Abstract
Objective
To systematically explore the pharmacological mechanism of Radix Paeoniae Rubra (RPR) against lung cancer (LC).
Methods
A network pharmacology approach, which involves active ingredients and target forecast, network construction, gene ontology and pathway enrichment, was employed in this research. In addition, the effect of Baicalein (BAI) in RPR on A549 cells was researched in vitro and in vivo.
Results
A total of 159 targets of the 29 active components in RPR were procured by pharmacokinetic parameters. The network analysis showed that β-sitosterol, baicalein, (+)-catechin, ellagic acid, stigmasterol, (2R, 3R)-4-methoxyl-distylin were the main ingredients and JUN, VEGFA, BCL2 were the hub targets of RPR in the treatment of LC. The functional enrichment analysis showed that RPR likely was useful to LC by regulating numerous pathways including Pathways in cancer, MAPK signaling pathway and so on. MTT results showed that 100μM, 200μM, 400μM of BAI had a time and dose-dependent inhibitory effect on A549 cells proliferation; Wound healing and transwell assays showed that 100μM, 200μM, 400μM of BAI could significantly restrain the migration and invasion of A549 cells; Flow cytometry assay results showed that 100μM, 200μM, 400μM of BAI could induce apoptosis of A549 cells. In vivo, BAI (50, 100 mg/kg) significantly inhibited tumor growth and promoted apoptosis of tumor cells compared with the control group.
Conclusion
BAI in RPR may exert anti-tumor effects by inhibiting the proliferation, migration and invasion of LC cells, and inducing the apoptosis of LC cells.
Keywords: network pharmacology, radix paeoniae rubra, lung neoplasms, baicalein
Introduction
Lung cancer (LC) is one of the most prevalent malignancies and the leading cause of cancer death. According to the latest global cancer statistics, there were approximately 18.1 million new cancer cases in 2018, of which 11.6% were LC.1 Effective therapies including surgery, radiotherapy, chemotherapy, targeted therapy, and immunotherapy have improved the prognosis and survival of LC.2,3 However, we still face with the challenge in cancer treatment such as multidrug resistance, which is closely related to the recurrence and metastasis of LC and mainly response for chemotherapy failure in a considerable number of patients.4,5 Therefore, there is a pressing necessity for more less noxious and effectual remedy of LC.
The Radix Paeoniae Rubra (RPR) is a Chinese herbal medicine extensively used in China. Many studies have shown that the main components of RPR are paeoniflorin, organic acids, hydroxy-4-acetylanisole, volatile oils and carbohydrate.6 It has been proved that RPR possesses high application value in liver protection,7 anti-inflammatory,8 cardiovascular system protection,9 antithrombosis,10 antiangiogenesis.11 Recent researches have confirmed that RPR and its active ingredients have anti-tumor effects,12,13 while the molecular mechanisms of RPR against LC remain unclear. Hence, it is expected that this research will provide RPR with a theoretical foundation in the remedy of LC.
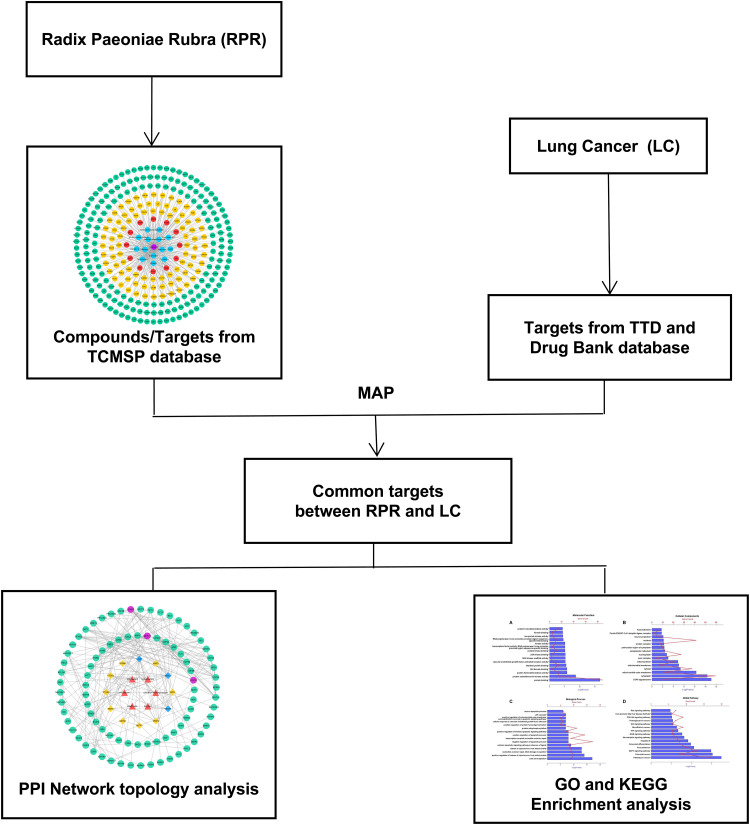
Network pharmacology studies the effects of drugs on diseases systematically from the disease-gene-target-drug interconnected perspective.14 Traditional Chinese medicine plays a therapeutic role of multiple components, multiple targets and multiple pathways. The current research used network pharmacology to probe the effective components of RPR and study its possible pharmacological mechanism for the treatment of LC. Flow diagram displayed in Figure 1.
Figure 1.
Network pharmacological flowchart of RPR against LC.
Abbreviations: RPR, Radix Paeoniae Rubra; LC, lung cancer.
Materials and Methods
Components and Targets of RPR
To collect the compounds and targets of RPR, we used the Traditional Chinese Medicine Systems Pharmacology Database (http://tcmspw.com/tcmspsearch.php, TCMSP).15 Active compounds were obtained from TCMSP database and screened with oral bioavailability (OB) ≥30%16 and drug likeness (DL)≥0.18%.17 Subsequently, the targets of the selected active ingredients were obtained from TCMSP.
