Abstract
The ability to respond quickly to salt stress can determine the tolerance level of a species. Here, we test how rapidly the roots of Calotropis procera react to high salinity conditions. In the first 24 h after saline exposure, the plants reduced stomatal conductance, increased CO2 assimilation, and water use efficiency. Thus, the root tissue showed an immediate increase in soluble sugars, free amino acid, and soluble protein contents. Twelve aquaporins showed differential gene expression in the roots of C. procera under salinity. Transcriptional upregulation was observed only after 2 h, with greater induction of CpTIP1.4 (fourfold). Transcriptional downregulation, in turn, occurred mainly after 8 h, with the largest associated with CpPIP1.2 (fourfold). C. procera plants responded quickly to high saline levels. Our results showed a strong stomatal control associated with high free amino acid and soluble sugar contents, regulated aquaporin expression in roots, and supported the high performance of the root system of C. procera under salinity. Moreover, this species was able to maintain a lower Na+/K+ ratio in the leaves compared to that of the roots of stressed plants. The first response of the root system, after immediate contact with saline solution, present an interesting scenario to discuss.
Supplementary Information
The online version contains supplementary material available at 10.1007/s12298-021-00957-9.
Keywords: Gas exchange, Root physiology, Salinity, Salt stress, Tonoplast intrinsic proteins
Introduction
Calotropis procera is a perennial Asian shrub with a significant adaptation to adverse climatic and poor soil conditions (Hassan et al. 2015). Among peculiar traits, it presents high photosynthetic performance, stomatal control, and water use efficiency under drought conditions (Rivas et al. 2017; 2020; Tezara et al. 2011;), and high metabolic performance under salinity (Mutwakil et al. 2017). Furthermore, although it originated in arid regions, this species is evergreen with a C3 photosynthetic metabolism. This set of characteristics makes it a relevant model for ecophysiological studies and an important source of gene pools to identify genes involved in tolerance to abiotic stresses. Among them, high salinity in the soil is one of the major abiotic stresses responsible for desertification processes worldwide, decreasing plant biomass production (Munns and Tester 2008).
The severity of the stress depends on the salt concentration to which the plant has been exposed. When plants are under very high salt concentrations (≥ 200 mM NaCl), it experiences more rapid and intense osmotic and ionic stresses, and exhibits greater susceptibility to plant cell plasmolysis (Shavrukov 2013). Plant water supply becomes precarious when roots are exposed to such high saline concentrations. Plant water relations depend on the root system. Therefore, root acclimatization responses to salt are essential to support water and nutrient conductivities (Rodríguez-Gamir et al. 2019) and avoid the accumulation of excess Na+ (Munns and Tester 2008). Thus, performance of roots under salinity by exploring their biochemical and molecular traits is unclear. Among the mechanisms triggered by roots related to salt tolerance, the performance of osmotic adjustment (Blum 2017; Slama et al. 2015;) through primary metabolite content (Sami et al. 2016) and the control of aquaporin (AQP) activity are worth highlighting (Kapilan et al. 2018; Rodríguez-Gamir et al. 2019). In all plant tissues, AQPs are intrinsic membrane proteins that act in the transport of cell water and small solutes, such as H2O2, ammonia, urea, glycerol, and others, in addition to gases and ions (Kapilan et al. 2018; Li et al. 2014). The increase in primary metabolite biosynthesis decreases the osmotic pressure and restores the plant water flow in relation to salinized soil (Blum et al. 2017). These compounds are associated with free radical elimination, cell structure protection, and maintenance of enzymatic activity (Sami et al. 2016; Mansour and Ali, 2017).
The AQPs family plays an important role in the water relations of the entire plant (Singh et al. 2020), which are largely regulated by salt concentrations (Liu et al. 2012) and drought (Li et al. 2014; Rodríguez-Gamir et al. 2019). Based on their phylogenetic distribution, plant AQPs are categorized into five main subfamilies (Bezerra-Neto et al. 2019): plasma membrane intrinsic proteins (PIP), nodulin 26-like intrinsic proteins, tonoplast intrinsic proteins (TIPs), small intrinsic proteins, and uncharacterized intrinsic proteins. These proteins support hydraulic regulation and nutrient transport (Li et al. 2014), stomatal conductance (Rodríguez-Gamir et al. 2019), mesophyll conductance, transpiration and photosynthesis (Moshelion et al. 2015), growth and plant development, and influence ionic homeostasis (An et al. 2017). The functional regulation and role of AQPS in plant growth, development, and physiology in response to abiotic stresses have been widely studied (Bezerra-Neto et al. 2019; Singh et al. 2020). Regarding AQPs, we focused on PIPs and TIPs transcriptional regulation. Most PIPs have been identified in plasma membranes, and they are generally located in organs characterized by a large water flux. TIPs, in turn, play a relevant role in intracellular water movement and non-limiting water flow through membranes (Kapilan et al. 2018; Singh et al. 2020).
This study evaluates the performance of the root system under exposure to high salinity. The main goal was to measure the root dynamics of carbon metabolism traits and the activity of aquaporins in the first 24 h under intense salinity stress. Thus, three questions were raised: (1) C. procera able to maintain water status and gas exchange during the first hours of salt exposure to roots?” (2) Are the roots of C. procera capable of producing rapid responses to adjust primary metabolites soon after exposure to high salinity? (3) How long does it take for C. procera roots to change aquaporin activity in response to high salinity? Considering these questions, there are two hypotheses: (1) the root system of C. procera presents biochemical responses that allow it to promote osmoregulation through the accumulation of organic solutes, and (2) under a high salt concentration, the upregulation of the gene expression of PIPs and TIPs occurs to promote the adjustment of plant water status and maintain water uptake by the root system.
Material and methods
Plant material and growth conditions
Calotropis procera (Aiton) W. T. Aiton (Apocynaceae) seeds were collected in the beach of Maria Farinha (municipality of Paulista) on the coast of Pernambuco state, Brazil (7° 50′ 32.9″ S, 34° 50′ 21.2″ W). The seed surface was sterilized by immersion in 0.5% sodium hypochlorite solution (v/v) for 5 min and washing with deionized water. The seeds were germinated in Petri dishes with wet filter paper and kept in a growth chamber (at 25 °C, 12 h photoperiod, and 70% relative humidity). After root emergence, the seedlings were transferred to pots containing 7 kg of washed sand and maintained in a greenhouse for 3 months. Throughout the experiment the plants grew in a greenhouse where the temperature ranged between 26 to 32 °C, full solar radiation within the environment. The plants were irrigated daily to keep the substrate fully hydrated. Every 15 days, the plants received a complete Hoagland solution.
After ninety days of emergence, the plants were exposed to two treatments: young plants watered daily with 300 mL deionized water (control), and young plants watered daily with 300 mL of 200 mM NaCl solution (salinity). The first watering was performed at 8:00 am in the morning and subsequent watering occurred after 24 h at the end of the daily gas exchange analysis.
The treatments (control and salinity) consisted of 28 plants each, with four time-points and seven replicates. To determine the collection time points of the plant material and the physiological measurements, the initial period of salt application (8:00 am) was used as a reference. The first day of the experiment included the time-points at 2 and 8 h of exposure to salt. Day 2 comprised the time points at 21 and 24 h of salt exposure.
