Abstract
Genetic heritability (h2) of alcohol use is reported to be greater in rural dwellers, distressed marriages, low socioeconomic status, in girls who are unmarried or lacking closeness with their parents or religious upbringing, in less-educated men, and in adolescents with peers using alcohol. However, these are all risk factors for heavy drinking, and the greater heritability could be due to quantile-dependent expressivity, i.e., h2 dependent upon whether the phenotype (alcohol intake) is high or low relative to its distribution. Quantile regression showed that h2 estimated from the offspring-parent regression slope increased significantly from lowest to highest gram/day of alcohol consumption (0.006±0.001 per percent, P=1.1x10−7). Heritability at the 90th percentile of the sample distribution (0.557±0.116) was 4.5-fold greater than at the 10th percentile (0.122±0.037). Heritabilities for intakes of other macronutrients were not quantile-dependent. Thus quantile-dependent expressivity may explain the higher estimated heritability associated with risk factors for high alcohol consumption.
Keywords: Alcohol, heritability, macronutrients, genetics, wine, beer, mixed drinks
Introduction
The inheritance of alcohol use has been studied extensively in twin pairs, family sets, and epidemiological cohorts (Rietschel & Treutlein 2013). Heritability (h2) for quantity consumed is estimated to be about 50% for adults (Dick et al. 2011, Hansell et al. 2008, Kendler et al. 2008, McGue 1999). Much of the genetic research has focused on problematic drinking such as alcohol use disorder (AUD) (American Psychiatric Association, 2013), which has a lifetime prevalence of 30% in the United States (Hasin et al. 2007), and which is also strongly heritable (h2 approximately 49%, Verhulst et al. 2015). Meta-analysis of 480,000 people aged 40 to 69 years of European descent identified 46 loci that account for 7% of the variance in the quantity of alcohol consumed (Evangelou 2019). Thus, similar to other complex traits, the polygenic inheritance of alcohol consumption appears to mostly involve a large number of genetic variants with small individual effects (Manolio et al. 2009).
Gene-environment interactions are thought to represent some of the missing heritability of alcohol intake, AUD, and other measures of alcohol use and misuse. For example, genetic heritability of alcohol use is purported to be greater in urban vs. rural areas (Davis et al. 2017, Legrand et al. 2008, Rose et al. 2001), in girls with less vs. greater parental closeness (Miles et al. 2005), in women lacking vs. having a religious upbringing (Koopmans et al. 1999), in unmarried vs. married women (Heath et al. 1989), in individuals with low vs. high socioeconomic status (Hamdi et al. 2015), in less vs. more educated men (Johnson et al. 2011), in individuals with vs. without distressed marriages (Jarnecke & South 2014), and in adolescents having peers and parents who drink (Dick et al. 2007). These interactions have been interpreted as behavioral or environmental restrictions that promote abstinence and restricted availability (social controls), and stressors that potentiate genetic liability (social triggers, Chartier et al. 2017).
Quantile-dependent expressivity (or penetrance) hypothesizes that the heritability of a trait (e.g., alcohol intake) depends upon whether the phenotype is high or low relative to its distribution (Williams 2012). We have shown that quantile-specific effects may play an important role in the genetics of body weight, lipoprotein concentrations, and coffee intake while not affecting other traits such as height (Williams 2012, 2020a-e). Prior studies assume that genetic and non-genetic factors affecting alcohol consumption in parents, offspring, and other related individuals apply equally over all quantiles of the phenotype distribution. However, urban dwelling, lack of parental closeness, non-religious upbringing, unmarried status, low socioeconomic status, low education, deviant peer group, and distressed marriages are all risk factors for higher consumption. We hypothesize that quantile-dependent expressivity may explain, at least in part, some of the gene-environment interactions observed for alcohol consumption and AUD because participants with and without these risk factors represent different portions of the intake distribution. Specifically, behavioral or environmental effects that tend to distinguish higher vs. lower alcohol consumption, dependence, or misuse could produce different h2 estimates implying genetic differences that might be simply explained by quantile-dependant expressivity.
It is not known whether the heritability of alcohol consumption, or of other nutrients, is quantile dependent. Therefore, we used self-reported food intake from the Harvard food frequency questionnaires completed by the Offspring and Third Generation Framingham Study cohorts (Kannel et al. 1979) to test whether spouse, offspring-parent and sibling resemblance increases significantly and substantially from the lowest to the highest consumption levels. Alcohol intake in particular is not normally distributed due to a sizable portion of abstainers and others that drink in excess. However, quantile regression is a robust technique for obtaining estimates of the regression slopes that does not require normality, and bootstrap resampling provides nonparametric estimates of statistical significance. The discussion examines the relevance of the findings to previously published gene-environment interactions of alcohol use and misuse. These include studies that used directly measured genotypes and alternative estimates of heritability, for the purpose of supporting its validity and demonstrating the concept’s potential to provide a common underlying basis for a diversity of environmental interactions.
Methods
Subjects
The data for the Framingham Offspring Cohort (the children of the Original Framingham Cohort and their spouses) and the Framingham Third Generation Cohort (children of the Framingham Offspring Cohort) were obtained from the National Institutes of Health FRAMCOHORT, GEN3, FRAMOFFSPRING Research Materials obtained from the National Heart Lung and Blood Institute (NHLBI) Biologic Specimen and Data Repository Information Coordinating Center. The Offspring Cohort consisted of 5,124 men and women who were originally recruited between 1971 and 1975 and subsequently examined at approximately 4-year intervals to investigate familial risk factors for cardiovascular disease (Kannel et al. 1979). Children of the Offspring Cohort were recruited to form the Third Generation Cohort (Splansky et al. 2007). The analyses presented in this report uses data from the 5th (1991–1995), 6th (1995–1998), 7th (1998–2001) and 8th (2005–2008) clinical exams from the Offspring Cohort and the 1st (2002–2005) and the 2nd (2008–2010) clinical exams from the Third Generation Cohort who were at least 16 years of age and self-identified as nonhispanic white. Participation rates were 74.1%, 68.9%, 69.1% and 59.0% for exams 5, 6, 7, and 8 of the Offspring Cohort, respectively (Wang et al. 2015).
A semi-quantitative, 126-item Willett food frequency questionnaire (FFQ) purple form was used to estimate alcohol intake in grams per day (g/d) and as a percent of total energy intake (%E) based on their food intake over the previous year (Rimm et al 1992). These questionnaires were mailed directly to study participants who returned them to the clinic staff for review at their clinical exam. Beer (glass, bottle or can), red wine (4 oz), white wine (4 oz), and liquor (1 drink or shot) were reported as: 1) never or less than once per month, 2) 1-3 per month, 3) 1 per week, 4) 2-4 per week, 5) 5-6 per week, 6) 1 per day, 7) 2-3 per day, 8) 4-5 per day, and 9) ≥6 per day. Categories were also used to record the consumptions of other foods. For each food item, if participants reported consuming ‘never or <1 serving/month’, their intake of that food item was coded as zero servings/week in the FFQ database. If participants reported consuming ‘1–3 servings/month’ or more of a particular item, their intake of that food item was converted to ‘servings/week ’ based on the mid-range value of the intake category. Questionnaires were excluded if more than 12 food items were left blank or the calculated energy intakes were <600 kcal/d or >4000 kcal/d (women) or >4200 kcal/d (men). Alcohol was computed as 10.8 g per 4-oz glass of wine, 13.2 g per 12-oz bottle of beer, and 15.1 g per mixed drink (Giovannucci et al. 1991). Alcohol intakes as percent of energy intake were reported to have intra-class correlation coefficients of 0.50 in 785 men and 0.62 in 949 women (Kimokoti et al. 2012). Somewhat weaker intraclass correlations were reported for male and female percent energy intake from total protein (0.32 and 0.34, respectively), total carbohydrate (0.34 and 0.29, respectively), total fat (0.23 and 0.19, respectively), saturated fat (0.31 and 0.21, respectively), monounsaturated fat (0.20 and 0.23, respectively), and polyunsaturated fat (0.13 and 0.18, respectively) (Kimokoti et al. 2012).
