Abstract
The pandemic outbreak of coronavirus disease 2019 (COVID‐19) across the world has led to millions of infection cases and caused a global public health crisis. Current research suggests that SARS‐CoV‐2 is a highly contagious coronavirus that spreads rapidly through communities. To understand the mechanisms of viral replication, it is imperative to investigate coronavirus viral replicase, a huge protein complex comprising up to 16 viral nonstructural and associated host proteins, which is the most promising antiviral target for inhibiting viral genome replication and transcription. Recently, several components of the viral replicase complex in SARS‐CoV‐2 have been solved to provide a basis for the design of new antiviral therapeutics. Here, we report the crystal structure of the SARS‐CoV‐2 nsp7+8 tetramer, which comprises two copies of each protein representing nsp7's full‐length and the C‐terminus of nsp8 owing to N‐terminus proteolysis during the process of crystallization. We also identified a long helical extension and highly flexible N‐terminal domain of nsp8, which is preferred for interacting with single‐stranded nucleic acids.
Keywords: COVID‐19, nsp7, nsp7+8 complex, nsp8, SARS‐CoV‐2
Short abstract
PDB Code(s): 7DCD;
1. INTRODUCTION
In early 2020, the COVID‐19 virus swept the world, creating a global pandemic greatest in risk since the influenza pandemic of 1918. 1 The virus has been identified as a new coronavirus named severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2), which is an enveloped, positive‐strand RNA (+RNA) virus belonging to the family, Coronaviridae, order Nidovirales, and realm Riboviria, with a large genome of approximately 29.8 kb in length. 2 , 3 , 4 , 5 Sequence analysis suggests that the SARS‐CoV‐2 genome is highly identical to that of bat coronavirus (BatCoV), which originated from infected bats. 4 , 6 The SARS‐CoV‐2 genome has been predicted to contain 14 functional open‐reading frames (ORFs), including ORF1ab, which occupies approximately two‐thirds of the 5′ end of this genome and encodes two polyproteins (pp1a and pp1ab) using a programmed ribosomal frameshift mechanism caused by a slippery sequence. 7 These two polyproteins contain two proteases, a papain‐like proteinase (PLpro, nsp3) and 3C‐like proteinase (3CLpro, nsp5), which autoproteolytically processes the polyproteins into 16 nonstructural proteins (nsp1‐16). 8 , 9 , 10 , 11 These nsps are believed to comprise the major components of the viral replication/transcription complex, a highly dynamic protein‐RNA complex that facilitates viral replication and transcription. Most of these nsps are well‐known types of enzymes, including a papain‐like protease 2 (PL2pro, nsp3), 12 a 3C‐like serine protease (Mpro, nsp5), 13 an RNA‐dependent RNA polymerase (RdRp, nsp12), an RNA helicase (nsp13), a guanine‐N7 methyltransferase (nsp14), an endoribonuclease (nsp15), and 2′O‐methyl‐transferase (2′O‐MTase, nsp16). 14 , 15 , 16 , 17 In addition, the replicase gene encodes several small associated proteins, including nsp1 and nsp7‐nsp11. 15 Proteins nsp7‐nsp10 are highly conserved among all coronaviruses (CoVs), but have no functional homologs outside of coronaviridae. 18 After the emergence of SARS‐CoV in 2003, there was an intense effort to characterize the complex structure of such nonstructural proteins. 19 Subsequently, the complex structures of nsp7+nsp8, nsp10+nsp14, nsp10+nsp16, and nsp12+nsp7+nsp8 of SARS‐CoV have been determined. 20 , 21 , 22 , 23 Very recently, several independent groups have reported the complex structure of nsp12 RNA‐dependent RNA polymerase with cofactors nsp7 and nsp8 from SARS‐CoV‐2 using cryo‐electron microscopy (cryo‐EM). 24 , 25 , 26
The hexadecameric structure of the nsp7 and nsp8 complex has been solved in SARS‐CoV. 20 The complex resembles a hollow cylinder with a central channel, which can encircle and bind RNA through a strong positive charge in the inner channel as exhibited in biochemical assays. Functional studies have revealed that nsp8 prefers the 5′‐(G/U)CC‐3′ trinucleotides on the RNA template to initiate the synthesis of complementary short oligonucleotides, and nsp8 is proposed to provide RNA primers required by nsp12 RdRp during replication and transcription. 27 Deletion of the nsp7 or nsp8 region results in impaired RNA synthesis and a lethal phenotype in MHV. 28 A report described that nsp8 co‐immunoprecipitates with nsp12, nsp9, nsp5, and helicase nsp13, and it also colocalizes with nsp7, nsp9, and nsp10. 19 , 29 Therefore, nsp7 and nsp8 seem to play an important role in the viral replication complex.
Herein, we determined the complex structure of nsp7 and nsp8 from SARS‐CoV‐2 using X‐ray crystallography. The 2.5 Å resolution structure is composed of a tetrameric complex of nsp7 and the C‐terminus of nsp8. The core architecture of the nsp7+8 tetramer is composed of two α‐helix bundles, which is highly conserved compared with the nsp7+8 hexadecameric structure from SARS‐CoV and the cryo‐EM structure of the nsp7+8+12 complex from SARS‐CoV‐2. We also confirmed that the N‐terminus of nsp8 is highly flexible with a positive electrostatic surface involved in nucleic acid interactions and complex formation with nsp7 and nsp12.
2. RESULTS
2.1. Overall structure of the SARS‐CoV‐2 nsp7+8 dimer
The full‐length recombinant proteins, GST‐tagged nsp7 (residues 1–84) and His6‐tagged nsp8 (residues 1–199) from SARS‐CoV‐2 were expressed in Escherichia coli (E. coli) BL21 (DE3). The nsp7+8 complex was prepared for crystallization with a stoichiometry ratio of 1:1 and further purified by size‐exclusion chromatography (SEC). The complex protein was screened for crystallization conditions, and a single crystal was grown for weeks into a proper size for X‐ray diffraction data collection (Figure S1D, S1E). The diffraction data were processed to a 2.5 Å resolution with the HKL2000 package. 30 The final nsp7+8 complex structure was resolved by molecular replacement in the P1 space group packing four nsp7+8 complexes into a unit cell. The N‐terminus of nsp8 (residues Ala1‐Ser77) could not be traced in the electron density map. The degradation of the nsp8 N‐terminus during the process of crystallization was identified by dissolving crystal in water and verified on SDS‐PAGE gel (Figure S1A‐C).
