Abstract
ADP-ribosylation is a protein post-translational modification that is critically involved in a wide array of biological processes connected to cell stress responses. Enzymes known as poly-ADP-ribose polymerases (PARPs) catalyze the addition of the ADP-ribose units to amino acids with various side chain chemistries. In particular, the PARP family member PARP1 is responsible for the modification of a large number of proteins and is involved in initiation of the DNA damage response, although the mechanisms through which PARP1 functions are still incompletely understood. The analysis of protein ADP-ribosylation is challenging because PARylation is a low-abundance, labile and heterogeneous protein modification. Recently, we developed an integrative proteomic platform for the site-specific analysis of protein ADP-ribosylation on Asp and Glu residues. Herein, we describe the method, and demonstrate its utility in quantitative characterization of the human Asp- and Glu-ADP-ribosylated proteome.
Keywords: ADP-ribosylation, PARP, DNA damage response, Cancer, NAD+ metabolism
1. Introduction
ADP-ribosylation is a protein post-translational modification (PTM) where either a single (mono-ADP-ribosylation) or multiple (up to 200) (poly-ADP-ribosylation) unit(s) of ADP-ribose is covalently attached to a target protein (D’Amours, Desnoyers, D’Silva, & Poirier, 1999a; Gibson & Kraus, 2012; Hottiger, 2015a). It has been reported that a number of enzymes, including poly-ADP-ribose polymerases (PARPs), bacterial toxins and several NAD+-dependent Sirtuins, could catalyze this reaction, where they transfer ADP-ribose molecules from the cofactor NAD+ to the acceptor protein (D’Amours, Desnoyers, D’Silva, & Poirier, 1999b; Gibson & Kraus, 2012; Hawse & Wolberger, 2009; Hottiger, 2015b; Krueger & Barbieri, 1995; Schreiber, 2006) (Figure 1). Among these enzymes, the PARP family is composed of 17 members, with PARP1, PARP2 and Tankyrases known to catalyze protein PARylation (Vyas et al., 2014). Other PARP enzymes possess either mono-ADP-ribosylation activity or no enzymatic activity (Collier, 2001; Vyas et al., 2014). PARP1 is arguably the best studied member of the PARP family. It is mainly localized in nucleus, and is known to play critical roles in regulating several cellular processes, including DNA damage response, RNA splicing, cell division, transcriptional regulation and apoptosis (Ali et al., 2012; Caldecott, Aoufouchi, Johnson, & Shall, 1996; Langelier, Planck, Roy, & Pascal, 2011a, 2012; Lautier, Lagueux, Thibodeau, Menard, & Poirier, 1993; C. Liu, Vyas, Kassab, Singh, & Yu, 2017; Ludwig, Behnke, Holtlund, & Hilz, 1988; Yamanaka, Penning, Willis, Wasson, & Carson, 1988). Under the basal state, PARP1 is often quiescent and there is generally a very low level of protein PARylation (Zhen, Zhang, & Yu, 2017). During genotoxic stress, PARP1 is rapidly recruited to DNA lesions and is responsible for the synthesis of more than 90% of the protein-linked PAR chains (Shieh et al., 1998). From a structural point of view, when DNA damage occurs, PARP1 recognizes DNA lesions by its Zinc Finger motifs (Caldecott et al., 1996; Langelier, Planck, Roy, & Pascal, 2011b). This binding event induces a dramatic conformational change of PARP1, resulting in a remodeling of an inhibitory motif near the catalytic domain (Ali et al., 2012; Langelier et al., 2012). The enzymatic activity of PARP1 is then stimulated to modify a large number of target proteins, including itself (Alvarez-Gonzalez & Althaus, 1989a; D’Amours et al., 1999a; Haince et al., 2008; M. Y. Kim, Mauro, Gevry, Lis, & Kraus, 2004; Kraus & Lis, 2003; Simonin, Poch, Delarue, & de Murcia, 1993; Wielckens, George, Pless, & Hilz, 1983).
Figure 1.
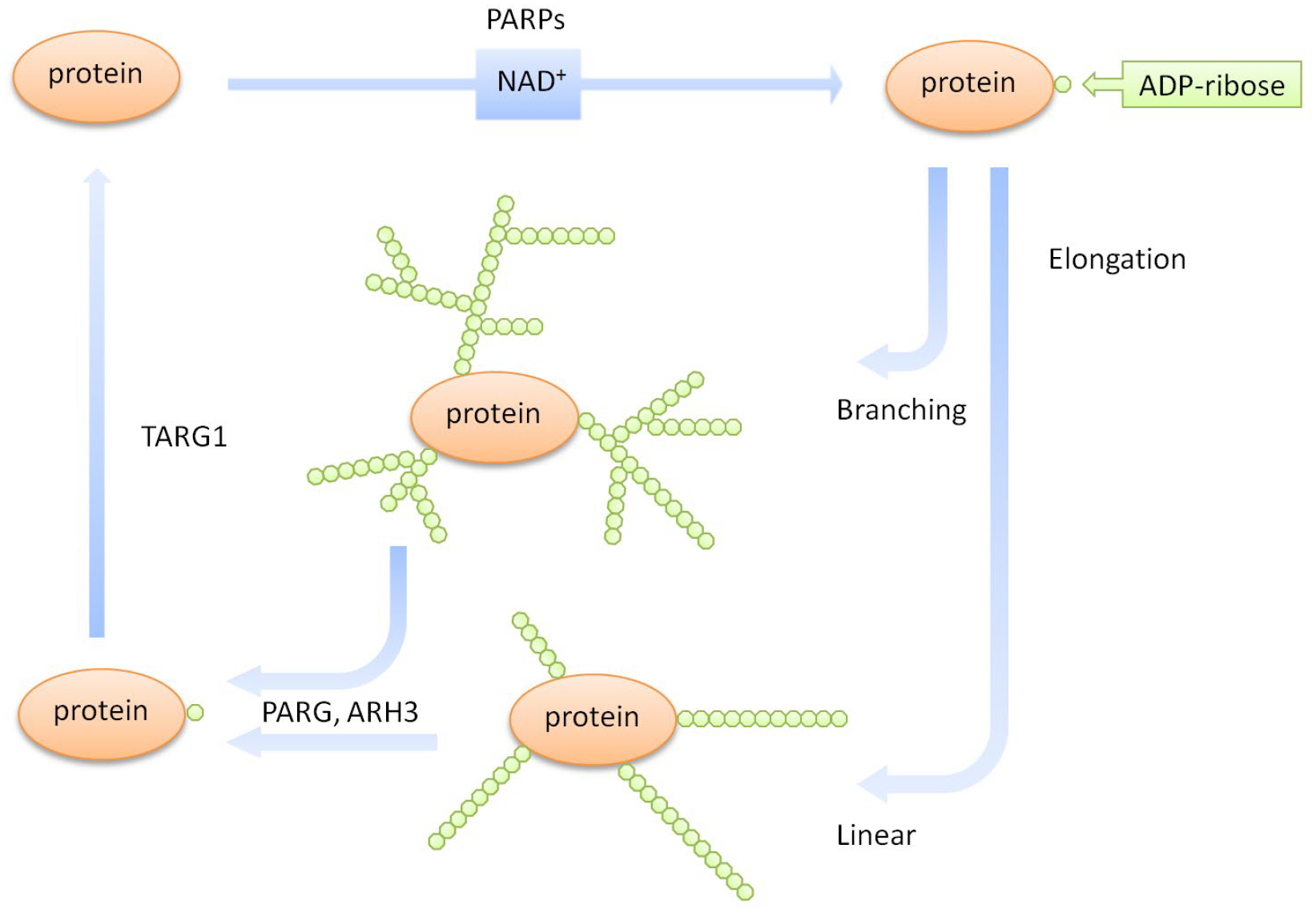
The chemistry of poly-ADP-ribose synthesis and degradation. ADP-ribosylation is catalyzed by PARPs using NAD+ as a cofactor. ADP-ribose monomers are joined in a linear and/or branched fashion to form a poly-ADP-ribose chain. PAR polymers are degraded by several enzymes, including PARG, ARH3, and TARG1.