Statistics
Age and sex adjustment was performed separately in the Offspring and Third Generation Cohorts using standard least-squares regression with the following independent variables: female (0,1), age, age2, female x age, and female x age2. Individual subject values were taken as the average of the residuals over all available visits. Offspring-parent correlations (rOP) and regression slopes (βOP) were computed using the parents and offspring from the Framingham Offspring and Framingham Third Generation Cohorts, respectively. They were computed by assigning a weight of one-half to the father-child pair and one-half to the mother-child pair (if both parents available), and assigning a weight of one to the parent-child pair if only one parent was available. Full sib correlations and regression slopes (βFS) were obtained by constructing all possible pairs using double entry (Karlin et al. 1981). The number of degrees of freedom for the standard error was Σki-2 for parent-offspring regression slopes and correlations, and Σ(ki-1) for sibship correlations and regression slopes, where ki is the number of offspring in family or sibship i, where the summation is taken over all i, i=1,…, N nuclear families. The log transformed alcohol intake was assigned a value of log(0.5) for abstainers.
Simultaneous quantile regression is a well-developed statistical procedure (Koenker & Hallock 2001, Gould 1992) that estimates the regression coefficients for multiple quantiles using linear programming to minimize the sum of asymmetrically weighted absolute residuals, and bootstrap resampling to estimate their corresponding variances and covariances (Gould 1992). Simultaneous quantile regression was performed using the sqreg command of Stata (version 11, StataCorp, College Station, TX) with one thousand bootstrap samples drawn to estimate the variance-covariance matrix for the 91 quantile regression coefficients between the 5th and 95th percentiles, and the post-estimation procedures (test and lincom) to test linear combinations of the slopes after estimation with Σki-2 degrees of freedom for offspring-parent regression slopes and Σ(ki-1) degrees of freedom for sibship correlations. Quantile-specific expressivity was assessed by: 1) estimating quantile-specific β-coefficient for the 5th, 6th,…, 95th percentiles of the sample distribution using simultaneous quantile regression (Figure 1, the <5th and >95th percentiles ignored because they were thought to be less stable); 2) plotting the quantile-specific β coefficient vs. the percentile of the trait distribution; and 3) testing whether the resulting graph is constant, or changes as a linear, quadratic, or cubic function of the percentile of the trait distribution using orthogonal polynomials (Winer et al. 1991). Heritability in the narrow sense (h2) was estimated as 2βOP/(1+rspouse) from offspring-parent regression slopes (βOP) and [(1+8 rspouse βFS)0.5-1]/2rspouse from full-sibs regression slopes (βFS) where rspouse is the spouse correlation (Falconer and Mackay, 1996)
Figure 1.

Increase in offspring alcohol intake per increase in their parent’s intake (g/d) for selected percentiles of the offspring’s distribution (i.e. offspring-parent slopes, upper panel), and offspring-parent slopes (vertical axis) plotted as a function of the percentiles of the offspring’s alcohol consumption (horizontal axis, lower panel). Shaded region designates the 95% confidence interval for the quantile-specific slopes. Parents and offspring data adjusted for sex, age, age2, sex x age, and sex x age2.
The analyses were approved by the Lawrence Berkeley National Laboratory Human Subjects Committee (HSC). The hypothesis tested is exploratory and not considered as part of the initial Framingham Study design.
Results
Estimated nutrient intakes were obtained for 1691 spouse-pairs and 2939 offspring (1691 with both parents, 1248 with one parent). There were 1684 families with more than one full sib (770 in the Offspring Cohort and 914 in the Third Generation Cohort). Their mean age and energy intakes averaged over all their available visits are displayed in Supplementary table 1, and deciles of alcohol intake are presented in Supplementary table 2. Spouse alcohol intakes were strongly correlated whether measured as g/d (rspouse=0.36), % calories consumed (rspouse=0.34), or g/kg of body weight (rspouse=0.35), and were more strongly correlated for g/d intake of alcohol from wine (rspouse=0.42), and liqueurs (rspouse=0.43) than beer (rspouse=0.09). The lower spouse concordance for beer is because substantially fewer women drank beer than other beverages.
Traditional offspring-parent regression analyses
As traditionally estimated by least squares regression, the offspring-parent regression slope (βOP±SE) was 0.157±0.019 for g/d of alcohol consumed, 0.129±0.016 for percent of calories consumed as alcohol, and 0.152±0.019 for g/d alcohol consumed divided by body weight (all P<10−15, Table 1). These correspond to heritabilities of 0.231±0.028, 0.193±0.024, and 0.224±0.028, respectively. As calculated from their traditional βOP’s presented in Table 1, the heritability for g/d alcohol intake was 0.203±0.042 from beer, 0.182±0.026 from wine and 0.077±0.016 from liqueurs. The heritability estimate for g/d of alcohol consumed was essentially unchanged when restricted to children with both parents (h2= 0.224±0.035, P=1.4x10−10).
Table 1.
Traditional and quantile regression analyses of alcohol intake
| Traditional regression analysis | Quantile regression analysis | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Classical regression slope, all observations (βFM, βOP βFS) |
Increase in slope per 1% increase in the percentile of the dependent variable’s distribution |
Difference in slope between the 90th and 10th percentiles |
|||||||
| Correlation | Linear effect | Nonlinear effects | |||||||
| Slope±SE | P | Slope±SE | Linear P | Quadratic P |
Cubic P | Difference±SE | P | ||
| Father-mother | |||||||||
| Alcohol g/d | 0.36 | 0.6839±0.0600 | <10−15 | 0.0094±0.0021 | 7.5x10−6 | 0.001 | 0.78 | 0.7402±0.2189 | 0.0007 |
| Alcohol %energy | 0.34 | 0.5555±0.0519 | <10−15 | 0.0073±0.0019 | 0.0001 | 0.001 | 0.99 | 0.5487±0.2353 | .02 |
| Alcohol (g/d)/kg | 0.35 | 0.5057±0.0466 | <10−15 | 0.0058±0.0012 | 2.1x10−6 | 2.2x10−8 | 0.06 | 0.3485±0.0903 | 0.0001 |
| Log alcohol (g/d) | 0.39 | 0.4505±0.0361 | <10−15 | −0.0048±0.0008 | 2.8x10−10 | <10−15 | 1.7x10−12 | −0.2029±0.0626 | 0.001 |
| Beer (g/d) | 0.09 | 0.3256±0.1266 | 0.01 | 0.0038±0.0046 | 0.41 | 0.32 | 0.27 | −0.1548±0.4565 | 0.73 |
| Wine (g/d) | 0.42 | 0.4530±0.0339 | <10−15 | 0.0132±0.0013 | <10−15 | 0.33 | 0.08 | 0.9629±0.1350 | 1x10−12 |
| Liquour (g/d) | 0.43 | 0.7160±0.0516 | <10−15 | 0.0195±0.0028 | 2.1x10−12 | 0.71 | 0.89 | 1.5961±0.3097 | 2.6 x10−7 |
| Offspring-parent | |||||||||
| Alcohol g/d | 0.15 | 0.1574±0.0193 | 4.4x10−16 | 0.0038±0.0007 | 1.1x10−7 | 0.004 | 0.66 | 0.2955±0.0804 | 0.0002 |
| Alcohol %energy | 0.15 | 0.1290±0.0161 | 8.9x10−16 | 0.0030±0.0006 | 2.7x10−7 | 0.14 | 0.80 | 0.2500±0.0537 | 3.2x10−6 |
| Alcohol (g/d)/kg | 0.15 | 0.1515±0.0186 | 4.4x10−16 | 0.0040±0.0007 | 1.2x10−9 | 0.05 | 0.70 | 0.3228±0.0524 | 7.0x10−10 |
| Log alcohol (g/d) | 0.20 | 0.1882±0.0174 | <10−16 | −0.0006±0.0004 | 0.09 | 6.8x10−14 | 4.1x10−8 | 0.0969±0.0272 | 0.001 |
| Beer (g/d) | 0.09 | 0.1106±0.0227 | 1.2x10−6 | 0.0030±0.0012 | 0.01 | 0.002 | 0.005 | 0.3063±0.1472 | 0.04 |
