Abstract
Deeper Rooting 1 (DRO1) gene identified from a major QTL on chromosome 9 increases the root growth angle (RGA) and thus facilitates survival under drought and hence is an excellent candidate for rice improvement. Twenty-four major Indian upland and lowland genotypes including the ‘yield under drought’ (DTY) QTL donors were subjected to allele mining of DRO1 (3058 bp) using four pairs of overlapping primers. A total of 216 and 52 SNPs were identified across all genotypes in the gene and coding region (756 bp) respectively with transversions 3.6 fold more common than transitions in the gene and 2.5 times in the CDS. In 251 amino acid long protein, substitutions were found in 19 positions, wherein change in position 92 was the most frequent. Based on allele mining, the 24 genotypes can be classified into 16 primary structure variants ranging from complete functional allele (Satti, IR36 and DTY 3.1 donor, IR81896-B-B-195) to truncated non-functional alleles in PMK2, IR64, IR20 and Swarna. All the DTY donors, other than IR81896-B-B-195, and most of the upland drought tolerant cultivars (Nagina 22, Vandana and Dhagaddeshi) had accumulated 6–19 SNPs and 4–8 amino acid substitutions resulting in substantial differences in their protein structure. The expression analysis revealed that all the genotypes showed upregulation under drought stress though the degree of upregulation varied among genotypes. The information on structural variations in DRO1 gene will be very useful for the breeders, especially in the light of recent breeding programmes on improving drought tolerance using several DTY donors and upland accessions.
Supplementary Information
The online version of this article (10.1007/s12298-021-00950-2).
Keywords: Rice, Drought stress, Root growth angle, DRO1, Allele mining
Introduction
Rice (Oryza sativa L.), an important cereal crop is consumed by more than 50% of humankind (Barah 2005). Rice also remains a means of livelihood to the rural population (Suhail et al. 2008). Hence, sustaining and improving rice production provides not only nourishment but also lifts the rural populations from poverty. Several factors impact sustainable rice production, of which, drought stress remains a more detrimental one (Serraj et al. 2009; Zhang et al. 2018). Drought stress in plants affects multiple vital physiological and biochemical processes such as structural and functional integrity of cell organelles and membrane, water relations, osmotic adjustment (OA), and plants’ mainstay activities, including nutrient uptake, photosynthesis and respiration, ultimately hampering the growth and productivity of crop plants (Benjamin and Nielsen, 2006; Blum, 2017). Plants combat drought stress or get acclimatized to it by means of alterations in structure (anatomy) and growth rate, enzymatic and non-enzymatic oxidative stress management systems and water relations (Duan et al. 2007; Prakash et al. 2016). The two major mechanisms which plants employ to survive through drought are dehydration avoidance and dehydration tolerance (Blum 2005). While early maturing varieties escape and thereby avoid drought, plants with dehydration avoidance mechanisms either minimise the transpirational loss of water or increase the amount of soil moisture extracted from deeper layers of soil when they have better root architecture (Redillas et al. 2012; Uga et al. 2013; Basu et al. 2016).
A number of genes and QTLs have been implicated in imparting drought tolerance to rice (Serraj et al. 2011; Swamy and Kumar 2013; Sahebi et al. 2018; Barik et al. 2019). These are the genes related to diverse plant functions including signalling (kinase) (Saijo et al. 2001; Liu et al. 2003), cell membrane integrity (LEA proteins) (Xu et al. 1996; Babu et al. 2004), carbohydrate metabolism (Jang et al. 2003), water uptake and its transport (aquaporins) (Martre et al. 2002) and oxidative stress management (Prakash et al. 2016). Of the various components of drought tolerance in rice, root architecture remains one of the most promising traits for analysing the differences among the rice genotypes in response to drought, though it has been less explored compared to the above ground changes (Gowda et al. 2011). There are four primary root traits related to drought tolerance, namely root length, volume, thickness and root growth angle (RGA; Uga et al. 2011). RGA is a key component root trait for drought tolerance as it determines the root depth. Deeper and profuse root systems help plants in surviving through the drought stress by extracting water from deeper soil layers. In upland rice, this type of root system has improved the drought tolerance ability of the plants by enhanced uptake of water (Price et al. 1999). Till now, 675 root QTLs and more than 85 genes related to 29 different root parameters have been reported in rice (Courtois et al. 2009; https://snp-seek.irri.org/) though no high yielding rice varieties (HYVs) introgressed with these QTLs/genes have yet been released for cultivation. Thus, the introduction of deep rooting characteristic in HYVs is an efficient way of improving drought tolerance in rice.
Three major QTLs controlling RGA have been reported in rice so far (Uga et al. 2011, 2013, 2015). Among these three, DEEPER ROOTING 1 (DRO1) remains the major one and this QTL has been fine mapped and the underlying gene, an early auxin responsive factor, has been cloned using IR64, a shallow rooted variety and Kinandang Patong (KP), a deeply rooted variety. It has also been shown that the other two QTLs, DRO2 and DRO3 interact with the DRO1 and thus function together. Moreover, a mega programme on QTL-Variety has transferred the major yield under drought QTLs in rice to major cultivated varieties (Singh et al. 2016; Kumar et al. 2014). In this context, we wanted to explore the donors of ‘yield under drought’ (DTY) QTLs and the major upland varieties of India which could be possible donors of DRO1 and a set of popular lowland varieties which could be possible recipients in drought tolerance breeding programmes of rice. The variation in the DRO1gene in the major Indian rice genotypes used in drought tolerant breeding programs and their relationship with root angle is not yet known. This study thus intends to determine the allelic variation in DRO1 across a set of rice genotypes with differential response to drought as well as those representing upland indica and ‘aus’ genotypes and lowland indica genotypes.
Materials and methods
Plant material
A set of 24 rice genotypes comprising of DTY donors from IRRI, upland rice genotypes of indica, aus and japonica types, popular lowland rice cultivars and a few landraces from India were used in the study. The name of the genotypes along with their response to drought stress, if known from the literature, is given in Table 1.
Table 1.
