Abstract
Salinity and submergence are two very prominent abiotic stress conditions affecting rice yield adversely in the coastal agro ecosystem. Marker Assisted Backcross Breeding (MABB) is an efficient and fast track molecular tool to incorporate a desired stress tolerant QTL/gene into an improved cultivar. The present study was carried out for the introgression of Saltol QTL responsible for salinity tolerance and Sub1 gene responsible for submergence tolerance into the high yielding rice variety Aiswarya independently through MABB. Final objective of the study is to develop dual stress tolerant (tolerance to salinity and submergence) Aiswarya rice variety by pyramiding the both target QTLs introgressed BC2F2 progenies having maximum background homozygosity. The donors of Saltol QTL and Sub1 gene used in the present study were FL478 and Swarna Sub1, respectively. Based on the background genome analysis of the introgressed plants, the plants with > 85–90% background similarity were selected for pyramiding of Saltol QTL and Sub1 gene into the elite background of rice variety Aiswarya. Those selected introgressed lines with Saltol QTL and Sub1 gene will be again crossed to pyramid both Saltol QTL and Sub1 gene into the rice variety Aiswarya. Such a mega rice variety pyramided with dual stress tolerant QTLs is the expected outcome of this study and can be recommended for cultivation in the flood prone saline coastal agroecosystem.
Keywords: Marker assisted breeding, Saltol QTL, Sub1 gene, SSR markers, Salinity tolerance, Submergence tolerance
Introduction
Rice, (Oryza sativa L.) is the most important food crop in India and is the staple food for half of the world’s population and providing 30–80% of the daily calories required in Asian population (Hossain and Narciso 2002). In developing countries, rice is the principal source of calories and nutrients. Various abiotic stresses such as drought, salinity, submergence, and extreme temperature cause a threatening impact on the growth and productivity of rice (Gregorio et al. 2013). Among these, salinity and submergence are the major issues faced by farmers of coastal agroecosystem. Farmers suffer from crop losses caused by the intrusion of saline water into rice fields and occasional flash floods during the monsoon season. Submergence stress due to unexpected heavy rains adversely affected rice crops and reduced the grain yield drastically (Sarkar and Bhattacharjee 2011). Rice can be susceptible to salinity at the seedling and reproductive stages (Lutts et al. 1995). The flowering stage is also a highly sensitive stage that is affected by salinity stress (Singh et al. 2007) and yield potential at the mature stage was affected due to salinity (Todaka et al. 2012). Salinity affects yield parameters such as panicle emergence, flowering, panicle length, spikelet number per panicle, and grain yield (Thomson et al. 2010). Submergence affects many plant physiological processes such as water absorption, respiration, photosynthetic activity and substantially reduce crop productivity (Fukao et al. 2011).
Thus, to tackle this abiotic stress induced yield reductions in stress fields; a permanent and sustainable approach is required. The most viable solution is to introgress the genes for abiotic stress tolerance into already developed improved high yielding rice cultivar through MABB. It has been identified that salinity tolerance is controlled by quantitative trait locus (QTL) on chromosome 1 which is responsible for low Na+ high K+ uptake (Mohammadi et al. 2008) and submergence tolerance is controlled by quantitative trait locus (QTL) on chromosome 9 (Neeraja et al. 2007). MABB is a precise and effective method to introgress the trait of interest without altering the essential characters of elite varieties. The objective of the present study was to improve the rice cultivar Aiswarya for salinity and submergence tolerance using the MABB strategy. In this paper, the parallel introgression of Saltol QTL and Sub1 gene through MABB strategy are discussed in detail which is a pre-requisite for pyramiding of these two abiotic stress tolerance QTLs, into the rice variety Aiswarya.
Materials and methods
Rice varieties – recurrent and donor parents
The saline tolerant rice breeding line FL-478, developed at IRRI (The International Rice Research Institute 1996) was used as the donor of Saltol QTL. The donor of Sub1 gene used was the Swarna Sub1, submergence tolerant rice variety developed at International Rice Research Institute, manila, Philippines. The recurrent parent used for improvement in this study is a high yielding rice cultivar, Aiswarya, developed at the Regional Agricultural Research Station, Pattambi, which is sensitive to salinity and submergence with an average yield of 6.3—6.5 tons/ha.
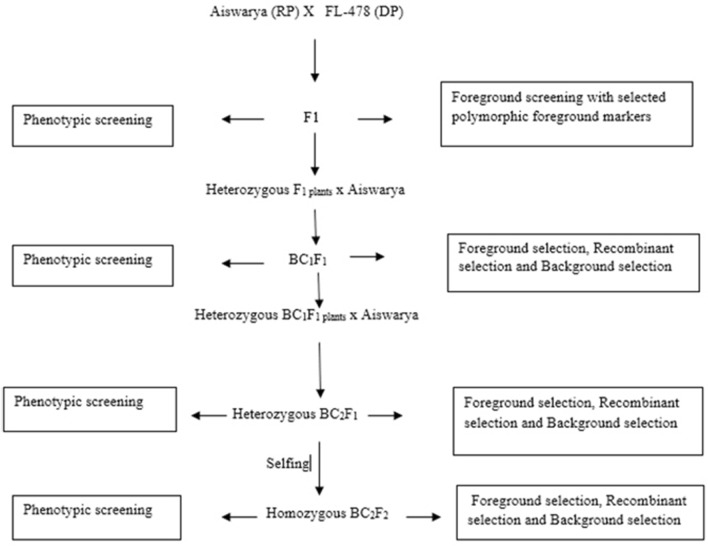
Breeding scheme for Saltol QTL introgression
The recurrent parent (RP) Aiswarya and donor parent (DP) FL-478 were crossed and F1 generation was raised. In case of Saltol QTL introgression, in each generation both phenotypic screening and genotypic screening were done, and the QTL introgressed plants were advanced into next generation. In the F1 generation genotypic screening was done in the phenotypically survived plants under saline condition and plants with heterozygous loci for the target trait alone were selected and backcrossed with the respective recurrent parent and raised BC1F1 generation. The heterozygous plants with the target locus in the BC1F1 generation were again selected for backcrossing with the respective recurrent parent to raise BC2F1 progenies. The heterozygous BC2F1 plants were selected to raise BC2F2 progenies by selfing and the homozygous plants in the target locus alone were selected from the selfed BC2F2 progenies (Fig. 1). After the Phenotyping of the BC2F2 progenies the plants with Score-3 (SES, IRRI) alone were selected for genotyping. The SES is a plant score of 1–9 spectrum based on morphological manifestation/stress symptoms under stress conditions where a lower SES score shows resistance and a higher score shows a sensitive response (Table 1).
Fig. 1.
Breeding scheme of Saltol QTL Introgression into mega rice variety Aiswarya, as a Recurrent Parent (RP) and Saltol- FL-478, as a Donor Parent (DP)
Table 1.
Standard evaluation system (SES) score for salinity tolerance (IRRI 1996)
| Score | Observation | Tolerance |
|---|---|---|
| 1 | Nearly normal growth, no leaf symptoms | Highly Tolerant |
| 3 | Nearly normal growth, but leaf tips or few leaves whitish and rolled | Tolerant |
| 5 | Growth severely retarded; most leaves rolled; only a few are elongating | Moderately Tolerant |
| 7 | Complete cessation of growth; most leaves dry; some plants dying | Susceptible |
| 9 | Almost all plants dead or dying | Highly Susceptible |
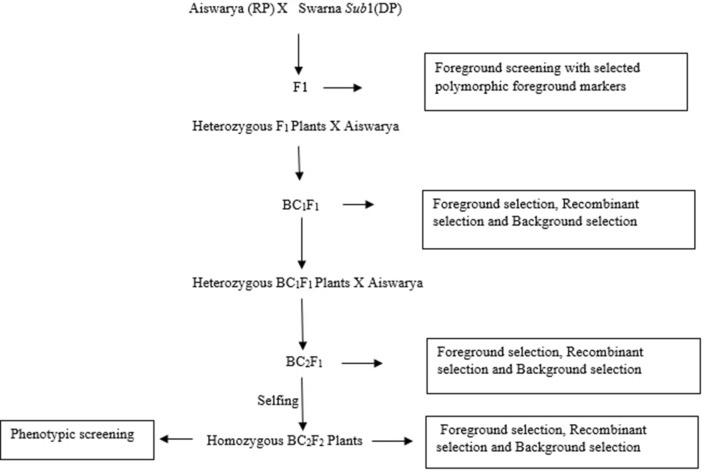
Breeding scheme for Sub 1 gene introgression
The recurrent parent (RP) Aiswarya and donor parent (DP) Swarna Sub1 were crossed and F1 generation was raised. In case of Sub1gene introgression the screening in each generation was done by genotyping and phenotypic screening was done in the final stage. In the F1 generation genotypic screening was done and plants with heterozygous loci for the target trait alone were selected and backcrossed with the respective recurrent parent and raised BC1F1 generation. The heterozygous plants with the target locus in the BC1F1 generation were again selected for backcrossing with the respective recurrent parent to raise BC2F1 progenies. The heterozygous BC2F1 plants were selected to raise BC2F2 progenies by selfing and the homozygous plants in the target locus alone were selected from the selfed BC2F2 progenies. After the Phenotyping of the BC2F2 progenies the plants with Score-3 (SES, IRRI) alone were selected (Fig. 2).