Physiological parameters
Foliar relative water content and soil electrical conductivity
Relative water content (RWC) was measured using a leaf disc collected at 5:00 am, 21 h after exposure to salt, according to the methodology of Barrs and Weatherley (1962). The RWC was calculated using the formula: RWC (%) = (FW − DW/TW − DW) × 100, where DW equals the dry weight, FW is the fresh weight, and TW equals the matter turgid weight. The RWC analysis was performed with three biologic repetitions (each treatment).
Sand samples of 25 g were collected and homogenized in 25 mL of deionized water. It was stirred three times for 2 min. The solution was kept immobile for 24 h and the supernatant was sampled to calculate electrical conductivity using a conductivity meter.
Gas exchange and chlorophyll contents
The stomatal conductance (gs), CO2 net assimilation (A), and transpiration (E) were measured in completely expanded and healthy leaves using a portable infrared gas exchange analyzer (ADC, LCi-Pro Model, Hoddesdon, UK). Photosynthetic photon flux density was determined by measuring the environmental incident radiation. The water use efficiency (WUE) was calculated from the A/E ratio. Gas exchange was determined with four repetitions (each treatment). The evaluations were performed in the morning (08:00–09:00 am), 24 h after the beginning of salt exposure. The vapor pressure deficit (VPD) was determined according to the method of Campbell and Norman (1998). In the morning, a VPD of 1.87 kPa was determined on the second day (24 h exposure to salt) of the experiment.
The chlorophyll contents were determined using leaves of C. procera collected 24 h after salt application. All samples were immediately frozen in liquid nitrogen and stored at − 80 °C. Chlorophyll a (CHLa) and chlorophyll b (CHLb) were quantified according to the method of Lichtenthaler and Buschmann (2001). The analyses were performed using a dual-beam spectrophotometer adjusted to the specific wavelength for each organic compound with three repetitions (each treatment).
Biochemical parameters
Proteins, amino acids, and sugars analysis
The analysis of the organic solutes was performed using the roots of C. procera collected at different time-points (2, 8, and 24 h) after salt application. All samples were immediately frozen in liquid nitrogen and stored at − 80 °C.
The collected roots were used to quantify soluble sugars (SS) (Dubois et al. 1956), free amino acids (FAA) (Moore and Stein 1948), and total soluble proteins (TSP) (Bradford 1976). The analyses were performed using a dual-beam spectrophotometer adjusted to the specific wavelength for each organic compound. The analysis of the organic solutes was carried out with three repetitions (each treatment).
Measurement of Na+and K+contents
To quantify the sodium and potassium contents, roots were collected after 24 h of exposure to salt. After drying in an oven (60 °C), 500 mg of plant material was digested in sulfuric acid P.A. (H2SO4) solution in a digester block at 350 °C for 84 min to obtain the sample extract (Thomas et al., 1967). Sodium and potassium contents were determined by flame emission photometry (DM-62, Digimed, São Paulo, Brazil). The Na+/K+ ratio was calculated using sodium and potassium content data. The analysis was conducted with four repetitions (each treatment).
Statistical analyses
Data were subjected to a test of normality (Shapiro–Wilk) and homogeneity of variance to determine if the prerequisites for parametric statistics used were met. The dataset that did not meet the requirements was subjected to non-parametric tests. Data on the sodium content, potassium content, Na+/K+ ratio, and biochemistry were subjected to a two-factor analysis of variance (ANOVA), where salt treatment versus exposure time to salinity or plant organ were analyzed. The differences in the values were submitted to Duncan test (p < 0.05). All data were analyzed using Statistica 8.0 (StatSoft. Inc., Tulsa, OK 74,104, USA).
Molecular analyses
Root material for AQP expression analysis
For the AQP gene expression study, root samples were collected at 2, 8, and 24 h (three replicates) after the salt exposure. All collected samples were immediately frozen in liquid nitrogen and stored at − 80 °C until RNA isolation.
Total RNA isolation and cDNA synthesis
Total RNA was isolated with the SV Total RNA Isolation System Kit (Promega, Fitchburg, WI, USA), according to the manufacturer's instructions. The integrity of RNA was verified by 1.5% (w/v) agarose gel electrophoresis and quantified by fluorometry (Qubit, Oregon, USA) and stored at − 80 °C until analysis. First-strand cDNA was synthesized from total RNA (1 μg) using GoScript™ Reverse Transcription System Oligo dT (Promega, Fitchburg WI, USA) according to the manufacturer's protocol.
Aquaporin selection and primer design
AQP protein seed sequences obtained from the Universal Protein (Uniprot) and National Center for Biotechnology Information (NCBI) databases were used for local BLAST (tBLASTn; e-value = ≤ 10–5) against C. procera leaf transcriptome generated by our group (Coêlho et al. 2019) and deposited in the NCBI database (accession number PRJNA508417). The retrieved sequences were translated by the ORF-Finder tool and submitted to the CD-Search domain recognition tool for full MIP domain selection. To determine the AQP nomenclature (Table A1), an alignment (MEGA 7 program) with C. procera and Arabidopsis thaliana AQP sequences was performed to construct a phylogenetic tree (Neighbor-Joining method, Bootstrap 1000 replications) (Tamura et al. 2007) (Fig. A1). The nomenclature was based on sequence homology (Johanson et al. 2001). The alignment was also used to confirm the presence and integrity of the NPA functional motifs and ar / R selective filter, both related to water transport (Wallace and Roberts 2004) (Fig. A2). The selected transcripts were coding six intrinsic plasma membrane proteins—PIPs (two PIP1 and four PIP2) and seven intrinsic tonoplast proteins—TIPs (four TIP1, two TIP2, and one TIP4). Primers were designed using the Primer3 tool (Rozen and Skaletsky 2000) with the following parameters: primer length of 18–22 bp, GC content of 45–55% (50% ideal), melting temperature (Tm) in a range of 58–62 °C (ideal 60 °C), and amplicon lengths of 100–200 bp. Peptidyl-prolyl cis-isomerase 23 (CYP23), Actin-104 (ACT104), and Ubiquitin carboxyl-terminal hydrolase 25 (UBP25) were used as reference genes (RGs) for data normalization, as recommended by Coêlho et al. (2019). The real-time quantitative PCR (qPCR) primer sequences and amplicon characteristics are described in Supplementary Table A1.
Table 1.
Relative water content (RWC, %), stomatal conductance (gs, mol m−2 s−1), CO2 assimilation (A, µmol m−2 s−1), water use efficiency (WUE, mmol m−2 s−1), chlorophyll a (CHLa, g.kg−1DW), chlorophyll b (CHLb, g.kg−1DW) in leaves of young C. procera plants under salinity conditions in a greenhouse (200 mM NaCl)
| Parameter | Treatment | |
|---|---|---|
| Control | 200 mM NaCl | |
| RWC | 65.6 ± 1.06 | 63.8 ± 1.07 |
| gs | 0.063 ± 0.003* | 0.050 ± 0.000 |
| A | 7.7 ± 0.1 | 9.5 ± 0.3* |
| WUE | 3.2 ± 0.1 | 5.6 ± 0.2* |
| CHLa | 3.6 ± 0.19 | 4.1 ± 0.24 |
| CHLb | 1.2 ± 0.08 | 1.2 ± 0.05 |
Values followed by * are different according to t-test (p < 0.05)
Real-time quantitative PCR
The qPCR analysis was performed using LineGene 9600 equipment (Bioer, Hangzhou, China). The reactions were prepared with the GoTaq® qPCR Master Mix (Promega, Fitchburg WI, USA) consisting of 5 μL of SYBR Green Super Mix (Applied Biosystems, Foster City CA, USA), 2 μL of diluted cDNA (1:10), and 0.3 μL of each primer pair (5 μM) in a final volume of 10 µL with sterile ddH2O. Non-template controls were included for each primer pair. qPCR amplification conditions were programmed to 95 °C for 2 min, followed by 35 cycles at 95 °C for 15 s, 62 °C for 1 min, and 72 °C for 15 s. The reactions were performed in triplicate.