Rice accessions used in the study
| S. no. | Name of the genotype | Response to drought stress | Ecotype (upland/ lowland) |
|---|---|---|---|
| 1 | IR 87,728–367-B-B | Drought tolerant; DTY-2.2 donor | Upland |
| 2 | IR 86,931-B-6 | Drought tolerant; DTY-3.2 donor | Upland |
| 3 | IR 8694-B-B-305 | Drought tolerant; DTY-1.1 donor | Upland |
| 4 | IR 87,728–75-B-B | Drought tolerant; DTY-2.2donor | Upland |
| 5 | IR 81,896-B-B-195 | Drought tolerant; DTY-3.1 donor | Upland |
| 6 | IR 87,728–59-B-B | Drought tolerant; DTY-9.1 donor | Upland |
| 7 | Azucena | Drought tolerant | Upland |
| 8 | Vandana | Drought tolerant | Upland |
| 9 | Dhagaddeshi | Drought tolerant | Upland |
| 10 | Rasi | Drought tolerant | Upland |
| 11 | Nagina 22 | Drought tolerant | Upland |
| 12 | PMK 2 | Drought tolerant | Upland |
| 13 | Bala | Drought sensitive | Upland |
| 14 | IR 20 | Drought sensitive | Lowland |
| 15 | CO 39 | Drought sensitive | Lowland |
| 16 | Swarna | Drought sensitive | Lowland |
| 17 | IR 64 | Drought sensitive | Lowland |
| 18 | Taipei 309 | Drought sensitive | Lowland |
| 19 | Satti | Drought sensitive | Upland |
| 20 | IR 36 | Drought sensitive | Lowland |
| 21 | IC 330,600 | Unknown; land race from Odisha, India | Unknown |
| 22 | Abor Red 4 | Unknown | Unknown |
| 23 | IC 526,266 | Unknown; land race from Andhra Pradesh, India | Unknown |
| 24 | Lalat | Drought tolerant | Upland |
Growth conditions and drought stress treatment
The 24 genotypes were grown in 6″ pots in soilrite (mixture of horticulture grade expanded perlite, Irish Peat moss and exfoliated vermiculite in equal ratio i.e., 1/3:1/3:1/3) in three replications each under two conditions, optimal water supply (WC) and water-deficit stress (WS) and maintained in the climate control glass house, ICAR-National Institute for Plant Biotechnology, New Delhi. For each treatment, three pots per replication were maintained @two plants per pot. So each genotype was represented by nine pots and 18 plants under well-watered treatment. A similar but separate set was used for water stress treatment. The seeds were pre-germinated in the laboratory in a petri dish and the germinated seeds were transferred to pots. The plants were irrigated regularly with half-strength MS medium. Irrigation was withheld under WS treatment 23 days after sowing (DAS). After seven days, i.e., on 30 DAS, from each replication three plants were used for phenotyping for relative water content (RWC), root length (cm) and root angle (°) while three plants were snap frozen and stored for allele mining and expression studies.
Morphological and physiological observations
RWC of the leaf samples was measured according to Barrs and Weatherley (1962). Roots were pulled from soilrite carefully and roots from both WC and WS treatment were scanned under Epson Perfection v700 Photo-Dual lens system root scanner. The images of the roots of each of the genotypes and root length (cm) and root angle (°) obtained via scanner were analysed. All the morphometric were analyzed in XL-stat.
DNA isolation and Primer designing for DRO1
Fresh young leaves from the 30 days old seedlings kept under WC were collected and kept at -80 °C till further use. DNA isolation was carried out by CTAB method (Doyle and Doyle, 1990) with some minor modifications. The full length gene sequence of DRO1 was downloaded from the ‘Rice Annotation Project Database’ and four pairs of overlapping primers (PP1 to PP4) covering the entire gene (3058 bp) (supplementary Figure 1), and with estimated amplicon length ranging from 904–944 bp, were designed using PRIMER 3 software (Supplementary Table 1). To ensure that single specific amplicons are obtained after amplification, primer sequences were searched in the rice genome (ensemble gramene) by BLAST and only those primers with single or minimum number of hits in the genome were chosen. PCR was carried out in 25 µl volume comprising of template DNA @ 30 ng/µl, 5 pmol of forward and reverse primers (Sigma Inc, India), 10 mMdNTPs (Takara) and 1 U of Taq DNA polymerase (Takara) and buffer containing MgCl2. The PCR products were examined in 2% agarose gel.
Sequencing of PCR amplified products
For removal of unused dNTPs and primers, purification using Exo-SAP (Exonuclease-Shrimp Alkaline Phosphatase) was carried out. 2 µl of Exo-SAP and 5 µl of PCR amplified products were kept for incubation at 37 °C for 15 min which were further incubated at 80 °C for 15 min. The purified products were subjected to (cycle) sequencing PCR using forward and reverse primers in separate reactions. Following this, purification of sequencing reaction products was carried out by addition of 10 µl EDTA (0.125 M) and 80 µl of ethanol with the total reaction volume being 10 µl and centrifugation at 3220 g. After discarding the supernatant, 70% ethanol washing was carried out twice and followed by centrifugation done at 3220 g for 30 min. 10 µl HiDiformamide was added to the purified product and incubated at 95 °C for 15 min for denaturation. For sequencing, denatured product was loaded in the sequencer maintained at ICAR-NIPB (DNA analyzer 3730xl, ABI, USA).
SNP analysis
The quality of the sequences was checked using the in-built software of the sequencer. Sequencing was repeated wherever the sequencing quality was below Phred score 20 and till complete sequence information was obtained for DRO1 in all the 24 genotypes. Contigs were made using CAP Contig Assembly in BioEdit software (Hall 1999). The consensus sequences were aligned with DRO1 sequence of Kinandang Patong (KP), used as the reference. Multiple sequence alignment of all the 24 genotypes was carried out using ClustalW (Larkin et al. 2007). CDS as well as amino acid sequences were aligned separately. Both SNP report and amino acid substitution report were generated from ClustalW.
RNA isolation and expression profiling by qRT-PCR
For RNA isolation, roots from WC treatment were cleaned in running water while the roots from WS treatment were cleaned with wet brush and immediately frozen in liquid N2. RNA isolation was carried out using AmbionPureLink® RNA Mini Kit following the manufacturer’s protocol. For determination of RNA quantity and quality, 260/280, 260/230 and concentration (ng/µl) values were taken using Nanodrop8000 spectrophotometer (Thermo Scientific, USA) and gel electrophoresis with formaldehyde agarose gel was carried out. For, cDNA conversion, first-strand synthesis kit (Thermo Fisher, USA) was used. qRT-PCR was then performed in three biological and three technical replications using a Roche LightCycler® 480 II machine facilitated with a 96 well plate system with the (VeriQuest SYBR® FAST) Master Mix reagent (Affymetrix, USA). The final reaction volume was 10 μl consisting of 2 μl cDNA having 50 ng cDNA, 0.5 μM of forward and reverse primer each and 5 µl of VeriQuest SYBR® FAST qPCR mastermix. For normalization, actin primers were used. The Pfaffl formula (Ratio = 2−ΔΔCt) was used to calculate the relative expression of DRO1 gene under WC and WS (Pfaffl et al. 2001).