Fig. 2.
Breeding scheme of Sub1gene Introgression into mega rice variety Aiswarya, as a Recurrent Parent (RP) and Swarna Sub1, as a Donor Parent (DP)
DNA isolation and PCR assay
The young leaf samples were collected, and genomic DNA was extracted using CTAB method (Doyle and Doyle 1987) The concentration of DNA was measured using Nano Drop 2000c (Thermo Scientific). Target gene sequence was amplified by PCR assay, in which the total volume of reaction is 20 μl containing 2μlDNA template, 10μlPCR Master Mix (Thermo Scientific) consist of 1XTaq Buffer, Taq DNA polymerase, MgCl2, 1 μl each of reverse and forward primer along with 6 μl water. To begin the PCR program the initial denaturation temperature was raised to 95°C then annealing temperature lowered to 55–65°C and extension temperature at 72°C. The cycle of changing temperature was then repeated up to 30–35 cycles.
Quality control with electrophoresis and DNA visualization
The DNA quality was tested by 8% Polyacrylamide gel electrophoresis (PAGE). Silver stain method were used for staining gels and were visualized using Gel Doc (Bio-Rad Gel Doc XR +). Selected SSR markers were used for foreground, recombinant and background selection.
Polymorphism assay and selection of SSR markers
Simple sequence repeats are co-dominant markers which occur at high frequency and they prominent throughout the genome. SSR markers shows a high level of polymorphism between the plant varieties. A total of 545 SSR markers were screened for polymorphism between the recurrent parent and donor parents, which is narrated in Table 10.
Table 10.
Genome wide SSR markers used for parental polymorphism assay
| Sl. No | Primer name | Chr. No | Sl. No | Primer name | Chr. No | Sl. No | Primer name | Chr. No |
|---|---|---|---|---|---|---|---|---|
| 1 | RM499 | 1 | 27 | RM140 | 1 | 53 | RM5536 | 1 |
| 2 | RM3252 | 1 | 28 | RM595 | 1 | 54 | RM315 | 1 |
| 3 | RM323 | 1 | 29 | RM129 | 1 | 55 | RM472 | 1 |
| 4 | RM476A | 1 | 30 | RM113 | 1 | 56 | RM431 | 1 |
| 5 | RM10209 | 1 | 31 | RM562 | 1 | 57 | RM3362 | 1 |
| 6 | RM1 | 1 | 32 | RM24 | 1 | 58 | RM6840 | 1 |
| 7 | RM220 | 1 | 33 | RM9 | 1 | 59 | RM165 | 1 |
| 8 | RM6289 | 1 | 34 | RM5 | 1 | 60 | OSR23 | 1 |
| 9 | RM283 | 1 | 35 | RM306 | 1 | 61 | RM104 | 1 |
| 10 | RM151 | 1 | 36 | RM488 | 1 | 62 | RM529 | 1 |
| 11 | RM576 | 1 | 37 | RM1196 | 1 | 63 | RM414 | 1 |
| 12 | RM490 | 1 | 38 | RM246 | 1 | 64 | RM14 | 1 |
| 13 | RM575 | 1 | 39 | RM237 | 1 | 65 | RM568 | 1 |
| 14 | RM259 | 1 | 40 | RM443 | 1 | 66 | RM3340 | 2 |
| 15 | RM35 | 1 | 41 | RM403 | 1 | 67 | OSR17 | 2 |
| 16 | RM243 | 1 | 42 | RM128 | 1 | 68 | RM154 | 2 |
| 17 | RM583 | 1 | 43 | RM543 | 1 | 69 | OSR14 | 2 |
| 18 | RM600 | 1 | 44 | RM302 | 1 | 70 | RM279 | 2 |
| 19 | RM578 | 1 | 45 | RM212 | 1 | 71 | RM279 | 2 |
| 20 | RM292 | 1 | 46 | RM102 | 1 | 72 | RM1075 | 2 |
| 21 | RM572 | 1 | 47 | RM476B | 1 | 73 | RM1347 | 2 |
| 22 | RM580 | 1 | 48 | RM486 | 1 | 74 | RM8 | 2 |
| 23 | RM312 | 1 | 49 | RM226 | 1 | 75 | RM555 | 2 |
| 24 | RM23 | 1 | 50 | RM265 | 1 | 76 | RM174 | 2 |
| 25 | RM81A | 1 | 51 | RM297 | 1 | 77 | RM1352 | 2 |
| 26 | RM238A | 1 | 52 | RM8236 | 1 | 78 | RM71 | 2 |
| 79 | RM1081 | 2 | 120 | RM406 | 2 | 161 | RM5959 | 3 |
| 80 | RM492 | 2 | 121 | RM213 | 2 | 162 | RM503 | 3 |
| 81 | RM5812 | 2 | 122 | RM482 | 2 | 163 | RM135 | 3 |
| 82 | RM452 | 2 | 123 | RM207 | 2 | 164 | RM426 | 3 |
| 83 | RM550 | 2 | 124 | RM266 | 2 | 165 | RM6135 | 3 |
| 84 | RM438 | 2 | 125 | RM498 | 2 | 166 | RM532 | 3 |
| 85 | RM5101 | 2 | 126 | RM535 | 2 | 167 | RM504 | 3 |
| 86 | RM4499 | 2 | 127 | RM7382 | 2 | 168 | RM1221 | 3 |
| 87 | RM465A | 2 | 128 | RM81B | 3 | 169 | RM203 | 3 |
| 88 | RM324 | 2 | 129 | RM60 | 3 | 170 | RM7324 | 3 |
| 89 | RM262 | 2 | 130 | RM132 | 3 | 171 | RM186 | 3 |
| 90 | RM561 | 2 | 131 | RM22 | 3 | 172 | RM55 | 3 |
| 91 | RM1303 | 2 | 132 | RM523 | 3 | 173 | RM168 | 3 |
| 92 | RM341 | 2 | 133 | RM569 | 3 | 174 | RM448 | 3 |
| 93 | RM5430 | 2 | 134 | RM231 | 3 | 175 | RM3583 | 3 |
| 94 | RM475 | 2 | 135 | RM175 | 3 | 176 | RM416 | 3 |
| 95 | RM3352 | 2 | 136 | RM489 | 3 | 177 | RM520 | 3 |
| 96 | RM1385 | 2 | 137 | RM545 | 3 | 178 | RM293 | 3 |
| 97 | RM1367 | 2 | 138 | RM36 | 3 | 179 | RM227 | 3 |
| 98 | RM3220 | 2 | 139 | OSR16 | 3 | 180 | RM468 | 3 |
| 99 | RM106 | 2 | 140 | RM517 | 3 | 181 | RM571 | 3 |
| 100 | RM263 | 2 | 141 | RM546 | 3 | 182 | RM422 | 3 |
| 101 | RM3275 | 2 | 142 | RM218 | 3 | 183 | RM422 | 3 |
| 102 | RM526 | 2 | 143 | RM3204 | 3 | 184 | RM143 | 3 |
| 103 | RM599 | 2 | 144 | RM232 | 3 | 185 | RM130 | 3 |
| 104 | RM5607 | 2 | 145 | RM251 | 3 | 186 | RM114 | 3 |
| 105 | RM221 | 2 | 146 | RM563 | 3 | 187 | RM514 | 3 |
| 106 | RM525 | 2 | 147 | RM5626 | 3 | 188 | RM570 | 3 |
| 107 | RM573 | 2 | 148 | RM554 | 3 | 189 | RM148 | 3 |
| 108 | RM1092 | 2 | 149 | RM282 | 3 | 190 | RM442 | 3 |
| 109 | RM5421 | 2 | 150 | RM338 | 3 | 191 | RM85 | 3 |
| 110 | RM318 | 2 | 151 | RM473D | 3 | 192 | RM307 | 4 |
| 111 | RM6 | 2 | 152 | RM2614 | 3 | 193 | RM2146 | 4 |
| 112 | RM3650 | 2 | 153 | RM156 | 3 | 194 | RM5414 | 4 |
| 113 | RM240 | 2 | 154 | RM411 | 3 | 195 | RM551 | 4 |
| 114 | RM7485 | 2 | 155 | RM487 | 3 | 196 | RM401 | 4 |
| 115 | RM425 | 2 | 156 | RM16 | 3 | 197 | RM537 | 4 |
| 116 | RM250 | 2 | 157 | RM347 | 3 | 198 | RM6770 | 4 |
| 117 | RM1063 | 2 | 158 | RM5172 | 3 | 199 | RM3471 | 4 |
| 118 | RM166 | 2 | 159 | RM6213 | 3 | 200 | RM5953 | 4 |
| 119 | RM208 | 2 | 160 | RM319 | 3 | 201 | RM335 | 4 |
| 202 | RM518 | 4 | 243 | RM413 | 5 | 284 | RM190 | 6 |
| 203 | RM3317 | 4 | 244 | RM194 | 5 | 285 | RM510 | 6 |
| 204 | RM261 | 4 | 245 | RM1089 | 5 | 286 | RM111 | 6 |
| 205 | RM456B | 4 | 246 | RM5994 | 5 | 287 | RM276 | 6 |
| 206 | RM185 | 4 | 247 | RM6229 | 5 | 288 | RM402 | 6 |
| 207 | RM177 | 4 | 248 | RM169 | 5 | 289 | RM6467 | 6 |
| 208 | RM471 | 4 | 249 | RM509 | 5 | 290 | RM527 | 6 |
| 209 | RM417 | 4 | 250 | RM1237 | 5 | 291 | RM5745 | 6 |
| 210 | RM1155 | 4 | 251 | RM598 | 5 | 292 | RM1161 | 6 |
| 211 | RM142 | 4 | 252 | RM465C | 5 | 293 | RM6818 | 6 |
| 212 | RM119 | 4 | 253 | RM249 | 5 | 294 | RM5753 | 6 |
| 213 | RM1136 | 4 | 254 | RM146 | 5 | 295 | RM541 | 6 |
| 214 | RM5320 | 4 | 255 | RM39 | 5 | 296 | RM1925 | 6 |
| 215 | RM273 | 4 | 256 | RM291 | 5 | 297 | RM454 | 6 |
| 216 | RM1703 | 4 | 257 | RM430 | 5 | 298 | RM162 | 6 |