LC Targets
LC-related targets were gathered from Therapeutic Target Database (https://db.idrblab.net/ttd/, TTD),18 DrugBank (DrugBank, https://www.drugbank.ca/).19 We extracted LC-related targets by searching these platforms with the keyword “lung cancer” and checked the extracted targets. Subsequently, the LC targets and the targets of RPR were further mapped to the UniProt (https://www.uniprot.org/) database for formalisation and standardized nomenclature.
Protein–Protein Interaction Data
The Protein–Protein Interaction (PPI) information were identified by the Search Tool for the Retrieval of Interacting Genes database (http://string-db.org/, STRING),20 which defines scores as low>0.15, medium>0.4, high>0.7. In present research, information with the scores>0.7 were selected for further study.
Network Construction and Analysis
The active compounds, targets and disease were inputted into Cytoscape software to build a network. Nodes were scored and ranked according to network features through Cytoscape’s plugin NetworkAnalyzer.21 For each node in the interaction network, degree was used to evaluate its topological parameter. The degree indicates the number of edges connected to the node. The greater the degree of a node means the more important the node is in the network.22 A disease-component-target gene network and protein–protein interaction network were constructed, in which high-degree nodes were analyzed as important nodes.23
Gene Ontology and Pathway Enrichment
The Database for Annotation, Visualization and Integrated Discovery (DAVID, https://david.ncifcrf.gov/), is a functional annotation tools which provides high-throughput gene functional analysis with comprehensive data.24 All the target genes were uploaded the DAVID database for Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment and gene ontology (GO) annotation. The pathway or gene functions with higher count were analyzed and P<0.01 was determined as the threshold value.
Cell Experiments
Based on the results of topological analysis, we selected some potential compounds of RPR to treat LC for in vitro experiments. The effect of RPR active ingredients such as baicalein (Herbpurify, Chengdu, China), (+)-catechin ((Herbpurify, Chengdu, China), paeoniflorin ((Herbpurify, Chengdu, China), and ellagic acid ((Herbpurify, Chengdu, China) on the proliferation of A549 cells (obtained from the American Type Culture Collection, USA) was observed by MTT assay. In MTT and scratch experiments, A549 cells were inoculated on 96-well plates and 6-well plates at a density of 4*103 and 5*105, respectively. In the transwell invasion and migration experiments, A549 cells were seeded in the upper chambers at a density of 106 and 5*105, respectively. The cells were stained with 4ʹ,6-diamidino-2-phenylindole (DAPI, BOSTER, Wuhan, China). Apoptosis was demonstrated and quantified by flow cytometry analysis using FITC Annexin V Apoptosis Detection Kit I (BD, America).
Animal Experiments
All animal studies were approved by the Experimental Animal Ethics Committee of Beijing Chinese Hospital of Traditional Chinese Medicine affiliated with Capital Medical University. A total of 18 Male BALB/c nude mice (5–6 weeks old, weighing 18–20g) were obtained from Beijing Vital River Laboratory Animal Technology Co. Ltd. [license No. SCXK (Beijing) 2016-0011]. A total of 5*106 A549 cells suspended in 100ul PBS were subcutaneously injected into the right flank of anesthetized BALB/c nude mice to establish the subcutaneous xenograft model. When the tumor volume was about 80 mm3, these mice were divided randomly into three groups: the control group (0.5% CMS-Na, N=6), high dose BAI group (dissolved in 0.5% CMC-Na, BAI-H, 100 mg/kg, N=6), low dose BAI group (dissolved in 0.5% CMC-Na, BAI-L, 50 mg/kg, N=6),25,26 and given intragastric administration of 0.2 mL 5 days a week. After 2 weeks, the mice were anesthetized by 1% pentobarbital sodium and sacrificed by decapitation.27 The tumors were removed for H&E staining (Hematoxylin and Eosin Staining Kit, Beyotime Biotechnology, China), TUNEL staining (TUNEL Assay Kit, Genepool, China) according to the manufacturer’s instructions.
Statistical Analysis
Statistical analysis was performed by GraphPad Prism (version 7, San Diego, CA), and P<0.05 was considered statistically significant.
Results
Component and Target of RPR and LC Targets
We obtained 29 compounds with better pharmacokinetic characteristics as active components of RPR in the TCMSP database with OB≥30 and DL≥0.18 as the screening conditions of major active ingredients, of which 14 active components have 101 target genes. A total number of 194 LC-related target genes were obtained from the TTD and DrugBank databases.
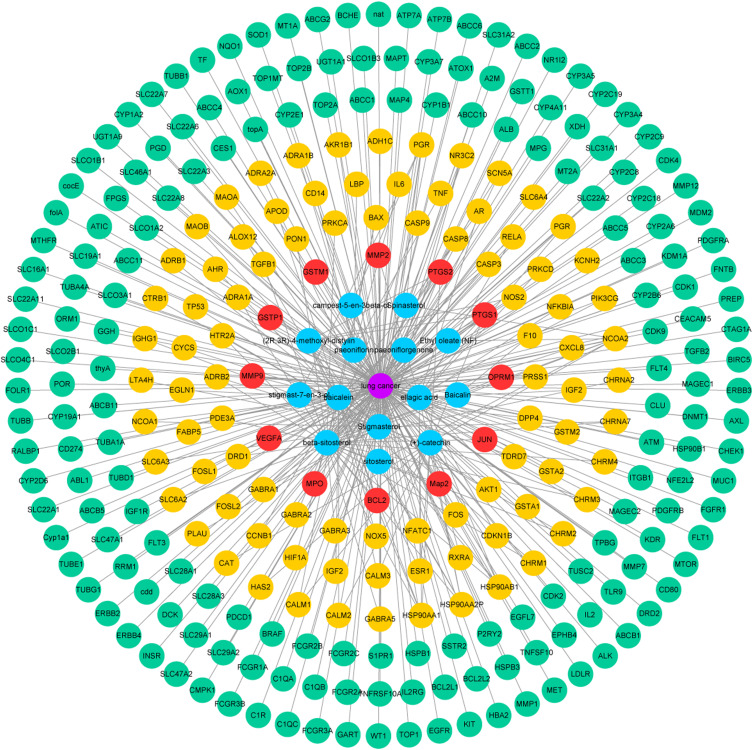
Compound-Disease-Target Protein Network
The compound-disease-target protein network is depicted in Figure 2, including 298 nodes and 353 edges. According to the topological analysis, the main active constituents of RPR were baicalein, β-sitosterol, stigmasterol, ellagic acid, (+)-catechin, (2R, 3R)-4-methoxyl-distylin, paeoniflorin, sitosterol, and spinasterol in the order of degree. The common targets between RPR and LC were BCL2, MMP9, VEGFA, JUN, MMP2, PTGS2, OPRM1, MPO, GSTP1, GSTM1, MAP2, PTGS1.