Protein modelling by I-TASSER
The amino acid sequences of the reference Kinandang Patong (KP) and two rice accessions, Nagina 22 and DTY 9.1 donor (IR 87728–59-B-B) which had different number of amino acid substitutions compared to the reference were submitted to I-TASSER (https://zhanglab.ccmb.med.umich.edu/I-TASSER; Zhang 2008; Roy et al. 2010) for protein structure models.
Results
Evaluation of rice genotypes for root angle and root length under WC and WS
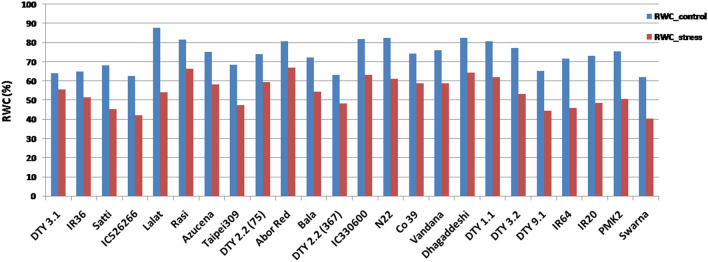
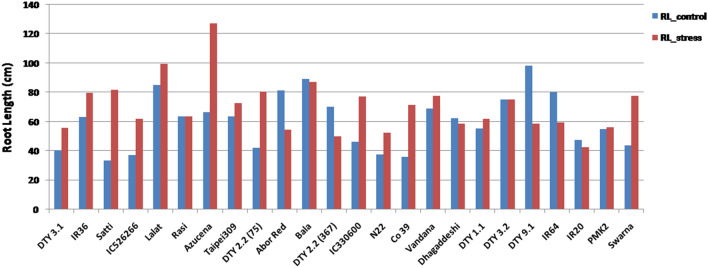
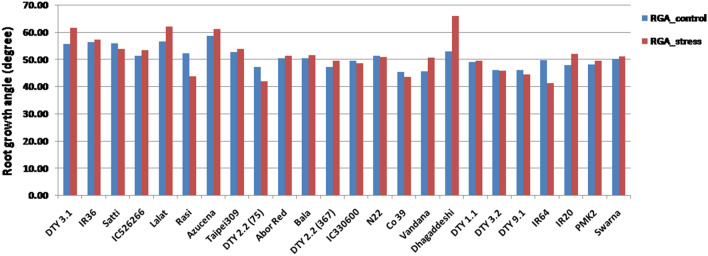
The RWC under WC varied from 61.94 to 87.86% while under WS, it had gone down to 39.36 to 66.93% implying that the stress condition was imposed appropriately and there was inherent variation among the genotypes for RWC (Fig. 1 and Table 2a). Lalat, followed by DTY 1.1 donor, Nagina 22, Dhagaddeshi, Rasi, Abor Red and Vandana had higher RWC (> 80%) while Swarna and IC526266 had the least RWC (< 62%) even under control conditions (Fig. 1). Similar trend was observed under WS too in these genotypes except Lalat. Lalat showed the highest % reduction in RWC between the treatments, WC and WS (38%). The co-efficient of variation (CV) for RWC was higher under WS compared to WC (14.46 against 10.18%; Table 2). Across all the genotypes, root length increased under WS compared to WC while CV did not (Fig. 2 and Table 2a). The mean root length across the rice genotypes was 60.07 cm under WC while it was 71.39 cm under WS (Table 2). Many genotypes, namely, Swarna, Satti, Azucena, Co39 and DTY 2.2 (75) showed more than 40% increase in root length under WS (Fig. 2).The range for RGA under WC was 45.6°–56.6° while under WS, it was 41.3° to 61.98° degrees. The standard deviations for RGA were nearly double (3.82 and 6.01) while the means were nearly equal (50.66 and 51.61) between the treatments, WC and WS respectively (Table 2a) suggesting that the RGA showed more variation under WS and it is a drought induced trait (Fig. 3). Pearson’s linear correlation analysis revealed that correlation between RL (root length) and RGA was not significant under any of the two treatments (Table 2b), while the correlation between WC and WS for RGA was significant (p < 0.01). Interestingly, RGA under WC showed significant correlation with RL under WS.
Fig. 1.
Relative water content (%) in the 24 rice genotypes under WC and WS
Table 2.
(a) Evaluation of rice genotypes under WC and WS for root traits. (b) Linear correlations among root angle and root length under WC and WS
| Descriptive statistics | RWC (%) | Root length (cm) | Root angle (°) | |||
|---|---|---|---|---|---|---|
| WC | WS | WC | WS | WC | WS | |
| (a) | ||||||
| Mean | 73.55 | 54.20 | 60.07 | 71.39 | 50.66 | 51.61 |
| Range | 61.94–87.86 | 39.36–66.93 | 33.43–98.40 | 42.38–127.06 | 45.60–56.63 | 41.26–61.98 |
| Standard deviation | 7.48 | 7.84 | 17.54 | 17.80 | 3.82 | 6.01 |
| CV (%) | 10.18 | 14.46 | 29.20 | 24.94 | 7.55 | 11.65 |
| RL-WC | RL-WS | RGA-WC | |
|---|---|---|---|
| (b) | |||
| RL-WS | 0.134 ns | ||
| RGA-WC | − 0.054 ns | 0.48** | |
| RGA-WS | 0.09 ns | 0.255 ns | 0.772** |
RL root length, RGA root growth angle, WC optimum water supply, WS water-deficit stress, ns non-significant
**p < 0.01
Fig. 2.
Root length (cm) across the study material under WC and WS regimes
Fig. 3.