| 217 | RM252 | 4 | 258 | RM473B | 5 | 299 | RM275 | 6 |
| 218 | RM456A | 4 | 259 | RM1090 | 5 | 300 | RM343 | 6 |
| 219 | RM3217 | 4 | 260 | RM440 | 5 | 301 | RM1340 | 6 |
| 220 | RM241 | 4 | 261 | RM459 | 5 | 302 | RM5814 | 6 |
| 221 | RM1100 | 4 | 262 | RM1187 | 5 | 303 | RM528 | 6 |
| 222 | RM451 | 4 | 263 | RM161 | 5 | 304 | RM1161 | 6 |
| 223 | RM470 | 4 | 264 | RM305 | 5 | 305 | RM1150 | 6 |
| 224 | RM303 | 4 | 265 | RM173 | 5 | 306 | RM30 | 6 |
| 225 | RM317 | 4 | 266 | RM534 | 5 | 307 | RM340 | 6 |
| 226 | RM5047 | 4 | 267 | RM188 | 5 | 308 | RM439 | 6 |
| 227 | RM1018 | 4 | 268 | RM538 | 5 | 309 | RM461 | 6 |
| 228 | RM255 | 4 | 269 | RM5770 | 5 | 310 | RM103 | 6 |
| 229 | RM348 | 4 | 270 | RM233B | 5 | 311 | RM494 | 6 |
| 230 | RM349 | 4 | 271 | RM6200 | 5 | 312 | RM295 | 7 |
| 231 | RM131 | 4 | 272 | RM421 | 5 | 313 | RM192 | 7 |
| 232 | RM124 | 4 | 273 | RM5968 | 5 | 314 | RM436 | 7 |
| 233 | RM127 | 4 | 274 | RM178 | 5 | 315 | RM427 | 7 |
| 234 | RM280 | 4 | 275 | RM26 | 5 | 316 | RM1132 | 7 |
| 235 | RM567 | 4 | 276 | RM274 | 5 | 317 | RM3555 | 7 |
| 236 | RM559 | 4 | 277 | RM87 | 5 | 318 | RM125 | 7 |
| 237 | RM153 | 5 | 278 | RM480 | 5 | 319 | RM1135 | 7 |
| 238 | RM507 | 5 | 279 | RM334 | 5 | 320 | RM180 | 7 |
| 239 | RM122 | 5 | 280 | RM133 | 6 | 321 | RM3484 | 7 |
| 240 | RM5693 | 5 | 281 | RM508 | 6 | 322 | RM6111 | 7 |
| 241 | RM5796 | 5 | 282 | RM197 | 6 | 323 | RM214 | 7 |
| 242 | RM17960 | 5 | 283 | RM469 | 6 | 324 | RM500 | 7 |
| 325 | RM445 | 7 | 366 | RM44 | 8 | 407 | RM219 | 9 |
| 326 | RM6776 | 7 | 367 | RM32 | 8 | 408 | RM524 | 9 |
| 327 | RM1362 | 7 | 368 | RM72 | 8 | 409 | RM342B | 9 |
| 328 | RM11 | 7 | 369 | RM88 | 8 | 410 | RM5526 | 9 |
| 329 | RM3859 | 7 | 370 | RM195 | 8 | 411 | RM105 | 9 |
| 330 | RM560 | 7 | 371 | RM330B | 8 | 412 | RM321 | 9 |
| 331 | RM182 | 7 | 372 | RM350 | 8 | 413 | RM1817 | 9 |
| 332 | RM336 | 7 | 373 | RM404 | 8 | 414 | RM2144 | 9 |
| 333 | RM351 | 7 | 374 | RM483 | 8 | 415 | RM409 | 9 |
| 334 | RM70 | 7 | 375 | RM1345 | 8 | 416 | RM460 | 9 |
| 335 | RM455 | 7 | 376 | RM137 | 8 | 417 | RM3912 | 9 |
| 336 | RM1085 | 7 | 377 | RM331 | 8 | 418 | RM566 | 9 |
| 337 | RM505 | 7 | 378 | RM1109 | 8 | 419 | RM4692 | 9 |
| 338 | RM1134 | 7 | 379 | RM1235 | 8 | 420 | RM434 | 9 |
| 339 | RM234 | 7 | 380 | RM223 | 8 | 421 | RM410 | 9 |
| 340 | RM18 | 7 | 381 | RM515 | 8 | 422 | RM2482 | 9 |
| 341 | RM47 | 7 | 382 | RM284 | 8 | 423 | RM257 | 9 |
| 342 | RM478 | 7 | 383 | RM6193 | 8 | 424 | RM1896 | 9 |
| 343 | RM118 | 7 | 384 | RM556 | 8 | 425 | RM3700 | 9 |
| 344 | RM134 | 7 | 385 | RM210 | 8 | 426 | RM242 | 9 |
| 345 | RM429 | 7 | 386 | RM531 | 8 | 427 | RM108 | 9 |
| 346 | RM1253 | 7 | 387 | RM419 | 8 | 428 | RM288 | 9 |
| 347 | RM5426 | 7 | 388 | RM256 | 8 | 429 | RM553 | 9 |
| 348 | RM172 | 7 | 389 | RM149 | 8 | 430 | RM278 | 9 |
| 349 | RM420 | 7 | 390 | RM308 | 8 | 431 | RM201 | 9 |
| 350 | RM2381 | 7 | 391 | RM230 | 8 | 432 | RM160 | 9 |
| 351 | RM248 | 7 | 392 | RM433 | 8 | 433 | RM107 | 9 |
| 352 | RM408 | 8 | 393 | RM1309 | 8 | 434 | RM328 | 9 |
| 353 | RM337 | 8 | 394 | RM502 | 8 | 435 | OSR28 | 9 |
| 354 | RM152 | 8 | 395 | RM458 | 8 | 436 | RM189 | 9 |
| 355 | RM1308 | 8 | 396 | RM477 | 8 | 437 | RM3164 | 9 |
| 356 | RM1376 | 8 | 397 | RM447 | 8 | 438 | RM2855 | 9 |
| 357 | RM3481 | 8 | 398 | RM281 | 8 | 439 | RM215 | 9 |
| 358 | RM3262 | 8 | 399 | RM264 | 8 | 440 | RM1026 | 9 |
| 359 | RM25 | 8 | 400 | RM41 | 9 | 441 | RM1328 | 9 |
| 360 | RM6471 | 8 | 401 | RM296 | 9 | 442 | RM 245 | 9 |
| 361 | RM5999 | 8 | 402 | RM1553 | 9 | 443 | RM205 | 9 |
| 362 | RM6032 | 8 | 403 | RM5799 | 9 | 444 | RM6364 | 10 |
| 363 | RM6008 | 8 | 404 | RM316 | 9 | 445 | RM474 | 10 |
| 364 | RM310 | 8 | 405 | RM464 | 9 | 446 | RM474 | 10 |
| 365 | RM547 | 8 | 406 | RM464 | 9 | 447 | RM330A | 10 |
| 448 | RM2504 | 10 | 489 | RM552 | 11 | 530 | RM277 | 12 |
| 449 | RM216 | 10 | 490 | RM116 | 11 | 531 | RM27973 | 12 |
| 450 | RM239 | 10 | 491 | RM120 | 11 | 532 | RM519 | 12 |
| 451 | RM311 | 10 | 492 | RM6115 | 11 | 533 | RM1246 | 12 |
| 452 | RM5689 | 10 | 493 | RM479 | 11 | 534 | RM313 | 12 |
| 453 | RM1375 | 10 | 494 | RM5731 | 11 | 535 | RM83 | 12 |
| 454 | RM467 | 10 | 495 | RM536 | 11 | 536 | RM463 | 12 |
| 455 | RM5629 | 10 | 496 | RM7303 | 11 | 537 | RM1986 | 12 |
| 456 | RM6100 | 10 | 497 | RM7120 | 11 | 538 | RM235 | 12 |
| 457 | RM596 | 10 | 498 | RM287 | 11 | 539 | RM270 | 12 |
| 458 | RM1108 | 10 | 499 | RM209 | 11 | 540 | RM1103 | 12 |
| 459 | RM184 | 10 | 500 | RM229 | 11 | 541 | RM2854 | 12 |
| 460 | RM271 | 10 | 501 | RM5961 | 11 | 542 | RM6396 | 12 |
| 461 | RM6128 | 10 | 502 | RM1341 | 11 | 543 | RM1296 | 12 |
| 462 | RM269 | 10 | 503 | RM457 | 11 | 544 | RM17 | 12 |
| 463 | RM258 | 10 | 504 | RM473E | 11 | 545 | RM1227 | 12 |
| 464 | RM5666 | 10 | 505 | RM6965 | 11 | |||
| 465 | RM1374 | 10 | 506 | RM206 | 11 | |||
| 466 | RM171 | 10 | 507 | RM254 | 11 | |||
| 467 | RM304 | 10 | 508 | RM456C | 11 | |||
| 468 | RM3123 | 10 | 509 | RM2136 | 11 | |||
| 469 | RM6016 | 10 | 510 | RM6094 | 11 | |||
| 470 | RM294A | 10 | 511 | RM224 | 11 | |||
| 471 | RM228 | 10 | 512 | RM139 | 11 | |||
| 472 | RM484 | 10 | 513 | RM144 | 11 | |||
| 473 | RM147 | 10 | 514 | RM27172 | 11 | |||
| 474 | RM333 | 10 | 515 | RM1880 | 12 | |||
| 475 | RM496 | 10 | 516 | RM2851 | 12 | |||
| 476 | RM590 | 10 | 517 | RM3483 | 12 | |||
| 477 | RM591 | 10 | 518 | RM19 | 12 | |||
| 478 | RM4B | 11 | 519 | RM453 | 12 | |||
| 479 | RM20B | 11 | 520 | RM247 | 12 | |||
| 480 | RM286 | 11 | 521 | RM117 | 12 | |||
| 481 | RM26652 | 11 | 522 | RM491 | 12 | |||
| 482 | RM7163 | 11 | 523 | RM512 | 12 | |||
| 483 | RM7173 | 11 | 524 | RM179 | 12 | |||
| 484 | RM1240 | 11 | 525 | RM415 | 12 | |||
| 485 | RM1124 | 11 | 526 | RM2529 | 12 | |||
| 486 | RM332 | 11 | 527 | RM1036 | 12 | |||
| 487 | RM5704 | 11 | 528 | RM3246 | 12 | |||
| 488 | RM167 | 11 | 529 | RM101 | 12 |
Foreground, recombinant and background selection for Saltol QTL
For the screening of Saltol QTL introgressed progenies of all generations, 7 foreground markers AP3206, SKC10, RM3412, RM8094, RM10713, RM10711, RM10772, 6 recombinant markers RM10793, RM493, RM10696, RM10701, RM1287, RM10694 and 545 genome wide SSR markers were screened between parents and then polymorphic markers were selected for genotypic screening of progenies of Aiswarya X FL-478.
