Abstract
By using complementary DNA sequences as surface ligands, we selectively allow two individual diffusing ‘dual-color’ carbon dots to interact in situ and in vitro. Spontaneous nanoscale oxidation of surface-abundant nitroso-/nitro- functionalities leads to two distinctly colored carbon dots (CD) which are isolated by polarity driven chromatographic separation. Green and red emitting carbon dots were decorated by complementary single stranded DNAs which produce a marked increase in the fluorescence emission of the respective carbon dots. Mutual colloidal interactions are achieved through hybridization of complementary DNA base pairs attached to the respective particles, resulting in quenching of their photoluminescence. The observed post-hybridization quenching is presumably due to a combined effect from an aggregation of CDs post duplex DNA formation and close proximity of multi-colored CDs, having overlapped spectral regions leading to a non-radiative energy transfer process possibly released as heat. This strategy may contribute to the rational design of mutually interacting carbon dots for a better control over the resulting assembly structure for studying different biological phenomenon including molecular cytogenetics. One of the newly synthesized CDs was successfully used to image intracellular location of GAPDH mRNA using event of change in fluorescence intensity (FI) of CDs. This selectivity was introduced by conjugating an oligonucleotide harboring complementary sequence to GAPDH mRNA. FI of this conjugated carbon dot, rCD-GAPDH was also found to decrease in presence of Ca2+ and varied in relation to H+ concentrations and could serve as a tool to quantify the intracellular concentrations of Ca2+ and pH value (H+) which can give important information about cell survival. Therefore, CD-oligonucleotide conjugates could serve as efficient probes for cellular events and interventions.
Keywords: Carbon dots, in-situ hybridization, luminescence quenching, intracellular recognitions, biosensing
Graphical Abstract
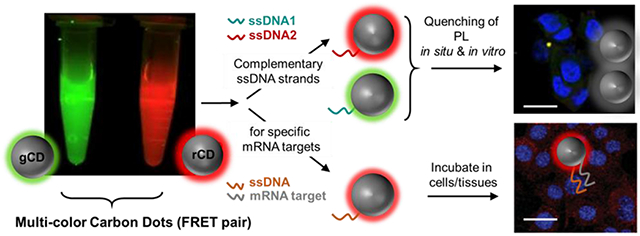
INTRODUCTION.
The ability to control colloidal particle assembly inspires fundamental interest as well as wide range of potential applications in areas of nanomechanics, electronics, biosensors, DNA scaffolds and drug delivery. [1–5] Highly selective bio-molecular interactions are utilized for manipulation of nano-assembly. Numerous biological complexes, e.g. peptides, proteins, and DNA have been utilized in the past as the components for these guided assembly systems. [6–9] DNA-DNA hybridization has long been exploited in the assembly of nanostructures. Most of these applications utilize complementary oligonucleotides tethered to metallic nanoparticles such as gold, silver and silica. [10–12] Additional sequences may be hybridized to these DNA-nanoparticle conjugates, or particles may be mutually hybridized with one another. For in situ hybridization-based applications, typically fluorescent dyes are used with these metallic particles; however, they suffer from inadequate signal-to-background, bleaching of fluorescence along the protocol time, diffusion of reporter molecules before reaching the site which can lead to orthogonal deposition of chemistries and lack of bio-neutrality. Fluorescent quantum dots conjugated with nucleotide probes have been used for countering some of these issues to identify genetic aberrances in morphologically preserved clinical tissue specimens. [13–15] Despite all these previous efforts, the quest to synthesize colloidal particles with photo-stability, narrow emission spectra, and biological inertness is still on. Carbon dots (CD) have emerged as one of the promising alternatives encompassing characteristics of easy synthesis, high photo-stability, narrow emission spectra, and tunability of photoluminescence, single source multi-color production and biological compatibility. [16–20] Mutually interacting multi-color CDs can provide systems for intercellular tracing. Here, we demonstrate that by using complementary DNA sequences as surface attached ligands, two individual diffusing ‘dual-color’ CDs can be selectively allowed to interact in situ and in vitro. Mutual colloidal interactions are realized through hybridization of complementary DNA sequences attached to the respective particles with anisotropic surface polarity (gCD-ssDNA1 and rCD-ssDNA2). (Figure SS1, SS2) Our results indicated that this interaction between ssDNA sequences in gCD-ssDNA1 and rCD-ssDNA2 leads to quenching of their photoluminescence presumably due to the non-radiative forster resonance energy transfer (FRET) occurring between the multi-colored carbon dots. We envision that this strategy could be used for biologically important application of detecting and targeting intracellular biomolecular targets including nucleic acids. Observations on changes in targeted biomolecules can be further utilized to provide vital information about their biological functions like cellular proliferation, metastasis, apoptosis, cell survival, etc. This hypothesis led to synthesis of new batch of CDs with targeting ability of GADPH RNA by introducing oligonucleotide sequences on the nanoscale surface of CDs.
RESULTS AND DISCUSSION
Synthesis and Physiochemical Characterization.
Spontaneous oxidation is a well-known phenomenon for carbohydrates. [21–22] However, nucleation of nano-sized materials from oxidization of sugar requires high temperature and pressure. [23] For preparing distinctly colored carbon dots, we hypothesized that the surface abundant oxidizable functionalities, such as nitroso benzene, can be subjected to assisted oxidation along with possibilities of spontaneous aerial oxidation of carbohydrate source. Consequently, this process will lead to anisotropical surface functionalities and in turn, polarity-driven wavelength tuning of their photoluminescence. In a typical synthetic process, nitroso (NO) or nitrobenzene (NO2) were used as a co-reactant with sucrose as the carbon source for hydrothermal (12 h) synthesis of carbon dots. With the increase in aerial oxidation, a more red-shifted emission peak is observed for CD-NO2 (Figure S1) as compared to CD-NO (Figure S1), probably due to increased oxidation owing to its more negative electrostatic potential. [31] The mass ratio of sucrose and nitrosobenzene (Ph-NO) / nitrobenzene (Ph-NO2) was kept at 1:1, and optimal temperature and reaction time were set at 180°C and 12h, respectively. (Fig. S2) CD-NO2 was selected for further fractionation via polarity-driven silica gel column chromatography [24–26] as it was found to give a significantly more red-shift in photoluminescence with increasing excitation wavelengths thereby implying that more red-emitting CDs were present. (Figure S3) The crude CD-NO2 samples were separated into fractions using column chromatography technique. Different fractions were collected and the ones which showed distinct green (gCD) and red (rCD) fluorescence emission characteristics were used for mutual interaction studies due to their overlapping absorption-emission characteristics. Obtained CDs were then re-dispersed in water (conc. = 1 mg/ml) and subjected to further physiochemical characterization studies.
Dynamic light scattering (DLS) studies indicated that gCD and rCD had hydrodynamic diameters between 15-30 nm and low polydispersity index (PDI) values. (Figure S4 and Table S1) The UV-Vis absorption of the separated fractions showed a typical n-π* transition at around 350 nm, typical for CDs (Figure 1a) [27] However, rCD exhibited lower energy absorption bands at around 550 nm, unlike gCD, indicating these fractions had different surface states. The PL emission spectra for gCD and rCD was collected and had an emission maximum at 510 nm and 595 nm, respectively. (Figure 1a). It was interesting to note that there was a spectral overlapping region between the emission region of gCD and the absorption spectrum of rCD, indicating plausible forster resonance energy transfer (FRET)-like behavior between the CDs. Such fractions with distinct green (gCD) and red (rCD) fluorescence emission characteristics (Figure 1b) can be used for mutual interaction studies. An elaborated study on morphological studies of these CDs by Transmission electron microscopy (TEM) and atomic force microscopy (AFM) of gCD, rCD, and gCD-ssDNA1 was conducted, which revealed spherical morphology of the nanoparticles (Figure S5).
Figure 1.
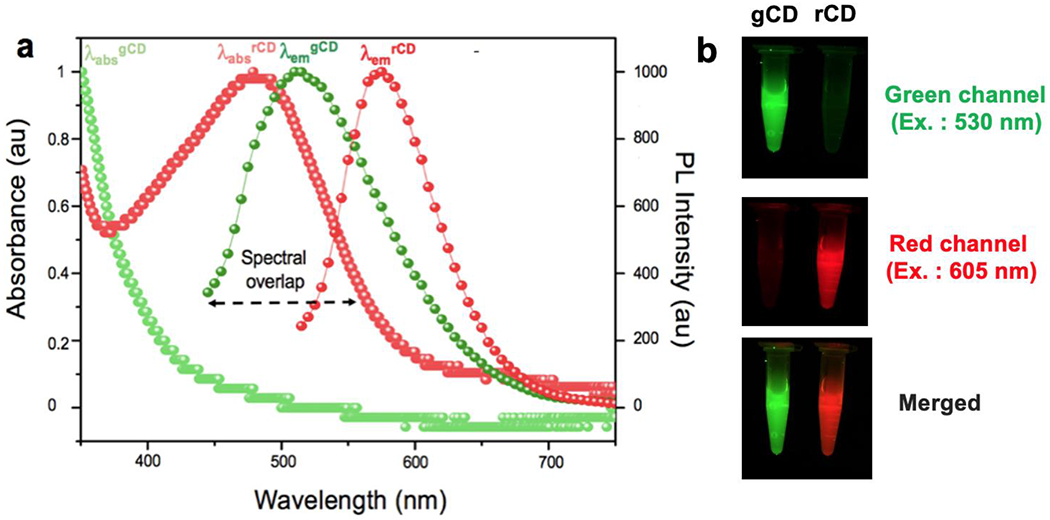
(a) UV-Vis absorption and corresponding PL spectra of gCD and rCD, with varying λex., (b) Geldock photoluminescence images of gCD and rCD at green channel (ex. 530nm), red channel (ex. 605nm) and merged image from both channels.
