Abstract
Pancreatic ductal adenocarcinoma (PDAC) is highly lethal. MUC4 (mucin4) is a heavily glycosylated protein aberrantly expressed in PDAC and promotes tumorigenesis via an unknown mechanism. To assess this, we genetically knocked out (KO) MUC4 in PDAC cells that did not express and did express truncated O-glycans (Tn/STn) using CRISPR/Cas9 technology. We found that MUC4 knockout cells possess less tumorigenicity in vitro and in vivo, which was further reduced in PDAC cells that express aberrant overexpression of truncated O-glycans. Also, MUC4KO cells showed a further reduction of epidermal growth factor receptors (ErbB) and their downstream signaling pathways in truncated O-glycan expressing PDAC cells. Tn-MUC4 specific 3B11 antibody inhibited MUC4-induced ErbB receptor and its downstream signaling cascades. MUC4 knockout differentially regulates apoptosis and cell cycle arrest in branched and truncated O-glycan expressing PDAC cells. Additionally, MUC4KO cells were found to be more sensitive to gemcitabine treatment. They possessed the upregulated expression of hENT1 and hCNT3 compared to parental cells, which were further affected in cells with aberrant O-glycosylation. Taken together, our results indicate that MUC4 enhances the malignant properties and gemcitabine resistance in PDAC tumors that aberrantly overexpress truncated O-glycans via altering ErbB/AKT signaling cascades and expression of nucleoside transporters, respectively.
Keywords: MUC4, pancreatic cancer, aberrant glycosylation, Tn-MUC4 antibody, nucleoside transporters
1. Introduction
Pancreatic ductal adenocarcinoma (PDAC) is the fourth leading cause of cancer-related death in the US, which accounts for 7% of all cancer deaths with a 5-year survival rate of 9%. An increased incidence of PDAC has been reported in the United States, with approximately 58,000 new cases reported in 2020 [1]. While emerging surgical technologies and adjuvant therapies have helped some patients, targeted therapies and chemotherapeutic regimens remain largely ineffective [2]. PDAC progression is characterized by mucin overexpression; MUC4 is one of the most differentially overexpressed mucins with clinicopathological significance during PDAC progression [3, 4]. MUC4 is not expressed in a healthy pancreas, whereas it is aberrantly overexpressed (70–80%) in pancreatic carcinogenesis [5, 6]. Studies have reported that MUC4 enhanced proliferative, invasiveness, migratory, and metastatic properties in pancreatic cancer but diminished cell-cell and cell-matrix interactions in several other epithelial cancers [7].
Numerous previous studies have revealed that the oncogenic potential of MUC4 may be mediated at least in part by direct interaction of MUC4 with epidermal growth factor (EGF)-like the domain of HER2/ErbB2 in several cancers [8–10]. ErbB2, in association with EGFR and ErbB3, triggers a rich network of oncogenic signaling cascades that control cell growth, differentiation, motility, and tumor metastasis. Studies have suggested that along with the ErbBs, the transmembrane subunit beta of MUC4 is a membrane partner of the domains of EGFs (EGF-1, −2, −3), which control the aggressive course of PDAC disease progression [11, 12]. It was recently demonstrated that the silencing of ErbB2 leads to increased sensitivity of pancreatic cancer cells to the chemotherapeutic drug gemcitabine (GEM) [13]. Also, the knockdown of MUC4 has been shown to regulate cellular sensitivity to GEM through the activation of nuclear transporters [14]. Human concentrative nucleoside transporter 1 and 3 (hCNT1 and hCNT3) and human equilibrative nucleoside transporter 1 (hENT1) are major transporter molecules that facilitate intracellular GEM uptake in pancreatic cancer cells [15]. Knockdown of MUC4 resulted in the activation of hENT1 and hCNT1/3, key components of GEM metabolism, and detoxifying channels via the NF-κB pathway [14]. Notably, 60–80% of PDAC cases are characterized by aberrant expression of truncated O-glycans, and studies have demonstrated that these aberrant O-glycans enhance the progression of PDAC [16, 17]. However, knowledge of MUC4 glycosylation and its role in pancreatic cancer tumorigenesis and drug resistance is limited. To determine the functional role of glycosylation of MUC4 in PDAC progression and drug resistance, we have genetically deleted MUC4 in isogenic PDAC cells with or without defective O-glycosylation using CRISPR/Cas9 genome editing technology. Here, we show evidence that the deletion of MUC4 in PDAC cells diminished tumor malignancies, which was further significantly affected by aberrant O-glycosylation. Genetic deletion of MUC4 reduced tumor growth via the modulation of ErbB receptors. Additionally, Tn-MUC4 specific antibody 3B11 treatment inhibits MUC4 mediated EGF receptor signaling cascades and induces cell death. We also show evidence that the deletion of MUC4 in PDAC cells causes GEM sensitivity through the regulation of nucleoside transporters hENT1 and hCNT3 expressions. For the first time, our results demonstrate the regulatory role of aberrant glycoforms of MUC4 during PDAC malignancy.
2. Material and Methods
2.1. Cell culture
Human PDAC cells T3M4 and Capan-2 [wildtype, WT; COSMC Knockout, SimpleCells (SC)] [18, 19] were maintained in Dulbecco’s Modified Eagle’s Medium (DMEM) (Hyclone, MA, USA) and Roswell Park Memorial Institute (RPMI) 1640 medium (Corning, NY, USA) respectively supplemented with 10% fetal bovine serum (FBS) (Corning, NY, USA) and 100 units/mL penicillin and 100 μg/mL streptomycin (Corning, NY, USA). All the cell lines were maintained at 37 °C with 5% CO2 in a humidified incubator.
2.2. Genetic deletion of MUC4 in PDAC cells
The MUC4 gene was genetically deleted in T3M4 WT and SC cells using CRISPR/Cas9 KO plasmid kits (Santa Cruz Biotechnology, CA, USA) as per the manufacturer’s instructions. Briefly, cells were transfected with a pool of three MUC4 guide RNA containing vectors (1. CGTTCTTATACCACGTTCCA; 2. CACCTTCGGCCGCTACTGCG; 3. TGTCGGTTGCCTGGGACGCC), which targets the MUC4 genome. After 48 h of transfection, the GFP positive cells were sorted using FACS cell sorting (BD FACS Aria II, UNMC Core Facility) in 96 well plates. MUC4 knockout clones (T3M4 WT-MUC4KO; 3B4 and 4D3; and T3M4 SC-MUC4KO; 1E11 and 5D10) were utilized for further studies.
2.3. Immunoblotting
The protein expression of MUC4 was analyzed by SDS-Agarose gel electrophoresis, as described previously [20]. Briefly, 70 μg of proteins were resolved on SDS (0.1%) - Agarose (1.5%) gel electrophoresis and transferred to the PVDF membrane through capillary transfer. For all other western blotting experiments, 4–20% SDS-PAGE was performed. Briefly, T3M4 WT, WT-MUC4KO (3B4, 4D3), T3M4 SC, SC-MUC4KO (1E11, 5D10) cells, as well as T3M4 WT and T3M4 SC cells were treated with various concentrations (2.5, 5.0, and 10 μg/ml) of Tn-MUC4 specific 3B11 antibody (a kind gift from Dr. Hans H. Wandall, University of Copenhagen, Denmark). For analyzing the effect of AKT inhibitor, T3M4 WT, and SC cells (1×106) were treated with 20 μM of LY294002 (Cell Signaling Technology, MA, USA) for 24 h. The cells were lysed in RIPA lysis buffer (Thermo Fisher Scientific, MA, USA) containing protease and phosphatase inhibitors (Roche, MO, USA) on ice for 20 min. The cells were then centrifuged at 12,500 X g for 15 min, and the supernatant was collected. Protein concentration was quantified using the BCA Protein Assay kit (Thermo Fisher Scientific, MA, USA) with bovine serum albumin (BSA) as a standard. Equal amounts of protein were resolved on 4–20% gradient SDS-PAGE gel (Bio-Rad, CA, USA) and transferred to 0.45 μm PVDF membranes (Millipore, MO, USA). After blocking in 5% nonfat skim milk for 1 h, the membranes were probed with specific primary antibodies overnight at 4 °C and secondary antibody at room temperature for 1 h. The following primary and secondary antibodies were diluted and used as per the manufacturer’s recommendations. Anti-phospho ErbB2 (Y1248), anti-ErbB2, anti-phospho ErbB3 (Y1289), anti-ErbB3, anti-ErbB1, anti-phospho AKT (S473), anti-AKT, anti-MMP-2, anti-Cyclin E1, anti-Cyclin B1, anti-hENT1, anti-hCNT3, anti-cytochrome c, anti-Mcl-1, anti-Bcl-2, anti-GAPDH, anti-β-actin (Cell Signaling Technology, MA, USA) anti-8G7, anti-phospho ErbB1 (Y1173) (Santa Cruz Biotechnology, CA, USA), α-tubulin (Developmental Studies Hybridoma Bank, IA, USA), and HRP conjugated horse anti-mouse IgG and horse anti-rabbit IgG (Cell Signaling Technology, MA, USA). The immunoreactivity was visualized using a western ECL detection kit (Bio-Rad, CA, USA).
2.4. Immunoprecipitation analysis
T3M4 WT and SC cells were lysed in non-denaturing RIPA buffer containing protease inhibitor. 500μg of proteins were incubated with either mouse anti-MUC4 (3μg/ml, 8G7) or Mouse IgG (3μg/ml; Jackson ImmunoResearch Laboratories, Inc, USA) overnight at 4°C. Lysates were then incubated with protein G Sepharose beads (GenScript, NJ, USA) for 2 h at room temperature. The beads were washed three times with RIPA buffer and boiled in SDS loading buffer (1X). Equal amounts of samples were subjected to 4–20% gradient (Bio-Rad, CA, USA) SDS-PAGE gel electrophoresis and transferred to PVDF membrane. The membranes were blocked with 5% skimmed milk and incubated with primary antibody rabbit anti-ErbB2 (Cell Signaling Technology, USA, 1:1000) and anti-mouse IgG (Jackson ImmunoResearch Laboratories, Inc, PA, USA, 1:4000) overnight. After incubating with respective secondary antibodies, the protein-antibody complex was detected using enhanced chemiluminescence (Bio-Rad, USA).