| Wine (g/d) | 0.13 | 0.1290±0.0184 | 2.4x10−12 | 0.0041±0.0011 | 0.0001 | 0.04 | 0.21 | 0.3088±0.1247 | 0.01 |
| Liquour (g/d) | 0.09 | 0.0551±0.0117 | 2.7x10−6 | 0.0017±0.0006 | 0.007 | 0.01 | 0.03 | 0.1382±0.0585 | 0.02 |
| Full-sib | |||||||||
| Alcohol g/d | 0.17 | 0.1653±0.0169 | <10−15 | 0.0044±0.0007 | 2.8x10−11 | 6.1x10−5 | 0.89 | 0.3602±0.0622 | 7.0x10−9 |
| Alcohol %energy | 0.16 | 0.1578±0.0169 | <10−15 | 0.0038±0.0007 | 1.1x10−8 | 0.12 | 0.11 | 0.2617±0.0586 | 8.0x10−6 |
| Alcohol (g/d)/kg | 0.16 | 0.1604±0.0170 | <10−15 | 0.0052±0.0007 | 3.6x10−13 | 0.002 | 0.44 | 0.3999±0.0833 | 1.6x10−6 |
| Log alcohol (g/d) | 0.20 | 0.2000±0.0168 | <10−15 | −0.0002±0.0004 | 0.65 | 2.0x10−14 | 0.001 | 0.0497±0.0398 | 0.21 |
| Beer (g/d) | 0.10 | 0.1034±0.0170 | 1.3x10−9 | 0.0026±0.001 | 0.008 | 0.001 | 0.01 | 0.2700±0.1102 | 0.01 |
| Wine (g/d) | 0.12 | 0.1174±0.0170 | 4.6x10−12 | 0.0047±0.0008 | 1.1x10−8 | 0.006 | 0.55 | 0.3680±0.0700 | 1.4x10−7 |
| Liquour (g/d) | 0.07 | 0.0667±0.0171 | 0.0001 | 0.0036±0.0011 | 0.0007 | 0.0029 | 0.009 | 0.3562±0.1000 | 0.0004 |
Quantile regression of offspring-parent alcohol intake
Figure 1 (upper panel) presents the offspring-parent regression slopes (βOP) for selected quantiles of the offsprings’ total alcohol intake (g/d) and their associated heritabilities. The slopes became progressively greater (i.e., steeper) with increasing quantiles of alcohol intake. These quantile-specific regression slopes were included with those of other quantiles to create the quantile-specific heritability function in the lower panel, i.e., where the offspring-parent slope (Y-axis) is plotted as a function of the quantile of the offsprings’ sample distribution (X-axis). Specifically, the Y-axis represents the slope at the 5th quantile of consumption, the 6th quantile of consumption,…, and the 95th quantiles of the offsprings’ alcohol consumption. The shaded area presents the 95% confidence intervals for the individual slopes at each quantile. The figure shows that each g/d increase in the parents’ alcohol intake was associated with an offspring increase of 0.083±0.025 g/d at the 10th percentile of the offsprings’ total alcohol intake (P=0.0008), 0.046±0.010 g/d at their 25th percentile (P=5.1x10−6), 0.130±0.025 g/d at the 50th percentile (P=3.6x10−7), 0.239±0.029 g/d at the 75th percentile (P=2.2x10−16), and 0.379±0.079 g/d at the 90th percentile (P=1.5x10−6). These correspond to h2 of 0.122, 0.068, 0.191, 0.351, and 0.557, respectively.
If heritability of total alcohol consumption was the same for all offspring quantiles as traditionally assumed, then the upper panel would display parallel regression lines, and the lower graph would present a simple horizontal line. In fact, the graph shows that the slope became progressively greater with increasing quantiles of its offsprings’ distribution, such that on average each 1-percent increase in the offspring distribution was associated with a 0.004±0.001 increase in slope and a 0.006±0.001 increase in heritability (P=1.1x10−7). The offspring-parent regression slope was 4.5-fold greater at the 90th than at the 10th percentile of offspring reported consumption. The difference in heritability between heavy (90th percentile) and light drinkers (i.e., 90th -10th percentile±SE: 0.435±0.118, P=0.0002) was nearly twice as great as the traditional heritability estimate calculated for the entire offspring sample (h2: 0.231). The offspring-parent slopes were significantly greater than zero at every percentile between the 11th and the 95th percentiles of the offspring’ alcohol intake. The heritabilities of g/d of alcohol intake by beverage type (from their βOP’s of Table 1) also increased with increasing percentiles of beer (0.006±0.002, P=0.01), wine (0.006±0.002, P=0.0001), and liquour consumption (0.002±0.001, P=0.007).
Full-sib regression analyses
The full-sib regression slopes (βFS) also increased significantly with increasing percentiles of the sibs’ distribution of alcohol intake (Table 1). Figure 2 shows that the graph of the full-sib slope for g/d intake vs. the quantile of sibs’ distribution is very similar to the offspring-parent slope, i.e., both show increasing concordance at the higher percentiles of their distribution, and a bump due to low consumers at the lower percentiles. The full-sib slope at the 90th percentile of the sibs' distribution (βFS±SE: 0.432±0.060) was 6-fold greater than the slope at the 10th percentile (0.072±0.024), and the difference between the 90th and 10th percentiles (0.360±0.062) was over twice as great as the traditional regression slope for the total sample (0.165±0.017, Table 1). The full-sib regression slope was significant (P<0.001) for all percentiles between the 11th and 95th percentiles of the sibs’ distribution. βFS for intakes of alcohol derived from beer, wine, and liqueurs also increased significantly with increasing percentiles of consumption (Table 1).
Figure 2.
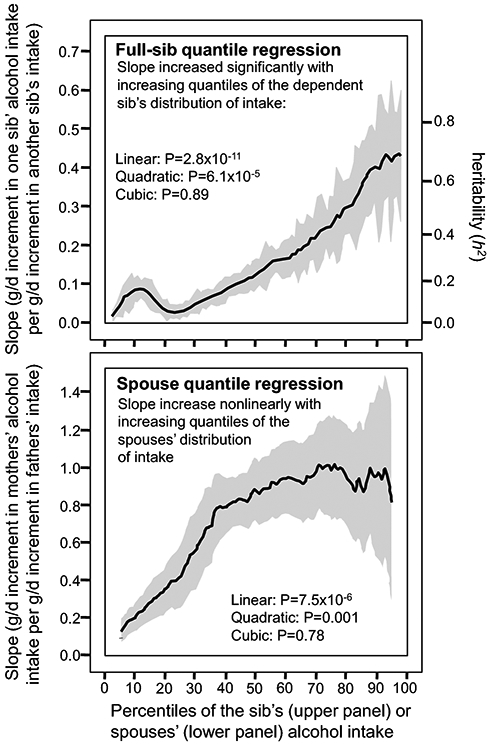
Full-sib regression slopes (vertical axis) by quantiles of the sib’s alcohol intake (horizontal axis, upper panel), and spouse regression slope of the mother (dependent variable) vs. father (independent variable, vertical axis) by quantiles of the mother’s alcohol intake (horizontal axis, lower panel). Shaded region designates the 95% confidence interval for the quantile-specific slopes. Spouse and sibling reported alcohol intake (g/d) adjusted for sex, age, age2, sex x age, and sex x age2.
Abstention
Quantile-dependent increases in heritability were confirmed when abstainers were excluded. One-hundred forty-nine fathers, 283 mothers, and 391 offspring reported not drinking. Parent and offspring abstinences were significantly associated (r=0.11, P<10−8) as were abstinences within sibpairs (r=0.10, P<10−8). Alcohol (g/d) consumption in 1910 offspring-parent pairs, where neither parents nor offspring abstained, correlated moderately between spouses (rspouse=0.38) with a heritability of h2=0.22, as traditionally estimated from the offspring-parent regression slope (βOP= 0.150±0.024, P=2.2x10−10). Quantile regression showed quantile-specific βOP increased 0.0035±0.0009 per percent increase in the population (P=4.0x10−10) and differed by 0.270±0.083 (P=0.001) between the 10th and 90th population percentile. Excluding abstainers left 4104 full siblings in 1915 sibships, βFS=0.175±0.021 (P=0.003), which corresponded to a heritability estimate of h2=0.314. Quantile regression showed quantile-specific βFS increased 0.0046±0.0009 per percent increase in the population (P=10−7), and differed by 0.362±0.135 (P=0.008) between the 10th and 90th population percentile. The bump in the lower percentiles of Figures 1 and 2 persisted when abstainers were excluded.
Standardized and log-transformed alcohol intake
Table 1 shows that βOP and βFS for alcohol intake divided by body weight and for alcohol intake as a percent of total energy intake were similar to those obtained for g/d. Table 1 and Supplementary figure 1 present the analyses of log transformed alcohol consumption. The spouse, offspring-parent, and sibling correlations are stronger than untransformed g/d consumed (Table 1). The curves relating log consumption between offspring and parents and among sibs exhibit substantial concavity suggesting substantially weaker heritability for higher and lower offspring consumption vs. intermediate offspring consumption.