Figure 2.
Compound-disease-target protein network. Purple circles represent LC. Blue circles represent active ingredients in RPR. Red circles represent common targets between ingredient targets from RPR and LC targets. Yellow circles represent targets of RPR. Green circles represent targets of LC.
Abbreviations: RPR, Radix Paeoniae Rubra; LC, lung cancer.
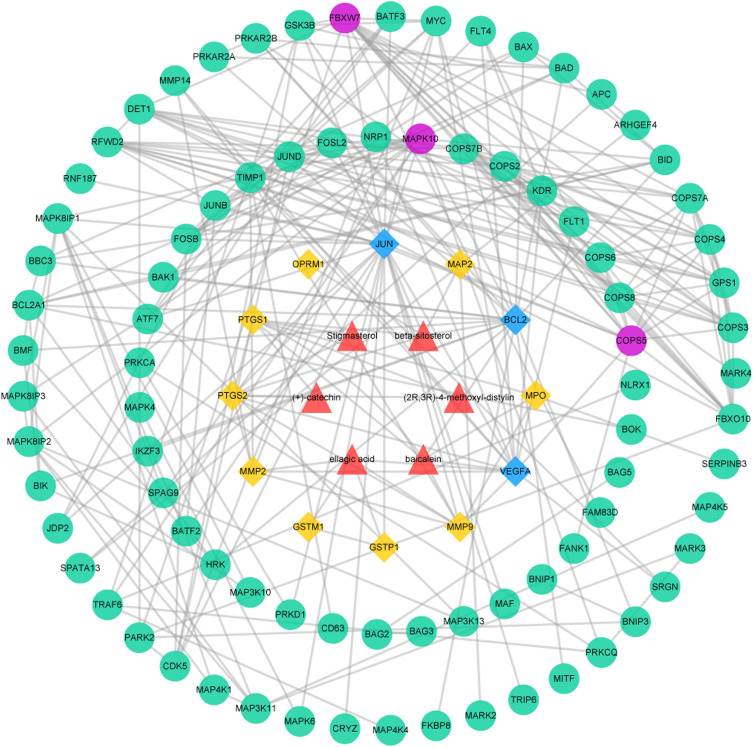
Protein–Protein Interaction Data
The PPI network was made up of the common targets and associated human proteins, as shown in Figure 3. The results of network analysis showed that the main active ingredients of RPR in the treatment of LC were baicalein, (+)-catechin, ellagic acid, stigmasterol, β-sitosterol, (2R, 3R)-4-methoxyl-distylin and the primary targets of RPR in the treatment of LC were JUN, VEGFA, BCL2.
Figure 3.
The common targets PPI network. Red triangles represent active ingredients in RPR. Yellow diamonds represent common targets from LC and active ingredients in RPR. Blue diamonds represent the main targets of common targets. Green circles represent associated human proteins that directly or indirectly interacted with common targets. Purple circles represent the main proteins of the associated human proteins.
Abbreviations: RPR, Radix Paeoniae Rubra; LC, lung cancer.
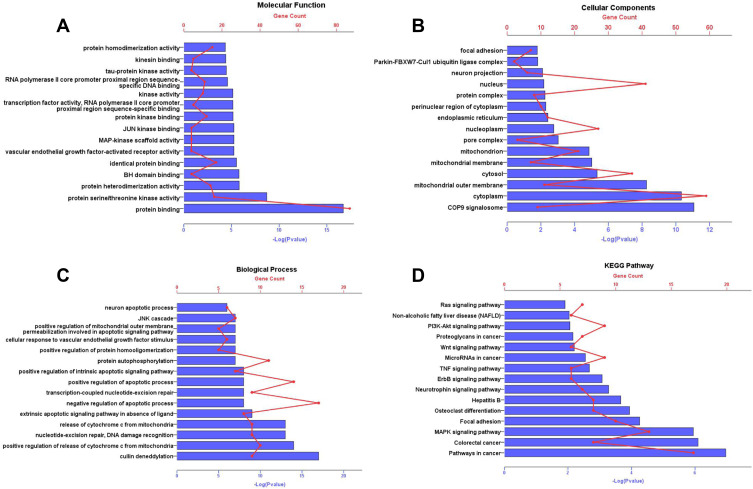
Gene Ontology and Pathway Enrichment
As shown in Figure 4, GO analysis results manifested that, in the molecular function category (Figure 4A), the common targets were chiefly enriched in protein binding. In the cell component category (Figure 4B), they were closely related to cytoplasm, nucleus, cytosol, nucleoplasm, mitochondrion. The related biological processes mainly included positive regulation of apoptotic process, negative regulation of apoptotic process and protein autophosphorylation (Figure 4C). KEGG results indicated that these targets were mainly enriched in MAPK signaling pathway, Pathways in cancer, Focal adhesion, PI3K-Akt signal pathway and so on (Figure 4D).