Protein PARylation is a dynamic modification. It has been shown that protein-linked PAR chains have a very short half-life, indicating the presence of an efficient mechanism for PAR-catabolism in cells (Alvarez-Gonzalez & Althaus, 1989b). It has been shown that PAR can be degraded by various enzymes, including PARG (Poly-ADP-ribose glycohydrolase), TARG1/C6orf130, ARH (ADP-ribosyl hydrolase) and proteins with the NUDIX (nucleoside diphosphates linked to moiety-X) domain and ENPP1 (ectonucleotide pyrophosphatase/phosphodiesterase 1) (Daniels, Thirawatananond, Ong, Gabelli, & Leung, 2015; Fontana et al., 2017; Niere et al., 2012; Sharifi et al., 2013; Slade et al., 2011). Among these proteins, PARG is probably one of the best studied and most dominant PAR-catabolizing enzyme. It is responsible for the hydrolysis of the endo- and exo-glycosidic linkages within PAR chains, generating free ADP-ribose monomers (Min & Wang, 2009). Although PARG is a very efficient enzyme that rapidly degrades PAR, it does not remove the terminal ADP-ribose unit that is linked to the side chain of an acceptor amino acid (Slade et al., 2011). The resulting MARylated proteins are instead digested by enzymes including TARG1 to reverse the mono-ADP-ribosylation (Sharifi et al., 2013). In addition to PARG and TARG1, recent studies have identified a number of other PAR-degrading enzymes (Daniels, Thirawatananond, et al., 2015; Fontana et al., 2017; Niere et al., 2012). For example, ARH3 is a Poly-ADP-ribose glycohydrolase that was shown to cleave the Serine-ADP-ribose bond (Fontana et al., 2017). In summary, these various PARPs and PAR-catabolizing enzymes function in concerted efforts to fine-tune cellular ADP-ribosylation homeostasis (Figure 1).
The biological function of protein ADP-ribosylation can be explored by studying how it affects the biochemical characteristics of the acceptor protein (Miyamoto, Kakizawa, & Hashizume, 1999; Rouleau, Patel, Hendzel, Kaufmann, & Poirier, 2010) (Figure 2). On one side, due to structural similarity between PAR and nucleic acids (both of them are bulky, charged, and flexible), PARylation can prevent a DNA- or RNA-binding protein from interacting with its nucleic acid targets (Ferro & Olivera, 1982a). In this case, it has been shown that automodified PARP1 is dissociated from DNA due to charge repulsion and steric hindrance (Ferro & Olivera, 1982b; Kanai et al., 2007; Mendoza-Alvarez & Alvarez-Gonzalez, 1993). On the other side, PAR chains can also serve as a scaffold to promote protein interactions (Gibson & Kraus, 2012). Recent studies have identified a number of PAR-binding motifs (PBZ), including WWE, PAR-binding zinc finger (PBZ), BRCA1 C terminus (BRCT), macrodomain, and oligonucleotide/oligosaccharide-binding (OB)-fold (Ahel et al., 2008; Aravind, 2001; Callow et al., 2011; Gagne et al., 2008; Gibson & Kraus, 2012; Han, Li, & Fu, 2011; Kang et al., 2011; Li, Lu, Yang, Wang, & Yu, 2013; Li & Yu, 2013; Masson et al., 1998; Wang et al., 2012; F. Zhang, Chen, Li, & Yu, 2014a, 2014b; F. Zhang, Shi, Chen, Bian, & Yu, 2015; Y. Zhang et al., 2011).
Figure 2.
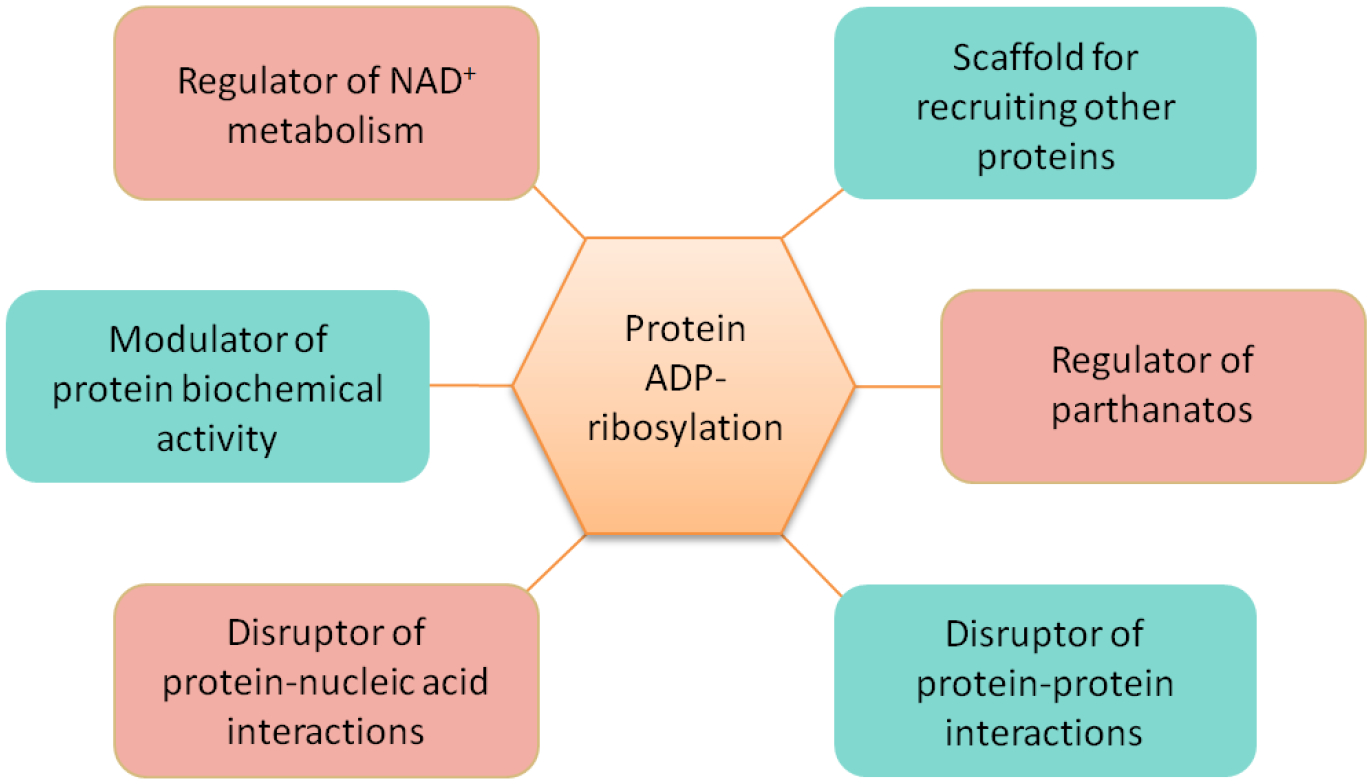
Cellular functions of protein ADP-ribosylation.
The tight link between PARP1 and DNA damage response provides the rationale for the clinical development of PARP1 inhibitors (Virag & Szabo, 2002). Indeed, four PARP1 inhibitors (Olaparib, Rucaparib, Niraparib and Talazoparib) have been approved by the FDA for the treatment of human malignancies (in particular, BRCA1/2mut ovarian and/or breast cancers) (Ledermann et al., 2012; Litton et al., 2018; Mirza et al., 2016; Swisher et al., 2017). Despite these very encouraging progresses in the clinic, the downstream signaling pathways of PARP1 and other PARP family members are still poorly characterized. Besides insights into the biological function of protein PARylation, approaches that allow unbiased and quantitative analysis of the ADP-ribosylated proteome would also greatly facilitate the mechanistic study of the PARP1 inhibitors (Bock, Todorova, & Chang, 2015; Gupte, Liu, & Kraus, 2017; Olsen & Mann, 2013). However, a number of technical challenges are known to be associated with the analysis of ADP-ribosylation (Y. Zhang, Wang, Ding, & Yu, 2013; Zhen & Yu, 2018; Zhen et al., 2017) (Figure 3). First, PARylation is a heterogeneous PTM without a defined mass shift for the modified amino acid residue. Second, PARylated proteins are usually of low abundance. Third, ADP-ribose moieties are linked to amino acids with distinct side chain chemistries. Fourth, the topological features of PAR polymers are highly complex, with chains elongated in both linear and branched manners (D’Amours et al., 1999a). Finally, the adenosine moiety, pyrophosphate bond, and the amino acid linkage are chemically labile. ADP-ribosylation is also a highly unstable PTM that readily decomposes under conventional tandem mass spectrometry (MS) conditions (Hengel & Goodlett, 2012; Matic, Ahel, & Hay, 2012). A number of approaches have been designed to overcome these difficulties (C. A. Vivelo & A. K. Leung, 2015). For example, affinity purification reagents, including the Af1521 macrodomain, 10H antibodies, ADP-ribose binding modules and boronate resins, have been employed to isolate and enrich PARylated proteins for their subsequent MS identification (Daniels, Ong, & Leung, 2015; Forst et al., 2013; Martello et al., 2016; Timinszky et al., 2009; C. A. Vivelo & A. K. L. Leung, 2015). More recently, various quantitative mass spectrometry approaches have been developed to address many technical challenges associated with the site-specific analysis of the PARylated proteome (C. A. Vivelo & A. K. Leung, 2015). For example, in order to tackle the heterogeneous nature of PARylation, Tao et al., used a PARP1 mutant (E988Q) that catalyzes only MARylation, but not PARylation. Using LC-MS experiments, they were able to identify a number of automodification sites on PARP1 (Tao, Gao, & Liu, 2009). Alternatively, PAR chains can also be digested by PARG, Nudix hydrolases and phosphodiesterases, which convert PARylated peptides into species that possess a defined mass addition to the modified amino acid residue (Chapman, Gagne, Poirier, & Goodlett, 2013; Syka, Coon, Schroeder, Shabanowitz, & Hunt, 2004). Finally, Leidecker et al., showed that ADP-ribose could be preserved using more gentle MS fragmentation techniques (e.g., ETD, electron transfer dissociation), and they reported Ser as a new acceptor amino acid of ADP-ribosylation (Leidecker et al., 2016).