Each nsp7+8 complex structure exhibits a heterodimer consisting of one copy of nsp7 (residues Met3–Asp75) and one copy of the nsp8 C‐terminus (residues Glu77‐Asn192, nsp8C) (Figure 1a). The nsp7 structure is composed of four α‐helices forming an antiparallel helix bundle with helices a1 (residues Ser4‐Gln19), a2 (residues Leu28‐Leu41), a3 (residues Thr45‐Ser61), and a4 (residues Ile68‐Met75) (Figure 1b). The four nsp7 monomers in the unit cell superimpose well (RMSD deviation <0.5 Å). A previous study reported that nsp7 in feline coronavirus adopts two different conformations in complex with nsp8. As seen in Figure S3B and S3C, the helix bundle conformation of nsp7 is highly conserved in SARS‐CoV‐2 replicase complexes, including within the structures of the nsp12+7+8 complex (PDB: 7C2K) and nsp12+13+7+8 complex (PDB: 6XEZ). We also compared our structure for SARS‐CoV‐2 nsp7 within the dimeric complex to the three structures of SARS‐CoV nsp7 with coordinates available in the PDB (2AHM, 6NUR, and 5F22). We noticed that the structure of SARS‐CoV‐2 nsp7 is highly conserved with that of SARS‐CoV nsp7 in complex with nsp8 (Figure S3D‐F). The RMS deviation for 62 Ca atoms between the two molecules (SARS‐CoV‐2 nsp7 and SARS‐CoV nsp7) is 0.583 Å. These data indicate that the structure of SARS‐Cov‐2 nsp7 is highly conserved with nsp7s of other CoVs, especially with the structure of SARS‐CoV nsp7, likely based on the high sequence identity between these CoVs (Figure S1F).
FIGURE 1.
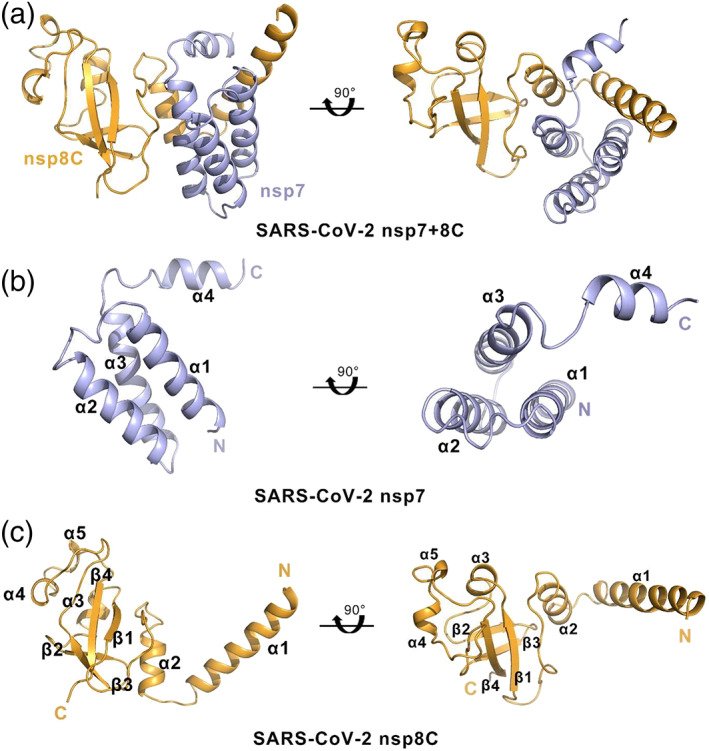
Overall structure of the SARS‐CoV‐2 nsp7+8C complex. a, The protomeric architecture of SARS‐CoV‐2 nsp7+8C. The nsp7 and nsp8C proteins are colored in light blue and orange, respectively. b, The four α helices of nsp7. c, The structural details of nsp8C. nsp8C contains five α helices and four β strands
SARS‐CoV nsp8 adopts two different conformations in the hexadecameric complex with nsp7. However, only one conformation of nsp8 was observed in our SARS‐CoV‐2 nsp7+8 complex. The nsp8 in our structure is incomplete owing to the degradation of the N‐terminus protein (residues Ala1‐Ser77) during crystallization. Therefore, we named nsp8C for an accurate description. nsp8C is composed of four α‐helices (α1, residues Asp78‐Arg 96; α2, residues Asp101‐Asn109; α3, residues Tyr135‐Lys139; α4, residues Lys169‐Glu171; α5, residues Ser177‐Asn179) packed against a four‐stranded antiparallel β‐sheet (b1, residues Leu128‐Ile132; b2, residues Thr146‐Tyr149; b3, residues Ala152‐Val160; b4, residues Leu184‐Leu189) is similar to the “golf club” nsp8 structures from SARS‐CoV and feline infectious peritonitis virus (FIPV) (Figure 1c). Superimposition of the SARS‐CoV‐2 nsp8C onto the SARS‐CoV nsp8 (PDB: 2AHM) and nsp8 in feline coronavirus (FCoV) nsp7+8 complex (PDB: 3UB0) indicates that the C‐terminal domains of nsp8 are highly conserved in coronaviruses, with RMSD values of 0.671 for 98 Cα atoms and 0.668 Å for 103 Cα atoms, respectively (Figure S3G, S3H). Compared with SARS‐CoV, there are five residues (Y15F, V132I, G136N, N145T, N193S) in SARS‐CoV‐2 that were mutated, four of which are located at the C‐terminus but had no significant effect on structure (Figure S1G).
nsp7 interacts tightly with nsp8C to yield a buried surface area of 1,467.4 Å2 (~26.4% of the whole surface area of nsp7), forming a heterodimer. The interaction interfaces between two proteins consist of a α‐helix bundle composed by five a‐helices (a1, a3, and a4 from nsp7; a1 and a2 from nsp8C) that interact with each other, which are predominant via hydrophobic interactions, along with nine hydrogen bonds and four salt bridges. The same interface was also observed in the hexadecameric complex of SARS‐CoV nsp7+8 (PDB: 2AHM), dimeric complex of SARS‐CoV nsp7+8C (PDB: 6NUR), and FCoV nsp7+8C trimeric complex (PDB: 3UB0), which indicates that the core structure of nsp7+8C is highly architecturally conserved among coronaviruses.
