Abstract
Pear fruit could be used as good medicine to relieve coughs, promote salivation, nourish lungs, and reduce the risk of many diseases for its phytochemical action. Lignin is a major secondary metabolite in Chinese pear fruit. Class III peroxidase (Class III PRX) is an important enzyme in the biosynthesis of lignin in plants. However, we poorly understand the role of PRXs in lignin biosynthesis in Chinese pear fruit. In our study, we cloned five PRXs from Chinese pear (Pyrus bretschneideri), namely PbPRX2, PbPRX22, PbPRX34, PbPRX64, and PbPRX75, which contained 978 bp encoded 326 amino acids (AA), 2607 bp encoded 869 AA, 972 bp encoded 324 AA, 687 bp encoded 229 AA, and 1020 bp encoded 340 AA, respectively. Enzyme activity analysis showed that four recombinant PbPRX proteins had catalytic activities for pyrogallol, guaiacol, ferulic acid, coniferyl alcohol, and sinapyl alcohol. Subcellular localization experiments showed that these genes were located in the cell wall or cell membrane. Enzyme activity and kinetics of PbPRX2 revealed its role in polymerization of lignin in Chinese pear fruit. The present study suggested that PbPRXs played critical roles in lignin biosynthesis in Chinese pear fruit.
Supplementary Information
The online version contains supplementary material available at 10.1007/s12298-021-00949-9.
Keywords: Chinese pear, PRX, Enzymatic, Subcellular localization
Introduction
Lignin, a complex aromatic heteropolymer, participates in water transport, mechanical support, control of fruit taste, and response to abiotic and biotic stresses (Cesarino 2019; Pomar et al. 2002). Lignin is derived mainly from sinapyl alcohol, coniferyl alcohol, and p-coumaryl alcohol (Ralph et al. 2019; Vanholme et al. 2010). The synthesis of lignin involves two steps, including monolignols synthesis and lignin polymerization (Ralph et al. 2019; Vanholme et al. 2010). Among them, Class III peroxidases (Class III PRXs) are unique to plants and play important roles in the polymerization of lignin (Fagerstedt et al. 2010; Pomar et al. 2002). The functions of PRX in the lignin biosynthesis pathway have been studied in many model plants, such as poplar (Populus trichocarpa), tobacco (Nicotiana tabacum), rice (Oryza sativa), and Arabidopsis (Arabidopsis thaliana) (Barceló and Aznar-Asensio 1999; Blee et al. 2003; Ehlting et al. 2005). These studies mainly focused on the localization of PRX protein in xylem, enzyme activity analysis, enzyme kinetic analysis, etc. (Barceló and Aznar-Asensio 1999; Blee et al. 2003; Ehlting et al. 2005). For example, Barceló and Aznar-Asensio found that a PRX protein was localized in the cell wall of Zinnia and have coniferyl alcohol oxidase activity (Barceló and Aznar-Asensio 1999). During the in vitro rooting of cherries, Dalet and Cornu found that the activity of PRX protein was positively correlated with the lignification of explants (Dalet and Cornu 1989). Sasaki et al. (2006) found that a PRX protein (CWPO-C) in poplar is located in the cell wall and can be combined with the substrate sinapyl alcohol to play an important role in the lignification process (Sasaki et al. 2006). Ren et al. (2014) identified the PRX family of poplar and analyzed the enzyme kinetics of 10 PRX proteins (Ren et al. 2014). Among them, 6 of the 10 PRX proteins had catalytic activity, and most of them had the highest catalytic activity for coniferol (Ren et al. 2014). Sinapyl alcohol, ferulic acid, and coniferyl alcohol are natural compounds that exist in plant cells (Cai et al. 2010; Ralph et al. 2019; Ren et al. 2014). Ferulic acid can increase the strength and rigidity of the cell wall by cross-linking hemicellulose, pentosan and arabinoxylan, and ultimately make the plant itself less susceptible to enzymatic hydrolysis when infected by foreign pathogens. Sinapyl alcohol and coniferyl alcohol are lignin monomers that involved in the formation of lignin polymers, so these two substances play a decisive role in the final biosynthesis of lignin.
The number of the PRX family have been identified in Chinese pear (Cao et al. 2016b), while the function of PRX was still unkonown, especially their role in the polymerization of lignin. In plants, PRXs can use H2O2 as an oxidant to produce lignin monomer phenoxy radicals, and finally form lignin polymers (Lewis and Yamamoto 1990). In A. thaliana, eight AtPRXs (At5g05340, At3g28200, At2g43480, At1g30870, At4g37520, At4g37530, At5g42180, and At2g37130) are involved in the synthesis and polymerization of lignin (Ehlting et al. 2005). In our previous research, the expression patterns of PbPRX2, PbPRX22, PbPRX34, PbPRX64, and PbPRX75 significantly correlated with the change in lignin content, indicating that these five genes play important roles in lignin polymerization during fruit development of Chinese pear (Cao et al. 2016b). In this study, we isolated and cloned 5 PRX genes, including PbPRX2, PbPRX22, PbPRX34, PbPRX64, and PbPRX75. The substrates of pyrogallol, guaiacol, ferulic acid, coniferyl alcohol, and sinapyl alcohol were used to detect the activity of PbPRX proteins. This study will help us understand the role of PbPRXs in the lignin polymerization of Chinese pear, and lay foundation for selecting suitable candidate genes to improve the quality of Chinese pear fruit at the molecular level in the future.
