Abstract
The majority of women with ovarian cancer are diagnosed with metastatic disease, therefore elucidating molecular events that contribute to successful metastatic dissemination may identify additional targets for therapeutic intervention and thereby positively impact survival. Using two human high grade serous ovarian cancer cell lines with inactive TP53 and multiple rounds of serial in vivo passaging, we generated sublines with significantly accelerated intra-peritoneal (IP) growth. Comparative analysis of the parental and IP sublines identified a common panel of differentially expressed genes. The most highly differentially expressed gene, upregulated by 60-65-fold in IP-selected sublines, was the type I transmembrane protein AMIGO2. As the role of AMIGO2 in ovarian cancer metastasis remains unexplored, CRISPR/Cas9 was used to reduce AMIGO2 expression, followed by in vitro and in vivo functional analyses. Knockdown of AMIGO2 modified the sphere-forming potential of ovarian cancer cells, reduced adhesion and invasion in vitro, and significantly attenuated IP metastasis. These data highlight AMIGO2 as a new target for a novel anti-metastatic therapeutic approach aimed at blocking cohesion, survival, and adhesion of metastatic tumorspheres.
Keywords: ovarian cancer; intra-peritoneal metastasis; AMIGO2; spheroid; multi-cellular aggregate; tumorsphere [deleted adhesion, invasion]
1. INTRODUCTION
Analysis of global cancer incidence and mortality data reveals that ovarian cancer is the 7th most common cancer diagnosis in women worldwide and the 8th most common cause of cancer death [1]. A worldwide increase in incidence of 55% is predicted by 2035 (371,000 new cases), accompanied by a predicted 67% increase in deaths (254,000 per year) [2]. While ovarian cancer incidence is much less common relative to other women’s malignancies such as breast cancer, endometrial cancer, and cervical cancer, the overall 5-year survival rate (46%) is substantially lower (87%, 79%, and 67%, respectively) [3]. This is predominantly due to the current inability to conduct population-level screening for early stage disease. For example, in the United States, most women are diagnosed with stage III or IV disease, resulting in a 39% and 17% 5-year survival, respectively [4]. The poor 5-year survival of women with ovarian cancer is attributed to the presence of widespread intra-peritoneal (IP) disease at the time of diagnosis. While most women initially respond to a treatment regimen consisting of surgical debulking and standard-of-care chemotherapy (platinum/taxane), most women ultimately develop recurrent metastatic disease [5, 6]. Elucidating molecular events that contribute to successful metastatic dissemination may identify additional targets for therapeutic intervention and thereby positively impact survival.
Ovarian cancer metastasis is initiated by exfoliation from the primary ovarian or fallopian tube tumor and direct extension into the peritoneal cavity [7, 8]. Hematogenous metastasis with peritoneal homing has also been reported [9, 10]. Matrix-detached cells survive in ascites fluid as single cells and as cell:cell adherent multi-cellular aggregates (MCAs, spheroids, tumorspheres) that subsequently contact the peritoneal mesothelium, induce mesothelial cell retraction, and adhere to the sub-mesothelial collagen-rich matrix wherein they anchor to generate widely disseminated secondary lesions [7, 11, 12]. MCAs exhibit enhanced anoikis-resistance and function as metastatic units, seeding secondary lesions within the peritoneal cavity.
Many factors have been identified that potentiate IP dissemination, including growth factors and bioactive lipids prevalent in ascites fluid [13-16]. Additional studies have used in vivo passaging approaches to generate metastatic variant sublines for comparative analyses. For example, using the human ovarian cancer line SKOV3, a variant designated SKOV3.ip1 was generated by growing tumor cells IP and isolating cells from ascites fluid [16]. Relative to parental SKOV3 cells, the SKOV3.ip1 subline had a higher rate of colony formation in soft agar, formed larger sub-cutaneous tumors, and resulted in decreased survival in mouse models. The enhanced malignancy was correlated with a 2-fold increase in expression of c-erbB-2/neu (HER2) [16]. Recently, however, comparative genomic profiling of human high grade serous ovarian carcinoma (HGSOC) tumors and ovarian cancer cell lines demonstrated that SKOV3 cells lack the TP53 mutations and copy number alterations that are commonly associated with HGSOC and are therefore no longer considered a strongly representative model of this disease [17-19]. A similar approach was taken to generate an aggressive subline of the murine ovarian cancer cell line ID8, syngeneic to C57Bl/6 mice [20]. Passaging of ID8 cells in vivo generated ID8-P1 cells, which exhibited more rapid in vivo growth and enhanced anoikis resistance relative to parental ID8 cells [20]. However, similar to SKOV3 cells, ID8 cells lack mutations in TP53 that are found in 95% of human HGSOCs [18, 21, 22].
In the current study, we employed two human ovarian cancer cell lines with mutational profiles characteristic of human HGSOC. We selected OVCAR5 (TP53 null due to a base insertion) and OVCAR8 (in frame TP53 deletion, KRAS G-T mutation) [19, 23, 24]. Three rounds of in vivo selection were used to generate ‘IP3’ sublines with significantly accelerated in vivo growth. Comparative RNASeq analysis identified a common panel of differentially expressed genes that may contribute to enhanced metastatic potential. CRISPR/Cas9-mediated editing of the most highly differentially expressed gene ‘amphoterin-induced gene and open reading frame 2’ (AMIGO2; adhesion molecule with IgG-like domain 2) substantially altered tumor cell behavior and significantly reduced both overall- and organ-specific tumor burden. Our results identify AMIGO2 as a regulator of ovarian cancer tumorsphere formation and subsequent metastatic dissemination, suggesting a novel target for anti-metastatic therapy.
2. MATERIALS AND METHODS
2.1. Cell Lines.
The human serous ovarian cancer cell lines OVCAR5 and OVCAR8 cells were obtained from American Type Culture Collection (Manassas, VA). Cell lines are routinely verified using short tandem repeat profiling (Genetica). Cells were cultured at 37 °C in Dulbecco's Modified Eagle Medium (DMEM) medium supplemented with 1% non-essential amino acids (Gibco) and 10% fetal bovine serum (FBS; Gibco) with 1% penicillin and streptomycin (Lonza). Red fluorescent protein (RFP) expressing cells were generated using an RFP lentiviral vector (GenTarget, San Diego, CA) with selection using a BD FACSAriaIII cell sorter.
2.2. In vivo passaging, tumor growth and image analysis.
All murine studies were conducted with the approval of the Notre Dame Institutional Animal Care and Use Committee (IACUC). To establish highly metastatic sublines of OVCAR5 and OVCAR8 cells, RFP-tagged cells (3x106 cells in 1 ml PBS) were injected into the peritoneal cavity (ip) of female athymic nude mice (nu/nu, Jackson Laboratory, n=4-6 per cohort). In vivo longitudinal imaging was performed under isoflurane anesthesia using a Bruker Xtreme or an IVIS Lumina II (Caliper Life Sciences) in vivo imaging system. Mice were observed weekly for signs of lethargy and for accumulation of ascites. At this time (7-9 weeks using parental OVCAR5 and OVCAR8 cells), mice were sacrificed using IACUC-approved procedures. In situ imaging of peritoneal tumor burden was performed following a midline incision to expose the peritoneal cavity. Peritoneal tumor nodules were collected, minced into ~5x5 mm pieces, and placed into culture dishes for 1h. After outgrowth, tissue was discarded and cells were sorted to collect RFP-expressing tumor cells, expanded in vitro (designated IP1), pooled, and again injected ip. Following tumor growth for 5-7 weeks (n=5-6 per cohort), tumor nodules were again collected, sorted, expanded (designated IP2), pooled, and re-injected. After 2-3 weeks, mice exhibited significant tumor burden as determined by in vivo imaging. Following in situ imaging and harvesting of peritoneal tumor nodules (designated IP3), abdominal organs were also collected and imaged ex vivo. Tumor burden was quantified using ImageJ as previously described [24].
2.3. Proliferation.
Proliferation was monitored by seeding cells (500-10,000 cells/well) in 96 well culture plates for 24-72h using the XTT Cell Proliferation Assay Kit (Cayman Chemical) according to manufacturer’s specifications. Absorbance was monitored at 450 nm using a Molecular Devices Spectramax microtitre plate reader.
2.4. Analysis of adhesion and invasion.
Cell adhesion to type I collagen-coated wells was assessed as previously described [24]. Invasion was evaluated using a Boyden chamber (8-um pore) coated with Matrigel (10 ug) as described [24]. Experiments were performed in triplicate.
2.5. Phosphokinase array.
A human phospho-kinase antibody array (R&D Systems, Minneapolis, MN, USA) was used according to manufacturer’s specifications. Briefly, duplicate lysates were incubated with array membranes overnight followed by detection of hybridization signal (chemiluminescence) using an ImageQuant LAS4000 biomolecular imager (GE Healthcare) and densitometric analysis using ImageJ.