Foreground, recombinant and background selection for Sub1 gene
For the screening of Sub1 gene introgression and background genome recovery 11 foreground markers which are IYT1, IYT3, AEX, Sub1A203, ART3, ART5, Sub1C173, Sub1AB1, Sub1BC1, Sub1BC2 and Sub1BC3, 16 recombinant markers RM316, RM219, RM464, RM23668, RM23679, RM8303, RM23770, RM23788, RM23778, RM23805, RM23887, RM23917, RM23922, RM23958, RM23928 and RM24005, and 545 genome wide markers were screened and evaluated. The markers which showed polymorphism between parents were selected for genotypic screening of all generations.
Analysis of molecular data
The Microsoft Excel sheet containing Saltol marker and Sub1 marker data was used and analysed in the Graphical Tools for Genotyper (GGT 2.0) (Ralph, 2008). GGT analysis software was used to measure recovery of parent genome in selected Introgressed progenies of Saltol X Sub1 lines. The alleles were scored for homozygous recipient allele, homozygous donor allele and heterozygous allele as ‘A’, ‘B’, ‘C’, respectively. So, the percentage of background genomic recovery were calculated.
Screening for salinity tolerance
Phenotypic screening at seedling stage for salinity and submergence was done. The salinity screening was done using the standard protocol SES developed at IRRI. The pre germinated seeds were sown in the holes on the float in distilled water for 3 days. FL-478 plants were used as the check and RP Aiswarya was used as susceptible check. On fourth day, distilled water was replaced with Yoshida nutrient solution having EC of 6 dS m−1 and on 7th day the Yoshida nutrient solution was replaced with fresh nutrient solution having EC of 12 dS m−1.The pH of the solution was maintained at 5.5 daily by adding either 1 N NaOH or 1 N HCL. The nutrient solution was replaced after every 7 days up to 21 days. The tolerance score was done using SES score developed at IRRI, 1996. Table 1 describes the SES score for salinity tolerance.
Screening for submergence tolerance
In screening for submergence tolerance, germinated seedlings of the selected progenies along with donor and recurrent parents were sown in pots. The experimental pots with fourteen-day old seedlings were completely submerged for 21 days in 1.5 m height tanks filled with water. The average depth of water was 90 cm and pH was 7.2. Control sets were also maintained for each of the varieties without any submergence treatment. After the completion of submergence treatment the plants were removed and 14 days after the completion of submergence treatment tolerance as per SES developed at IRRI. Table 2 describes the SES score for submergence tolerance.
Table 2.
Standard evaluation system (SES) score for submergence tolerance (IRRI 1996)
| Score | Percentage survival | Tolerance |
|---|---|---|
| 1 | 100% | Minor visible symptoms |
| 3 | 95–99% | Some visible symptoms |
| 5 | 75–94% | Moderately injury |
| 7 | 50–54% | Severe injury |
| 9 | 0–94% | Complete death |
Results and discussion
The MABB approach was used to introgress the Saltol QTL and Sub 1 genes into Aiswarya rice variety and is an effective and appropriate method for introgressing our gene of interest and recovering the maximum genome background of recurrent parents in progenies (Collard and Mackill 2007). The highly tolerant F1 plants were selected based on phenotyping and genotyped with selected foreground markers for confirmation, and the tolerant lines with heterozygous locus were alone selected. The selected tolerant lines in F1 and BC1F1 generations were backcrossed with recurrent parent and genotyped with foreground, recombinant and background primers. At last from the selfed BC2F2 lines having the salinity and submergence tolerance score similar to FL-478 and Swarna Sub1 with maximum recipient genotype recovery could be identified.
Parental Polymorphism Assay of Foreground, Recombinant and Background Markers
Total 545 SSR markers covering 12 chromosomes were used for the parental polymorphic screening between recurrent parent and donor parent. For salinity screening 88 polymorphic markers and for submergence screening 90 polymorphic markers were selected and used for screening of every F1, BC1F1, BC2F1 and BC2F2 generation. Foreground markers are used for efficient selection of target locus and these markers are tightly linked with the target gene (Hospital and Charcosset 1997). The recombinant markers that flank a target gene are used for eliminating the undesirable gene as quick as possible and linkage drag can be minimized (Hasan et al. 2015). Background selection involves selecting progenies with the maximum proportion of RP genome, and these markers are unlinked to the target locus. Genetic analysis of the salt tolerant rice varieties using SSR markers through molecular characterization is done (John and Shylaraj.2017). The result was verified with some other previous reports of the polymorphic markers between the parents in target region of Saltol (Niones 2004). Within the Sub1 cluster the diagnostic markers developed and microsatellite markers along the Sub1 region has been reported (Neeraja et al. 2007). Sub1 QTL was introgressed into the most popular rice variety, Jyothi, from the donor parent Swarna-Sub1 for submergence tolerance (John and Shylaraj 2017). Number of Polymorphic markers screened and used for parental screening are narrated in Table 3 below. Tables 4,5,6,7,8,9 describes the name of selected foreground, recombinant and background SSR markers. Total 545 genomewide SSR markers screened for parental polymorphism in both salinity and submergence were narrated in Table 10.
Table 3.
Polymorphic markers screened and markers used for parental screening
| SSR markers screened for polymorphism | Polymorphic markers selected | ||||
|---|---|---|---|---|---|
| Salinity | Submergence | Salinity | Submergence | ||
| 1 | Foreground markers | 7 | 11 | 3 | 3 |
| 2 | Recombinant markers | 6 | 16 | 2 | 3 |
| 3 | Background markers | 545 | 545 | 88 | 90 |
Table 4.
Saltol SSR markers used for foreground selection
| Primer | Sequence | Distance (mb) |
|---|---|---|
| AP206 F | TTCTCATCGCACCATCTCTG | 11.2 |
| AP206R | GGAGGAGGAGAGGAAGAAG | |
| SKC10F | ATAGGGGATATTGGCTGCAC | 11.2 |
| SKC10R | CAACCAAGCGTGACTAAAAAGA | |
| RM3412F | TGATGGATCTCTGAGGTGTAAAGAGC | 11.6 |
| RM3412R | TGCACTAATCTTTCTGCCACAGC |
Table 5.