2.5. Immunofluorescence analysis
Immunofluorescence staining of PDAC cells was carried out as described previously [20]. Briefly, cells were washed with PBS and blocked with 1% BSA. Cells were incubated with mouse anti-core 1 synthase (C1GALT1) (1:200) (Santa Cruz Biotechnology, CA, USA), mouse anti-STn (TKH2) (1:200), and mAb 8G7 (1:200) for 2 h at room temperature. After washing, the cells were incubated with Alexa Fluor 647-conjugated goat anti-mouse IgG and Alexa Fluor 488-conjugated goat anti-mouse IgG (1:500) secondary antibodies (Jackson ImmunoResearch Laboratories, Inc, PA, USA). For co-localization studies, T3M4 and Capan-2 (WT and SC) cells were processed as described above and incubated with rabbit anti-ErbB2 (1:200) and mouse anti-MUC4 (8G7) (1:200) for 2 h at room temperature. After washing, the cells were incubated with Alexa Fluor 488-conjugated goat anti-rabbit IgG (1:500) and Alexa Fluor 647-conjugated goat anti-mouse IgG (1:500) secondary antibodies for 1 h. Cells were washed and mounted with Vectashield mounting medium with DAPI (Vector Laboratories, CA, USA). Immunofluorescence images were captured using confocal laser scanning microscopy at the UNMC Core Facility.
2.6. Cell proliferation assay
T3M4 WT, WT-MUC4KO (3B4, 4D3), T3M4 SC, and SC-MUC4KO (1E11, 5D10) cells (1×103) were plated in a 96-well plate. After 24, 48, 72, 96, and 120 hours of incubation, 1/10 volume of Alamar blue reagent (Invitrogen, CA, USA) was added and incubated for 1–3 h in the dark at 37 °C. Plates were washed, and the fluorescence intensity was determined at excitation/emission 560nm/590nm using a Spectramax M5e (Molecular Devices, CA, USA) fluorescence reader.
2.7. Cell cycle analysis
Cells were grown to a confluence of 70–80% and were harvested, washed with PBS, and counted. 1×106 cells were fixed in ice-cold ethanol at 4 °C for 1 h. After fixation, cells were washed with PBS, re-suspended in PBS containing RNase A (100 μg/mL; Thermo Fisher Scientific, MA, USA) propidium iodide (PI; 50 μg/mL, Invitrogen, CA, USA) and 0.1% Triton X 100, then incubated for another 30 min at 37 °C in the dark. Data of cell-cycle phase distribution was acquired by caliber flow cytometry (BD, UNMC Core Facility) and analyzed by ModFit LT software.
2.8. Cell migration assay
The tumor cell migration assay was performed as described previously [20]. Briefly, T3M4 WT, WT-MUC4KO (4D3), T3M4 SC, and SC-MUC4KO (5D10) cells (0.5×106) were plated on the top chamber of a non-coated 24-well plate inserts (Corning, NY, USA). After 24 h, the cells that did not migrate through the membrane pores were removed with a cotton swab. The migrated cells were fixed and stained with a Diff-Quick cell stain kit (Siemens Healthcare Diagnostic, PA, USA). The cells were counted under a light microscope.
2.9. Cell invasion assay
The tumor cell invasion assay was performed as described previously [20]. Briefly, T3M4 WT, WT-MUC4KO (4D3), T3M4 SC, and SC-MUC4KO (5D10) cells (0.5×106) were seeded on Matrigel-coated invasion membranes (BD Biosciences, CA, USA) in serum-free medium and incubated for 24 h at 37 °C. After 24 h, cells on the upper chamber were removed, and the invaded cells were fixed and stained with Diff-Quick cell stain kit. The cells were counted under a light microscope.
2.10. Clonogenic assay
T3M4 WT, WT-MUC4KO (3B4 and 4D3), T3M4 SC, and SC-MUC4KO (1E11 and 5D10) cells (500 cells) were seeded in 6-well plates with a low attachment surface (Corning, NY, USA). Colonies were allowed to grow for 8–12 days, with a medium change every 3 days. Colonies were fixed with paraformaldehyde for 15 min, stained with 0.5% crystal violet for 2 h at room temperature. The colony number was determined in triplicate after capturing the image in a 10X bright field microscope.
2.11. Apoptosis assay
The apoptosis index was analyzed by using an annexin V/FITC apoptosis detection kit (BD Biosciences, CA, USA), according to the manufacturer’s instructions and as described by Moon et al. (2018) [21]. Briefly, T3M4 WT, WT-MUC4KO (3B4 and 4D3), T3M4 SC, and SC-MUC4KO (1E11 and 5D10) cells were collected, washed with ice-cold PBS, and then re-suspended in 100 μL (1 × 105 cells) of binding buffer containing 5 μL FITC‐conjugated annexin V and 5 μL propidium iodide (PI). After incubation for 20 min at room temperature in the dark, the cells were analyzed by flow cytometry using BD FACS LSR II (UNMC Core Facility). Cell populations in different quadrants were calculated, and statistical significance was analyzed.
2.12. Live/dead cell assay
T3M4 and Capan-2 cells were treated with anti-Tn-MUC4 antibody 3B11 (5 μg/ml) for 24 h and processed for the live/dead cell assay as per the manufacturer’s instruction (Invitrogen, CA, USA). Briefly, cells were washed well with cell-culture grade PBS. 20 μl ethidium homodimer-1 (EthD-1, 2 mM) was dissolved in 10 ml PBS. To this solution, 5 μl of calcein acetoxymethyl ester (Calcein-AM, 4 mM) was added. 150 μl of this combined reagent was added to the grown cells on a coverslip and incubated for 30–45 minutes. The numbers of live and dead cells were detected with confocal microscopy (UNMC Core Facility). The ratio of dead cells to total cells was calculated for quantitative comparisons.
2.13. Gemcitabine cytotoxicity assay
T3M4 WT, WT-MUC4KO, T3M4 SC, and SC-MUC4KO cells (1×104) were plated in a 96-well plate. After 24 h, cells were treated with 15 nM of gemcitabine and incubated for another 72 h. After the incubation period, 1/10 volume of Alamar blue reagent was added and incubated for 2 h in the dark at 37 °C. Fluorescence intensity was determined at excitation/emission 560nm/590nm using a Spectramax M5e (Molecular Devices, Sunnyvale, CA, USA) fluorescence reader.
2.14. Real-time PCR analysis
Total RNA was extracted from T3M4 WT, WT-MUC4KO, T3M4 SC, and SC-MUC4KO cells using an RNA isolation kit (Qiagen CA, USA) according to the manufacturer’s recommended protocols. A total of 1.0 μg of RNA was used to synthesize cDNA using the Verso cDNA synthesis kit (Thermo Fisher Scientific, MA, USA). cDNA was amplified with the SYBR™ Green PCR master mix (Applied Biosystems, MA, USA). The following primers were used for the analysis, hENT1: sense (5′−3′) CAGGCAAAGAGGAATCTGGA, anti-sense (5′−3′) GGCCCAACCAGTCAAAGATA; hCNT3: sense (5′−3′) GGGTCCCTAGGAATCGTGATC anti-sense (5′−3′) CGAGGCGATATCACGCTTTC; MUC4: sense (5′−3′) GCCCAAGCTACAGTGTGACTCA anti-sense (5′−3′) ATGGTGCCGTTGTAATTTGTTGT; GAPDH: sense (5′−3′) TCGACAGTCAGCCGCATCTTCTTT anti-sense (5′−3′) ACCAAATCCGTTGACTCCGACCTT. The comparative threshold cycle (CT) method was used to analyze the PCR data.
2.15. Orthotopic pancreatic tumor model
All the athymic nu/nu mice (Crl: NU-Foxn1nu) were purchased from Charles River Laboratories, MA, USA, housed under standard recommended housing conditions at the UNMC Comparative Medicine animal facility. All the animal procedures involved in this study were approved by the UNMC Institutional Animal Care and Use Committees (IACUC). T3M4 WT, WT-MUC4KO (4D3), T3M4 SC, and SC-MUC4KO (5D10) cells (3×105/30 μl PBS) were orthotopically implanted into the pancreas of mice (n=7). After 28 days, the animals were euthanized, and tumor weight and volume, and metastasis to various organs were analyzed as described previously [18].
2.16. Immunohistochemistry
The standard immunohistochemistry protocol was used for the analysis of protein expression of orthotopic mouse tumor tissue samples. Briefly, after deparaffinized the slides in xylene, the sections were rehydrated with a descending series of alcohol and quenched with H2O2. Citrate buffer (pH = 6.0) was used for antigen retrieval. After blocking with a universal blocker (Thermo Fisher Scientific, MA, USA), the slides were incubated with antibodies p-ErbB2 (1:50), p-AKT (1:50), hENT1 (1:100), hCNT3 (1:100), and cleaved caspase3 (1:50) overnight at 4 °C. The slides were washed and incubated with HRP conjugated respective secondary antibodies (Dako, CA, USA) and incubated for 1 h at room temperature. The antigen-antibody complex was developed using 3,3’-diaminobenzidine tetrahydrochloride (DAB, Vector laboratories, CA, USA) substrate and then counterstained with hematoxylin. All the slides were dehydrated with an ascending series of alcohol, and after xylene washes, the slides were mounted with the coverslip. The protein expression was scored by a pathologist. The histological scoring was performed based on stain proportion (0–100%) and intensity (0-negligible, 1-low, 2-moderate, 3-high). The histoscore was generated by multiplying the stain proportion score (1=<5%, 2=5–25%, 3=26–50%, 4=51–75%, 5=>75%) with the intensity score (0 – 3) to obtain values between 0–15.
2.17. Statistical analysis
For statistical analyses, an unpaired Student’s t-test was used to compare two groups. One-way ANOVA with Dunnett’s multiple comparisons test was used to compare more than two groups. Analyses were performed with GraphPad Prism 8.4.2 (San Diego, CA, USA). A p-value of less than 0.05 was considered statistically significant.