Figure 3.
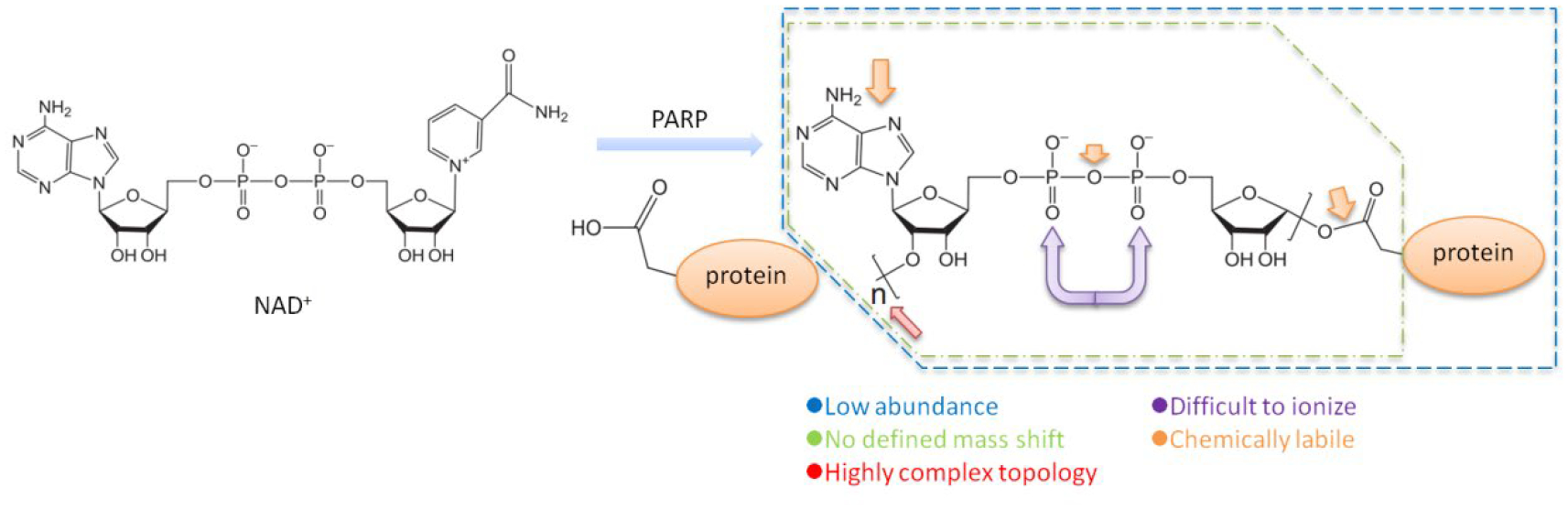
Challenges for the study of PARylation.
We recently developed an integrated strategy for the site-specific characterization of the Asp- and Glu-ADP-ribosylated proteome (Y. Zhang et al., 2013) (Figure 4). In this protocol, the PARylated peptides are enriched using boronate affinity chromatography based on the specific interaction between boron and the 1,2-cis-diol moiety in ADP-ribose (X. C. Liu & Scouten, 2000). PARylated peptides are then eluted by NH2OH treatment, during which ADP-ribosylated Asp and Glu residues are converted into hydroxamic acids (Moss, Yost, & Stanley, 1983). This mass tag produces a defined mass shift (+15.0109 Da) that is highly stable and is amenable to MS analysis using conventional fragmentation methods (e.g., collision-induced dissociation, CID). This chapter describes the protocol (including sample preparation, data acquisition and bioinformatics) that is used to identify and quantify the Asp- and Glu-ADP-ribosylated proteome in cells.
Figure 4.
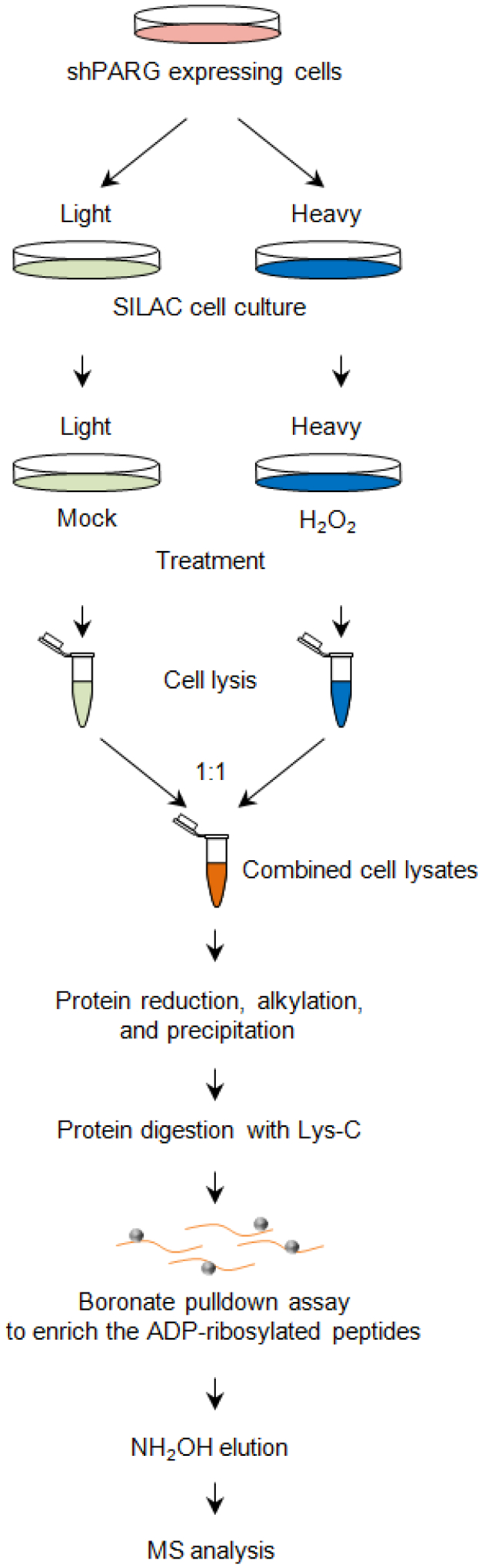
Workflow for the site-specific characterization of protein Asp- and Glu-ADP-ribosylation by quantitative mass spectrometry.