Other macronutrients
Supplementary table 3 shows that quantile-specific concordance among family members was not characteristic of other macronutrient intakes. Specifically, the percent intakes of protein, fat (total, saturated, monounsaturated, polyunsaturated), and carbohydrates exhibited moderate offspring-parent (0.07 to 0.16) and sibling correlations (0.10 to 0.17), but little evidence of quantile-specific expressivity, i.e., the slopes were generally the same (i.e., regression lines parallel) for all percentiles of the offspring distribution.
Discussion
Our analyses suggest that the heritability of alcohol consumption increases with increasing percentiles of its population distribution. Heritability (h2=2βPO/(1+rspouse)) at the 90th percentile of the population distribution was 4.5-fold greater than h2 at the 10th percentile. The alcohol results were replicated for both offspring-parent and full-sib estimates of heritability. Important caveats to our analyses are: 1) h2 lacks the specificity of directly measured genotypes even if it is a more inclusive genetic measure; and 2) the formula used for h2 probably do not adequately represent the true complexity of alcohol genetics and shared environmental effects. To addresses these concerns, the discussion to follow re-evaluates published studies from the perspective of quantile-dependent expressivity. These are studies that differed from our analyses in that they measured genetic variants directly, or they used different estimates of heritability. They expand the analyses to include examples of both alcohol consumption and dependence, where the genetic effect size (i.e., difference between genotypes) increases in association with an increase in the phenotypic mean, per Figures 1 and 2.
Quantile-dependent expressivity is intended to provide a perspective to alcohol’s gene-environment interactions that complements their more traditional interpretations, i.e. as specific behavioral or environmental restrictions that promote abstinence and restricted availability (social controls), and stressors that potentiate genetic liability (social triggers, Chartier et al. 2017). Quantile-dependent expressivity considers these restrictions and stressors as affecting overall average consumption, which in turn affects the expressivity of the genetic variants, as has been shown for other traits. Specifically, it is hypothesized that the environment affects alcohol consumption, which in turn affects the phenotypic expression of genetic trait (quantile-specific expressivity: environment→ phenotype→genetic expression) rather than the more traditional interpretation of gene-environment interaction (environment→genetic expression→phenotype).
D4 dopamine receptor gene (DRD4)
For example, Hutchison et al. (2003) reported that the DRD4 variable number of tandem repeat (VNTR) polymorphism affected the change in alcohol craving when participants viewed, poured, smelled, and tasted their favorite beer. Specifically, providing beer produced a greater urge to drink in carriers of the L-allele (i.e., ≥7 tandem repeats) than in SS homozygotes (containing <7 tandem repeats, Figure 3A histogram). Alternatively, the line-graph in Figures 3A (derived from their figure 1) suggests that the genotype-specific increases were possibly the effects of quantile-dependent expressivity in association with the higher average craving when beer was provided. In particular, the smaller genetic effect size at lower (pre-beer) than higher (post-beer) average craving requires that the craving trajectories cannot move in parallel for the two genotypes when average craving increased. Subtracting pre-exposure from the post-exposure craving will necessarily require a greater craving increase in for the genotype with the higher pre-exposure craving (LL/LS genotypes) vis-à-vis that with lower exposure craving (SS genotype).
Figure 3.

Traditional perspective (histogram inserts of mean changes or differences by genotype) vs. quantile-dependent expressivity interpretation (line graphs showing larger genetic effect size when average levels were high vs. lower), for: A) Hutchison et al. 2003 report (derived from controls in their figure 1) on alcohol craving before and after viewing, pouring, smelling, and tasting their favorite beer in 21 participants homozygous for <7 tandem repeats in their D4 dopamine receptor gene polymorphism and 9 participants with one or both alleles having ≥7 tandem repeats; B) Meyer et al. 2015 report on the effect of childhood adversity on the maximum number of drinks in a 24-hour period (Maxdrinks) during the period of lifetime heaviest drinking by 601 GG homozygotes and 542 A-allele carriers of the ADH1B-rs1229984 polymorphism (Pinteraction<0.01); C) the aforementioned analysis for DSM-Alcohol Use Disorder (AUD) severity (Pinteraction<0.01); D) Schacht et al.’s 2017 report on the effect of 4-month μ-opioid antagonist naltrexone on percent heavy drinking days in 33 AA homozygotes and 33 G-allele carriers of the μ-opioid receptor (OPRM1) gene.
Alcohol dehydrogenase 1B (ADH1B)
The same principle applies when comparing conditions of low and high alcohol consumption or use disorder (AUD) cross-sectionally. The A-allele of the ADH1B rs1229984 polymorphism encodes a histidine for arginine48 substitution (ADH1B*2) that causes alcohol dehydrogenase 1B activity and alcohol clearance to increase substantially. Increase ethanol-acetaldehyde conversion causes a buildup of acetaldehyde, which results in flushing and other unpleasant effects that discourage drinking. Meyers et al. (2015) reported that childhood adversity significantly increased both the maximum drinks/day (Maxdrinks, P<0.001) and AUD severity (P<0.0001) in Israeli Jews, and that the magnitude of the increase was significantly reduced in carriers of the A-allele vis-à-vis GG-homozygotes for both Maxdrinks (Figure 3B histogram, Pinteraction<0.01), and AUD severity (Figure 3C histogram, Pinteraction<0.01). Alternatively, the line-graphs of Figures 3B and 3C suggest that the genotype-specific differences were possibly the effects of quantile-dependent expressivity in association with the higher average Maxdrinks and AUD of childhood adversity. In particular, the smaller genetic effect size at lower (non-adverse) than higher (adverse) average Maxdrinks requires that the differences cannot move in parallel for the two genotypes when average consumption is increased. Subtracting the non-adverse from the adverse values will necessarily require a greater increase in Maxdrinks for the genotype with the higher non-adverse value (ADH1B*2 carriers) vis-à-vis the genotype with lower non-adverse value (ADH1B*1). The same goes for AUD.
In another study, Chartier et al. (2016) reported that religious involvement significantly decreased both maximum drinks consumed and alcohol dependence symptoms (P<0.0001), and that ADH1B-rs2066702, ADH1C-rs698, and ADH4-rs1042364 risk variants had significantly weaker associations with both maximum drinks (Pinteraction≤0.03) and alcohol dependence (Pinteraction≤0.005) when there was greater religious involvement, i.e., smaller genetic effect size at lower average consumption and dependence per quantile-dependent expressivity.
Mu-opioid receptor gene (OPRM1)
Naltrexone is an opioid receptor antagonist that modulates alcohol consumption by reducing dopamine release in the medial forebrain and nucleus accumbens (Hartwell et al. 2020). Although individual studies have yielded mixed results, meta-analyses of seven randomized trials suggest that the non-synonymous Asn40Asp substitution (A118G) in OPRM1 rs1799971 modifies naltrexone effect on drinks per day (P=0.02) (Hartwell, 2020). For example, the histogram in Figure 3D presents Schacht et al’s finding that the naltrexone-placebo difference in the proportion of heavy drinking days was greater the carriers of the G allele than AA homozygotes. However, the associated line graph suggests that the genetic effect size was smaller on naltrexone when heavy drinking averaged less (15%), than on placebo, when heavy drinking averaged more (23.7%).
α2 subunit of the GABAA receptor (GABRA2)
Our analysis of the drunkenness frequency data presented by Dick et al. (2014) suggests a highly significant concordance between average frequency and the estimated genotype difference between AA and G-allele carriers of the GABRA2 rs279858 polymorphism in young adults who were ≥18 years old. From their figures 1 and 2, the provided means (±SE) and estimated genotype differences for drunk days/year were, respectively: 18.9±2.4 and −2.7 at eighteen, 32.5±3.5 and 13.0 at nineteen, 33.4±3.3 and 11.3 at twenty, 29.6±2.8 and 9.0 at twenty-one, 28.0±3.0 and 7.4 at twenty-two, 27.3±3.6 and 5.1 at twenty-three, 27.2±4.9 and 3.7 at twenty-four, and 19.1±3.8 and 1.6 at twenty-five (regression of genetic effect size vs. mean, slope±SE: 0.90±0.13, P=0.0006).