Figure 4.
GO annotation and KEGG pathway enrichment analysis for associated human proteins that directly or indirectly interacted with common targets. The y-axis shows significantly enriched Molecular Function, Cellular Components, Biological Process, KEGG pathways of the targets. The x-axis shows the enrichment scores of these terms or the counts of targets. P value<0.01. (A) Molecular Function; (B) Cellular Components; (C) Biological Process; (D) KEGG pathways.
Abbreviations: GO, gene ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes.
Cell Experiments
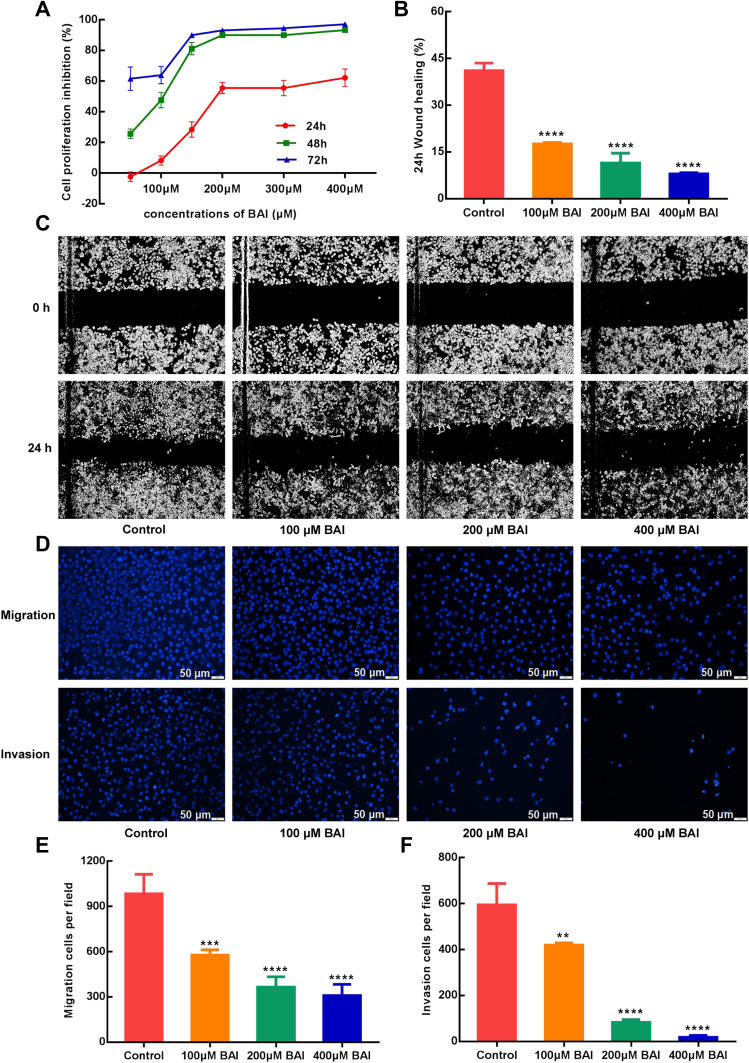
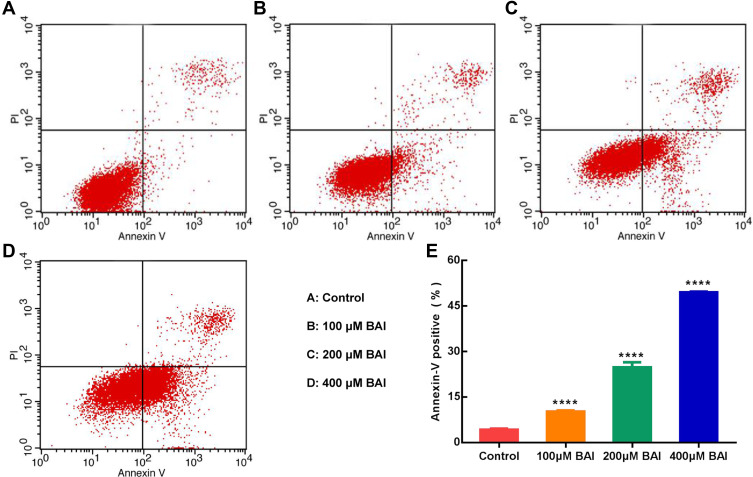
In vitro, the results of MTT assay showed that (+)-catechin (maximum concentration 800 μM) and paeoniflorin (maximum concentration 800 μM) had no obvious anti-tumor effect on A549 cells in 24, 48 and 72 h and the inhibition rate of 200 μM ellagic acid on A549 cells was only about 35% in 48 h. However, baicalein (BAI) inhibited the proliferation of A549 cells in a time and dose-dependent manner, as the IC50 of BAI at 24, 48 and 72 h were 217.5 μM, 98.43 μM, and 91.05 μM, respectively (Figure 5A). Based on the above results, we further conducted in vitro experiments for verifying the effect of BAI on the migration, invasion and apoptosis of A549 cells. Scratch test results showed that 100 μM, 200 μM, 400 μM of BAI could prevent the migration of A549 cells significantly and the percent of wound healing was 17.55%±0.34%, 11.45%±2.24%, 7.80%±0.27%, respectively (Figure 5B–C). Transwell experiments also showed that 100 μM, 200 μM, 400 μM of BAI prevented the migration and invasion of A549 cells in comparison with the control group. The count of migrated A549 cells was 577.50±35.11, 365.80±67.82, 309.00±74.35 VS 984.33±129.11 and the number of invasion was 420.33±8.51, 82.67±12.22, 18.34±8.02 VS 594.33±93.33 (Figure 5D–F). Flow cytometry assay results indicated that BAI could induce the apoptosis of A549 cells in a dose-dependent manner. The apoptotic rate (including early and late stage of apoptosis) was 10.29%±0.27%, 24.77%±1.72%, 49.5%±0.28%, respectively (Figure 6A–E).
