Key Points
Extracellular rhERAPs trigger innate immunity.
Anti–HIV-1 activity of extracellular ERAPs relies on monocyte/macrophage activation.
Abstract
Recombinant human (rh) ERAP2-treated PBMCs are less susceptible to in vitro HIV-1 infection even when CD8+ T cells are depleted. We therefore investigated whether ERAP2 can trigger other immunocompetent cells, boosting their antiviral potential. To this end, human monocyte-derived macrophages (MDMs) differentiated from PBMCs of 15 healthy donors were in vitro HIV-1 infected in the presence/absence of 100 ng/ml of rhERAP2, rhERAP1, or rhERAP1+rhERAP2. Notably, rhERAP2 treatment resulted in a 7-fold reduction of HIV-1 replication in MDMs (p < 0.05). This antiviral activity was associated with an increased mRNA expression of CD80, IL-1β, IL-18, and TNF-α (p < 0.01 for cytokine) in in vitro ERAP2-treated HIV-1–infected MDMs and a greater release of IL-1β, TNF-α, IL-6, and IL-8 (p < 0.01 for each cytokine). The rhERAPs addition also induced the functional inflammasome activation by ASC speck formation in monocytes (p < 0.01) and in THP1-derived macrophages (p < 0.01) as well as a rise in the percentage of activated classical (CD14+CD16−HLA-DRII+CCR7+) and intermediate (CD14++CD16+HLA-DRII+CCR7+) monocytes (p < 0.02). Finally, THP-1–derived macrophages showed an increased phagocytosis following all ERAPs treatments. The discovery that ERAPs are able to trigger several antiviral mechanisms in monocyte/macrophages suggests that their anti-HIV potential is not limited to their canonical role in Ag presentation and CD8+ T cell activation. These findings pose the premise to further investigate the role of ERAPs in both innate and adaptive immunostimulatory pathways and suggest their potential use in novel preventive and therapeutic approaches against HIV-1 infection.
Introduction
Endoplasmic reticulum (ER) aminopeptidase (ERAP) 1 and 2 are two closely related IFN-γ–inducible, ubiquitously expressed, ER-localized aminopeptidases that play a major role in the antigenic peptide editing quality control. By trimming the N-termini of precursor peptides generated in the cytosol by the proteasome, ERAPs influence the development of Ag repertoire that will be presented in MHC class I, manipulating both CD8+ T lymphocyte– (1) and NK-mediated immune responses (2–4). The role of ERAPs in shaping the quality and the potency of immune responses has been shown in a number of autoimmune and inflammatory diseases by several independent groups (5–8). Besides this canonical function, ERAPs can nevertheless display additional unconventional roles whose relevance should not be undervalued. First of all, enzymatically active ERAP1 has been found in the secretome of RAW264.7 murine macrophage cell lines upon both inflammatory stimuli (9) and increased cytokine-induced intracellular Ca2+ levels (10). Furthermore, a subset of secreted ERAP1 was found to be bound to exosomes released from LPS/IFN-γ–treated murine macrophages (11). Secreted ERAP1 mediates a number of functions, including 1) the activation of inflammatory processes by enhancing phagocytoses and NO synthesis from macrophages (9–11); 2) the trimming of peptides with N-terminal arginine residues, favoring NO production (12); 3) blood pressure regulation (13); 4) increased inflammatory cytokine production in human PBMCs through NOD-like receptor, pyrin domain-containing (NLRP) 3 activation (14); and 5) cytokine receptor shedding (15, 16).
The role played by the highly homologous ERAP2 in both Ag presentation and innate modulation has not been as extensively characterized as for ERAP1. This lack of information is mainly due to the ERAP2 absence in rodent genomes (17). In the field of Ag presentation, ERAP2 acts on different substrates compared with ERAP1 and shows particular preference for arginine residues (18, 19). Only recently, the release of ERAP2 in the secretome of IFN-γ/LPS–stimulated monocyte-derived macrophages (MDMs) has been documented (20), but its biological function in this milieu has only partially been clarified. Based on the results so far obtained, it has been speculated that secreted ERAP2 may re-enter the cell and relocalize into the ER, thus strengthening CD8+ T cell activation. Supporting this assumption are data showing that the addition of recombinant human (rh) ERAP2 to cell culture upregulated CD8+ activated lymphocytes (CD25+HLA−DRII+) and effector memory/terminally differentiated CD8+ T cells, resulting in a more efficient in vitro control of HIV infection/replication (20). However, in the same study, it was demonstrated that HIV replication was partially reduced by the rhERAP2 addition even in CD8+ T cell–depleted cultural conditions, suggesting that effector mechanisms or cellular types other than CD8+ T cells may be modulated by ERAP2. Hence, considering the high structural and functional homology between the two ERAP isoforms, the current study aims to verify if extracellular ERAP2 could have direct or indirect effects on natural immunity as previously ascertained for ERAP1. To this end, we investigated monocyte/macrophage activation by analyzing inflammasome activation, phagocytosis, and monocyte differentiation. Furthermore, as ERAP1 and ERAP2 can heterodimerize at an intracellular level and generate final peptides at a faster rate than their individual counterparts (21), we performed these same analyses using a combination of the two isoforms of ERAPs. The in vitro HIV infection model was used as several studies have demonstrated a correlation between both susceptibility (20, 22–25) and progression (26–28) of HIV infection and ERAPs.
Materials and Methods
Healthy control blood samples and PBMC isolation
Thirty milliliters of whole blood were collected in EDTA-containing vacutainer tubes (Becton Dickinson, Rutherford, NJ) from 15 healthy controls (HCs). PBMCs were separated on lymphocyte separation medium (Cedarlane Laboratories, Hornby, ON, Canada) and washed twice in PBS. Leukocyte viability was determined using a Bio-Rad TC20 Automated Cell Counter (Bio-Rad Laboratories).
The Ethical Committee of the Fondazione IRCCS Ca’ Granda Ospedale Maggiore Policlinico (Milan, Italy) approved the study (Protocol No. 0028257). All the donors signed an informed consent form in accordance with the Declaration of Helsinki.
MDMs differentiation
The percentage of CD14+ monocytes was determined in PBMCs isolated from 15 HCs by flow cytometer analyses. Monocytes were isolated via plastic adherence and MDMs were generated as previously described (29), incubating 1 × 106 monocytes for 5 d in RPMI 1640 with 20% of FBS (EuroClone, Milan, Italy) and 100 ng/ml M-CSF (R&D Systems, Minneapolis, MN). MDM differentiation was assessed by optical microscope observation (ZOE Fluorescent Cell Imager; Bio-Rad Laboratories) (Supplemental Fig. 1).
PBMCs and MDMs in vitro HIV-1 infection assay
PBMCs from five HC and MDMs from 15 HCs were cultured in RPMI 1640 containing 20% FBS in the presence/absence of 100 ng/ml of rhERAP1, rhERAP2, or rhERAP1+rhERAP2 for 3 h (R&D Systems Code 2334-ZN and 3830ZN, respectively; R&D Systems). The dose of rh proteins to be used in this experimental setting was assessed in a previous work (20). Moreover, in the Human Protein Atlas database (https://www.proteinatlas.org/humanproteome/blood/proteins+detected+in+ms), ERAP1 (27 μg/l) and ERAP2 (7.4 μg/l) human plasma concentration detected by mass spectrometry are reported. Although the concentration of rhERAPs employed in our assay is somewhat higher (3.7-fold for ERAP1 and 13.5-fold for ERAP2), we did not observe any toxic effect on the treated cells in a previous study (20).
After incubation, cells were in vitro HIV-1Ba-L infected (0.5 ng/1 × 106cells) at 37°C and 5% CO2. After 24 h, PBMCs and the attached macrophages were washed twice with PBS and cultured in complete medium containing 20% FBS for 6 d. Postinfection, half of the medium was changed every 2 d, and rhERAPs were added in the corresponding cell cultures. Only for PBMCs, IL-2 (15 ng/ml) (R&D Systems) was added as well. Six days postinfection, viral Ag replication was assessed in the supernatant of PBMCs and MDMs by p24 Ag ELISA (Cell Biolabs, San Diego, CA). Cytokine production was evaluated in the supernatant of infected MDMs by multiplex ELISA (Bio-Rad Laboratories). Gene expression analyses was performed on MDMs.