Root growth angle across the study material under well watered and water deficit regimes
Allele mining for DRO1 gene in rice
Single nucleotide polymorphisms (SNPs) in DRO1 in the experimental material
The complete gene had 216 SNPs (7%) and 239 InDels whereas CDS of 756 bp of DRO1 had 52 SNPs (7%) and no InDels with respect to KP genotype (as reference) from which the functional DRO1 gene was mapped and functionally validated. Out of the 216 SNPs in the complete gene, there were 180 transversions and 50 transitions, thus yielding 3.6 times more transversions than transitions. Interestingly, 14 positions had both transitions and transversions as well as InDels thus yielding tetra allelic variations across the accessions. The maximum number of changes were harboured by IC526266, Azucena, IC330600, Rasi and PMK2 having 190, 174, 143, 143 and 138 changes respectively. Along with the higher number of SNP changes, Azucena also had the longest InDel region of 44 nucleotides (Supplementary Table 2). Among the 52 SNPs of CDS, 15 were transitions while 37 were transversions with nearly 2.5 fold higher transversions (Supplementary Table 2). In CDS also, a triallelic SNP was found at position 375 (A to G and A to T changes) and hence 51 positions harboured 52 SNPs. The SNP positions which had mutations in high frequency were 400, 401 and 402 (in 10 genotypes) followed by SNP 274 (9 genotypes) and SNPs 91 and 93 (8 genotypes) (Supplementary Figure 2).The genotype-wise comparison of SNPs in the DRO1 gene revealed that DTY 3.1 had the least number of variation (1 SNP) followed by DTY 2.2 donor while DTY 9.1 had the highest number of SNPs (19 SNPs). Other DTY donors, 3.2 and 1.1 also had high number of SNPs, 16 and 15 respectively (Supplementary Table 3). In fact, only DTY 3.1 and 2.2 donors (IR81896-B-B-195 and IR 87728–75-B-B) had minimal number of SNPs.
Amino acid substitutions in DRO1 in the experimental material
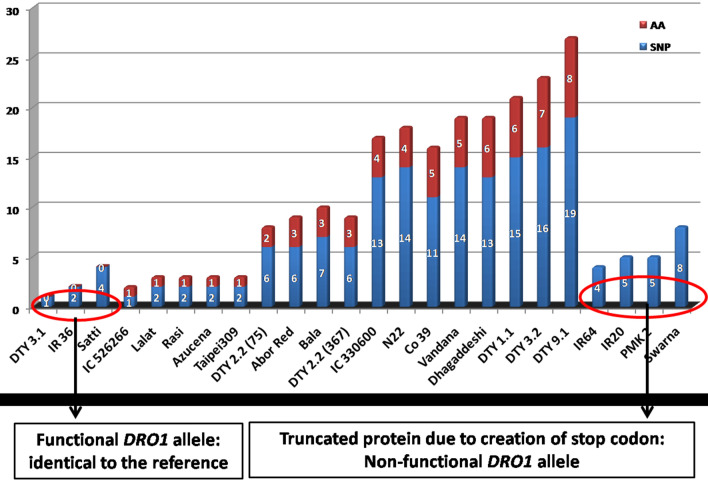
Across the gene, there were 19 amino acid changes in the 24 genotypes (Supplementary Figure 3) (Fig. 4). The reference position 92 showed the highest number of amino acid substitutions (9 genotypes; phenylalanine to isoleucine) followed by amino acid substitution of asparagine to histidine in position 31 in 8 genotypes (Supplementary Table 3 and supplementary Figure 3). From allele mining for DRO1, the entire study material could be clubbed into 16 different primary structure variants, covering the variation from complete functional allele (Satti, IR36 and IR81896-B-B-195) to truncated non-functional alleles in PMK2, IR64, IR20 and Swarna owing to two transversions at positions 475 (A to T) and 477 (C to A) changing the reading frame from AGC to TGA i.e., serine to stop codon (Fig. 4 and Supplementary Table 4). The genotypes that harboured maximum amino acid substitutions were donors of QTLs, DTY 9.1, 3.2, 1.1, and two drought tolerant accessions Dhagaddeshi followed by Vandana and a popular variety Co39 (Fig. 4). Genotypes that harboured a few amino acid changes included IC 526266 (31; glutamic acid to aspartic acid), Lalat, Azucena, Rasi and Taipei 309 (39; asparagine to histidine) all with single amino acid substitution, followed by DTY 2.2 (75) with two amino acid substitutions.
Fig. 4.
Allelic variation in the DRO1 gene in the rice accessions studied
Expression analysis of DRO1 in the study material under WC and WS
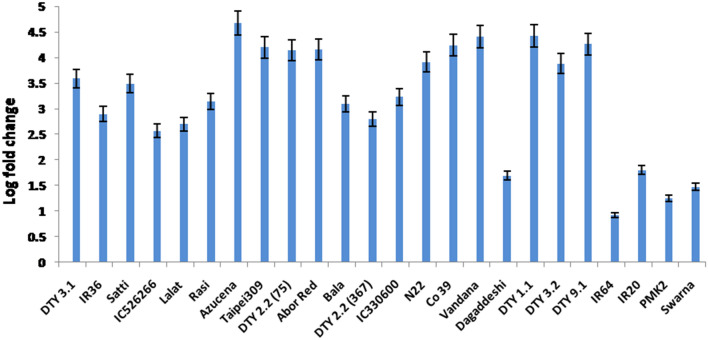
A pair of primers specific to DRO1 gene (F: 5′-3′: TAGTTCCAGAACCAAGGAACAC; and R: 5′-3′: CTTCCAGTCCAAATGCCTCTTA) and actin (F: 5′-3′: CTGGGTTCGCCGGAGATGAT; R: 5′-3′: TGAGATCACGCCCAGCAAGG) were designed for expression analysis. Compared to WC conditions, the expression of DRO1 was upregulated under WS across all the 24 genotypes tested suggesting that cis-element control in all the rice accessions, irrespective of their lineage, was similar. This also suggested that the structural variation in the alleles of DRO1 is more important than their transcriptional control as the RGA of these genotypes varied (Figs. 3 and 5). In genotypes, IR64, IR20, Dhagaddeshi, PMK2 and Swarna, the upregulation was less than two fold while in the popular drought tolerant japonica variety, Azucena, and another japonica genotype Taipei 309, the upregulation was nearly fivefold and fourfold respectively. Dhagaddeshi and PMK2, well-known drought tolerant genotypes showed least upregulation, in fact, on par with well-known drought sensitive genotypes such as IR64, IR20 and Swarna which harboured a non-functional allele.