2. Materials
2.1. Cell line
HEK293TD (packaging cell line)
2.2. Plasmids
VSVG (envelope plasmid)
Δ8.9 (packaging plasmid)
pLKO.1-puro-shPARG (Sigma)
2.3. Reagents
Dialyzed fetal bovine serum (FBS) (Fisher Scientific)
Light lysine (12C614N2) (Sigma)
Light arginine (12C614N4) (Sigma)
Heavy lysine (13C615N2) (Sigma)
Heavy arginine (13C615N4) (Sigma)
SILAC medium (Thermo Scientific)
DMEM (Thermo Scientific)
Lipofectamine 2000 (Invitrogen)
Opti-MEM (Gibco)
Polybrene (Sigma), 8 mg/ml
Puromycin (Sigma), 2 mg/ml, sterilized through a 0.22-μm filter
Anti-PARG antibody (Millipore Sigma)
BCA protein assay kit (Fisher Scientific)
1 M Dithiothreitol (DTT) (Fisher Scientific)
0.5 M Iodoacetamide (IAA) (Sigma)
Methanol, HPLC grade (Fisher Scientific)
Chloroform, HPLC grade (Sigma)
Water, HPLC grade (Fisher Scientific)
Lysyl endopeptidase (Lys-C) (Wako Chemicals), 10 AU resuspended in 50 mM acetic acid (for a 2 μg/μl stock) and stored at −80 °C
Trypsin (Thermo Scientific), MS grade, 1 μg/μl, stored at −80 °C
m-Aminophenylboronic acid-agarose beads (Sigma)
2.4. Solutions
Phosphate-buffered saline (PBS): 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4 (pH 7.4), sterilized by autoclaving
0.25% Trypsin-EDTA solution
2 M Hydrogen peroxide (Fisher Scientific), freshly-made
SDS lysis buffer: 1% SDS, 10 mM HEPES (pH 7.0), 2 mM MgCl2, 500 U universal nuclease (Sigma)
200 mM HEPES (Fisher Scientific) (pH 8.5)
0.5 M NH2OH (Sigma) in 200 mM HEPES buffer (pH 8.5)
Boronate bead wash buffer: 1% SDS, 200 mM HEPES (pH 8.5)
200 mM HEPES (pH 8.8)
SDS wash buffer: 1% SDS, 200 mM HEPES (pH 8.5), 150 mM NaCl
HEPES wash buffer: 200 mM HEPES (pH 8.5), 150 mM NaCl
2 M NH2OH in 200 mM HEPES buffer (pH 8.5)
20% Trifluoroacetic acid (TFA) (Fisher Scientific)
0.1% TFA
0.1% formic acid (FA) (Fisher Scientific) in 40% acetonitrile (ACN) (Fisher Scientific)
15.0.1% FA in H2O
HPLC solvent A: 0.1% FA in H2O
HPLC solvent B: 90% ACN, 0.1% FA in H2O
2.5. Equipment
LTQ-Velos Pro Orbitrap Mass Spectrometer
Evolution 60S UV-Visible Spectrophotometer
Thermo EASY-nLC 1200 Liquid Chromatography System
Hand-pulled fused silica microcapillary column (0.075 mm ID × 150 mm) filled with reverse-phase Magic C18AQ beads, 3 μm, 200 Å
10-cc Syringes
OASIS HLB Extraction Cartridges, 10 mg
Vacuum manifold
Standard lab incubator
Vari mix platform rocker
Rotamix rotator
Vortex mixer
Microcentrifuge
Vacufuge
3. Protocols
Our recently developed approach overcomes different aspects of abovementioned technical challenges for the study of Asp- and Glu-ADP-ribosylation. ADP-ribosylation of these acidic residues have been shown to represent the major form of cellular ADP-ribosylation, at least under oxidative stress conditions (Adamietz & Hilz, 1976). The ester bond between Asp/Glu and ADP-ribose is sensitive to nucleophilic attack by NH2OH, and this reaction converts an ADP-ribosylated-D/E residue into a hydroxamic acid derivative (with an addition of 15.0109 Da) (Moss et al., 1983). This stable mass tag can be readily pinpointed by conventional tandem mass spectrometry (MS) experiments (e.g., collision-induced dissociation). The workflow for this protocol is shown in Figure 4.
3.1. Preparation of shPARG-expressing cells
Grow HEK293TD cells to ~80% confluency. Co-transfect the HEK293TD cells with the packaging vector (Δ8.9), envelope vector (VSVG), and pLKO.1-puro-shPARG (see Note 1) at a ratio of 3:3:4 with Lipofectamine 2000 (see Note 2).
At 48h after transfection, remove the virus-containing supernatants and filter the viral supernatants through 0.45-μm filters to remove cell debris. Add fresh medium to the cells. Infect the target cells (e.g., HCT116) with the virus-containing medium, which includes the filtered virus supernatants, 8 μg/ml polybrene, and fresh growth medium (see Note 3).
After 24h, remove the virus-containing supernatants from the HEK293TD cells and filter the viral supernatants through 0.45-μm filters to remove cell debris. Re-infect the target cells with the second batch of virus.
After 48h, select for the cells that stably express the shPARG construct by the addition of puromycin to 2 μg/ml (see Note 4). Grow cells for another 2 days. To confirm the efficiency of PARG depletion, analyze the cell lysates by immuno-blotting with the anti-PARG antibody.
3.2. SILAC cell culture
Separate the shPARG-expressing cells into two aliquots. Grow one sample in the light Lys/Arg SILAC medium and one sample in the heavy Lys/Arg SILAC medium. Both SILAC media contain 10% dialyzed FBS. Passage the cells every 2 days for at least five generations. Cells with an incorporation rate of over 97% for the heavy amino acids are used for the following experiment.
3.3. Sample preparation for mass spectrometry analysis
Grow the SILAC-labeled cells to ~80% confluency in culture dishes. Treat the heavy and light cells with the appropriate stimuli (see Note 5). After the treatment, discard the culture medium and wash each dish with 10 ml ice-cold PBS twice and remove the remaining PBS.
Lyse the cells with 1 ml SDS lysis buffer per dish (see Note 6). Incubate the dishes for 10 min on a platform rocker at room temperature. Collect cell lysates in 15-ml centrifuge tubes. Determine protein concentrations of each sample with a BCA protein assay. Combine 25 mg of heavy lysate with 25 mg of light lysate for each experimental condition.
To reduce the disulfide bonds, add DTT (final concentration of 3 mM) to the lysates, vortex well, and incubate for 20 min at room temperature. Alkylate the cysteines by adding IAA (to a final concentration of 50 mM), vortex well and incubate for 20 min in the dark.
To precipitate the proteins, add 4 volumes of methanol to each tube and vortex well. Add 1 volume of chloroform (relative to the original lysate volume) and vortex well. Finally, add 3 volumes of water (relative to the original lysate volume), vortex well, and centrifuge at the maximum speed of the microcentrifuge (i.e., 7830 rpm) for 15 min. Carefully collect the protein layer. Wash the pellets with methanol (4 volumes relative to the initial volume of the lysate sample), and centrifuge at the maximum speed of the microcentrifuge (i.e., 7830 rpm) for 5 min and remove the methanol completely.
3.4. Boronate bead-based pulldown assay
Dissolve the protein pellets in the SDS lysis buffer, mix well, and sonicate to solubilize the proteins completely (see Note 7). Digest the proteins by addition of a 2 μg/μl stock of Lys-C (use a 1:100 enzyme : protein substrate ratio). Incubate the samples by rotating for 1.5 h at room temperature.
Prepare the boronate beads (200 μl beads per 150-mm dish of cells). Add 1 ml of the bead mixture to the necessary number of Eppendorf tubes. Centrifuge beads at 3500 rpm in a microfuge for 2 min, and remove the buffer completely. Wash each tube of the boronate beads with 1 ml of 200 mM HEPES (pH 8.5) twice. After each wash, centrifuge the tubes at 3500 rpm for 2 min and remove the buffer completely. Prepare a solution of 0.5 M NH2OH in 200 mM HEPES (pH 8.5) and add 1 ml to each tube of beads. Incubate at room temperature for 5 min. Centrifuge the tubes at 3500 rpm for 2 min and remove the buffer completely. Then wash three times with 1 ml 200 mM HEPES (pH 8.5) and once with 1 ml boronate bead wash buffer. Centrifuge the tubes at 3500 rpm for 2 min and remove the solution completely after each wash (see Note 8). Add 1 ml boronate bead wash buffer for 200 μl of beads to resuspend the boronate beads.
After the 1.5-h digestion of the proteins with Lys-C, adjust the pH of the cell lysate samples to 8.5 with 200 mM HEPES (pH 8.8) (see Note 9). Add 500μl of bead suspension to each sample, and rotate end-to-end at room temperature for 1 h. Then add the other half of the pre-washed beads, and rotate end-to-end at room temperature for 1 h.
Centrifuge samples at 3500 rpm for 2 min and transfer each sample to a new 2-ml Eppendorf tube. Wash the beads with 1 mL SDS wash buffer for seven times, with the HEPES wash buffer for ten times, and with 200 mM HEPES (pH 8.5) once. After each wash, centrifuge the beads at 3500 rpm for 2 min, and remove the buffer completely. Resuspend the beads in 1.3 ml of 2 M NH2OH solution (in 200 mM HEPES buffer, pH 8.5) containing 1 μl Lys-C and 1μl trypsin (see Note 10). Rotate end-to-end overnight at room temperature.
Centrifuge at 3500 rpm for 2 min, and transfer the supernatants into new 2-ml tubes. Incubate the beads twice with 1 ml 200 mM HEPES buffer (pH 8.5) with end-to-end rotation at room temperature for 10 min. Centrifuge the beads at 3500 rpm for 2 min, and combine the supernatants. Adjust the pH values of the combined supernatants to between pH 2 and pH 3 with 20% TFA, and mix well. Use OASIS HLB-cartridges to desalt the eluted peptides. Lyophilize the eluates completely, and dissolve each sample in 10 μl of 0.1% FA.
3.5. Mass spectrometry analysis
Use a hand-pulled fused silica microcapillary column to separate the peptides. Use 75-μm ID × 15 cm analytical columns (New Objective) packed with Maccel C18 3-μm, 200-Å beads (The Nest Group). Elute with a 75-min linear gradient ranging from 7% to 32% ACN in 0.1% FA at a flow rate of 300 nl/min. Analyze the samples on an LTQ-Velos Pro Orbitrap mass spectrometer or any mass spectrometers with conventional CID capabilities (Olsen et al., 2009). The isolation window and the minimal signal threshold for MS/MS experiments should be set to be 2 Th and 500 counts, respectively. The AGC for the Orbitrap (MS1) and the ion trap (MS2) is set to be 1,000,000 and 7,500, respectively. Only peptides with +2 or higher charge states are selected for MS2 experiments. The normalized collision energy is set to be 35 eV, with a minimal signal threshold of 500. Dynamic exclusion is enabled with an exclusion duration of 60 sec. The ReAdW.exe programs should be used to convert the raw files into the mzXML format (https://sourceforge.net/projects/sashimi/files/).