ζ-potential studies performed on these CDs revealed a negative ζ-potential. (Figure S6) [28] The quantum yields (Q.Y.) were calculated using quinine bisulfate in 0.1 M H2SO4 and Fluorescein isothiocynate (FITC) in 0.1mg/ml water as reference and were found to be 21% for gCD and 18% for rCD, respectively (Table S2). [29, 30] Additionally, these CDs were having high brightness values too after computing the full brightness (FB) values from previous literature reports. (Table S3) [31–35] Compositional characterization of CD-NO2 was studied by multiple analytical and spectroscopic techniques, including Fourier transform-infrared (FT-IR) spectroscopy (Figure S7), X-ray photoelectron spectroscopy (XPS) (Figure S8, S9, Table S4) supporting the successful synthesis. Emission spectra of gCD and rCD were carried out at different excitation wavelengths including 400, 450 and 500 nm. (Figure S10) These excitation wavelengths were chosen based on absorption pattern of gCD and rCD. Around 400 nm wavelength, a cross over in absorption spectra of gCD and rCD spectra were recorded, however, minimum overlap between both was recorded at 450 nm. At 500 nm, only rCD was found to absorb and overlap with emission spectra of gCD revealing an emission maximum at 510 nm. rCD had an emission maxima at ~ 600 nm.
FT-IR spectra for CD-NO and CD-NO2 revealed that the CDs possessed abundant hydrophilic groups such as O-H (3300 cm−1), and COOH (~1750 cm−1) on their surfaces, thereby ensuring good solubility in water. (Figure S7) [36] Asymmetric N=O stretch at ~ 1500 cm−1 and symmetric N=O stretch at ~1350 cm−1 was observed in both CDs. XPS measurements were performed on CD-NO2 to elucidate the bonding states of carbon, nitrogen and oxygen in CDs. This technique was used to provide an insight into the core structure of CDs with C1s, N1s, and O1s bands at 285 eV, 401 eV and 532.5 eV, respectively. (Figure S8) The high resolution C1s spectra were de-convoluted into 4 different signals corresponding to carbon atoms in sp2 C-C/ sp3 C-C (~ 284.8 eV), C-O/C-N/C=N (~286.5 eV), C=O (288.5 eV) and COOH (291.7 eV). [36] O1s and N1s peaks were further de-convolved to generate different signals. (Figure S9A, B, C)
Ethidium Bromide Intercalation Studies.
Whether two species will co-assemble depends on the chemical, physical and structural complementarity of the interacting components. In our study, the success of two particles to interact would rely on the ability of nucleotide probes binding to another complementary nucleotide sequence in intracellular space. We showed the duplex formation between gCD-ssDNA1 and rCD-ssDNA2 is feasible in situ and intracellularly. The process was thoroughly studied through spectroscopy, interaction with DNA duplex intercalators, flow assisted cell assay, and confocal experiments. Our results indicated that formation of duplex between gCD and rCD leads to co-localization of particles in intracellular space and in turn diminish luminescence as suggested by spectroscopy, flow cytometry and confocal studies. (Fig. 1– Fig. 6, Fig. S16) For luminescence hybridization process, isolated particles (gCD and rCD) with different emission properties at different excitation wavelength (Figure S10), were subjected to carbodiimide-mediated conjugation with complementary deoxyribonucleic acid single strands to generate, gCD-ssDNA1 and rCD-ssDNA2, respectively. UV-Vis absorption spectroscopy and ethidium bromide (EtBr) intercalation experiments was carried out to confirm the successful conjugation of single strand-DNA (ss-DNA) probes onto the surface of CD, and in situ formation of double-stranded DNA (ds-DNA) when gCD-ssDNA1 and rCD-ssDNA2 were mixed. UV-Vis spectroscopy of gCD-ssDNA1, rCD-ssDNA2 or their mixture showed a characteristic oligonucleotide absorption peak at around 260 nm (Figure S11) and is in accordance to earlier literature report. [37] DLS measurements performed on gCD-ssDNA1 and rCD-ssDNA2 revealed an increase in their hydrodynamic diameter compared to either gCD or rCD, respectively without any significant changes in their PDI values (Figure 2a and Table S1). To further demonstrate that ds-DNA was formed when gCD-ssDNA1 and rCD-ssDNA2 were mixed, ethidium bromide (EtBr) intercalation experiment was conducted.
Figure 6.
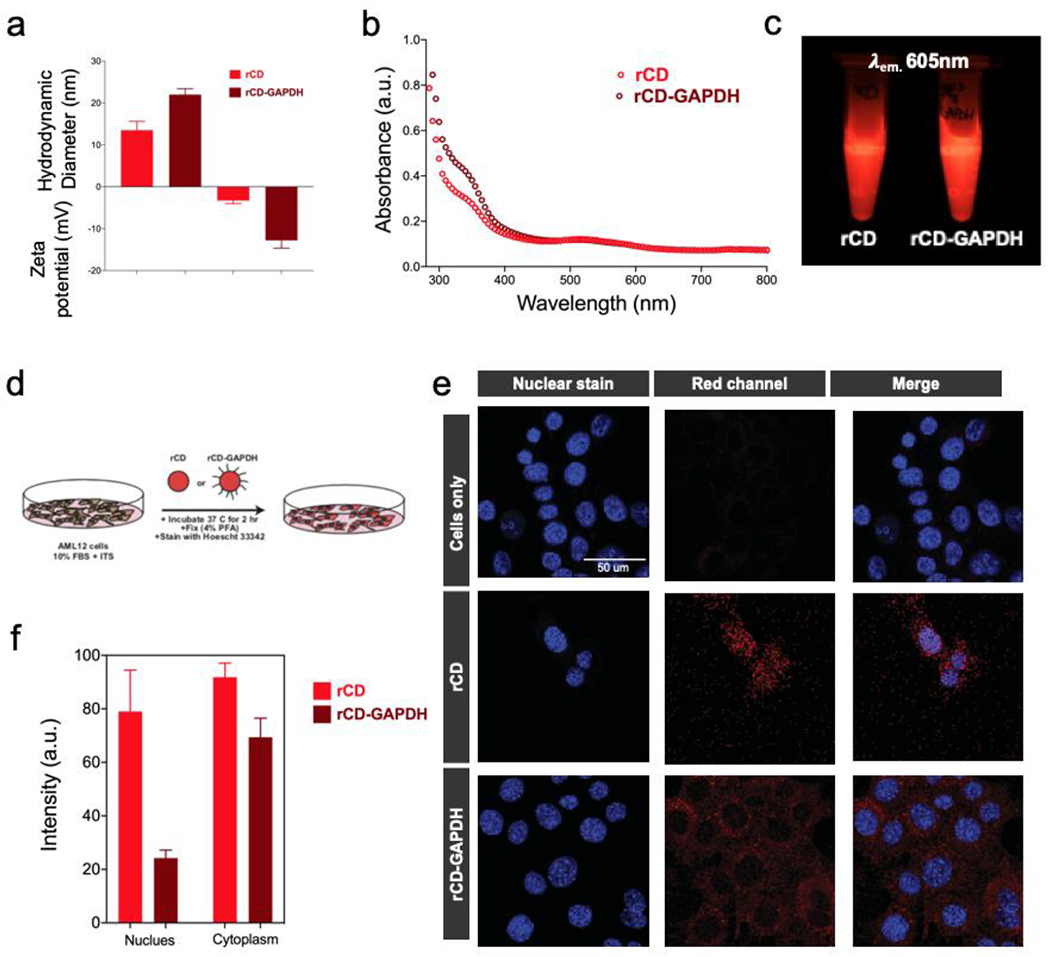
(a) Hydrodynamic diameter measurements, zeta potential measurements and (b) UV-Vis absorbance spectroscopy measurements for rCD and rCD-GAPDH. (c) Photographic presentation of rCD and rCD-GAPDH samples under exposure of light at 605 nm. (d) Representation of culture conditions used in cell experiment for confocal studies. Confocal intracellular imaging of CD treated cells and quantifications. (e) Localization of rCD and rCD-GAPDH in intracellular space and (f) respective differential intensity of CD fluorescence originated from nuclear and cytoplasmic regions.
Figure 2.
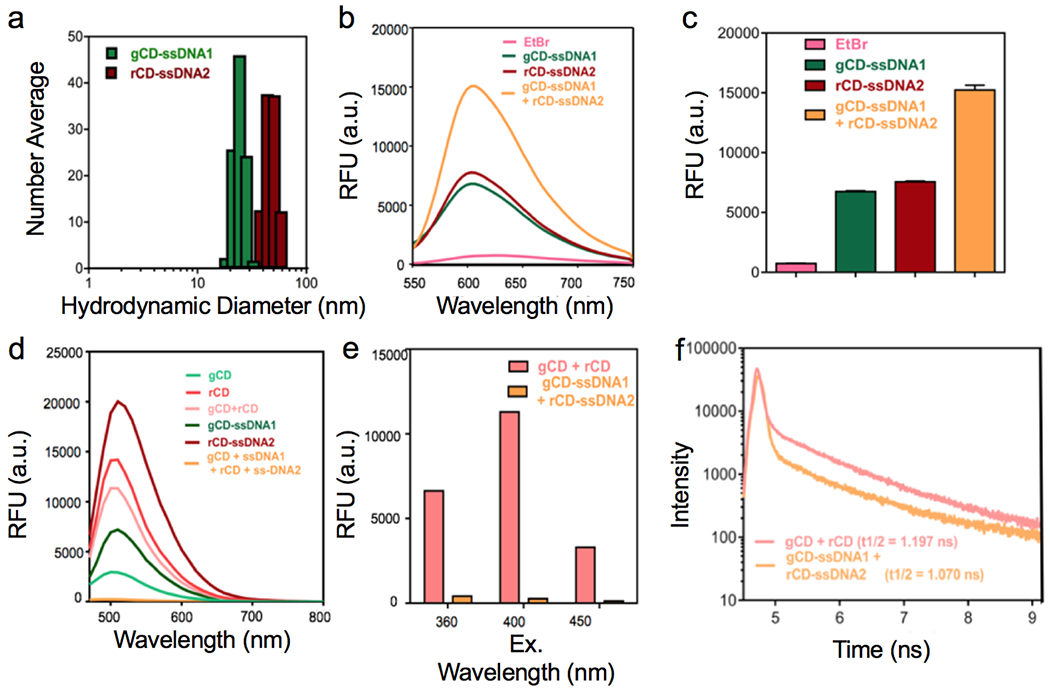
(a, b) Ethidium bromide intercalation study to confirm the formation of ds-DNA and PL intensity comparison between different samples. (c) Emission spectra profiles of different samples at λex. = 400 nm, (d, e) Corresponding emission spectra profiles comparison for gCD + rCD and gCD-ssDNA1 + rCD-ssDNA2 at different excitation wavelengths, (f) Time-resolved PL study performed on gCD + rCD and gCD-ssDNA1 and rCD-ssDNA2 mixtures, at excitation wavelength of 510 nm.