Fig. 5.
Expression analysis of DRO1 in WS with WC as control in the study material
Prediction of protein structure
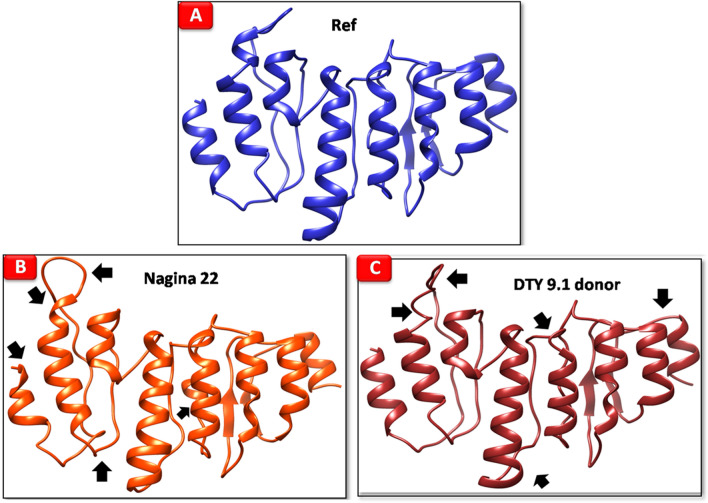
Significant differences were observed between the protein model of reference genotype KP and the two genotypes tested (Fig. 6). The reference genotype showed a mixed α-β structure having α-helix along with two twisted β-sheets. In Nagina 22, and DTY 9.1 donor (IR 87728–59-B-B), the differences were observed in the α-helical configurations, wherein loop formation was not similar to the reference.
Fig. 6.
Protein models of reference genotype, Nagina 22, DTY 2.2 (75) donorand DTY 9.1 donor (predicted by I-TASSER)
Discussion
Drought causes several changes at the physiological, metabolic and molecular level and thus hampers the normal growth and development process of plants (Zu et al. 2017). As response to drought stress in a species varies across the genotypes and which is strongly affected by environment and G x E interaction, the utility of physiological traits as an indirect selection criteria (surrogate) would be crucial in supplementing the yield-based selection procedures (Lonbani and Arzani, 2011). Drought tolerance is indicated by high water status (high leaf water potential and relative water content), better osmotic adjustment, greater membrane stability and high proline content (Larkunthod et al. 2018). Zu et al. 2017 have developed a new method called ‘Drought Tolerance Degree’ or DTD method for efficiently evaluating the DTD of upland rice cultivars. DTD values are correlated with several physiological traits such as, water potential, proline, chlorophyll content and fluorescence, malondialdehyde content, survival rate and yield per plant. The role of grain carbon isotope discrimination has been established as an indirect selection criterion for evaluating drought tolerance and osmotic stress tolerance of crops (Yasir et al. 2019; Faseela et al. 2019). Similarly, different morpho-physiological indicators have also been used for evaluation of degree of tolerance aganist salinity stress in rice seedlings (Abdelhamid 2020; Rasel et al. 2020).
Root growth angle (RGA) has been explored considerably in rice with three major QTLs and several minor QTLs reported, of which one major QTL is dissected to identify the DRO1 gene (Uga et al. 2011, 2013, 2015; Kitomi et al. 2015). Other than characterization of a few upland varieties, that too mostly japonica, the rest of the germplasm, especially the ones relevant to drought tolerance breeding in the indica rice growing regions, remain unexplored for RGA (Kato et al. 2006; Kitomi et al. 2015).
Reduction in growth and reproductive parameters is a common feature under stress (Pandit et al. 2013; Lima et al. 2015; Tiwari et al. 2016). However, root length is an exception and always increases under drought stress compared to ambient conditions (Liu et al. 2004; Lima et al. 2015; Xu et al. 2015). In the present study also, increase in root length across all the 24 genotypes was observed in WS conditions compared to WC (Fig. 2). Though most of the linear correlations between root length and RGA under the two water regimes were not significant, the correlation between RGA in WS and WC was significant (Table 2). This suggested that the constitutive genetic determinants for RGA are stronger and remain the same irrespective of the growth conditions as observed in a study with 12 upland rice genotypes (Kato et al. 2006). Importantly, based on the experimentation, the study material could be classified into two groups, namely, wide or deep RGA genotypes (9 rice accessions; Azucena, DTY 3.1, IR36, Satti, IC526266, Lalat, Rasi, Nagina22 and Dhagaddeshi) and narrow or shallow RGA genotypes (15 accessions). Further we have tried to correlate the % reduction in RWC and % increase in RGA and RL under water stress with the allelic groups by assigning a drought tolerance score based on the performance of the reference set of 24 genotypes used in the study (supplementary table 5). For example, for RWC where low % reduction is the favourable drought tolerance response, those genotypes with % reduction value less than the mean (m) and standard deviation (sd) of the 24 genotypes were assigned a score of 1; those between this value (m-sd) and mean were assigned a score of 0.75; those between mean and (m + sd) were assigned a score 0.5 and those greater than the m + sd were assigned a score of 0.25. The same pattern of scoring was followed for RGA and RL too with the difference being there higher increase got higher scores (i.e., > m + sd got the score of 1). The cumulative score over the three traits was finally compared with the allelic groups. The cumulative score though did not have one-on-one relationship with the allelic groups, the functional groups with no amino acid substitutions had higher scores (2.25–2.75) except in case of Satti. Similarly, all the stop codon groups had a lower cumulative score ranging from 0.75 to 1.75. Thus RGA, RL and RWC do not explain the drought tolerance nature of a genotype in its entirety. They are only some components of drought tolerance; nevertheless important to improve drought tolerance.
Gene rich regions in the indica subspecies are known to harbour comparatively higher variation than japonica (Zhang et al. 2011). The study material had two accessions from japonica, one from aus and the rest 21 from indica including some of the landraces from different parts of India (Table 1; Tiwari et al. 2015). In both the CDS and complete gene 7% of SNPs with an additional 7% of InDels in the latter. Considering the number of genotypes involved in the study, the frequency of SNPs obtained is quite high (Bao et al. 2006; Dixit et al. 2013). This could be because rice has been largely adapted to irrigated ecosystem with hardly any need for deep root systems and hence genes like DRO1 are not important for survival and could accumulate more mutations. As against the well accepted notion that transitions are more frequent than transversions, especially in exonic regions across species (Tsaftaris and Polidoros, 2000), we found more transversions (nearly > two fold) than transitions as reported in a few studies (Dixit et al. 2013).