The melting curves were generated by varying the amplification temperature from 65–95 °C. Amplification efficiency (E) was determined for each primer from a standard curve, and generated after serial cDNA dilutions (1:10, 1:100, 1:1000, and 1:10,000) in triplicate and calculated using the equation: E = 10 (− 1/slope of the standard curve) (Rasmussen 2001). The inclinations of the standard curve line in the range from − 3.58 to − 3.10 were considered acceptable for the trial (Biassoni 2014). These line slope values correlated with amplification efficiencies between 90 (E = 1.9) and 110% (E = 2.1).
The Rest2009 software package (REST Standard mode) was used to calculate and analyze the relative expression of the target transcripts. Relative expression was calculated using the formula: E (ΔCq aquaporin)/ E (ΔCq RG), where E represents the average efficiency for each gene, ΔCq is the difference between mean Cq-value of a control sample and the mean Cq-value of treated sample. Such analysis was based on paired comparisons (of target transcript and reference genes under salinity conditions and controls) using randomization and bootstrapping—Pair-wise Fixed Reallocation Randomization Test© (Pfaffl et al., 2002). Hypothesis testing (p < 0.05) was applied to determine if differences in the expression of target transcripts under control and treated conditions were significant.
Results
Foliar relative water content and soil electrical conductivity
Leaf RWC analysis revealed no significant difference after 24 h of salinity in relation to the control treatment. The average RWC values were 66 and 64% for the control and salinity treatments, respectively (Table 1). After 24 h under saline stress the soil electrical conductivity was 0.36 ± 0.02 and 0.98 ± 0.15 mS cm−1, for the control and saline treatment, respectively.
Sodium and potassium content and Na+/K+ ratio
The application of saline solution resulted in an increase of Na+ ions in the roots, stems, and leaves. In relation to the control, the greatest increase occurred in the roots (Fig. 1A). On the other hand, C. procera showed the ability to maintain K+ ion content in the three measured organs (Fig. 1B). The Na+/K+ ratio remained unchanged in the leaves, however lower than root content values (Fig. 1C).
Fig. 1.
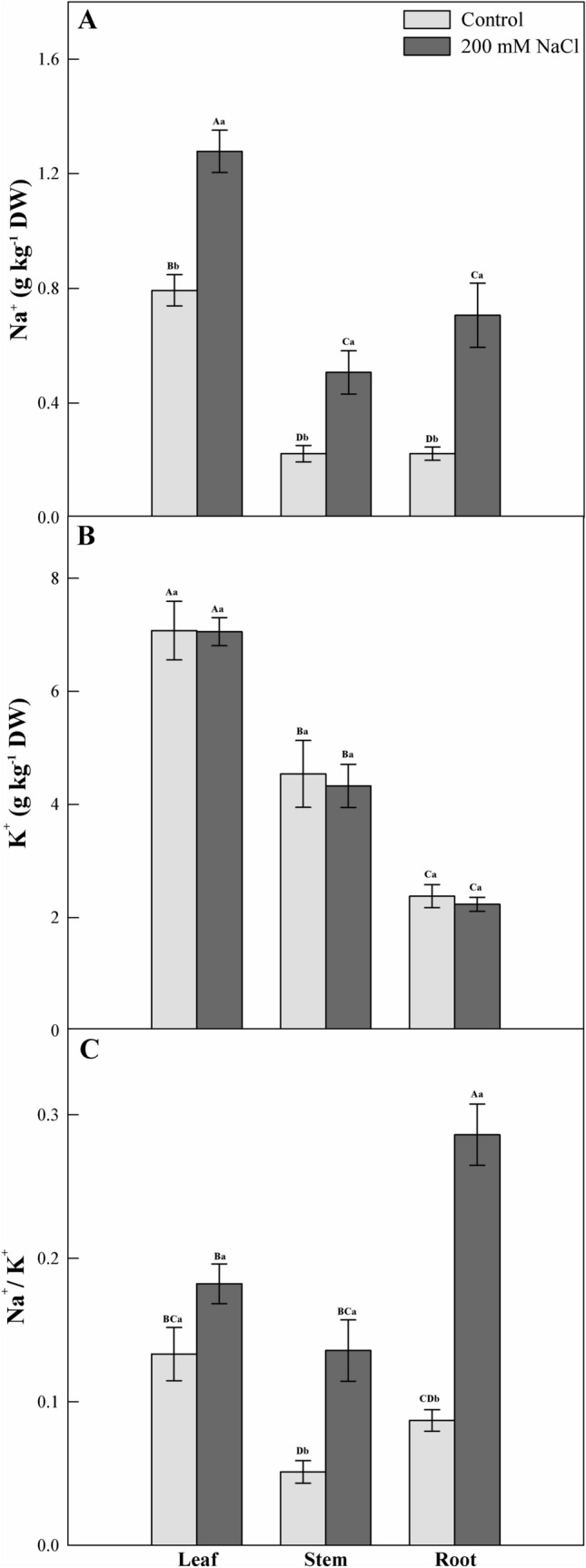
Sodium and potassium content and Na+/K+ ratio in the leaf, stem and root of young plants of Calotropis procera 24 h after exposure to salinity (200 mM NaCl) in a greenhouse assay. Each bar represents (n = 4 ± SE), when followed by same uppercase letters did not differ between treatments among different organs, and same lowercase letters did not differ between treatments at the same organ (p < 0.05)
Gas exchange and chlorophyll contents
Stomatal conductance (gs) was reduced in the salinity group compared to that in the control group after 24 h of exposure to salt (21%) (Table 1). The CO2 assimilation (A) increased by 23% (Table 1) in the plants under salinity. Also the water use efficiency (WUE) increased by 74% in salt-exposed plants compared to control plants (Table 1). Furthermore, plants exposed to salt showed no reduction in chlorophyll a (CHLa) and chlorophyll b (CHLb) after 24 h (Table 1).
Root sugars, amino acid, and protein contents
Salt application promoted rapid changes in the dynamics of root primary metabolism in C. procera (Fig. 2). Plants subjected to salinity treatment showed a 1.6-fold increase in soluble sugars (SS) after 2 h of exposure to salt. However, SS decreased (24–24%) after 8 and 24 h of exposure to salt (Fig. 2 A).
Fig. 2.