3. Results
3.1. Knockout of MUC4 decreases the tumorigenic potential of PDAC cells
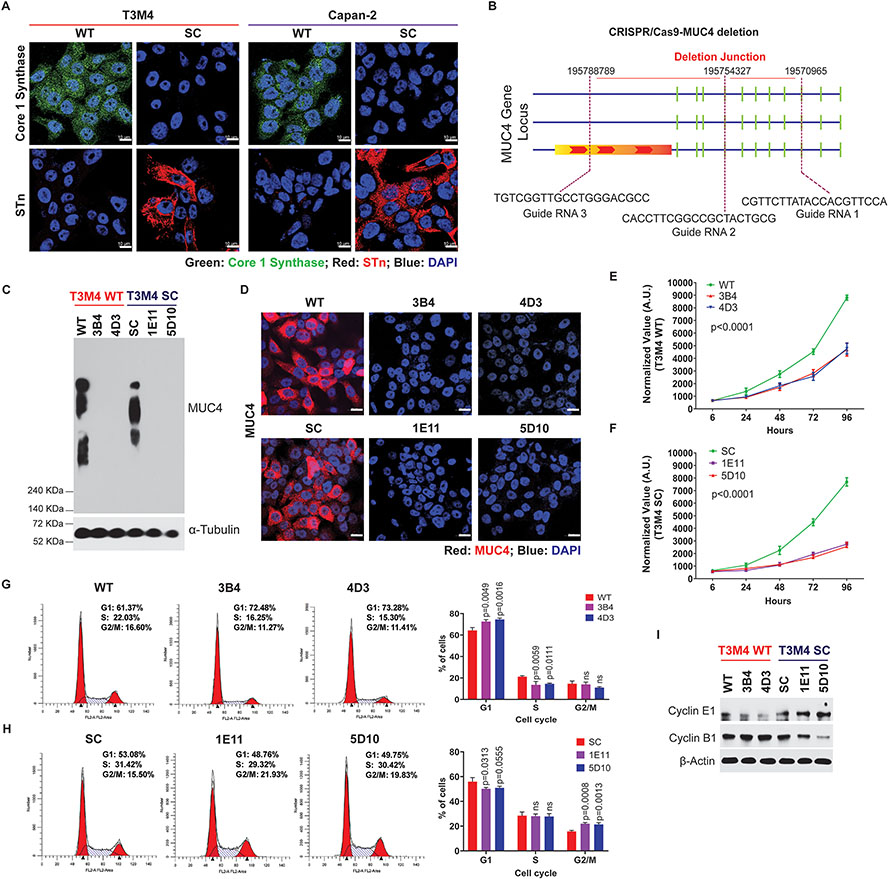
Induced aberrant overexpression of truncated O-glycans by genetic deletion of COSMC enhances the tumorigenic potential of PDAC cells with an unknown mechanism (18). Similarly, we found reduced expression of Core 1 synthase and increased expression of STn antigens in COSMC Knockout T3M4, and Capan-2 cells (SimpleCells, SC) compared to parental (wildtype, WT) cells (Figure 1A). Previous studies have shown a close association between MUC4 and PDAC disease progression, metastasis, and reduced overall survival [22–24]. To decipher the oncogenic potential of aberrant glycoforms of MUC4 in detail, we have disrupted MUC4 in T3M4 WT (wildtype, which expresses branched O-glycans) and SC (SimpleCells, which expresses truncated O-glycans (Tn/STn antigens) defective O-glycosylation using CRISPR/Cas9 genome editing technology. CRISPR/Cas9 deletion construct and the deletion site on the MUC4 gene is represented in Figure 1B. Genetic deletion of MUC4 in T3M4 WT (T3M4 WT-MUC4KO) and T3M4 SC (T3M4 WT-MUC4KO) cells was confirmed by qRT-PCR (Supplementary Figure 1A and 1B), western blotting (Figure 1C), and immunofluorescence (Figure 1D). To analyze the tumorigenic potential of MUC4 in WT and SC cells, we performed cell proliferation analyses. We found that the genetic deletion of MUC4 exhibited a remarkable reduction in the rate of cell proliferation in both T3M4 WT (Figure 1E) (overall p<0.0001; WT vs 3B4, WT vs 4D3) as well as T3M4 SC (Figure 1F) (overall p<0.0001; SC vs 1E11, SC vs 5D10) cells. Because we found a significant difference in the cell proliferation rate, we analyzed the proportion of cells in the various phases of the cell cycle. It was found that genetic deletion of MUC4 in T3M4 WT cells resulted in cell cycle arrest in the G1 phase (Figure 1G; WT vs 3B4, p=0.0049; WT vs 4D3, p=0.0016), whereas deletion of MUC4 in T3M4 SC cells resulted in cell cycle arrest in the G2/M phase (Figure 1H; SC vs 1E11, p=0.0008; SC vs 5D10, p=0.0013). This result was further confirmed by the immunoblot analysis of cell cycle markers. We found that G1 phase marker Cyclin E1 was significantly downregulated in T3M4 WT-MUC4KO clones (3B4 and 4D3) compared to T3M4 WT cells, whereas there was no change in T3M4 SC-MUC4KO clones (Figure 1I). Expectedly, the G2/M phase cell cycle marker Cyclin B1 showed no difference between T3M4 WT and WT-MUC4KO clones; however, reduced expression of Cyclin B1 was observed in SC-MUC4KO clones (1E11 and 5D10) as compared to T3M4 SC cells (Figure 1I).
Figure 1. Genetic deletion of MUC4 reduces PDAC cell tumorigenicity.
A. Immunofluorescence analysis of core 1 synthase and STn in T3M4 and Capan-2 (WT and SC) cells. Scale bar (10μm). B. Schematic representation of MUC4 deletion using the CRISPR-Cas9 MUC4 gene construct. C. Immunoprobing of MUC4 in T3M4 WT, WT-MUC4KO (3B4 and 4D3), and T3M4 SC, SC-MUC4KO (1E11 and 5D10) cells. D. Immunofluorescence analysis of MUC4 in T3M4 WT, WT-MUC4KO (3B4 and 4D3), and T3M4 SC, SC-MUC4KO (1E11 and 5D10) cells. Scale bar (10μm). E and F Alamar blue cell proliferation assays for T3M4 WT, WT-MUC4KO (3B4 and 4D3), and T3M4 SC, SC-MUC4KO (1E11 and 5D10) cells at different time points (6, 24, 48, 72, and 96 h) (mean ± SD, n=3). G and H Cell cycle analysis of T3M4 WT, WT-MUC4KO (3B4 and 4D3), and T3M4 SC, SC-MUC4KO (1E11 and 5D10) cells by FACS (mean ± SD, n=3). I. Western blot analysis of Cyclin E1 and Cyclin B1 in T3M4 WT, WT-MUC4KO (3B4 and 4D3), SC, SC-MUC4KO (1E11 and 5D10) cells. β-actin acts as a loading control. [ns = not significant].
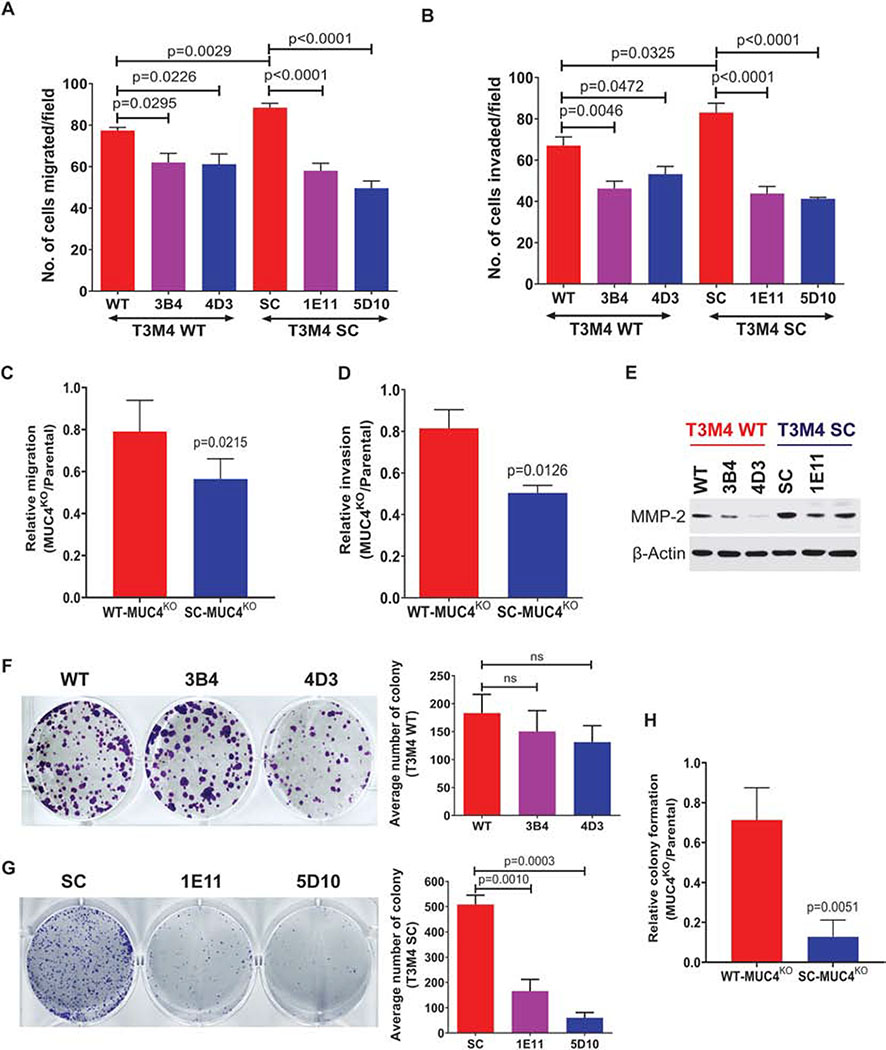
Next, we examined the rate of cell motility of PDAC cells with or without MUC4. The in vitro migration assay showed that T3M4 WT-MUC4KO (3B4, p=0.0295; 4D3, p=0.0226) and T3M4 SC-MUC4KO (1E11, p<0.0001; 5D10, p<0.0001) clones significantly reduced tumor cell migration as compared to parental cells (Figure 2A). Similarly, T3M4 WT-MUC4KO clones (3B4, p=0.0046; 4D3, p=0.0472) and T3M4 SC-MUC4KO clones (1E11, p<0.0001; 5D10, p<0.0001) exhibited significant reduction in tumor cell invasion through Matrigel compared to parental cells (Figure 2B). Next, we measured the relative (KO clone/parental cells) tumor cell migration and invasion between WT-MUC4KO and SC-MUC4KO clones. MUC4KO in SC cells showed a significantly reduced migration (p=0.0215) and invasion (p=0.0126) compared to MUC4KO in WT cells (Figure 2C and 2D, respectively). Tumor cells promote tumor invasiveness through the upregulation of matrix metalloproteases (MMPs). Among MMPs, MMP-2 and MMP-9 are most relevant to tumor invasion [25]. We found that the expression of MMP-2 was downregulated in WT- and SC-MUC4KO clones (Figure 2E). Next, we determined the colony-forming ability of these MUC4KO clones by colony formation assay. We found a decreased colony formation in WT-MUC4KO clones (3B4 and 4D3) compared to WT cells; however, this was not statistically significant (Figure 2F). However, significantly less colonies were observed in T3M4 SC-MUC4KO (1E11, p=0.0010; 5D10, p=0.0003) clones compared to T3M4 SC cells (Figure 2G). Also, we detected a significantly reduced colony formation in the SC-MUC4KO clone (p=0.0051) compared to the WT-MUC4KO clone (Figure 2H).