Despite the huge number of SNPs in the gene, there were only a total of 19 amino acid substitutions which is suggestive of their common lineage or ancestry. For example, among the DTY donors, only DTY 3.1 had the complete functional allele; while DTY donors 3.2 and 9.1 had glutamic acid to aspartic acid substitution at position 125, Nagina 22 and DTY 1.1 donor had glutamic acid to alanine change. Since, Nagina 22 is the donor of DTY 1.1, this could be expected (Singh et al. 2016). Though the expression analyses showed that most of the DTY donors and upland genotypes like Vandana showed enhanced expression of DRO1, the alleles were quite different across these genotypes harbouring substantial variations in the primary and secondary structure of proteins (Figs. 5 and 6).
Conclusion
The current report on DRO1 allele mining can serve as a guideline for designing markers for selecting for functional DRO1 in breeding programmes aimed at enhancing drought tolerance. Further, the on-going drought tolerance breeding programmes in rice across globe can benefit from the information generated here on designing their breeding schemes.
Supplementary information
Acknowledgements
The first author acknowledges the financial support in the form of fellowship from Post-Graduate School, ICAR-Indian Agricultural Research Institute, New Delhi during her Master of science program.
Author contributions
BKS and RK carried out the experiment and compiled the results, BKS prepared the manuscript, MD and AS supervised drought stress and expression studies respectively, NKS supplied the seed material, AKS involved in conceptualization of experiment and manuscript writing and editing and SVA conceptualized, supervised the whole experiment, edited and finalized the manuscript. All the authors read and approved the manuscript.
Funding
The corresponding author acknowledges the institute funding from ICAR-National Institute for Plant Biotechnology through Research project proposal (RPP) mode.
Compliance with ethical standards
Conflicts of interest
The authors declare no conflicting interests for publishing it to the journal ‘Physiology and Molecular Biology of Plants’.
Consent for publication
All the authors give their consent to publish the research article to the journal ‘Physiology and Molecular Biology of Plants’.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Bablee Kumari Singh and M. K. Ramkumar have contributed equally for this work.
References
- Abdelhamid MT (2020) New approaches for improving salt stress tolerance in rice. In: Roychoudhury A (ed), rice research for quality improvement. Genomics and genetic engineering. Doi:10.1007/978-981-15-4120-9_10
- Babu RC, Zhang JX, Blum A, Ho THD, Wu R, Nguyen HT. HVA1, a LEA gene from barley confers dehydration tolerance in transgenic rice (Oryza sativa L.) via cell membrane protection. Plant Sci. 2004;166:855–862. doi: 10.1016/j.plantsci.2003.11.023. [DOI] [Google Scholar]
- Bao JS, Corke H, Sun M. Nucleotide diversity in starch synthaseIIa and validation of single nucleotide polymorphisms in relation to starch gelatinization temperature and other physicochemical properties in rice (Oryza sativa L.) Theor Appl Genet. 2006;113:1171–1183. doi: 10.1007/s00122-006-0355-6. [DOI] [PubMed] [Google Scholar]
- Barah BC (2005) Dynamics of rice economy in india: emerging scenario and policy options. Department of Economic Analysis and Research National bank for agriculture and rural development Mumbai Occasional Paper 47.
- Barik SR, Pandit E, PradhanSK MSP, Mohapatra T. Genetic mapping of morpho-physiological traits involved during reproductive stage drought tolerance in rice. PLoS ONE. 2019;14(12):e0214979. doi: 10.1371/journal.pone.0214979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrs HD, Weatherly PE. A re-examination of the relative turgidity technique for estimating water deficits in leaves. Aust J Biol Sci. 1962;15:413–428. doi: 10.1071/BI9620413. [DOI] [Google Scholar]
- Basu S, Ramegowda V, Kumar A, Pereira A. Plant adaptation to drought stress. F1000 Res. 2016;5:1554–1563. doi: 10.12688/f1000research.7678.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamin JG, Nielsen DC. Water deficit effects on root distribution of soybean, field pea and chickpea. Field Crops Res. 2006;97:248–253. doi: 10.1016/j.fcr.2005.10.005. [DOI] [Google Scholar]
- Blum A. Drought resistance, water-use efficiency, and yield potential-are they compatible, dissonant, or mutually exclusive? Aust J Agric Res. 2005;56:1159–1168. doi: 10.1071/AR05069. [DOI] [Google Scholar]
- Blum A. Osmotic adjustment is a prime drought stress adaptive engine in support of plant production. Plant Cell Environ. 2017;40(1):4–10. doi: 10.1111/pce.12800. [DOI] [PubMed] [Google Scholar]
- Courtois B, Ahmadi N, Khowaja F, Price AH, Rami JF, Frouin J, Ruiz HC, M, Rice root genetic architecture: Meta-analysis from a drought QTL database. Rice. 2009;2:115–128. doi: 10.1007/s12284-009-9028-9. [DOI] [Google Scholar]
- Dixit N, Dokku P, Amitha Mithra SV, Parida SK, Singh NK, Mohapatra T. Haplotype structure in grain weight gene GW2 and its association with grain characteristics in rice. Euphytica. 2013;192(1):55–61. doi: 10.1007/s10681-012-0852-4. [DOI] [Google Scholar]
- Doyle JJ, Doyle JL. Isolation of plant DNA from fresh tissue. Focus. 1990;12(1):13–15. [Google Scholar]
- Duan B, Yang Y, Lu Y, Korpelainen H, Berninger F, Li C. Interactions between drought stress, ABA and genotypes in Piceaasperata. J Exp Bot. 2007;58:3025–3036. doi: 10.1093/jxb/erm160. [DOI] [PubMed] [Google Scholar]