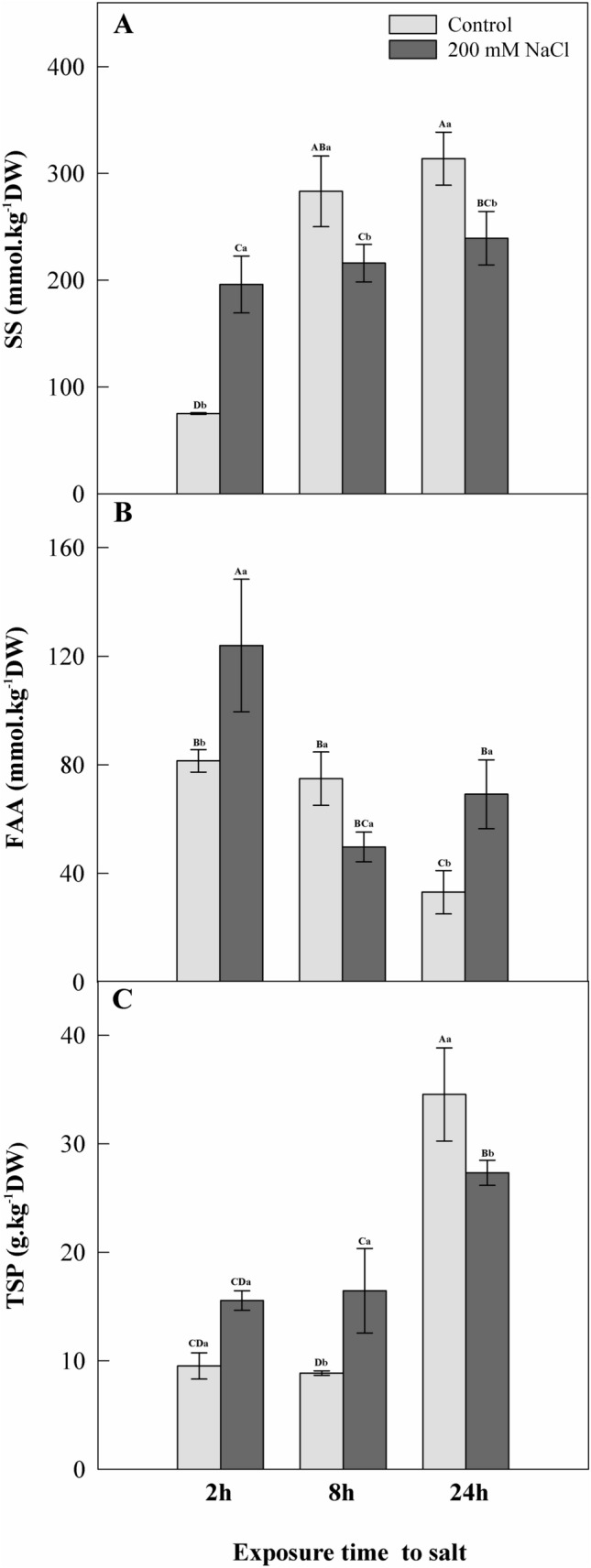
Root content of A soluble sugars (SS), B free amino-acids (FAA) and C total soluble proteins (TSP) in roots of young plants of Calotropis procera under salinity conditions (200 mM NaCl) in a greenhouse assay. Each bar represents (n = 3 ± SE), when followed by same lowercase letters did not differ between treatments within each exposure time-point to salinity and same uppercase letters did not differ between treatments between exposure time-points to salinity (p < 0.05)
Solutes involved in nitrogen metabolism showed fast changes in the dynamics of the root system of plants exposed to salt. Increased FAA (52%) was also observed after 2 h of salt addition (Fig. 2 B). TSP increased by 85% in stressed plants compared to control plants after 8 h of exposure to salt (Fig. 2 C). Compared to the control plants, plants under saline treatment increased FAA content (109%) and decreased TSP (21%) after 24 h of exposure to salt (Fig. 2 B,C).
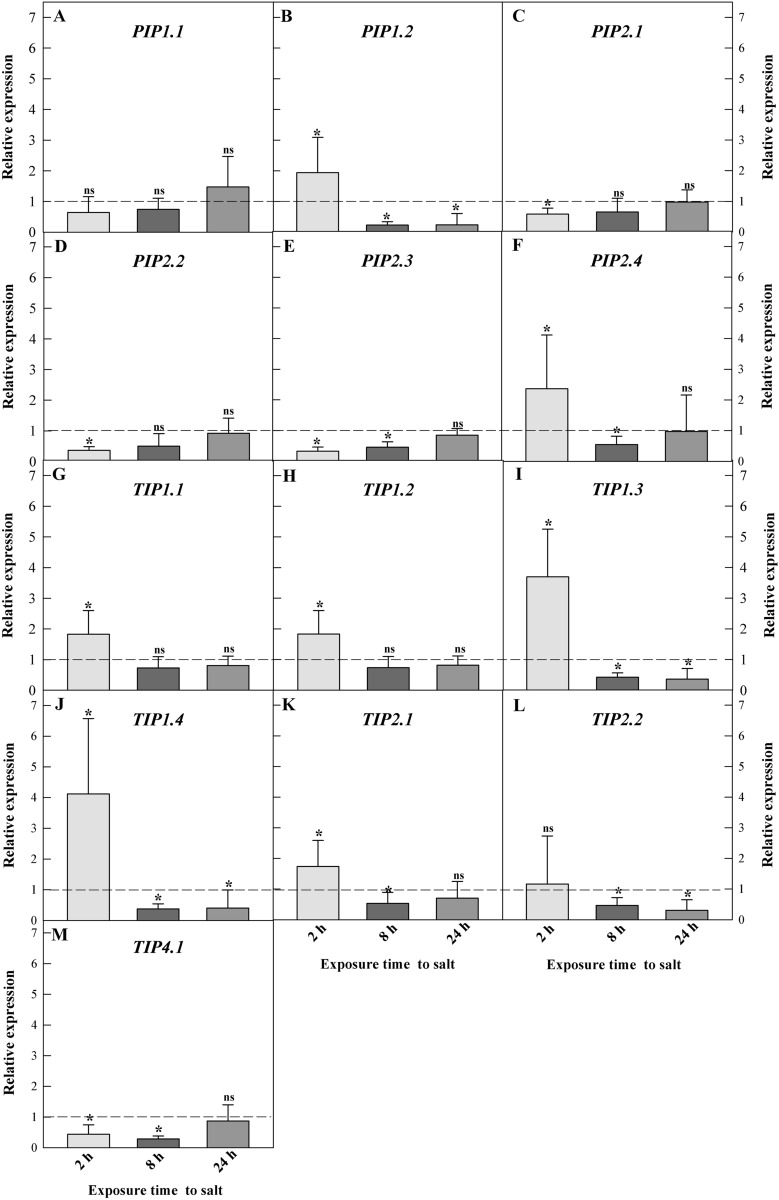
Gene expression of AQPs in the root system
Salt promoted rapid changes in CpPIP and CpTIP gene expression dynamics in C. procera roots. The CpTIP1.4 isoform was highly upregulated (fourfold), whereas CpPIP1.2 was the most downregulated (fourfold) (Fig. 3 BJ). Plants subjected to salinity treatment showed upregulation of CpPIP1.2, CpPIP2.4 (Fig. 3 BF), CpTIP1.1, CpTIP1.2, CpTIP1.3, CpTIP1.4, and CpTIP2.1 (Fig. 3 G-K), and downregulation of CpPIP2.1, CpPIP2.2, CpPIP2.3 (Fig. 3 E-C), and CpTIP4.1 (Fig. 3 M) after 2 h of exposure to NaCl. Salinity promoted downregulation of CpPIP1.2, CpPIP2.3, CpPIP2.4 (Fig. 3 BEF), CpTIP1.3, CpTIP1.4, CpTIP2.1, CpTIP2.2 (Fig. 3 I-L), and CpTIP4.1 isoform (Fig. 3 M) at 8 h. When the plants were exposed for 24 h to NaCl, the CpPIP1.2 (Fig. 3 B), CpTIP1.3, CpTIP1.4, and CpTIP2.2 (Fig. 3 IJL) isoforms were downregulated.