2.6. RNASeq and qRT-PCR.
Pooled cultures of OVCAR5 and OVCAR8 parental and IP3 variants were analyzed by Drop-seq sequencing analysis as described [25]. Briefly, cells were loaded onto a microfluidic device fabricated using the CAD file from http://mccarrolllab.org/dropseq/ at ~100 cells/μl. Barcoded microbeads (Chemgenes, MACOSKO201110) and single cell suspensions in lysis buffer were encapsulated into droplets to compartmentalize RNA capture, harvested, and subjected to reverse transcription with Maxima H Minus Reverse Transcriptase (Thermo Fisher Scientific, EP0752). cDNA was amplified and PCR products purified and quantified on a BioAnalyzer (Agilent) prior to fragmenting and amplifying for sequencing using a Nextera XT DNA sample preparation kit (Illumina). Libraries were sequenced using an Illumina HiSeq 2500. Base calling was performed using Illumina real time analysis, demultiplexed and converted to Fastq files that were processed using a modified bulk RNA-seq data workflow with the DESeq2 R package to identify differential gene expression. Differentially expressed genes have a p<0.01. Quantitative real-time PCR (qRT-PCR) was used to validate differential expression. Total RNAs were isolated using RNeasy plus mini kit (Qiagen, Valencia, CA), resuspended in RNase-free water and RNA concentrations were determined using NanoDrop One Microvolume UV-Vis Spectrophotometer (Thermo Fisher Scientific). Total RNA (1μg) was reverse-transcribed to cDNA using QuantiTect Reverse Transcription Kit (Qiagen). Primer sequences are shown in Suppl. Table 1. qRT-PCR was performed on StepOnePlus Real-Time PCR System (Thermo Fisher Scientific) using synthesized cDNA (40x dilution), primers (200 nM) and iTaq Universal SYBR Green Supermix (BioRad, Hercules, CA). Conditions for qRT-PCR were: initial 95°C for 1 min, and then 40 cycles of 95°C for 15 sec and 60°C for 1 min with real-time data acquisition, followed by melt curve detection at 0.5°C per 10-sec increment from 60°C to 95°C. The specificity of the qPCR reaction was verified by both melting curve analysis and agarose gel electrophoresis of the amplification products. Relative gene expression was determined with the comparative delta-delta Ct method and normalized using HPRT1 and HSPCB reference genes.
2.7. Plasminogen activation assay.
Net plasminogen activator activity in conditioned media from OVCAR5 and OVCAR8 parental and IP3 cells was quantified using a coupled assay to monitor plasminogen activation and the resulting plasmin hydrolysis of a colorimetric plasmin substrate (Val-Leu-Lys-p-nitroanilide, VLKpNA) as previously described [26]. Assays were performed in triplicate.
2.8. CRISPR/Cas9-mediated AMIGO2 gene editing.
Based on qRT-PCR validation of RNASeq expression data, the AMIGO2 gene was targeted for further analysis. CRISPR/Cas9-mediated AMIGO2 gene editing was performed on the OVCAR5-IP3 cell line with Synthego (Menlo Park, CA) synthetic single guide RNAs (sgRNAs). In general, four sgRNAs targeting exon3 of AMIGO2 were designed using Synthego CRISPR Design Tool (https://design.synthego.com) and synthesized with modifications (Suppl. Table 2) to minimize off-target effects [27, 28]. Each sgRNA was complexed with S. pyogenes Cas9 2NLS nuclease (Synthego) at a molar ratio of 1.3:1 (sgRNA: Cas9) to form ribonucleoproteins (RNPs) for 10 min at room temperature. Delivery in the RNP format restricts Cas9 expression to a brief time window to reduce off target editing [28-31]. The resulting sgRNA/Cas9 RNP complexes were individually delivered into the OVCAR5-IP3 cell line (1.2 x 104 cells/well in 24-well plate) using Lipofectamine CRISPRMAX Cas9 transfection reagent (Thermo Fisher Scientific, Waltham, MA) following the manufacturer's protocol. Post lipofection (48h), cells were split into two wells on duplicate 24-well plates. One plate was used for clonal expansion and the other plate was used to determine the editing efficiency. Cells were directly lysed in the well and genomic DNA was extracted using QuickExtract DNA Extraction Solution (Lucigen, Middleton, WI). The extracted genomic DNA immediately proceeded to PCR using AmpliTaq Gold 360 master mix (Thermo Fisher Scientific) with locus-specific primers (Suppl. Table 3). The amplification conditions were as follows: 95°C for 10 min, 40 cycles of 95°C for 30 sec, 55°C for 30 sec, 72°C for 1 min, and then 72°C for 7 min. The PCR products were run on a 1% agarose gel to verify amplification of a single band of the correct size. Sanger sequencing was then performed through the Genomics & Bioinformatics Core Facility at University of Notre Dame with sequencing primers (Suppl. Table 3). Sanger trace data was analyzed for CRISPR edits using the Synthego ICE (Inference of CRISPR Edits) tool (https://ice.synthego.com). Based on the highest ICE Score (the indel frequency represents the overall CRISPR editing efficiency) and KO Score (frameshift- or a large indel in protein-coding region represents potential knockout efficiency), two of the sgRNA-edited cell pools (sgRNA2 and sgRNA3) were selected for single cell sorting via BD FACSAria III Cell Sorter into three 96-well plates per sgRNA at an average density of 0.5 cells/well. Cells were monitored over time and in total 72 single live cell clones (36 clones from each sgRNA) were further expanded (Suppl. Table 4). Screening for AMIGO2 knockdown/knockout cell clones was individually performed by genomic PCR, Sanger sequencing of target regions and ICE analysis as described above. Sanger sequencing verified the presence of indel mutations.
2.9. Western blotting.
Western blotting was used to confirm AMIGO2 protein knockdown in selected clones as previously described [24]. Membranes were incubated overnight at 4 °C with anti-AMIGO2 antibody (ab179747, 1:500; Abcam). The immunoreactive proteins were visualized with horseradish peroxidase-conjugated goat anti-rabbit IgG (A6667, 1:3,000; MilliporeSigma) followed by enhanced chemiluminescent developing with SuperSignal West Dura extended duration substrate (Thermo Fisher Scientific, Kalamazoo, MI) using an ImageQuant LAS 4000 biomolecular imager (GE Healthcare). Protein blots were then stripped and reblotted with anti-β-actin-peroxidase antibody (A3854, clone AC-15, 1:100,000; MilliporeSigma) to ensure that similar amounts of protein were present in each lane. The assay was repeated in three independent biological replicates.
2.10. Aldehyde dehydrogenase assays.
The ALDEFLUOR assay kit (STEMCELL Technologies) was used according to manufacturer’s specifications. Briefly, cells (2x105) were incubated with Aldefluor reagent containing ALDH substrate (Bodipy-Aminoacetaldehyde) +/− the ALDH1 inhibitor diethylaminobenzaldehyde (DEAB). Biological duplicates were performed. Samples were incubated at 37 °C for 35-55 prior to analysis using the green fluorescence channel (515-535 nm) in a Cytomics FC500 flow cytometer (Beckman Coulter, Brea, CA). Data were analyzed using FlowJo Flow Cytometry Data Analysis Software (Tree Star, Ashland, OR). ALDH activity was also evaluated using a colorimetric assay (Abcam assay kit ab155893). Experiments were performed in triplicate.
2.11. Sphere-forming assay.
MCAs (spheroids, tumorspheres) were generated using a hanging drop method. Specifically, parental OVCAR5 cells, OVCAR5-IP3, or OVCAR5-IP3-AMIGO2-KD cells were counted and re-suspended at 1.5 x 104 and 5 x 104 cells/ml in culture media, and 20 μl droplets (300 or 1,000 cells per droplet, designated MCA300 or MCA1000) were individually seeded on a culture dish lid. The lid was inverted gently and MCAs were cultured as hanging drops at 37°C for 2 days to allow sphere formation. In some experiments, OVCAR5-IP3 cells were incubated in the presence of anti-AMIGO2 antibody (1:50, Santa Cruz sc 373699) during the MCA formation process. The resulting MCAs were imaged with Echo Revolve hybrid microscopy. MCA area (μm2) was measured using ImageJ and statistical analysis was conducted using one-way ANOVA. Sphere formation assay was repeated three times with ten technical replicates per assay.
2.12. In vivo comparison of OVCAR5-IP3-AMIGO2-KD tumor growth.
RFP-tagged cells (3x106 cells in 1 ml PBS) were injected into the peritoneal cavity (ip) of female athymic nude mice (nu/nu, Jackson Laboratory, n=5 per cohort). In vivo longitudinal imaging was performed as described in 2.2 above. After 3 weeks, mice were sacrificed and in situ imaging of peritoneal tumor burden and ex vivo imaging of peritoneal organs was performed as described in 2.2 above. Tumor burden was quantified using ImageJ [24].