Materials and method
Cloning of PbPRX genes
The RNAprep pure Plant Kit (Tiangen, Beijing, China) was used to extract total RNA from Chinese pear fruits. The First Strand cDNA synthesis Kit (Takara, Dalian, China) was used to synthesize the first-strand complementary DNA (cDNA). The Primer (version 5.0) was used to design the specific primers of target genes. Using specific primers (Table S1), we isolated and cloned 5 PbPRX genes from the Chinese pear fruit cDNA library.
Multiple sequence alignment
The ExPASy ProtParam (Gasteiger et al. 2005) was used to predict the theoretical molecular weight and isoelectric point of PbPRX2, PbPRX22, PbPRX34, PbPRX64, and PbPRX75 genes. In recent years, researchers have analyzed the crystal structure of PRX proteins (Østergaard et al. 2000; Schuller et al. 1996) for comprehensive and systematic study of their functions and structures. The MAFFT software (Katoh et al. 2005) was used for multiple sequence alignments of these five proteins and other related proteins, and the ESpript online tool (Gouet et al. 2005) was used for visualization.
Purification of PbPRX proteins
The open reading frames (ORFs) of PbPRX2, PbPRX22, PbPRX34, PbPRX64, and PbPRX75 were subcloned into pMAL-c2X vector, respectively, and transformed into E. coli BL21 (GENERAL BIOL, Hefei, China) competent cells. Strains with completely correct sequencing were cultured at 37 °C until the OD600 reached 0.6, IPTG was added to it to make the final concentration reach 1.0 mmol/L, and then the protein expression was induced overnight at 28 °C. The bacterial pellet were collected by centrifugation at 4 °C in a cryocentrifuge tobe (5000 rpm/min, 10 min), the supernatant was removed, and a suitable buffer solution was added to suspend the bacterial pellet. After the suspended cells were broken by ultrasonication, the supernatant was centrifuged and diluted with buffer solution, and finally the expressed protein was purified by affinity chromatography, as described by Han et al. (2017).
Analyses of enzymatic activity and enzyme kinetics of PbPRXs
The catalytic activity of PbPRX proteins for pyrogallol and guaiacol was determined by the method of Kvaratskhelia et al. (1997). The catalytic activity of PbPRX proteins for coniferyl alcohol and sinapyl alcohol was determined by the method of (Barceló and Aznar-Asensio 1999). The catalytic activity of PbPRX proteins for ferulic acid was determined by the method of Sanchez et al. (1996). All the above reactions were carried out at room temperature, and the protein concentration was measured using A280. The kinetic analyses of PbPRXs proteins was performed according to the method of Ren et al. (2014).
Analyses of subcellular localization of PbPRXs
The ORFs of PbPRX2, PbPRX22, PbPRX34, PbPRX64, and PbPRX75 were subcloned into pCambia1304 vector by using GenRec Assembly Master Mix Kit (GENERAL BIOL, Hefei, China), respectively. The constructed pPbPRX-GFP vectors were electroporated into Agrobacterium tumefaciens EHA105 (GENERAL BIOL, Hefei, China) by using a Gene Pulser Xcell (BIO-RAD, USA). As described by Cao et al. (2016a, b), the suspensions were infiltrated into the tobacco leaves. The laser scanning microscopy (CarlZeiss LSM710, Germany) was used to observe the expressed PbPRX-GFPs.
Results
Characteristics of PbPRX genes
Five PbPRX (PbPRX2, PbPRX22, PbPRX34, PbPRX64, and PbPRX75) genes were cloned from the Chinese pear fruit. Their full-length of cDNAs are 978 bp, 2607 bp, 972 bp, 687 bp, and 1020 bp, respectively, encoding 326 amino acids (AA), 869 AA, 324 AA, 229 AA, and 340 AA (Table 1). The ExPASy ProtParam was used to predict their molecular weights and isoelectric points. The molecular weights of PbPRX2, PbPRX22, PbPRX34, PbPRX64, and PbPRX75 were 35.27 kilodaltons (kDa), 98.12 kDa, 34.41 kDa, 25.11 kDa, and 37.26 kDa, respectively, and the isoelectric points were 8.82, 6.33, 5.88, 8.93, and 5.36, respectively (Table 1).
Table 1.