Figure 2. Genetic deletion of MUC4 reduces PDAC cell migration and invasion.
A. Transwell migration assay in T3M4 WT, WT-MUC4KO (3B4 and 4D3), T3M4 SC and SC-MUC4KO (1E11 and 5D10) cells (mean ± SEM, n=3). B. Transwell invasion assay in T3M4 WT, WT-MUC4KO (3B4 and 4D3), T3M4 SC and SC-MUC4KO (1E11 and 5D10) cells (mean ± SEM, n=3) C and D Relative (MUC4KO/parental) migration and invasion of WT-MUC4KO (4D3) and SC-MUC4KO (5D10) clones. (mean ± SEM, n=3). E. Western blot analysis of MMP-2 in T3M4 WT, WT-MUC4KO (3B4 and 4D3), SC, SC-MUC4KO (1E11 and 5D10) cells. β-actin acts as a loading control. F and G Colony formation assay and its graphical representation in T3M4 WT, WT-MUC4KO (3B4 and 4D3), T3M4 SC, and SC-MUC4KO (1E11 and 5D10) cells (mean ± SEM, n=3). [ns = not significant]. H. Relative (MUC4KO/parental) colony formation of WT-MUC4KO (4D3) and SC-MUC4KO (5D10) clones. (mean ± SEM, n=3).
3.2. Knockout of Tn/STn-MUC4 reduces tumor growth, proliferation, and angiogenesis
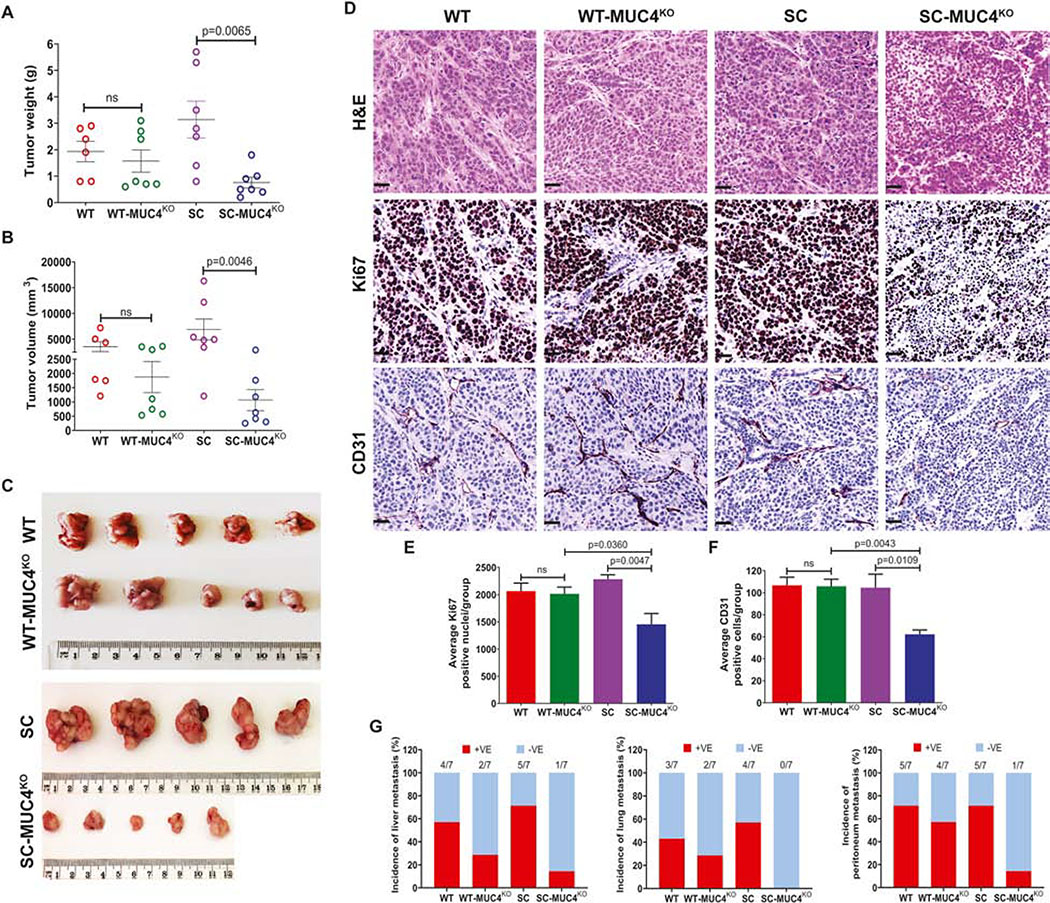
Next, we evaluated the biological impact of MUC4 on in vivo tumor growth and metastasis in the murine xenograft model. The mice with WT-MUC4KO clone (4D3) implanted in them exhibited a decreased tumor weight, and volume with no significant difference from the mice with T3M4 WT implanted cells (Figure 3A and 3B). Interestingly, SC-MUC4KO clone (5D10) implanted in tumor-bearing animals showed a significantly reduced tumor progression by weight (p=0.0065) and volume (p=0.0046) as compared to T3M4 SC cells implanted tumors (Figure 3A and 3B). The representative tumor images are given in Figure 3C. Additionally, we observed a significant downregulation of tumor proliferation marker Ki67 (p=0.0047) and angiogenesis marker CD31 (p=0.0109) in tumors of SC-MUC4KO implanted clone. In contrast, no significant change in the expression of Ki67 and CD31 between tumors of WT and WT-MUC4KO implanted clones (Figure 3D, 3E, and 3F). Additionally, SC-MUC4KO clone implanted tumors showed a significantly reduced Ki67 (p=0.0360) and CD31 (p=0.0043) expression compared to WT-MUC4KO clone implanted tumors (Figure 3E and 3F, respectively). Also, we observed a reduced percentage of tumor metastasis to liver, lung, and peritoneum in MUC4KO tumor-bearing animals (Figure 3G).
Figure 3. MUC4 deletion affects in vivo tumorigenesis.
A and B Quantification of tumor weight (g) and tumor volume (mm3) in T3M4 WT, WT-MUC4KO (4D3), SC, SC-MUC4KO (5D10) cells derived orthotopic tumors (n=7). C. Representative tumor images from the above-mentioned tumor models. D. Immunohistochemical analysis of H&E, Ki67 and CD31 in the above-mentioned tumor tissues. E. Average Ki67 positive cells (mean ± SD, n=5). F. Average CD31 positive cells (mean ± SD, n=5). G. Percentage of tumor metastasis (liver, lung, and peritoneum) in T3M4 WT, WT-MUC4KO (4D3), SC, SC-MUC4KO (5D10) cells implanted orthotopic tumor bearing-animals (n=7). [ns = not significant].