Figure 5.
BAI inhibit the proliferation, migration and invasion of A549 cells. MTT assays show that BAI significantly inhibits the proliferation of A549 cells at 24, 48 and 72 h (A). Wound-healing assays show that BAI (100 μM/200 μM/400 μM) significantly inhibits the migration of A549 cells at 24 h (B and C). Transwell assays show that BAI (100 μM/200 μM/400 μM) significantly inhibits the migration and invasion of A549 cells at 24h (D/E/F). ****p<0.0001, ***p<0.001, **p<0.01 versus Control cells.
Abbreviation: BAI, Baicalein.
Figure 6.
BAI induces the apoptosis of A549 cells. Annexin V/PI staining shows that BAI (100 μM/200 μM/400 μM) induces the apoptosis of A549 cells at 24 h (E). The apoptotic rates of A549 cells (early apoptosis and late apoptosis) are 4.21%±0.34%, 10.29%±0.27%, 24.77%±1.72%, 49.5%±0.28% in the Control (A), 100 μM BAI (B), 200 μM BAI (C), 400 μM BAI (D), respectively. ****p<0.0001 versus Control cells. BAI, Baicalein.
Animal Experiments
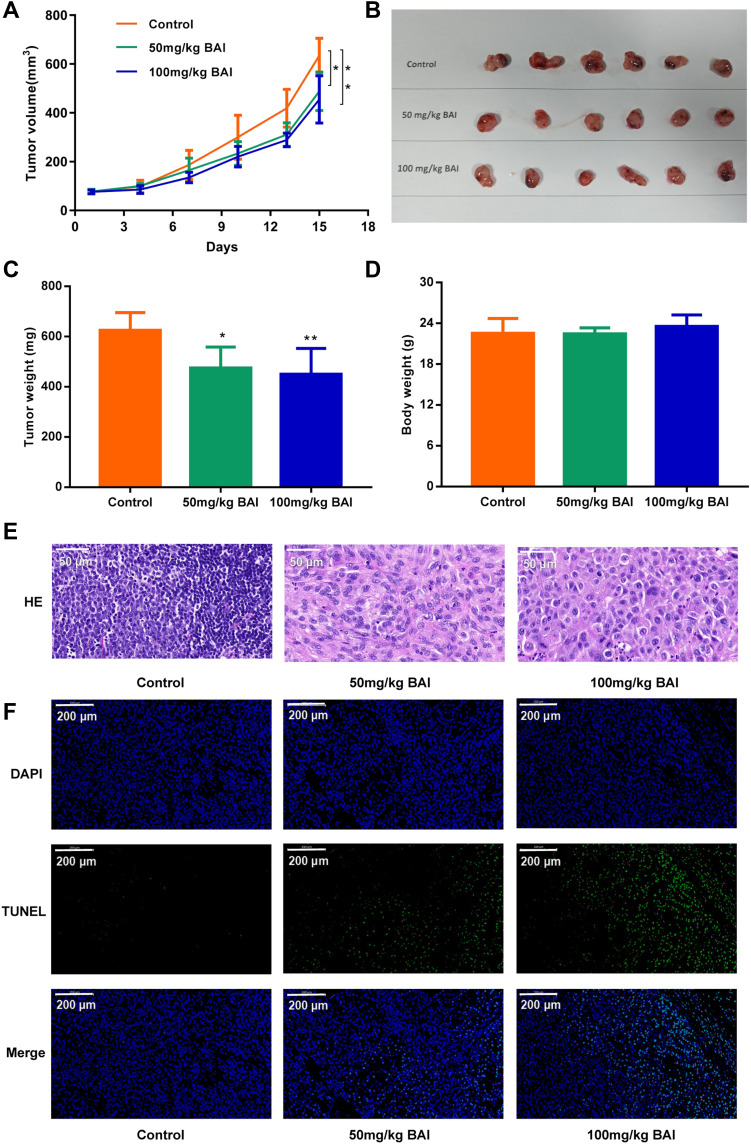
As shown in Figure 7A–C, the tumors volume and weight in BAI treatment (50, 100 mg/kg) group were lower than the control group (P<0.05). There was no statistical difference in body weight between the BAI treatment group and the control group (Figure 7D). H&E staining revealed that BAI treatment showed significant morphological changes compared to the control group (Figure 7E). These results suggested that BAI significantly inhibited tumors growth. In addition, TUNEL staining showed that the number of green-stained positive cells was significantly increased after treatment with BAI (50, 100 mg/kg) compared with the control group, indicating that BAI treatment (50, 100 mg/kg) significantly promoted apoptosis of tumor cells (Figure 7F).
Figure 7.
BAI inhibits the growth of subcutaneous xenograft tumor of A549. The subcutaneous xenograft models were randomly divided into 3 groups (n=6): Control group (0.5% CMC-Na), BAI-L (50 mg/kg) and BAI-H (100 mg/kg) (A). Tumor volume (B). Tumor image (C). Tumor weight (D). Body weight (E). The morphology of tumor was determined by H&E staining (scale bar, 50μm) (F). Apoptosis ratio was determined by TUNEL assay (scale bar, 200μm) (*p < 0.01, **p <0.001 vs control group).
Discussion and Conclusions
The RPR was reported that its active constituents could play an important role in inhibiting LC,28 gastric carcinoma,29 breast carcinoma,30 pancreatic carcinoma,31 and intestinal carcinoma.32 However, previous studies mainly focused on the pharmaceutical effect of single component rather than the complex ingredients in RPR, which limits our understanding of RPR without a holistic view. The network pharmacological method has similar characteristics to the multiple components, multiple targets and multiple pathway effects of traditional Chinese medicine. With the development of multidisciplinary pharmacology, network pharmacology offers a new direction for interpreting the complex pharmacological mechanism of Chinese herb.