Saltol Polymorphic SSR markers used for Recombinant selection
| Primer | Sequence | Distance (mb) |
|---|---|---|
| RM493 F | GTACGTAAACGCGGAAGGTGACG | 12.3 |
| RM493R | CGACGTACGAGATGCCGATCC | |
| RM10696F | CCTTCGACTCCATGAAACAAACG | 10.6 |
| RM10696R | TCTCTTTGCCCTAACCCTATGTCC |
Table 6.
Saltol Polymorphic SSR markers used for Background selection
| Chr. No | SSR markers | Position (cM) | Chr.No | SSR markers | Position (cM) | Chr.No | SSR markers | Position (cM) |
|---|---|---|---|---|---|---|---|---|
| 1 | 1196 | 0 | 4 | RM 537 | 0 | 6 | RM 439 | 117 |
| 1 | RM 10209 | 18.8 | 4 | RM 3471 | 16.7 | 7 | RM 295 | 0 |
| 1 | RM 600 | 61.30 | 4 | RM 3317 | 32.1 | 7 | RM 3859 | 42.4 |
| 1 | RM595 | 78.40 | 4 | RM471 | 53.8 | 7 | RM 214 | 61.5 |
| 1 | RM 246 | 115.2 | 4 | RM 401 | 63.4 | 7 | RM 11 | 93.8 |
| 1 | RM 212 | 148.7 | 4 | RM 142 | 100.2 | 7 | RM 118 | 130.5 |
| 1 | RM 472 | 171.6 | 4 | RM 273 | 116.8 | 7 | RM 420 | 144.6 |
| 2 | RM 174 | 33.8 | 4 | RM 153 | 0.5 | 8 | RM 408 | 0 |
| 2 | RM 1347 | 26.6 | 4 | RM17960 | 18.8 | 8 | RM1235 | 12.8 |
| 2 | RM 71 | 49.8 | 4 | RM 1089 | 37.2 | 8 | RM 1376 | 15.2 |
| 2 | RM 1352 | 48.1 | 5 | RM 169 | 57.9 | 8 | RM547 | 27.3 |
| 2 | RM 5430 | 11.5 | 5 | RM 6229 | 63.6 | 8 | RM25 | 59.6 |
| 2 | RM 1303 | 81.7 | 5 | RM 1187 | 96.1 | 8 | RM149 | 122.1 |
| 2 | RM 526 | 136.3 | 5 | RM249 | 122.1 | 8 | RM 447 | 131.3 |
| 2 | RM240 | 158 | 5 | RM5770 | 103.9 | 9 | RM 1328 | 0 |
| 2 | RM 498 | 194.6 | 5 | RM334 | 141.8 | 9 | RM 316 | 1.5 |
| 3 | RM 175 | 18.9 | 6 | RM508 | 0 | 9 | RM 3912 | 46.3 |
| 3 | RM545 | 35.3 | 6 | RM469 | 2.8 | 9 | RM 1553 | 68.2 |
| 3 | RM 227 | 120.1 | 6 | RM 197 | 15.1 | 9 | RM 189 | 90.7 |
| 3 | RM 168 | 138.6 | 6 | RM 276 | 30.8 | 9 | RM 205 | 114.7 |
| 3 | RM6135 | 158.2 | 6 | RM 527 | 59.9 | 10 | RM 214 | 27.8 |
| 3 | RM 85 | 179.7 | 6 | RM 454 | 86.5 | 10 | RM 2504 | 35.9 |
| 3 | RM7324 | 166.4 | 6 | RM 1340 | 82.9 | 10 | RM 311 | 46.4 |
| 3 | RM520 | 191.6 | 6 | RM 30 | 115.7 | 10 | RM184 | 79.3 |
| 10 | RM 294A | 109.4 | 11 | RM 6965 | 101.9 | 12 | RM 1105 | 91.4 |
| 11 | RM 332 | 12.3 | 11 | RM 206 | 104.2 | 12 | RM 1296 | 108.2 |
| 11 | RM 536 | 42 | 11 | RM 144 | 121.4 | 12 | RM 17 | 109.1 |
| 11 | RM 7120 | 56.2 | 12 | RM1328 | 0 | 12 | RM 1227 | 130.8 |
| 11 | RM 229 | 66.5 | 12 | RM 19 | 20.9 | |||
| 11 | RM 209 | 84.7 | 12 | RM 27973 | 57.6 |
Table 7.
Submergence SSR markers used for foreground selection
| Primer | Sequence | Distance (mb) |
|---|---|---|
| SUB1C173F | AACGCCAAGACCAACTTCC | Exon of Sub1C |
| SUB1CI73R | AGGAGGCTGTCCATCAGGT | |
| ART5F | CAGGGAAAGAGATGGTGGA | Sub1C promoter |
| ART5R | TTGGCCCTAGGTTGTTTCAG | |
| Sub1BC2F | AAAACAATGGTTCCATACGAGAC | Between Sub1B and C |
| Sub1BC2R | GCCTATCAATGCGTGCTCTT |
Table 8.
Submergence SSR markers used for recombinant selection
| Primer | Sequence | Distance (mb) |
|---|---|---|
| RM8303F | AGGGGAGAGGACACACACAC | 2.3 |
| RM8303R | GGATCCTCCTGCAAAATCAA | |
| RM23770F | GACCTTGTCCAGAGTGATTTTG | 3.7 |
| RM23770R | ATTTGAGAATAACTTTTCCTACTTCG | |
| RM23958F | GAGACAGATGTGTACGGTTTGGTG | 7.9 |
| RM23958R | TTGACAAGGGAATTGAAGGAGAAG |
Table 9.
Submergence Polymorphic SSR markers used for Background selection
| Chr. No | SSR Marker | Position (cM) | Chr. No | SSR Marker | Position (cM) | Chr. No | SSR Marker | Position (cM) |
|---|---|---|---|---|---|---|---|---|
| 1 | RM 3252 | 0.0 | 4 | RM 261 | 31.1 | 6 | RM 340 | 137.5 |
| 1 | RM 595 | 78.4 | 4 | RM 401 | 63.4 | 7 | RM 295 | 0.0 |
| 1 | RM1196 | 107.5 | 4 | RM 273 | 116.8 | 7 | RM 3859 | 42.4 |
| 1 | RM246 | 115.2 | 4 | RM 142 | 100.2 | 7 | RM 125 | 44.4 |
| 1 | RM443 | 122.7 | 4 | RM 559 | 173.3 | 7 | RM 214 | 61.5 |
| 2 | RM7382 | 0.0 | 4 | RM 1155 | 58.9 | 7 | RM 11 | 93.8 |
| 2 | RM 1347 | 26.6 | 5 | RM 153 | 0.0 | 7 | RM 1362 | 116.1 |
| 2 | RM324 | 66.0 | 5 | RM 169 | 57.9 | 7 | RM 420 | 144.6 |
| 2 | RM 5430 | 111.5 | 5 | RM 188 | 108.7 | 8 | RM 337 | 0.1 |
| 2 | RM7485 | 145.7 | 5 | RM 334 | 11.7 | 8 | RM 1235 | 12.8 |
| 2 | RM 240 | 158.0 | 5 | RM 6467 | 143.8 | 8 | RM 1376 | 15.2 |
| 3 | RM 175 | 18.9 | 5 | RM 30 | 174.6 | 8 | RM 547 | 27.3 |
| 3 | RM 545 | 35.3 | 6 | RM 469 | 2.8 | 8 | RM 3481 | 44.7 |
| 3 | RM 282 | 61.5 | 6 | RM 402 | 3.2 | 8 | RM 310 | 57.0 |
| 3 | RM5626 | 122.4 | 6 | RM 197 | 15.1 | 8 | RM 25 | 59.6 |
| 3 | RM 118 | 138.6 | 6 | RM 276 | 30.8 | 8 | RM 433 | 87.2 |
| 3 | RM 227 | 229.7 | 6 | RM 454 | 86.5 | 8 | RM 149 | 122.1 |
| 4 | RM 551 | 0.0 | 6 | RM 1340 | 82.9 | 8 | RM 447 | 131.3 |
| 8 | RM 264 | 138.2 | 10 | RM 1374 | 72.8 | 11 | RM 206 | 104.2 |
| 9 | RM 1328 | 0.0 | 10 | RM 184 | 79.3 | 11 | RM 144 | 121.4 |
| 9 | RM 316 | 1.5 | 10 | RM 6016 | 83.0 | 11 | RM 27172 | 123.3 |
| 9 | RM 3912 | 46.3 | 10 | RM 294A | 109.4 | 11 | RM 2136 | 136.8 |
| 9 | RM 3164 | 72.1 | 11 | RM 332 | 12.3 | 12 | RM 247 | 12.0 |
| 9 | RM 328 | 82.4 | 11 | RM 287 | 57.4 | 12 | RM 19 | 20.9 |
| 9 | RM 245 | 112.3 | 11 | RM 7303 | 64.2 | 12 | RM 3246 | 48.2 |
| 10 | RM 216 | 27.8 | 11 | RM 229 | 66.5 | 12 | RM 27973 | 57.6 |
| 10 | RM 2504 | 35.9 | 11 | RM 1341 | 80.2 | 12 | RM 235 | 82.0 |
| 10 | RM 596 | 34.8 | 11 | RM 5961 | 79.9 | 12 | RM 1103 | 91.4 |
| 10 | RM 311 | 46.4 | 11 | RM 209 | 84.7 | 12 | RM 17 | 109.1 |
| 10 | RM 1108 | 55.3 | 11 | RM 6965 | 101.9 | 12 | RM 1227 | 130.8 |
Genotyping of F1 generation
Saltol F1 generation
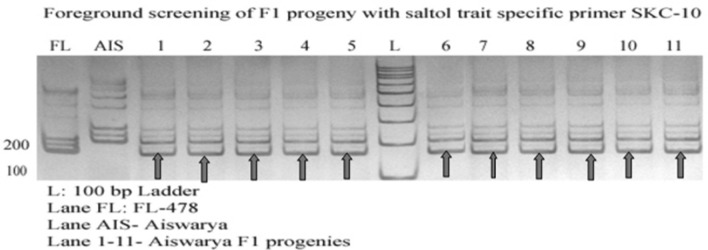
For the foreground selection of Saltol F1 generation, 35 / 55 plants were heterozygous for AP3206, SKC-10 and RM3412 foreground markers. After that recombinant selection was done with RM493 and RM10696. Recently the highly salt tolerant rice variety FL478 has been used to transfer Saltol QTL into the high yielding grown cultivars ASS996 using MABC strategy in Vietnam (Huyen et al. 2012). Figure 3 shows the genotypic screening with SKC10 marker.