2.13. Statistical analysis.
Statistical analyses (Student’s t test, Mann-Whitney U test, ANOVA) were performed using Sigmaplot.
3. RESULTS
3.1. In vivo passaging enhances tumor growth kinetics.
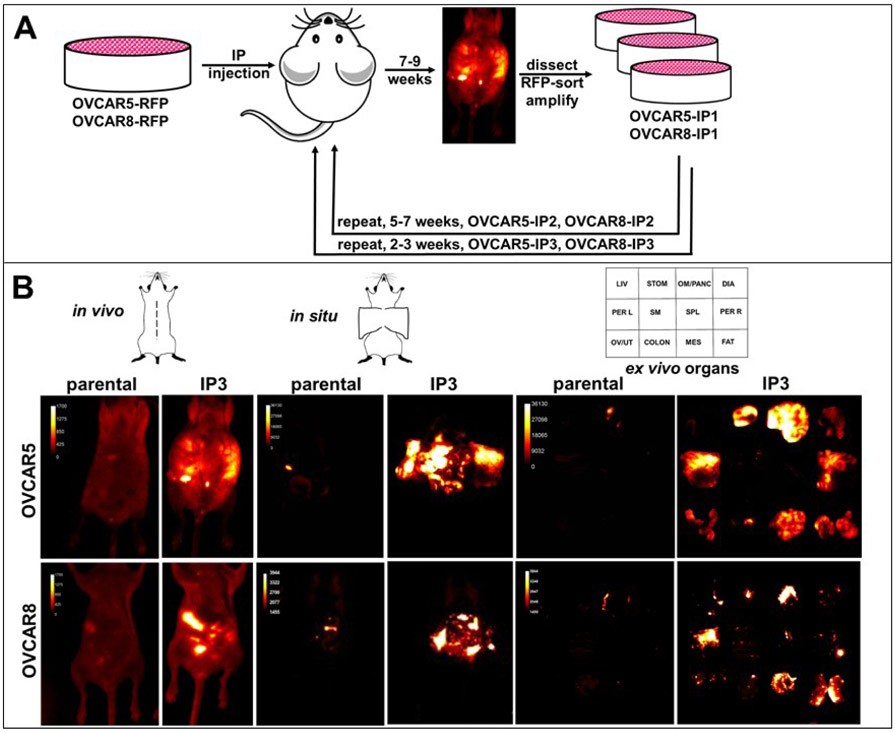
To evaluate the effect of in vivo passaging on tumor growth rate, two human ovarian cancer cell lines (OVCAR5, OVCAR8) tagged with red fluorescent protein (RFP) were subjected to three rounds of in vivo intraperitoneal (IP) passaging (Fig. 1A). Following initial IP injection, both parental lines grew as widely disseminated metastatic nodules, frequently accompanied by accumulation of malignant ascites. Animals were monitored for tumor formation via longitudinal in vivo fluorescent imaging and by observation of abdominal distension and weight changes. Animals injected with parental OVCAR5 and OVCAR8 cells were sacrificed at 7-9 weeks, when they exhibited significant tumor burden, according to institutional IACUC guidelines. After a second round of in vivo passaging (designated IP2, Fig. 1A), mice exhibited significant tumor burden at 5-7 weeks. Following the third round of in vivo passaging (designated IP3 cells), rapid tumor growth necessitated humane sacrifice at 2-3 weeks. Direct comparison of the in vivo growth of parental vs IP3 cells is shown in Fig. 1B. Following midline dissection, peritoneal organs were imaged in situ. Organs were also dissected and imaged ex vivo. Whereas the in vivo passaged IP3 cells exhibit widely disseminated metastatic lesions on the omentum, peritoneum, mesentery and adipose tissue at 2.5 weeks, little tumor burden is apparent at this time point in mice injected with parental cells.
Figure 1. Serial in vivo passaging generates aggressive sublines of OVCAR5 and OVCAR8.
(A) Cells tagged with RFP (3x106 in 1 ml PBS) were injected IP into female nu/nu mice and tumor burden monitored longitudinally by in vivo fluorescent imaging. When animals displayed significant tumor burden, they were sacrificed, tumor nodules dissected and cultured. Two additional rounds of in vivo selection were performed to generate the sublines OVCAR5-IP3 and OVCAR8-IP3 (n=4-6 per cohort). (B) Fluorescent images of tumors were obtained of the whole body (in vivo), midline dissected mice (in situ), or ex vivo organs (ex vivo). The template depicts the position of individual organs.
In both OVCAR5 and OVCAR8 cell lines, IP3 cells exhibited significant increases in adhesion to type I collagen, a key component of the peritoneal extracellular matrix (1.7-2.5-fold change, Fig. 2A, B). Matrix invasive activity was also significantly increased in the IP3 sublines (2-7 fold, Fig. 2C, D). Growth of parental and IP3 cell lines was also evaluated using the XTT Cell Proliferation Assay kit. In contrast to results observed in vivo (Fig. 1) no differences in in vitro proliferation between parental and IP3 cells were detected in either cell line (data not shown). Moreover, a phosphokinase array showed no significant differences in detection of phosphorylated kinases between parental and IP3 sublines (Suppl. Fig. 1).
Figure 2. In vitro comparison of parental and IP3 cell lines.
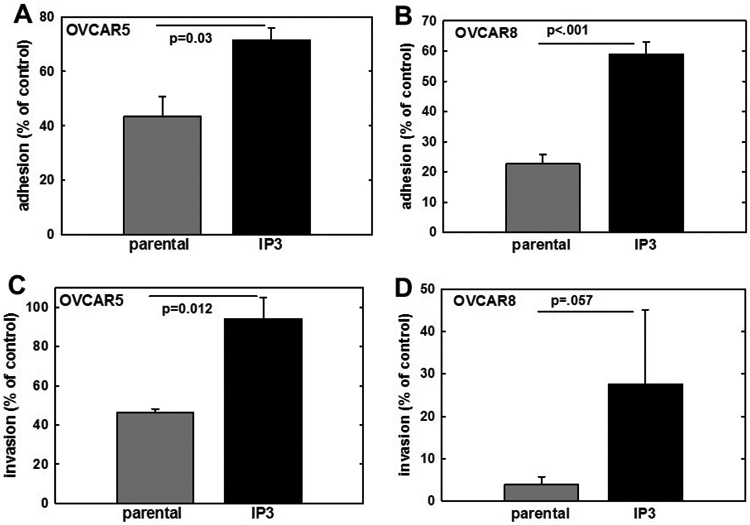
(A, B) Analysis of adhesion. Parental OVCAR5 or OVCAR8, respectively, or their matched IP3 sublines were assayed for adhesion to type I collagen-coated wells. Cells (1x105) were added to wells and allowed to adhere for 45 min, followed by washing to remove non-adherent cells and enumeration of adherent cells (minimum of 20 high-powered fields per condition). Adhesion is shown as % of control adhesion (24h time point). (C,D) Analysis of invasion. Parental OVCAR5 or OVCAR8, respectively, or their matched IP3 sublines, were assayed for invasive activity using a Boyden chamber (8-um pore) coated with Matrigel (10 ug). Cells (2x105) were added to the chamber in serum-free medium for 24-72h. At the termination of the assay, non-invading cells were removed from the top of the filter using a cotton swab. Diff-Quik was used to fix and stain filters and migrating cells adherent to the bottom of the filter were enumerated by counting a minimum of 20 high-powered fields. All experiments were performed in at least triplicate and results show mean +/− SEM.