- Faseela P, Sinisha AK, Brestic M, Puthur JT. Chlorophyll a fluorescence parameters as indicators of a particular abiotic stress in rice. Photosynthetica. 2019;57(SI):108–115. [Google Scholar]
- Gowda VRP, Henry A, Yamauchi A, Shashidhar HE, Serraj R. Root biology and genetic improvement for drought avoidance in rice. Field Crops Res. 2011;122:1–13. doi: 10.1016/j.fcr.2011.03.001. [DOI] [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids SympSer. 1999;41:95–98. [Google Scholar]
- Jang IC, Oh SJ, Seo JS, Choi WB, Song SI, Kim CH, Kim YS, Seo HS, DoChoi Y, Nahm BH, Kim JK. Expression of a bifunctional fusion of the Escherichia coli genes for trehalose-6-phosphatesynthase and trehalose-6-phosphate phosphatase in transgenic rice plants increases trehalose accumulation and abiotic stress tolerance without stunting growth. Plant Physiol. 2003;131:516–524. doi: 10.1104/pp.007237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato Y, Abe J, Kamoshita A, Yamagishi J. Genotypic variation in root growth angle in rice (Oryza sativa L.) and its association with deep root development in upland fields with different water regimes. Plant Soil. 2006;287:117–129. doi: 10.1007/s11104-006-9008-4. [DOI] [Google Scholar]
- Kitomi Y, Kanno N, Kawai S, Mizubayashi T, Fukuoka S, Uga Y. QTLs underlying natural variation of root growth angle among rice cultivars with the same functional allele of DEEPER ROOTING 1. Rice. 2015;8:16. doi: 10.1186/s12284-015-0049-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Dixit S, Ram T, Yadaw RB, Mishra KK, Mandal NP. Breeding high-yielding drought-tolerant rice genetic variations and conventional and molecular approaches. J Exp Bot. 2014;65(21):6265–6278. doi: 10.1093/jxb/eru363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23(21):2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Larkunthod P, Nounjan N, Siangliw JL, Toojinda T, Sanitchon J, Boonrat J, Theerakulpisut P. Physiological responses under drought stress of improved drought—tolerant rice lines and their parents. Not Bot Horti Agrobo. 2018;46(2):679–687. doi: 10.15835/nbha46211188. [DOI] [Google Scholar]
- Lima JM, Nath M, Dokku P, Raman KV, Kulkarni KP, Vishwakarma C, Sahoo SP, Mohapatra UB, AmithaMithra SV, Chinnusamy V, Robin S, Sarla N, Seshashayee M, Singh K, Singh AK, Singh NK, Sharma RP, Mohapatra T. Physiological, anatomical and transcriptional alterations in a rice mutant leading to enhanced water stress tolerance. AoB Plants. 2015;7:plv023. doi: 10.1093/aobpla/plv023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Xu ZH, Luo D, Xue HW. Roles of OsCKI1, a rice casein kinase i, in root development and plant hormone sensitivity. Plant J. 2003;36:189–202. doi: 10.1046/j.1365-313x.2003.01866.x. [DOI] [PubMed] [Google Scholar]
- Liu HS, Li FM, Xu H. Deficiency of water can enhance root respiration rate of drought-sensitive but not drought-tolerant spring wheat. Agr Water Manage. 2004;64:41–48. doi: 10.1016/S0378-3774(03)00143-4. [DOI] [Google Scholar]
- Lonbani M, Arzani A. Morpho-physiological traits associated with terminal drought stress tolerance in triticale and wheat. Agron Res. 2011;9(1–2):315–329. [Google Scholar]
- Martre P, Morillon R, Barrieu F, North GB, Nobel PS, Chrispeels MJ. Plasma membrane aquaporins play a significant role during recovery from water deficit. Plant Physiol. 2002;130:2101–2110. doi: 10.1104/pp.009019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandit A, Rai V, Bal S, Sinha S, Kumar V, Chauhan M, Gautam RK, Singh R, Sharma PC, Singh AK, Gaikwad K, Sharma TR, Mohapatra T, Singh NK. Combining QTL mapping and transcriptome profiling of bulked RILs for identification of functional polymorphism for salt tolerance genes in rice (Oryza sativa L.) Mol Genet Genomics. 2013;284:121–136. doi: 10.1007/s00438-010-0551-6. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucl Acids Res. 2001;29:2002–2007. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash C, Mithra SVA, Singh PK, Mohapatra T, Singh NK. Unraveling the molecular basis of oxidative stress management in a drought tolerant rice genotype nagina 22. BMC Genomics. 2016;17:774–787. doi: 10.1186/s12864-016-3131-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AH, Courtois B. Mapping QTLs associated with drought resistance in rice: progress, problems and prospects. Plant Growth Regul. 1999;29:123–133. doi: 10.1023/A:1006255832479. [DOI] [Google Scholar]
- Rasel M, Tahjib-Ul-Arif M, Hossain MA, Hassan L, Farzana S, Brestic M. Screening of salt-tolerant rice landraces by seedling stage phenotyping and dissecting biochemical determinants of tolerance mechanism. J Plant Growth Regul. 2020 doi: 10.1007/s00344-020-10235-9. [DOI] [Google Scholar]
- Redillas MC, Jeong JS, Kim YS, Jung H, Bang SW, Choi YD, Ha SH, Reuzeau C, Kim JK. The overexpression of OSNAC9 alters the root architecture of rice plants enhancing drought resistance and grain yield under field conditions. Plant Biotechnol J. 2012;10:792–805. doi: 10.1111/j.1467-7652.2012.00697.x. [DOI] [PubMed] [Google Scholar]
- Roy A, Kucukural A, Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc. 2010;5(4):725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahebi M, Hanafi MM, Rafii YY, Mahmud TMM, Azizi P, Osman M, Abiri R, Taheri S, Kalhori N, Shabanimofrad M, Miah G, Atabaki N. Improvement of drought tolerance in rice (Oryza sativa L.): genetics, genomic tools, and the WRKY gene family. BioMed Res Int. 2018 doi: 10.1155/2018/3158474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saijo Y, Kinoshita N, Ishiyama K, Hata S, Kyozuka J, Hayakawa T, Nakamura T, Shimamoto K, YamayaT IK. A Ca2+-dependent protein kinase that endows rice plants with cold- and saltstresstolerance functions in vascular bundles. Plant and Cell Physiol. 2001;42:1228–1233. doi: 10.1093/pcp/pce158. [DOI] [PubMed] [Google Scholar]