Fig. 3.
Genotypic screening of Saltol trait specific F1 progenies with foreground marker SKC-10 (Aiswarya X FL 478)
Sub1 F1 generation
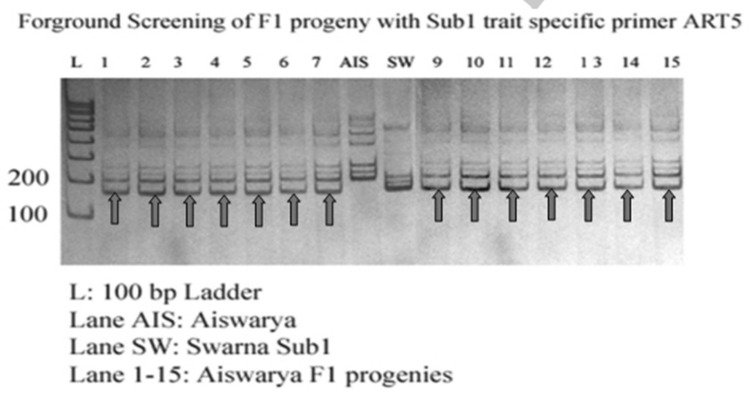
For the selection of Sub1 F1 generation of the 48 plants, 26 plants were heterozygous for ART5, SUIBC2 And SUB1C173 foreground markers and recombinant selection carried out with RM8303, RM23770 and RM24005. The successful introgression of sub1 from donor rice variety IR64 into popular rice variety AS996 through MABC using ART5 and SC3 has been done in Vietnam (Cuc et al. 2012). After selection of heterozygous plants, they were backcrossed with the respective recurrent parents to develop BC1F1 plants. Figure 4 shows the Genotypic screening of Sub1 F1 progenies with foreground marker ART5.
Fig. 4.
Genotypic screening of Sub1 trait specific F1 progenies with foreground marker ART5 (Aiswarya X Swarna Sub1)
Genotyping of BC1F1 generation
Saltol BC1F1 generation
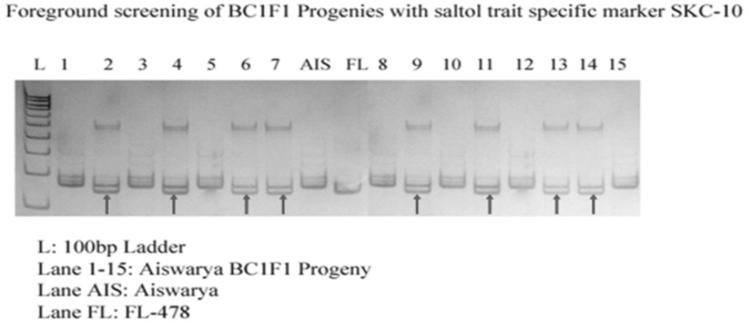
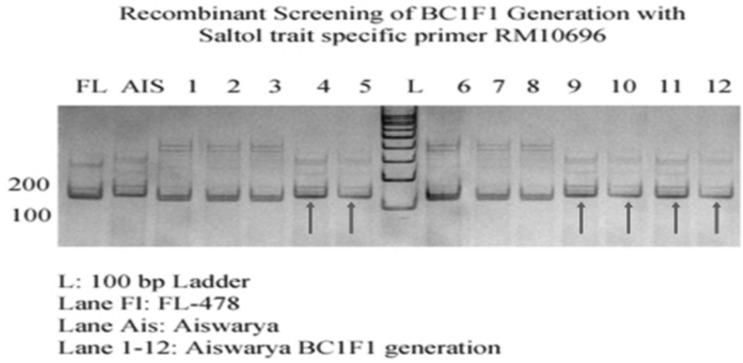
In 90 plants of BC1F1 the 58 plants were Saltol introgressed plants, confirmed with AP3206, SKC-10 and RM3412 foreground markers and Recombinant selection was carried out on 58 Saltol introgressed plants and 66 Sub1 introgressed plants. Background analysis was done of Saltol introgressed plants with selected SSR markers and indicated a recovery of 64%. The Introgression of Saltol QTL into an elite rice variety Jyothi is done using marker assisted selection (Rohini and Shylaraj 2017). Seventeen Saltol BC1F1 recombinant plants were further used for backcross breeding. Figure 5 shows the Genotypic screening of Saltol BC1F1 progenies with foreground marker SKC-10 and Fig. 6 shows the Genotypic screening of Saltol BC1F1 progenies with recombinant marker RM10696.
Fig. 5.
Genotypic screening of Saltol trait specific BC1F1 progenies with foreground marker SKC-10
Fig. 6.
Genotypic screening of Saltol trait specific BC1F1 progenies with recombinant marker RM10696
Sub1 BC1F1 generation
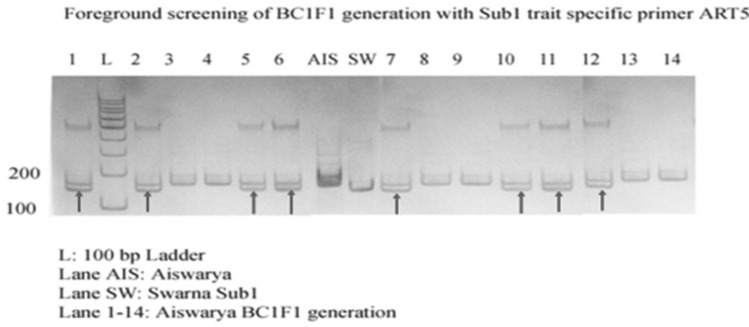
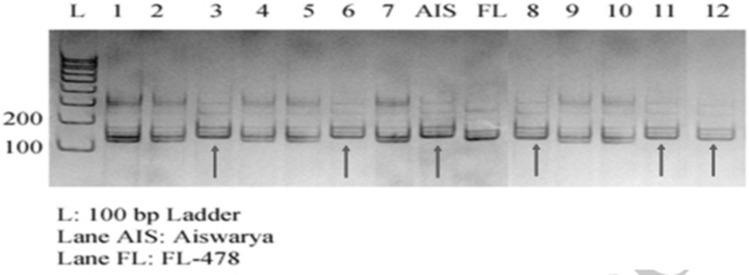
In 94 BC1F1 plants, 66 plants were Sub1 introgressed plants, confirmed with ART5, SUIBC2 And SUB1C173 and Recombinant selection was carried out on 58 Saltol introgressed plants and 66 Sub1 introgressed plants. Background analysis was done in Sub1 introgressed plants with selected SSR markers and indicated a recovery of 67%. Nineteen Sub1 BC1F1 recombinant plants were further used for backcross breeding. Figure 7 shows the Genotypic screening of Sub1 BC1F1 progenies with foreground marker ART5 and Fig. 8 shows the Genotypic screening of Sub1 BC1F1 progenies with recombinant marker RM8303.
Fig. 7.
Genotypic screening of Sub1 trait specific BC1F1 progenies with foreground marker ART5
Fig. 8.