The basic information of fivePbPRXs genes
| Gene name | Accession number | Molecular weight (kD) of proteins | cDNA length (bp) | ORF length (bp) | Size (Amino acids) | pI |
|---|---|---|---|---|---|---|
| PbPRX2 | Pbr035186.1 | 35.27 | 978 | 978 | 326 | 8.82 |
| PbPRX22 | Pbr013845.1 | 98.12 | 2607 | 2607 | 869 | 6.33 |
| PbPRX34 | Pbr020590.1 | 34.41 | 972 | 972 | 324 | 5.88 |
| PbPRX64 | Pbr039193.1 | 25.11 | 687 | 687 | 229 | 8.93 |
| PbPRX75 | Pbr007872.1 | 37.26 | 1020 | 1020 | 340 | 5.36 |
Determination of enzyme activity of PbPRX proteins
After induction and fragmentation, the fusion proteins of pMAL-PbPRX2, pMAL-PbPRX22, pMAL-PbPRX34, pMAL-PbPRX64, and pMAL-PbPRX75 were purified. Among them, pMAL-PbPRX22 was mainly obtained in the precipitate, so we only studied the four proteins PbPRX2, PbPRX34, PbRX64, and PbPRX75 in further experiments. These four purified proteins had certain catalytic activities for pyrogallol, guaiacol, ferulic acid, coniferyl alcohol, and sinapyl alcohol. Among them, PbPRX2 showed higher activity for both sinapyl alcohol and coniferyl alcohol, compared with the other three PbPRX proteins (Table 2). These data suggested that PbPRX2 might play a decisive role in the lignin biosynthesis of Chinese pear fruit.
Table 2.
Specific activities of the Pyrus bretschneideri PbPRX proteins for five substrates
| Specific activity (μmol/min per mg) | |||||
|---|---|---|---|---|---|
| Pyrogallol | Guaiacol | Ferulic acid | Coniferyl alcohol | Sinapyl alcohol | |
| PbPRX2 | 1108.25 ± 21.33 | 426.76 ± 23.69 | 1311.23 ± 28.27 | 5003.13 ± 9.65 | 2004.21 ± 10.54 |
| PbPRX34 | 1321.62 ± 19.24 | 428.13 ± 9.32 | 2142.27 ± 32.14 | 3002.93 ± 110.32 | 111.81 ± 5.32 |
| PbPRX64 | 634.21 ± 12.33 | 233.18 ± 9.87 | 634.23 ± 19.42 | 3303.21 ± 9.32 | 80.32 ± 1.22 |
| PbPRX75 | 719.11 ± 19.29 | 215.24 ± 4.97 | 691.75 ± 23.65 | 2331.31 ± 132.10 | 47.31 ± 4.52 |
Enzyme kinetic analyses of PbPRXs
Using pyrogallol, ferulic acid, and coniferyl alcohol as substrates, the kinetic constants of the four purified PbPRX proteins (PbPRX2, PbPRX34, PbRX64, and PbPRX75) were tested. Compared with pyrogallol, these four PbPRX proteins have better kcat/Km (higher catalytic efficiency), bette kcat (higher conversion number), and bette 1/Km (higher affinity) for ferulic acid and coniferyl alcohol (Table 3).