In current study, network pharmacological analysis found that the main active ingredients of RPR against LC were baicalein, (+)-catechin, ellagic acid, β-sitosterol, stigmasterol, (2R,3R)-4-methoxyl-distylin. JUN, VEGFA, BCL2 were identified as the key potential therapeutic targets of RPR in treating LC. JUN is a member of the AP-1 transcription factor family and regulates the expression of many genes involved in extracellular matrix degradation, angiogenesis, and cell migration. One study has shown that down-regulation of JUN and AP-1 expression can inhibit apoptosis of lung adenocarcinoma cells.33 Vascular endothelial growth factor A (VEGFA) plays a critical role in neoplasm angiogenesis. When VEGFA specifically binds to its receptor, it participates in tumor invasion and metastasis by stimulating tumor vascular endothelial cell neoplasia.34 BCL2 is an anti-apoptotic gene. Apoptosis is a form of autonomic and orderly death of cells regulated by a series of related genes. The development of tumors is often accompanied by inhibition of expression of pro-apoptotic proteins and inhibitors of apoptosis-inhibiting proteins. Induction of tumor cell apoptosis is the most important mechanism of action of most anti-tumor drugs. Xu et al confirmed that total paeony glycoside could inhibit Bcl-2 and Bcl-xL expression and promote Bax expression to induce apoptosis of chronic granulocyte leukemia K562 cells.35
GO enrichment analysis found that the main biological processes related to this research were cell apoptosis and protein phosphorylation. Cancer is a complicated disease involving multiple genes regulation. Abnormal growth, differentiation and apoptosis of cancer cells are often accompanied with aberrant protein phosphorylation in cell signaling pathways. Chen et al analyzed the expression of phosphorylated proteins in serum from breast cancer patients compared to healthy donors. They found that phosphorylated proteins were closely related to breast cancer.36 RPR exerts inhibitory influence on tumor development by regulating several signaling pathways. The MAPK signal pathway is composed of four signal families, ERK, JNK, ERK5, and the p38 signaling family. Studies have shown that abnormal or excessive activation of MAPK signal pathway plays an influential role in malignant transformation of cells. ERK, JNK, and P38 signal pathways have been shown to be bound up with tumorigenesis.37,38 The PI3K-Akt signaling pathway is closely related to the proliferation, apoptosis and invasion of multiple cancer cells. The dysregulation of PI3K-Akt signal pathway is mainly due to excessive activation of Akt.39,40 Zhang et al found that reducing the phosphorylation of PI3K and AKT and restraining activation of PI3K-AKT pathway could restrain the proliferation and promote the apoptosis of LC cells.41
In vitro, we selected baicalein, paeoniflorin, ellagic acid, (+)-catechin of the active ingredients in RPR to observe the inhibitory effect on A549 cells. Paeoniflorin has been proven with anti-tumor effects in nasopharyngeal carcinoma cells via downregulation of NEDD4.42 Ellagic acid was reported to induce autophagy and exhibit anti-tumor effects in lung adenocarcinoma HOP62 and H1975 cells.43 Payen et al reported that (+)-catechin: lysine 1:2 could suppress cervix carcinoma cell migration in vitro and have antimetastatic effect in mouse melanoma models.44 However, paeoniflorin, ellagic acid, (+)-catechin had no significant anti-tumor effect on A549 cells in our present study. Baicalein has been proven to have a variety of anti-tumor effects, such as undifferentiated thyroid cancer cells,45 breast cancer46 and bladder cancer.47 The present results showed that 100 μM, 200 μM, 400 μM of BAI could significantly restrain the proliferation, migration and invasion of A549 cells. In vivo, BAI (50, 100 mg/kg) also significantly inhibited the growth of subcutaneous xenograft tumor of A549 cells. According to PPI network analysis and GO analysis, the anti-tumor effect of RPR was closely related to the induction of tumor cell apoptosis. Flow cytometry assay results also indicated that BAI could induce the apoptosis of A549 cells. TUNEL staining showed that BAI (50, 100 mg/kg) significantly promoted apoptosis of tumor cells compared with the control group. However, there are some limitations in predicting effective components and targets of LC through network pharmacology based on existing databases. Different databases may predict different results, so we should choose multiple databases as comprehensive as possible when making network predictions. In addition, LC also has multiple cell lines, so multiple cells can be tested in parallel in future experiments.
Acknowledgments
The research was supported by the National Natural Science Foundation of China (No.81473643) and Beijing Natural Science Foundation (7202065).
Ethics Approval
All animal studies were approved by the Experimental Animal Ethics Committee of Beijing Chinese Hospital of Traditional Chinese Medicine affiliated to Capital Medical University and were perform in accordance with the recommended procedures of the National Institutes of Health guide for the welfare and treatment of the laboratory animals.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest for this work.