The aldehyde dehydrogenase gene (ALDH2) exception
Quantile-dependent expressivity is probably a gene-specific property. The ALDH2*2 allele of the mitochondrial ALDH2 rs671 polymorphism causes acetaldehyde to accumulate from drinking. This causes carriers of the ALDH2*2 allele to experience dysphoric symptoms, which limits their drinking and AUD risk relative to homozygotes for the fully active allele (ALDH2*1). Several studies report that the genetic effect size of the ALDH2 polymorphism is greater under conditions generally associated with lower (not greater) consumption. Irons et al. (2012) reported that high parental alcohol use and misuse reduced ALDH2*2’s protective effect, whilst low parental alcohol use and misuse enhanced ALDH2*2’s protective effect. Higuchi et al. (1994) also reported reduced protective effects of ALDH2*2 genotype as exposure to alcohol increased. Bujarski et al. (2015) reported significantly greater (not less) alcohol intake in ALDH2*2 carriers than ALDH1*1 homozygotes in Asian-American young adults with family history of alcohol problems and who drank more, but not in those without such history who drank less (Pinteraction<0.01). Thus the genetic effect size may be greater at lower percentiles of the alcohol use distribution, however, O'Shea et al. (2017) did report that both genetic effect size (P=0.01) and average consumption (P=0.001) increase with the proportion of friends who get intoxicated. These ALDH2*2-allele results may not be particularly relevant to the Framingham analysis of Figures 1 and 2 given the allele is found almost exclusively in northeast Asians. Moreover, the proportion of the total variance explained by any particular SNP is generally too small to noticeably affect overall h2 (Evangelou 2019).
The modifying effects of the serotonin transporter gene (Covault et al. 2007, Nilsson et al. 2005), the dopamine D2 receptor gene (Pieters et al. 2012, van der Zwaluw et al. 2010), and the Monoamine oxidase A gene (Nilsson et al. 2007) on alcohol consumption and the environment are discussed as supplementary material.
There are multiple twin studies of alcohol consumption and dependence using alternative estimates of heritability that are consistent with quantile-dependent expressivity.
Alcohol-specific and general externalizing genetic risk factors.
Kendler et al. (2011) reported that peer group deviance, alcohol availability, and low pro-social behavior interacted significantly with inherited risk factors in affecting maximal yearly alcohol consumption in boys 12-14 years old (Figures 4A-4C histograms). Alternatively, the line graphs of Figures 4A-4C suggest a larger genetic effect size when average intake was greater at high (+1 SD) vs. low (−1 SD) peer group deviance, high (+1 SD) vs. low alcohol availability (−1 SD), and low (−1 SD) vs. high (+1 SD) pro-social activity levels, consistent with quantile-dependent expressivity. The larger genetic effect size when adolescence maximal alcohol consumption was high vs. low mandates unequal effects by genotypes (represented by the nonparallel lines).
Figure 4.

Traditional perspective (histogram inserts of drinking differences between environments by tertiles of inherited risk score) vs. quantile-dependent expressivity interpretation (line graphs showing larger inherited effect when average levels were high vs. lower) for Kendler et al’s 2011 report on estimated maximum yearly alcohol consumed (log transformed) in boys aged 12–14 by: A) peer group deviance (PGD) and low (−1 SD), intermediate (mean), and high inherited risk (+1 SD) for alcohol use disorders (Pinteraction<0.0001); B) alcohol (alc) availability and low (−1 SD), intermediate (mean), and high inherited risk (+1 SD) for externalizing disorders (Pinteraction<0.0001); and C) pro-social activity level (PAL) and low (- SD), intermediate (mean), and high (+1 SD) inherited risk for externalizing disorders (Pinteraction <0.0001). Derived from their figures 2-4. IRS: inherited risk score, SD: standard deviation.
Parental closeness
Miles et al. (2005) reported a greater genetic effect on alcohol use as estimated by the difference in correlations between MZ and DZ twins (0.97 vs. 0.67, p < .001) in families with low parental closeness vis-à-vis a smaller genetic effect in families with strong parental closeness (MZ-DZ correlations: 0.79 vs. 0.73, p = 0.24), consistent with lower intake of the close parent households and quantile-dependent expressivity.
Parental drinking
Cleveland et al. (2003) reported that parental drinking was positively correlated with adolescent drinking (r=0.14, P<0.05), and correspondingly, the heritability of adolescent drinking was greater in families with drinking parents than in families with parents who drank little or abstained entirely (h2=0.57 vs. 0.34, computed among sibs).
Peer deviance, stressful life events, parental knowledge
Dick et al. (2007) reported that adolescent alcohol use was positively correlated with friends’ alcohol use (r=0.48), and consistent with quantile-dependent expressivity, genetic influences on the adolescent’s alcohol use were greater when more peers used alcohol. Cooke et al. (2015) report that the frequency of alcohol intake increased in association with greater peer deviance (r=0.44) and stressful life events (r=0.11) and less parental knowledge (r=−0.31, P<0.01 for all) in 803 MZ (rMZ=0.71) and 825 DZ twin pairs (rDZ=0.49). Consistent with quantile-dependent expressivity, the additive genetic variance increased significantly with increasing peer deviance and decreasing parental monitoring, but did not change significantly with the number of potentially stressful life events (i.e., the environmental factor showing the weakest effect on frequency of alcohol use).
Marriage
Marriage is reported to moderate the relative importance of genetic effects on alcohol consumption, consistent with quantile-dependent expressivity. Heath et al. (1989) reported that genetic differences in young female twins (≤30 years) accounted for a greater percentage of the variance in alcohol consumption if single than married (60% vs. 31%) consistent with the higher alcohol intake of the single women (6.43±0.35 vs. 4.17±0.21 drinks/wk). In older twins, the genetic contribution to variations in their alcohol intake was 76% if single vs. 59% if married, which again corresponded with higher average alcohol intake when single (6.74±0.55 vs. 6.23±0.26 drinks/wk).
Rural vs. Urban dwelling
Studies show greater heritability of alcohol use or involvement in urban than rural residents (Davis et al. 2017, Legrand et al. 2008, Rose et al. 2001). The difference in heritability has been attributed to a restrictive rural environment that does not allow for the expression of a genetic predisposition versus a more permissive or enriching urban environment that allows for its expression. Rural existence may also engender a greater respect towards community monitoring, greater religiousness, and more poverty than urban existence. The difficulty with these explanations, and one based on quantile-dependent expressivity, is that only one of the four studies shows greater alcohol use in urban than rural adolescents.
The exception is Davis et al.’s (2017) analysis of the 1962 National Merit Twin Study, in which alcohol involvement was greater in urban than rural college-bound adolescent females (1.44 vs. 0.84) and males (1.76 vs. 1.49). Heritability for alcohol involvement was also greater in urban than rural females (0.44 vs. 0.02), consistent with quantile-dependent expressivity, but not in males (0.07 vs. 0.66).
The other studies report minor regional differences in average consumption levels. For example, Rose et al. (2001) hypothesized that the smaller genetic effect in rural drinking frequency they observed was due to the limited number of shops selling alcohol in the sparsely populated rural areas. However, rural and urban drinking frequency did not differ in their sample. Legrand et al. (2008) also reported significantly smaller genetic effects in rural than urban male (3% vs. 49%) and female twins (3% vs. 19%). Again, there was little difference in mean rural vs. urban consumption scores (males: 3.4±0.24 vs. 3.3±0.16, respectively; females: 2.7±0.16 vs. 2.8±0.16, respectively). Rural areas include a lower proportion of young adults, sell less alcohol, and experience less migration than urban areas in Finland. Dick et al. (2001) reported that the heritability of alcohol intake was lower for regions with the least vs. most young adults (i.e., 13% vs. 63%), with the lowest vs. highest alcohol sales (males: 32% vs. 49%; females: 25% vs. 61%), and with the lowest vs. highest migration (16% vs. 60%). In contrast to these large differences in heritability, the frequency of alcohol use was only weakly related to differences in the proportion of young adults (r=0.04), money spent in alcohol sales (r=0.08), and migration rates across regions (r=0.03).
Age
The genetic influences affecting alcohol intake and behavior increase with adolescent age in accordance with increasing mean intake, consistent with quantile-dependent expressivity. Rose et al. (2001) reported that the proportion of the genetic variance in drinking frequency increased from 33% at age 16, 49% at 17 and 50% at 18 in their twin study as abstinence fell by a factor of 2.5 or more and the proportion of the sample drinking ≥twice monthly more than doubled.
Additional exceptions
Sartor et al. (2014) reported that AUD symptoms showed smaller (not larger) genotype differences for the ADH1B-rs1229984 polymorphism in European–American men experiencing childhood adversity than those who did not, despite their higher average AUD symptoms. Davis et al. (2018) reported substantially smaller (not larger) genetic influences in adolescent twins having the highest vs. the lowest family income (2% vs. 50%) despite a weak concordance between income and alcohol involvement r=0.09, P=0.002).