Table 3.
Kinetic constants of the Pyrus bretschneideri PbPRXs proteins for three substrates
| Ferulic acid | Pyrogallol | Coniferyl alcohol | |||||||
|---|---|---|---|---|---|---|---|---|---|
| 1/Km | Kcat | Kcat/Km | 1/Km | Kcat | Kcat/Km | 1/Km | Kcat | Kcat/Km | |
| (mM−1) | (S−1) | (mM−1S−1) | (mM−1) | (S−1) | (mM−1S−1) | (mM−1) | (S−1) | (mM−1S−1) | |
| PbPRX2 | 2.11 | 12,543.24 | 26,466.24 | 0.13 | 698.32 | 90.78 | 4.3 | 997.35 | 4288.61 |
| PbPRX34 | 3.12 | 12,501.29 | 39,004.02 | 0.21 | 832.22 | 174.77 | 2.23 | 9975.19 | 22,244.67 |
| PbPRX64 | 2.22 | 5229.13 | 11,608.67 | 0.32 | 312.25 | 99.92 | 3.16 | 4543.56 | 14,357.65 |
| PbPRX75 | 4.45 | 2176.45 | 9685.21 | 3.56 | 74.32 | 264.58 | 4.09 | 8114.43 | 33,188.02 |
Multiple sequence alignment analyses of PbPRX proteins
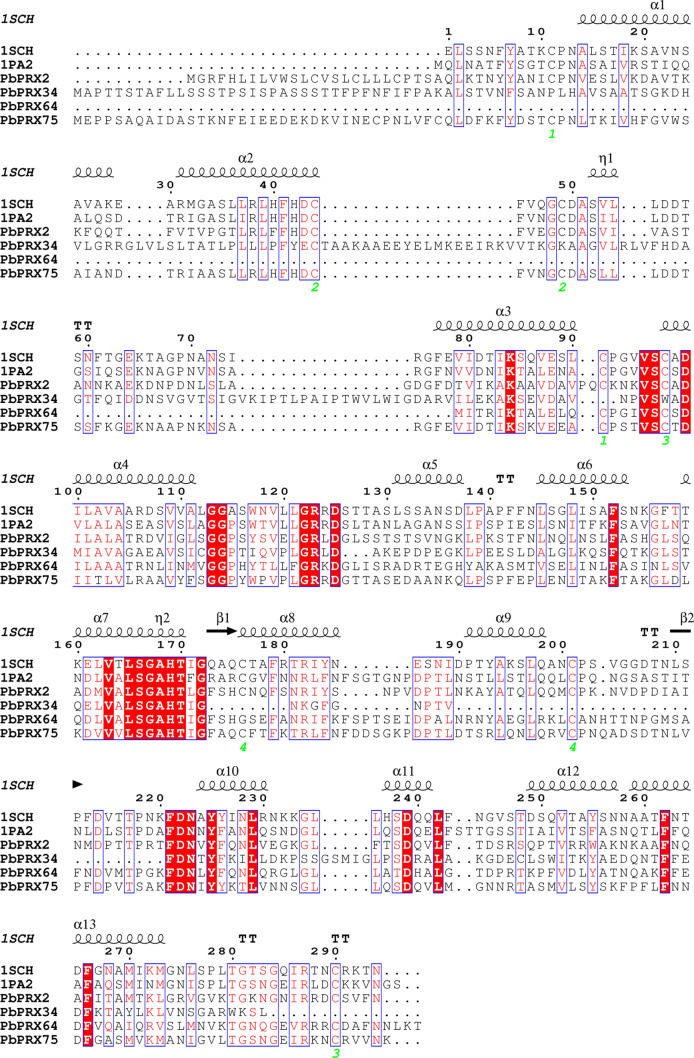
In this study, the amino acid sequences of the two PRX proteins, viz. 1SCH (Schuller et al. 1996) and 1PA2 (Østergaard et al. 2000) were used as templates to analyze the secondary structure of PbPRX2, PbPRX34, PbRX64, and PbPRX75 proteins. As shown in Fig. 1, the typical conserved domains of peroxidase existed in these four proteins of Chinese pear. The most conserved regions in these PbPRX proteins mainly include residues used to maintain the folding and catalytic activity of peroxidase. For example, the eight Cys residues involved in the four disulfides are highly conserved in all PRX tested.
Fig. 1.
Structural alignment of PbPRX2, PbPRX34, PbRX64 and PbPRX75 with two PRXs from the research collaboratory for structural bioinformatics PDB
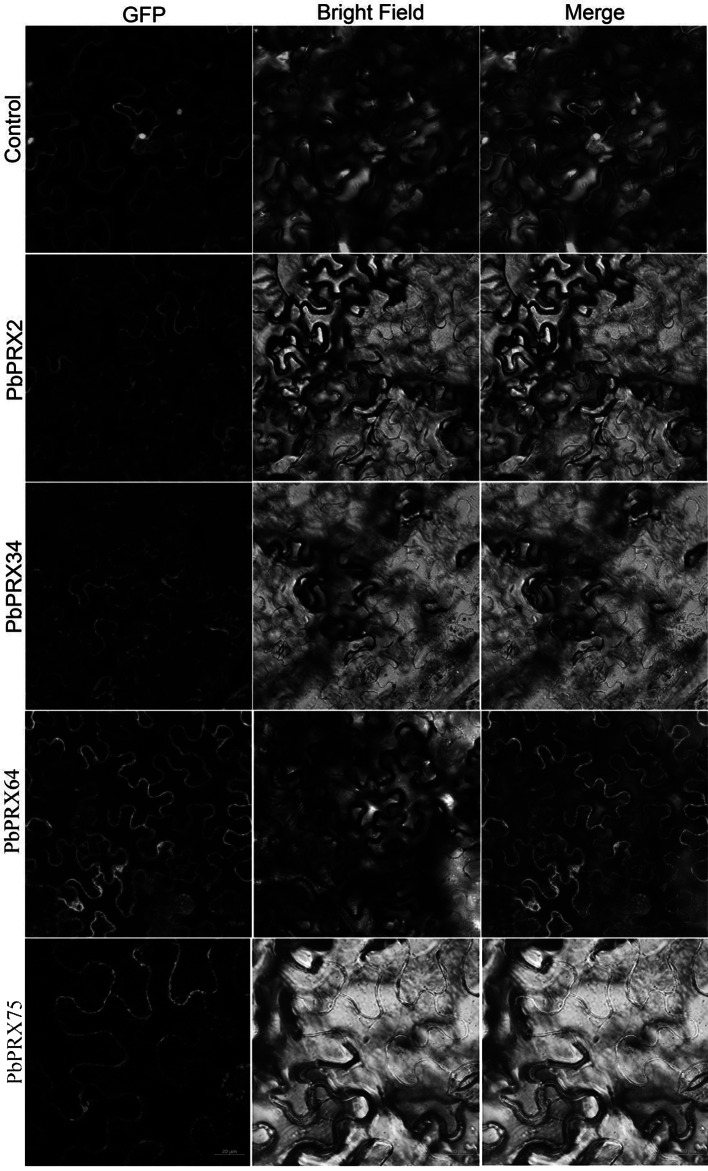
Subcellular localization of PbPRX proteins
To determine the subcellular localization of PbPRX2, PbPRX34, PbRX64, and PbPRX75, we constructed pPbPRX-GFP expression vectors and then transformed them into tobacco, respectively. The green fluorescence signals from the expressed fusion PbPRX2-GFP, PbPRX34-GFP, PbPRX64-GFP, and PbPRX75-GFP proteins were specifically distributed on the cell membrane or cell wall, as shown in Fig. 2. However, the green fluorescence from the expressed GFP alone was distributed on the whole cell, which means that it was a constitutive expression pattern.