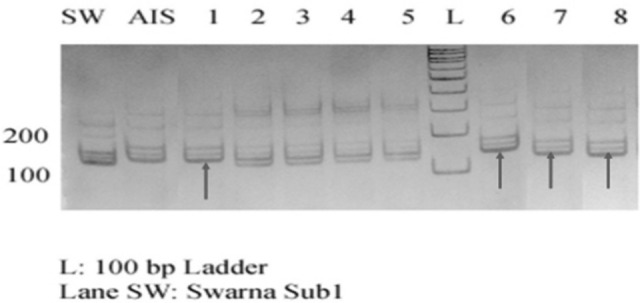
Genotypic screening of Sub1 trait specific BC1F1 progenies with recombinant marker RM8303
Genotyping of BC2F1 and BC2F2 generation
Saltol BC2F1 and BC2F2 generation
In BC2F1 generation 10 plants with Saltol introgressed were phenotypically and genotypically analyzed and indicated a recovery of 75% to 82%. After that, all the selected BC2F1 plants were selfed and BC2F2 generation were obtained. On the basis of phenotypic and genotypic screening, the best four plants with maximum genome recovery of 89% to 92% were selected. Figure 9 shows the Genotypic screening of Saltol BC2F2 progenies with foreground marker SKC-10. Figure 10 shows the Genotypic screening of Saltol trait specific BC2F2 progenies with recombinant marker RM10696. The Fig. 11 shows the graphical representation of selected best plant with 92% background genomic recovery.
Fig. 9.
Genotypic screening of Saltol trait specific BC2F2 progenies with foreground marker SKC-10
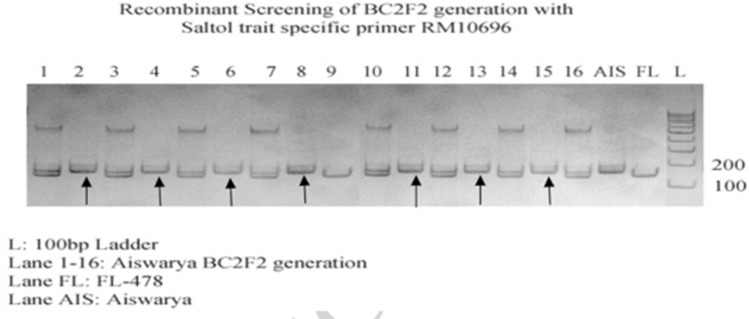
Fig. 10.
Genotypic screening of Saltol trait specific BC2F2 progenies with recombinant marker RM10696
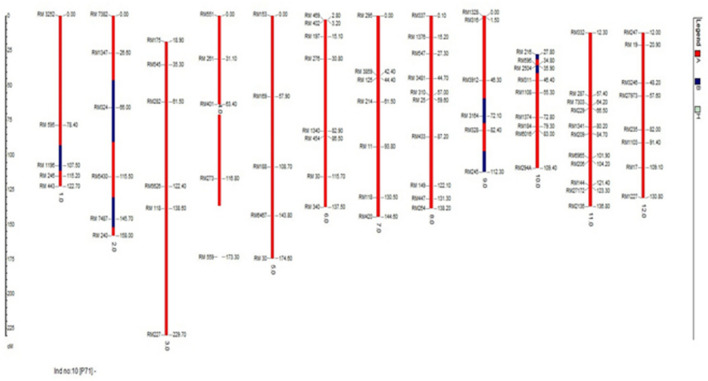
Fig. 11.
Graphical presentation of the best BC2F2 plant with Saltol QTL introgression (A: Homozygous to the recipient genome B: Homozygous to the donor genome H: Heterozygous region)
Sub1 BC2F1 and BC2F2 generation
In BC2F1 generation 12 plants with Sub1 introgressed were phenotypically and genotypically analyzed and indicated a recovery of 70% to 80%. After that, all the selected BC2F1 plants were selfed and BC2F2 generation produced. On the basis of phenotypic and genotypic screening, the best four plants with maximum genome recovery of 85% to 93% were selected. Figure 12 shows the Genotypic screening of Sub1 BC2F2 progenies with foreground marker ART5. Figure 13 shows the Genotypic screening of Sub1 BC2F2 progenies with recombinant marker RM8303. The Fig. 14 shows the graphical presentation of best selected plant with 93% background genomic recovery.
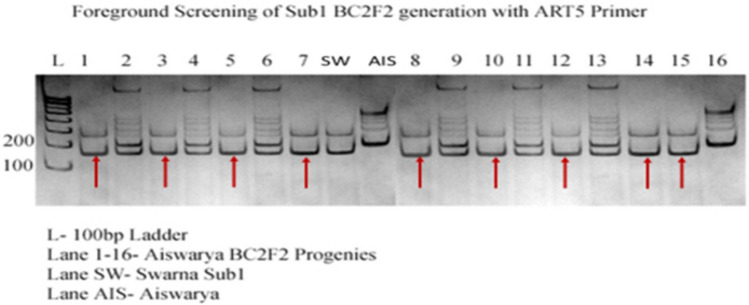
Fig. 12.
Genotypic screening of Sub1 trait specific BC2F2 progenies with foreground marker ART5
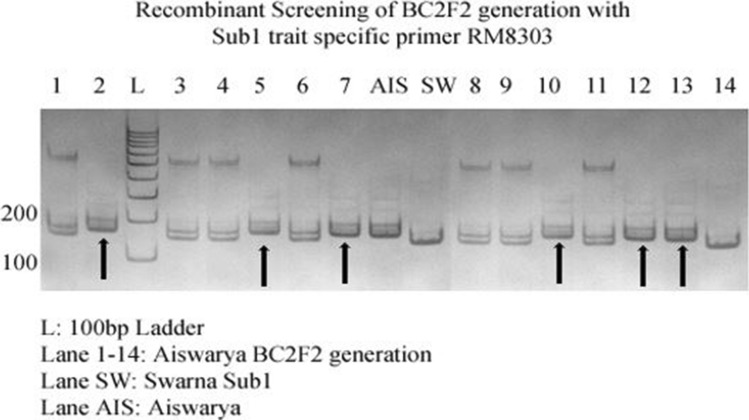
Fig. 13.
Genotypic screening of Sub1 trait specific BC2F2 progenies with recombinant marker RM8303
Fig. 14.
Graphical presentation of the best BC2F2 plant with Sub 1 gene introgression (A: Homozygous to the recipient genome B: Homozygous to the donor genome H: Heterozygous region)
Phenotypic Screening for Salinity tolerance and submergence tolerance
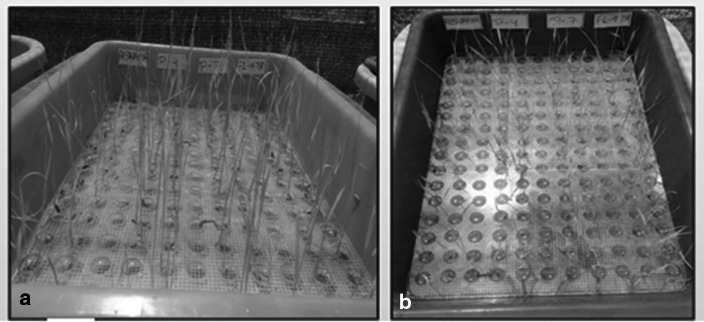
The selected Saltol BC2F2 lines showed salinity tolerance at seedling stage under salt stress. The salinity tolerance score of these lines were similar to the donor variety FL-478. A salinity tolerant Q5DB was developed using marker assisted selection, which was controlled by a major Saltol QTL (Huyen et al. 2013) and the selected Sub1 introgressed BC2F2 lines showed submergence tolerance and recovery after submergence of 21 days so the tolerance score of these lines were similar to the donor variety Swarna Sub1. A single major quantitative trait locus (QTL) SUBMERGENCE 1 (SUB1) near the centromere of chromosome 9 in the rice genome along with a number of minor QTLs linked to submergence tolerant had been identified (Xu et al. 2006). Based on these results the introgression of Saltol and Sub1 QTL in these lines was confirmed. Figure 15 shows the phenotypic screening of selected progenies for salinity tolerance and Fig. 16 shows the phenotypic screening of submergence tolerance.
Fig. 15.
Phenotypic screening for salinity screening. a Control set of progenies without salinity stress b progenies with salinity stress
Fig. 16.

Phenotypic screening of submergence tolerance
Outcome of the study
The present study could develop 5 best lines of Saltol QTL introgressed Aiswarya with 85%–92% of background homozygosity of Aiswarya and 4 best lines of Sub1 gene introgressed lines of Aiswarya with 88%–93% of background homozygosity of Aiswarya. These lines can be released for cultivation in the saline prone coastal areas as well as in submergence prone coastal areas. Further these lines can be used to pyramid both the abiotic stresses into the variety Aiswarya to develop a dual stress tolerant rice variety.
Conclusion
This work aimed to develop a Saltol QTL and Sub1 QTL introgressed Aiswarya variety by using Marker Assisted Backcross Breeding. This study can make a good impact on rice breeding through the development of a salinity and submergence tolerant variety Aiswarya which can be cultivated throughout Kerala. Climatic changes and environmental issues harming rice production, which is the most important food crop in India (Kurokawa et al. 2018). Abiotic stresses include drought, salinity, submergence, heat, cold etc. disapprovingly impends crop production and causes yield loss in low land areas and coastal areas (Pareek et al. 2010). Due to unexpected linkage drag it is difficult to use conventional breeding method to introgress these tolerant genes into high yielding varieties (Jeung et al. 2005). Hence using molecular breeding is a good option which did not affect grain quality and yield. A rice cultivar OMCS2000 developed with improved salt tolerant genes using maker assisted selection (Lang et al. 2008). Salt tolerance is controlled by many eleven QTL which are located on 5 chromosomes from the population of Nona Bokra and Koshihikari (Ren et al., 2005). 10.000 hectares of rice were submerged by 2011 floods, due to which faced a severe economic loss and loss of rice productivity (MARD 2011). Approximately 40.000 km2, a huge amount of area will be disappeared if sea level rise is by 1 m (Khanh et al. 2013).