Fig. 4.
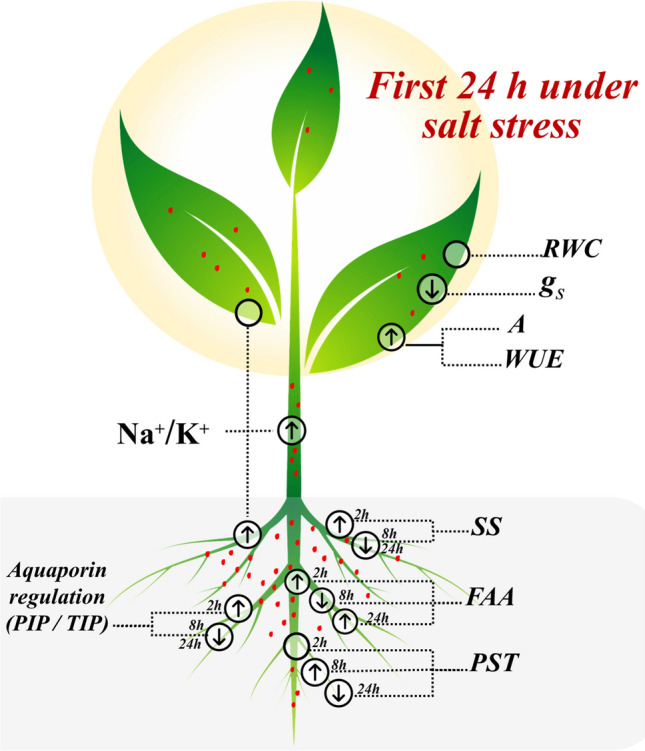
Major traits that helped the Calotropis procera plants tolerate salinity (200 mM) by controlling changes in physiological and biochemical parameters. The arrows indicate the trait behavior during the saline treatment, without change (), increasing (↑) or decreasing (↓) its activity or content in the first 24 h under salt stress. The red dots represent the proportion of the Na+/K+ ratio in the three measured tissues. Relative water content (RWC), stomatal conductance (gs), CO2 assimilation (A), water use efficiency (WUE), soluble sugars (SS), free amino acids (FAA), protein soluble total (PST), plasma membrane proteins (PIP) and intrinsic tonoplast proteins (TIP)
Fig. 3.
Relative expression‡ of aquaporins in roots of young Calotropis procera plants under salinity conditions (200 mM NaCl) in a greenhouse assay. A–F Intrinsic plasma membrane proteins—PIPs. G–M Tonoplast intrinsic proteins—TIPs (–) Dashed line indicates the limit between the positive (above the line) and negative (below the line) regulation of expression. Values followed by * indicate significant differences between treatments (Control and Salinity) by the Fixed Paired Reallocation Randomization test (p < 0.05). ‡ Relative expression was calculated using the formula: E (ΔCq aquaporin)/ E (ΔCq reference genes), where E represents the average efficiency for each transcript (aquaporin and reference genes), ΔCq is the difference between mean Cq-value of each control sample and the mean Cq-value of each treated sample
Salinity (200 mM, NaCl) promoted a rapid change in CpPIP and CpTIP expression levels in C. procera roots. Among the seven CpTIP isoforms, five (CpTIPs 1.1, 1.2, 1.3, 1.4, and 2.1) were significantly upregulated after 2 h of salinity, three of these (CpTIPs 1.3, 1.4, and 2.1) were repressed during the longest periods of salt exposure (8 and 24 h).
CpPIP 1.2 and CpPIP 2.4 were also upregulated at 2 h, although the expression was repressed after 8 h. On the other hand, three CpPIPs (2.1, 2.2, and 2.3) were downregulated in the first analysis time (2 h).
Discussion
Our results suggest that C. procera under salinity stress has fast modulation of root aquaporin activity. Moreover, the increase in CO2 assimilation, even with reduced stomatal conductance, promotes greater WUE in the first 24 h, soluble sugar and protein contents in the first 2 and 8 h of exposure to salt. This species also has an outstanding ability to maintain a higher Na+/K+ ratio in the root system and stem, alleviating the ionic imbalance in the leaf tissue.
Under salt exposure, Na+ accumulation in plant tissues is unavoidable. However, the ability to avoid targeting some tissues may represent a key factor in salinity tolerance. In this regard, several tolerance mechanisms are triggered by the selectivity of Na+ uptake by roots. Thus, the preferential xylem was loaded over K+ instead of Na+, and sodium was removed from the xylem by roots, stems, petiole, and leaf sheaths (Munns and Tester 2008). C. procera was able to keep K+ concentrations significantly unchanged, contributing to metabolism protection. Thus, a balance of K+ and Na+ concentrations act to combat ionic toxicity. Ions of K+ are essential for preventing Na+ entry, promoting the activation of numerous enzymes, stabilizing protein synthesis, and maintaining homeostasis of cytosolic pH (Munns and Tester 2008; Dreyer and Uozum 2011).
The first question was, “is C. procera able to maintain water status and gas exchange during the first hours of salt exposure to roots?” During exposure to salinity, changes in water status are common (Munns and Tester 2008), however small changes in leaf RWC probably occur due to mechanisms of WUE, such as stomatal control (Rodríguez-Gamir et al. 2019), osmotic adjustment (Blum 2017) and transcriptional reprogramming of AQPs (Moshelion et al. 2015).
Indeed, the ability to maintain water status is crucial for maintaining photosynthetic activity under salinity (Munns and Tester 2008). In our study, C. procera had robust photosynthetic performance in the face of higher stomatal resistance; thus, WUE increased under salinity. Opposite responses of A and gs presuppose the influence of non-stomatal factors on photosynthetic rates. The species studied here showed similar behavior in comparison with those in previous studies under water deficit (Rivas et al. 2017). This work argues that the high photosynthesis rate of C. procera was supported by an increased mesophilic conductance and electron transport under water deficit. Stomatal control is a plant acclimatization response to low water availability, favoring WUE in C. procera (Rivas et al. 2017). The high CO2 assimilation in C. procera at the onset of salinity and increased root soluble sugars could support water uptake.
The second question, was “are roots of C. procera capable of producing quick responses to adjust primary metabolites soon after exposure to high salinity?” Increased contents of organic solutes such as soluble sugars, amino acids, and proteins can play a role in osmoregulation contributing to salt tolerance in plants (Blum 2017; Sami et al. 2016). Under salinity conditions, C. procera exhibited a quick changes in the dynamics of root organic solutes. After the first two hours under salinity stress, the plants showed the highest SS and FAA contents. The quick responses of these metabolites suggest plasticity and contribute to the clarification of the salinity tolerance of this species (Slama et al. 2015).
Rapid exposure to salinity is not common in the wild, where salinity occurs progressively. However, studies such as ours can demonstrate the ability of a species to use efficient mechanisms very quickly. Amino acids were strongly regulated under exposure to salt. A recent study on C. procera metabolic and transcriptomic responses to saline conditions showed evident participation of different amino acids, as an acclimatization response over time (Mutwakil et al. 2017).