References
- 1.Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 2.Maemondo M, Inoue A, Kobayashi K, et al. Gefitinib or chemotherapy for non–small-cell lung cancer with mutated EGFR. N Engl J Med. 2010;362(25):2380–2388. doi: 10.1056/NEJMoa0909530 [DOI] [PubMed] [Google Scholar]
- 3.Arrieta O, Barron F, Ramirez-Tirado LA, et al. Efficacy and safety of pembrolizumab plus docetaxel vs docetaxel alone in patients with previously treated advanced non-small cell lung cancer: the PROLUNG Phase 2 randomized clinical trial. JAMA Oncol. 2020;6(6):1–9. doi: 10.1001/jamaoncol.2020.0409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gao J, Chen L, Qi R, et al. Simultaneous delivery of gene and chemotherapeutics via copolymeric micellar nanoparticles to overcome multiple drug resistance to promote synergistic tumor suppression. J Biomater Appl. 2019;34(1):130–140. doi: 10.1177/0885328219839254 [DOI] [PubMed] [Google Scholar]
- 5.Sameiyan E, Hayes AW, Karimi G. The effect of medicinal plants on multiple drug resistance through autophagy: a review of in vitro studies. Eur J Pharmacol. 2019;852:244–253. doi: 10.1016/j.ejphar.2019.04.001 [DOI] [PubMed] [Google Scholar]
- 6.Li SL, Song JZ, Choi FF, et al. Chemical profiling of Radix Paeoniae evaluated by ultra-performance liquid chromatography/photo-diode-array/quadrupole time-of-flight mass spectrometry. J Pharm Biomed Anal. 2009;49(2):253–266. doi: 10.1016/j.jpba.2008.11.007 [DOI] [PubMed] [Google Scholar]
- 7.Wang R, Xiong AZ, Teng ZQ, et al. Radix paeoniae rubra and radix paeoniae alba attenuate CCl4-induced acute liver injury: an ultra-performance liquid chromatography-mass spectrometry (UPLC-MS) based metabolomic approach for the pharmacodynamic study of Traditional Chinese Medicines (TCMs). Int J Mol Sci. 2012;13(11):14634–14647. doi: 10.3390/ijms131114634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tu J, Guo Y, Hong W, et al. The regulatory effects of paeoniflorin and its derivative paeoniflorin-6′-o-benzene sulfonate CP-25 on inflammation and immune diseases. Front Pharmacol. 2019;10:57. doi: 10.3389/fphar.2019.00057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu X, Chen K, Zhuang Y, et al. Paeoniflorin improves pressure overload-induced cardiac remodeling by modulating the MAPK signaling pathway in spontaneously hypertensive rats. Biomed Pharmacother. 2019;111:695–704. doi: 10.1016/j.biopha.2018.12.090 [DOI] [PubMed] [Google Scholar]
- 10.Xie P, Cui L, Shan Y, et al. Antithrombotic effect and mechanism of radix paeoniae rubra. Biomed Res Int. 2017. doi: 10.1155/2017/9475074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abd El-Aal NF, Abdelbary EH. Paeoniflorin in experimental BALB/c mansoniasis: a novel anti-angiogenic therapy. Exp Parasitol. 2019;197:85–92. doi: 10.1016/j.exppara.2018.11.002 [DOI] [PubMed] [Google Scholar]
- 12.Yang J, Ren Y, Lou Z, et al. Paeoniflorin inhibits the growth of bladder carcinoma via deactivation of STAT3. Acta Pharm. 2018;68(2):211–222. doi: 10.2478/acph-2018-0013 [DOI] [PubMed] [Google Scholar]
- 13.Yue M, Li S, Yan G, et al. Paeoniflorin inhibits cell growth and induces cell cycle arrest through inhibition of FoxM1 in colorectal cancer cells. Cell Cycle. 2018;17(2):240–249. doi: 10.1080/15384101.2017.1407892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xue J, Shi Y, Li C, et al. Network pharmacology-based prediction of the active ingredients, potential targets, and signaling pathways in compound Lian-Ge granules for treatment of diabetes. J Cell Biochem. 2019;120(4):6431–6440. doi: 10.1002/jcb.27933 [DOI] [PubMed] [Google Scholar]
- 15.Ru J, Li P, Wang J, et al. TCMSP: a database of systems pharmacology for drug discovery from herbal medicines. J Cheminform. 2014;6(1):1–6. doi: 10.1186/1758-2946-6-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu X, Zhang W, Huang C, et al. A novel chemometric method for the prediction of human oral bioavailability. Int J Mol Sci. 2012;13(6):6964–6982. doi: 10.3390/ijms13066964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zheng C, Pei T, Huang C, et al. A novel systems pharmacology platform to dissect action mechanisms of traditional Chinese medicines for bovine viral diarrhea disease. Eur J Pharm Sci. 2016;94:33–45. doi: 10.1016/j.ejps.2016.05.018 [DOI] [PubMed] [Google Scholar]
- 18.Li YH, Yu CY, Li XX, et al. Therapeutic target database update 2018: enriched resource for facilitating bench-to-clinic research of targeted therapeutics. Nucleic Acids Res. 2018;46(D1):D1121–D1127. doi: 10.1093/nar/gkx1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wishart DS, Feunang YD, Guo AC, et al. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 2018;46(D1):D1074–D1082. doi: 10.1093/nar/gkx1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Szklarczyk D, Morris JH, Cook H, et al. The STRING database in 2017: quality-controlled protein–protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45(D1):D362–D368. doi: 10.1093/nar/gkw937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Assenov Y, Ramirez F, Schelhorn SE, et al. Computing topological parameters of biological networks. Bioinformatics. 2008;24(2):282–284. doi: 10.1093/bioinformatics/btm554 [DOI] [PubMed] [Google Scholar]
- 22.Jeong H, Mason SP, Barabási AL, Oltvai ZN. Lethality and centrality in protein networks. Nature. 2001;411(6833):41–42. doi: 10.1038/35075138 [DOI] [PubMed] [Google Scholar]
- 23.Xiong Y, Yang Y, Xiong W, et al. Network pharmacology-based research on the active component and mechanism of the antihepatoma effect of Rubia cordifolia L. J Cell Biochem. 2019;120(8):12461–12472. doi: 10.1002/jcb.28513 [DOI] [PubMed] [Google Scholar]