2.2. Structural basis for SARS‐CoV‐2 nsp7+8 tetramer
In order to obtain more information about the interfaces and likely biological assemblies of the nsp7+8 complex, we analyzed the crystal structure using the online programs, PISA (Protein, Interfaces, Structures and Assemblies) 31 and EPPIC (Evolutionary Protein–Protein Interface Classifier). 32 The results suggested that nsp7+8 forms a stable symmetric tetramer during crystal packing. The average total surface area buried of the tetramer was 7,790 Å2. The analysis of the crystal structure showed that a tetramer with a “butterfly” shape was present in the unit cell (Figure 2a). It is formed by the association of two nsp7+8 dimers, which contact each other with the two helix bundles. According to the PISA calculation, the formation of the tetramer is mainly because of an “α‐helix bands” hydrophobic core between nsp7 (α1 and α2) and nsp8 (α1). There are two hydrogen bonds between residues Met94 of nsp8 and Ser4 of nsp7. In addition, Cys8 between the two nsp7 residues forms a disulfide bond to stabilize the tetramer (Figure 2b).
FIGURE 2.
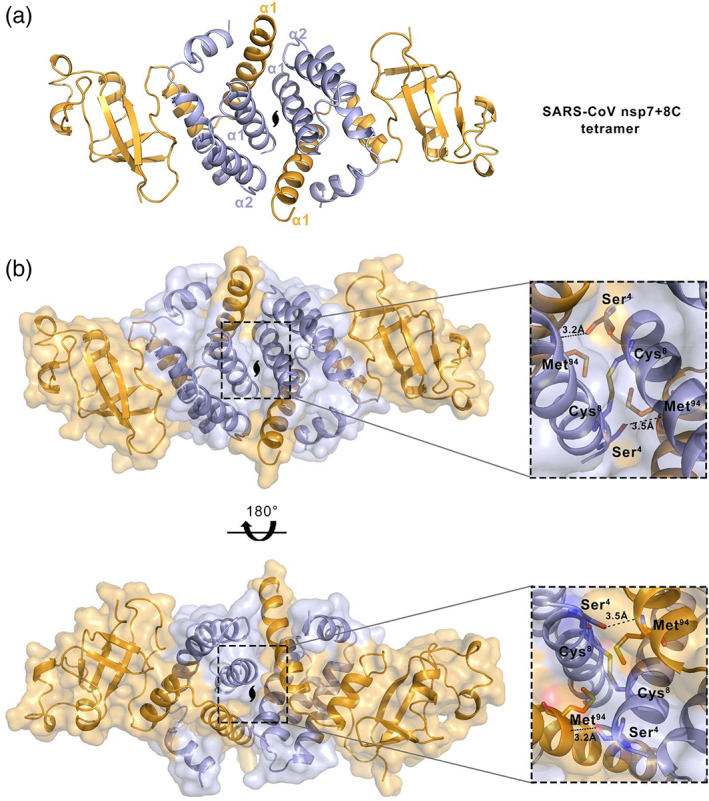
The teteramer structure and interface details of SARS‐CoV‐2 nsp7+8C. a, The overall teteramer structure of SARS‐CoV‐2 nsp7+8C. There is a central symmetry between the two nsp7+8C molecules protomerically. b, The interface of the SARS‐CoV‐2 nsp7+8C teteramer. The interactional amino acid residues and distances are represented by abbreviated letters and numbers
There was also the question of whether the quaternary structure of the SARS‐CoV‐2 nsp7+8C tetramer exists in solution. First, we performed chemical cross‐linking analysis. After treatment with different concentrations (0.005%, 0.01%, and 0.02%) of glutaraldehyde, isolated nsp7, nsp8, and nsp8C displayed cross‐linked homopolymers in addition to their monomers (Figure S2B‐E). When the nsp7+8C complex was cross‐linked by glutaraldehyde, we detected the nsp7+8C heterodimer (24 kDa) and heterotetramer (48 kDa) (Figure S2F). We also applied analytical SEC to determine the solution states of the nsp7, nsp8, nsp8C, nsp7+8, and nsp7+8C complexes. As shown in Figure S2A, the nsp7+8 and nsp7+8C complexes eluted with a retention volume of 14.75 ml and 15.12 ml, respectively, representing a tetramer with estimated molecular mass (MM) of 58 kDa and 49 kDa, respectively, which corresponded reasonably well to the MM indicated by cross‐linking. nsp7 and nsp8 had retention volumes of 16.95 ml and 15.28 ml, respectively. The estimated MMs calculated from the retention volumes were also in agreement with the MMs of their cross‐linked tetramer. Consequently, these data indicate that the tetrameric model of nsp7+8C is a feasible biological assembly.