3.2. Differential gene expression in IP-passaged aggressive ovarian cancer sublines.
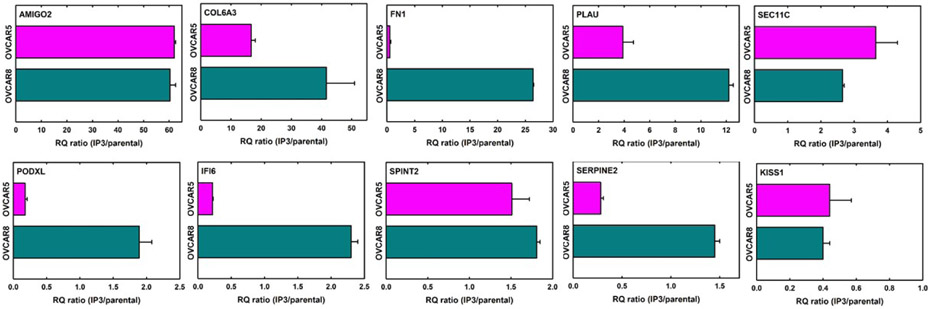
To identify genes, the expression of which may contribute to enhanced in vivo metastatic seeding, matched parental and IP3 sublines were evaluated using RNASeq. The top 20 genes differentially expressed in both cell lines are listed in Table 1. To validate the RNASeq data, 10 of these genes were chosen and expression quantified by qRT-PCR. Results are shown as the ratio of expression in IP3/parental line (Fig. 3). The gene AMIGO2, an Ig-like superfamily member, was upregulated 60-62-fold in the IP3 sublines from both OVCAR5 and OVCAR8, while the metastasis suppressor KISS1 was downregulated 14-25 fold in both IP3 cell lines. Proteases including PLAU and SEC11C were upregulated, while the protease inhibitor SERPINE2 was downregulated in IP3 sublines. Increased PLAU (urinary type plasminogen activator, or uPA) expression was confirmed using a coupled colorimetric assay to detect uPA-mediated conversion of plasminogen to plasmin. A 10-12-fold increase in uPA activity was detected in the conditioned medium of both IP3 sublines (Suppl. Fig. 2). Expression of the extracellular matrix protein COL6A3 was also significantly enhanced in both IP3 sublines, while FN1 expression increased only in OVCAR8-IP3 cells. Differential expression of PODXL, IFI6, and SPINT2 was also validated.
Table 1. Top 20 differentially expressed genes from RNASeq.
Listed are the normalized counts of the top 20 differentially expressed genes with p<0.01, ranked by p value. The 10 genes designated in bold were subsequently validated by qRT-PCR (Fig. 3). Values shown in italics depict decreased expression in the IP3 cell line relative to the parental. ND – not detected, therefore no ratio is calculated.
| Gene | Gene name(s) | OVCAR5 | OVCAR5IP3 | 5IP3/parental | OVCAR8 | OVCAR8IP3 | 8IP3/parental |
|---|---|---|---|---|---|---|---|
| PLAU | urinary-type plasminogen activator, urokinase | 51.03 | 334.11 | 6.55 | 16.57 | 155.89 | 9.41 |
| KISS1 | Kiss-1 metastasis suppressor | 31.90 | 1.16 | 0.04 | 39.46 | 2.57 | 0.07 |
| TAOK1 | serine-threonine protein kinase | 2055.71 | 80.33 | 0.04 | 70.24 | 55.17 | 0.79 |
| SPOCK1 | testican-1 proteoglycan | 194.57 | 12.81 | 0.07 | 26.83 | 3.85 | 0.14 |
| COL6A3 | α3 chain of type VI collagen | 4.78 | 61.70 | 12.91 | 1.58 | 24.38 | 15.43 |
| SEC11C | signal peptidase complex catalytic subunit | 28.71 | 102.44 | 3.57 | 29.20 | 89.17 | 3.05 |
| SERPINE2 | serine protease inhibitor serpin E2, protease nexin 1 | 307.80 | 46.57 | 0.15 | 74.97 | 42.98 | 0.57 |
| IFI44 | interferon induced protein 44 | 6.38 | 3.99 | 0.62 | 168.89 | 1.28 | 0.01 |
| SDPR | serum deprivation response protein, cavin-2 | 271.12 | 20.95 | 0.08 | 199.67 | 94.95 | 0.48 |
| PODXL | podocalyxin-like protein 1 | 146.72 | 23.28 | 0.16 | 48.93 | 23.09 | 0.48 |
| GAL | galanin and GMAP prepropeptide | 35.09 | 9.31 | 0.27 | 64.71 | 12.19 | 0.19 |
| TENM2 | teneurin transmembrane protein 2 | ND | 8.15 | --- | ND | 49.37 | --- |
| FN1 | fibronectin 1 | 17.54 | 23.29 | 1.33 | 21.31 | 214.91 | 10.08 |
| TUBB2A | tubulin beta 2A | 157.89 | 18.62 | 0.12 | 84.44 | 47.78 | 0.57 |
| AMIGO2 | adhesion molecule with Ig-Like domain 2 | ND | 53.55 | --- | ND | 6.41 | --- |
| SPINT2 | serine peptidase inhibitor kunitz type 2, hepatocyte growth factor activator inhibitor type 2 (HAI-2) | 41.47 | 165.31 | 3.99 | 49.72 | 94.30 | 1.90 |
| IFI6 | interferon inducible protein 6 | 38.27 | 13.97 | 0.37 | 65.50 | 13.47 | 0.21 |
| PSMD2 | proteasome 26S subunit non-ATPase-2 | 244.00 | 403.95 | 1.66 | 247.81 | 704.38 | 2.84 |
| IL32 | interleukin 32 | 51.03 | 2.32 | 0.05 | 41.83 | 14.11 | 0.34 |
| TGM2 | transglutaminase-2 | 141.94 | 16.30 | 0.11 | 32.36 | 25.02 | 0.77 |
Figure 3. Differential gene expression between parental and IP3 cell lines.
From the list of top 20 differentially expressed genes shown in Table 1, qPCR validation was performed to verify differential expression of 10 genes. Total RNAs were isolated from cells, quantified, and reverse transcribed using 1 ug RNA. qRT-PCR was performed using synthesized cDNA (40x dilution), primers (200 nM, Supplemental Table 1) and iTaq Universal SYBR Green Supermix (BioRad, Hercules, CA). qRT-PCR conditions were as specified in Materials and Methods. Relative gene expression was determined with the comparative delta-delta Ct method and normalized using HPRT1 and HSPCB reference genes. Panels depict the expression ratio of IP3 to corresponding parental cell line. Experiments were performed in at least triplicate and results show mean +/− SEM.
3.3. CRISPR/Cas9-mediated knockdown of AMIGO2 modulates MCA dynamics and aggressive in vivo growth of OVCAR5-IP3.
Based on RNASeq identification and qRT-PCR validation data showing a high degree of upregulation in both IP3 sublines (60-62-fold, Fig. 3) relative to the respective parental lines, the gene AMIGO2 was chosen for further analysis. As the overall expression of AMIGO2 by OVCAR5-IP3 cells was 3-fold higher than that of OVCAR8-IP3 cells (data not shown), OVCAR5-IP3 cells were selected for CRISPR/Cas9-mediated editing of AMIGO2 gene expression. Four different single guide RNAs (sgRNAs) targeting exon3 of AMIGO2 were selected using the Synthego CRISPR Design Tool based on the lowest off-target potentials (Suppl. Fig.3A, B). The assembled sgRNA/Cas9 RNP complexes were individually introduced into OVCAR5-IP3 cells and genomic PCR was performed to verify amplification of a single product (Suppl. Fig.4A). Genome editing efficiency of the four sgRNA-edited cell pools was measured by Sanger sequencing and Inference of CRISPR Edits (ICE) analysis (Suppl. Fig.4B-D). According to the highest ICE Score and KO Score, AMIGO2 sgRNA2- and sgRNA3- edited cell pools were selected for single cell FACS sorting (Suppl. Table 4). The resulting 72 single cell clones were individually screened through Sanger sequencing and ICE analysis. Clones that displayed the highest ICE and KO Score were further expanded. A ~50% decrease in AMIGO2 expression was observed in two clones (clone 2.6 and 2.23) using qPCR (Fig. 4A), suggesting knockdown of one AMIGO2 allele. This result was confirmed by immunoblotting, showing a similar reduction in AMIGO2 protein levels (Fig. 4B). Together these results indicate that CRISPR/Cas9 system effectively mediated the knockdown of the AMIGO2 gene in OVCAR5-IP3 cells (designated OVCAR5-IP3-AMIGO2-KD).
Figure 4. Validation of CRISPR/Cas9-mediated AMIGO2 knockdown.