3.3. MUC4 enhances pancreatic cancer malignancy through the activation of EGF receptors
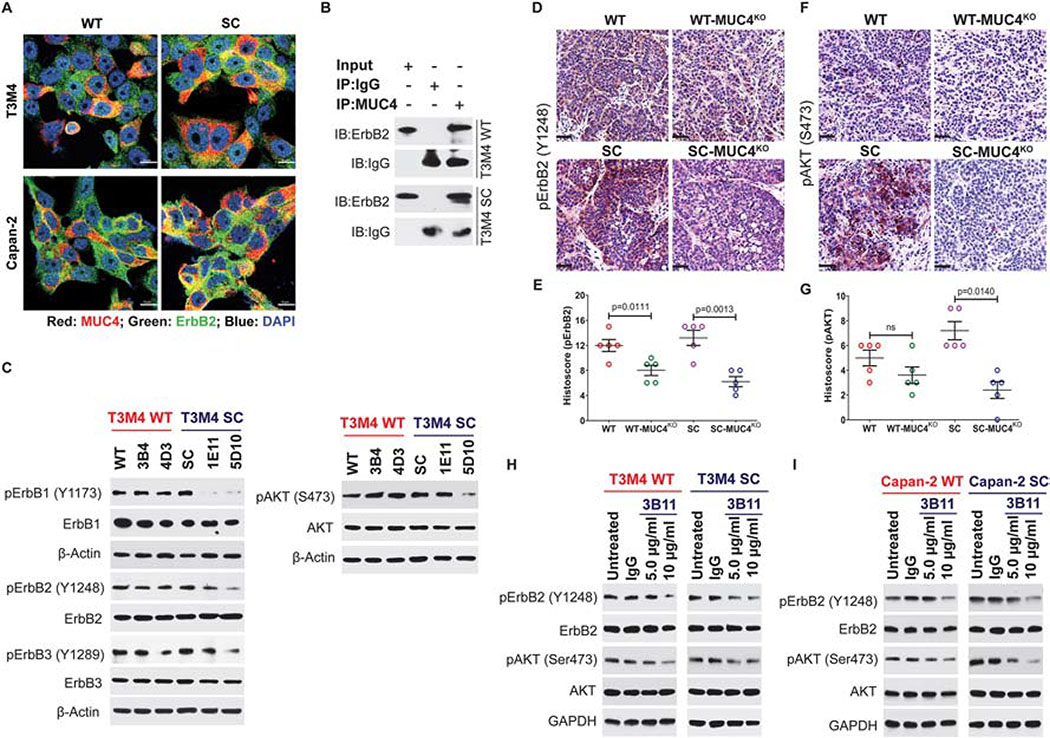
Several studies have shown that mucin-type glycoproteins contain EGF-like domains that bind ErbB receptors [26, 27] (including FGFR1 [28]), stabilize the expression of growth factor receptors, and activate downstream oncogenic cell signaling pathways. In this study, we examined the expression and subcellular localization of MUC4 and ErbB2 using co-localization confocal microscopy. Both MUC4 (red) and ErbB2 (green) are predominantly expressed in both cell membrane and cytoplasm and found increased col-localization (yellow) of these proteins in T3M4 SC and Capan-2 SC cells (Figure 4A). We further validated these MUC4-ErbB2 interactions by Immunoprecipitation. We detected increased interaction between MUC4, and ErbB2 in T3M4 SC cells as compared to T3M4 WT cells (Figure 4B). Our previous studies demonstrated that the cancer-specific truncation of mucin-type O-glycosylation enhances epidermal growth factor (EGF) receptor family signaling in PDAC cells [18]. Hence, we explored MUC4 promoted ErbB mediated oncogenic signaling in PDAC cells that express truncated O-glycans. We found downregulation of the phosphorylation of ErbB1 (Y1173), ErbB2 (Y1248), ErbB3 (Y1289), and AKT (S473) in T3M4 SC-MUC4KO clones (1E11 and 5D10) compared to SC cells (Figure 4C). However, there was not a difference in the expression of phosphorylated ErbB1, ErbB2, ErbB3, and AKT between T3M4 WT-MUC4KO clones (3B4 and 4D3) and parental WT cells (Figure 4C). To confirm these findings, we performed immunohistochemical analysis of phosphorylated ErbB2 in tumors derived from T3M4 WT, WT-MUC4KO, T3M4 SC, and SC-MUC4KO cells and found that MUC4KO clones (WT and SC) implanted tumors exhibited less expression of p-ErbB2 (p=0.0111 and p=0.0013, respectively) compared to their parental cells (Figure 4D and 4E). As one of the oncogenic drivers, ErbB2 can directly or indirectly bind PI3K and transmit signals down the PI3K/AKT pathway to facilitate tumor progression. Hence, we analyzed MUC4-ErbB2 mediated phosphorylation of AKT in orthotopic tumor tissues. A significantly reduced expression of p-AKT was detected in tumors of SC-MUC4KO implanted clones compared to SC cells derived tumors (p=0.0140, Figure 4F and 4G). However, we observed a reduced expression of p-AKT with no significant difference in WT-MUC4KO cells derived tumors than WT tumors (Figure 4F and 4G). These results support the hypothesis that the oncogenic potential of MUC4 is significantly affected by aberrant O-glycosylation.
Figure 4. MUC4 mediates PDAC tumorigenicity through the regulation of ErbB2.
A. Co-localization of MUC4 (red) and ErbB2 (green) in T3M4 and Capan-2 (WT and SC) cells. Representative images are shown (n=3). Scale bar 10 μm. B. Immunoprecipitation (IP) of T3M4 (WT and SC) cell lysates with MUC4 (mAb 8G7) and mouse IgG (isotype control), and immunoprobed (IB) with anti-ErbB2 and anti-mouse IgG antibodies. C. Western blot analysis of phospho-ErbB1 (Y1173), ErbB1, phospho-ErbB2 (Y1248), ErbB2, phospho-ErbB3 (Y1289), and ErbB3, phospho-AKT (S473) and AKT in T3M4 WT, WT-MUC4KO, SC, and SC-MUC4KO cell lysate. β-actin acts as a loading control. IHC analysis of p-ErbB2 (D and E) and p-AKT (S473) (F and G) in T3M4 WT, WT-MUC4KO, SC, and SC-MUC4KO cells implanted orthotopic tumor tissues (n=5). H. Western blot analysis of p-ErbB2, ErbB2, p-AKT, and AKT in T3M4 WT and SC cells treated with varying doses of anti-Tn-MUC4 mAb 3B11 (5 and 10 μg/ml). I. Western blot analysis of p-ErbB2, ErbB2, p-AKT, and AKT in Capan-2 WT and SC cells treated with varying doses of anti-Tn-MUC4 mAb 3B11 (5 and 10 μg/ml) for 24 h. Treatment of PDAC cells with mouse IgG (5 μg/ml) served as an isotype control. GAPDH act as a loading control. [ns = not significant].
To further validate the aberrant glycoforms of MUC4 mediated PDAC tumor malignancy, we have treated isogenic PDAC cells T3M4 WT and T3M4 SC with anti-Tn-MUC4 specific mAb 3B11. mAb 3B11 treatment induces a dose-dependent reduction of p-ErbB2 (Y1248) and p-AKT (S473) in both WT and SC cells as compared to untreated, and IgG treated control cells (Figure 4H). Interestingly, we observed a similar result in mAb 3B11-treated Capan-2 WT and SC cells (Figure 4I). Treatment of both T3M4 WT as well as SC cells with AKT inhibitor (LY294002) showed a reduced expression of p-AKT (S473) (Supplementary Figure 1C). Expression of p-AKT was significantly high in SC cells compared to WT cells and confirmed the hypothesis that MUC4 enhances tumor malignancy through ErbB2/AKT signaling axis in truncated O-glycan expressing PDAC cells.
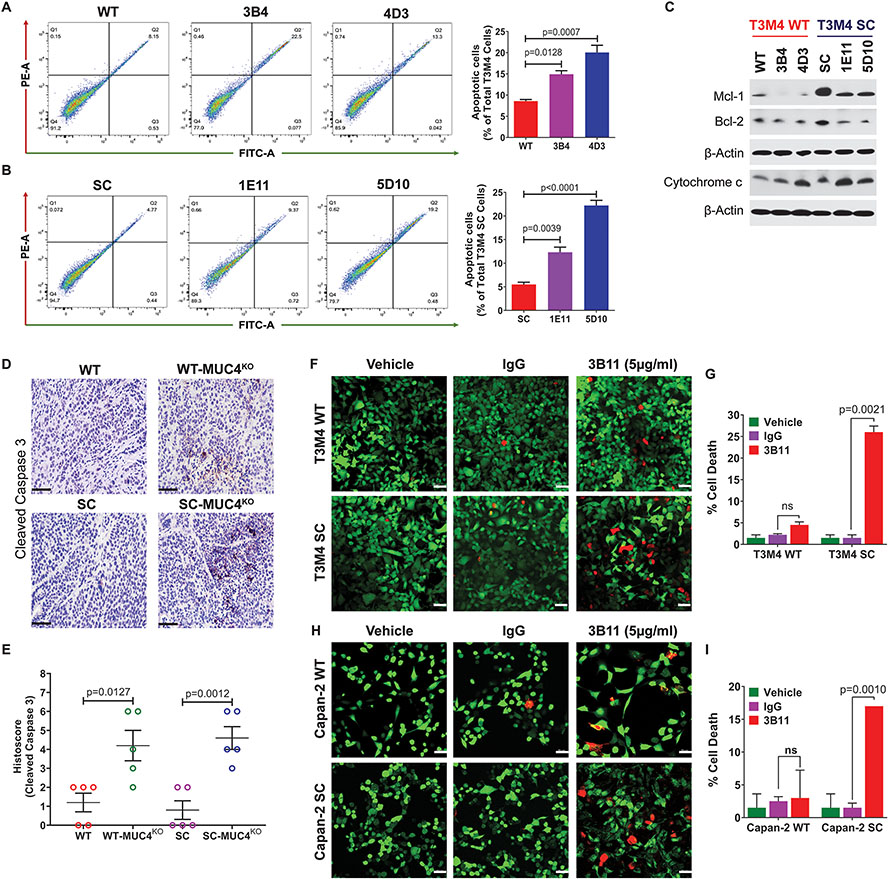
3.4. Knockout of MUC4 promotes apoptosis in PDAC cells
It is well documented that MUC4 signals through ErbB2 to promote tumor cell survival [29]. The knockdown of MUC4 was reported to increase the Bax/BclXL ratio, suggesting more susceptibility to apoptosis [14]. Therefore, we sought to analyze the MUC4 induced apoptotic index in PDAC cells with or without aberrant O-glycosylation. We found that genetic deletion of MUC4 in both WT and SC cells significantly induced apoptosis. WT-MUC4KO clones exhibited a three-fold higher number of cells in the apoptotic quadrant than their parental cells (3B4, p=0.0128; 4D3, p=0.0007, Figure 5A). Similarly, SC-MUC4KO clones exhibited a significantly increased number of apoptotic cells than SC cells (1E11, p=0.0039; 5D10, p<0.0001, Figure 5B). This result was further confirmed with the expression of pro- and anti-apoptotic markers in parental versus KO clones. MUC4KO clones showed decreased expressions of anti-apoptotic markers Mcl-1 and Bcl-2 with concomitant increased expression of pro-apoptotic marker cytochrome c, suggesting a higher susceptibility to induce apoptosis in MUC4KO clones (Figure 5C). To further confirm these results, we performed immunohistochemical analysis of cleaved caspase 3 in tumor tissues of WT, WT-MUC4KO, SC, and SC-MUC4KO implanted cells. We detected increased expression of cleaved caspase 3 in SC-MUC4KO (p=0.0012) and WT-MUC4KO (p=0.0127) clone implanted tumor tissues compared to their parental cells implanted tumors (Figure 5D and 5E). To further validate the MUC4 induced apoptosis, we have measured the status of apoptosis in isogeneic T3M4 and Capan-2 (WT and SC) cells treated with mAb 3B11 (5 μg/ml) for 24 h using a Live/Dead cell assay. Significantly increased cell death was observed in mAb 3B11 treated T3M4 SC and Capan-2 SC cells as compared to IgG control cells (Figure 5F and 5G and Figure 5H and 5I, respectively). However, WT cells of both cell lines did not show a significant increase in cell death upon incubation with mAb 3B11. These results indicate T3M4 SC and Capan-2 SC cells are more susceptible to cell death with the treatment of anti-Tn-MUC4 specific mAb 3B11 compared to WT cells, which further confirm the role of aberrant glycoforms of MUC4 on the regulation of apoptosis in PDAC cells.