2.3. Nucleic acid‐binding domain located in N‐terminus of nsp8
Based on our knowledge, the N‐terminus of nsp8 is highly dynamic in solution and adopts diverse confirmation depending on the assembly elements in the complex. We calculated several full‐length SARS‐CoV‐2 nsp8 models based on the structures of nsp8 from the SARS‐CoV nsp7‐8 hexadecamer (PDB: 2AHM) using SWISS‐MODEL. The models present an N‐terminal domain containing a long α‐helix or several short helices. The surface charge distribution pattern of nsp8 in SARS‐CoV‐2 reveals a highly positive charge region located on the N‐terminus, indicating a potential nucleic acid‐binding region (Figure 3a, b). Therefore, surface plasmon resonance (SPR) was performed to investigate whether the nsp8 and nsp7+8 complexes could bind single‐stranded nucleic acids. The binding curves of nucleic acids with various binding partners were studied (Figure 3c, d) and the interaction of the nsp8 wild‐type to ssRNA showed a KD of 5.20 μM while the interaction of the nsp7+8 to ssRNA was 11.5 μM. Furthermore, the electrophoretic mobility shift assay (EMSA) approach was employed to identify the nucleic acid binding with the wild‐type nsp8, specifically two truncated versions nsp8ΔN and nsp8ΔC, respectively. The mobility of the nucleic acid reduced in the lanes of the nsp8 wild‐type and nsp7+8 complex indicates that the specificity of the interaction between protein and nucleic acid, nsp8ΔC or nsp7+nsp8ΔC, binds slightly to the nucleic acid, while there is no obvious combination between free nsp7, nsp8ΔN, or nsp7+nsp8ΔN and nucleic acid (Figure 3e, f). This implies that full‐length nsp8 is needed to stabilize its binding to nucleic acids. Our data support that the N‐terminus of nsp8 among coronaviruses is a nucleic acid binding motif. Without the existence of nucleic acid, the N‐terminus of nsp8 is highly unstable and easily degradable in vitro. Very recently, studies on the complex of SARS‐CoV‐2 nsp12+7+8 complex resolved by cryo‐EM showed that the N‐terminus of nsp8 plays pivotal roles in RNA interaction. 24 , 25
FIGURE 3.
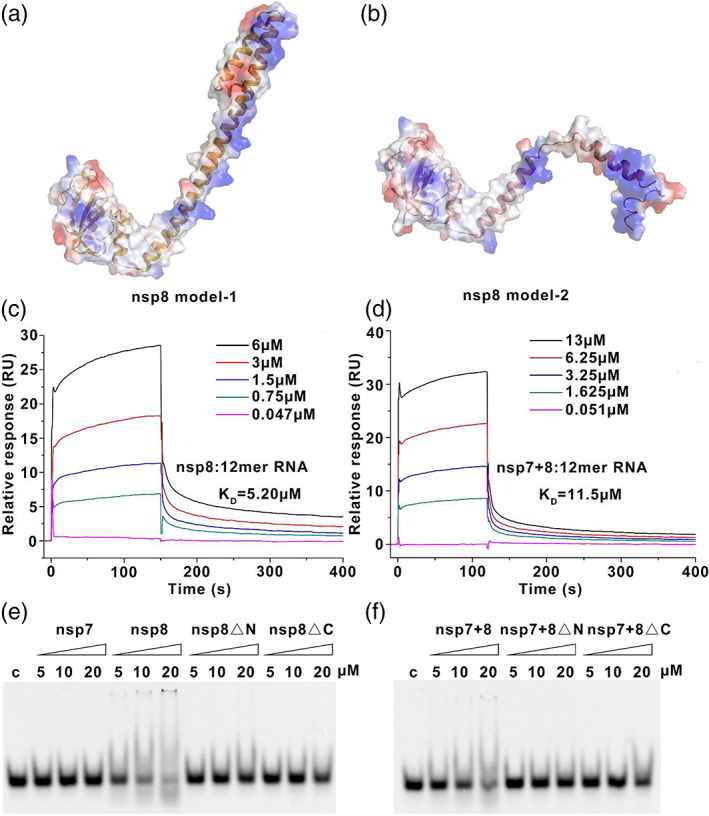
The nsp8 is responsible for nucleic acid binding. a‐b, The full‐length nsp8 SARS‐CoV‐2 model show as cartoon and surface (blue, positive charge (+60 kBT); red, negative charge (−60 kBT)). c‐d, SPR sensorgram showing the binding kinetics for the SARS‐CoV‐2 nsp7+8 complex and 12‐mer ssRNA. Dates are shown as color lines. (e‐f) Single‐stranded (ss) RNA‐binding abilities of SARS‐CoV‐2 nsp7+8 as determined by electrophoretic mobility shift assay. The state of the SARS‐CoV‐2 nsp7+8 complex and the concentration of proteins used in the reaction system are indicated above the gel
3. DISCUSSION
Although our crystal of nsp7+8 complex was obtained and diffraction resolution could be up to 2.5 Å, the crystal had a few flaws when collecting the diffraction data, which resulting the data was processed through P1 space group instead of higher symmetry space group. Compared with structures of SARS‐CoV nsp7+8 hexadecamer complex and the heterotrimeric complex of FCoV nsp7+8, the nsp7+8C structure of SARS‐CoV‐2 forms a heterotetramer consisting with two of nsp7 molecules and two nsp8 C‐terminus. Considering the N‐terminus of nsp8 is highly conserved in coronaviruses and is vital in multimeric assembly of the nsp7+8 complex in SARS‐CoV and FCoV, we analyzed the solution state of the SARS‐CoV‐2 full‐length nsp7+8 complex in multiple ways. The SARS‐CoV‐2 nsp7 and nsp8 were mixed at different ratios before loading to the SEC column, but the nsp7+8 complex always eluted with a retention volume of 14.75 ml with an estimated MM of 58 kDa. Chemical cross‐linking also indicated that the 2:2 stoichiometry of nsp7:nsp8 observed in our crystal structure exists in solution, as well. Based on these results, SARS‐CoV‐2 nsp7 and nsp8 can form a stable tetrameric conformation in the solution state.