Assortative mating
Spousal drinking habits are known to correlate due to phenotypic assortment (Ask et al. 2012, Reynolds et al. 2006) and cohabitation (Ask et al. 2012, Grant et al. 2007), and to a lesser extent social homogamy (Reynolds et al. 2006, Maes et al. 1998). Quantile regression showed that the wifes’ alcohol intake was moderately related to that of the husband’s. Moreover, the effect of the husband’s intake was greater at higher percentiles of the wife’s distribution. Similar results were obtained for wine and liquour but not beer because beer was primarily consumed by men, and probably in the company of men. This suggests that the traditional spouse correlation may underestimate the importance of spouse interaction at the highest consumption levels, i.e., that portion of the distribution having the highest priority for behavioral interventions. The Framingham family sets provide no insight on the degree to which spouse concordance was the consequence of assortative mating, social homogamy, or marital contagion.
Log transformation
Alcohol consumption is often logarithmically transformed because of its right-skewed distribution (van Beek et al. 2014, Heath et al. 1989, Schumann et al. 2011). This transformation appears to be made purely to conform to the statistical requirements of the model (normality) and not for theoretical considerations of inherited drinking. Quantile regression provides a robust approach to estimating βOP and βFS that does not require normality, however, the slopes will be affected by data transformations. The log transformation has the effect of changing the quantile-specific graphs of the offspring-parent and sibling slopes from a slightly convex increasing (Figures 1) or linearly increasing function (Figure 2) to a strongly concave function (Supplementary figure 1) that may be less easily interpreted than the untransformed results. We would argue that untransformed alcohol intake (i.e., g/d or as percent calories) is a more natural metric than their log transformation, and untransformed intake has been used in twin and other studies of consumption (Kendler et al. 2010).
Other macronutrients
The heritabilities we estimated for intakes of protein, fat and carbohydrates are consistent with those published by others (Hasselbalch et al. 2008) and showed no evidence for quantile-specific inheritance. The genetic regulation of alcohol intake may be behaviorally driven whereas other macronutrients may be driven by nutritional requirements.
Limitations
The analysis of the Framingham family sets is based on self-reported alcohol consumption that is subject to recall bias and under-reporting by heavy drinkers. The gene-environment examples cited above include a variety of metrics including drinking frequency, intoxication, and dependence that may involve different genetic influences than usual intake. Nonetheless, self-reported measures of alcohol consumption, including drink frequency and regular quantity at the time of heaviest drinking, have been shown to correlate strongly with the genetic risk for alcohol use disorders such as dependence. Drinkers at higher percentiles of the alcohol consumption distribution may represent a different phenotype (alcohol use disorder) that has a higher heritability. Our analyses do not control for gene-environment correlation, when a larger genetic effect results in the greater active or passive selection of environmental conditions that increase consumption average. Supplementary table 1 shows that the Framingham sample were middle-aged and older, which is not when alcohol intake is at its peak. As evident from the exceptions noted, not all published reports fit the quantile-dependent expressivity model, just as not all published reports provide consistent evidence for the same gene-environment interactions. This inconsistency probably reflects type I and type II statistical errors, study and population differences, and heterogeneity in gene-environment interactions.
In conclusion, the heritability of alcohol consumption, but not other nutrients, depends upon whether average consumption is high or low relative to the population distribution. The many published examples of gene-environment interactions that are consistent with quantile-dependent expressivity support its validity and utility. Previously published gene-environment interactions involving parental drinking, parental knowledge, parental control, good family relations, peer deviance, stressful life events, and marriage suggest practical approaches for limiting alcohol misuse. Quantile-dependent expressivity provides a conceptual framework for interpreting these interactions that has been shown to be apropos to simpler phenotypes, including cholesterol, triglycerides, body weight, postprandial lipemia, and pulmonary function.
Supplementary Material
Acknowledgement
The data were obtained from the National Institutes of Health FRAMCOHORT, GEN3, FRAMOFFSPRING Research Materials obtained from the NHLBI Biologic Specimen and Data Repository Information Coordinating Center. The author (PTW) was responsible for the project conception, development of overall research plan, analyzing data including performed the statistical analysis, and wrote the paper. The sole author had responsibility for all parts of the manuscript. There are no conflicts of interest to report. The Framingham Heart Study was conducted and supported by the National Heart, Lung, and Blood Institute (NHLBI) in collaboration with Boston University (Contract No. N01-HC-25195 and HHSN268201500001I). Funding support for the Framingham Food Frequency Questionnaire dataset was provided by ARS Contract #53- 3k06-5-10, ARS Agreement #’s 58-1950-9-001, 58-1950-4-401 and 58-1950-7-707. This manuscript was not prepared in collaboration with investigators of the Framingham Heart Study and does not necessarily reflect the opinions or views of the Framingham Heart Study, FRAMCOHORT, GEN3, FRAMOFFSPRING, Boston University, or NHLBI.
This research was supported by grant R21ES020700 from the National Institute of Environmental Health Sciences, and an unrestricted gift from HOKA ONE ONE.
Footnotes
Conflicts of interest: None to report.
Publisher's Disclaimer: This Author Accepted Manuscript is a PDF file of an unedited peer-reviewed manuscript that has been accepted for publication but has not been copyedited or corrected. The official version of record that is published in the journal is kept up to date and so may therefore differ from this version.
Literature cited
- American Psychiatric Association (2013) Diagnostic and Statistical Manual of Mental Disorders. 5. Arlington, VA: American Psychiatric Publishing. [Google Scholar]
- Ask H, Rognmo K, Torvik FA, Røysamb E, Tambs K (2012) Non-random mating and convergence over time for alcohol consumption, smoking, and exercise: the Nord-Trøndelag Health Study. Behav Genet. 42:354–65. [DOI] [PubMed] [Google Scholar]
- Bujarski S, Lau AS, Lee SS, Ray LA (2015) Genetic and environmental predictors of alcohol use in Asian American young adults. J Stud Alcohol Drugs. 76:690–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chartier KG, Dick DM, Almasy L, Chan G, Aliev F, Schuckit MA, Scott DM, Kramer J, Bucholz KK, Bierut LJ, Nurnberger J Jr, Porjesz B, Hesselbrock VM (2016) Interactions between alcohol metabolism genes and religious involvement in association with maximum drinks and alcohol dependence symptoms. J Stud Alcohol Drugs. 77:393–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chartier KG, Karriker-Jaffe KJ, Cummings CR, Kendler KS. (2017) Review: Environmental influences on alcohol use: Informing research on the joint effects of genes and the environment in diverse U.S. populations. Am J Addict. 26:446–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleveland HH, Wiebe RP (2003) The moderation of genetic and shared-environmental influences on adolescent drinking by levels of parental drinking. J Stud Alcohol. 64:182–94. [DOI] [PubMed] [Google Scholar]
- Cooke ME, Meyers JL, Latvala A, Korhonen T, Rose RJ, Kaprio J, Salvatore JE, Dick DM (2015) Gene-environment interaction effects of peer deviance, parental knowledge and stressful life events on adolescent alcohol use. Twin Res Hum Genet. 18:507–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covault J, Tennen H, Armeli S, Conner TS, Herman AI, Cillessen AH, Kranzler HR (2007) Interactive effects of the serotonin transporter 5-HTTLPR polymorphism and stressful life events on college student drinking and drug use. Biol Psychiatry. 61:609–16. [DOI] [PubMed] [Google Scholar]