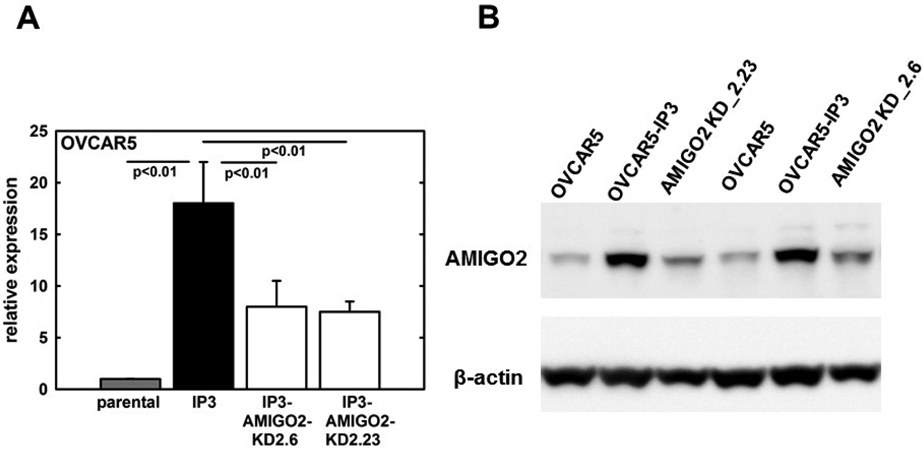
(A) qRT-PCR analysis for relative expression levels of AMIGO2 mRNA in OVCAR5 parental, OVCAR5-IP3 cells, and two OVCAR5-IP3-AMIGO2-KD clones 2.6 and 2.23. Experiments were performed in at least triplicate and results show mean +/− SEM. (B) Western blot analysis for relative expression of AMIGO2 protein. Lysates were electrophoresed in 9% SDS-polyarylamide gels and electroblotted to Immobilon. After blocking, blots were incubated with anti-AMIGO2 (1:500 dilution) overnight, washed, and incubated with an HRP-conjugated secondary antibody (1:3,000) and developed using SuperSignal West Dura substrate. Blots were then stripped and blotted with anti-b-actin-peroxidase antibody (1:100,000) as a loading control. Blots were performed in triplicate and a representative example is shown.
A variety of assays was then performed to evaluate potential mechanisms by which acquisition of AMIGO2 expression may alter pro-metastatic behaviors. To determine whether AMIGO2-expressing OVCAR5-IP3 cells have enhanced cancer stem cell-like or progenitor features [32], ALDH activity was assessed in OVCAR5 parental, -IP3, and –IP3-AMIGO2-KD cells using the flow cytometry-based ALDEFLUOR assay kit. Low levels of ALDH activity were detected using ALDEFLUOR and no differences were detected between parental OVCAR5 and OVCAR5-IP3-AMIGO2-KD cells (Suppl. Fig. 5A). To enhance the sensitivity of detection, a colorimetric assay was used to examine ALDH activity in cell lysates. Although overall activity levels were low, results show a 2-fold increase in ALDH activity when comparing –IP3 cells to OVCAR5 parental cells (Suppl. Fig. 5B). However, relative to –IP3 cells, ALDH activity was not significantly decreased in OVCAR5-IP3-AMIGO2-KD cells (Suppl. Fig. 5B), suggesting that factor(s) other than AMIGO2 contribute to the enhanced ALDH activity observed in OVCAR5-IP3 cells. Knockdown of AMIGO2 also abrogated the enhanced collagen adhesion (Suppl. Fig. 5C) and invasion (Suppl. Fig. 5D) observed in OVCAR5-IP3 cells in vitro.
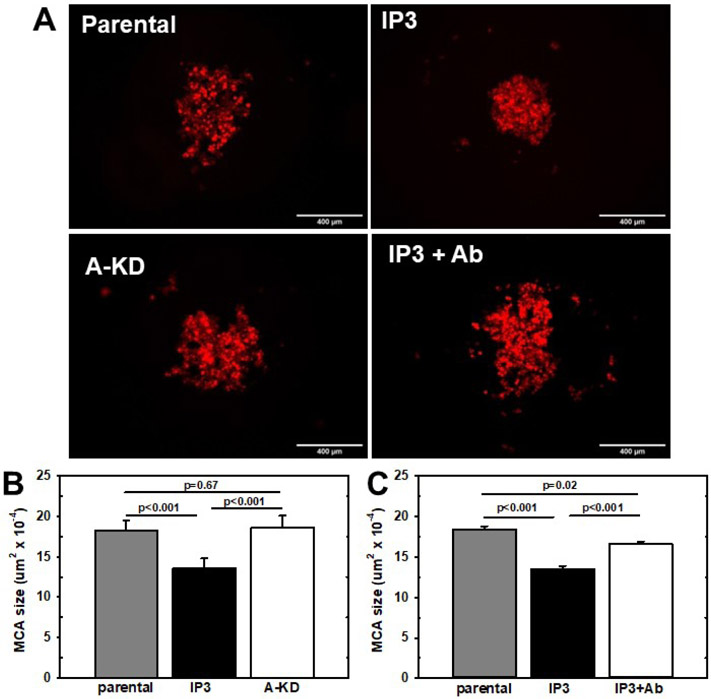
As multi-cellular aggregates (MCAs) or tumorspheres ranging in size from 50-400 um in diameter are commonly found in ascites of women with ovarian cancer, survive in suspension, and are key unit of metastatic dissemination [32-35], MCA dynamics were investigated. Cells were seeded in hanging drops at two different densities to generate small or large aggregates. MCA formation was monitored by phase-contrast or fluorescence microscopy and MCA area analyzed with ImageJ. Relative to parental OVCAR5 cells, a significant compaction of both small and large MCAs was observed with OVCAR5-IP3 cells (Fig. 5A,B; Suppl. Fig. 6A,B). This enhanced spheroid compaction was not observed with OVCAR5-IP3-AMIGO2-KD cells or in the presence of AMIGO2 blocking antibodies (Fig 5A,C; Suppl. Fig. 6A,C).
Figure 5. AMIGO2 regulates multicellular aggregate dynamics.
(A) MCAs were generated via the hanging drop method by seeding cells at a density of 300 cells per 20 μl droplet onto the lid of culture dishes. Dishes were gently inverted and allowed to aggregate for 2 days. MCAs were imaged using Echo Revolve hybrid microscopy in bright field and fluorescence modes. Shown are parental OVCAR5 cells, OVCAR5-IP3, OVCAR5-IP3-AMIGO2-KD or OVCAR5-IP plus anti-AMIGO2 antibody as indicated. (B,C) MCA areas (μm2) were measured using ImageJ and statistical analysis was performed using one-way ANOVA. Experiments were performed in triplicate.
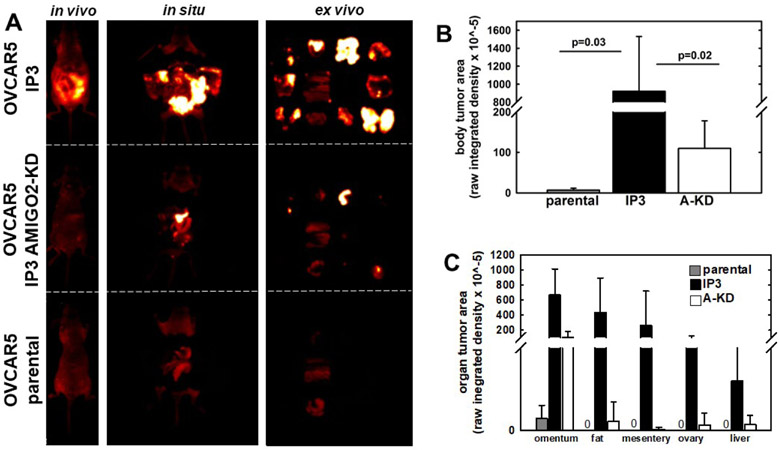
Figure 6. Downregulation of AMIGO2 expression reduces IP metastatic burden.
(A) OVCAR5 parental, OVCAR5-IP3, or OVCAR5-IP3-AMIGO2-KD cells (3x106 in 1 ml PBS) were injected IP into female nu/nu mice and tumor burden monitored longitudinally by in vivo fluorescent imaging (n=5 per cohort). After 3 weeks, fluorescent images were obtained of the whole body (in vivo), midline dissected mice (in situ), or ex vivo organs (ex vivo). Representative images are shown to illustrate regions quantified in B and C below. (B) Quantitation of whole body tumor burden. Fluorescence imaging was performed in situ following a midline incision to expose the peritoneal cavity. (C) Quantitation of organ-specific tumor burden. Following in situ imaging, peritoneal tumor nodules were harvested and imaged ex vivo, positioned as on the template shown in Figure 1. Tumor burden was quantified using ImageJ.
To assess the effects of AMIGO2 knockdown and the resulting enhancement of tumorsphere compaction on in vivo metastatic success, mice were injected IP with OVCAR5 parental, -IP3, or -IP3-AMIGO2-KD cells and animals were monitored for tumor formation using longitudinal in vivo fluorescent imaging (Fig. 6A). After 3 weeks, animals were sacrificed and tumor burden imaged in vivo. Following midline dissection, peritoneal organs were imaged in situ. Organs were also dissected and imaged ex vivo. Quantitation of either whole body or organ-specific tumor burden shows a substantial increase in metastatic burden in OVCAR5-IP3 relative to parental OVCAR5 cells, that is significantly abrogated in OVCAR5-IP3-AMIGO2-KD cells (Fig. 6B,C).