The alignment of the structures of all three nsp7+8 complexes (2AHM, 6NUR, and 5F22) in SARS coronaviruses reveals a conserved six‐helical bundle contact surface, which is the major interface that forms the nsp7+8C tetramer. The structure of the SARS‐CoV nsp7+nsp8 hexadecamer can be regarded as a supercomplex assembled from four nsp7+8 tetramers, which are tightly associated with the long α‐helix at the N‐terminal of nsp8. The proteolysis product of SARS‐CoV nsp8, which loses the N‐terminal long α‐helix, only forms a tetrameric architecture similar to the structure of SARS‐CoV‐2 nsp7+8 tetramer (Figure S4A, S5). However, it should be noted that the nsp12+nsp7+nsp8 complex in SARS coronaviruses represents the minimal RNA polymerase complex required for viral genomic transcription and replication. There are two cryo‐EM structures of RdRp (nsp12) from SARS‐CoV‐2 bound to nsp7 and nsp8 co‐factors, providing a view of the overall architecture of the super complex. 24 , 25 However, no hexadecameric or tetrameric forms of nsp7+8 observed in these structures indicated that nsp7+8 may perform diverse biological functions through differential assembly conformations (Figure S4B, S4C). Very recently, Quan et al. also described the SARS‐CoV‐2 polymerase‐RNA complex structure and proposed that the hexadecameric nsp7+8 primase complex is dissociated by half after primer synthesis and delivers the nucleic acid to the polymerase‐RNA complex directly. 24 The crystal structure of the nsp7+8 complex from SARS‐CoV‐2 reported here suggests that the core architecture of the nsp7+8 complex plays a pivotal role in RNA‐dependent RNA polymerase assembly.
4. METHODS
4.1. Plasmid construction, protein purification, and crystallization
The SARS‐CoV‐2 nsp7 (genome nucleotides 11,578‐11,826; Gene ID: 43740578) and nsp8 (genome nucleotides 11,827‐12,420; Gene ID: 43740578) genes were synthesized and subcloned into the pGEX‐6p‐1 [using BamHI and XhoI restriction enzymes (Takara)] and pET‐DUET1 [using BamHI and HindIII restriction enzymes (Takara)] vectors, respectively. Recombinant proteins nsp7 and nsp8, as well as two truncated nsp8 versions, nsp8△C (residues 1–78) and nsp8△N (residues 77–198) which subcloned into the pET‐DUET1 (Table S1), were expressed in Escherichia coli (E. coli) BL21 (DE3). The cells were harvested after 18 h induced by 0.5 mM isopropyl β‐D‐1‐thiogalactopyranoside (IPTG) at 16°C. Next, the cell pellet was resuspended into buffer with 20 mM Tris pH 8.0 and 150 mM NaCl before the homogenization process. After centrifugation to remove cell debris, the Glutathione S‐transferase (GST) tag labeled nsp7 protein was purified with a glutathione affinity column (Qiagen, Hilden, Germany) and the GST tag was released by incubation with PreScission Protease at a final concentration of 0.05 mg/ml in solution. The nsp8 and truncations were purified by Ni‐NTA affinity chromatography (Qiagen, Hilden, Germany) following Resource Q (nsp8 and nsp△N) or Resource S (nsp8△C) chromatography (Cytiva). The final protein samples were concentrated to 1 ml in Amicon Ultra concentrators (cut‐off size of 3 kDa; Millipore) and loaded onto a Superdex™ 200 column (GE Healthcare, USA) for further purification and characterization, with a buffer of 20 mM Tris pH 8.0, 300 mM NaCl, 1 mM dithiothreitol (DTT), and 10% glycerol (v/v). The purity of the nsp7 and nsp8 proteins reached above 95%, as confirmed by SDS‐PAGE. Nsp8 or nsp8△N was then mixed with 1 M excess of nsp7 and passed over the Superdex™ 200 column in the same buffer. Fractions of the nsp7+nsp8 complex were prepared with 1:1 stoichiometry and concentrated to 20 mg/ml for crystallization. Protein quantity was assessed using absorbance readings at 280 nm (Thermo NanoDrop 2000, USA). The nsp7+nsp8 crystals were initially grown at 20°C for 7 days by using the hanging‐drop vapor‐diffusion method with a mixture of 1 μL protein and 1 μL reservoir solution. One single protein crystal with high X‐ray diffraction quality was obtained in 200 mM NaCl, 100 mM Tris–HCl, pH 8.2 and 25% PEG3350. The single crystal was stored in liquid nitrogen for data collection.
4.2. Data collection, processing, and structure determination
The diffraction data of nsp7+nsp8 complex crystal were collected at 100 K in at the SSRF Beamline BL18U1 (Shanghai, China) at a wavelength of 0.97930 Å. Data were processed and scaled using the HKL2000 package. 30 The nsp7+nsp8 structure was solved by molecular replacement using Phaser in the CCP4 package with the initial searching coordinates of SARS‐CoV nsp7 and nsp8 (PDB: 2AHM). 33 Cycles of refinement and model building were carried out with the REFMAC5, Phenix, and COOT software programs. 34 , 35 Model geometry was verified using the SWISS model. 36 Structural figures were drawn using PyMOL. 37 The data collection and refinement statistics are shown in Table 1.
TABLE 1.
SARS‐CoV‐2 nsp7+8 data collection and refinement statistics
| SARS‐CoV‐2 nsp7+8 | |
|---|---|
| Data collection | |
| Resolution (Å) | 2.57 |
| Space group | P1 |
| Unit‐cell parameters (Å, °) | 43.031 42.99100.63 90.012 90.035 72.613 |
| Resolution range (Å) | 41.06–2.57 (2,662–2.57) |
| R merge a (%) | 13.8 (23.6) |
| R pim b (%) | 7.5 (12.7) |
| Average I/σ(I) | 11.6 (4.5) |
| No. of unique reflections | 20,426 (1981) |
| Completeness (%) | 93.14 (89.76) |
| Solvent content (%) | 50 |
| Molecules per asymmetric unit | 8 |
| Refinement | |
| Rwork/Rfree | 0.25/0.35 |
| Ramachandran favored (%) | 87.92 |
| Ramachandran outliers (%) | 0.67 |
| No. of atoms | |
| Protein | 5,809 |
| Water | 75 |
| Wilson B value | 43.68 |
| Root‐mean‐square deviations | |
| Bond length (Å) | 0.010 |
| Bond angle (°) | 1.239 |
R merge = ∑hkl∑i∣Ii(hkl) – 〈I(hkl)〉∣/∑hkl∑iIi(hkl), where Ii(hkl) is an individual intensity measurement and 〈I(hkl)〉 is the average intensity for all i reflections.