Figure 5. Genetic deletion of MUC4 induces apoptosis in PDAC cells.
A and B Apoptosis analysis using Annexin V/FITC in T3M4 WT, WT-MUC4KO (3B4 and 4D3), T3M4 SC, and SC-MUC4KO (1E11 and 5D10) cells by FACS (mean ± SD, n=3). C. Western blotting of Mcl-1, Bcl-2, and cytochrome c in T3M4 WT, WT-MUC4KO (3B4 and 4D3), T3M4 SC, SC-MUC4KO (1E11 and 5D10) cells. β-actin acts as a loading control. D. Immunohistochemical analysis of cleaved caspase 3 in T3M4 WT, WT-MUC4KO (4D3), T3M4 SC, and SC-MUC4KO (5D10) cells implanted xenograft tumors. E. Average IHC histoscore of cleaved caspase 3 (n=5). Live/Dead cell analysis of anti-Tn-MUC4 mAb 3B11 (5 μg/ml, for 24 h) treated T3M4 WT and SC cells (F and G), and Capan-2 WT and SC cells (H and I) (mean ± SD, n=3). [ns = not significant].
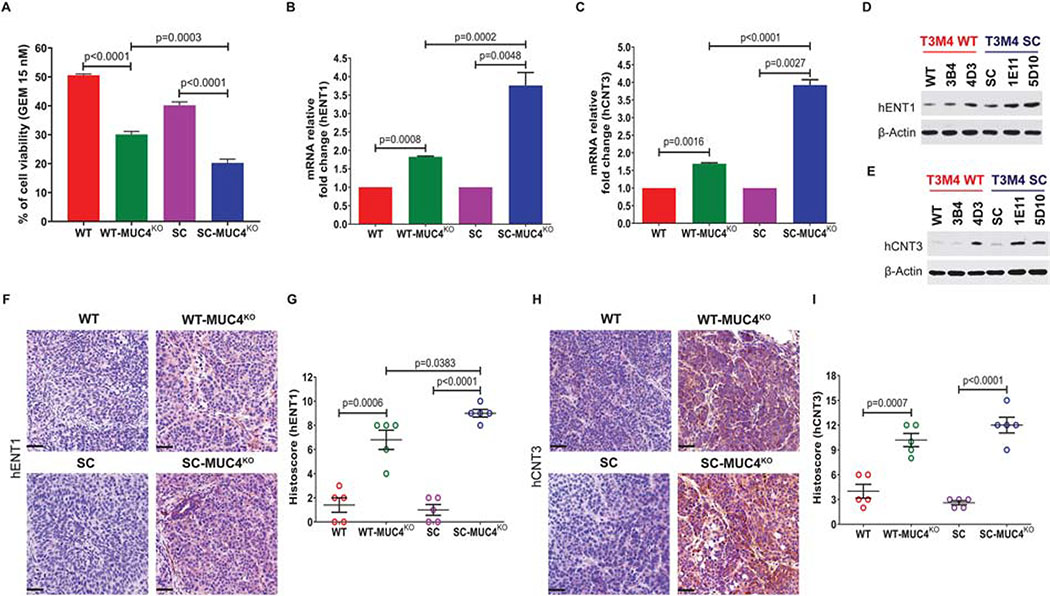
3.5. Knockout of MUC4 induces cellular sensitivity to gemcitabine and alters gemcitabine (GEM) metabolism markers in PDAC cells
Previous MUC4 knockdown studies have demonstrated that MUC4 can affect chemoresistance [30, 31]. Here, we utilized genetic knockout models of MUC4 with and without aberrant glycosylation to determine the MUC4-mediated drug resistance. Treatment of 15 nM of GEM resulted in a significant decrease in the cell viability in the T3M4 WT-MUC4KO clone (p<0.0001, Figure 6A). Interestingly, treatment of the same concentration of GEM resulted in a further reduction of cell viability of the T3M4 SC-MUC4KO clone (p<0.0001, Figure 6A). However, the SC-MUC4KO clone showed a significantly reduced cell viability (p=0.0003) compared WT-MUC4KO clone (Figure 6A). One of the mechanisms of action of the chemotherapeutic drug GEM in human pancreatic cancer cells is the induction of apoptosis [32]. The observed increased sensitivity to GEM in MUC4KO clones was accompanied by the induction of apoptosis. The efficacy of GEM depends on the expression of nucleoside transporters (hENT1, hCNT1, and hCNT3) that help to transport and incorporate GEM into the DNA. We sought to study the mechanism behind MUC4-mediated drug resistance and found that genetic deletion of MUC4 alters the expressions of hENT1, as well as hCNT3, in PDAC cells. qPCR analysis of mRNA levels of hENT1 showed that both WT-MUC4KO clone (p=0.0008) and SC-MUC4KO clone (p=0.0048) enhanced the expression of hENT1 (Figure 6B). Similarly, mRNA expression of hCNT3 was also upregulated in WT-MUC4KO clone (p=0.0016) and SC-MUC4KO clone (p=0.0027) compared to their respective parental cells (Figure 6C). Additionally, the SC-MUC4KO clone showed a significantly increased hENT1 (p=0.0002) and hCNT3 (p<0.0001) expression compared WT-MUC4KO clone (Figure 6B and 6C, respectively). Further, western blot analysis revealed that protein expressions of hENT1 and hCNT3 were significantly upregulated in T3M4 WT-MUC4KO and T3M4 SC-MUC4KO clones compared to T3M4 WT/SC cells (Figure 6D and 6E). These observations were further confirmed with the immunohistochemical analysis of hENT1 and hCNT3 in T3M4 WT and SC cells with or without MUC4 implanted tumor tissues. Protein expression of hENT1 was significantly upregulated in both WT-MUC4KO (p=0.0006) and SC-MUC4KO clones (p<0.0001) implanted tumors (Figure 6F and 6G). Similarly, protein expression of hCNT3 was also significantly high in WT-MUC4KO (p=0.0007) and SC-MUC4KO clones (p<0.0001) implanted tumor tissues (Figure 6H and 6I). However, the SC-MUC4KO clone showed a significantly increased hENT1 (p=0.0383) expression compared to WT-MUC4KO clone implanted tumors (Figure 6G). Also, we observed increased expression of hCNT3 in SC-MUC4KO compared to WT-MUC4KO clone implanted tumors (Figure 6I). These results further confirm that MUC4-mediated drug resistance is enhanced by aberrant glycosylation.
Figure 6. Genetic deletion of MUC4 induces sensitivity to gemcitabine in PDAC cells.
A. Cytotoxicity assay of gemcitabine in T3M4 WT, WT-MUC4KO (4D3), T3M4 SC and SC-MUC4KO (5D10) cells (mean ± SD, n=5). B and C qRT-PCR analysis of mRNA expression of hENT1 and hCNT3 in T3M4 WT, WT-MUC4KO (4D3), T3M4 SC and SC-MUC4KO (5D10) cells (mean ± SD, n=3). D and E Western blotting of protein expression of hENT1 and hCNT3 in T3M4 WT, WT-MUC4KO (3B4 and 4D3), SC, SC-MUC4KO (1E11 and 5D10). β-actin acts as a loading control. Immunohistochemical analysis of hENT1 (F and G) and hCNT3 (H and I) in T3M4 WT, WT-MUC4KO (4D3), T3M4 SC, and SC-MUC4KO (5D10) cells implanted xenograft tumors (mean ± SD, n=5).
Discussion
Pancreatic cancer malignancy is highly lethal due to asymptomatic characteristics, poor resectability, early metastasis, lack of consistent biomarkers, limited efficacies of current treatment modalities, and high recurrence rate. The development of resistance to chemotherapeutics is yet another attribute of pancreatic malignancy. Previous studies showed the significant role of MUC4 in pancreatic cancer cell motility, sustained growth, activation of oncogenic signaling cascades, and local and distant metastases [5, 33, 34]. Our previous studies demonstrated that induced expression of aberrant O-glycans enhanced PDAC tumor growth via promoting the epithelial-to-mesenchymal transition [18, 35]. Approximately 80% of the human PDAC tumors express truncated O-glycans [17]. However, how MUC4 glycoprotein affects tumorigenic functions of cells with and without aberrant O-glycosylation is poorly explored in PDAC. Here, we show evidence that the genetic deletion of MUC4 significantly inhibited the malignant potential of PDAC cells, which is further affected in cells that aberrantly overexpress truncated O-glycans. More specifically, we showed that aberrant glycoforms of MUC4 differentially regulate the cell cycle and induce apoptosis in PDAC cells. Importantly, MUC4 with truncated O-glycans enhanced PDAC tumorigenesis through the activation of EGF receptors. Additionally, MUC4 is involved in PDAC chemoresistance by altering the expression of nucleoside transporters.