- Davis CN, Natta SS, Slutske WS (2017) Moderation of genetic influences on alcohol involvement by rural residency among adolescents: results from the 1962 National Merit Twin Study. Behav Genet. 47:587–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis CN, Slutske WS (2018) Socioeconomic status and adolescent alcohol involvement: evidence for a gene-environment interaction. J Stud Alcohol Drugs. 79:725–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick DM, Rose RJ, Viken RJ, Kaprio J, Koskenvuo M (2001) Exploring gene-environment interactions: socioregional moderation of alcohol use. J Abnorm Psychol. 110:625–32. [DOI] [PubMed] [Google Scholar]
- Dick DM, Pagan JL, Holliday C, Viken R, Pulkkinen L, Kaprio J, Rose RJ (2007) Gender differences in friends' influences on adolescent drinking: a genetic epidemiological study. Alcohol Clin Exp Res. 31:2012–9. [DOI] [PubMed] [Google Scholar]
- Dick DM, Meyers JL, Rose RJ, Kaprio J, Kendler KS (2011) Measures of current alcohol consumption and problems: Two independent twin studies suggest a complex genetic architecture. Alcoholism: Clinical and Experimental Research 35:2152–2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick DM, Cho SB, Latendresse SJ, Aliev F, Nurnberger JI Jr, Edenberg HJ, Schuckit M, Hesselbrock VM, Porjesz B, Bucholz K, Wang JC, Goate A, Kramer JR, Kuperman S (2014) Genetic influences on alcohol use across stages of development: GABRA2 and longitudinal trajectories of drunkenness from adolescence to young adulthood. Addict Biol. 19:1055–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evangelou E, Gao H, Chu C, Ntritsos G, Blakeley P, Butts AR, Pazoki R, Suzuki H, Koskeridis F, Yiorkas AM, Karaman I, Elliott J, Luo Q, Aeschbacher S, Bartz TM, Baumeister SE, Braund PS, Brown MR, Brody JA, Clarke TK, Dimou N, Faul JD, Homuth G, Jackson AU, Kentistou KA, Joshi PK, Lemaitre RN, Lind PA, Lyytikäinen LP, Mangino M, Milaneschi Y, Nelson CP, Nolte IM, Perälä MM, Polasek O, Porteous D, Ratliff SM, Smith JA, Stančáková A, Teumer A, Tuominen S, Thériault S, Vangipurapu J, Whitfield JB, Wood A, Yao J, Yu B, Zhao W, Arking DE, Auvinen J, Liu C, Mäannikkö M, Risch L, Rotter JI, Snieder H, Veijola J, Blakemore AI, Boehnke M, Campbell H, Conen D, Eriksson JG, Grabe HJ, Guo X, van der Harst P, Hartman CA, Hayward C, Heath AC, Jarvelin MR, Kähönen M, Kardia SLR, Kühne M, Kuusisto J, Laakso M, Lahti J, Lehtimäki T, McIntosh AM, Mohlke KL, Morrison AC, Martin NG, Oldehinkel AJ, Penninx BWJH, Psaty BM, Raitakari OT, Rudan I, Samani NJ, Scott LJ, Spector TD, Verweij N, Weir DR, Wilson JF, Levy D, Tzoulaki I, Bell JD, Matthews PM, Rothenfluh A, Desrivières S, Schumann G, Elliott P (2019) New alcohol-related genes suggest shared genetic mechanisms with neuropsychiatric disorders. Nat Hum Behav. 3:950–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falconer DS, Mackay TFC. (1996) Introduction to Quantitative Genetics (fourth ed.) Longmans Green, Harlow, Essex, UK [Google Scholar]
- Giovannucci E, Colditz G, Stampfer MJ, Rimm EB, Litin L, Sampson L, Willett WC (1991) The assessment of alcohol consumption by a simple self-administered questionnaire. Am J Epidemiol. 133:810–817. [DOI] [PubMed] [Google Scholar]
- Gould WW (1992) Quantile regression with bootstrapped standard errors. Stata Technical Bulletin. 9:19–21. [Google Scholar]
- Grant JD, Heath AC, Bucholz KK, Madden PA, Agrawal A, Statham DJ, Martin NG (2007) Spousal concordance for alcohol dependence: evidence for assortative mating or spousal interaction effects? Alcohol Clin Exp Res. 31:717–28. [DOI] [PubMed] [Google Scholar]
- Hamdi NR, Krueger RF, South SC (2015) Socioeconomic status moderates genetic and environmental effects on the amount of alcohol use. Alcohol Clin Exp Res. 39:603–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansell NK, Agrawal A, Whitfield JB, Morley KI, Zhu G, Lind PA, Pergadia ML, Madden PA, Todd RD, Heath AC, Martin NG (2008) Long-term stability and heritability of telephone interview measures of alcohol consumption and dependence. Twin Res Hum Genet. 11:287–305. [DOI] [PubMed] [Google Scholar]
- Hartwell EE, Feinn R, Morris PE, Gelernter J, Krystal J, Arias AJ, Hoffman M, Petrakis I, Gueorguieva R, Schacht JP, Oslin D, Anton RF, Kranzler HR (2020) Systematic review and meta-analysis of the moderating effect of rs1799971 in OPRM1, the mu-opioid receptor gene, on response to naltrexone treatment of alcohol use disorder. Addiction. 2020 (in press) doi: 10.1111/add.14975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasselbalch AL, Heitmann BL, Kyvik KO, Sørensen T (2008) Studies of twins indicate that genetics influence dietary intake. J Nutr. 2008;138:2406–12. [DOI] [PubMed] [Google Scholar]
- Hasin DS, Stinson FS, Ogburn E, Grant BF (2007) Prevalence, correlates, disability, and comorbidity of DSM-IV alcohol abuse and dependence in the United States: results from the National Epidemiologic Survey on Alcohol and Related Conditions. Archives of General Psychiatry. 64:830–842 [DOI] [PubMed] [Google Scholar]
- Heath AC, Jardine R, Martin NG (1989) Interactive effects of genotype and social environment on alcohol consumption in female twins. J Stud Alcohol. 50:38–48. [DOI] [PubMed] [Google Scholar]
- Higuchi S, Matsushita S, Imazeki H, Kinoshita T, Takagi S, Kono H (1994) Aldehyde dehydrogenase genotypes in Japanese alcoholics. Lancet 1994;343:741–742. [DOI] [PubMed] [Google Scholar]
- Hutchison KE, Wooden A, Swift RM, Smolen A, McGeary J, Adler L, Paris L (2003) Olanzapine reduces craving for alcohol: a DRD4 VNTR polymorphism by pharmacotherapy interaction. Neuropsychopharmacology 28:1882–8. [DOI] [PubMed] [Google Scholar]
- Irons DE, Iacono WG, Oetting WS, McGue M (2012) Developmental trajectory and environmental moderation of the effect of ALDH2 polymorphism on alcohol use. Alcohol Clin Exp Res. 36:1882–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson C, Henriksen L, Dickinson D (1999) Alcohol-specific socialization, parenting behaviors and alcohol use by children. J Stud Alcohol 60:362–367. [DOI] [PubMed] [Google Scholar]
- Jarnecke AM, South SC (2014) Genetic and environmental influences on alcohol use problems: moderation by romantic partner support, but not family or friend support. Alcohol Clin Exp Res. 38:367–75. [DOI] [PubMed] [Google Scholar]
- Johnson W, Kyvik KO, Mortensen EL, Skytthe A, Batty GD, Deary IJ (2011) Does education confer a culture of healthy behavior? Smoking and drinking patterns in Danish twins. Am J Epidemiol. 173:55–63. [DOI] [PubMed] [Google Scholar]
- Kannel WB, Feinleib M, McNamara PM, Garrison RJ, Castelli WP (1979) An investigation of coronary heart disease in families. The Framingham offspring study. Am J Epidemiol. 110:281–90. [DOI] [PubMed] [Google Scholar]
- Karlin S, Cameron EC, Williams PT (1981) Sibling and parent--offspring correlation estimation with variable family size. Proc Natl Acad Sci U S A. 78:2664–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendler KS, Schmitt E, Aggen SH, Prescott CA (2008) Genetic and environmental influences on alcohol, caffeine, cannabis, and nicotine use from early adolescence to middle adulthood. Arch Gen Psychiatry. 65:674–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendler KS, Myers J, Dick D, Prescott CA (2010) The relationship between genetic influences on alcohol dependence and on patterns of alcohol consumption. Alcohol Clin Exp Res. 34:1058–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendler KS, Gardner C, Dick DM (2011) Predicting alcohol consumption in adolescence from alcohol-specific and general externalizing genetic risk factors, key environmental exposures and their interaction. Psychol Med 41:1507–1516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimokoti RW, Newby PK, Gona P, Zhu L, Campbell WR, D'Agostino RB, Millen BE. (2012) Stability of the Framingham Nutritional Risk Score and its component nutrients over 8 years: the Framingham Nutrition Studies. Eur J Clin Nutr. 66:336–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenker R, Hallock KF (2001) Quantile regression. J Economic Perspectives. 15:143–56. [Google Scholar]