MDM gene expression analysis
RNA extraction and retrotranscription were performed as previously described (30). cDNA quantification (Bio-Rad Laboratories) was assessed by using a real-time PCR (CFX96 connect; Bio-Rad Laboratories) and a SYBR Green PCR Mix (Bio-Rad Laboratories), and all reactions were run in duplicate. The results are presented as the average of the relative expression units to the GAPDH and β-actin reference genes calculated by the 2−ΔΔCt equation using the CFX manager 3.1 (Bio-Rad Laboratories). Reactions were performed according to the following thermal profile: initial denaturation (95°C, 15 min) followed by 40 cycles of 15 s at 95°C (denaturation) and 20 s at 60°C (annealing) and 20 s at 72°C (extension). Melting curve analysis was also analyzed for amplicon identification. Cycle threshold values of 35 or higher were excluded from the analyses. Each experiment was run in duplicate.
Cytokine and chemokine measurement by multiplex assay
The concentration of 27 cytokines/chemokines was assessed in the supernatants of HIV-infected MDMs in the presence/absence of rhERAPs by using immunoassays formatted on magnetic beads (Bio-Rad Laboratories), according to manufacturer’s protocol via Luminex 100 technology (Luminex, Austin, TX). Supernatants were obtained 6 d post–MDMs HIV infection. Each experiment was run in duplicate.
Cell cultures: human monocytic THP-1 cell line, PBMCs, and MDMs
The human monocytic cell line THP-1 was provided by Istituto Zooprofilattico Sperimentale della Lombardia e dell’Emilia Romagna (Brescia, Italy) and maintained in RPMI 1640 supplemented with 10% FBS, 2 mM l-glutamine, and 1% penicillin (Invitrogen, Paisley, U.K.) (medium) at 37°C in a humidified 5% CO2 atmosphere. To differentiate these cells into macrophages, THP-1 human monocytes were seeded in six-well plates at a density of 1 × 106 cells/well in medium containing 50 nM of phorbol 12-myristate 13-acetate (PMA) (Sigma-Aldrich, St. Louis, MO) and incubated for 12 h at 37°C in 5% CO2.
For ASC speck formation, THP-1–derived macrophages (duplicate) and PBMCs were resuspended at 1 × 106 cells/ml and were either 1) cultured in medium alone (unstimulated), 2) stimulated with the NLRP3 agonist nigericin (5 μM for 1 h) (positive control), 3) primed 2 h with LPS (1 μg/ml) (Sigma-Aldrich), or 4) primed 2 h with LPS and stimulated with 100 ng/ml of rhERAP1, rhERAP2, or rhERAP1+rhERAP2 for 22 h at 37°C in a humidified 5% CO2 atmosphere. LPS preincubation is required because neither NLRP3 nor pro–IL-1β are constitutively expressed and require transcriptional induction (31, 32).
For monocyte activation, uninfected and HIV-1–infected PBMCs were cultured in medium alone or stimulated with rhERAP1 (100 ng/ml), rhERAP2 (100 ng/ml), or a combo of ERAP1+rhERAP2 (100 ng/ml) for 24 h at 37°C in a humidified 5% CO2 atmosphere. For phagocytoses assay, THP-1–derived macrophages were cultured analogously.
Intracellular inflammasome protein staining
THP-1–derived macrophages (1 × 106) or monocytes, stimulated as described above, were fixed with 100 μl of paraformaldehyde (PFA) (1%) for 10 min, permeabilized with 100 μl of Saponin (0.1%) (Life Science VWR, Lutterworth, U.K.), and stained with PE anti-human ASC (clone HASC-71, isotype mouse IgG1; BioLegend, San Diego, CA) for 1 h at room temperature. Cells were then washed with PBS, centrifuged at 1500 rpm for 10 min, resuspended in 50 μl of PBS, and analyzed by Amnis FlowSight Imaging Flow Cytometer (Luminex).
Immunofluorescent staining
Monocyte subsets and MHC class II expression.
Uninfected and HIV-1–infected PBMCs cultured in medium alone or stimulated with rhERAPs were stained with CD14 PE-Cy7 (Isotype IgG2a Mouse, Clone RMO52; Beckman Coulter), CD16 PE-Cy5 (Isotype IgG1 Mouse, Clone 3G8; Beckman Coulter), CCR7 FITC (Isotype IgG2a Mouse, Clone G043H7; BioLegend), and HLA-DRII ECD (Isotype IgG2b Mouse, Clone B8.12.2; Beckman Coulter)–specific mAbs for 30 min at room temperature in the dark. Cells were then washed, fixed in 1% PFA in PBS, and acquired by flow cytometry.
Flow cytometry analysis
Analyses were performed by using a Beckman Coulter Gallios Flow Cytometer equipped with two lasers operating at 488 and 638 nm, respectively, interfaced with Gallios software and analyzed with Kaluza v 1.2. Two hundred thousand events were acquired and gated on CD14+ for monocyte cells, considering isotype background.
Data were collected using linear amplifiers for forward and side scatter and logarithmic amplifiers for fluorescences (FL) 1, FL2, FL3, FL4, and FL5. Samples were first run using isotype control or single fluorochrome-stained preparations for color compensation. Rainbow Calibration Particles (Spherotech, Lake Forest, IL) were used to standardize flow cytometry results.
Gating strategies were as follows (Supplemental Fig. 2): an initial assessment of monocyte population was performed using a forward scatter plot/side scatter plot. A first gate was set around the monocytes. The monocyte population was gated on a side scatter/CD14 scatter plot and on CD14+ to identify the monocyte population; another two gates were set on a CD14/CD16 scatter plot to distinguish the classical monocytes (CD14+/CD16−) from intermediate (CD14++/CD16+) and nonclassical monocytes (CD14+/CD16++).
The percentage of CCR7+ and HLA-DRII+ was then calculated on classical, intermediate, and nonclassical monocytes. Samples were first run using isotype controls or single fluorochrome-stained preparations for color compensation. Rainbow Calibration Particles (Spherotech) were used to standardize flow cytometry results.
Phagocytosis assay
THP-1–derived macrophages (1 × 106), stimulated as described above, were cultured with latex beads coated with rabbit IgG–FITC complex (Cayman Chemical) to a final dilution of 1:100 for 1 h. Medium containing 0.05% Trypsin–EDTA (Code ECB3052D; EuroClone) was added for 10 min at 37°C in 5% CO2. Cells were then resuspended in RPMI 1640 supplemented with 10% FBS and centrifuged at 1500 rpm for 10 min. Pellets were fixed with 0.1% PFA for 10 min, washed, resuspended in 50 μl PBS, and analyzed by Amnis FlowSight Imaging Flow Cytometer (Luminex). Each experiment was run in duplicate.
Amnis FlowSight imaging analysis
Phagocytosis and ASC speck formation were analyzed by Amnis FlowSight Imaging Flow Cytometer (Luminex), an imaging flow cytometer equipped with two lasers operating at 488 and 642 nm, 2 cameras, and 12 standard detection channels that merge flow cytometry and high-resolution microscopy. The machine simultaneously produces side scatter (darkfield) images, one or two transmitted light (brightfield [BF]) images, and up to 10 channels of FL imagery of every cell. FlowSight using the INSPIRE system, acquires 2000 cells/s, and operates with a 1-μm pixel size (∼×20 magnification), allowing visualization of FL from the membrane, cytoplasm, or nucleus. The IDEAS image analysis software allows quantification of cellular morphology and FL at different cellular localizations by defining specific cellular regions (masks) and mathematical expressions that uses image pixel data or masks (features).