The premise behind this study was that EtBr will selectively bind with ds-DNA and result in an increase in the fluorescence intensity. Fluorescence intensity of EtBr is known to drop in hydrophilic environment. However, this effect is significantly less pronounced for ss-DNAs. [38] When either gCD-ssDNA1 or rCD-ssDNA2 were incubated with EtBr, it increased their fluorescence intensity compared to just EtBr. However, the effect of increase in fluorescence intensity was more pronounced when mixture was incubated with EtBr. This indicates the formation of duplex ds-DNA when two surfaces bound complementary ss-DNA conjugated on gCD and rCD (Figure 2b, 2c) mutually interact and can bind to the complementary nucleotide sequence. It was found that only a duplex DNA of plasmid (pDNA) was able to show migrated DNA band but gCD-ssDNA and rCD-ssDNA did not. It further indicates that the single stranded nature of ssDNA conjugated to CDs which could not intercalate EtBr to generate any visible band (Figure S12).
Photoluminescence and Time-Resolved Photoluminescence Studies.
To probe this further, photoluminescence (PL) spectra of samples at same concentration (1mg/ml) was recorded at different excitation wavelengths, i.e. 400 nm (Figure 2d, 2e), 360 nm, and 450 nm. (Figure S13) For mixture of gCD and rCD, it revealed an intermediate PL intensity respect to PL of individual gCD or rCD. Interestingly, conjugation of a ss-DNA strand on gCD or rCD results in an increase in PL intensity presumably due to the presence of nucleotides on the surface of the particles. In situ mixing of gCD-ssDNA1 and rCD-ssDNA2 resulted in a remarkable quenching in PL intensity. Probably, addition of oligonucleotide units (containing nitrogen bases with inherent fluorescence properties) on CDs improving overall photoluminescence intensity. This photoluminescence increment could be reduced to a significant level upon synthesis of complementary duplex between gCD-ssDNA1 and rCD-ssDNA. [39,40] Oligonucleotides with multiple fluorophoric nitrogen bases add up their individual fluorescence to intrinsic PL of CDs after conjugation. This added emission intensity increases overall emission intensity of CD-ssDNAs and showed up in PL studies. [41, 42] Furthermore, time-resolved photoluminescence (TRPL) study was performed on mixture of gCD and rCD & gCD1-ssDNA1 and rCD-ssDNA2 mixtures, at an excitation wavelength of 510 nm. It was observed that once the gCD-ssDNA1 and rCD-ssDNA2 were mixed, it leads to sharp fall in half-life value as compared to the case when just gCD and rCD were mixed, due to quenching effect, leading to more non-radiative transition and ultimately lowering the half-life value. (Figure 2f, Figure S14 and Table S5) This observation could be due to formation of aggregated structure comprising of numerous well-matched duplex ds-DNA CDs as a result of mixing of CDs with complementary sequences coupled by non-radiative FRET occurring between gCD-ssDNA1 and rCD-DNA2, possibly releasing the energy as heat. We suggest that both these processes are presumably contributing simultaneously towards the PL quenching effect observed. [43–45] As the distance between rCD and gCD would not be changing during FRET process due to duplex formation between ssDNA1 and ssDNA2 conjugated to these nanoparticles, an effective fluorescent distance of donors and acceptors when the transfer efficiency is 50% (R0) would not be possible to calculate in this case while distance between donors and acceptor of fluorescence (R) was ~20 nm. [46]
Recovery of Emission Intensity in Presence of Endonucleases.
The formation of ds-DNA duplex resulting from the close proximity of gCD and rCD, lead to non-radiative energy transfer and in turn quenching of the fluorescence. We next attempted to recover the quenched fluorescence. To demonstrate this, we used endonuclease (DNase I) at a high concentration of 1U/mL to trigger the cleavage of the DNA linkers thereby allowing the release of gCD and rCD from each other and resulting in the recovery of luminescence. [47–49] In Fig. 3a, b, after addition of 1U/ml of DNase I to gCD-ssDNA1 + rCD-ssDNA2 mixture, the emission intensity was significantly increased over the course of time (0.5h to 24h) and was able to recover emission intensity nearly equal to the scenario where gCD and rCD were mixed without any ss-DNA probes conjugated to their surface. We anticipate that such recovery of luminescence in presence of nucleases would have significant applications in bio-sensing, etc. [50]
Figure 3.
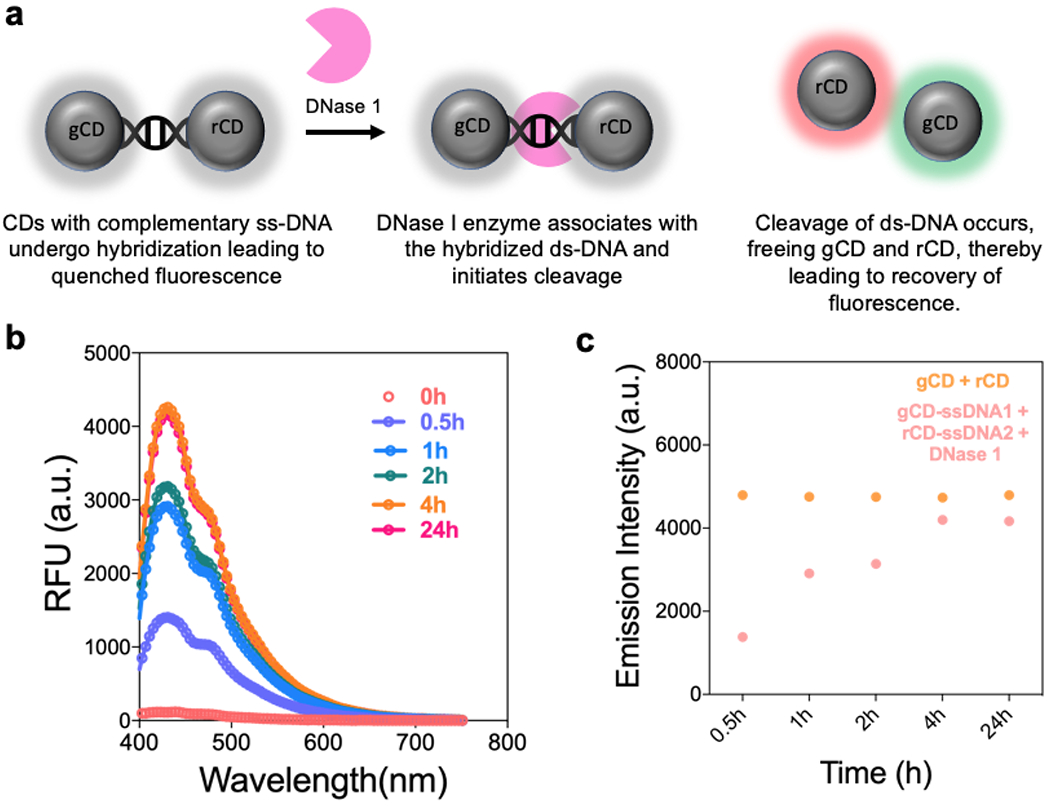
(a) Schematic representation of the recovery of luminescence of gCD-ssDNA1 and rCD-ssDNA2 in presence of endonuclease enzyme, DNase 1. (b) Fluorescence emission intensity profile of gCD-ssDNA1 + rCD-ssDNA2 without (t = 0) and with 1U/mL DNase I as a function of incubation time varying from 0.5h to 24h. (c) Emission intensity comparison at 430 nm for gCD + rCD and gCD-ssDNA1 + rCD-ssDNA2 (in presence of DNase I) as a function of time.
Cellular Toxicity Studies and Mechanism of Cellular Entry.
Metabolic processes of cells are known to have redox activities in mitochondria. A large number of live cells account for higher metabolic activity and directly correlates with less cytostatic and cytotoxic effects of threated agents. Mitochondrial reduction of MTT reagent in cell population with and without treatments can indicate on cytotoxicity efficiencies of CDs. MTT assay for evaluation of cytotoxic effects of different CDs samples in MCF-7 breast cancer cells at various concentrations revealed that none of the CD or mixed CD suspensions with or without conjugated ssDNA sequence showed any significant level of cytotoxicity as such (Figure S15). We also performed MTT assay of these probes in a healthy cell line, AML12 (mouse cell line of hepatocyte lineage). It did not reveal any cytotoxic effect even at increased concentrations of the probes (Figure S16).
Cellular entry of nanoparticles occurs through different pathways and can be verified by decrease in after-effect of cellular entry viz. nanoparticle mediated cell toxicity. [49] Cellular entry of gCD-ssDNA1 and rCD-ssDNA2 were verified by blocking different cellular entry pathways using different pharmacological manipulators or inhibitors. Cells were pre-incubated with various inhibitors before allowing the internalization of effective dose of CDs via lipid raft, metabolic, energy, clathrin, and/or caveolar mediated pathways. A 25% (v/v of 1 mg/mL stock solution in reconstituted DMEM medium) concentration of CDs was used for the study (Figure 4). It was found that among different used inhibitors including sodium azide (NaN3) and 2-deoxyglucose (DOG), chlorpromazine (CPM), nystatin, dynasore hydrate, and Methyl-β-cyclodextrin, nystatin and dynasore hydrate were found to playing major role in cellular entry of gCD-ssDNA1 and rCD-ssDNA2. Dynasore, an inhibitor of the dynamin GTPase, is known to inhibit dynamin-dependent endocytosis in cells [51] while nystatin, a sterol-binding agent disassembles caveolae in the membrane causing clathrin-independent inhibition of endocytosis. [52] Blocking of these two pathways resulted in increasing the cell viability to significant levels indicating the major role of dynamin-dependent and lipid-raft mediated cellular entry of both gCD-ssDNA1 and rCD-ssDNA2. It was interesting to note that in a healthy cell line (AML12), it revealed that probes used a combination of cholesterol-dependent and lipid-raft mediated endocytic pathways to internalize which was different when compared to their entry in MCF-7 cells (Figure S17). Furthermore, cellular entry of rCD-ssDNA2 was found to be more affected by these inhibitors compared to gCD-ssDNA1 in both MCF-7 and AML12 cells. This could be an after effect of higher cellular entry of rCD-ssDNA2 when compared to gCD-ssDNA1, probably, due to a more oxidized surface of rCD-ssDNA2 allows higher cellular entry and in turn gets affected more with inhibitor-based blocking of endocytosis. [53–55]
Figure 4.