4. DISCUSSION
The vast majority of women with ovarian cancer present at diagnosis with stage III or IV disease, characterized by numerous widespread IP metastatic lesions. Malignant ascites, present at diagnosis in one third of ovarian cancer patients and in the majority of women with recurrent cancer, contains cells shed from the primary tumor. These anchorage-independent cells are present as individual cells and as multi-cellular aggregates (MCAs, or spheroids). Both MCAs and single cells can adhere to peritoneal mesothelial cells, induce mesothelial cell retraction, and anchor in the sub-mesothelial collagen-rich matrix forming metastatic lesions throughout the peritoneal cavity [7, 12, 33-38]. As a molecular-level understanding of factors that regulate metastatic dissemination is necessary for the development of anti-metastatic therapeutics, experiments in this study were designed to identify gene products that functionally contribute to metastasis. Using two distinct human ovarian cancer cell lines and three rounds of in vivo selection, we identified a common panel of differentially expressed genes in the highly metastatic OVCAR5 and OVCAR8 ‘IP3’ sublines. Functional validation focused on the most highly differentially expressed gene, AMIGO2.
AMIGO2 is a member of a family of three structurally homologous type I transmembrane proteins originally cloned from neurons treated with the neurite-promoting protein amphoterin [39]. The extracellular domain contains multiple leucine-rich repeats thought to be active in protein-protein interaction and an immunoglobulin domain. AMIGO family members exhibit both homophilic and heterophilic binding interactions and have been proposed as a novel family of cell adhesion molecules that participate in neurite outgrowth [39]. Although functional studies are limited, emerging data suggest a role for AMIGO2 in cancer as well. AMIGO2 expression was identified in gastric adenocarcinoma, wherein it was referred to as DEGA/AMIGO2 [40]. Antisense downregulation of DEGA/AMIGO2 in gastric adenocarcinoma cells decreased cell adhesion and invasion in vitro and tumor growth in a subcutaneous injection model [40].
Subsequent studies showed upregulation of AMIGO2 in stage IV colorectal cancers relative to stage I [41] and in liver metastatic fibrosarcoma cells [42]. AMIGO2 was also identified as a breast cancer cell invasion driver in a novel model system using keratin-14 reporter cells coupled to a suicide gene used to identify the contribution of genes in the basal cell population to breast cancer progression. Knockdown of AMIGO2 dramatically decreased invasion of 4T1 cells through a collagen/Matrigel matrix [43]. While functional data on AMIGO2 expression in ovarian cancer are lacking, it was recently reported that AMIGO2 is modulated by sustained focal adhesion kinase signaling in a cellular model of adhesion-independent ovarian tumorspheres [44].
In the current study, AMIGO2 was highly overexpressed (60-fold) in two sublines generated from two distinct human parental cells repeatedly selected in vivo for enhanced IP metastatic potential. CRISPR/Cas9-mediated knockdown of AMIGO2 inhibited both cell adhesion and invasion in vitro and substantially reduced overall- and organ-specific metastasis in vivo. Despite the use of multiple distinct sgRNAs, resulting clones showed an incomplete knockout, retaining approximately 50% expression of the AMIGO2 gene. Similar results were observed by western blotting of AMIGO2 protein levels. The most likely explanation for these results is that AMIGO2 may regulate cell survival, such that complete knockout resulted in a lethal phenotype. To this end, it is interesting to note that AMIGO2 expression has been linked to survival in endothelial cells [45] and melanoma [46]. MCAs (tumorspheres or spheroids) are prevalent in ovarian cancer patient ascites and exhibit a significant survival advantage relative to single cells [12, 32, 35]. Our results demonstrate that expression of AMIGO2 enhances spheroid compaction of OVCAR5-IP3 cells, an effect that is significantly reduced in AMIGO2-knockdown clones. Spheroid compaction is associated with a more invasive subpopulation of ovarian cancer cells with enhanced survival characteristics [12, 36]. Consistent with this observation, IP injected OVCAR5-IP3 cells survive in suspension in vivo, seeding substantially more robust metastatic tumor growth relative to OVCAR5-IP3-AMIGO2-KD cells. This observation is supported by a recent study identifying AMIGO2 as a pro-survival gene in melanoma [46]. Taken together, our results support a model wherein the cell-cell adhesion properties of AMIGO2-expressing cells promote formation of compact spheroids, thereby enhancing survival of detached clusters and promoting subsequent IP seeding of multiple metastatic cell clusters.
In addition to AMIGO2, several other genes of interest were differentially regulated in the ‘IP3’ sublines. For example, a 7-9-fold increase in PLAU expression was shown by qRT-PCR, which translated into a 10-12-fold increase in uPA activity. Moreover, a 2-7-fold downregulation of the protease inhibitor SERPINE2, that inhibits uPA activity, was also observed. Numerous studies have demonstrated that increased uPA expression accompanies malignant transformation in ovarian cancer and support the consideration of uPA as a therapeutic target [47-49]. In contrast to SERPINE2, expression of the protease inhibitor SPINT2 (hepatocyte growth factor inhibitor-2 or HAI-2) was upregulated 2-4 fold in IP3 cells. HAI-2 inhibits the serine protease matriptase, expression of which has been linked to defects in epidermal barrier function and to epidermal carcinogenesis [50, 51]. While this result was unexpected, it is interesting to note that a highly related protein HAI-1 (encoded by SPINT1) was recently identified as one of three core proteins in an 11-protein biomarker panel for detection of ovarian cancer from plasma samples [52]. Upregulation of extracellular matrix (ECM) proteins including COL6A3 (13-15 fold) and FN1 (10-fold) was also observed. These results are consistent with emerging data showing ECM-associated molecular signatures that correlate with disease severity and overall survival in ovarian cancer patients [53, 54]. We also observed downregulation of KISS1 (14-25-fold), a metastasis-suppressor that inhibits migration, growth in soft agar, and in vivo metastatic colonization of ovarian cancer [55, 56].
In summary, we have generated highly aggressive sublines of human ovarian cancer cells by serial in vivo passaging and have identified genes related to ovarian cancer progression and metastasis. Functional validation studies support a role for AMIGO2 in regulation of ovarian cancer cell adhesion, tumorsphere compaction, and IP metastatic success. Together these data suggest that AMIGO2 may represent a new target for a novel anti-metastatic therapeutic approach aimed at blocking cohesion, survival and adhesion of metastatic spheroids. To this end, it is interesting to note that the AMIGO2 C-terminal region has been shown to interact with 3-phosphoinositide-dependent kinase 1 (PDK1) in endothelial cells, leading to Akt activation and a pro-survival phenotype [45]. Moreover, a 9 amino acid peptide (representative of amino acids 464-475 in AMIGO2) conjugated to a cell-penetrating peptide inhibited the AMIGO2-PDK1 interaction and subsequent Akt activation in these cells [45]. Based on these observations, future research investigating the AMIGO2-PDK1-Akt axis in ovarian cancer and its potential as a therapeutic target to block tumorsphere survival and metastatic implantation is warranted.