Phagocytosis was evaluated by analyzing the internalization feature using a mask representing the whole cell, defined by the BF image, and an internal mask defined by eroding the whole cell mask to eliminate the fluorescent signal coming from the latex beads rabbit IgG–FITC complex attached to the cell surface, thus measuring only the internalized part. The internalization feature was first used to calculate the ratio of the intensity of latex beads rabbit IgG–FITC complex inside the cell/total FAM intensity outside the cell. Higher internalization scores (IS) indicate a greater concentration of latex beads rabbit IgG–FITC complex inside the cell. THP-derived macrophages with an IS >2 were defined as those with a functional phagocytic activity. The inflammasome assembly was determined by ASC speck formation using a threshold mask plotting Max Pixel MC (Ch03) versus area threshold (M03, Ch03, and Ch70) that defined cell with ASC speck from cells with an ASC diffuse pattern. This mask allows us to separate within the population of ASC fluorescent cells those with small area and high maximum pixel (ASC speck) from those with large area and low maximum pixel (ASC diffuse) (33).
Statistical analyses
Data were analyzed using Student t or ANOVA test by GraphPad Prism version 5 (GraphPad Software, La Jolla, CA), and p values ≤0.05 were considered to be significant.
Results
rhERAPs induce the differentiation and activation of monocytes
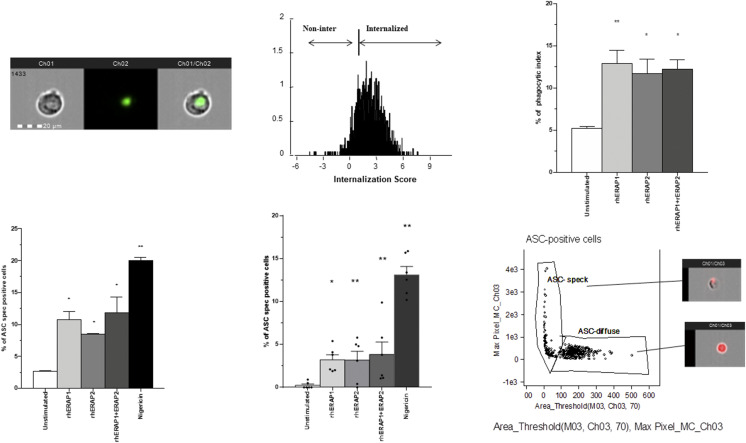
We first investigated the effect exerted by ERAPs on monocyte differentiation and activation. To this end, PBMCs were stimulated with rhERAPs and differentiation/activation subset into classical, intermediate, and nonclassical monocytes and were evaluated. Notably, we observed an increase in the percentage of activated (CCR7+DRII+) total monocytes following any rhERAP treatment, with the strongest effect being associated with the use of rhERAP2 (p < 0.01) (Fig. 1). Moreover, when we evaluated monocyte subsets, we found that the addition of rhERAP2+rhERAP1 resulted in the activation (CCR7+DRII+) of all three subtypes of monocytes: classical monocytes (CD14++ CD16−) (p < 0.05), intermediate monocytes (CD14++CD16+) (p < 0.05), and nonclassical monocytes (CD14+CD16++) (p < 0.05) (Fig. 1). Notably, rhERAP2 addition significantly activated classical (p < 0.01) and intermediate monocytes (p < 0.05) (Fig. 1), whereas rhERAP1 treatment induced only the activation of classical monocytes (p < 0.05) (Fig. 1). As classical and intermediate monocytes are responsible for the induction of a robust innate inflammatory immune response, these results suggest that rhERAPs treatment favors the establishment of a prompt and potent immune response, which could protect against infection.
FIGURE 1.
rhERAPs stimulation favors monocyte activation and differentiation in specific subsets. PBMCs isolated from 15 HCs were stimulated with 100 ng/ml of rhERAP1, rhERAP2, or rhERAP1+rhERAP2 for 24 h. The percentage of total CD14+ CD16+), classical (CD14++CD16−), intermediate (CD14++CD16+), and nonclassical (CD14+CD16++) monocytes expressing the activation markers DRII and CCR7 was assessed by flow cytometry. Mean values and SEM are shown. *p < 0.05, **p < 0.01.
rhERAPs stimulation triggers the phagocytic activity of THP-1–derived macrophages
To elucidate the pathophysiological relevance of extracellular rhERAP protein addition, we next measured the phagocytic activity of rhERAPs-treated THP-1–derived macrophages. Data revealed that the ability of THP-1–derived macrophages to phagocytose latex beads coated with fluorescently labeled rabbit IgG significantly increased following rhERAPs addition (Fig. 2A). In particular, as shown in Fig. 2B, a moderate rate of phagocytosis was seen in untreated cells. However, rhERAP1, rhERAP2, and rhERAP1+rhERAP2 induced a substantial increase (nearby 2-fold) of this parameter (rhERAP1: p < 0.01; rhERAP2: p < 0.05; and rhERAP1+rhERAP2: p < 0.05). These results suggest that extracellular ERAPs are also responsible for the enhancement of THP-1–derived macrophage phagocytosis.
FIGURE 2.
rhERAPs stimulation resulted into increased phagocytosis in THP-1–derived macrophages into inflammasome ASC speck assembly. (A) Representative images captured by Amnis FlowSight Cytometry of THP-1–derived macrophage incubated with latex beads rabbit IgG–FITC complex are shown. The first column shows BF image of macrophages incubated with latex beads rabbit IgG–FITC complex, the second column shows image related to latex beads rabbit IgG–FITC complex FL, and the third column shows merged latex beads rabbit IgG–FITC complex FL with BF (left); IS calculated on latex beads rabbit IgG–FITC complex positive macrophage by IDEAS software is shown (right). Scale bar, 20 μm. (B) THP-1–derived macrophages were incubated with or without 100 ng/ml of rhERAP1, rhERAP2, or rhERAP1+rhERAP2 for 3 h. Latex beads rabbit IgG–FITC complex was added directly to the culture medium at a 1:200 dilution and incubated at 37°C for 2 h and immediately analyzed by FlowSight Amnis. The phagocytic activity of THP-1–derived macrophages incubated in the absence of rhERAPs treatment was measured as a control (C–E). NLRP3/ASC speck colocalization was analyzed by Amnis FlowSight Cytometry. 1 × 106 LPS-primed (1 μg/ml) THP-1–derived macrophages (C) and PBMCs (six HCs) (D) were stimulated for 24 h with rhERAP1, rhERAP2, or rhERAP1+rhERAP2 (100 ng/ml), and results were summarized as the percentage of positive cells for NLRP3/ASC speck formation. (E) Representative image of nigericin-stimulated THP-1–derived macrophage identifying ASC speck. ASC-positive cells are plotted on Max Pixel MC (Ch03) versus area threshold (M03, Ch03, and Ch70) scatter plot. This mask allows us to discriminate between cells characterized by speck formation, in which a functional inflammasome complex is assembled, and cells with an ASC diffuse pattern. Mean values and SEM are shown. *p < 0.05, **p < 0.01.
ASC speck formation in LPS-primed and rhERAPs-stimulated THP-1–derived macrophages and monocytes
As rhERAPs-treated PBMCs significantly increased IL-1β release as well as IL-18 and CASP1 expression and rhERAP1 has already been proven to induce NLRP3 activation (14), we wondered whether all rhERAPs might activate specific innate immune sensors, responsible for IL-1β processing and release, a mechanism orchestrated by inflammasome.
Apoptosis-associated speck-like protein containing CARD-speck formation, the hallmark of inflammasome activation, was analyzed by FlowSight Amnis in LPS-primed and rhERAPs-stimulated THP-1–derived macrophages as well as in CD14+ cells labeled from HC PBMCs. Results showed that the percentage of ASC speck–positive THP-1–derived macrophages was significantly increased following nigericin (positive control) stimulation (p < 0.05) (Fig. 2C). Remarkably, THP-1–derived macrophages primed with LPS and stimulated with all rhERAPs showed a significant increase of ASC speck assembly compared with the unstimulated condition (p < 0.05 for all comparisons) (Fig. 2C).