Aberrant glycosylation of proteins such as MUC1, MUC4, and MUC16 are expected to affect tumorigenic functions by modifying the extracellular domain. Structural changes in the glycoproteins regulate cellular functions either alone or through interaction with other receptors or ligands [24]. For example, aberrant expression of Core 1 β1, 3-galactosyltransferase (C1GALT1) enhanced breast cancer malignancy through activating the MUC1-C/β-catenin signaling pathway [36]. Also, studies have shown that MUC1 can escalate the expression of truncated glycans during tumorigenesis [37]. Though MUC4 has been shown to promote tumor invasion and metastasis through the interactions with ErbB2, ErbB3, and extracellular matrix (ECM) proteins [38]; however, the biological consequences of aberrant O-glycosylation of MUC4 and their effect on PDAC tumorigenicity is poorly understood. Our results indicate that MUC4 significantly enhances the tumorigenic properties of PDAC cells that aberrantly express truncated O-glycans. The deletion of genes is one of the reliable approaches for functional analyses. To study the effect of MUC4 during PDAC progression, we deleted MUC4 in PDAC cells that express fully branched and truncated O-glycans. We found that genetic deletion of MUC4 significantly affected cell proliferation, invasion, migration, and colony formation in truncated O-glycan expressing PDAC cells. Decreased proliferation in MUC4 deleted cells is associated with cell cycle arrest. Interestingly, MUC4 regulates cell cycle in a glycosylation dependent manner; MUC4 knockout in PDAC cells with defective O-glycosylation showed cell cycle arrest at the G2/M phase, as evidenced by decreased expression of cyclin B1, whereas MUC4 deletion in branched O-glycan expressing (WT) PDAC cells resulted in cell cycle arrest at the G1 phase. Phosphorylation of Cyclin B1 by Cdk1 facilitates its translocation to the nucleus and coordinates entry into mitosis [39]. MUC4 deletion in truncated O-glycan expressing PDAC cells, therefore, significantly prevents entry into mitosis and thereby inhibits tumor cell proliferation. However, further evaluations are warranted to define the mechanisms by which aberrant O-glycosylation induces cell cycle arrest at the G2/M phase. Additionally, the deletion of MUC4 in truncated O-glycan expressing PDAC cells significantly reduced in vivo tumor growth, proliferation, and angiogenesis in an orthotopic tumor model. These findings support the phenomenon of aberrant glycosylation-mediated tumor progression [18, 40, 41]. Here, we provide evidence that MUC4 significantly affects the tumorigenic properties of PDAC cells that aberrantly overexpress truncated O-glycans.
Another important observation in the present study was diminished expressions of phosphorylated EGF receptors in MUC4 deleted clones compared to parental cells, which was affected by aberrant O-glycosylation. This hypothesis was further confirmed in truncated O-glycan expressing PDAC cells treated with monoclonal antibody 3B11 that binds specifically with Tn-MUC4 and significantly downregulated the activation of ErbB2 and its downstream AKT signaling cascades. Overexpression of the ErbB family of receptors has been shown to correlate with tumor aggressiveness during PDAC progression [42]. Moreover, MUC4 was shown to act as an intramembrane ligand for ErbB2 and encourages its dimerization with other ErbB receptors [43, 44]. In our study, we found that the interaction of MUC4 with ErbB2 in PDAC is highly influenced by aberrant O-glycosylation and represents one of the major mechanisms behind tumor aggressiveness. Also, our results demonstrating the therapeutic potential of the Tn-MUC4-specific monoclonal antibody against MUC4 driven EGF signaling mediated PDAC progression. The heavily glycosylated extracellular domain of MUC4 may have the capacity to bind to various lectins and receptors that describe the oncogenic potential of MUC4, which should be investigated in the future.
We also show that loss of MUC4 is sufficient to modify the expression of apoptotic proteins and promote apoptosis by upregulating the expressions of pro-apoptotic proteins and downregulating the expressions of anti-apoptotic proteins. The development of pancreatic cell resistance to gemcitabine is one of the major hurdles in the treatment options for PDAC [45]. Several factors are responsible for the sensitivity to gemcitabine, particularly nucleoside transporters [46]. hENT1, hCNT1, and hCNT3 are the major molecules that facilitate cellular uptake of GEM [15]. Here, we found that loss of MUC4 significantly modified and upregulated the hENT1 and hCNT3 transporters in PDAC tumors that aberrantly overexpress truncated O-glycans. These results indicate that MUC4 with aberrant O-glycosylation may be a potential predictive marker in pancreatic cancer drug resistance.
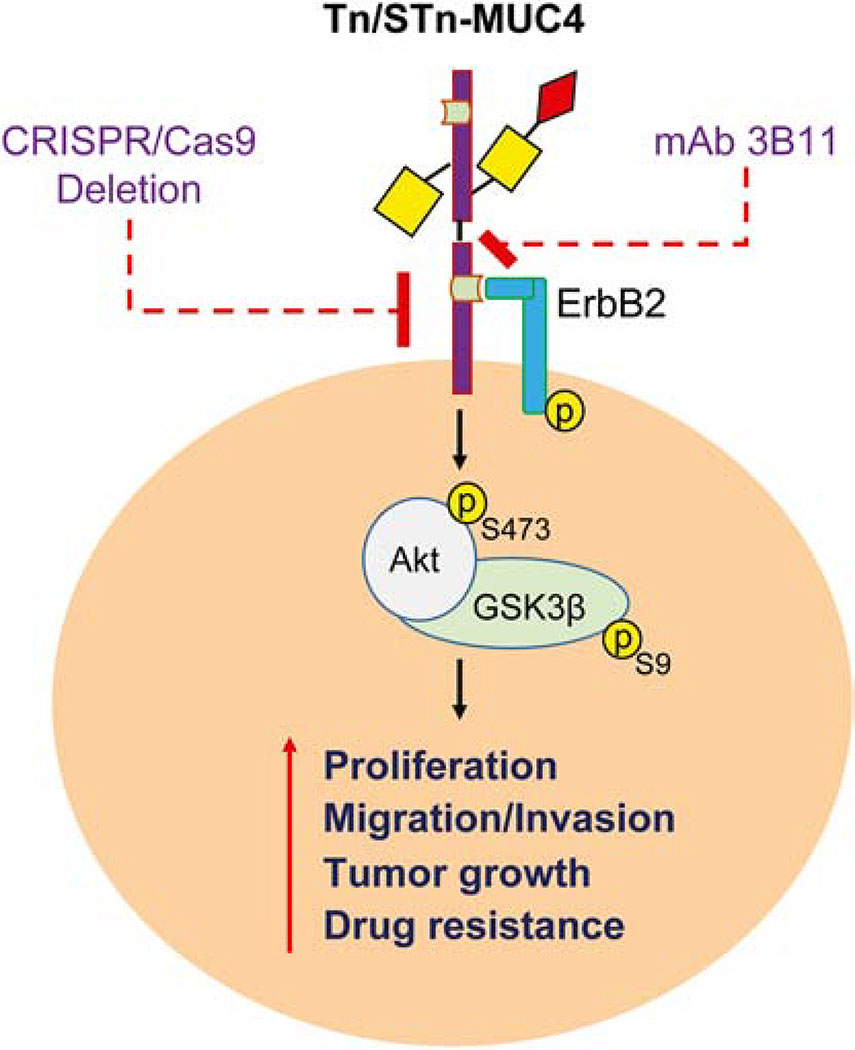
In summary, this study provide evidence for the first time on the potential oncogenic role of MUC4 that enhances tumor cell growth, invasion, migration, colony formation, and in vivo tumor growth of PDAC cells that possess aberrant O-glycosylation, which is the most common characteristic features of PDAC tumors. Also, this study show that the genetic deletion of MUC4 prevents PDAC progression through the downregulation EGF receptors and their downstream signaling pathways, which is further regulated by aberrant O-glycosylation. Additionally, this study found that loss of MUC4 facilitates the upregulation of nuclear transporters hENT1 and hCNT3 in truncated O-glycan expressing PDAC tumors (Figure 7). Hence, MUC4 can be considered as a potential predictive biomarker for PDAC progression as well as drug resistance in PDAC tumors that aberrantly overexpress truncated O-glycans.
Figure 7. Aberrant glycosylation of MUC4 promotes PDAC malignancy.
Genetic deletion of MUC4 in truncated O-glycan expressing PDAC cells alters PDAC malignancy and drug sensitivity. Tn-MUC4 specific antibody 3B11 inhibits MUC4 driven ErbB/AKT mediated PDAC tumorigenesis.
Supplementary Material
Highlights.
MUC4 alter in vitro and in vivo tumorigenic properties of truncated O-glycan expressing PDAC cells via ErbB/AKT signaling
mAb 3B11 binds with aberrant glycoforms of MUC4 (Tn/STn-MUC4) and diminishes ErbB mediated oncogenic signaling
MUC4 alter drug sensitivity in truncated O-glycan expressing PDAC via nucleoside transporters
Acknowledgment
This work was supported, in part, by the National Cancer Institute at the National Institutes of Health R01 CA208108, NE-DHHS/LB506, and Fred & Pamela Buffett Cancer Center start-up funds to (P.R). The European Research Council (ERC Consolidator Grant, EU project 772735), the Danish National Research Foundation Grant DNRF107, and the NEYE FOUNDATION to HHW. We thank Jeffrey Patterson (Research Editorial Office, at UNMC) for editing this manuscript.