Search the MS/MS spectra against the human Uniprot protein database (or the protein sequence database appropriate for the samples) and its reversed complement using the Sequest (Rev28) algorithm. Search parameters should allow for dynamic modifications of 15.0109 Da to aspartic acid and glutamic acid, a static modification of 57.02146 Da on cysteine, and a variable modification of 15.994915 Da on methionine. The stable isotopes on arginine and lysine should be set as 10.00827 Da and 8.01420 Da, respectively.
Filter the search results to include <1% matches to the reverse database by the linear discriminator function using parameters including Xcorr, dCN, missed cleavage, charge state (exclude 1+ peptides), mass accuracy, peptide length, and fraction of ions matched to MS/MS spectra as previously described (Huttlin et al., 2010). Use appropriate algorithms to assess the localization of ADP-ribosylation sites. For example, the ModScore evaluate site-specific fragment ions and the localized sites have scores of ≥ 13 (P ≤ 0.05) (W. Kim et al., 2011).
4. Notes
This protocol can be used with any cell lines that can be infected with lentiviruses.
A decrease in the activity of PARG stabilizes PARylated proteins (Gagne et al., 2008; Kawamitsu et al., 1984; Mortusewicz, Fouquerel, Ame, Leonhardt, & Schreiber, 2011; Petesch & Lis, 2012). Here we used an shRNA targeting PARG. Other approaches to inactivate the function of PARG have been described (Hengel, Shaffer, Nunn, & Goodlett, 2009; Laing, Koch-Nolte, Haag, & Buck, 2011; Margarit, Davidson, Frego, & Stebbins, 2006; Messner et al., 2010; Mueller-Dieckmann et al., 2006; Oka, Kato, & Moss, 2006; Rosenthal et al., 2011; Tao et al., 2009). For example, (1) levels of endogenous PARylated proteins can be increased by pre-treating the cells with the PARG inhibitor (PDD00017273 (Gravells, Grant, Smith, James, & Bryant, 2017; James et al., 2016)) before the activation of PARylation; (2) a PARG inhibitor ADP-HPD can be added to the lysis buffer (Slama et al., 1995), or (3) siRNA can be used to inhibit PARG expression (Jungmichel et al., 2013a; Y. Zhang et al., 2013).
Polybrene is used to increase the infection efficiency.
Cell lines differ in their sensitivity to puromycin; therefore, the optimal concentration of puromycin should be determined in advance for the cell line used.
H2O2 is a genotoxic agent that is known to activate PARP1. Other treatment conditions could be applied to perturb the PARylated proteome. For example, specific PARP1 inhibitors could be used to assess how the global PARylated proteome responds to PARP1 inhibition.
A recent study showed that PARP1 could be activated by sheared DNA generated during cell lysis, leading to the artificial formation of PARylated proteins (Jungmichel et al., 2013a). Therefore, a denaturing buffer (e.g., the SDS buffer) should be used to inactivate PARP (and also PAR-degrading enzymes) during cell lysis. Alternatively, PARP1 inhibitors (and also PARG inhibitors) could be added to the lysis buffer to prevent non-physiological PARylation (Jungmichel et al., 2013b).
The water in the water bath should be changed several times during the sonication to ensure that an appropriate temperature is maintained.
Complete removal of the residual NH2OH is necessary for efficient peptide binding.
The boronate affinity enrichment is optimal at pH 8.5.
After the treatment with NH2OH, the resulting moiety is small and does not suppress the ionization of the modified peptides.
5. Summary and Perspectives
Recent advances in mass spectrometry-based proteomic technologies have allowed the characterization of the ADP-ribosylated proteome in a global, quantitative and site-specific manner (Chapman et al., 2013; Ogata, Ueda, & Hayaishi, 1980; Ogata, Ueda, Kagamiyama, & Hayaishi, 1980; Tao et al., 2009; Y. Zhang et al., 2013). These progresses have greatly facilitated the study of this PTM, leading to a fundamental understanding of the functional role this PTM in many pathophysiological processes (Y. Zhang et al., 2013). In particular, proteome-wide studies of PARylation have shown that in addition to DNA damage response, PARylated proteins and PAR-binding proteins are involved in a wide variety of cellular processes linked to cell stress responses, including transcription control, RNA metabolism, and epigenetic regulation (Cohen-Armon et al., 2007; Gibson et al., 2016).
Despite these abovementioned progresses, it is important to characterize PARylation-mediated signaling events under both stressed as well as unstressed conditions. Furthermore, many key questions still remain in the field of PARP and ADP-ribosylation biology: (1) How to define the specificity of PARPs and PAR degrading enzymes? (2) What are the biological roles of a specific ADP-ribosylated site? (3) How does ADP-ribosylation influence protein functions? (4) What are the signaling networks downstream of PARPs? (5) What are the potential functions of specific PAR chain topologies (e.g., lengths, linear vs. branched, etc.). It is expected that new approaches for characterization of the ADP-ribosylated proteome will provide the cornerstone to address these questions and to dissect the functional role of this critically important PTM.
Table 1.
FDA-approved PARP1 inhibitors.
| PARP1 inhibitors | Year of FDA Approval | Disease Indication |
|---|---|---|
| Olaparib (AZD-22S1) | 2014 | Ovarian cancer (BRCA mutated) |
| 2016 | ATM-mutated, castration-resistant prostate cancer (BRCA mutated) | |
| 2018 | HER2-negative, advanced breast cancer (germline BRCA) | |
| Rucaparib (AG-014699) | 2016 | Ovarian cancer (BRCA mutated) |
| Niraparib (MK-4827) | 2017 | Platinum-sensitive, relapsed ovarian cancer in post-chemotherapy maintenance |
| Talazoparib (BMN-673) | 2018 | Germline BRCA-mutated, HER2-negative locally advanced or metastatic breast cancer |
Olaparib has never been approved for prostate cancer.
Acknowledgments
This work was supported in part by grants from the Welch Foundation (I-1800 to Y.Y.) and NIH (GM122932 to Y.Y.).