- Serraj R, Kumar A, McNally KL, Slamet-Loedin I, Bruskiewich R, Mauleon R, Cairns J, Hijmans RJ. Improvement of drought resistance in rice. AdvAgron. 2009;103:41–98. [Google Scholar]
- Serraj R, McNally KL, Slamet-Loedin I, Kohli A, HaefeleSM AG, Kumar A. Drought resistance improvement in rice: an integrated genetic andresource management strategy. Plant Prod Sci. 2011;14:1–14. doi: 10.1626/pps.14.1. [DOI] [Google Scholar]
- Singh R, Singh Y, Xalaxo S, Verulkar S, Yadav N, Singh S, Singh N, Prasad KSN, Kondayya K, Rao PVR, Rani MG, Anuradha T, Suraynarayana Y, Sharma PC, Krishnamurthy SL, Sharma SK, Dwivedi JL, Singh AK, Singh PK, Nilanjay SNK, Kumar R, Chetia SK, Ahmad T, Rai M, Perraju P, Pande A, Singh DN, Mandal NP, Reddy JN, Singh ON, Katara JL, Marandi B, Swain P, Sarkar RK, Singh DP, Mohapatra T, Padmawathi G, Ram T, Kathiresan RM, Paramsivam K, Nadarajan S, Thirumeni S, Nagarajan M, Singh AK, Vikram P, Kumar A, Septiningshih E, Singh US, Ismail AM, Mackill D, Singh NK. From QTL to variety-harnessing the benefits of QTLs for drought, flood and salt tolerance in mega rice varieties of India through a multi-institutional network. Plant Sci. 2016;242:278–287. doi: 10.1016/j.plantsci.2015.08.008. [DOI] [PubMed] [Google Scholar]
- Suhail AJ, Ahmad M, Asghar M, Tzyyib MM. Determination of economic threshold level for the chemical control of rice stems borers (Sciropophaga incertules WIk. and Sciropophaga innotata WIk.) Pak Entomol. 2008;30(2):175–178. [Google Scholar]
- Swamy BPM, Kumar A. Genomics-based precision breeding approaches to improve drought tolerance in rice. Biotechnol Adv. 2013;31:1308–1318. doi: 10.1016/j.biotechadv.2013.05.004. [DOI] [PubMed] [Google Scholar]
- Tiwari KK, Singh A, Pattnaik S, Sandhu M, Kaur S, Jain S, Tiwari S, Mehrotra S, Anumalla M, Samal R, Bhardwaj J, Dubey N, Sahu V, Kharshing GA, Zeliang PK, Sreenivasan K, Kumar P, Parida SK, Mithra SVA, Rai V, Tyagi W, Agrawal PK, Rao AR, Pattanayak A, Chandel G, Singh AK, Bisht IS, Bhat KV, Rao GJN, Khurana JP, Singh NK, Mohapatra T. Identification of a diverse mini-core panel of Indian rice germplasm based on genotyping using microsatellite markers. Plant Breed. 2015;134(2):164–171. doi: 10.1111/pbr.12252. [DOI] [Google Scholar]
- Tiwari S, Lata C, Chauhan PS, Nautiyal CS. Pseudomonas putida attunes morpho physiological, biochemical and molecular responses in Cicer arietinum L. during drought stress and recovery. Plant Physiol Biochem. 2016;99:108–117. doi: 10.1016/j.plaphy.2015.11.001. [DOI] [PubMed] [Google Scholar]
- Tsaftaris AS, Polidoros AN. DNA methylation and plant breeding. Plant Breed Rev. 2000;18:87–176. doi: 10.1002/9780470650158.ch3. [DOI] [Google Scholar]
- Uga Y, Okuno K, Yano M. Dro1, a major QTL involved in deep rooting of rice under upland field conditions. JExp Bot. 2011;62(8):2485–2494. doi: 10.1093/jxb/erq429. [DOI] [PubMed] [Google Scholar]
- Uga Y, Sugimoto K, Ogawa S, Rane S, Ishitani M, Hara N, Kitomi Y, Inukai Y, Ono K, Kanno N, Inoue H, Takehisa H, Motoyama R, Nagamura Y, Wu J, Matsumoto T, Takai T, Okuno K, Yano M. Control of root system architecture by DEEPER ROOTING 1 increases rice yield under drought conditions. Nat Genet. 2013;45(9):1097–1102. doi: 10.1038/ng.2725. [DOI] [PubMed] [Google Scholar]
- Uga Y, Kitomi Y, Yamamoto E, Kanno N, Kawai S, Mizubayashi T, Fukuoka S. A QTL for root growth angle on rice chromosome 7 is involved in the genetic pathway of DEEPER ROOTING 1. Rice. 2015;8:8. doi: 10.1186/s12284-015-0044-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Duan X, Wang B, Hong B, Ho TH, Wu R. Expression of a late embryogenesis abundant (LEA) protein gene, HvA1, from barley confers tolerance to drought and salinity in transgenic rice. Plant Physiol. 1996;110:249–257. doi: 10.1104/pp.110.1.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Cui K, Xu A, Nie L, Huang J, Peng S. Drought stress condition increases root to shoot ratio via alteration of carbohydrate partitioning and enzymatic activity in rice seedlings. Acta Physiol Plant. 2015;37:9. doi: 10.1007/s11738-014-1760-0. [DOI] [Google Scholar]
- Yasir TA, Wasaya A, Hussain M, Ijaz M, Farooq M, Farooq O, Nawaz A, Hu YG. Evaluation of physiological markers for assessing drought tolerance and yield potential in bread wheat. Physiol Mol Biol Plants. 2019;25(5):1163–1174. doi: 10.1007/s12298-019-00694-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. I-TASSER server for protein 3D structure determination. BMC Bioinformatics. 2008;9:40–47. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Li J, Li X, Liu X, Zhao X, Lu Y. Population structure and genetic diversity in a rice core collection (Oryza sativa L.) investigated with SSR markers. PLoSONE. 2011;6(12):e27565. doi: 10.1371/journal.pone.0027565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Zhang S, Cheng M, Jiang H, Zhang X, Peng C, Lu X, Zhang M, Jin J. Effect of drought on agronomic traits of rice and wheat: a meta-analysis. Int J Environ Res Public Health. 2018;15(5):839–853. doi: 10.3390/ijerph15050839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zu X, Lu Y, Wang Q, Chu P, Miao W, Wang H, La H. A new method for evaluating the drought tolerance of upland rice cultivars. Crop J. 2017;5:488–498. doi: 10.1016/j.cj.2017.05.002. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.