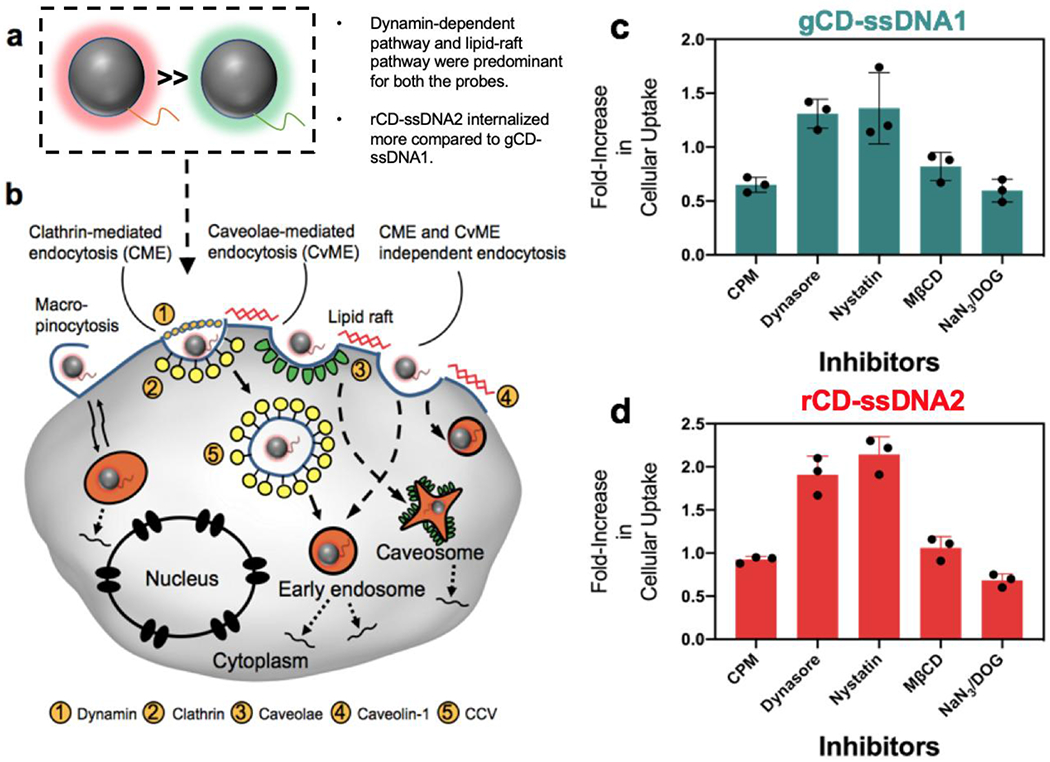
(a) Schematic representation gCD-ssDNA1 and rCD-ssDNA2. (b) Various endocytic pathways for intracellular delivery of CDs. (c, d) Cellular internalization studies of gCD and rCD. MCF-7 cells were pre-incubated with inhibitors reconstituted in culture medium having inhibitors sodium azide (NaN3) and 2-deoxyglucose (DOG), chlorpromazine (CPM), nystatin, dynasore hydrate, and Methyl-β-cyclodextrin (MβCD), respectively. Cells were post incubated with CDs for 48h before performing MTT assay.
Evaluation of Duplex Formation via Confocal Microscopy Imaging and Flow Cytometry.
Confocal laser scanning microscopy (CLSM) was further performed on a model cell system to investigate the duplex formation and intracellular quenching once gCD-ssDNA1 and rCD-ssDNA2 were co-localized. All samples (at conc. of 1 mg/mL) were incubated with breast cancer MCF-7 cells for 24h (80% confluency), followed by fixing them after 4h of post incubation and consequently, being imaged (Figure 5a, b). A significant decrease in CD emission was obtained in cells when they were incubated with gCD-ssDNA1 or rCD-ssDNA2, compared to cells just incubated with gCD or rCD. Interestingly, there was a drastic decrease in emission when cells were co-incubated with gCD-ssDNA1 and rCD-ssDNA2 mixture, (Figure 5b) compared to incubations with mixture of gCD and rCD. This effect could be due to hybridization of complementary DNA base pairs attached to the respective particles formed ds-DNA complex leading to a possible aggregation. Formation of the ds-DNA complex could also allow interaction between the two CDs (gCD and rCD) due to close proximity, resulting in a non-radiative energy transfer process leading to quenching. [41–44] Respective green and red channels were visibly following the trend of increased emission intensity for ssDNA conjugated CDs and decreased emission intensity for intracellularly mixed CD-ssDNAs, which remained viable in the merged images as well (Figure 5a). These changes were further quantified by calculating emission intensities from various confocal images. A ratio of merged emission intensities and emission in individual red and green channels revealed an around four-fold decrease in emission intensity for gCD-ssDNA1 duplex with rCD-ssDNA2 compared to mixture of gCD and rCD (Figure 5b). This experiment evaluates the feasibility of using this strategy for both qualitative and quantitative intracellular studies. These results were further corroborated quantitatively with flow assisted cell assay results. MCF-7 cells untreated and treated with gCD and rCD with or without conjugated ssDNA were flow sorted (Guava EasyCyte Plus). For each sample, data from 5000 single cell events were collected within 1 min at green channel (λex: 525nm) and red channel (λem: 633nm) to conclude that cells with co-incubation of gCD and rCD show only a minor drop (~10%) in high emission cell populations comprising of M2 histogram marker region compared to cells incubated to rCD particles while co-incubation of gCD-ssDNA1 and rCD-ssDNA2 exhibited a significant drop (~34%) in fluorescence in the same histogram marker region. (Figure S16a, b and Table S6) Cells are known to have some background fluorescence due to presence of aromatic amino acids which shows up in flow cytometry analysis. Any additional auto-fluorescent or photoluminescent agent can add up the internal fluorescence to treated cells and will contribute to the higher fluorescence during flow cytometric analysis. Similar technique was used to verify the PL of CDs in treated cells (Figure S18a). It was noticed that addition of g-CD and r-CD improved the overall population of cells with additional fluorescence which was not reduced to any significant extent after incubating both the CDs together. Incubation of cells with ssDNA conjugated g- and r-CD were found to increase the base fluorescence of cells to greater extent and reduced to significant level on co-incubation (Figure S18b). These results corroborate to cellular internalization of various CD species. [56–58]
Figure 5.
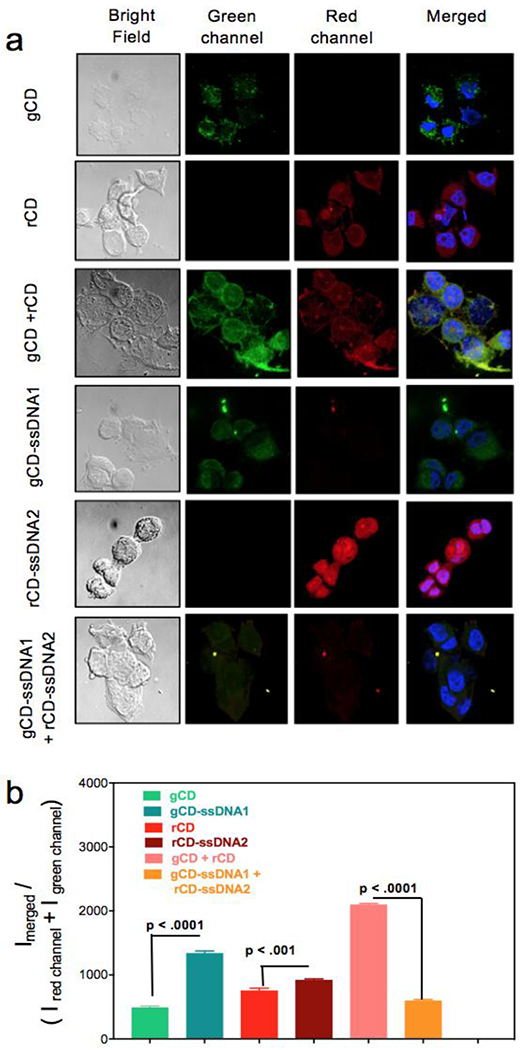
(a) CLSM images of MCF-7 cells incubated with gCD, rCD, gCD + rCD, gCD-ssDNA1, rCD-ssDNA2 and gCD-ssDNA1 + rCD-ssDNA2. Four different cell panels were acquired, bright field, green channel (for gCD), red channel (for rCD) and overlapped green and blue channels with blue from DAPI. (b) Semi-quantitative fluorescence intensity calculations were performed with help from ImageJ [35] and corroborated with the PL experiments.
Recognition of Essential Intracellular Proteins using DNA conjugated Carbon Dots.
Finally, these multi-color CDs, owing to several advantages such as good signal sensitivity, fluorescence stability and ability to be multiplexed with various intracellular targets could be utilized to provide vital information about biological functions like cellular proliferation, metastasis, apoptosis, etc. similar to quantum dots [59–61]. Here, we have utilized one of the CDs to target an intracellular mRNA coding for Glyceraldheyde-3-phosphate dehydrogenase (GAPDH) protein. It has been long established that GAPDH is an important enzyme for energy metabolism and production of ATP and pyruvate via the anaerobic glycolysis in the cytoplasm. [62] However, it has been shown that an increased GAPDH expression and enzymatic function is associated with cell proliferation and tumorigenesis, and conditions such as oxidative stress diminish the catalytic activity of GAPDH and eventually leads to cellular aging and apoptosis. Here, we designed a 22nt long probe (5’-GTGAGTGGAGTCATACTGGAACA-3’) targeting GAPDH mRNA. In addition, the probe also harbors two 18-carbon spacers at the 3’-end to maximize distance to the curvature of CD, thereby avoiding possible physical hindrance to hybridization. This probe was conjugated with rCD and is referred to as rCD-GAPDH in future experiments.
Hydrodynamic diameter and zeta potential measurements (Figure 6a, b) revealed an increase in size and zeta potential upon conjugation of the GAPDH probe onto rCD. UV-Vis absorbance studies revealed an absorption peak plateauing at wavelength ~ 360nm, with lower energy absorption bands at around ~500 nm, remaining the same for both rCD and rCD-GAPDH. A clear difference in improved FI of rCD could be seen in photographic presentations of rCD and rCD-GAPDH samples under exposure of light at 605 nm as captured using Geldock luminescence imaging system (Figure 6c).