To verify if such phenomenon is not limited to cell lines, experiments were replicated on PBMCs labeled with CD14 fluorophore to distinguish monocytes, the main inflammasome-expressing cells (34). Results confirmed that rhERAPs addition to cell cultures induces cytoplasmic diffuse ASC molecules to cluster into ASC speck (Fig. 2D). The increase was statistically significant in all the rhERAPs-treated cultures (rhERAP1: p < 0.02; rhERAP2: p < 0.01; and rhERAP1+rhERAP2: p < 0.01). Representative images are provided in Fig. 2E.
rhERAPs decrease in vitro HIV-1 replication in MDMs
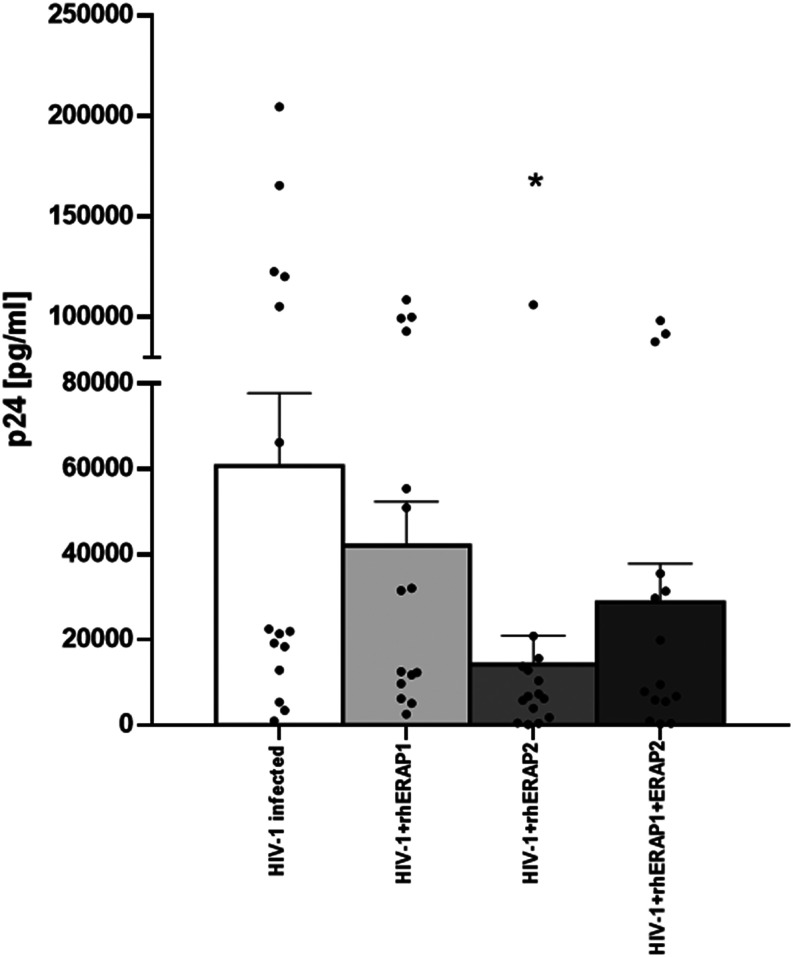
Because the addition of rhERAP2 reduces in vitro HIV-1 replication in PBMCs and the mechanism of action is only partially dependent on an acquired immune response (20), we decided to verify if rhERAPs could exert the same effect on HIV-1–infected MDMs. To this end, MDMs differentiated from monocytes were in vitro HIV-1 infected in the absence/presence of rhERAP1, rhERAP2, or rhERAP1+rhERAP2. As shown in Fig. 3, viral replication in MDMs was reduced in all cases and reached statistical significance in the presence of rhERAP2 (p < 0.05). These results suggest that rhERAPs, in particular rhERAP2, confer protection from HIV-1 infection by modulating innate immune responses.
FIGURE 3.
Susceptibility to in vitro HIV-1 infection was reduced in MDMs treated with rhERAPs. MDMs from 15 HCs were treated with 100 ng/ml of rhERAP1, rhERAP2, or rhERAP1+rhERAP2 and in vitro HIV infected with a R5 HIV1Ba-L. P24 concentration was measured by ELISA in 6 d post–in vitro HIV-1–infected supernatants. Mean values and SEM are shown. *p < 0.05.
Modulation of the expression of immune effectors genes by rhERAPs in in vitro HIV-1–infected MDMs
To verify whether the antiviral activity exerted by rhERAPs on MDMs relies on the activation of a natural immune response, we evaluated the mRNA expression of involved key players by real-time PCR. Analyses were performed on 6 d post–in vitro HIV-1–infected MDMs, which were treated/untreated with rhERAPs. Notably, the addition of different rhERAPs to cell cultures resulted in the upregulation of the same genes, which are implicated in the acquired and innate immune responses (Fig. 4); again, rhERAP2 seems to display a more consistent effect on immune activation. In particular, following rhERAP2 stimulation, a high transcription rate was observed for CD80 (p < 0.01), IFN-α (p < 0.05), TAP (p < 0.05), IL-1β (p < 0.01), IL-18 (p < 0.01), IL-8 (p < 0.01), CASP1 (p < 0.05), NPLR3 (p < 0.05), TNF-α (p < 0.05), and IFN-γ (p < 0.05) (Fig. 4).
FIGURE 4.
mRNA expression of cytokines and activation markers was increased in in vitro rhERAPs-treated HIV-1–infected MDMs. Following 6 d in vitro HIV infection, mRNA expression of several cytokines and immune determinants was assessed by real-time PCR in rhERAPs-stimulated MDMs differentiated from 15 HCs. Mean values and SEM are shown. *p < 0.05, **p < 0.01.
Modulation of cytokine and chemokine production by rhERAPs in in vitro HIV-1–infected MDMs
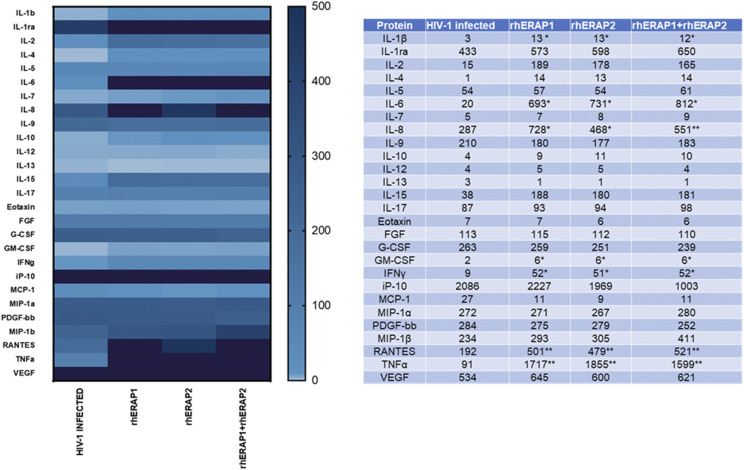
To further elucidate the antiviral mechanism elicited by rhERAPs in in vitro HIV-1–infected MDMs, we next assessed the production of 27 cytokines and chemokines related to immune response activation. Again, all rhERAPs significantly induced the production of key cytokines involved in several aspects of macrophage activation (Fig. 5). In particular, rhERAP2 significantly increased the secretion of proinflammatory cytokines, including IL-1β (p < 0.05), IL-6 (p < 0.05), IL-8 (p < 0.05), IFN-γ (p < 0.05), and TNF-α (p < 0.05) as well as of chemokines like RANTES (p < 0.01). These data mirror the results obtained by gene expression analyses, suggesting that the protection conferred by rhERAPs is correlated with an increased expression and production of several cytokines.
FIGURE 5.