Soluble sugars are another important class of metabolites recognized for having numerous functions, mediating vital physiological processes in plants (Sami et al. 2016). The solutes involved in carbon metabolism significantly increased as an initial response to salt exposure. Accumulation of carbohydrates can alleviate the negative effects of excess salt by acting on osmotic homeostasis, eliminating free radicals and stabilizing membranes (Sami et al. 2016). Additionally, sugars are related to stress signaling by modulating gene expression (Martínez-Noël and Tognetti 2018), reducing Na+ and increasing K+ uptake under salinity (Sami et al. 2016). Another important aspect of sugar metabolism is its role in sensing embolism in xylem vessels. When there is difficulty in the uptake of water from the soil, for example, under water deficit or high salinity conditions, the risk of embolism in xylem vessels increases and thus further damages the water supply for shoots. The mechanism by which plants can perceive embolism has been investigated. Sugars play an important role in triggering the chain of events that end by refilling xylem vessels (Secchi and Zwieniecki 2011). Indeed, C. procera plants were able to maintain RWC even under salinity conditions.
The third question was “how long does it take for C. procera roots to change AQP activity in response to salinity?” Once solutes have accumulated and a favorable water potential gradient has been created for hydraulic conductivity in the whole plant, it is necessary to facilitate the water flow through the tissues. Water flow is controlled by the activity of AQPs, and this mechanism is essential for growth in saline environments because these proteins play an important physiological role in water status, controlling hydraulic conductivity in the whole plant, water uptake of roots from the soil solution, and stomatal conductance (Li et al. 2014; Moshelion et al. 2015).
AQPs from C. procera roots showed a differential response to salinity. Five cpTIP isoforms were up-regulated and five were down-regulated. TIP members are located mainly on the vacuole membrane (tonoplast) and show high activity in water transport (Maurel et al. 2008; Wallace and Roberts 2004). In addition to water, TIPs are capable of transporting glycerol, urea, ammonia, and CO2. The overexpression of TIPs has been associated with increased salt tolerance in several plant species. Overexpression of PgTIP1 in soybeans promotes the upregulation of NHX, SOS1, CAT1, and APX1, contributing to ion compartmentalization in the vacuole and antioxidant defense under salinity (An et al. 2017). Furthermore, transformed Arabidopsis plants overexpressing SlTIP2.2 show increased activity of antioxidant enzymes (CAT, SOD, POD), K+/Na+ ratio, and tolerance to salt stress (Xin et al. 2014).
CpPIP isoforms were also upregulated in C. procera roots, but to a lesser extent than CpTIP1 isoforms. The PIP subfamily acts on water transport with high efficiency, controlling almost the entire membrane water permeability potential by the members PIP1 and PIP2 (Bellati et al. 2010). On the other hand, when the expression of PIPs is inhibited, there is a reduction in water transport in roots, hydraulic conductivity, and leaf water potential, and a delay in plant recovery after water deficit. Additionally, the overexpression of GmPIP1.6 in roots confers tolerance to salt in soybeans by promoting the maintenance of hydraulic conductivity, increased CO2 assimilation, and growth (Zhou et al. 2014). Indeed, the overexpression of PIP2.7 induces a six-fold increase in the hydraulic conductivity of roots in A. thaliana conditioned to salt stress (Pou et al. 2016).
The AQP isoforms in C. procera had a transient regulation in relation to exposure time to NaCl. Temporal variation in the expression of AQPs was also observed in rice roots under salt stress (Li et al., 2014). The transcriptional and post-transcriptional regulation of AQP isoforms is complex mechanisms (Kapilan et al. 2018), although, among the AQPs studied here, it was possible to trace a pattern of temporal expression, with upregulation at 2 h and downregulation after 8 and 24 h of exposure to salinity.
Previous studies have shown the importance of inducing AQPs to maintain the water status of plant species (Pou et al. 2016; Zhou et al. 2014;) by the coordinated expression of PIPs and TIPs. This is a key point for improving cell water flow and water uptake in roots (Maurel et al. 2015). Indeed, tomato plants that maintain PIP1 abundance are more tolerant to salt stress than those genotypes with a decreased AQP presence (Han et al. 2020). On the other hand, the downregulation of AQPs negatively influences hydraulic conductivity and stomatal conductance, thus relieving the transpiration rate, which in turn favors the conservation of water in shoots (Moshelion et al. 2015; Skorupa-Kłaput et al. 2015). These proteins also play a role in the hydraulic control of xylem parenchyma cells, highlighting their participation in maintaining xylem transport capacity and restoring vessel functionality after embolism events (Secchi et al. 2017).
At a high salt concentration (200 mM, NaCl), there was upregulation of CpPIPs and CpTIPs (especially CpPIP2.4, CpTIP1.4, and CpTIP1.3), possibly indicating its involvement in adjusting cell water flow (vacuole-cytosol-apoplast) and increasing water uptake by the root system (Maurel et al. 2015). On the other hand, we propose that the downregulation of some AQPs, especially CpPIP1.2, CpTIP4.1, and CpTIP1.4 at 8 h, and CpPIP1.2, CpTIP2.2, and CpTIP1.3, after 24 h of exposure to NaCl, can be a strategy used by C. procera to prevent excess salt contact with the root system, which quickly reaches the shoots (Skorupa-Kłaput et al. 2015). Therefore, previous studies have argued that lower aquaporin activity can be offset by an increase in the number of proteins in the plasma membrane. Thus, the intensity of uptake water is increased without greater activity in each of the channels and the dilution of NaCl within the root cells is maintained (Muries et al. 2011). Most studies do not measure protein synthesis and expression at the same time, but those who did found varied responses, which may be antagonistic or not. This would be due to the stress intensity and duration imposed, as well as the species studied, mainly under saline stress (Yepes-Molina et al. 2020). Under another abiotic stress, such as caused by low temperature in corn plants, the recovery of root hydraulic conductivity is pointed out by the authors due to the greater activity and/or abundance of aquaporins (Aroca et al. 2005).
The plasticity in modulating the primary metabolism and gene expression of AQPs supports the robustness of the C. procera root system in response to osmotic and ionic stresses generated by salt stress (Munns and Tester 2008; Shavrukov 2013). Such tolerance mechanisms can act locally to protect cellular processes and maintain the activity of the root system through osmoregulation, detoxification of reactive oxygen species, and storage of cytosolic sodium (An et al. 2017; Han et al. 2020; Mansour and Ali 2017; Sami et al. 2016). It can regulate the water homeostasis of the entire plant (Maurel et al. 2015; Pou et al. 2016; Rodríguez-Gamir et al. 2019).
In summary, root physiological responses of C. procera under salinity conditions show that high salt concentrations cause a quick response detectable by analyzing physiological parameters of young plants under greenhouse conditions. Exposure to high NaCl concentrations (200 mM) did not affect the photosynthesis and shoot water status in a short time (24 h exposed to salt). Our first hypothesis was that C. procera root system would show biochemical responses that allowed it to promote osmoregulation through the accumulation of organic solutes. Quick changes in the roots of treated plants suggested that C. procera metabolites, such as soluble sugars and free amino acids, might be involved in osmoregulation to tolerate osmotic stress. The second hypothesis was that under high salt conditions, the upregulation of the gene expression of PIPs and TIPs would occur to promote the adjustment of plant water status and maintain the water uptake by the root system, maintaining leaf RWC. At the beginning of salinity exposure, there was no difference in RWC between the treated and control plants. The temporal modulation of CpPIP and CpTIP expressions support the plasticity of the root system in water uptake to mitigate the decreasing water availability in the soil and cellular dehydration generated by the salinized soil. Finally, we concluded that strong stomatal control associated with high amino acid and soluble sugar contents, as well as the regulation of AQP expressions in roots, support the high performance of the root system and alleviates C. procera leaf metabolism.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
This work was supported by the National Council for Scientific and Technological Development (CNPq) [CNPq-470247/2013-4; 310871/2014-0; and 433931/2018-3]. This study was financed in part by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior – Brasil (CAPES) (Finance Code 001) for scholarship to M.R.C. M.G.S. and A.M.B.I. recognize CNPq for fellowships and financial support. A.M.B.I. recognizes the CAPES BioComputacional Program [88882.160046 / 2013-01] for financial support.