Fig. 2.
Subcellular localization of PbPRX from Chinese pear (Pyrus bretschneideri)
Discussion
Lignin is a kind of biological macromolecule with complex and extremely stable structure in plants (Hatakeyama and Hatakeyama 2009; Martínez et al. 2008; Ralph et al. 2019). Lignin is mainly composed of three structural units, i.e., hydroxyphenyl propane, guaiacyl propane, and syringyl propane (Ralph et al. 2019; Vanholme et al. 2010). S-lignin (syringyl lignin) consists of syringyl structural units, while H-lignin (hydroxyphenyl lignin) is composed of p hydroxyphenylpropane units, and G-lignin (guaiacyl lignin) consists of guaiacyl units (Cai et al. 2010; Ralph et al. 2019). The stone cells of pear fruit, which have adversely affected pear quality and flavor, are mainly composed of lignin (Cai et al. 2010; Rogers and Campbell 2004). Therefore, lignin content is one of the important factors affecting the fruit quality of Chinese pear (Jin et al. 2013; Yan et al. 2014). The core lignification gene families in Chinese pear, including C4H, CSE, COMT, 4CL, CCR, HCT, CCoAOMT, C3H, PAL, F5H, and CAD, have been studied in previously published articles (Cao et al. 2016a, 2019; Cheng et al. 2017; Ding et al. 2020; Li et al. 2021). For the polymerization of lignin, although Cao et al. (2016a, b) also identified the number of the PbPRX family in Chinese pear, but the function of PRXs still remains underexplored.
PRXs play an important role in various physiological processes of plants, such as cell wall structure, response to biotic and abiotic stresses, and biosynthesis of secondary metabolites (Blee et al. 2003; Fagerstedt et al. 2010; Fernández-Fueyo et al. 2014; Hoffmann et al. 2020; Kidwai et al. 2020; Miller et al. 2007). PRXs usually exist in the form of the gene family in plants. For example, Wang et al. (2015) identified 119 ZmPRXs in maize (Zea mays), Li et al. (2020) cloned CsPRXs in tea (Camellia sinensis), and Yan et al. (2019) identified 374 TaPRXs in wheat (Triticum aestivum) (Li et al. 2020; Wang et al. 2015; Yan et al. 2019). PRXs are very important enzymes in plants that mainly catalyze the polymerization of lignin monomers in the last step of the lignin metabolism pathway (Davin et al. 2008). In our study, we cloned five PbPRXs (PbPRX2, PbPRX22, PbPRX34, PbPRX64, and PbPRX75), and found that four (PbPRX2, PbPRX34, PbPRX64, and PbPRX75) of these genes may be involved in the lignin biosynthesis in Chinese pear fruit. Sequence alignment analysis revealed that these five PbPRX proteins have typical conserved domains of peroxidase (Cosio and Dunand 2009; Passardi et al. 2004; Teixeira et al. 2004). Ren et al., (2014) found that PRX proteins are mainly located in the cell membrane or cell wall (Ren et al. 2014), which was also confirmed in our results (Fig. 2).
PRX proteins have catalytic activity for a variety of substrates, such as pyrogallol, guaiacol, ferulic acid, coniferyl alcohol, and sinapyl alcohol (Kokkinakis and Brooks 1979; Lai et al. 2006; Ren et al. 2014; Wariishi and Gold 1989). To study the catalytic activity of PbPRX2, PbPRX34, PbPRX64, and PbPRX75 for these substrates, these proteins were purified, and their enzyme activities and enzyme kinetic constants were analyzed. Enzyme activity analysis showed that PbPRX2 has high catalytic activity for both coniferyl alcohol and sinapyl alcohol. Previous studies have shown that coniferyl alcohol and sinapyl alcohol are precursors for the synthesis of lignin monomers (Amthor 2003; del Río et al. 2020; Jin et al. 2013; Vanholme et al. 2019; Yan et al. 2014), which suggested that PbPRX2 may play a vital role in the lignin biosynthesis of Chinese pear fruit. Indeed, Koutaniemi et al., (2005) found that one gene PaPRX5 encoded protein preferred coniferyl alcohol and contributed to the lignin biosynthesis in Norway spruce (Picea abies) (Koutaniemi et al. 2005).