- 24.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- 25.Deng X, Liu J, Liu L, Sun X, Huang J, Dong J. Drp1-mediated mitochondrial fission contributes to baicalein-induced apoptosis and autophagy in lung cancer via activation of AMPK signaling pathway. Int J Biol Sci. 2020;16(8):1403–1416. doi: 10.7150/ijbs.41768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu L, Zhang M, Wang X, et al. Baicalein enhances the antitumor efficacy of docetaxel on nonsmall cell lung cancer in a β-catenin-dependent manner. Phytother Res. 2020;34(1):104–117. PMID: 31515889. doi: 10.1002/ptr.6501. [DOI] [PubMed] [Google Scholar]
- 27.Ma Y, Li G, Sun X, et al. Huayu Wan prevents lewis lung cancer metastasis in mice via the platelet pathway. Evid Based Complement Alternat Med. 2020;2020:1306207. doi: 10.1155/2020/1306207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu Q, Chen GL, Li YJ, et al. Paeoniflorin inhibits macrophage-mediated lung cancer metastasis. Chin J Nat Med. 2015;13(12):925–932. doi: 10.1016/S1875-5364(15)30098-4 [DOI] [PubMed] [Google Scholar]
- 29.Shin EJ, Choi H, Sung MJ, et al. Anti-tumour effects of beta-sitosterol are mediated by AMPK/PTEN/HSP90 axis in AGS human gastric adenocarcinoma cells and xenograft mouse models. Biochem Pharmacol. 2018;152:60–70. doi: 10.1016/j.bcp.2018.03.010 [DOI] [PubMed] [Google Scholar]
- 30.Yan W, Ma X, Zhao X, et al. Baicalein induces apoptosis and autophagy of breast cancer cells via inhibiting PI3K/AKT pathway in vivo and vitro. Drug Des Devel Ther. 2018;12:3961–3972. doi: 10.2147/DDDT.S181939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li Y, Gong L, Qi R, et al. Paeoniflorin suppresses pancreatic cancer cell growth by upregulating HTRA3 expression. Drug Des Devel Ther. 2017;11:2481–2491. doi: 10.2147/DDDT.S134518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang J, Li L, Wu W, et al. Anti-tumor effects of paeoniflorin on epithelial-to-mesenchymal transition in human colorectal cancer cells. Med Sci Monit. 2018;24:6405–6413. doi: 10.12659/MSM.912227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nagesh R, Kiran Kumar KM, Naveen Kumar M, et al. Stress activated p38 MAPK regulates cell cycle via AP-1 factors in areca extract exposed human lung epithelial cells. Cytotechnology. 2019;71(2):507–520. doi: 10.1007/s10616-019-00297-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Claesson-Welsh L, Welsh M. VEGFA and tumour angiogenesis. J Intern Med. 2013;273(2):114–127. doi: 10.1111/joim.12019 [DOI] [PubMed] [Google Scholar]
- 35.Xu HY, Chen ZW, Wu YM. Antitumor activity of total paeony glycoside against human chronic Antitumor activity of total paeony glycoside against human chronic myelocytic leukemia K562 cell lines in vitro and in vivo. Med Oncol. 2012;29(2):1137–1147. doi: 10.1007/s12032-011-9909-9 [DOI] [PubMed] [Google Scholar]
- 36.Chen IH, Xue L, Hsu CC, et al. Phosphoproteins in extracellular vesicles as candidate markers for breast cancer. Proc Natl Acad Sci U S A. 2017;114(12):3175–3180. doi: 10.1073/pnas.1618088114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Burotto M, Chiou VL, Lee J, et al. The MAPK pathway across different malignancies: a new perspective. Cancer. 2014;120(22):3446–3456. doi: 10.1002/cncr.28864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim EK, Choi E. Compromised MAPK signaling in human diseases: an update. Arch Toxicol. 2015;89(6):867–882. doi: 10.1007/s00204-015-1472-2 [DOI] [PubMed] [Google Scholar]
- 39.Pal I, Mandal M. PI3K and Akt as molecular targets for cancer therapy: current clinical outcomes. Acta Pharmacol Sin. 2012;33(12):1441–1458. doi: 10.1038/aps.2012.72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Noorolyai S, Shajari N, Baghbani E, et al. The relation between PI3K/AKT signalling pathway and cancer. Gene. 2019;698:120–128. doi: 10.1016/j.gene.2019.02.076 [DOI] [PubMed] [Google Scholar]
- 41.Zhang L, Liu B. Sapylin inhibits lung cancer cell proliferation and promotes apoptosis by attenuating PI3K/AKT signaling. J Cell Biochem. 2019;1–9. doi: 10.1002/jcb.28729 [DOI] [PubMed] [Google Scholar]
- 42.Chen Y, Zhang R, Zhao W, et al. Paeoniflorin exhibits antitumor effects in nasopharyngeal carcinoma cells through downregulation of NEDD4. Am J Transl Res. 2019;11(12):7579–7590. [PMC free article] [PubMed] [Google Scholar]
- 43.Duan J, Zhan JC, Wang GZ, et al. The red wine component ellagic acid induces autophagy and exhibits anti-lung cancer activity in vitro and in vivo. J Cell Mol Med. 2019;23(1):143–154. doi: 10.1111/jcmm.13899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Payen VL, Porporato PE, Danhier P, et al. (+)-Catechin in a 1:2 complex with lysine inhibits cancer cell migration and metastatic take in mice. Front Pharmacol. 2017;8:869. doi: 10.3389/fphar.2017.00869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang M, Qiu S, Qin J. Baicalein induced apoptosis and autophagy of undifferentiated thyroid cancer cells by the ERK/PI3K/Akt pathway. Am J Transl Res. 2019;11(6):3341–3352. [PMC free article] [PubMed] [Google Scholar]
- 46.Nguyen L, Song YW, Cho SK. Retraction note: baicalein inhibits epithelial to mesenchymal transition via downregulation of Cyr61 and LOXL-2 in MDA-MB231 breast cancer cells. Mol Cells. 2019;42(4):377. doi: 10.14348/molcells.2019.3243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jiang L, Song H, Guo H, et al. Baicalein inhibits proliferation and migration of bladder cancer cell line T24 by down-regulation of microRNA-106. Biomed Pharmacother. 2018;107:1583–1590. doi: 10.1016/j.biopha.2018.08.107. [DOI] [PubMed] [Google Scholar]