Author contributions
MRC and RR conducted the experiment and performed the measurements. MRC analyzed the data and wrote the initial draft first version of the manuscript. MS is the advisor of MRC and participated in the planning of the study, data analysis, and writing of the manuscript. JRCF-N, VP, and AMB-I participated in the molecular data analysis, discussions about results, and helped in the writing of the final manuscript.
Compliance with ethical standards
Conflict of interest
No conflict of interest exists in the submission of this manuscript, and the manuscript has been approved by all authors for publication.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- An J, Hu Z, Che B, Chen H, Yu B, Cai W. Heterologous expression of Panax ginseng PgTIP1 confers enhanced salt tolerance of soybean cotyledon hairy roots, composite, and whole plants. Front Plant Sci. 2017;8:1232. doi: 10.3389/fpls.2017.01232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aroca R, Amoedo G, Fernandéz-Ilescas S, Herman EM, Chaumont F, Chrispeels M. The role of aquaporins and membrane damage in chilling and hydrogen peroxide induced changes in the hydraulic conductance of maize roots. Plant Physiol. 2005;137:341–353. doi: 10.1104/pp.104.051045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrs HD, Weatherley PE. A re-examination of the relative turgidity technique for estimating water deficits in leaves. Aust J Biol Sci. 1962;15:413–428. doi: 10.1071/bi9620413. [DOI] [Google Scholar]
- Bellati J, Alleva K, Soto G, Vitali V, Josefkowicz C, Amoedo G. Intracellular pH sensing is altered by plasma membrane PIP aquaporin co-expression. Plant Mol Biol. 2010;74:105–118. doi: 10.1007/BF00018060. [DOI] [PubMed] [Google Scholar]
- Bezerra-Neto P, Araújo FC, Ferreira-Neto JRC, Silva MD, Pandolfi V, Aburjaile FF, Sakamoto T, Silva RLO, Kido EA, Amorim LLB, Ortega JM, Benko-Iseppon AM. Plant Aquaporins: diversity, evolution and biotechnological applications. Curr Protein Pept Sci. 2019;20:368–395. doi: 10.1007/s11103-010-9658-8. [DOI] [PubMed] [Google Scholar]
- Biassoni R. Quantitative Real-Time PCR: Methods and Protocols. In: Biassoni R, Raso A, editors. Methods in Molecular Biology. New York: Springer; 2014. p. 1160. [Google Scholar]
- Blum A. Osmotic adjustment is a prime drought stress adaptive engine in support of plant production. Plant Cell Environ. 2017;40:4–10. doi: 10.1111/pce.12800. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Campbell GS, Norman JM. An introduction to environmental biophysics. New York: Springer; 1998. [Google Scholar]
- Coêlho MRV, Rivas R, Ferreira-Neto JCR, Pandolfi V, Bezerra-Neto JP, Benko-Iseppon AM, Santos MG. Reference genes selection for Calotropis procera under different salt stress conditions. PLoS ONE. 2019;14:e0215729. doi: 10.1371/journal.pone.0215729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyer I, Uozumi N. Potassium channels in plant cells. FEBS J. 2011;278:4293–4303. doi: 10.1111/j.1742-4658.2011.08371.x. [DOI] [PubMed] [Google Scholar]
- Dubois M, Gilles KA, Hamilton JK, Rebers PA, Smith F. Colorimetric method for determination of sugars and related substances. Anal Chem. 1956;28:350–356. doi: 10.1021/ac60111a017. [DOI] [Google Scholar]
- Flexas J, Diaz-Espejo A, Conesa MA, Coopman RE, Douthe C, Gago J, Gallé A, Galmés J, Medrano H, Ribas-Carbo M, Tomàs M, Niinemets U. Mesophyll conductance to CO2 and Rubisco as targets for improving intrinsic water use efficiency in C3 plants. Plant Cell Environ. 2016;39:965–982. doi: 10.1111/pce.12622. [DOI] [PubMed] [Google Scholar]
- Han W, Jia J, Hu Y, Liu J, Guo J, Shi Yu, Huo H, Gong H. Maintenance of root water uptake contributes to salt-tolerance of a wild tomato species under salt stress. Arch Agron Soil Sci. 2020 doi: 10.1080/03650340.2020.1720911. [DOI] [Google Scholar]
- Hassan LM, Galal TM, Farahat EA, El-Midany MM. The biology of Calotropis procera (Aiton) W.T. Trees. 2015;29:311–320. doi: 10.1007/s00468-015-1158-7. [DOI] [Google Scholar]
- Johanson U, Karisson M, Johansson I, Gustavsson S, Sjovall S, Fraysse L, Weig AR, Kjellbom P. The Complete set of genes encoding major intrinsic proteins in Arabidopsis provides a framework for a new nomenclature for major intrinsic proteins in plants. Plant Physiol. 2001;126:1358–1369. doi: 10.1104/pp.126.4.1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapilan R, Vaziri M, Zwiazek JJ. Regulation of aquaporins in plants under stress. Biol Res. 2018 doi: 10.1186/s40659-018-0152-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Santoni V, Maurel C. Plant aquaporins: Roles in plant physiology. Biochimica et Biophysica Acta (BBA) - General Subjects. Aquaporins. 2014;1840:1574–1582. doi: 10.1016/j.bbagen.2013.11.004. [DOI] [PubMed] [Google Scholar]
- Lichtenthaler H, Buschmann C (2001) Chlorophylls and carotenoids: measurement and characterization by UV‐VIS spectroscopy. In: Current protocols in food analytical chemistry, pp 1–8. Doi: 10.1002/0471142913.faf0403s01
- Liu C, Li C, Liang D, Wei Z, Zhou S, Wang R, Ma F. Differential expression of ion transporters and aquaporins in leaves may contribute to different salt tolerance in Malus species. Plant Physiol Biochem. 2012;58:159–165. doi: 10.1016/j.plaphy.2012.06.019. [DOI] [PubMed] [Google Scholar]
- Mansour MMF, Ali EF. Evaluation of proline functions in saline conditions. Phytochemistry. 2017;140:52–68. doi: 10.1016/j.phytochem.2017.04.016. [DOI] [PubMed] [Google Scholar]
- Martínez-Noël GMA, Tognetti JA. Sugar signaling under abiotic stress in plants. In: Ahmad P, Ahanger MA, Singh VP, Tripathi DK, Alam P, Alyemeni MN, editors. Plant metabolites and regulation under environmental stress. New York: Academic Press; 2018. pp. 397–406. [Google Scholar]
- Maurel C, Verdouck L, Luu DT, Santoni V. Plant aquaporins: membrane channels with multiple integrated functions. Ann Rev Plant Biol. 2008;59:595–624. doi: 10.1146/annurev.arplant.59.032607.092734. [DOI] [PubMed] [Google Scholar]
- Maurel C, Boursiac Y, Luu DT, Santoni V, Shahzad Z, Verdoucq L. Aquaporins in plants. Physiol Rev. 2015;95:1321–1358. doi: 10.1152/physrev.00008.2015. [DOI] [PubMed] [Google Scholar]
- Moore S, Stein WH. Photometric ninhydrin method for use in the chromatography of amino acids. J Biol Chem. 1948;176:367–388. doi: 10.1016/S0021-9258(18)51034-6. [DOI] [PubMed] [Google Scholar]