Supplementary Material
Highlights:
Most women with ovarian cancer are diagnosed with diffuse intra-peritoneal (IP) metastases and succumb to metastatic burden
Ovarian cancer cells selected in vivo for IP metastatic aggressiveness display altered gene expression
The adhesion molecule AMIGO2 is upregulated in IP metastatic sublines and modulates ovarian cancer tumorsphere compaction
AMIGO2 represents a novel anti-metastatic target in ovarian cancer
5. ACKNOWLEDGEMENTS:
This work was supported in part by Research Grants RO1 CA109545 (M.S.S.), RO1 CA194697 (S.Z.) and KO1 CA218305 (T.S.H.) from the National Institutes of Health, National Cancer Institute; by the Leo and Anne Albert Charitable Trust (M.S.S.); and the Walther Cancer Foundation Interdisciplinary Interface Training Program (E.I.H.). (E.I.H.) is a previous fellow of the Chemistry-Biochemistry-Biology Interface (CBBI) Program at the University of Notre Dame, supported by training grant T32GM075762 from the National Institute of General Medical Sciences. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of General Medical Sciences or the National Institutes of Health. (E.I.H.) is currently supported by F99 AG068527 from the National Institutes of Health, National Institute on Aging.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
CONFLICTS OF INTEREST: The authors have no conflicts of interest to declare.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
REFERENCES
- [1].Reid F, Bhatla N, Oza AM, Blank SV, Cohen R, Adams T, Benites A, Gardiner D, Gregory S, Suzuki M, Jones A, The World Ovarian Cancer Coalition Every Woman Study: identifying challenges and opportunities to improve survival and quality of life, Int J Gynecol Cancer, (2020). [DOI] [PubMed] [Google Scholar]
- [2].Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F, Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012, Int J Cancer, 136 (2015) E359–386. [DOI] [PubMed] [Google Scholar]
- [3].Quaresma M, Coleman MP, Rachet B, 40-year trends in an index of survival for all cancers combined and survival adjusted for age and sex for each cancer in England and Wales, 1971-2011: a population-based study, Lancet, 385 (2015) 1206–1218. [DOI] [PubMed] [Google Scholar]
- [4].Urban RR, He H, Alfonso R, Hardesty MM, Gray HJ, Goff BA, Ovarian cancer outcomes: Predictors of early death, Gynecol Oncol, 140 (2016) 474–480. [DOI] [PubMed] [Google Scholar]
- [5].Howlader N, Noone AM, Krapcho M, Miller D, Brest A, Yu M, Ruhl J, Tatalovich Z, Mariotto A, Lewis DR, Chen HS, Feuer EJ, Cronin KA, (eds). SEER Cancer Statistics Review, 1975-2017, National Cancer Institute. Bethesda, MD, https://seer.cancer.gov/csr/1975_2017/, based on November 2019. SEER data submission, posted to the SEER web site, April 2020. [Google Scholar]
- [6].Marcus CS, Maxwell GL, Darcy KM, Hamilton CA, McGuire WP, Current approaches and challenges in managing and monitoring treatment response in ovarian cancer, J Cancer, 5 (2014) 25–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Lengyel E, Ovarian cancer development and metastasis, Am J Pathol, 177 (2010) 1053–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Lengyel E, Burdette JE, Kenny HA, Matei D, Pilrose J, Haluska P, Nephew KP, Hales DB, Stack MS, Epithelial ovarian cancer experimental models, Oncogene, 33 (2014) 3619–3633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Coffman LG, Burgos-Ojeda D, Wu R, Cho K, Bai S, Buckanovich RJ, New models of hematogenous ovarian cancer metastasis demonstrate preferential spread to the ovary and a requirement for the ovary for abdominal dissemination, Transl Res, 175 (2016) 92–102 e102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Pradeep S, Kim SW, Wu SY, Nishimura M, Chaluvally-Raghavan P, Miyake T, Pecot CV, Kim SJ, Choi HJ, Bischoff FZ, Mayer JA, Huang L, Nick AM, Hall CS, Rodriguez-Aguayo C, Zand B, Dalton HJ, Arumugam T, Lee HJ, Han HD, Cho MS, Rupaimoole R, Mangala LS, Sehgal V, Oh SC, Liu J, Lee JS, Coleman RL, Ram P, Lopez-Berestein G, Fidler IJ, Sood AK, Hematogenous metastasis of ovarian cancer: rethinking mode of spread, Cancer Cell, 26 (2014) 77–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Klymenko Y, Kim O, Stack MS, Complex Determinants of Epithelial: Mesenchymal Phenotypic Plasticity in Ovarian Cancer, Cancers (Basel), 9 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Al Habyan S, Kalos C, Szymborski J, McCaffrey L, Multicellular detachment generates metastatic spheroids during intra-abdominal dissemination in epithelial ovarian cancer, Oncogene, 37 (2018) 5127–5135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Mills GB, Moolenaar WH, The emerging role of lysophosphatidic acid in cancer, Nat Rev Cancer, 3 (2003) 582–591. [DOI] [PubMed] [Google Scholar]
- [14].Ahmed N, Stenvers KL, Getting to know ovarian cancer ascites: opportunities for targeted therapy-based translational research, Front Oncol, 3 (2013) 256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Kipps E, Tan DS, Kaye SB, Meeting the challenge of ascites in ovarian cancer: new avenues for therapy and research, Nat Rev Cancer, 13 (2013) 273–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Yu D, Wolf JK, Scanlon M, Price JE, Hung MC, Enhanced c-erbB-2/neu expression in human ovarian cancer cells correlates with more severe malignancy that can be suppressed by E1A, Cancer Res, 53 (1993) 891–898. [PubMed] [Google Scholar]
- [17].Domcke S, Sinha R, Levine DA, Sander C, Schultz N, Evaluating cell lines as tumour models by comparison of genomic profiles, Nat Commun, 4 (2013) 2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Cancer N Genome Atlas Research, Integrated genomic analyses of ovarian carcinoma, Nature, 474 (2011) 609–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Mitra AK, Davis DA, Tomar S, Roy L, Gurler H, Xie J, Lantvit DD, Cardenas H, Fang F, Liu Y, Loughran E, Yang J, Sharon Stack M, Emerson RE, Cowden Dahl KD, M VB, Nephew KP, Matei D, Burdette JE, In vivo tumor growth of high-grade serous ovarian cancer cell lines, Gynecol Oncol, 138 (2015) 372–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Cai Q, Fan Q, Buechlein A, Miller D, Nephew KP, Liu S, Wan J, Xu Y, Changes in mRNA/protein expression and signaling pathways in in vivo passaged mouse ovarian cancer cells, PLoS One, 13 (2018) e0197404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Walton J, Blagih J, Ennis D, Leung E, Dowson S, Farquharson M, Tookman LA, Orange C, Athineos D, Mason S, Stevenson D, Blyth K, Strathdee D, Balkwill FR, Vousden K, Lockley M, McNeish IA, CRISPR/Cas9-Mediated Trp53 and Brca2 Knockout to Generate Improved Murine Models of Ovarian High-Grade Serous Carcinoma, Cancer Res, 76 (2016) 6118–6129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, Wilson CJ, Lehar J, Kryukov GV, Sonkin D, Reddy A, Liu M, Murray L, Berger MF, Monahan JE, Morais P, Meltzer J, Korejwa A, Jane-Valbuena J, Mapa FA, Thibault J, Bric-Furlong E, Raman P, Shipway A, Engels IH, Cheng J, Yu GK, Yu J, Aspesi P Jr., de Silva M, Jagtap K, Jones MD, Wang L, Hatton C, Palescandolo E, Gupta S, Mahan S, Sougnez C, Onofrio RC, Liefeld T, MacConaill L, Winckler W, Reich M, Li N, Mesirov JP, Gabriel SB, Getz G, Ardlie K, Chan V, Myer VE, Weber BL, Porter J, Warmuth M, Finan P, Harris JL, Meyerson M, Golub TR, Morrissey MP, Sellers WR, Schlegel R, Garraway LA, The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity, Nature, 483 (2012) 603–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Bykov VJ, Issaeva N, Selivanova G, Wiman KG, Mutant p53-dependent growth suppression distinguishes PRIMA-1 from known anticancer drugs: a statistical analysis of information in the National Cancer Institute database, Carcinogenesis, 23 (2002) 2011–2018. [DOI] [PubMed] [Google Scholar]
- [24].Asem M, Young AM, Oyama C, Claure De La Zerda A, Liu Y, Yang J, Hilliard TS, Johnson J, Harper EI, Guldner I, Zhang S, Page-Mayberry T, Kaliney WJ, Stack MS, Host Wnt5a Potentiates Microenvironmental Regulation of Ovarian Cancer Metastasis, Cancer Res, 80 (2020) 1156–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Wang Q, Guldner IH, Golomb SM, Sun L, Harris JA, Lu X, Zhang S, Single-cell profiling guided combinatorial immunotherapy for fast-evolving CDK4/6 inhibitor-resistant HER2-positive breast cancer, Nat Commun, 10 (2019) 3817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Stack MS, Gray RD, Pizzo SV, Modulation of murine B16F10 melanoma plasminogen activator production by a synthetic peptide derived from the laminin A chain, Cancer Res, 53 (1993) 1998–2004. [PubMed] [Google Scholar]