Confocal imaging of rCDs with or without conjugated GAPDH oligonucleotide sequence revealed differential pattern of cellular localization. While rCD were found to localize equivalently to both cytoplasmic and nuclear spaces, the rCD-GAPDH probe showed much higher cytoplasmic localization in AML12 mouse cells (hepatocyte lineage) (Figure 6d, e). The increased preference for cytoplasmic localization of rCD-GAPDH probe reflects selective binding to cytoplasmic GAPDH mRNA (Figure 6f). These results reveal the efficiency of rCD-GAPDH probe to work qualitatively and quantitatively in cytoplasmic and intranuclear space.
Additionally, we attempted to show that our probe (rCD-GAPDH) could be eventually used for biological sensing as it was responsive to both, changes in pH and presence of Ca2+ ions. Intracellular pH plays an important role in the pathophysiological processes of cells, and fluorescence probes responsive to pH could serve as a valuable tool for assessing the pH of intact cells. Furthermore, presence of excessive Ca2+ in the cells activates a cascade of cytotoxic events which could eventually lead to activation of enzymes which causes breakdowns of lipids, proteins, etc. Hence, detecting the presence of excess Ca2+ could prove to a valuable asset for a fluorescence probe. Both rCD and rCD-GAPDH were re-dispersed in pH solutions ranging from pH 2 to pH 12 and corresponding area spectrum (Figure S19a) and FI (Figure S19b, c) were collected at 590nm emission after 530nm excitation. It revealed that for rCD, lowering the pH to 2 leads to an increase in fluorescence intensity whereas increasing the pH to 12 leads to a drastic drop in the fluorescence intensity. Surprisingly, for rCD-GAPDH, it leads to significant drop in FI at both high and low pH values, while being the highest at pH 7. Separately, we observed that upon increasing the concentration of Ca2+ ions from 1μM to 200 μM, the fluorescence intensity of rCD-GAPDH probe dropped gradually (Figure S20). Subsequently, limit of detection (LOD) was calculated to be 3.85μM, according to an equation 3σ/S, where σ is the standard deviation of the baseline signal (n=3) and S is the slope of the calibration plot. [63] We further attempted to demonstrate the same effect by spiking Ca2+ ions in AML12 cells and incubating them with rCD-GAPDH and collecting the fluorescence intensity values. A similar trend was observed, highlighting the usefulness of these probes in monitoring Ca2+ ions in the cells (Figure S21). During apoptosis, the concentration of Ca2+ ions increases drastically from 0.1μM to 100/1000-folds (100 μM). [64] Hence, incubation of rCD-GAPDH probe to these apoptotic cells would show a drop-in fluorescence intensity, something that could also be visualized via confocal microscopy. Hence, our probe could also be used to sense excessive Ca2+ as well the pH variation in the cells, which are closely related. [66–68]
CONCLUSIONS
Intracellular machinery and biochemical processes play very important role in homeostasis of living beings including humans. It becomes more important to study these intracellular machineries’ and biochemical processes in disease condition. A better understanding of such events can eventually lead to therapeutic interventions. Intracellular imaging might be answer to such a process but requires intracellularly interacting nature of probes, long lifetimes and better biocompatibilities. Though some of the photo-emitting nanoscale entities are highly resistant to metabolic degradation and maintain their fluorescent properties after chemical modification if required, existing key difficulties are because of their specificity and cytotoxicity for living-cell imaging. Carbon nanodots have emerged as new class of auto-emission particles having easy and economical controlled syntheses, surface chemistries, optical properties and biologically safe entity. A DNA sequence conjugated CD may be helpful in designing intracellularly interacting CDs conjugated with complementary sequence. Here, intracellular hybridization process was achieved by using gCD-ssDNA1 and rCD-ssDNA2 with ssDNA1 and ssDNA2 being complementary sections of single strand DNA sequences. Their intracellular interaction was confirmed through changes in spectroscopic patterns, interaction with DNA duplex intercalators, flow assisted cell assay and confocal studies. It was found that gCD-ssDNA1 and rCD-ssDNA2 can mutually co-localize to form duplex which results in a luminescence quenching.
As a model biochemical process for intracellular investigation study, we chose one of the biologically important proteins GAPDH which is known to get involved in process of glycolysis. Location and quantification of GAPDH synthesis in intracellular space can be vital information about the downstream events including stage of cell growth. Our rCD-GAPDH oligo conjugated probe was able to bind GAPDH mRNA sequence and in-turn decrease the FI of conjugated carbon dot which was further decreased in presence of Ca2+ and very high or very low H+ concentrations. This further signifies the probable possibility of quantifying the local concentrations of Ca2+ and pH value, which controls many of the cell survival events including activation of enzymes that break down proteins, lipids and nucleic acid. Thus, newly developed oligo-conjugated CDs could be highly efficient and selective probe for intracellular imaging for biological events These results can contribute to the rational design of mutually interacting carbon dots for a better control over the resulting assembly structure. Our findings offer control of spherical carbon dots as building blocks for the assembly of sophisticated nano- and macroscopic materials and provide a framework to understand complementarity and directional interactions in DNA-mediated nanoparticle assembly. The chemistry we report here for mutually interacting DNA/carbon dots may open up novel ways to many applications proposed or pursued in the past.
METHODS
Materials:
Unless otherwise stated, all chemicals were purchased from Aldrich Chemical Co. (St. Louis, MO) and used as received. Modified DNA sequences were purchased from Integrated DNA Technologies Inc. 8180 N. McCormick Blvd. Skokie, Illinois 60076, USA.
Synthesis of small molecule passivated carbon nanoparticles:
First, sucrose (250mg) and nitrobenzene (NO2)/ nitrosobenzene (NO) (250mg) was dissolved in 5ml of ethanol (200 proof, UPS specs, Decon Labs, Inc., PA, USA); then, the solution was transferred into a PTFE-lined stainless-steel autoclave. After being heated at 180 °C for 12h (to form CD-NO2 and CD-NO, respectively) and then cooled to room temperature, the obtained solution was filtered using a 0.22-micron filter. Afterwards, these were freeze-dried and suspended in water for use. The synthetic conditions for preparation of CD-NO and CD-NO2 were the same except the pre-cursor molecules for passivation, which were NO and NO2, respectively.
Dynamic light scattering:
The hydrodynamic size distribution of the nanoparticles was determined by dynamic light scattering measurements on Malvern Zetasizer ZS90 instrument (Malvern Instruments Ltd, United Kingdom) at a fixed angle of 90°. A 50 μL of particle suspension was mixed with 950 μL of nanopure water to run the samples in DLS machine. A photomultiplier aperture of 400 nm was used and the incident laser power was so adjusted to obtain a photon counting rate between 200 and 300 kcps. Measurements for which the measured and calculated baselines of the intensity autocorrelation function were within 0.1% range were used for diameter values. All measurements were carried out in a triplet of thirteen consecutive measurements.
Zeta potential measurements:
Zeta potential (ζ) values for each nanoparticle were determined using a Malvern Zetasizer (Malvern Instruments Ltd, United Kingdom) of Nano series. The experiments were performed at 25°C and pH 7 at the light scattering mode in the phase analysis light scattering (PALS) mode following solution equilibration at 25 °C. Calculation of ζ from the measured nanoparticle electrophoretic mobility (μ) employed the Smoluchowski equation: μ = εζ/η where ε and η are the dielectric constant and the absolute viscosity of the medium, respectively. Measurements of ζ were reproducible within ±4 mV of the mean value having three trials/determinations, with 15 data accumulations in each trial.
Column chromatography:
Different colored fractions of as-prepared CD-NO2 were critically separated using a normal-phase silica gel (230-400 mesh size) by column chromatography technique using a mixture of ethyl acetate and ethanol. Obtained fractions differed in terms of their polarity. For complete separation of different colored fractions, it took 2 days due to slow flow rate maintained while separating the fractions to avoid possible cross-contamination amongst CDs with different polarities. The polarity of the eluting solvent was increased gradually along the process.
UV-Vis and photoluminescence measurements:
Ultraviolet-visible (UV-Vis) absorbance of CDs was recorded on GENESYSTM 10S UV-Vis Spectrophotometer (Thermo Scientific, MA, USA) using quartz cuvettes. Absorbance spectra were collected at an interval of 1 nm scanning from 250–800 nm. For fluorescence measurements, the emission spectra of CDs were obtained using TECAN Infinite F200 PRO using a Corning® 384 Well Optical Imaging Flat Clear Bottom Black Polystyrene TC-Treated microplates. Measurements were done at different excitation-emission wavelengths, with gain set to 100 for all measurements throughout this work. Furthermore, integration time per each well was set as 20 μs, and the temperature was kept at 25°C.
Transmission electron microscopy:
CDs were mildly vortexed prior to samples preparation for TEM imaging. Ten microliters of CD sample were drop cast on a 200-mesh copper grid. After waiting for 1 min, the excess fluid was absorbed by lint-free Kimwipes. The transmission electron micrographs were acquired on a JOEL 2100 Cryo TEM machine and imaged by Gatan UltraScan 2k X 2k CCD camera.
FT-IR measurements
An 200μL aqueous suspension of the nanoparticle (CD-NO2) was dried onto a MirrIR IR-reflective glass slide (Kevley Technologies, Chesterland, Ohio, USA) for Fourier Transform Infrared (FT-IR) measurements using a Nicolet Nexus 670 FT-IR (Fredrick Seitz Material Research Laboratories (FSMRL), Urbana, Illinois, USA). For each measurement 100 × 100 μm images were collected at 1 cm−1 spectral resolution with 64 scans per pixel and a 25 × 25 μm pixel size and individual spectra were corrected for atmospheric contributions. Prior to running the measurements, a background measurement was performed, and background corrections were done on subsequent measurements.
X-ray photoelectron spectroscopy:
X-ray photoelectron spectroscopy (XPS) measurements for CD-NO2 was obtained after deposition of 20 μL sample on a glass surface, leading to a thick vacuum dried. It was then run using Physical Electronics PHI 5400 spectrometer with Al Kα (1486.6 eV) radiation. The spectrum was referenced to the adventitious C 1s feature at 285.0 eV and remaining peak assignments were followed from references.