Conclusion
In our study, we cloned PbPRX2, PbPRX22, PbPRX34, PbPRX64, and PbPRX75 genes from Chinese pear fruit. Subsequently, we performed sequence alignment, enzyme activity, enzyme kinetics, and subcellular localization analyses. Finally, we found that PbPRX2 played a key role in the polymerization of lignin in Chinese pear fruit. Our results provided candidate PbPRX genes for improving pear fruit quality at the molecular level in the future.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
We are grateful the supported by the Outstanding youth of the Education Department of Hunan Province (20B624), and the Talent Research Start-up Fund of Central South University of Forestry and Technology (2019YJ012).
Abbreviations
- Class III PRX
Class III peroxidase
- AA
Amino acids
- ORFs
Open reading frames
- cDNA
Complementary DNA
- kDa
Kilodaltons
- S-lignin
Syringyl lignin
- H-lignin
Hydroxyphenyl lignin
- G-lignin
Guaiacyl lignin
Funding
This work was supported by the Outstanding youth of the Education Department of Hunan Province (20B624), and the Talent Research Start-up Fund of Central South University of Forestry and Technology (2019YJ012).
Compliance with ethical standards
Conflict of interest
The authors declare no competing financial interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Xi Zhu and Lan Jiang have contributed equally to this work.
References
- Amthor JS. Efficiency of lignin biosynthesis: a quantitative analysis. Ann Bot. 2003;91:673–695. doi: 10.1093/aob/mcg073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barceló AR, Aznar-Asensio G. Coniferyl alcohol oxidase activity of a cell-wall-located class III peroxidase. Funct Plant Biol. 1999;26:411–419. [Google Scholar]
- Blee KA, Choi JW, O'Connell AP, Schuch W, Lewis NG, Bolwell GP. A lignin-specific peroxidase in tobacco whose antisense suppression leads to vascular tissue modification. Phytochemistry. 2003;64:163–176. doi: 10.1016/s0031-9422(03)00212-7. [DOI] [PubMed] [Google Scholar]
- Cai Y, Li G, Nie J, Lin Y, Nie F, Zhang J, Xu Y. Study of the structure and biosynthetic pathway of lignin in stone cells of pear. Sci Hortic. 2010;125:374–379. [Google Scholar]
- Cao Y, Han Y, Li D, Lin Y, Cai Y. Systematic analysis of the 4-Coumarate: coenzyme a ligase (4CL) related genes and expression profiling during fruit development in the Chinese pear. Genes. 2016;7:89. doi: 10.3390/genes7100089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Han Y, Meng D, Li D, Jin Q, Lin Y, Cai Y. Structural, evolutionary, and functional analysis of the class III peroxidase gene family in Chinese pear (Pyrus bretschneideri) Front Plant Sci. 2016;7:1874. doi: 10.3389/fpls.2016.01874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Li X, Jiang L. Integrative analysis of the core fruit lignification toolbox in pear reveals targets for fruit quality bioengineering. Biomolecules. 2019;9:504. doi: 10.3390/biom9090504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesarino I. Structural features and regulation of lignin deposited upon biotic and abiotic stresses. Curr Opin Biotechnol. 2019;56:209–214. doi: 10.1016/j.copbio.2018.12.012. [DOI] [PubMed] [Google Scholar]
- Cheng X, et al. Characterization and analysis of CCR and CAD gene families at the whole-genome level for lignin synthesis of stone cells in pear (Pyrus bretschneideri) fruit. Biol Open. 2017;6:1602–1613. doi: 10.1242/bio.026997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosio C, Dunand C. Specific functions of individual class III peroxidase genes. J Exp Bot. 2009;60:391–408. doi: 10.1093/jxb/ern318. [DOI] [PubMed] [Google Scholar]
- Dalet F, Cornu D. Lignification level and peroxidase activity during in vitro rooting of Prunus avium. Can J Bot. 1989;67:2182–2186. [Google Scholar]
- Davin LB, Jourdes M, Patten AM, Kim K-W, Vassão DG, Lewis NG. Dissection of lignin macromolecular configuration and assembly: comparison to related biochemical processes in allyl/propenyl phenol and lignan biosynthesis. Nat Prod Rep. 2008;25:1015–1090. doi: 10.1039/b510386j. [DOI] [PubMed] [Google Scholar]
- del Río JC, Rencoret J, Gutiérrez A, Elder T, Kim H, Ralph J. Lignin monomers from beyond the canonical monolignol biosynthetic pathway: another brick in the wall. ACS Sustain Chem Eng. 2020;8:4997–5012. [Google Scholar]