References
- Adamietz P, & Hilz H (1976). Poly(adenosine diphosphate ribose) is covalently linked to nuclear proteins by two types of bonds. Hoppe Seylers Z Physiol Chem, 357(4), 527–534. [DOI] [PubMed] [Google Scholar]
- Ahel I, Ahel D, Matsusaka T, Clark AJ, Pines J, Boulton SJ, & West SC (2008). Poly(ADP-ribose)-binding zinc finger motifs in DNA repair/checkpoint proteins. Nature, 451(7174), 81–85. doi: nature06420 [pii] 10.1038/nature06420 [DOI] [PubMed] [Google Scholar]
- Ali AAE, Timinszky G, Arribas-Bosacoma R, Kozlowski M, Hassa PO, Hassler M, … Oliver AW (2012). The zinc-finger domains of PARP1 cooperate to recognize DNA strand breaks. Nat Struct Mol Biol, 19(7), 685–692. doi: 10.1038/nsmb.2335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Gonzalez R, & Althaus FR (1989a). Poly(ADP-ribose) catabolism in mammalian cells exposed to DNA-damaging agents. Mutation Research, 218(2), 67–74. [DOI] [PubMed] [Google Scholar]
- Alvarez-Gonzalez R, & Althaus FR (1989b). Poly(ADP-ribose) catabolism in mammalian cells exposed to DNA-damaging agents. Mutat Res, 218(2), 67–74. [DOI] [PubMed] [Google Scholar]
- Aravind L (2001). The WWE domain: a common interaction module in protein ubiquitination and ADP ribosylation. Trends Biochem Sci, 26(5), 273–275. doi: S0968–0004(01)01787-X [pii] [DOI] [PubMed] [Google Scholar]
- Bock FJ, Todorova TT, & Chang P (2015). RNA Regulation by Poly(ADP-Ribose) Polymerases. Molecular Cell, 58(6), 959–969. doi: 10.1016/j.molcel.2015.01.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldecott KW, Aoufouchi S, Johnson P, & Shall S (1996). XRCC1 polypeptide interacts with DNA polymerase beta and possibly poly (ADP-ribose) polymerase, and DNA ligase III is a novel molecular ‘nick-sensor’ in vitro. Nucleic Acids Res, 24(22), 4387–4394. doi: 10.1093/nar/24.22.4387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callow MG, Tran H, Phu L, Lau T, Lee J, Sandoval WN, … Costa M (2011). Ubiquitin ligase RNF146 regulates tankyrase and Axin to promote Wnt signaling. PLoS One, 6(7), e22595. doi: 10.1371/journal.pone.0022595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman JD, Gagne JP, Poirier GG, & Goodlett DR (2013). Mapping PARP-1 auto-ADP-ribosylation sites by liquid chromatography-tandem mass spectrometry. J Proteome Res, 12(4), 1868–1880. doi: 10.1021/pr301219h [DOI] [PubMed] [Google Scholar]
- Cohen-Armon M, Visochek L, Rozensal D, Kalal A, Geistrikh I, Klein R, … Seger R (2007). DNA-independent PARP-1 activation by phosphorylated ERK2 increases Elk1 activity: a link to histone acetylation. Mol Cell, 25(2), 297–308. doi: 10.1016/j.molcel.2006.12.012 [DOI] [PubMed] [Google Scholar]
- Collier RJ (2001). Understanding the mode of action of diphtheria toxin: a perspective on progress during the 20th century. Toxicon, 39(11), 1793–1803. [DOI] [PubMed] [Google Scholar]
- D’Amours D, Desnoyers S, D’Silva I, & Poirier GG (1999a). Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem J, 342 (Pt 2), 249–268. [PMC free article] [PubMed] [Google Scholar]
- D’Amours D, Desnoyers S, D’Silva I, & Poirier GG (1999b). Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochemical Journal, 342, 249–268. [PMC free article] [PubMed] [Google Scholar]
- Daniels CM, Ong SE, & Leung AK (2015). The Promise of Proteomics for the Study of ADP-Ribosylation. Molecular Cell, 58(6), 911–924. doi: 10.1016/j.molcel.2015.06.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels CM, Thirawatananond P, Ong SE, Gabelli SB, & Leung AK (2015). Nudix hydrolases degrade protein-conjugated ADP-ribose. Sci Rep, 5, 18271. doi: 10.1038/srep18271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferro AM, & Olivera BM (1982a). Poly(ADP-ribosylation) in vitro. Reaction parameters and enzyme mechanism. J Biol Chem, 257(13), 7808–7813. [PubMed] [Google Scholar]
- Ferro AM, & Olivera BM (1982b). Poly(ADP-ribosylation) in vitro. Reaction parameters and enzyme mechanism. Journal of Biological Chemistry, 257(13), 7808–7813. [PubMed] [Google Scholar]
- Fontana P, Bonfiglio JJ, Palazzo L, Bartlett E, Matic I, & Ahel I (2017). Serine ADP-ribosylation reversal by the hydrolase ARH3. Elife, 6. doi: 10.7554/eLife.28533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forst AH, Karlberg T, Herzog N, Thorsell AG, Gross A, Feijs KL, … Luscher B (2013). Recognition of mono-ADP-ribosylated ARTD10 substrates by ARTD8 macrodomains. Structure, 21(3), 462–475. doi: 10.1016/j.str.2012.12.019 [DOI] [PubMed] [Google Scholar]
- Gagne JP, Isabelle M, Lo KS, Bourassa S, Hendzel MJ, Dawson VL, … Poirier GG (2008). Proteome-wide identification of poly(ADP-ribose) binding proteins and poly(ADP-ribose)-associated protein complexes. Nucleic Acids Res, 36(22), 6959–6976. doi: 10.1093/nar/gkn771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson BA, & Kraus WL (2012). New insights into the molecular and cellular functions of poly(ADP-ribose) and PARPs. Nat Rev Mol Cell Biol, 13(7), 411–424. doi: 10.1038/nrm3376 [DOI] [PubMed] [Google Scholar]
- Gibson BA, Zhang Y, Jiang H, Hussey KM, Shrimp JH, Lin H, … Kraus WL (2016). Chemical genetic discovery of PARP targets reveals a role for PARP-1 in transcription elongation. Science, 353(6294), 45–50. doi: 10.1126/science.aaf7865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravells P, Grant E, Smith KM, James DI, & Bryant HE (2017). Specific killing of DNA damage-response deficient cells with inhibitors of poly(ADP-ribose) glycohydrolase. DNA Repair (Amst), 52, 81–91. doi: 10.1016/j.dnarep.2017.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupte R, Liu Z, & Kraus WL (2017). PARPs and ADP-ribosylation: recent advances linking molecular functions to biological outcomes. Genes and Development, 31(2), 101–126. doi: 10.1101/gad.291518.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haince JF, McDonald D, Rodrigue A, Dery U, Masson JY, Hendzel MJ, & Poirier GG (2008). PARP1-dependent kinetics of recruitment of MRE11 and NBS1 proteins to multiple DNA damage sites. Journal of Biological Chemistry, 283(2), 1197–1208. doi: 10.1074/jbc.M706734200 [DOI] [PubMed] [Google Scholar]
- Han W, Li X, & Fu X (2011). The macro domain protein family: structure, functions, and their potential therapeutic implications. Mutat Res, 727(3), 86–103. doi: S1383–5742(11)00005–6 [pii] 10.1016/j.mrrev.2011.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawse WF, & Wolberger C (2009). Structure-based mechanism of ADP-ribosylation by sirtuins. Journal of Biological Chemistry, 284(48), 33654–33661. doi: 10.1074/jbc.M109.024521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hengel SM, & Goodlett DR (2012). A Review of Tandem Mass Spectrometry Characterization of Adenosine Diphosphate-Ribosylated Peptides. Int J Mass Spectrom, 312, 114–121. doi: 10.1016/j.ijms.2011.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hengel SM, Shaffer SA, Nunn BL, & Goodlett DR (2009). Tandem mass spectrometry investigation of ADP-ribosylated kemptide. Journal of the American Society for Mass Spectrometry, 20(3), 477–483. doi: 10.1016/j.jasms.2008.10.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hottiger MO (2015a). Nuclear ADP-Ribosylation and Its Role in Chromatin Plasticity, Cell Differentiation, and Epigenetics. Annu Rev Biochem, 84, 227–263. doi: 10.1146/annurev-biochem-060614-034506 [DOI] [PubMed] [Google Scholar]
- Hottiger MO (2015b). Nuclear ADP-Ribosylation and Its Role in Chromatin Plasticity, Cell Differentiation, and Epigenetics. Annual Review of Biochemistry, 84, 227–263. doi: 10.1146/annurev-biochem-060614-034506 [DOI] [PubMed] [Google Scholar]
- Huttlin EL, Jedrychowski MP, Elias JE, Goswami T, Rad R, Beausoleil SA, … Gygi SP (2010). A tissue-specific atlas of mouse protein phosphorylation and expression. Cell, 143(7), 1174–1189. doi: 10.1016/j.cell.2010.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- James DI, Smith KM, Jordan AM, Fairweather EE, Griffiths LA, Hamilton NS, … Ogilvie DJ (2016). First-in-Class Chemical Probes against Poly(ADP-ribose) Glycohydrolase (PARG) Inhibit DNA Repair with Differential Pharmacology to Olaparib. ACS Chem Biol, 11(11), 3179–3190. doi: 10.1021/acschembio.6b00609 [DOI] [PubMed] [Google Scholar]