Surface conjugation of ssDNA1 and ssDNA2 on gCD and rCD:
A carbodiimide coupling using (1-Ethyl-3-(3-dimethylaminopropyl)carbodiimide, EDC, 0.5 μMol) and N-Hydroxysuccinimide (NHS, 0.5 μMol) procedure was used to conjugate ssDNA1 (5’-ACCTTCGGGTTTT-3’-O-CH2-CHOH-CH2-O-CH2-CH2-CH2-NH2, 0.5 μMol) and ssDNA2 (H2N-C6H14-O-PO2-O-5’-TGGAAGCCCAAAA-3’, 0.5 μMol) to gCD (1 mg/mL) and rCD (1 mg/mL), respectively. An around 2% surface -COOH groups on CD were employed to generate amide bond formation between CD and ssDNA entities. Conjugations were performed in water medium (0.5 mL) at room temperature (RT) by stirring for overnight. Conjugated particles were characterized by change in photo-properties with ethidium bromide intercalation study which confirms the formation of ds-DNA (Figure 2a–f) which is only possible for DNA conjugated CDs, a simple mixing of unconjugated one of both the CDs will not lead to these results.
MTT assay:
MCF-7 cells (10,000) were plated in 96 well plate for 24 h to achieve a ~80% confluence before performing MTT based cell viability assay. Cells were incubated with various CDs at a concentration of 10 % (v/v) having overall CD concentration 1 mg/mL for 48 h. All treatments were performed in triplicates. At the end of the incubation, 20 μL of MTT solution (5 mg/mL) were added to each well of growing cells. Cells were further incubated for 4 h. The entire spent medium was replaced with 200 μL of DMSO and mixed at RT for 5 sec before performing absorption studies at 592 nm in a plate reader. Cell viability was calculated by using following formulae:
Inhibitor study:
MCF-7 cells (10 × 103) were grown in 96 well plates for 24 h before incubation with various cell internalization inhibitors. Inhibitors were prepared with reconstituted medium having inhibitors sodium azide (NaN3) and 2-deoxyglucose (DOG), chlorpromazine (CPM), nystatin, dynasore hydrate, Methyl-β-cyclodextrin and cholesterol at a concentration of 10 × 10−6 M, 50 × 10−6 M, 28 × 10−6 M, 180 × 10−9 M, 80 × 10−6 M, 2 × 10−6 and 1 × 10−6 M, respectively. Cells were incubated with inhibitors for 1h at ambient condition. Inhibitors were replaced with CDs at a concentration of 25% v/v for 48h. All the additions were performed in triplicate. Cells only with inhibitor treatments were considered as negative controls whereas cells treated with only CDs (CD-NO and CD-NO2) without any pre-inhibitor treatment were used as positive controls. The cytotoxic effect of CDs was investigated with an MTT assay (Sigma-Aldrich, MO, USA) as described above.
Ethidium bromide intercalation experiments:
CDs and ssDNA-CDs were mixed in equal amount of 50 μg before adding ethidium bromide before performing fluorescence emission experiment. Samples were excited at 400 nm and emission was reported at 605 nm. Fluorescence intensity from free EtBr and intercalated EtBr were compared and increase in EtBr on intercalation was compared to conclude DNA duplex formation.
Recovery of emission intensity in presence of endonucleases:
A stock solution of DNase I was prepared in DNase buffer (25% Glycerin in H2O) and stored at −20°C in the refrigerator for further use. Activated DNase I (concentration = 1U/mL) was incubated with gCD-ssDNA1 + rCD-ssDNA2 for different time (0-24h) at 37°C. Then the emission spectra were recorded on the spectrofluorometer excited at 365 nm. The control experiment was carried out with gCD + rCD in absence of DNase I.
Confocal imaging:
To verify the intracellular pairing of ssDNA conjugated CDs in intracellular space, fluorescence confocal studies were performed. Cells (MCF-7, 0.4 x 106) were grown on glass coverslips (15 mm) for 24 h before treating with 1 mg/mL of CD or CD-ssDNA particles along with co-incubated gCD and rCD and gCD-ssDNA and rCD-ssDNA for 4h. At the end of incubation cells were fixed with 4% paraformaldehyde for 10 min and washed with DPBS for 2 times. Cells were mounted on DAPI containing mounting medium and used for confocal studies. Cells were excited at 488 and 555nm for the experiment
Flow cytometry:
To further support the process of intracellular pairing of ssDNA conjugated CDs in intracellular space, FACS studies were performed. Cells (MCF-7, 60,000) were grown in 24 well plate for 24 h before treating with 1 mg/mL of CD or CDssDNA particles along with co-incubated gCD and rCD and gCD-ssDNA and rCDssDNA for 4h. At the end of incubation cells were trypsinized and collected in 0.2% FBS containing DPBS. Samples were analyzed using a Guava EasyCyte Plus Flow cytometer. For each sample, data from 5000 single cell events were collected for 1 minute, in triplicates, at green channel (λex = 525 nm) and red channel (λex = 633 nm).
Quantum yield calculations:
Quinine sulfate in 0.1 M H2SO4 solution was used as a standard. The quantum yield of the green and red fraction was calculated using the following equation:
where ∅ is the quantum yield, I is the measured integrated emission intensity, μ is the refractive index of the solvent, and A is the absorbance. The subscript R refers to the standard, Quinine sulfate (QS) in 0.1 M H2SO4 or Fluorescein isothiocynate (FITC) in 0.1mg/ml water. The absorbance of each of the samples including references were taken at 365 nm (QS) or 470 nm (FITC) and correspondingly, area of photoluminescence emission intensity was calculated after the samples were excited at 400 nm and repeated in triplicates. (Table S2).
PL lifetime experiments:
PL lifetime experiments were performed for CDs at an excitation wavelength of 510 nm. A single-photon-counting setup at MRL, UIUC facility was used. The experiment was performed with the lifetime of emitting states with single photon sensitivity and <1 ns temporal resolution at a temperature of 273 K. A particle concentration of 1 mg/mL was used for the experiment in a volume of 200 μL loaded in a quartz cuvette.
Gel electrophoresis:
A 1% agarose gel was used to run gCD-ssDNA (200 μg) and rCD-ssDNA (200 μg) along with a plasmid DNA duplex PBR3322 (200 ng). Gel was stained with 1 μg/mL of EtBr for 5 min and washed in 1× TAE buffer for 3 min before imaging by Gel Doc.
Cell Culturing and Confocal Experiments for intracellular imaging of rCD-GAPDH and differential location of GAPDH mRNA:
Cells (AML12) (standard conditions DMEM supplemented with 10% FBS and ITS) were grown on glass fluori-dishes for 24 h before treating with 1 mg/mL of rCD or rCD-GAPDH particles for 2h. At the end of incubation, cells were fixed with 4% paraformaldehyde for 10 min and washed with PBS 3 times. Cells were mounted on Hoescht 33342 containing mounting medium and used for confocal imaging on Zeiss LSM710 microscope.
pH experiments and calcium ion incubation experiments:
Solutions having different pH were prepared in acetate buffer. Fluorescence spectra of rCD and rCD-GAPDH probes at different pH were recorded using an Infinite 200 PRO multimode microplate reader (TECAN, NC, USA) at excitation of 530 nm, emission of 590 nm, and gain 100.
For calcium ion experiments, different concentration of Ca2+ were prepared by taking calcium chloride (CaCl2) ranging from 1μM to 200μM. For detecting Ca2+ in solutions, different concentration of Ca2+ ranging from 1μM to 200μM were incubated to 200μL of rCD-GAPDH solution in a well plate. Subsequent fluorescence intensities were collected using the microplate reader at excitation of 530 nm, emission of 590 nm, and gain 100. Similar protocol was followed for AML12 cells were cultured so that they were confluent and then plated on a well plate (1000 per well, counted via flow cytometry). Subsequently, Ca2+ ions were added/spiked and incubated with rCD-GAPDH probe and allowed to incubate at 37°C for 30 minutes. Subsequently, fluorescence spectra were recorded using the microplate reader in triplicates.
Supplementary Material
Scheme 1.
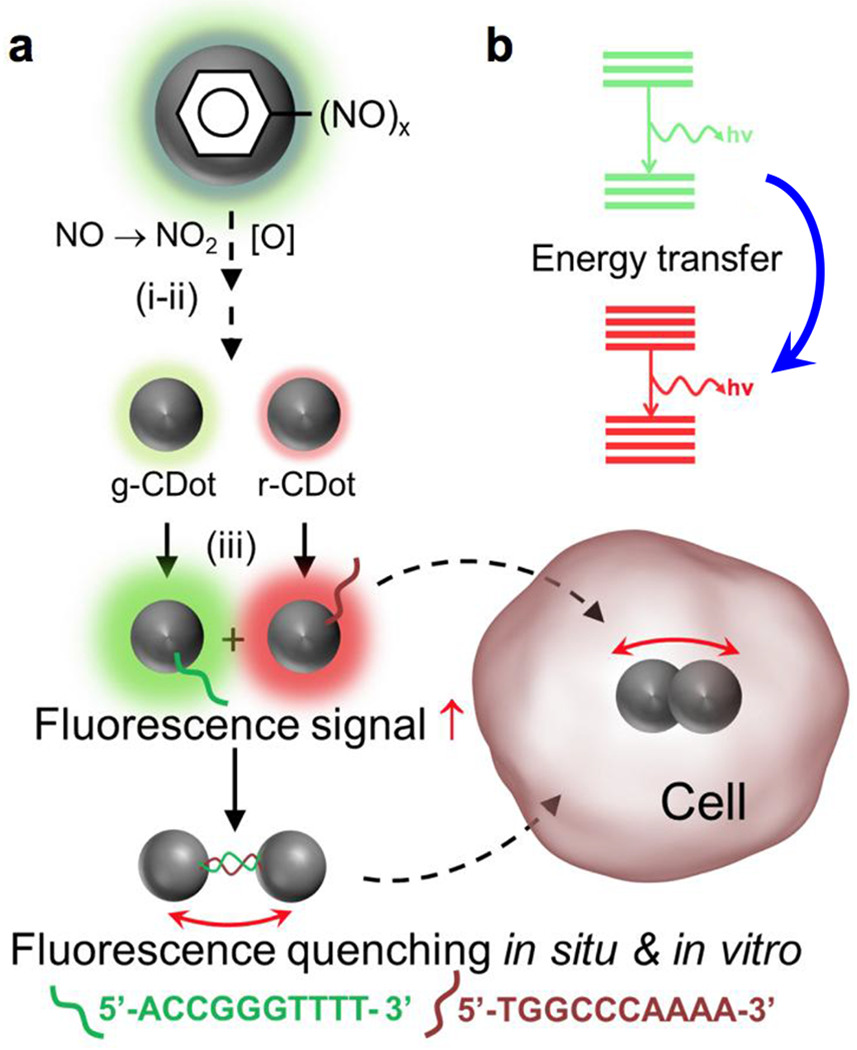
Anisotropic complementarity in DNA-DNA hybridization directed assembly. (a) spontaneous nanoscale oxidation of nitroso carbon dots leads to multicolored particles (i) and separated by chromatography (ii); surface attachment of two complementary ssDNA; (b) quenching through proximity driven energy transfer between two systems. Red arrow (↑) indicates fluorescence intensity enhancement; Double headed arrow (↔) indicates hybridization between two complementary ssDNA.