R pim is approximately estimated by multiplying the R merge value by the factor [1/(N − 1)]1/2, where N is the overall redundancy of the data set.
4.3. Cross‐linking assay by glutaraldehyde
Cross‐linking was carried out by incubating 1 mg/ml of wildtype SARS‐CoV‐2 nsp7, nsp8, nsp8C, nsp7+8, and nsp7+8C in 50 mM HEPES pH 7.5, 150 mM NaCl, and 10% glycerol (v/v). Different final concentrations of glutaraldehyde (0.005%, 0.01%, and 0.02% (v/v)) were utilized at 25°C for 5, 10, and 15 min. The reaction was quenched by adding 1 M Tris pH 8.0 to a final concentration of 0.3 M and analysis was carried out by 14% SDS‐PAGE with detection by Coomassie Brilliant Blue staining.
4.4. Analytic size‐exclusion chromatography
SEC analysis was conducted with evaluation of the oligomeric state of the nsp7, nsp8, nsp8C, nsp7+nsp8 and nsp7+nsp8C truncated forms using a Superdex™ 200 10/300 GL column (Cytiva). Commercial Gel Filtration Standard (Bio‐Rad, #1511901) was used as molecular weight standard. The column was equilibrated with elution buffer in 20 mM Tris (pH 8.0), 300 mM NaCl, 1 mM DTT, and 10% glycerol (v/v) at a flow rate of 0.3 ml/min followed by protein samples at a concentration of ~1 mg/ml at 4°C.
4.5. Electrophoretic mobility‐shift assay
FAM‐labeled 12‐mer ssRNA oligonucleotide (5′FAM‐GCTTTGATTTCG‐3′) was used for the electrophoretic mobility‐shift assay. Initially, 2 nmol DNA was incubated with different concentrations of nsp7+nsp8 complex along with free nsp7, nsp8, and truncated nsp8 forms in 10 mM HEPES pH 8.0, 50 mM KCl, 1 mM EDTA, 0.05% Triton‐X‐100, and 5% Glycerol for 30 min at 4°C, followed by the addition of 10 × loading buffer (250 mM Tris–HCl pH 7.9, 40% glycerol) to the mixture. Samples were then run on 6.5% non‐denaturing TBE polyacrylamide gels for 30 min at a voltage of 100 V, and the results were determined with a Bio‐Rad ChemiDoc Touch Imaging System (Bio‐Rad).
4.6. Surface plasmon resonance analysis
The interaction between nsp8 or nsp7+nsp8 complex proteins with 12‐mer ssRNA was monitored by SPR using a BIAcore 8 K (GE Healthcare) carried out at 16°C in running buffer composed of 50 mM HEPES pH 7.5, 150 mM NaCl, and 0.05% Tween 20. The serial concentrations of nsp8 or nsp7+nsp8 complex proteins were 0.375, 0.75, 1.5, 3, and 6 mM when testing interactions with 12‐mer ssRNA, respectively. The biotin‐labeled 12‐mer ssRNA was immobilized to a sensor chip SA to a level of ~200 RUs using a BIAcore 8 K in the same running buffer described earlier. The resulting data were fitted to a 1:1 banding model, and the binding affinity was calculated using BIAcore 8 K Evaluation Software (GE Healthcare).
4.7. Protein data Bank accession code
The structure factor and atomic coordinate have been deposited in the Protein Data Bank with the PDB ID code: 7DCD.
AUTHOR CONTRIBUTIONS
Changhui Zhang: Investigation; writing‐original draft. Li Li: Visualization; writing‐original draft. Jun He: Formal analysis; writing‐original draft. Cheng Chen: Formal analysis; writing‐original draft. Dan Su: Conceptualization; project administration; writing‐original draft.
CONFLICT OF INTEREST
The authors declare that they have no conflicts of interest surrounding the contents of this article.
Supporting information
Supplementary Figure 1 SARS‐CoV‐2 nsp7+8 expression and purification. (A) SDS‐PAGE analysis of the nsp7+8 protein complex. (B) SDS‐PAGE analysis (cropped from different parts of the same gel) of the crystals of the nsp7+8 complex (de‐nsp8 refers to degraded nsp8, M refers to marker). (C) SDS‐PAGE analysis of the nsp7+8C protein complex. (D) The crystal for X‐ray date collection of SARS‐CoV‐2 nsp7+8C after crystallization conditions optimized. (E) X‐ray diffraction pattern of nsp7+8C for structure analysis. (F‐G) Sequence alignment of CoV nsp7 and nsp8 homologs. Comparison of the SARS‐CoV, HCoV‐229E, and FCoV sequences with the SARS‐CoV‐2 sequence. Identical residues are highlighted with a red background, and conserved residues are shown in red font. The table was produced with ESPript 3.0.
Supplementary Figure 2. Information on the SARS‐CoV‐2 nsp7+8 complex. (A) The SEC of SARS‐CoV‐2 nsp7+nsp8 and nsp7+nsp8C with a Superdex200™ 10/300 column. (B‐F) The cross‐linking assays for nsp7, nsp8, nsp8C, nsp7+8, and nsp7+8C.
Supplementary Figure 3. Superimposition of SARS‐CoV‐2 nsp7 and nsp8 with the SARS‐CoV nsp7 and nsp8 structure. (A‐C) Superimposition of SARS‐CoV‐2 nsp7 with other structures in different assembly forms (PDB: 7C2K and 6XEZ). (D‐F) Superimposition of SARS‐CoV‐2 nsp7 with other nsp7 structures for SARS‐CoV (PDB: 2AHM, 5F22 and 6NUR). (G‐H) Superimposition of SARS‐CoV‐2 nsp8C with SARS‐CoV and FCoV nsp8.