- Moshelion M, Halperin O, Wallach R, Oren R, Way D. Role of aquaporins in determining transpiration and photosynthesis in water-stressed plants: crop water-use efficiency, growth and yield. Plant Cell Environ. 2015;38:1785–1793. doi: 10.1111/pce.12410. [DOI] [PubMed] [Google Scholar]
- Munns R, Tester M. Mechanisms of salinity tolerance. Annu Rev Plant Biol. 2008;59:651–681. doi: 10.1146/annurev.arplant.59.032607.092911. [DOI] [PubMed] [Google Scholar]
- Muries B, Faize M, Carvajal M, Martínez-Ballesta MC. Identification and differential induction of the expression of aquaporins by salinity in broccoli plants. Mol BioSyst. 2011;7:1322–1335. doi: 10.1039/c0mb00285b. [DOI] [PubMed] [Google Scholar]
- Mutwakil MZ, Hajrah NH, Atef A, Edris S, Sabir MJ, Al-Ghamdi AK, Sabir MJSM, Nelson C, Makki RM, Ali HM, El-Domyati FM, Al-Hajar ASM, Gloaguen Y, Al-Zahrani HS, Sabir JSM, Jansen RK, Bahieldin A, Hall N. Transcriptomic and metabolic responses of Calotropis procera to salt and drought stress. BMC Plant Biol. 2017;17:231. doi: 10.1186/s12870-017-1155-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW, Horgan GW, Dempfle L. Relative expression software tool (REST©) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucl Acids Res. 2002;30:e36. doi: 10.1093/nar/30.9.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pou A, Jeanguenin L, Milhiet T, Batoko H, Chaumont F, Hachez C. Salinity-mediated transcriptional and post-translational regulation of the Arabidopsis aquaporin PIP2;7. Plant Mol Biol. 2016;92:731–744. doi: 10.1007/s11103-016-0542-z. [DOI] [PubMed] [Google Scholar]
- Rasmussen R (2001) Quantification on the LightCycler. In: Meuer S, Wittwer C, Nakagawara K-I (Eds) Rapid cycle real-time PCR: methods and applications. Springer, Berlin, pp 21–34. Doi: 10.1007/978-3-642-59524-0_3
- Rivas R, Barros V, Falcão H, Frosi G, Arruda E, Santos M. Ecophysiological traits of invasive C3 species Calotropis procera to maintain high photosynthetic performance under high VPD and low soil water balance in semi-arid and seacoast zones. Front Plant Sci. 2020;11:717. doi: 10.3389/fpls.2020.00717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas R, Frosi G, Ramos DG, Pereira S, Benko-Iseppon AM, Santos MG. Photosynthetic limitation and mechanisms of photoprotection under drought and recovery of Calotropis procera, an evergreen C3 from arid regions. Plant Physiol Biochem. 2017;118:589–599. doi: 10.1016/j.plaphy.2017.07.026. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Gamir J, Xue J, Clearwater MJ, Meason DF, Clinton PW, Domec JC. Aquaporin regulation in roots controls plant hydraulic conductance, stomatal conductance, and leaf water potential in Pinus radiata under water stress. Plant Cell Environment. 2019;42:717–729. doi: 10.1111/pce.13460. [DOI] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. In: Krawetz S, Misener S, editors. Bioinformatics methods and protocols: methods in molecular biology. Totowa: Humana Press; 2000. pp. 365–386. [DOI] [PubMed] [Google Scholar]
- Sami F, Yusuf M, Faizan M, Faraz A, Hayat S. Role of sugars under abiotic stress. Plant Physiol Biochem. 2016;109:54–61. doi: 10.1016/j.plaphy.2016.09.005. [DOI] [PubMed] [Google Scholar]
- Secchi F, Zwieniecki MA. Sensing embolism in xylem vessels: the role of sucrose as a trigger for refilling. Plant, Cell Environ. 2011;34:514–524. doi: 10.1111/j.1365-3040.2010.02259.x. [DOI] [PubMed] [Google Scholar]
- Secchi F, Pagliarani C, Zwieniecki MA. The functional role of xylem parenchyma cells and aquaporins during recovery from severe water stress. Plant Cell Environ. 2017;40:858–871. doi: 10.1111/j.1365-3040.2010.02259.x. [DOI] [PubMed] [Google Scholar]
- Shavrukov Y. Salt stress or salt shock: which genes are we studying? J Exp Bot. 2013;64:119–127. doi: 10.1093/jxb/ers316. [DOI] [PubMed] [Google Scholar]
- Singh RK, Deshmukh R, Muthamilarasan M, Rani R, Prasad M. Versatile roles of aquaporin in physiological processes and stress tolerance in plants. Plant Physiol Biochem. 2020;149:178–189. doi: 10.1016/j.plaphy.2020.02.009. [DOI] [PubMed] [Google Scholar]
- Skorupa-Klaput M, Szczepanek J, Kurnik K, Tretyn A, Tyburski J. The expression patterns of plasma membrane aquaporins in leaves of sugar beet and its halophyte relative, Beta vulgaris ssp. maritima, in response to salt stress. Biologia. 2015;70:467–477. doi: 10.1515/biolog-2015-0056. [DOI] [Google Scholar]
- Slama I, Abdelly C, Bouchereau A, Flowers T, Sauvoré A. Diversity, distribution and roles of osmoprotective compounds accumulated in halophytes under abiotic stress. Ann Bot. 2015;115:433–447. doi: 10.1093/aob/mcu239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Tezara W, Colombo R, Coronel I, Marín O. Water relations and photosynthetic capacity of two species of Calotropis in a tropical semi-arid ecosystem. Annals Botany. 2011;107:397–405. doi: 10.1093/aob/mcq245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas RL, Sheard RW, Moyer JR. Comparison of conventional and automated procedures for nitrogen, phosphorus, and potassium analysis of plant material using a single digestion. Agron J. 1967;59:240–243. doi: 10.2134/agronj1967.00021962005900030010x. [DOI] [Google Scholar]
- Wallace IS, Roberts DM. Homology modeling of representative subfamilies of Arabidopsis major intrinsic proteins. Classification based on the aromatic/arginine selectivity filter. Plant Physiol. 2004;135:1059–1068. doi: 10.1104/pp.103.033415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin S, Yu G, Sun L, Qiang X, Xu N, Cheng X. Expression of tomato SlTIP2;2 enhances the tolerance to salt stress in the transgenic Arabidopsis and interacts with target proteins. J Plant Res. 2014;127:695–708. doi: 10.1007/s10265-014-0658-7. [DOI] [PubMed] [Google Scholar]
- Yepes-Molina L, Barzana G, Carvajal M. Controversial regulation of gene expression and protein transduction of aquaporins under drought and salinity stress. Plants. 2020;9:1662. doi: 10.3390/plants9121662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Wang C, Liu R, Han Q, Vandeleur RK, Du J, Tyerman S. Constitutive overexpression of soybean plasma membrane intrinsic protein GmPIP1;6 confers salt tolerance. BMC Plant Biol. 2014;14:181. doi: 10.1186/1471-2229-14-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.