- Jungmichel S, Rosenthal F, Altmeyer M, Lukas J, Hottiger MO, & Nielsen ML (2013a). Proteome-wide identification of poly(ADP-Ribosyl)ation targets in different genotoxic stress responses. Mol Cell, 52(2), 272–285. doi: 10.1016/j.molcel.2013.08.026 [DOI] [PubMed] [Google Scholar]
- Jungmichel S, Rosenthal F, Altmeyer M, Lukas J, Hottiger MO, & Nielsen ML (2013b). Proteome-wide identification of poly(ADP-Ribosyl)ation targets in different genotoxic stress responses. Molecular Cell, 52(2), 272–285. doi: 10.1016/j.molcel.2013.08.026 [DOI] [PubMed] [Google Scholar]
- Kanai M, Hanashiro K, Kim SH, Hanai S, Boulares AH, Miwa M, & Fukasawa K (2007). Inhibition of Crm1-p53 interaction and nuclear export of p53 by poly(ADP-ribosyl)ation. Nat Cell Biol, 9(10), 1175–1183. doi: 10.1038/ncb1638 [DOI] [PubMed] [Google Scholar]
- Kang HC, Lee YI, Shin JH, Andrabi SA, Chi Z, Gagne JP, … Dawson TM (2011). Iduna is a poly(ADP-ribose) (PAR)-dependent E3 ubiquitin ligase that regulates DNA damage. Proc Natl Acad Sci U S A, 108(34), 14103–14108. doi: 10.1073/pnas.1108799108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamitsu H, Hoshino H, Okada H, Miwa M, Momoi H, & Sugimura T (1984). Monoclonal antibodies to poly(adenosine diphosphate ribose) recognize different structures. Biochemistry, 23(16), 3771–3777. [DOI] [PubMed] [Google Scholar]
- Kim MY, Mauro S, Gevry N, Lis JT, & Kraus WL (2004). NAD+-dependent modulation of chromatin structure and transcription by nucleosome binding properties of PARP-1. Cell, 119(6), 803–814. doi: 10.1016/j.cell.2004.11.002 [DOI] [PubMed] [Google Scholar]
- Kim W, Bennett EJ, Huttlin EL, Guo A, Li J, Possemato A, … Gygi SP (2011). Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol Cell, 44(2), 325–340. doi: 10.1016/j.molcel.2011.08.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraus WL, & Lis JT (2003). PARP goes transcription. Cell, 113(6), 677–683. [DOI] [PubMed] [Google Scholar]
- Krueger KM, & Barbieri JT (1995). The family of bacterial ADP-ribosylating exotoxins. Clinical Microbiology Reviews, 8(1), 34–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laing S, Koch-Nolte F, Haag F, & Buck F (2011). Strategies for the identification of arginine ADP-ribosylation sites. J Proteomics, 75(1), 169–176. doi: 10.1016/j.jprot.2011.07.003 [DOI] [PubMed] [Google Scholar]
- Langelier MF, Planck JL, Roy S, & Pascal JM (2011a). Crystal structures of poly(ADP-ribose) polymerase-1 (PARP-1) zinc fingers bound to DNA: structural and functional insights into DNA-dependent PARP-1 activity. Journal of Biological Chemistry, 286(12), 10690–10701. doi: 10.1074/jbc.M110.202507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langelier MF, Planck JL, Roy S, & Pascal JM (2011b). Crystal structures of poly(ADP-ribose) polymerase-1 (PARP-1) zinc fingers bound to DNA: structural and functional insights into DNA-dependent PARP-1 activity. J Biol Chem, 286(12), 10690–10701. doi: 10.1074/jbc.M110.202507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langelier MF, Planck JL, Roy S, & Pascal JM (2012). Structural basis for DNA damage-dependent poly(ADP-ribosyl)ation by human PARP-1. Science, 336(6082), 728–732. doi: 10.1126/science.1216338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lautier D, Lagueux J, Thibodeau J, Menard L, & Poirier GG (1993). Molecular and biochemical features of poly (ADP-ribose) metabolism. Molecular and Cellular Biochemistry, 122(2), 171–193. [DOI] [PubMed] [Google Scholar]
- Ledermann J, Harter P, Gourley C, Friedlander M, Vergote I, Rustin G, … Matulonis U (2012). Olaparib maintenance therapy in platinum-sensitive relapsed ovarian cancer. N Engl J Med, 366(15), 1382–1392. doi: 10.1056/NEJMoa1105535 [DOI] [PubMed] [Google Scholar]
- Leidecker O, Bonfiglio JJ, Colby T, Zhang Q, Atanassov I, Zaja R, … Matic I (2016). Serine is a new target residue for endogenous ADP-ribosylation on histones. Nat Chem Biol, 12(12), 998–1000. doi: 10.1038/nchembio.2180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Lu LY, Yang CY, Wang S, & Yu X (2013). The FHA and BRCT domains recognize ADP-ribosylation during DNA damage response. Genes Dev, 27(16), 1752–1768. doi: 27/16/1752 [pii] 10.1101/gad.226357.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, & Yu X (2013). Function of BRCA1 in the DNA damage response is mediated by ADP-ribosylation. Cancer Cell, 23(5), 693–704. doi: S1535–6108(13)00132–3 [pii] 10.1016/j.ccr.2013.03.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litton JK, Rugo HS, Ettl J, Hurvitz SA, Goncalves A, Lee KH, … Blum JL (2018). Talazoparib in Patients with Advanced Breast Cancer and a Germline BRCA Mutation. N Engl J Med, 379(8), 753–763. doi: 10.1056/NEJMoa1802905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Vyas A, Kassab MA, Singh AK, & Yu X (2017). The role of poly ADP-ribosylation in the first wave of DNA damage response. Nucleic Acids Res, 45(14), 8129–8141. doi: 10.1093/nar/gkx565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XC, & Scouten WH (2000). Boronate affinity chromatography. Methods Mol Biol, 147, 119–128. doi: 10.1385/1-59259-041-1:119 [DOI] [PubMed] [Google Scholar]
- Ludwig A, Behnke B, Holtlund J, & Hilz H (1988). Immunoquantitation and size determination of intrinsic poly(ADP-ribose) polymerase from acid precipitates. An analysis of the in vivo status in mammalian species and in lower eukaryotes. Journal of Biological Chemistry, 263(15), 6993–6999. [PubMed] [Google Scholar]
- Margarit SM, Davidson W, Frego L, & Stebbins CE (2006). A steric antagonism of actin polymerization by a salmonella virulence protein. Structure, 14(8), 1219–1229. doi: 10.1016/j.str.2006.05.022 [DOI] [PubMed] [Google Scholar]
- Martello R, Leutert M, Jungmichel S, Bilan V, Larsen SC, Young C, … Nielsen ML (2016). Proteome-wide identification of the endogenous ADP-ribosylome of mammalian cells and tissue. Nat Commun, 7, 12917. doi: 10.1038/ncomms12917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masson M, Niedergang C, Schreiber V, Muller S, Menissier-de Murcia J, & de Murcia G (1998). XRCC1 is specifically associated with poly(ADP-ribose) polymerase and negatively regulates its activity following DNA damage. Molecular and Cellular Biology, 18(6), 3563–3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matic I, Ahel I, & Hay RT (2012). Reanalysis of phosphoproteomics data uncovers ADP-ribosylation sites. Nat Methods, 9(8), 771–772. doi: 10.1038/nmeth.2106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza-Alvarez H, & Alvarez-Gonzalez R (1993). Poly(ADP-ribose) polymerase is a catalytic dimer and the automodification reaction is intermolecular. Journal of Biological Chemistry, 268(30), 22575–22580. [PubMed] [Google Scholar]
- Messner S, Altmeyer M, Zhao H, Pozivil A, Roschitzki B, Gehrig P, … Hottiger MO (2010). PARP1 ADP-ribosylates lysine residues of the core histone tails. Nucleic Acids Res, 38(19), 6350–6362. doi: 10.1093/nar/gkq463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min W, & Wang ZQ (2009). Poly (ADP-ribose) glycohydrolase (PARG) and its therapeutic potential. Front Biosci (Landmark Ed), 14, 1619–1626. [DOI] [PubMed] [Google Scholar]
- Mirza MR, Monk BJ, Herrstedt J, Oza AM, Mahner S, Redondo A, … Investigators, E.-O. N. (2016). Niraparib Maintenance Therapy in Platinum-Sensitive, Recurrent Ovarian Cancer. N Engl J Med, 375(22), 2154–2164. doi: 10.1056/NEJMoa1611310 [DOI] [PubMed] [Google Scholar]
- Miyamoto T, Kakizawa T, & Hashizume K (1999). Inhibition of nuclear receptor signalling by poly(ADP-ribose) polymerase. Mol Cell Biol, 19(4), 2644–2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortusewicz O, Fouquerel E, Ame JC, Leonhardt H, & Schreiber V (2011). PARG is recruited to DNA damage sites through poly(ADP-ribose)- and PCNA-dependent mechanisms. Nucleic Acids Res, 39(12), 5045–5056. doi: 10.1093/nar/gkr099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss J, Yost DA, & Stanley SJ (1983). Amino acid-specific ADP-ribosylation. J Biol Chem, 258(10), 6466–6470. [PubMed] [Google Scholar]