Supplementary Figure 4. Superimposition of the SARS‐CoV‐2 nsp7+nsp8C complex with other nsp7+nsp8 complexes. (A) Superimposition of the SARS‐CoV‐2 nsp7+nsp8C complex with the SARS‐CoV nsp7+nsp8 complex (PDB: 2AHM). (B) SARS‐CoV nsp7+8+12 complex (PDB: 6NUR). (C) FCoV nsp7+8 complex (PDB: 3UB0).
Supplementary Figure 5. The structure of the SARS‐CoV nsp7+nsp8 complex. (A) The octamer structure of the SARS‐CoV nsp7+nsp8 complex from the hexadecameric supercomplex (PDB: 2AHM). (B) The structure of the SARS‐CoV nsp7+nsp8 complex without the N‐terminal a‐helix forms a tetrameric architecture.
Supplementary Figure 6. Uncropped gels used for the indicated figures.
Supplementary Table 1. The primers used to construct the two truncated nsp8 versions.
ACKNOWLEDGMENTS
We appreciate the staff of the State Key Laboratory of Biotherapy, Sichuan University, during the period of the COVID‐19 epidemic. X‐ray diffraction experiments were carried out at the Shanghai Synchrotron Radiation Facility (BSRF) BL18U1. We further thank the Beamline staff for approving our urgent proposal submission for this COVID‐19 research project. Moreover, we are grateful to Dr. Guangwen Lu for providing us with the associated plasmids. This work was supported by grants from the National Key Research and Development Program of China (2017YFA0505903), the National Natural Science Foundation of China (2018YFC1312300), the Special Research Fund on COVID‐19 of Sichuan Province (2020YFS0010), and the Urgent Project on COVID‐19 of West China Hospital, Sichuan University (HX‐2019‐nCoV‐044).
Zhang C, Li L, He J, Chen C, Su D. Nonstructural protein 7 and 8 complexes of SARS‐CoV‐2. Protein Science. 2021;30:873–881. 10.1002/pro.4046
Funding information Special Research Fund on COVID‐19 of Sichuan Province, Grant/Award Number: 2020YFS0010; National Key Research and Development Program of China, Grant/Award Number: 2017YFA0505903; National Natural Science Foundation of China, Grant/Award Number: 2018YFC1312300; The Urgent Project on COVID‐19 of West China Hospital, Sichuan University, Grant/Award Number: HX‐2019‐nCoV‐044
REFERENCES
- 1. Kaul D. An overview of coronaviruses including the SARS‐2 coronavirus ‐ molecular biology, epidemiology and clinical implications. Curr Med Res Pract. 2020;10:54–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zhu N, Zhang D, Wang W, et al. China novel coronavirus I, research T. a novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Coronaviridae Study Group of the International Committee on Taxonomy of . V. the species severe acute respiratory syndrome‐related coronavirus: Classifying 2019‐nCoV and naming it SARS‐CoV‐2. Nat Microbiol. 2020;5:536–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lu R, Zhao X, Li J, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. Lancet. 2020;395:565–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chan JF, Kok KH, Zhu Z, et al. Genomic characterization of the 2019 novel human‐pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg Microbes Infect. 2020;9:221–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zhou P, Yang XL, Wang XG, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Graham RL, Sparks JS, Eckerle LD, Sims AC, Denison MR. SARS coronavirus replicase proteins in pathogenesis. Virus Res. 2008;133:88–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hui DS, E IA, Madani TA, et al. The continuing 2019‐nCoV epidemic threat of novel coronaviruses to global health ‐ the latest 2019 novel coronavirus outbreak in Wuhan. China Int J Infect Dis. 2020;91:264–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Brian DA, Baric RS. Coronavirus genome structure and replication. Curr Top Microbiol Immunol. 2005;287:1–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kim D, Lee JY, Yang JS, Kim JW, Kim VN, Chang H. The architecture of SARS‐CoV‐2 transcriptome. Cell. 2020;181:914–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Snijder EJ, Bredenbeek PJ, Dobbe JC, et al. Unique and conserved features of genome and proteome of SARS‐coronavirus, an early split‐off from the coronavirus group 2 lineage. J Mol Biol. 2003;331:991–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Shin D, Mukherjee R, Grewe D, et al. Van der Heden van Noort GJ, Ovaa H, Muller S, Knobeloch KP, Rajalingam K, Schulman BA, Cinatl J, hummer G, Ciesek S, Dikic I. papain‐like protease regulates SARS‐CoV‐2 viral spread and innate immunity. Nature. 2020;587:657–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhang L, Lin D, Sun X, et al. Crystal structure of SARS‐CoV‐2 main protease provides a basis for design of improved alpha‐ketoamide inhibitors. Science. 2020;368:409–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ahn DG, Choi JK, Taylor DR, Oh JW. Biochemical characterization of a recombinant SARS coronavirus nsp12 RNA‐dependent RNA polymerase capable of copying viral RNA templates. Arch Virol. 2012;157:2095–2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Thiel V, Ivanov KA, Putics A, et al. Mechanisms and enzymes involved in SARS coronavirus genome expression. J Gen Virol. 2003;84:2305–2315. [DOI] [PubMed] [Google Scholar]