ACKNOWLEDGMENT
We would like to thank Drs. H. Walukiewicz, R. T. Haasch for help with fluorescence, measurements and XPS studies, respectively. We thank Frederick Seitz Materials Research Laboratory and Beckman Institute for providing access to characterization techniques. Financial support from University of Illinois and National Institute of Health (R01HL126845 to A.K.) is gratefully acknowledged. NIH Tissue Microenvironment training program (T32-EB019944) to S.B.
Footnotes
Supporting Information. A listing of the contents of each file supplied as Supporting Information should be included. For instructions on what should be included in the Supporting Information as well as how to prepare this material for publications, refer to the journal’s Instructions for Authors.
REFERENCES
- (1).Yang B; Sharp JS; Smith MI Shear Banding in Drying Films of Colloidal Nanoparticles. ACS Nano 2015, 9 (4), 4077–4084, [DOI] [PubMed] [Google Scholar]
- (2).Talapin VD; Lee J-S; Kovalenko MV; Shevchenko EV Prospects of Colloidal Nanocrystals for Electronic and Optoelectronic Applications. Chem. Rev 2010, 110 (1), 389–458. [DOI] [PubMed] [Google Scholar]
- (3).Zhu J; Heram MC Assembly and Electronic Applications of Colloidal Nanomaterials. Adv. Mater 2017, 29, 1603895. [DOI] [PubMed] [Google Scholar]
- (4).Puchner EM; Kufer SK; Strackharn M; Stah SW; Gaub HE Nanoparticle Self-Assembly on a DNA-Scaffold Written by Single-Molecule Cut-and-Paste. Nano Lett. 2008, 8 (11), 3692–3695. [DOI] [PubMed] [Google Scholar]
- (5).Colombo M; Fiandra L; Alessio G; Mazzucchelli S; Nebuloni M; Palma C.De; Kantner K; Pelaz B; Rotem R; Corsi F; Parak WJ; Prosperi D Tumour Homing and Therapeutic Effect of Colloidal Nanoparticles Depend on the Number of Attached Antibodies. Nat Commun. 2016, 7, 13818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Gurunatha KL; Fournier AC; Urvoas A; Valerio-Lepiniec M; Marchi V; Minard P; Dujardin E Nanoparticles Self-Assembly Driven by High Affinity Repeat Protein Pairing. ACS Nano 2016, 10, 3176–3185. [DOI] [PubMed] [Google Scholar]
- (7).Lalander CH; Zheng Y; Dhuey S; Cabrini S; Bach U DNA-Directed Self-Assembly of Gold Nanoparticles onto Nanopatterned Surfaces: Controlled Placement of Individual Nanoparticles into Regular Arrays. ACS Nano 2010, 4, 6153–6161. [DOI] [PubMed] [Google Scholar]
- (8).Bromleya KM; Morrisa RJ; Hobley L; Brandania G; Gillespieb RMC; McCluskeya M; Zachariaec M; Marenduzzoa D; Stanley-Wallb NR; MacPhee CE Interfacial Self-Assembly of a Bacterial Hydrophobin. Proc Natl Acad Sci U S A 2014, 112, 5419–5424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Shim M; Guyot-Sionnest P N-Type Colloidal Semiconductor Nanocrystals. Nature 2000, 407, 981–983. [DOI] [PubMed] [Google Scholar]
- (10).Lee O-S; Prytkova TR; Schatz GC Using DNA to Link Gold Nanoparticles, Polymers, and Molecules: A Theoretical Perspective. J Phys Chem Lett. 2010, 1(12), 1781–1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Nicewarner Pena SR; Raina S; Goodrich GP; Fedoroff NV, Keating CD Hybridization and Enzymatic Extension of Au Nanoparticle-Bound Oligonucleotides. J. Am. Chem. Soc 2002, 124 (25), 7314–7323. [DOI] [PubMed] [Google Scholar]
- (12).Fu Y; Lakowicz JR Enhanced Fluorescence of Cy5-Labeled DNA Tethered to Silver Island Films: Fluorescence Images and Time-Resolved Studies Using Single-Molecule Spectroscopy. Anal. Chem 2006, 78, 6238–6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Wang Y; Hu R; Lin G; Roy I; Yong K-T Functionalized Quantum Dots for Biosensing and Bioimaging and Concerns on Toxicity. ACS Appl. Mater. Interfaces 2013, 5(8), 2786–2799 [DOI] [PubMed] [Google Scholar]
- (14).Pathak S; Choi S-K; Arnheim N; Thompson ME Hydroxylated Quantum Dots as Luminescent Probes in In Situ Hybridization. J. Am. Chem. Soc 2001, 123 (17), 4103–4104. [DOI] [PubMed] [Google Scholar]
- (15).Wu S-M; Zhao X; Zhang Z-L; Xie H-Y; Tian Z-Q; Peng J; Lu Z-X; Pang D-W; Xie Z-X Quantum-Dot-Labeled DNA Probes for Fluorescence In Situ Hybdrization (FISH) in the Microorganism Escherichia Coli. ChemPhysChem, 7, 1062–1067. Doi: 10.1002/cphc.200500608 [DOI] [PubMed] [Google Scholar]
- (16).Jiang K; Sun S; Zhang L; Lu Y; Wu A; Cai C Lin H Red, Green, and Blue Luminescence by Carbon Dots: Full-Color Emission Tuning and Multicolor Cellular Imaging. Angew. Chem. Int. Ed 2015, 54, 5360–5363. [DOI] [PubMed] [Google Scholar]
- (17).Ding H; Yu S-B; Wei J-S; Xiong H-M Full-Color Light Emitting Carbon Dots with a Surface-State-Controlled Luminescence Mechanism. ACS Nano 2016, 10 (1), 484–491. [DOI] [PubMed] [Google Scholar]
- (18).Hola K; Sudolska M; Kalytchuk S; Nachtigallova D; Rogach AL; Otyepka M; Zboril R Graphitic Nitrogen Triggers Red Fluorescence in Carbon Dots. ACS Nano 2017, 11 (2), 12402–12410. [DOI] [PubMed] [Google Scholar]
- (19).Ding H; Wei J-S; Zhong N; Gao Q-Y, Xiong H-M Highly Efficient Red-Emitting Carbon Dots with Gram-Scale Yield for Bioimaging. Langmuir 2017, 33, 12635–12642. [DOI] [PubMed] [Google Scholar]
- (20).Srivastava I; Misra SK; Ostadhossein F; Daza E; Singh J; Pan D Surface Chemistry of Carbon Nanoparticles Functionally Select Their Uptake in Various Stages of Cancer Cells. Nano Research 2017, 10 (10), 3269–3284. [Google Scholar]
- (21).Mathews AP Laboratory of Biochemistry and Pharmacology, University of Chicago. January 24, 1909. [Google Scholar]
- (22).Tcyrulnikov NA; Tikhomirova AA; Tcyrulnikov S; Wilson RM Spontaneous Aerobic Oxidation of 1,1,2,2-Tetrakis (N-methylpyridin-4-ium) ethane Ioidie to the Alkene and the Epoxide. Org. Lett 2018, 20 (5), pp 1279–1282. [DOI] [PubMed] [Google Scholar]
- (23).Gautier M, Rohani V, Fulcheri L & Trelles JP Influence of Temperature and Pressure on Carbon Black Size Distribution During Allothermal Cracking of Methane. Aerosol Sci. Techn. 2016, 50 (1), 26. [Google Scholar]
- (24).Choi Y; Kang B; Lee J; Kim GT; Kang H; Lee BR; Kim H; Shim S-H; Lee G; Kwon O-H, Kim B-S Integrative Approach towards Uncovering the Origin of Photoluminescence in Dual Heteroatom-Doped Carbon Nanodots. Chem. Mater 2016, 28, 6840–6847. [Google Scholar]
- (25).Srivastava I, Sar D, Mukherjee P, Schwartz-Duval AS, Huang Z, Jaramillo C, Civantos A, Tripathi I, Allain JP, Bhargava R, Pan D Enzyme-Catalyzed Biodegradation of Carbon Dots follow Sequential Oxidation in a Time-Dependent Manner. Nanoscale 2019, 11, 8226–8236. [DOI] [PubMed] [Google Scholar]
- (26).Sharma A; Gadly T; Neogy S; Ghosh SK; Kumbhakar M Molecular Origin and Self-Assembly of Fluorescent Carbon Nanodots in Polar Solvents. J. Phys. Chem. Lett 2017, 8, 1044–1052. [DOI] [PubMed] [Google Scholar]
- (27).Srivastava IS, Khamo J, Pandit S, Fathi P, Huang X, Cao A, Haasch R, Nie S, Zhang K, Pan D Influence of Electron Donor and Electron Acceptor on the Photophysical Properties of Carbon Dots: A Comparative Investigation at the Bulk-State and Single-Particle Level. Adv. Funct. Mater 2019, doi: 10.1002/adfm.201902466. [DOI] [Google Scholar]
- (28).Misra SK; Srivastava I; Khamo JS.; Krishnamurthy VV; Sar D; Schwartz-Duval a. S.; Soares JANT; Zhang K; Pan D. Carbon Dots with Induced Surface Oxidation Permits Imaging at Single-Particle Level for Intracellular Studies. Nanoscale, 2018, doi: 10.1039/c8nr04065f [DOI] [PubMed] [Google Scholar]
- (29).Wurth C; Grabolle M; Pauli J; Spieles M; Resch-Genger U Relative and absolute determination of fluorescence quantum yields of transparent samples. Nat Protoc. 2013, 8, 1535–1550. [DOI] [PubMed] [Google Scholar]
- (30).Misra SK; Srivastava I; Tripathi I; Daza E; Ostadhossein F; Pan D Macromolecularly “Caged” Carbon Nanoparticles for Intracellular Trafficking via Switchable Photoluminescence. J. Am. Chem. Soc 2017, 139, 1746–1749. [DOI] [PubMed] [Google Scholar]
- (31).Domljanovic I; Ulven ER; Ulven T; Thomsen RP; Okholm AH; Kjems J; Voss A; Taskova M; Astakhova K Dihydropyridine Fluorophores Allow for Specific Detection of Human Antibodies in Serum. ACS Omega 2018, 3, 7580–7586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Taskova M; Astakhova K Fluorescent Oligonucleotides with Bis(prop-2-yn-1-yloxy)butane-1,3-diol Scaffold Rapidly Detect Disease-Associated Nucleic Acids. Bioconjugate Chem. 2019, 30, 3007–3012. [DOI] [PubMed] [Google Scholar]