- Ding B, Hu C, Feng X, Cui T, Liu Y, Li L. Systematic analysis of the OFP genes in six Rosaceae genomes and their roles in stress response in Chinese pear (Pyrus bretschneideri) Physiol Mol Biol Plants. 2020;26:2085–2094. doi: 10.1007/s12298-020-00866-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlting J, et al. Global transcript profiling of primary stems from Arabidopsis thaliana identifies candidate genes for missing links in lignin biosynthesis and transcriptional regulators of fiber differentiation. Plant J. 2005;42:618–640. doi: 10.1111/j.1365-313X.2005.02403.x. [DOI] [PubMed] [Google Scholar]
- Fagerstedt KV, Kukkola EM, Koistinen VVT, Takahashi J, Marjamaa K. Cell wall lignin is polymerised by class III secretable plant peroxidases in Norway spruce. J Integr Plant Biol. 2010;52:186–194. doi: 10.1111/j.1744-7909.2010.00928.x. [DOI] [PubMed] [Google Scholar]
- Fernández-Fueyo E, Ruiz-Dueñas FJ, Martínez MJ, Romero A, Hammel KE, Medrano FJ, Martínez AT. Ligninolytic peroxidase genes in the oyster mushroom genome: heterologous expression, molecular structure, catalytic and stability properties, and lignin-degrading ability. Biotechnol Biofuels. 2014;7:2. doi: 10.1186/1754-6834-7-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasteiger E, Hoogland C, Gattiker A, Duvaud SE, Wilkins MR, Appel RD, Bairoch A. Protein identification and analysis tools on the ExPASy server. Proteom Protoc Handb. 2005;112:571–607. [Google Scholar]
- Gouet P, Robert X, Courcelle E. ESPript/ENDscript: sequence and 3D information from protein structures. Acta Crystallogr. 2005;61:42–43. [Google Scholar]
- Han Y, et al. Functional analysis of two flavanone-3-hydroxylase genes from Camellia sinensis: a critical role in flavonoid accumulation. Genes. 2017;8:300. doi: 10.3390/genes8110300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatakeyama H, Hatakeyama T. Biopolymers. Berlin: Springer; 2009. Lignin structure, properties, and applications; pp. 1–63. [Google Scholar]
- Hoffmann N, Benske A, Betz H, Schuetz M, Samuels AL. Laccases and peroxidases co-localize in lignified secondary cell walls throughout stem development. Plant Physiol. 2020;184:806–822. doi: 10.1104/pp.20.00473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Q, Yan C, Qiu J, Zhang N, Lin Y, Cai Y. Structural characterization and deposition of stone cell lignin in Dangshan Su pear. Sci Hortic. 2013;155:123–130. [Google Scholar]
- Katoh K, Kuma K-I, Toh H, Miyata T. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucl Acids Res. 2005;33:511–518. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidwai M, Ahmad IZ, Chakrabarty D. Class III peroxidase: an indispensable enzyme for biotic/abiotic stress tolerance and a potent candidate for crop improvement. Plant Cell Rep. 2020;39:1381–1393. doi: 10.1007/s00299-020-02588-y. [DOI] [PubMed] [Google Scholar]
- Kokkinakis DM, Brooks JL. Tomato peroxidase: purification, characterization, and catalytic properties. Plant Physiol. 1979;63:93–99. doi: 10.1104/pp.63.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koutaniemi S, et al. Characterization of basic p-coumaryl and coniferyl alcohol oxidizing peroxidases from a lignin-forming Picea abies suspension culture. Plant Mol Biol. 2005;58:141–157. doi: 10.1007/s11103-005-5345-6. [DOI] [PubMed] [Google Scholar]
- Kvaratskhelia M, Winkel C, Thorneley RN. Purification and characterization of a novel class III peroxidase isoenzyme from tea leaves. Plant Physiol. 1997;114:1237–1245. doi: 10.1104/pp.114.4.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai L-S, Wang D-J, Chang C-T, Wang C-H. Catalytic characteristics of peroxidase from wheat grass. J Agric Food Chem. 2006;54:8611–8616. doi: 10.1021/jf060888w. [DOI] [PubMed] [Google Scholar]
- Lewis NG, Yamamoto E. Lignin: occurrence, biogenesis and biodegradation. Annu Rev Plant Biol. 1990;41:455–496. doi: 10.1146/annurev.pp.41.060190.002323. [DOI] [PubMed] [Google Scholar]
- Li G, et al. Comparative genomic analysis of superoxide dismutase (SOD) genes in three Rosaceae species and expression analysis in Pyrus bretschneideri. Physiol Mol Biol Plants. 2021;27:39–52. doi: 10.1007/s12298-021-00926-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li HJ, Wang HB, Chen Y, Ma QP, Zhao Z, Li XH, Chen X. Isolation and expression profiles of class III PRX gene family under drought stress in Camellia sinensis. Biol Plant. 2020;64:280–288. [Google Scholar]