In this study we used the MABB breeding method to introgress the Saltol gene and Sub1 gene into a popular rice variety Aiswarya by phenotypic and genotypic selection. Using SSR markers the Saltol gene and Sub1 gene introgression was confirmed. Our results accomplish that a salinity tolerant gene (Saltol) from the donor parent FL478 was successfully transferred into Aiswarya and submergence tolerant gene Sub1 from the donor parent Swarna Sub1 was successfully transferred into Aiswarya independently. The future line of work of the study is to pyramid both these abiotic stresses into the rice variety Aiswarya to make it dual stress tolerant variety. Better profitability can be achieved directly benefiting the farmers by increasing their harvest in saline and flood affected lands. Due to this sole reason this work will have a significant impact on the socio-economic factors of low-lying areas and will lead to upliftment of farmers of these regions.
Acknowledgement
The authors are grateful to the funding agency, Department of Science and Technology, Government of India for the Inspire Fellowship and Kerala Agriculture University for providing the necessary lab facilities to carry out the study.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Collard CY, Mackill J (2007) Marker-assisted selection: an approach for precision plant breeding in the twenty-first century Phil Trans R Soc B 36310.1098/rstb.2007.2170 [DOI] [PMC free article] [PubMed]
- Cuc LM, Huyen LTN, Hien PTM, Hang VTT, Dam NQ, Mui PT, Quang VD, Ismail AM, Ham LH. Application of marker assisted backcrossing to introgress the submergence tolerance QTLSUB1 into the Vietnam elite rice variety-AS996. Am J Plant Sci. 2012;3:528–536. doi: 10.4236/ajps.2012.34063. [DOI] [Google Scholar]
- Doyle JJ, Doyle JL. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 1987;19:11–15. [Google Scholar]
- Fukao T, El Y, Bailey-Serres J. The submergence tolerance regulator sub1a mediates crosstalk between submergence and drought tolerance in rice. Plant Cell. 2011;23:412–427. doi: 10.1105/tpc.110.080325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregorio GB, Islam MR, Vergara GV, Thirumeni S. Recent Advances in rice science to design salinity and other abiotic stress tolerant rice varieties. SABRAO J Breed Genet. 2013;45:31–41. [Google Scholar]
- Hasan MM, Rafii MY, Ismail MR, et al. Marker-assisted backcrossing: a useful method for rice improvement. Biotechnol Equip. 2015;29:237–254. doi: 10.1080/13102818.2014.995920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hospital F, Charcosset A. Marker-Assisted Introgression of Quantitative Trait Loci. Genetics. 1997;147:1469–1485. doi: 10.1093/genetics/147.3.1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain M, Narciso J. Global Rice Market: Trends and Perspectives. The Bangladesh Dev Stud. 2002;28:1–28. [Google Scholar]
- Huyen LTN, Cuc LM, Ismail AM, Ham LH. Introgression the salinity tolerance QTLs Saltol into AS996, the elite rice variety of Vietnam. Am J Plant Sci. 2012;3:981–987. doi: 10.4236/ajps.2012.37116. [DOI] [Google Scholar]
- Huyen LTN, Cuc LM, Ham LH, Khanh TD. Introgression the SALTOL QTL into Q5DB, The Elite Variety of Vietnam Using Marker-Assisted-Selection (MAS) Am J BioSci. 2013;1:80–84. doi: 10.11648/j.ajbio.20130104.15. [DOI] [Google Scholar]
- IRRI (1996). Standard evaluation system for rice. The International Rice Research Institute, Manila, Philippines. pp 44
- Jeung JU, Hwang HG, Moon HP, Jena KK. Fingerprinting temperate japonica and tropical Indica rice genotypes by comparative analysis of DNA markers. Euphytica. 2005;146:239–251. doi: 10.1007/s10681-005-9022-2. [DOI] [Google Scholar]
- John D, Shylaraj KS. Introgression of Sub1 QTL into an elite rice (Oryza sativa L.) variety Jyothi through Marker Assisted Backcross Breeding. J Tropical Agric. 2017;55:1–11. [Google Scholar]
- John D, Rohini PS, Shylaraj KS. Genetic analysis of the salt tolerant rice varieties of Kerala through molecular characterization using SSR markers. Green Farming. 2017;8:1012–1017. [Google Scholar]
- Khanh TD, Linh LH, Ham LH, Xuan TD. Rapid and high-precision marker assisted backcrossing to introgress the SUB1 QTL into the Vietnamese elite rice variety. J Plant Bred Crop Sci. 2013;5:26–33. doi: 10.5897/JPBCS12.052. [DOI] [Google Scholar]
- Kurokawa Y, Nagai K, Huan PD, Shimazaki K, Qu H, Mori Y, Toda Y, Kuroha T, Hayashi N, Aiga S. Rice leaf hydrophobicity and gas films are conferred by a wax synthesis gene (LGF1) and contribute to flood tolerance. New Phytol. 2018;218:1558–1569. doi: 10.1111/nph.15070. [DOI] [PubMed] [Google Scholar]
- Lang NT, Buu BC, Ismail AM. Molecular mapping and marker assisted selection for salt tolerance in rice (Oryza sativa L.) Omonrice. 2008;16:0–56. [Google Scholar]
- Lutts S, Kinet JM, Bouharmont J. Changes in plant response to NaCl during development of rice (Oryza sativa L.) varieties differing in salinity resistance. J Exp Bot. 1995;46:1843–1852. doi: 10.1093/jxb/46.12.1843. [DOI] [Google Scholar]
- [MARD] Ministry of Agriculture and Rural Development (2011). Vietnam News Agency report, Nov 12.
- Mohammadi N, Arzani A, Rezai AM, Singh RK, Gregorio GB. Assessment of rice genotypes for salt tolerance using microsatellite markers associated with the saltol. QTL Afr J Biotech. 2008;7:9. [Google Scholar]
- Neeraja C, Maghirang Rodriguez R, Pamplona A, Heuer S, Collard B, Septiningsih E. A marker-assisted backcross approach for developing submergence-tolerant rice cultivars. Theor Appl Genet. 2007;115:767–776. doi: 10.1007/s00122-007-0607-0. [DOI] [PubMed] [Google Scholar]
- Niones, JM (2004) Fine mapping of the salinity tolerance gene on chromosome 1 of rice (Oryza sativa L.) using near- isogenic lines. MS dissertation. University of the Philippines Los Baños. pp.78
- Pareek A, Sopory SK, Bohnert HJ, Govindjee (2010) Abiotic stress adaptation in plants. Physiol mol genomic found. 10.1007/978-90-481-3112-9
- Ralph VB. GGT 2.0: Versatile software for visualization and analysis of genetic data. J Hered. 2008;99:232–236. doi: 10.1093/jhered/esm109. [DOI] [PubMed] [Google Scholar]
- Ren ZH, Gao JP, Li LG, Cai XL, Huang W, Chao DY, Zhu MZ, Wang ZY, Luan S, Lina HX. Rice quantitative trait locus for salt tolerance encodes a sodium transporter. Nat Genet. 2005;37:1141–1146. doi: 10.1038/ng1643. [DOI] [PubMed] [Google Scholar]
- Rohini PC, Shylaraj KS. Introgression of Saltol QTL into an elite rice variety Jyothi through marker assisted selection. Green Farming. 2017;8:512–517. [Google Scholar]
- Sarkar RK, Bhattacharjee B. Rice Genotypes with SUB1 QTL Differ in Submergence Tolerance, Elongation Ability during Submergence and Re-generation Growth at Re-emergence. Rice (NY) 2011;295(5):7. doi: 10.1007/s12284-011-9065-z. [DOI] [Google Scholar]
- Singh RK, Glein B, Gregorio RK. QTL mapping for salinity tolerance in rice. Physiol Mol Biol Plants. 2007;13:87–99. [Google Scholar]
- Thomson MJ, Ocampo DE, Egdane J, et al. Characterizing the Saltol quantitative trait locus for salinity tolerance in rice. Rice. 2010;3:148–160. doi: 10.1007/s12284-010-9053-8. [DOI] [Google Scholar]
- Todaka D, Nakashima K, Shinozaki K. Toward understanding transcriptional regulatory networks in abiotic stress responses and tolerance in rice. Rice. 2012;5:6. doi: 10.1186/1939-8433-5-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K, Xu X, Fukao T, Canlas P, Maghirang Rodriguez R, Heuer S, Ismail AM, Serres B, Ronald J, Mackill DJ. Sub1A is an ethylene-response-factor-like gene that confers submergence tolerance to rice. Nature. 2006;442:705–708. doi: 10.1038/nature04920. [DOI] [PubMed] [Google Scholar]