The secretion of 27 cytokines/chemokines that are part of the inflammatory response is upregulated in in vitro rhERAPs-treated HIV-infected MDMs. The production of 27 cytokines/chemokines regulating immune response activation was significantly increased in in vitro HIV-infected MDMs isolated from 15 HCs following rhERAPs treatment. Cytokine production (picograms per milliliter) is shown as a color scale from white to black (0 to +500). Mean values and SEM are shown. *p < 0.05, **p < 0.01.
Differentiation and activation of monocytes in rhERAPs-treated HIV-1–infected PBMCs
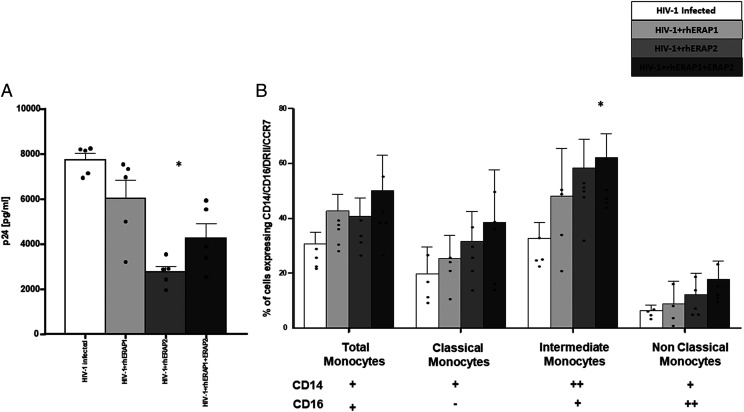
To confirm that the addition of rhERAPs reduces in vitro HIV-1 infection, PBMCs isolated from five HCs were in vitro infected with HIV-1Bal. Results showed that 6 d after in vitro HIV-1 infection, p24 levels were significantly lower in PBMCs treated with rhERAP2 and rhERAP1+rhERAP2 (p < 0.05). To verify if such protective effect could be secondary to rhERAP-induced monocyte differentiation and activation, 3 d post–in vitro infection PBMCs were labeled so as to analyze the different monocyte subsets.
Of note, in HIV-infected rhERAP-treated PBMCs, we observed an increase in the percentage of 1) total activated monocytes (p < 0.05), 2) classical monocytes following rhERAP2 addition (p < 0.05), and 3) classical and intermediate monocytes following rhERAP1+rhERAP2 treatment (p < 0.05). The induction of these inflammatory monocyte subsets by rhERAPs treatment could explain the suppression of viral replication in in vitro HIV infection assay (Fig. 6).
FIGURE 6.
rhERAPs stimulation favors monocyte activation and differentiation in specific subsets in an in vitro HIV-1 infection assay. (A) PBMCs from five HCs were treated with 100 ng/ml of rhERAP1, rhERAP2, or rhERAP1+rhERAP2 and in vitro HIV-infected with a R5 HIV-1Ba-L. P24 concentration was measured by ELISA in 6 d post–in vitro HIV-1–infected supernatants. Mean values and SEM are shown. *p < 0.05. (B) In the same cell cultures, 3 d post–in vitro HIV-1 infection, the percentage of total (CD14+ CD16+), classical (CD14++CD16−), intermediate (CD14++CD16+), and nonclassical (CD14+CD16++) monocytes expressing the activation markers DRII and CCR7 was assessed by flow cytometry. Mean values and SEM are shown. *p < 0.05.
Discussion
As clearly implied by their acronyms, ERAP1 and ERAP2 are primarily located in the ER, where they participate in the generation of the antigenic repertoire to be presented in association with MHC class I molecules. Therefore, they do play a key role in the selection, modulation, and activation of specific CD8+ T cell clones that can result in a strengthened or weakened immune response. Several studies, however, reported that besides acting as Ag-processing enzymes in the ER, ERAPs may be secreted into the extracellular milieu, where they display various pathophysiological functions. Such roles have been extensively investigated and reviewed by Tsujimoto et al. (35) for ERAP1. The finding that also the evolutionary younger sister ERAP2 may be released in the extracellular environment in response to inflammatory stimuli is far more recent (20, 36), and its role in this atypical context is almost entirely to be explored. In this report, for the first time, to our knowledge, we document ERAP2 extracellular ability to trigger natural immunity, besides an acquired one, at multiple levels.
First, the addition of rhERAPs to cell cultures significantly modified the distribution of monocyte subsets and in general increased their activation, as demonstrated by DRII and CCR7 analyses. Monocytes are, indeed, heterogeneous and comprise at least three distinct subsets of mononuclear phagocytes: the classical CD14+CD16−, intermediate CD14++CD16+, and nonclassical CD14+CD16++ subsets, although new ones have been recently described (37). Each subset shows functional differences in inflammatory (38, 39), migratory, and phagocytic capabilities (38) and then display a different impact on both homeostasis and disease progression. Classical monocytes are critical for the initial inflammatory response (40), and they can differentiate into macrophages in tissue (41) and show enhanced phagocytic activity (42). Intermediate monocytes are described as the major source of proinflammatory cytokines upon stimulation (43, 44). Conversely, nonclassical monocytes are widely viewed as anti-inflammatory as they maintain vascular homeostasis (45). The observation that ERAPs are able to regulate monocyte differentiation/activation is fairly recent. Of note, all rhERAPs treatments determined a rise in the percentage of total monocytes, suggesting that these aminopeptidases directly influence their expansion and activation. Even more important, following rhERAP stimulation, we observed a skew toward the percentage of activated classical and intermediate monocytes compared with the nonclassical ones. We can, thus, assume that following ERAPs secretion or addition, monocytes, which have demonstrated extensive plasticity, adjust their functional phenotype so as to generate a robust innate inflammatory immune response. These results allow also to speculate on a possible use of rhERAPs to establish a prompt and fortified immune response driven by monocyte elicitation.
Second, in our experimental setting, rhERAPs functionally activated the monocyte/macrophage lineage, as demonstrated by both increased phagocytosis and inflammasome assembly. In fact, this is not the first insight that ERAP1 stimulates macrophage phagocytosis. In 2011, Goto et al. (9) first reported that the addition of recombinant wild-type, but not inactive mutant, ERAP1 to culture medium enhanced phagocytosis in RAW264.7 murine cells, a property shared by other aminopeptidases (P-LAP and laeverin/APQ). The same feature has been further detailed in more recent studies (11, 14), leading the authors to suppose that both secreted and exosome-associated ERAP1 produce phagocytosis-enhancing peptide(s) via cleavage of substrate(s) in the extracellular space (9, 11). Remarkably, our results extend this property even to rhERAP2 and the combo of the two aminopeptidases in THP-1–derived macrophages human cell lines. We can, therefore, infer that phagocytosis-enhancing activity is a characteristic commonly shared by different aminopeptidases (9), which could be exploited as therapeutic intervention strategy in several pathological conditions showing reduced phagocytic functions. Likewise, caspase1/NLRP3 inflammasome pathway activation by extracellular ERAP1 has been previously documented by means of IL-1β secretion and caspase-1 production (14). Our data not only confirm these findings but move a step forward by pointing at rhERAP2 as another inflammasome-activating enzyme. Moreover, our data show for the first time, to our knowledge, that rhERAPs stimulation is able to induce cytoplasmic dispersed ASC molecules to cluster in one condensed micrometric-sized ASC speck complex, the hallmark of inflammasome activation. Although an excessive NLRP3 inflammasome activation has been implicated in a variety of human diseases, including Alzheimer disease (46, 47), multiple sclerosis (48), metabolic dysfunctions (49, 50), infectious diseases, and tumorigenesis (51, 52), ASC speck assembly is highly beneficial to the well-being of the host, and it is indispensable for the onset of a correct and protective immune response. As a matter of fact, by producing IL-1β inflammatory cytokine and inducing pyroptosis, the primary function of NLRP3 inflammasome is to protect the host from invading microorganisms (53). Notably, NLRP3-induced IL-1β production was also demonstrated to boost T cell function in patients receiving chemotherapy (54). Hence, we could hypothesize to take advantage of rhERAPs controlled induction of inflammasome in all the physiological and pathological circumstances requiring its intervention.