Abbreviations
- BSA
bovine serum albumin
- DAPI
4′,6-diamidino-2-phenylindole
- EGFR
epidermal growth factor receptor
- FBS
fetal bovine serum
- FITC
fluorescein isothiocyanate
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- hENT1
human equilibrative nucleoside transporter 1
- hCNT3
human concentrative nucleoside transporter 1
- mAb
monoclonal antibody
- MUC4
mucin4
- MMP
matrix metalloproteases
- PDAC
pancreatic ductal adenocarcinoma
- PI
propidium iodide
Footnotes
Conflict of Interest
HC is a shareholder with GO-Therapeutics, and HHW is a shareholder and has a consultancy arrangement with GO-Therapeutics.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.American cancer society. Cancer facts & figures. 2020 [DOI] [PMC free article] [PubMed]
- 2.Ryan DP, Hong TS, Bardeesy N. Pancreatic adenocarcinoma. N Engl J Med. 2014;371(11):1039–1049. [DOI] [PubMed] [Google Scholar]
- 3.Hollingsworth MA, Swanson BJ. Mucins in cancer: protection and control of the cell surface. Nat Rev Cancer. 2004;4(1):45–60. [DOI] [PubMed] [Google Scholar]
- 4.Ansari D, Bauden MP, Sasor A, Gundewar C, Andersson R. Analysis of MUC4 expression in human pancreatic cancer xenografts in immunodeficient mice. Anticancer Res. 2014;34(8):3905–3910. [PubMed] [Google Scholar]
- 5.Andrianifahanana M, Moniaux N, Schmied BM, et al. Mucin (MUC) gene expression in human pancreatic adenocarcinoma and chronic pancreatitis: a potential role of MUC4 as a tumor marker of diagnostic significance. Clin Cancer Res. 2001;7(12):4033–4040. [PubMed] [Google Scholar]
- 6.Lahdaoui F, Delpu Y, Vincent A, et al. miR-219–1-3p is a negative regulator of the mucin MUC4 expression and is a tumor suppressor in pancreatic cancer. Oncogene. 2015;34(6):780–788. [DOI] [PubMed] [Google Scholar]
- 7.Xia P, Choi AH, Deng Z, et al. Cell membrane-anchored MUC4 promotes tumorigenicity in epithelial carcinomas. Oncotarget. 2017;8(8):14147–14157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karg A, Dinç ZA, Başok O, et al. MUC4 expression and its relation to ErbB2 expression, apoptosis, proliferation, differentiation, and tumor stage in non-small cell lung cancer (NSCLC). Pathol Res Pract. 2006;202(8):577–583. [DOI] [PubMed] [Google Scholar]
- 9.Funes M, Miller JK, Lai C, et al. The mucin Muc4 potentiates neuregulin signaling by increasing the cell-surface populations of ErbB2 and ErbB3. J Biol Chem. 2006;281(28):19310–19319. [DOI] [PubMed] [Google Scholar]
- 10.Ramsauer VP, Pino V, Farooq A, et al. Muc4-ErbB2 complex formation and signaling in polarized CACO-2 epithelial cells indicate that Muc4 acts as an unorthodox ligand for ErbB2. Mol Biol Cell. 2006;17(7):2931–2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jonckheere N, Skrypek N, Merlin J, et al. The mucin MUC4 and its membrane partner ErbB2 regulate biological properties of human CAPAN-2 pancreatic cancer cells via different signalling pathways. PLoS One. 2012;7(2):e32232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carraway KL, Theodoropoulos G, Kozloski GA, et al. Muc4/MUC4 functions and regulation in cancer. Future Oncol. 2009;5(10):1631–1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Skrypek N, Vasseur R, Vincent A, et al. The oncogenic receptor ErbB2 modulates gemcitabine and irinotecan/SN-38 chemoresistance of human pancreatic cancer cells via hCNT1 transporter and multidrug-resistance associated protein MRP-2. Oncotarget. 2015;6(13):10853–10867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Skrypek N, Duchêne B, Hebbar M, et al. The MUC4 mucin mediates gemcitabine resistance of human pancreatic cancer cells via the Concentrative Nucleoside Transporter family. Oncogene. 2013;32(13):1714–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Damaraju VL, Damaraju S, Young JD, et al. Nucleoside anticancer drugs: the role of nucleoside transporters in resistance to cancer chemotherapy. Oncogene. 2003;22(47):7524–7536. [DOI] [PubMed] [Google Scholar]
- 16.Pinho SS, Reis CA. Glycosylation in cancer: mechanisms and clinical implications. Nat Rev Cancer. 2015;15(9):540–555. [DOI] [PubMed] [Google Scholar]
- 17.Julien S, Videira PA, Delannoy P. Sialyl-tn in cancer: (how) did we miss the target?. Biomolecules. 2012;2(4):435–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Radhakrishnan P, Dabelsteen S, Madsen FB, et al. Immature truncated O-glycophenotype of cancer directly induces oncogenic features. Proc Natl Acad Sci U S A. 2014;111(39):E4066–E4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Steentoft C, Vakhrushev SY, Joshi HJ, et al. Precision mapping of the human O-GalNAc glycoproteome through SimpleCell technology. EMBO J. 2013;32(10):1478–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Radhakrishnan P, Grandgenett PM, Mohr AM, et al. Expression of core 3 synthase in human pancreatic cancer cells suppresses tumor growth and metastasis. Int J Cancer. 2013;133(12):2824–2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moon JS, Ko HM, Park JI, et al. Inhibition of human mesenchymal stem cell proliferation via Wnt signaling activation. J Cell Biochem. 2018;119(2):1670–1678. [DOI] [PubMed] [Google Scholar]
- 22.Singh AP, Moniaux N, Chauhan SC, et al. Inhibition of MUC4 expression suppresses pancreatic tumor cell growth and metastasis. Cancer Res. 2004;64(2):622–630. [DOI] [PubMed] [Google Scholar]
- 23.Saitou M, Goto M, Horinouchi M, et al. MUC4 expression is a novel prognostic factor in patients with invasive ductal carcinoma of the pancreas. J Clin Pathol. 2005;58(8):845–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Remmers N, Anderson JM, Linde EM, et al. Aberrant expression of mucin core proteins and o-linked glycans associated with progression of pancreatic cancer. Clin Cancer Res. 2013;19(8):1981–1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnsen M, Lund LR, Rømer J, et al. Cancer invasion and tissue remodeling: common themes in proteolytic matrix degradation. Curr Opin Cell Biol. 1998;10(5):667–671. [DOI] [PubMed] [Google Scholar]
- 26.Carraway KL 3rd, Rossi EA, Komatsu M, et al. An intramembrane modulator of the ErbB2 receptor tyrosine kinase that potentiates neuregulin signaling. J Biol Chem. 1999;274(9):5263–5266. [DOI] [PubMed] [Google Scholar]
- 27.Chaturvedi P, Singh AP, Chakraborty S, et al. MUC4 mucin interacts with and stabilizes the HER2 oncoprotein in human pancreatic cancer cells [published correction appears in Cancer Res. 2008 May 1;68(9):3550]. Cancer Res. 2008;68(7):2065–2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rachagani S, Macha MA, Ponnusamy MP, et al. MUC4 potentiates invasion and metastasis of pancreatic cancer cells through stabilization of fibroblast growth factor receptor 1. Carcinogenesis. 2012;33(10):1953–1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Workman HC, Sweeney C, Carraway KL 3rd. The membrane mucin Muc4 inhibits apoptosis induced by multiple insults via ErbB2-dependent and ErbB2-independent mechanisms. Cancer Res. 2009;69(7):2845–2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bafna S, Kaur S, Momi N, et al. Pancreatic cancer cells resistance to gemcitabine: the role of MUC4 mucin. Br J Cancer. 2009;101(7):1155–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kalra AV, Campbell RB. Mucin overexpression limits the effectiveness of 5-FU by reducing intracellular drug uptake and antineoplastic drug effects in pancreatic tumours. Eur J Cancer. 2009;45(1):164–173. [DOI] [PubMed] [Google Scholar]
- 32.Hill R, Rabb M, Madureira PA, et al. Gemcitabine-mediated tumour regression and p53-dependent gene expression: implications for colon and pancreatic cancer therapy. Cell Death Dis. 2013;4(9):e791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhu Y, Zhang JJ, Zhu R, et al. The increase in the expression and hypomethylation of MUC4 gene with the progression of pancreatic ductal adenocarcinoma. Med Oncol. 2011;28 Suppl 1:S175–S184. [DOI] [PubMed] [Google Scholar]
- 34.Swartz MJ, Batra SK, Varshney GC, et al. MUC4 expression increases progressively in pancreatic intraepithelial neoplasia. Am J Clin Pathol. 2002;117(5):791–796. [DOI] [PubMed] [Google Scholar]
- 35.Thomas D, Sagar S, Caffrey T, et al. Truncated O-glycans promote epithelial-to-mesenchymal transition and stemness properties of pancreatic cancer cells. J Cell Mol Med. 2019;23(10):6885–6896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chou CH, Huang MJ, Chen CH, et al. Up-regulation of C1GALT1 promotes breast cancer cell growth through MUC1-C signaling pathway. Oncotarget. 2015;6(8):6123–6135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Solatycka A, Owczarek T, Piller F, et al. MUC1 in human and murine mammary carcinoma cells decreases the expression of core 2 β1,6-N-acetylglucosaminyltransferase and β-galactoside α2,3-sialyltransferase. Glycobiology. 2012;22(8):1042–1054. [DOI] [PubMed] [Google Scholar]
- 38.Senapati S, Gnanapragassam VS, Moniaux N, et al. Role of MUC4-NIDO domain in the MUC4-mediated metastasis of pancreatic cancer cells. Oncogene. 2012;31(28):3346–3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gavet O, Pines J. Progressive activation of CyclinB1-Cdk1 coordinates entry to mitosis. Dev Cell. 2010;18(4):533–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu YM, Liu CH, Huang MJ, et al. C1GALT1 enhances proliferation of hepatocellular carcinoma cells via modulating MET glycosylation and dimerization. Cancer Res. 2013;73(17):5580–5590. [DOI] [PubMed] [Google Scholar]
- 41.Bergstrom K, Liu X, Zhao Y, et al. Defective Intestinal Mucin-Type O-Glycosylation Causes Spontaneous Colitis-Associated Cancer in Mice. Gastroenterology. 2016;151(1):152–164.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meyers N, Gérard C, Lemaigre FP, et al. Differential impact of the ERBB receptors EGFR and ERBB2 on the initiation of precursor lesions of pancreatic ductal adenocarcinoma. Sci Rep. 2020;10(1):5241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Komatsu M, Jepson S, Arango ME, et al. Muc4/sialomucin complex, an intramembrane modulator of ErbB2/HER2/Neu, potentiates primary tumor growth and suppresses apoptosis in a xenotransplanted tumor. Oncogene. 2001;20(4):461–470. [DOI] [PubMed] [Google Scholar]
- 44.Carraway KL, Carvajal ME, Li P, et al. ErbB2 and its ligand Muc4 (sialomucin complex) in rat lacrimal gland. Adv Exp Med Biol. 2002;506(Pt A):289–295. [DOI] [PubMed] [Google Scholar]
- 45.Grasso C, Jansen G, Giovannetti E. Drug resistance in pancreatic cancer: Impact of altered energy metabolism. Crit Rev Oncol Hematol. 2017;114:139–152. doi: 10.1016/j.critrevonc.2017.03.026. [DOI] [PubMed] [Google Scholar]
- 46.Mackey JR, Mani RS, Selner M, et al. Functional nucleoside transporters are required for gemcitabine influx and manifestation of toxicity in cancer cell lines. Cancer Res. 1998;58(19):4349–4357. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.