- Koopmans JR, Slutske WS, van Baal GC, Boomsma DI (1999) The influence of religion on alcohol use initiation: evidence for genotype X environment interaction. Behav Genet. 29:445–53. [DOI] [PubMed] [Google Scholar]
- Legrand LN, Keyes M, McGue M, Iacono WG, Krueger RF (2008) Rural environments reduce the genetic influence on adolescent substance use and rule-breaking behavior. Psychol Med. 38:1341–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maes HH, Neale MC, Kendler KS, Hewitt JK, Silberg JL, Foley DL, Meyer JM, Rutter M, Simonoff E, Pickles A, Eaves LJ (1998) Assortative mating for major psychiatric diagnoses in two population-based samples. Psychol Med. 28:1389–401. [DOI] [PubMed] [Google Scholar]
- Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, McCarthy MI, Ramos EM, Cardon LR, Chakravarti A, Cho JH, Guttmacher AE, Kong A, Kruglyak L, Mardis E, Rotimi CN, Slatkin M, Valle D, Whittemore AS, Boehnke M, Clark AG, Eichler EE, Gibson G, Haines JL, Mackay TF, McCarroll SA, Visscher PM (2009) Finding the missing heritability of complex diseases. Nature. 461:747–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGue M (1999). Behavioral genetic models of alcoholism and drinking. In Leonard KE & Blane HT (Eds.), Psychological theories of drinking and alcoholism (2nd ed., pp. 372–421). New York, NY [Google Scholar]
- Meyers JL, Shmulewitz D, Wall MM, Keyes KM, Aharonovich E, Spivak B, Weizman A, Frisch A, Edenberg HJ, Gelernter J, Grant BF, Hasin D (2015) Childhood adversity moderates the effect of ADH1B on risk for alcohol-related phenotypes in Jewish Israeli drinkers. Addict Biol. 20:205–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miles DR, Silberg JL, Pickens RW, Eaves LJ (2005) Familial influences on alcohol use in adolescent female twins: testing for genetic and environmental interactions. J Stud Alcohol 66:445–51. [DOI] [PubMed] [Google Scholar]
- Nilsson KW, Sjöberg RL, Damberg M, Alm PO, Ohrvik J, Leppert J, Lindström L, Oreland L (2005) Role of the serotonin transporter gene and family function in adolescent alcohol consumption. Alcohol Clin Exp Res. 2005 29:564–70. [DOI] [PubMed] [Google Scholar]
- Nilsson KW, Sjöberg RL, Wargelius HL, Leppert J, Lindström L, Oreland L (2007) The monoamine oxidase A (MAO-A) gene, family function and maltreatment as predictors of destructive behaviour during male adolescent alcohol consumption. Addiction. 102:389–98. [DOI] [PubMed] [Google Scholar]
- O'Shea T, Thomas N, Webb BT, Dick DM, Kendler KS, Chartier KG (2017) ALDH2*2 and peer drinking in East Asian college students. Am J Drug Alcohol Abuse. 43:678–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds CA, Barlow T, Pedersen NL (2006) Alcohol, tobacco and caffeine use: spouse similarity processes. Behav Genet. 36:201–15. [DOI] [PubMed] [Google Scholar]
- Rietschel M, Treutlein J (2013) The genetics of alcohol dependence. Ann N Y Acad Sci. 1282:39–70. [DOI] [PubMed] [Google Scholar]
- Rimm EB, Giovannucci EL, Stampfer MJ, Colditz GA, Litin LB, Willett WC (1992) Reproducibility and validity of an expanded self-administered semiquantitative food frequency questionnaire among male health professionals. Am J Epidemiol. 135:1114–26 [DOI] [PubMed] [Google Scholar]
- Rose RJ, Dick DM, Viken RJ, Kaprio J (2001) Gene-environment interaction in patterns of adolescent drinking: regional residency moderates longitudinal influences on alcohol use. Alcohol Clin Exp Res. 25:637–43. [PubMed] [Google Scholar]
- Sartor CE, Wang Z, Xu K, Kranzler HR, Gelernter J (2014) The joint effects of ADH1B variants and childhood adversity on alcohol related phenotypes in African-American and European-American women and men. Alcohol Clin Exp Res. 38:2907–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schacht JP, Randall PK, Latham PK, Voronin KE, Book SW, Myrick H, Anton RF (2017) Predictors of naltrexone response in a randomized trial: reward-related brain activation, OPRM1 genotype, and smoking status. Neuropsychopharmacology. 42:2640–2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumann G, Coin LJ, Lourdusamy A, Charoen P, Berger KH, Stacey D, Desrivières S, Aliev FA, Khan AA, Amin N, Aulchenko YS, Bakalkin G, Bakker SJ, Balkau B,Beulens JW, Bilbao A, de Boer RA, Beury D, Bots ML, Breetvelt EJ, Cauchi S, Cavalcanti-Proença C, Chambers JC, Clarke TK, Dahmen N, de Geus EJ, Dick D, Ducci F, Easton A, Edenberg HJ, Esko T, Fernández-Medarde A, Foroud T, Freimer NB, Girault JA, Grobbee DE, Guarrera S, Gudbjartsson DF, Hartikainen AL, Heath AC, Hesselbrock V, Hofman A, Hottenga JJ, Isohanni MK, Kaprio J, Khaw KT, Kuehnel B, Laitinen J, Lobbens S, Luan J, Mangino M, Maroteaux M, Matullo G, McCarthy MI, Mueller C, Navis G, Numans ME, Núñez A, Nyholt DR, Onland-Moret CN, Oostra BA, O'Reilly PF, Palkovits M, Penninx BW, Polidoro S, Pouta A, Prokopenko I, Ricceri F, Santos E, Smit JH, Soranzo N, Song K, Sovio U, Stumvoll M, Surakk I, Thorgeirsson TE, Thorsteinsdottir U, Troakes C, Tyrfingsson T, Tönjes A, Uiterwaal CS, Uitterlinden AG, van der Harst P, van der Schouw YT, Staehlin O, Vogelzangs N, Vollenweider P, Waeber G, Wareham NJ, Waterworth DM, Whitfield JB, Wichmann EH, Willemsen G, Witteman JC, Yuan X, Zhai G, Zhao JH, Zhang W, Martin NG, Metspalu A, Doering A, Scott J, Spector TD, Loos RJ, Boomsma DI, Mooser V, Peltonen L, Stefansson K, van Duijn CM, Vineis P, Sommer WH, Kooner JS, Spanagel R, Heberlein UA, Jarvelin MR, Elliott P (2011) Genome-wide association and genetic functional studies identify autism susceptibility candidate 2 gene (AUTS2) in the regulation of alcohol consumption. Proc Natl Acad Sci U S A. 108:7119–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Splansky GL, Corey D, Yang Q, Atwood LD, Cupples LA, Benjamin EJ, D'Agostino RB Sr, Fox CS, Larson MG, Murabito JM, O'Donnell CJ, Vasan RS, Wolf PA, Levy D (2007) The Third Generation Cohort of the National Heart, Lung, and Blood Institute's Framingham Heart Study: design, recruitment, and initial examination. Am J Epidemiol. 165:1328–35. [DOI] [PubMed] [Google Scholar]
- van Beek JH, de Moor MH, Geels LM, Willemsen G, Boomsma DI (2014) Explaining individual differences in alcohol intake in adults: evidence for genetic and cultural transmission? J Stud Alcohol Drugs. 75:201–10. [DOI] [PubMed] [Google Scholar]
- van der Zwaluw CS, Engels RC, Vermulst AA, Franke B, Buitelaar J, Verkes RJ, Scholte RH (2010) Interaction between dopamine D2 receptor genotype and parental rule-setting in adolescent alcohol use: evidence for a gene-parenting interaction. Mol Psychiatry.15:727–35. [DOI] [PubMed] [Google Scholar]
- Verhulst B, Neale MC, Kendler KS (2015) The heritability of alcohol use disorders: a meta-analysis of twin and adoption studies. Psychol Med. 45:1061–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Fox CS, Troy LM, Mckeown NM, Jacques PF (2015) Longitudinal association of dairy consumption with the changes in blood pressure and the risk of incident hypertension: the Framingham Heart Study. Br J Nutr. 114:1887–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams PT (2012) Quantile-specific penetrance of genes affecting lipoproteins, adiposity and height. PLoS One. 2012;7(1):e28764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams PT (2020a) Quantile-specific heritability may account for gene-environment interactions involving coffee consumption. Behavioral Genetics 50:119–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams PT (2020b) Quantile-dependent expressivity of postprandial lipemia. PLoS One 15:e0229495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams PT (2020c) Gene-environment interactions due to quantile-specific heritability of triglyceride and VLDL concentrations. Sci Rep. 10:4486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams PT (2020d) Spirometric traits show quantile-dependent heritability, which may contribute to their gene-environment interactions with smoking and pollution. PeerJ. 8:e9145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams PT. (2020e) Quantile-specific heritability of high-density lipoproteins with implications for precision medicine. J Clin Lipidol (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winer BJ, Brown DR, Michels KM. 1991. Statistical principles in experimental design. Third edition. McGraw-Hill; New York. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