- Martínez ÁT, Rencoret J, Marques G, Gutiérrez A, Ibarra D, Jiménez-Barbero J, José C. Monolignol acylation and lignin structure in some nonwoody plants: a 2D NMR study. Phytochemistry. 2008;69:2831–2843. doi: 10.1016/j.phytochem.2008.09.005. [DOI] [PubMed] [Google Scholar]
- Miller G, Suzuki N, Rizhsky L, Hegie A, Koussevitzky S, Mittler R. Double mutants deficient in cytosolic and thylakoid ascorbate peroxidase reveal a complex mode of interaction between reactive oxygen species, plant development, and response to abiotic stresses. Plant Physiol. 2007;144:1777–1785. doi: 10.1104/pp.107.101436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Østergaard L, et al. Arabidopsis ATP A2 peroxidase. Expression and high-resolution structure of a plant peroxidase with implications for lignification. Plant Mol Biol. 2000;44:231–243. doi: 10.1023/a:1006442618860. [DOI] [PubMed] [Google Scholar]
- Passardi F, Longet D, Penel C, Dunand C. The class III peroxidase multigenic family in rice and its evolution in land plants. Phytochemistry. 2004;65:1879–1893. doi: 10.1016/j.phytochem.2004.06.023. [DOI] [PubMed] [Google Scholar]
- Pomar F, Caballero N, MA P, Ros Barceló A. H2O2 generation during the auto-oxidation of coniferyl alcohol drives the oxidase activity of a highly conserved class III peroxidase involved in lignin biosynthesis. FEBS Lett. 2002;529:198–202. doi: 10.1016/s0014-5793(02)03339-2. [DOI] [PubMed] [Google Scholar]
- Ralph J, Lapierre C, Boerjan W. Lignin structure and its engineering. Curr Opin Biotechnol. 2019;56:240–249. doi: 10.1016/j.copbio.2019.02.019. [DOI] [PubMed] [Google Scholar]
- Ren L-L, Liu Y-J, Liu H-J, Qian T-T, Qi L-W, Wang X-R, Zeng Q-Y. Subcellular relocalization and positive selection play key roles in the retention of duplicate genes of Populus class III peroxidase family. Plant Cell. 2014;26:2404–2419. doi: 10.1105/tpc.114.124750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers LA, Campbell MM. The genetic control of lignin deposition during plant growth and development. New Phytol. 2004;164:17–30. doi: 10.1111/j.1469-8137.2004.01143.x. [DOI] [PubMed] [Google Scholar]
- Sanchez M, Pena MJ, Revilla G, Zarra I. Changes in dehydrodiferulic acids and peroxidase activity against ferulic acid associated with cell walls during growth of Pinus pinaster hypocotyl. Plant Physiol. 1996;111:941–946. doi: 10.1104/pp.111.3.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki S, Baba KI, Nishida T, Tsutsumi Y, Kondo R. The cationic cell-wall-peroxidase having oxidation ability for polymeric substrate participates in the late stage of lignification of Populus alba L. Plant Mol Biol. 2006;62:797–807. doi: 10.1007/s11103-006-9057-3. [DOI] [PubMed] [Google Scholar]
- Schuller DJ, Ban N, van Huystee RB, McPherson A, Poulos TL. The crystal structure of peanut peroxidase. Structure. 1996;4:311–321. doi: 10.1016/s0969-2126(96)00035-4. [DOI] [PubMed] [Google Scholar]
- Teixeira FK, Menezes-Benavente L, Margis R, Margis-Pinheiro M. Analysis of the molecular evolutionary history of the ascorbate peroxidase gene family: inferences from the rice genome. J Mol Evol. 2004;59:761–770. doi: 10.1007/s00239-004-2666-z. [DOI] [PubMed] [Google Scholar]
- Vanholme R, De Meester B, Ralph J, Boerjan W. Lignin biosynthesis and its integration into metabolism. Curr Opin Biotechnol. 2019;56:230–239. doi: 10.1016/j.copbio.2019.02.018. [DOI] [PubMed] [Google Scholar]
- Vanholme R, Demedts B, Morreel K, Ralph J, Boerjan W. Lignin biosynthesis and structure. Plant Physiol. 2010;153:895–905. doi: 10.1104/pp.110.155119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Wang Q, Zhao Y, Han G, Zhu S. Systematic analysis of maize class III peroxidase gene family reveals a conserved subfamily involved in abiotic stress response. Gene. 2015;566:95–108. doi: 10.1016/j.gene.2015.04.041. [DOI] [PubMed] [Google Scholar]
- Wariishi H, Gold MH. Lignin peroxidase compound III: formation, inactivation, and conversion to the native enzyme. FEBS Lett. 1989;243:165–168. [Google Scholar]
- Yan C, Yin M, Zhang N, Jin Q, Fang Z, Lin Y, Cai Y. Stone cell distribution and lignin structure in various pear varieties. Sci Hortic. 2014;174:142–150. [Google Scholar]
- Yan J, et al. Genome-wide and evolutionary analysis of the class III peroxidase gene family in wheat and Aegilops tauschii reveals that some members are involved in stress responses. BMC Genomics. 2019;20:666. doi: 10.1186/s12864-019-6006-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.