Interestingly, all the above-described properties perfectly justify the reduced susceptibility to in vitro HIV infection registered in rhERAPs-treated MDMs and PBMCs. Thus far, it has been reported that rhERAP2 addition to PBMC cell culture supernatant reduces in vitro HIV replication with a mechanism that is only partially dependent on CD8+ T cell activation (20). Data in this study suggest another defensive strategy triggered by rhERAP2 to contain viral replication mediated by the induction of several determinants intervening in natural immunity mainly mediated by monocytes activation and differentiation and inflammatory response. Indeed, the addition of rhERAPs to infected PBMCs results in an increase of both classical and intermediate monocytes, suggesting that the reduction of viral replication is at least partially mediated by the induction of a prompt and defensive innate response. Moreover, mRNA expression of cytokines (IL-1β, IL-8, IL-18, TNF-α, IFN-α, and IFN-γ), inflammasome components (CASP1 and NLRP3), activation markers (CD80), and Ag presentation machinery (TAP) and the production of several proinflammatory cytokines (IL-1β, IL-6, IL-8, IFN-γ, TNF-α, and RANTES) were, indeed, upregulated in HIV-infected MDMs following rhERAPs addition, suggesting their intervention in the orchestration of biological defense response to viral infection/replication. IL-1β increase was observed both at mRNA and protein level and was likely derived by rhERAPs-induced activation of inflammasome, a data corroborated by the higher mRNA expression of CASP1 following rhERAP addition. Although most of the studies reported in the literature establish a negative correlation between HIV infection and inflammosome complex assembly in both acute and chronic infection (51, 55, 56), it is also possible that inflammasome plays a protective role in the first steps of HIV-1 infection, as previously proposed by others (55). Actually, a plethora of studies correlate natural resistance to HIV infection in HIV-exposed seronegative individuals with the induction of an inflammatory process directing host immune response against the virus and/or disease progression (57–60). According to these assumptions, immune activation in itself is not always sufficient to favor HIV infection but rather could result in protection against HIV, possibly hampering viral replication and spreading and reducing the chance of an overt infection. Furthermore, immune activation could result in the elicitation of a more robust adaptive antiviral immune response. In this perspective, rhERAPs, by triggering inflammasome assembly, phagocytoses, and monocyte differentiation toward an inflammatory profile, could be considered a therapeutic strategy to enhance the natural protective resources during primary exposure to the virus, as observed in HIV-exposed seronegative individuals.
Analyses were performed stimulating cell cultures also with the combo of the two enzymes. Indeed, it was recently demonstrated that ERAP1 and ERAP2 can heterodimerize at an intracellular level, creating complexes with superior peptide-trimming efficacy (21, 61). Unexpectedly, the addition of rhERAP1 and rhERAP2 to the same cell culture did not result in an increased natural immune response in terms of monocyte/macrophage activation. However, at present, several questions concerning rhERAPs behavior in the extracellular milieu are still unanswered: 1) may the two aminopeptidases dimerize at an extracellular level, 2) which substrate(s) is recognized by the two enzymes, 3) is the quantity of rhERAPs added to the cell culture supernatant sufficient to saturate the system, and 4) may the rhERAPs re-enter the cells and relocalize into the ER, thus modulating CD8+ T cell activation? Answering these questions will allow us to deepen the role of ERAP1 and ERAP2 in the secretome so as to better exploit their functions in different experimental, physiological, and pathological settings.
Growing evidence shows that proteins can have multiple functions according to cellular localization. Considering that ERAPs may bind different substrates sharing specific amino acidic characteristics, it is conceivable that, depending on their subcellular localization, they may change binding partners and thus display multifunctional activities, with roles in both an innate and acquired immune system as well as inflammatory responses, which are crucial in the biological defense systems. Aside from Ag presentation, ERAP1 modulates angiogenesis, blood pressure control, cytokine receptor shedding, phagocytosis, NO synthesis, inflammasome activation, and TNF-α and IL-1β production, all of which apparently depend on its localization in the cells. The results reported in this study suggest that also ERAP2 may be considered as a “moonlighting protein,” acting as a final processing enzyme of MHC class I–presented Ags as is the case in peptides in the ER as well as a regulatory protein binding to several substrates involved in the natural immune response in the extracellular milieu. Further studies will clarify the processes and conditions that regulate the intracellular and extracellular location of the enzyme and the substrates recognized in the different settings.
Supplementary Material
The online version of this article contains supplemental material.
- BF
- brightfield
- ER
- endoplasmic reticulum
- ERAP
- ER aminopeptidase
- FL
- fluorescence
- HC
- healthy control
- IS
- internalization score
- MDM
- monocyte-derived macrophage
- NLRP
- NOD-like receptor, pyrin domain-containing
- PFA
- paraformaldehyde
- rh
- recombinant human.
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Shastri, N., Nagarajan N., Lind K. C., Kanaseki T.. 2014. Monitoring peptide processing for MHC class I molecules in the endoplasmic reticulum. Curr. Opin. Immunol. 26: 123–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cifaldi, L., Romania P., Falco M., Lorenzi S., Meazza R., Petrini S., Andreani M., Pende D., Locatelli F., Fruci D.. 2015. ERAP1 regulates natural killer cell function by controlling the engagement of inhibitory receptors. Cancer Res. 75: 824–834. [DOI] [PubMed] [Google Scholar]
- 3.Wilczyńska, K., Wiśniewski A., Malinowski A., Barcz E., Wilczyński J. R., Kuśnierczyk P., Nowak I.. 2019. ERAP, KIR and HLA-C gene interaction in susceptibility to recurrent spontaneous abortion in the Polish population. Hum. Immunol. 80: 344–348. [DOI] [PubMed] [Google Scholar]
- 4.Cifaldi, L., Lo Monaco E., Forloni M., Giorda E., Lorenzi S., Petrini S., Tremante E., Pende D., Locatelli F., Giacomini P., Fruci D.. 2011. Natural killer cells efficiently reject lymphoma silenced for the endoplasmic reticulum aminopeptidase associated with antigen processing. Cancer Res. 71: 1597–1606. [DOI] [PubMed] [Google Scholar]
- 5.Agrawal, N., Brown M. A.. 2014. Genetic associations and functional characterization of M1 aminopeptidases and immune-mediated diseases. Genes Immun. 15: 521–527. [DOI] [PubMed] [Google Scholar]
- 6.Reeves, E., James E.. 2018. The role of polymorphic ERAP1 in autoinflammatory disease. Biosci. Rep. 38: BSR20171503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hanson, A. L., Morton C. J., Parker M. W., Bessette D., Kenna T. J.. 2019. The genetics, structure and function of the M1 aminopeptidase oxytocinase subfamily and their therapeutic potential in immune-mediated disease. Hum. Immunol. 80: 281–289. [DOI] [PubMed] [Google Scholar]
- 8.Compagnone, M., Cifaldi L., Fruci D.. 2019. Regulation of ERAP1 and ERAP2 genes and their disfunction in human cancer. Hum. Immunol. 80: 318–324. [DOI] [PubMed] [Google Scholar]