- [27].Hendel A, Bak RO, Clark JT, Kennedy AB, Ryan DE, Roy S, Steinfeld I, Lunstad BD, Kaiser RJ, Wilkens AB, Bacchetta R, Tsalenko A, Dellinger D, Bruhn L, Porteus MH, Chemically modified guide RNAs enhance CRISPR-Cas genome editing in human primary cells, Nat Biotechnol, 33 (2015) 985–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Cameron P, Fuller CK, Donohoue PD, Jones BN, Thompson MS, Carter MM, Gradia S, Vidal B, Garner E, Slorach EM, Lau E, Banh LM, Lied AM, Edwards LS, Settle AH, Capurso D, Llaca V, Deschamps S, Cigan M, Young JK, May AP, Mapping the genomic landscape of CRISPR-Cas9 cleavage, Nat Methods, 14 (2017) 600–606. [DOI] [PubMed] [Google Scholar]
- [29].Liang X, Potter J, Kumar S, Zou Y, Quintanilla R, Sridharan M, Carte J, Chen W, Roark N, Ranganathan S, Ravinder N, Chesnut JD, Rapid and highly efficient mammalian cell engineering via Cas9 protein transfection, J Biotechnol, 208 (2015) 44–53. [DOI] [PubMed] [Google Scholar]
- [30].Kim S, Kim D, Cho SW, Kim J, Kim JS, Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins, Genome Res, 24 (2014) 1012–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Vakulskas CA, Behlke MA, Evaluation and Reduction of CRISPR Off-Target Cleavage Events, Nucleic Acid Ther, 29 (2019) 167–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Liao J, Qian F, Tchabo N, Mhawech-Fauceglia P, Beck A, Qian Z, Wang X, Huss WJ, Lele SB, Morrison CD, Odunsi K, Ovarian cancer spheroid cells with stem cell-like properties contribute to tumor generation, metastasis and chemotherapy resistance through hypoxia-resistant metabolism, PLoS One, 9 (2014) e84941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Burleson KM, Casey RC, Skubitz KM, Pambuccian SE, Oegema TR Jr., Skubitz AP, Ovarian carcinoma ascites spheroids adhere to extracellular matrix components and mesothelial cell monolayers, Gynecol Oncol, 93 (2004) 170–181. [DOI] [PubMed] [Google Scholar]
- [34].L'Esperance S, Bachvarova M, Tetu B, Mes-Masson AM, Bachvarov D, Global gene expression analysis of early response to chemotherapy treatment in ovarian cancer spheroids, BMC Genomics, 9 (2008) 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Shield K, Ackland ML, Ahmed N, Rice GE, Multicellular spheroids in ovarian cancer metastases: Biology and pathology, Gynecol Oncol, 113 (2009) 143–148. [DOI] [PubMed] [Google Scholar]
- [36].Sodek KL, Ringuette MJ, Brown TJ, Compact spheroid formation by ovarian cancer cells is associated with contractile behavior and an invasive phenotype, Int J Cancer, 124 (2009) 2060–2070. [DOI] [PubMed] [Google Scholar]
- [37].Klymenko Y, Kim O, Loughran E, Yang J, Lombard R, Alber M, Stack MS, Cadherin composition and multicellular aggregate invasion in organotypic models of epithelial ovarian cancer intraperitoneal metastasis, Oncogene, 36 (2017) 5840–5851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Klymenko Y, Johnson J, Bos B, Lombard R, Campbell L, Loughran E, Stack MS, Heterogeneous Cadherin Expression and Multicellular Aggregate Dynamics in Ovarian Cancer Dissemination, Neoplasia, 19 (2017) 549–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Kuja-Panula J, Kiiltomaki M, Yamashiro T, Rouhiainen A, Rauvala H, AMIGO, a transmembrane protein implicated in axon tract development, defines a novel protein family with leucine-rich repeats, J Cell Biol, 160 (2003) 963–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Rabenau KE, O'Toole JM, Bassi R, Kotanides H, Witte L, Ludwig DL, Pereira DS, DEGA/AMIGO-2, a leucine-rich repeat family member, differentially expressed in human gastric adenocarcinoma: effects on ploidy, chromosomal stability, cell adhesion/migration and tumorigenicity, Oncogene, 23 (2004) 5056–5067. [DOI] [PubMed] [Google Scholar]
- [41].Huo T, Canepa R, Sura A, Modave F, Gong Y, Colorectal cancer stages transcriptome analysis, PLoS One, 12 (2017) e0188697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Kanda Y, Osaki M, Onuma K, Sonoda A, Kobayashi M, Hamada J, Nicolson GL, Ochiya T, Okada F, Amigo2-upregulation in Tumour Cells Facilitates Their Attachment to Liver Endothelial Cells Resulting in Liver Metastases, Sci Rep, 7 (2017) 43567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Sonzogni O, Haynes J, Seifried LA, Kamel YM, Huang K, BeGora MD, Yeung FA, Robert-Tissot C, Heng YJ, Yuan X, Wulf GM, Kron KJ, Wagenblast E, Lupien M, Kislinger T, Hannon GJ, Muthuswamy SK, Reporters to mark and eliminate basal or luminal epithelial cells in culture and in vivo, PLoS Biol, 16 (2018) e2004049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Diaz Osterman CJ, Ozmadenci D, Kleinschmidt EG, Taylor KN, Barrie AM, Jiang S, Bean LM, Sulzmaier FJ, Jean C, Tancioni I, Anderson K, Uryu S, Cordasco EA, Li J, Chen XL, Fu G, Ojalill M, Rappu P, Heino J, Mark AM, Xu G, Fisch KM, Kolev VN, Weaver DT, Pachter JA, Gyorffy B, McHale MT, Connolly DC, Molinolo A, Stupack DG, Schlaepfer DD, FAK activity sustains intrinsic and acquired ovarian cancer resistance to platinum chemotherapy, Elife, 8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Park H, Lee S, Shrestha P, Kim J, Park JA, Ko Y, Ban YH, Park DY, Ha SJ, Koh GY, Hong VS, Mochizuki N, Kim YM, Lee W, Kwon YG, AMIGO2, a novel membrane anchor of PDK1, controls cell survival and angiogenesis via Akt activation, J Cell Biol, 211 (2015) 619–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Fontanals-Cirera B, Hasson D, Vardabasso C, Di Micco R, Agrawal P, Chowdhury A, Gantz M, de Pablos-Aragoneses A, Morgenstern A, Wu P, Filipescu D, Valle-Garcia D, Darvishian F, Roe JS, Davies MA, Vakoc CR, Hernando E, Bernstein E, Harnessing BET Inhibitor Sensitivity Reveals AMIGO2 as a Melanoma Survival Gene, Mol Cell, 68 (2017) 731–744 e739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Young TN, Rodriguez GC, Moser TL, Bast RC Jr., Pizzo SV, Stack MS, Coordinate expression of urinary-type plasminogen activator and its receptor accompanies malignant transformation of the ovarian surface epithelium, Am J Obstet Gynecol, 170 (1994) 1285–1296. [DOI] [PubMed] [Google Scholar]
- [48].Schmalfeldt B, Prechtel D, Harting K, Spathe K, Rutke S, Konik E, Fridman R, Berger U, Schmitt M, Kuhn W, Lengyel E, Increased expression of matrix metalloproteinases (MMP)-2, MMP-9, and the urokinase-type plasminogen activator is associated with progression from benign to advanced ovarian cancer, Clin Cancer Res, 7 (2001) 2396–2404. [PubMed] [Google Scholar]
- [49].Zhang Y, Kenny HA, Swindell EP, Mitra AK, Hankins PL, Ahn RW, Gwin K, Mazar AP, O'Halloran TV, Lengyel E, Urokinase plasminogen activator system-targeted delivery of nanobins as a novel ovarian cancer therapy, Mol Cancer Ther, 12 (2013) 2628–2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Sales KU, Friis S, Abusleme L, Moutsopoulos NM, Bugge TH, Matriptase promotes inflammatory cell accumulation and progression of established epidermal tumors, Oncogene, 34 (2015) 4664–4672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Szabo R, Hobson JP, List K, Molinolo A, Lin CY, Bugge TH, Potent inhibition and global co-localization implicate the transmembrane Kunitz-type serine protease inhibitor hepatocyte growth factor activator inhibitor-2 in the regulation of epithelial matriptase activity, J Biol Chem, 283 (2008) 29495–29504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Enroth S, Berggrund M, Lycke M, Broberg J, Lundberg M, Assarsson E, Olovsson M, Stalberg K, Sundfeldt K, Gyllensten U, High throughput proteomics identifies a high-accuracy 11 plasma protein biomarker signature for ovarian cancer, Commun Biol, 2 (2019) 221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Pearce OMT, Delaine-Smith RM, Maniati E, Nichols S, Wang J, Bohm S, Rajeeve V, Ullah D, Chakravarty P, Jones RR, Montfort A, Dowe T, Gribben J, Jones JL, Kocher HM, Serody JS, Vincent BG, Connelly J, Brenton JD, Chelala C, Cutillas PR, Lockley M, Bessant C, Knight MM, Balkwill FR, Deconstruction of a Metastatic Tumor Microenvironment Reveals a Common Matrix Response in Human Cancers, Cancer Discov, 8 (2018) 304–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Cheon DJ, Tong Y, Sim MS, Dering J, Berel D, Cui X, Lester J, Beach JA, Tighiouart M, Walts AE, Karlan BY, Orsulic S, A collagen-remodeling gene signature regulated by TGF-beta signaling is associated with metastasis and poor survival in serous ovarian cancer, Clin Cancer Res, 20 (2014) 711–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Ji K, Ye L, Mason MD, Jiang WG, The Kiss-1/Kiss-1R complex as a negative regulator of cell motility and cancer metastasis (Review), Int J Mol Med, 32 (2013) 747–754. [DOI] [PubMed] [Google Scholar]
- [56].Jiang Y, Berk M, Singh LS, Tan H, Yin L, Powell CT, Xu Y, KiSS1 suppresses metastasis in human ovarian cancer via inhibition of protein kinase C alpha, Clin Exp Metastasis, 22 (2005) 369–376. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.