- 16. te Velthuis AJ, Arnold JJ, Cameron CE, van den Worm SH, Snijder EJ. The RNA polymerase activity of SARS‐coronavirus nsp12 is primer dependent. Nucleic Acids Res. 2010;38:203–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Subissi L, Imbert I, Ferron F, et al. SARS‐CoV ORF1b‐encoded nonstructural proteins 12‐16: Replicative enzymes as antiviral targets. Antiviral Res. 2014;101:122–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Johnson MA, Jaudzems K, Wuthrich K. NMR atructure of the SARS‐CoV nonstructural protein 7 in solution at pH 6.5. J Mol Biol. 2010;402:619–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. von Brunn A, Teepe C, Simpson JC, et al. Analysis of intraviral protein‐protein interactions of the SARS coronavirus ORFeome. PLoS One. 2007;2:e459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhai Y, Sun F, Li X, et al. Insights into SARS‐CoV transcription and replication from the structure of the nsp7‐nsp8 hexadecamer. Nat Struct Mol Biol. 2005;12:980–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ma Y, Wu L, Shaw N, et al. Structural basis and functional analysis of the SARS coronavirus nsp14‐nsp10 complex. Proc Natl Acad Sci U S A. 2015;112:9436–9441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chen Y, Su C, Ke M, et al. Biochemical and structural insights into the mechanisms of SARS coronavirus RNA ribose 2′‐O‐methylation by nsp16/nsp10 protein complex. PLoS Pathog. 2011;7:e1002294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kirchdoerfer RN, Ward AB. Structure of the SARS‐CoV nsp12 polymerase bound to nsp7 and nsp8 co‐factors. Nat Commun. 2019;10:2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wang Q, Wu J, Wang H, et al. Structural basis for RNA replication by the SARS‐CoV‐2 polymerase. Cell. 2020;182:417–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hillen HS, Kokic G, Farnung L, Dienemann C, Tegunov D, Cramer P. Structure of replicating SARS‐CoV‐2 polymerase. Nature. 2020;584:154–156. [DOI] [PubMed] [Google Scholar]
- 26. Yin W, Mao C, Luan X, et al. Structural basis for inhibition of the RNA‐dependent RNA polymerase from SARS‐CoV‐2 by remdesivir. Science. 2020;368:1499–1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Imbert I, Guillemot JC, Bourhis JM, et al. A second, non‐canonical RNA‐dependent RNA polymerase in SARS coronavirus. EMBO J. 2006;25:4933–4942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Deming DJ, Graham RL, Denison MR, Baric RS. Processing of open reading frame 1a replicase proteins nsp7 to nsp10 in murine hepatitis virus strain A59 replication. J Virol. 2007;81:10280–10291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pan J, Peng X, Gao Y, et al. Genome‐wide analysis of protein‐protein interactions and involvement of viral proteins in SARS‐CoV replication. PLoS One. 2008;3:e3299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Otwinowski Z, Minor W. Processing of X‐ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. [DOI] [PubMed] [Google Scholar]
- 31. Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. [DOI] [PubMed] [Google Scholar]
- 32. Scharer MA, Grutter MG, Capitani G. CRK: An evolutionary approach for distinguishing biologically relevant interfaces from crystal contacts. Proteins. 2010;78:2707–2713. [DOI] [PubMed] [Google Scholar]
- 33. Winn MD, Ballard CC, Cowtan KD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. 2011;67:235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Adams PD, Afonine PV, Bunkoczi G, et al. A comprehensive python‐based system for macromolecular structure solution. Acta Crystallogr. 2010;66:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. McCoy AJ, Grosse‐Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Cryst. 2007;40:658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Waterhouse A, Bertoni M, Bienert S, et al. SWISS‐MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46:W296–W303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rigsby RE, Parker AB. Using the PyMOL application to reinforce visual understanding of protein structure. Biochem Mol Biol Educ. 2016;44:433–437. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1 SARS‐CoV‐2 nsp7+8 expression and purification. (A) SDS‐PAGE analysis of the nsp7+8 protein complex. (B) SDS‐PAGE analysis (cropped from different parts of the same gel) of the crystals of the nsp7+8 complex (de‐nsp8 refers to degraded nsp8, M refers to marker). (C) SDS‐PAGE analysis of the nsp7+8C protein complex. (D) The crystal for X‐ray date collection of SARS‐CoV‐2 nsp7+8C after crystallization conditions optimized. (E) X‐ray diffraction pattern of nsp7+8C for structure analysis. (F‐G) Sequence alignment of CoV nsp7 and nsp8 homologs. Comparison of the SARS‐CoV, HCoV‐229E, and FCoV sequences with the SARS‐CoV‐2 sequence. Identical residues are highlighted with a red background, and conserved residues are shown in red font. The table was produced with ESPript 3.0.
Supplementary Figure 2. Information on the SARS‐CoV‐2 nsp7+8 complex. (A) The SEC of SARS‐CoV‐2 nsp7+nsp8 and nsp7+nsp8C with a Superdex200™ 10/300 column. (B‐F) The cross‐linking assays for nsp7, nsp8, nsp8C, nsp7+8, and nsp7+8C.
Supplementary Figure 3. Superimposition of SARS‐CoV‐2 nsp7 and nsp8 with the SARS‐CoV nsp7 and nsp8 structure. (A‐C) Superimposition of SARS‐CoV‐2 nsp7 with other structures in different assembly forms (PDB: 7C2K and 6XEZ). (D‐F) Superimposition of SARS‐CoV‐2 nsp7 with other nsp7 structures for SARS‐CoV (PDB: 2AHM, 5F22 and 6NUR). (G‐H) Superimposition of SARS‐CoV‐2 nsp8C with SARS‐CoV and FCoV nsp8.
Supplementary Figure 4. Superimposition of the SARS‐CoV‐2 nsp7+nsp8C complex with other nsp7+nsp8 complexes. (A) Superimposition of the SARS‐CoV‐2 nsp7+nsp8C complex with the SARS‐CoV nsp7+nsp8 complex (PDB: 2AHM). (B) SARS‐CoV nsp7+8+12 complex (PDB: 6NUR). (C) FCoV nsp7+8 complex (PDB: 3UB0).
Supplementary Figure 5. The structure of the SARS‐CoV nsp7+nsp8 complex. (A) The octamer structure of the SARS‐CoV nsp7+nsp8 complex from the hexadecameric supercomplex (PDB: 2AHM). (B) The structure of the SARS‐CoV nsp7+nsp8 complex without the N‐terminal a‐helix forms a tetrameric architecture.
Supplementary Figure 6. Uncropped gels used for the indicated figures.
Supplementary Table 1. The primers used to construct the two truncated nsp8 versions.