- (33).Mulberg MW; Taskova M; Thomsen RP; Okholm AH; Kjems J; Astakhova K New Fluorescent Nanoparticles for Ultrasensitive Detection of Nucleic Acids by Optical Methods. ChemBioChem 2017, 18, 1599–1603. [DOI] [PubMed] [Google Scholar]
- (34).Domljanovic I; Carstens A; Okholm A; Kjems J; Nielsen CT; Heegaard NHH; Astakhova K Complexes of DNA with fluorescent dyes are effective reagents for detection of autoimmune antibodies. Sci. Rep 2017, 19, 1925–1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Banerjee A; Pons T; Lequeux N; Dubertret. Quantum dots–DNA bioconjugates: synthesis to applications. Interface Focus 2019, 6, 20160064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Srivastava I, Misra SK, Tripathi I, Schwartz-Duval AS, Pan D In Situ Time-Dependent and Progressive Oxidation of Reduced State Functionalities at the Nanoscale of Carbon Nanoparticles for Polarity-Driven Multiscale Near-Infrared Imaging. Adv. Biosys 2018, 2, 1800009. [Google Scholar]
- (37).Guo CX; Xie J; Wang B; Zheng X; Yang HB; Li CM A New Class of Fluorescent-Dots: Long Luminescent Lifetime Bio-Dots Self-Assembled from DNA at Low Temperatures. Sci. Rep 2013. 3. 2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Vardevanyan PO; Antonyan AP; Parsadanyan MA; Davtyan HG; Karapetyan AT The Binding of Ethidium Bromide with DNA: Interaction with Single- and Double-stranded Structures. Experimental and Molecular Medicine 2003, 35 (6), 527–533. [DOI] [PubMed] [Google Scholar]
- (39).Mladenova P, Udono H, Angelov A, Lukanov A Carbon Nanodots Coated with Oligonucleotides as Fluorescent Hybridization Probes for DNA Microarray. Journal of Mining and Geological sciences 2017, 60, Part II. [Google Scholar]
- (40).Shao S, Fan J, Xue B, Yuwen L, Liu X, Pan D, Fan C, and Wang L DNA-Conjugated Quantum Dot Nanoprobe for High-Sensitivity Fluorescent Detection of DNA and micro-RNA. ACS Appl. Mater. Interfaces 2014, 6 (2), 1152–1157. [DOI] [PubMed] [Google Scholar]
- (41).Onidas D, Markovitsi D, Marguet S, Sharonov A, Gustavsson TJ Phys. Chem. B, 2002, 106 (43), pp 11367–11374 [Google Scholar]
- (42).Onidas D, Markovitsi D, Marguet S, Sharonov A, Gustavsson T Fluorescence Properties of DNA Nucleosides and Nucleotides: A Refined Steady-State and Femtosecond Investigation. J. Phys. Chem. B 2002, 106 (43), 11367–11374. [Google Scholar]
- (43).Geddes CD, Lakowicz JR Metal-Enhanced Fluorescence. J/ Fluor. 2002, 12 (2), 121–129. [Google Scholar]
- (44).Zhou D; Li D; Jing P; Zhai Y; Shen D; Qu, Songnan; Rogach, A. L. Conquering Aggregation-Induced Solid-State Luminescence Quenching of Carbon Dots through a Carbon Dots-Triggered Silica Gelation Process. Chem. Mater 2017, 29, 4, 1779–1787. [Google Scholar]
- (45).Zhang J; Smaga LP; Satyavolu NSR; Chan J; Lu Y DNA Aptamer-Based Activable Probes for Photoacoustic Imaging in Living Mice. J. Am. Chem. Soc 2017, 139, 17225–17228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Shi J; Tian F; Lyu J; Yang M Nanoparticle based fluorescence resonance energy transfer (FRET) for biosensing applications, J. Mater. Chem. B, 2015, 3, 6989. [DOI] [PubMed] [Google Scholar]
- (47).Graff BM; Bloom BP; Wierzbinski E; Waldeck DH Electron Transfer in Nanoparticle Dyads Assembled on a Colloidal Template. J. Am. Chem. Soc 2016, 138, 13260–13270. [DOI] [PubMed] [Google Scholar]
- (48).Seferos DS; Prigodich AE; Giljohann DA; Patel PC; Mirkin CA Nano Lett., 2009, 9(1), 308–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Zagorovsky K; Chou LYT; Chan WCW Controlling DNA-Nanoparticle Serum Interactions. Proc. Natl Acad Sci USA 2016, 113 (48), 13600–13605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Xu X; Han MS; Mirkin CA A Gold-Nanoparticle-Based Real-Time Colorimetric Screening Method for Endonuclease Activity and Inhibition. Angew Chem Int Ed Engl. 2007, 46(19), 3468–70. [DOI] [PubMed] [Google Scholar]
- (51).Vercauteren D; Vandenbroucke RE; Jones AT; Rejman J; Demeester J; De Smedt SC; Sanders NN; Braeckmans K The Use of Inhibitors to Study Endocytic Pathways of Gene Carriers: Optimization and Pitfalls. Mol. Ther 2010, 18, 561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Kirchhausen T; Macia E; Pelish HE Use of Dynasore, the Small Molecule Inhibitor of Dynamin, in the Regulation of Endocytosis. Methods Enzymol. 2008, 438, 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Sigismund S; Woelk T; Puri C; Maspero E; Tacchetti C; Transidico P; Di Fiore PP; Polo S Clathrin-Independent Endocytosis of Ubiquitinated Cargos. Proc. Natl. Acad. Sci. USA 2005, 102, 2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Zhang S, Gao H, and Bao G Physical Principles of Nanoparticles Cellular Endocytosis. ACS Nano 2015, 9 (9), 8655–8671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Kampert T, Misra SK, Srivastava I, Tripathi I, Pan D Phenotypically Screened Carbon Nanoparticles for Enhanced Combinatorial Therapy in Triple Negative Breast Cancer. Cell. Mol. Bioeng 2017, 9, 371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Hildebrandt N; Spillmann CM; Algar WR; Pons T; Stewart MH; Oh E; Susumu K; Diaz SA; Delehanty JB; Medintz IL Energy Transfer with Semiconducting Quantum Dot Bioconjugates: A Versatile Platform for Biosensing, Energy Harvesting, and Other Developing Applications. Chem. Rev 2017, 117, 536–711. [DOI] [PubMed] [Google Scholar]
- (57).Giljohann DA; Seferos DS; Patel PC; Millstone JE; Rosi NL; Mirkin CA Oligonucleotide Loading Determines Cellular Uptake of DNA-Modified Gold Nanoparticles. Nano Lett. 2007, 7 (12), 3818–3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Rogers WB; Crocker JC Direct Measurements of DNA-Mediated Colloidal Interactions and their Quantitative Modeling. Proc Natl Acad Sci USA 2011, 108, 15687–15692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Saurabh S; Beck LE; Maji S; Baty CJ; Wang Y; Yan Q; Watkins SC; Bruchez MP Multiplexed Modular Genetic Targeting of Quantum Dots. ACS Nano 2014, 8, 111380–11146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Ma L; Tu C; Le P; Chittor S; Sung JL; Zahid MU; Teng KW; Ge P; Selvin PR; Smith AM Multidentate Polymer Coatings for Compact and Homogeneous Quantum Dots with Efficient Bioconjugation. J. Am. Chem. Soc 2016, 138, 3382–3384. [DOI] [PubMed] [Google Scholar]
- (61).Naganbabu M; Perkins LA; Wang Y; Kurish J; Schmidt BF; Bruchez MP Multiexcitation Fluorogenic Labeling of Surface, Intracellular, and Total Protein Pools in Living Cells. Bioconjugate Chem. 2016, 27, 1525–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Zhang JY, Zhang F, Hong CQ, Giuliano AE, Cui XJ, Zhou GJ, Zhang GJ, Cui YK Critical Protein GAPDH and its Regulatory Mechanisms in Cancer Cells. Cancer Biol Med. 2015, 12, 10–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Yue J; Li L; Cao L; Zan M; Yang D; Wang Z; Chang Z; Mei Q; Miao P; Dong W-F Two-Step Hydrothermal Preparation of Carbon Dots for Calcium Detection. ACS Appl. Mater. Interfaces 2019, 11, 44566–44572. [DOI] [PubMed] [Google Scholar]
- (64).Milo R; Philips R Cell Biology by the Numbers, 1st ed.; Garland Science: New York, 2016 [Google Scholar]
- (65).Mari Y; Katnik C; Cuevas J ASIC1a Cannels are Activated by Endogenous Protons CDuring Ischemia and Contribute to Synergistic Potentiation of Intracellular Ca2+ Overload During Ischemia and Acidosis. Cell Calcium 2010, 48, 70–82 [DOI] [PubMed] [Google Scholar]
- (66).Liu Z; Pei H; Zhang L; Tian Y Mitochondria-Targeted DNA Nanoprobe for Real-Time Imaging and Simultaneous Quantification of Ca2+ and pH in Neurons. ACS Nano 2018, 12, 12357–12368. [DOI] [PubMed] [Google Scholar]
- (67).Xiong Z; Zhu X; Chu X; Minami M; Hey J; Wei W; MacDonald JF; Wemmie JA; Price MP; Welsh MJ, Simon RP Neuroprotection in Ischemia: Blocking Calcium-Permeable Acid-Sensing Ion Channels. Cell 2004, 118, 687–698. [DOI] [PubMed] [Google Scholar]
- (68).Wang Y; Zeng W; Xiao X; Huang Y; Song X; Yu Z; Tang D; Dong X; Zhu M; Xu T Intracellular ASIC1a Regulates Mitochondrial Permeability Transition-Dependent Neuronal Death. Cell Death Differ. 2013, 20, 1359–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