- 9.Goto, Y., Ogawa K., Hattori A., Tsujimoto M.. 2011. Secretion of endoplasmic reticulum aminopeptidase 1 is involved in the activation of macrophages induced by lipopolysaccharide and interferon-γ. J. Biol. Chem. 286: 21906–21914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goto, Y., Ogawa K., Nakamura T. J., Hattori A., Tsujimoto M.. 2014. TLR-mediated secretion of endoplasmic reticulum aminopeptidase 1 from macrophages. J. Immunol. 192: 4443–4452. [DOI] [PubMed] [Google Scholar]
- 11.Goto, Y., Ogawa Y., Tsumoto H., Miura Y., Nakamura T. J., Ogawa K., Akimoto Y., Kawakami H., Endo T., Yanoshita R., Tsujimoto M.. 2018. Contribution of the exosome-associated form of secreted endoplasmic reticulum aminopeptidase 1 to exosome-mediated macrophage activation. Biochim. Biophys. Acta Mol. Cell Res. 1865: 874–888. [DOI] [PubMed] [Google Scholar]
- 12.Goto, Y., Ogawa K., Nakamura T. J., Hattori A., Tsujimoto M.. 2015. Substrate-dependent nitric oxide synthesis by secreted endoplasmic reticulum aminopeptidase 1 in macrophages. J. Biochem. 157: 439–449. [DOI] [PubMed] [Google Scholar]
- 13.Goto, Y., Hattori A., Ishii Y., Tsujimoto M.. 2006. Reduced activity of the hypertension-associated Lys528Arg mutant of human adipocyte-derived leucine aminopeptidase (A-LAP)/ER-aminopeptidase-1. FEBS Lett. 580: 1833–1838. [DOI] [PubMed] [Google Scholar]
- 14.Aldhamen, Y. A., Pepelyayeva Y., Rastall D. P. W., Seregin S. S., Zervoudi E., Koumantou D., Aylsworth C. F., Quiroga D., Godbehere S., Georgiadis D., et al. 2015. Autoimmune disease-associated variants of extracellular endoplasmic reticulum aminopeptidase 1 induce altered innate immune responses by human immune cells. J. Innate Immun. 7: 275–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Adamik, B., Islam A., Rouhani F. N., Hawari F. I., Zhang J., Levine S. J.. 2008. An association between RBMX, a heterogeneous nuclear ribonucleoprotein, and ARTS-1 regulates extracellular TNFR1 release. Biochem. Biophys. Res. Commun. 371: 505–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cui, X., Hawari F., Alsaaty S., Lawrence M., Combs C. A., Geng W., Rouhani F. N., Miskinis D., Levine S. J.. 2002. Identification of ARTS-1 as a novel TNFR1-binding protein that promotes TNFR1 ectodomain shedding. J. Clin. Invest. 110: 515–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Andrés, A. M., Dennis M. Y., Kretzschmar W. W., Cannons J. L., Lee-Lin S.-Q., Hurle B., Schwartzberg P. L., Williamson S. H., Bustamante C. D., Nielsen R., et al. NISC Comparative Sequencing Program . 2010. Balancing selection maintains a form of ERAP2 that undergoes nonsense-mediated decay and affects antigen presentation. PLoS Genet. 6: e1001157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zervoudi, E., Papakyriakou A., Georgiadou D., Evnouchidou I., Gajda A., Poreba M., Salvesen G. S., Drag M., Hattori A., Swevers L., et al. 2011. Probing the S1 specificity pocket of the aminopeptidases that generate antigenic peptides. Biochem. J. 435: 411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.López de Castro, J. A. 2018. How ERAP1 and ERAP2 shape the peptidomes of disease-associated MHC-I proteins. Front. Immunol. 9: 2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saulle, I., Ibba S. V., Torretta E., Vittori C., Fenizia C., Piancone F., Minisci D., Lori E. M., Trabattoni D., Gelfi C., et al. 2019. Endoplasmic reticulum associated aminopeptidase 2 (ERAP2) is released in the secretome of activated MDMs and reduces in vitro HIV-1 infection. Front. Immunol. 10: 1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen, H., Li L., Weimershaus M., Evnouchidou I., van Endert P., Bouvier M.. 2016. ERAP1-ERAP2 dimers trim MHC I-bound precursor peptides; implications for understanding peptide editing. Sci. Rep. 6: 28902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saulle, I., Vicentini C., Clerici M., Biasin M.. 2020. An overview on ERAP roles in infectious diseases. Cells 9: 720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Biasin, M., Sironi M., Saulle I., de Luca M., la Rosa F., Cagliani R., Forni D., Agliardi C., lo Caputo S., Mazzotta F., et al. 2013. Endoplasmic reticulum aminopeptidase 2 haplotypes play a role in modulating susceptibility to HIV infection. AIDS 27: 1697–1706. [DOI] [PubMed] [Google Scholar]
- 24.Tenzer, S., Wee E., Burgevin A., Stewart-Jones G., Friis L., Lamberth K., Chang C. H., Harndahl M., Weimershaus M., Gerstoft J., et al. 2009. Antigen processing influences HIV-specific cytotoxic T lymphocyte immunodominance. Nat. Immunol. 10: 636–646. [DOI] [PubMed] [Google Scholar]
- 25.Saulle, I., Vanetti C., Goglia S., Vicentini C., Tombetti E., Garziano M., Clerici M., Biasin M.. 2020. A new ERAP2/iso3 isoform expression is triggered by different microbial stimuli in human cells. Could it play a role in the modulation of SARS-CoV-2 infection? Cells 9: 1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Draenert, R., Le Gall S., Pfafferott K. J., Leslie A. J., Chetty P., Brander C., Holmes E. C., Chang S.-C., Feeney M. E., Addo M. M., et al. 2004. Immune selection for altered antigen processing leads to cytotoxic T lymphocyte escape in chronic HIV-1 infection. J. Exp. Med. 199: 905–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carr, D. F., Bourgeois S., Chaponda M., Takeshita L. Y., Morris A. P., Castro E. M. C., Alfirevic A., Jones A. R., Rigden D. J., Haldenby S., et al. 2017. Genome-wide association study of nevirapine hypersensitivity in a sub-Saharan African HIV-infected population. J. Antimicrob. Chemother. 72: 1152–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lori, E. M., Cozzi-Lepri A., Tavelli A., Mercurio V., Ibba S. V., Lo Caputo S., Castelli F., Castagna A., Gori A., Marchetti G., et al. 2020. Evaluation of the effect of protective genetic variants on cART success in HIV-1-infected patients. J. Biol. Regul. Homeost. Agents 34: 1553–1559. [DOI] [PubMed] [Google Scholar]
- 29.Merlini, E., Tincati C., Biasin M., Saulle I., Cazzaniga F. A., d’Arminio Monforte A., Cappione A. J. I. III, Snyder-Cappione J., Clerici M., Marchetti G. C.. 2016. Stimulation of PBMC and monocyte-derived macrophages via toll-like receptor activates innate immune pathways in HIV-infected patients on virally suppressive combination antiretroviral therapy. Front. Immunol. 7: 614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ortega, P. A. S., Saulle I., Mercurio V., Ibba S. V., Lori E. M., Fenizia C., Masetti M., Trabattoni D., Caputo S. L., Vichi F., et al. 2018. Interleukin 21 (IL-21)/microRNA-29 (miR-29) axis is associated with natural resistance to HIV-1 infection. AIDS 32: 2453–2461. [DOI] [PubMed] [Google Scholar]
- 31.Martinon, F., Pétrilli V., Mayor A., Tardivel A., Tschopp J.. 2006. Gout-associated uric acid crystals activate the NALP3 inflammasome. Nature 440: 237–241. [DOI] [PubMed] [Google Scholar]
- 32.Halle, A., Hornung V., Petzold G. C., Stewart C. R., Monks B. G., Reinheckel T., Fitzgerald K. A., Latz E., Moore K. J., Golenbock D. T.. 2008. The NALP3 inflammasome is involved in the innate immune response to amyloid-β. Nat. Immunol. 9: 857–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cossarizza, A., Chang H.-D., Radbruch A., Acs A., Adam D., Adam-Klages S., Agace W. W., Aghaeepour N., Akdis M., Allez M., et al. 2019. Guidelines for the use of flow cytometry and cell sorting in immunological studies (second edition). Eur. J. Immunol. 49: 1457–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Awad, F., Assrawi E., Jumeau C., Georgin-Lavialle S., Cobret L., Duquesnoy P., Piterboth W., Thomas L., Stankovic-Stojanovic K., Louvrier C., et al. 2017. Impact of human monocyte and macrophage polarization on NLR expression and NLRP3 inflammasome activation. PLoS One 12: e0175336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tsujimoto, M., Aoki K., Ohnishi A., Goto Y.. 2020. Endoplasmic reticulum aminopeptidase 1 beyond antigenic peptide-processing enzyme in the endoplasmic reticulum. Biol. Pharm. Bull. 43: 207–214. [DOI] [PubMed] [Google Scholar]