- Mueller-Dieckmann C, Kernstock S, Lisurek M, von Kries JP, Haag F, Weiss MS, & Koch-Nolte F (2006). The structure of human ADP-ribosylhydrolase 3 (ARH3) provides insights into the reversibility of protein ADP-ribosylation. Proc Natl Acad Sci U S A, 103(41), 15026–15031. doi: 10.1073/pnas.0606762103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niere M, Mashimo M, Agledal L, Dolle C, Kasamatsu A, Kato J, … Ziegler M (2012). ADP-ribosylhydrolase 3 (ARH3), not poly(ADP-ribose) glycohydrolase (PARG) isoforms, is responsible for degradation of mitochondrial matrix-associated poly(ADP-ribose). J Biol Chem, 287(20), 16088–16102. doi: 10.1074/jbc.M112.349183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata N, Ueda K, & Hayaishi O (1980). ADP-ribosylation of histone H2B. Identification of glutamic acid residue 2 as the modification site. J Biol Chem, 255(16), 7610–7615. [PubMed] [Google Scholar]
- Ogata N, Ueda K, Kagamiyama H, & Hayaishi O (1980). ADP-ribosylation of histone H1. Identification of glutamic acid residues 2, 14, and the COOH-terminal lysine residue as modification sites. J Biol Chem, 255(16), 7616–7620. [PubMed] [Google Scholar]
- Oka S, Kato J, & Moss J (2006). Identification and characterization of a mammalian 39-kDa poly(ADP-ribose) glycohydrolase. J Biol Chem, 281(2), 705–713. doi: 10.1074/jbc.M510290200 [DOI] [PubMed] [Google Scholar]
- Olsen JV, & Mann M (2013). Status of large-scale analysis of post-translational modifications by mass spectrometry. Mol Cell Proteomics, 12(12), 3444–3452. doi: 10.1074/mcp.O113.034181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen JV, Schwartz JC, Griep-Raming J, Nielsen ML, Damoc E, Denisov E, … Horning S (2009). A dual pressure linear ion trap Orbitrap instrument with very high sequencing speed. Mol Cell Proteomics, 8(12), 2759–2769. doi: 10.1074/mcp.M900375-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petesch SJ, & Lis JT (2012). Activator-induced spread of poly(ADP-ribose) polymerase promotes nucleosome loss at Hsp70. Mol Cell, 45(1), 64–74. doi: 10.1016/j.molcel.2011.11.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenthal F, Messner S, Roschitzki B, Gehrig P, Nanni P, & Hottiger MO (2011). Identification of distinct amino acids as ADP-ribose acceptor sites by mass spectrometry. Methods in Molecular Biology, 780, 57–66. doi: 10.1007/978-1-61779-270-0_4 [DOI] [PubMed] [Google Scholar]
- Rouleau M, Patel A, Hendzel MJ, Kaufmann SH, & Poirier GG (2010). PARP inhibition: PARP1 and beyond. Nat Rev Cancer, 10(4), 293–301. doi: 10.1038/nrc2812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber V (2006). [Present state of endocrinology in the Czech Republic]. Cas Lek Cesk, 145(8), 595–596. [PubMed] [Google Scholar]
- Sharifi R, Morra R, Appel CD, Tallis M, Chioza B, Jankevicius G, … Ahel I (2013). Deficiency of terminal ADP-ribose protein glycohydrolase TARG1/C6orf130 in neurodegenerative disease. EMBO J, 32(9), 1225–1237. doi: 10.1038/emboj.2013.51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shieh WM, Ame JC, Wilson MV, Wang ZQ, Koh DW, Jacobson MK, & Jacobson EL (1998). Poly(ADP-ribose) polymerase null mouse cells synthesize ADP-ribose polymers. J Biol Chem, 273(46), 30069–30072. [DOI] [PubMed] [Google Scholar]
- Simonin F, Poch O, Delarue M, & de Murcia G (1993). Identification of potential active-site residues in the human poly(ADP-ribose) polymerase. Journal of Biological Chemistry, 268(12), 8529–8535. [PubMed] [Google Scholar]
- Slade D, Dunstan MS, Barkauskaite E, Weston R, Lafite P, Dixon N, … Ahel I (2011). The structure and catalytic mechanism of a poly(ADP-ribose) glycohydrolase. Nature, 477(7366), 616–620. doi: 10.1038/nature10404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slama JT, Aboul-Ela N, Goli DM, Cheesman BV, Simmons AM, & Jacobson MK (1995). Specific inhibition of poly(ADP-ribose) glycohydrolase by adenosine diphosphate (hydroxymethyl)pyrrolidinediol. J Med Chem, 38(2), 389–393. [DOI] [PubMed] [Google Scholar]
- Swisher EM, Lin KK, Oza AM, Scott CL, Giordano H, Sun J, … McNeish IA (2017). Rucaparib in relapsed, platinum-sensitive high-grade ovarian carcinoma (ARIEL2 Part 1): an international, multicentre, open-label, phase 2 trial. Lancet Oncol, 18(1), 75–87. doi: 10.1016/S1470-2045(16)30559-9 [DOI] [PubMed] [Google Scholar]
- Syka JE, Coon JJ, Schroeder MJ, Shabanowitz J, & Hunt DF (2004). Peptide and protein sequence analysis by electron transfer dissociation mass spectrometry. Proc Natl Acad Sci U S A, 101(26), 9528–9533. doi: 10.1073/pnas.0402700101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Z, Gao P, & Liu HW (2009). Identification of the ADP-ribosylation sites in the PARP-1 automodification domain: analysis and implications. J Am Chem Soc, 131(40), 14258–14260. doi: 10.1021/ja906135d [DOI] [PubMed] [Google Scholar]
- Timinszky G, Till S, Hassa PO, Hothorn M, Kustatscher G, Nijmeijer B, … Ladurner AG (2009). A macrodomain-containing histone rearranges chromatin upon sensing PARP1 activation. Nat Struct Mol Biol, 16(9), 923–929. doi: 10.1038/nsmb.1664 [DOI] [PubMed] [Google Scholar]
- Virag L, & Szabo C (2002). The therapeutic potential of poly(ADP-ribose) polymerase inhibitors. Pharmacol Rev, 54(3), 375–429. [DOI] [PubMed] [Google Scholar]
- Vivelo CA, & Leung AK (2015). Proteomics approaches to identify mono-(ADP-ribosyl)ated and poly(ADP-ribosyl)ated proteins. Proteomics, 15(2–3), 203–217. doi: 10.1002/pmic.201400217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivelo CA, & Leung AKL (2015). Proteomics approaches to identify mono-(ADP-ribosyl)ated and poly(ADP-ribosyl)ated proteins. Proteomics, 15(2–3), 203–217. doi: 10.1002/pmic.201400217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vyas S, Matic I, Uchima L, Rood J, Zaja R, Hay RT, … Chang P (2014). Family-wide analysis of poly(ADP-ribose) polymerase activity. Nat Commun, 5, 4426. doi: 10.1038/ncomms5426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Michaud GA, Cheng Z, Zhang Y, Hinds TR, Fan E, … Xu W (2012). Recognition of the iso-ADP-ribose moiety in poly(ADP-ribose) by WWE domains suggests a general mechanism for poly(ADP-ribosyl)ation-dependent ubiquitination. Genes Dev, 26(3), 235–240. doi: gad.182618.111 [pii] 10.1101/gad.182618.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wielckens K, George E, Pless T, & Hilz H (1983). Stimulation of poly(ADP-ribosyl)ation during Ehrlich ascites tumor cell “starvation” and suppression of concomitant DNA fragmentation by benzamide. Journal of Biological Chemistry, 258(7), 4098–4104. [PubMed] [Google Scholar]
- Yamanaka H, Penning CA, Willis EH, Wasson DB, & Carson DA (1988). Characterization of human poly(ADP-ribose) polymerase with autoantibodies. Journal of Biological Chemistry, 263(8), 3879–3883. [PubMed] [Google Scholar]
- Zhang F, Chen Y, Li M, & Yu X (2014a). The oligonucleotide/oligosaccharide-binding fold motif is a poly(ADP-ribose)-binding domain that mediates DNA damage response. Proc Natl Acad Sci U S A, 111(20), 7278–7283. doi: 1318367111 [pii] 10.1073/pnas.1318367111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Chen Y, Li M, & Yu X (2014b). The oligonucleotide/oligosaccharide-binding fold motif is a poly(ADP-ribose)-binding domain that mediates DNA damage response. Proceedings of the National Academy of Sciences of the United States of America, 111(20), 7278–7283. doi: 10.1073/pnas.1318367111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Shi J, Chen SH, Bian C, & Yu X (2015). The PIN domain of EXO1 recognizes poly(ADP-ribose) in DNA damage response. Nucleic Acids Res, 43(22), 10782–10794. doi: gkv939 [pii] 10.1093/nar/gkv939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu S, Mickanin C, Feng Y, Charlat O, Michaud GA, … Cong F (2011). RNF146 is a poly(ADP-ribose)-directed E3 ligase that regulates axin degradation and Wnt signalling. Nat Cell Biol, 13(5), 623–629. doi: 10.1038/ncb2222 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Wang J, Ding M, & Yu Y (2013). Site-specific characterization of the Asp- and Glu-ADP-ribosylated proteome. Nat Methods, 10(10), 981–984. doi: 10.1038/nmeth.2603 [DOI] [PubMed] [Google Scholar]
- Zhen Y, & Yu Y (2018). Proteomic Analysis of the Downstream Signaling Network of PARP1. Biochemistry, 57(4), 429–440. doi: 10.1021/acs.biochem.7b01022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhen Y, Zhang Y, & Yu Y (2017). A Cell-Line-Specific Atlas of PARP-Mediated Protein Asp/Glu-ADP-Ribosylation in Breast Cancer. Cell Rep, 21(8), 2326–2337. doi: 10.1016/j.celrep.2017.10.106 [DOI] [PMC free article] [PubMed] [Google Scholar]