- 36.Wu, C.-C., Lin J.-D., Chen J.-T., Chang C.-M., Weng H.-F., Hsueh C., Chien H.-P., Yu J.-S.. 2018. Integrated analysis of fine-needle-aspiration cystic fluid proteome, cancer cell secretome, and public transcriptome datasets for papillary thyroid cancer biomarker discovery. Oncotarget 9: 12079–12100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Merah-Mourah, F., Cohen S. O., Charron D., Mooney N., Haziot A.. 2020. Identification of novel human monocyte subsets and evidence for phenotypic groups defined by interindividual variations of expression of adhesion molecules. Sci. Rep. 10: 4397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cros, J., Cagnard N., Woollard K., Patey N., Zhang S.-Y., Senechal B., Puel A., Biswas S. K., Moshous D., Picard C., et al. 2010. Human CD14dim monocytes patrol and sense nucleic acids and viruses via TLR7 and TLR8 receptors. Immunity 33: 375–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Belge, K.-U., Dayyani F., Horelt A., Siedlar M., Frankenberger M., Frankenberger B., Espevik T., Ziegler-Heitbrock L.. 2002. The proinflammatory CD14+CD16+DR++ monocytes are a major source of TNF. J. Immunol. 168: 3536–3542. [DOI] [PubMed] [Google Scholar]
- 40.Chimen, M., Yates C. M., McGettrick H. M., Ward L. S. C., Harrison M. J., Apta B., Dib L. H., Imhof B. A., Harrison P., Nash G. B., Rainger G. E.. 2017. Monocyte subsets coregulate inflammatory responses by integrated signaling through TNF and IL-6 at the endothelial cell interface. J. Immunol. 198: 2834–2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gross, L. Z. F., Sacerdoti M., Piiper A., Zeuzem S., Leroux A. E., Biondi R. M.. 2020. ACE2, the receptor that enables infection by SARS-CoV-2: biochemistry, structure, allostery and evaluation of the potential development of ACE2 modulators. ChemMedChem 15: 1682–1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kapellos, T. S., Bonaguro L., Gemünd I., Reusch N., Saglam A., Hinkley E. R., Schultze J. L.. 2019. Human monocyte subsets and phenotypes in major chronic inflammatory diseases. Front. Immunol. 10: 2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tolouei Semnani, R., Moore V., Bennuru S., McDonald-Fleming R., Ganesan S., Cotton R., Anuradha R., Babu S., Nutman T. B.. 2014. Human monocyte subsets at homeostasis and their perturbation in numbers and function in filarial infection. Infect. Immun. 82: 4438–4446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rossol, M., Kraus S., Pierer M., Baerwald C., Wagner U.. 2012. The CD14(bright) CD16+ monocyte subset is expanded in rheumatoid arthritis and promotes expansion of the Th17 cell population. Arthritis Rheum. 64: 671–677. [DOI] [PubMed] [Google Scholar]
- 45.Narasimhan, P. B., Marcovecchio P., Hamers A. A. J., Hedrick C. C.. 2019. Nonclassical monocytes in health and disease. Annu. Rev. Immunol. 37: 439–456. [DOI] [PubMed] [Google Scholar]
- 46.Saresella, M., La Rosa F., Piancone F., Zoppis M., Marventano I., Calabrese E., Rainone V., Nemni R., Mancuso R., Clerici M.. 2016. The NLRP3 and NLRP1 inflammasomes are activated in Alzheimer’s disease. Mol. Neurodegener. 11: 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.La Rosa, F., Saresella M., Marventano I., Piancone F., Ripamonti E., Al-Daghri N., Bazzini C., Zoia C. P., Conti E., Ferrarese C., Clerici M.. 2019. Stavudine reduces NLRP3 inflammasome activation and modulates amyloid-β autophagy. J. Alzheimers Dis. 72: 401–412. [DOI] [PubMed] [Google Scholar]
- 48.Deerhake, M. E., Biswas D. D., Barclay W. E., Shinohara M. L.. 2019. Pattern recognition receptors in multiple sclerosis and its animal models. Front. Immunol. 10: 2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu, K. K.-L., Cheung S. W.-M., Cheng K. K.-Y.. 2020. NLRP3 inflammasome activation in adipose tissues and its implications on metabolic diseases. Int. J. Mol. Sci. 21: 4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rainone, V., Schneider L., Saulle I., Ricci C., Biasin M., Al-Daghri N. M., Giani E., Zuccotti G. V., Clerici M., Trabattoni D.. 2016. Upregulation of inflammasome activity and increased gut permeability are associated with obesity in children and adolescents. Int. J. Obes. 40: 1026–1033. [DOI] [PubMed] [Google Scholar]
- 51.Bandera, A., Masetti M., Fabbiani M., Biasin M., Muscatello A., Squillace N., Clerici M., Gori A., Trabattoni D.. 2018. The NLRP3 inflammasome is upregulated in HIV-infected antiretroviral therapy-treated individuals with defective immune recovery. Front. Immunol. 9: 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tartey, S., Kanneganti T.-D.. 2019. Differential role of the NLRP3 inflammasome in infection and tumorigenesis. Immunology 156: 329–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Franchi, L., Muñoz-Planillo R., Núñez G.. 2012. Sensing and reacting to microbes through the inflammasomes. Nat. Immunol. 13: 325–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Antonopoulos, C., Dubyak G. R.. 2014. Chemotherapy engages multiple pathways leading to IL-1β production by myeloid leukocytes. OncoImmunology 3: e27499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pontillo, A., Santillo B. T., Duarte A. J., Oshiro T. M.. 2013. Differential inflammasome expression and IL-1β secretion in monocyte-derived dendritic cells differentiated with IL-4 or IFN-α. AIDS Res. Ther. 10: 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Feria, M. G., Taborda N. A., Hernandez J. C., Rugeles M. T.. 2018. HIV replication is associated to inflammasomes activation, IL-1β, IL-18 and caspase-1 expression in GALT and peripheral blood. PLoS One 13: e0192845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fenizia, C., Saulle I., Clerici M., Biasin M.. 2020. Genetic and epigenetic regulation of natural resistance to HIV-1 infection: new approaches to unveil the HESN secret. Expert Rev. Clin. Immunol. 16: 429–445. [DOI] [PubMed] [Google Scholar]
- 58.Fenizia, C., Rossignol J.-F., Clerici M., Biasin M.. 2018. Genetic and immune determinants of immune activation in HIV-exposed seronegative individuals and their role in protection against HIV infection. Infect. Genet. Evol. 66: 325–334. [DOI] [PubMed] [Google Scholar]
- 59.Tomescu, C., Seaton K. E., Smith P., Taylor M., Tomaras G. D., Metzger D. S., Montaner L. J.. 2015. Innate activation of MDC and NK cells in high-risk HIV-1-exposed seronegative IV-drug users who share needles when compared with low-risk nonsharing IV-drug user controls. J. Acquir. Immune Defic. Syndr. 68: 264–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Saulle, I., Biasin M., Gnudi F., Rainone V., Ibba S. V., Lo Caputo S., Mazzotta F., Trabattoni D., Clerici M.. 2016. Short communication: immune activation is present in HIV-1-exposed seronegative individuals and is independent of microbial translocation. AIDS Res. Hum. Retroviruses 32: 129–133. [DOI] [PubMed] [Google Scholar]
- 61.Saveanu, L., Carroll O., Lindo V., Del Val M., Lopez D., Lepelletier Y., Greer F., Schomburg L., Fruci D., Niedermann G., van Endert P. M.. 2005. Concerted peptide trimming by human ERAP1 and ERAP2 aminopeptidase complexes in the endoplasmic reticulum. Nat. Immunol. 6: 689–697. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.