Abstract
Most acute promyelocytic leukemias (APL) are characterized by reciprocal translocations t(15;17)(q22;21), which results in the fusion of the promyelocytic leukemia protein (PML) gene at 15q22 with retinoic acid receptor a (RARα) gene at 17q21. However, several complex variant translocations also have been reported. Here, we report a 62-year-old man with typical morphology and clinical features of APL with a complex karyotype including add(11)(p15) and t(13;20)(q12;q11.2) without typical t(15;17) assayed by the G-banding analysis. The fluorescence in situ hybridization with a PML/RARα dual-color DNA probe showed an atypical fusion signal, quantitative real-time polymerase chain reaction analysis showed PML/RARα fusion transcripts, and NGS detected FLT3, WT1, and KRAS mutations. The patient achieved complete remission after treatment with conventional chemotherapy combined with all-trans retinoic acid (ATRA) and arsenic trioxide (ATO). Although the mechanism of this kind of cryptic variant remains unknown, we conclude that the cryptic PML/RARα fusion with add(11)(p15) and t(13;20)(q12;q11.2) seems not to alter the effectiveness of chemotherapy combined with ATRA and ATO.
Keywords: Cryptic t(15;17), acute promyelocytic leukemia, complex karyotype, PML/RARα fusion
INTRODUCTION
Acute promyelocytic leukemia (APL) is classified as M3 subtype of acute myeloid leukemia (AML) according to French–American–British (FAB) classification systems, which accounts for approximately 4% to 20% of adult AML cases [1-3]. Typical APL is caused by the t(15;17)(q24.1;q21.2) translocation which leads to a promyelocytic leukemia protein (PML) gene and retinoic acid receptor a (RARα) gene rearrangement on der(15)and a RARα/PML rearrangement on der(17) at the molecular level. PML/RARα and RARα/PML were found in, respectively, 100% and 67% of patients with positive t(15;17). The PML/RARa fusion protein maintains the ligand-binding domain of RARa and the functional domain of PML, which obstructs the transcription of genes to drive leukemogenesis by deregulating differentiation and self-renewal of myeloid progenitors [4].
In some APL cases, the classic translocation is undetectable, which is considered as masked or cryptic t(15;17). These cases have been detected as submicroscopic insertions of PML or RARα or as more complex rearrangements such as three- or four-way translocations [5-8]. Here, we report a patient with typical morphology and clinical features of APL with a complex karyotype including add(11)(p15) and t(13;20)(q12;q11.2) but masked t(15;17) based on the conventional chromosome (CC) analysis. The patient achieved complete remission with induction treatment of chemotherapy combined with all-trans retinoic acid (ATRA) and arsenic trioxide (ATO).
CASE REPORT
A 62-year-old male presented with pharyngalgia, fatigue, and gum bleeding for 1 week. On admission, peripheral blood counts showed pancytopenia: Hemoglobin 78g/L platelet count 3×109/L, leucocytes 2.53×109/L with 75.6% neutrophils, and 15.3% lymphocytes. Abnormal coagulation screening tests showed prothrombin time 15.6s (normal 9.6–13.7s), activated partial thromboplastin time 25.1s (normal 20-40s), fibrinogen degradation products 168.98mg/L (normal <0-5 mg/L), and D-dimer > 20000 mg/L FEU (normal < 500mg/L FEU). The bone marrow (BM) was hypercellular and packed with 95.2% abnormal promyelocytes that were strongly positive for myeloperoxidase, leading to the morphological diagnosis of APL (Figure 1). The flow cytometry of BM specimens demonstrated that the blasts accounted for 87.27% which were positive for CD117, CD33, myeloperoxidase (MPO), CD13, CD58, CD38, and CD81. Based on the diagnosis and for the patient’s severely compromised overall condition, therapy with ATRA combined with ATO was initiated, followed by cytarabine (Ara-C) and daunorubicin (DNR)-based induction chemotherapy.
FIGURE 1.
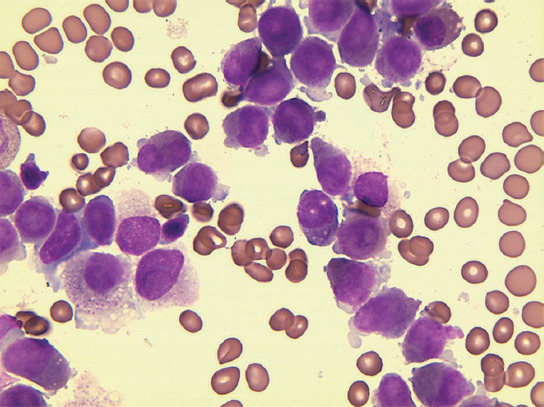
Bone marrow smear of the patient on admission.
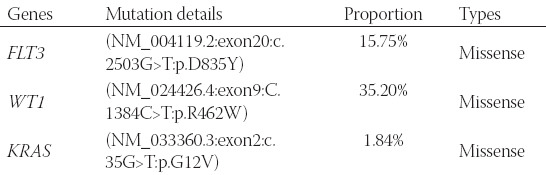
Subsequently, the fluorescence in situ hybridization (FISH) analysis with a probe specific for the PML locus at 15q22 and the RARα locus at 17q21 was performed according to standard procedures. The atypical PML/ RARα fusion fluorescence signal, two red signals (PML), one green signal (RARα) and one yellow signal (PML/RARα fusion) or two red signals (PML), two green signals (RARα), and one yellow signal (2R1G1Y/2R2G1Y), was revealed in 91% (182/200) interphase cells (Figure 2). Quantitative real-time polymerase chain reaction (RT-qPCR) analysis showed that the major PML/RARα transcript harbored the three type breakpoints. The copy number of PML/RARα longtype was 19500, the varianttype was 11100, and the shorttype was 610. The longtype breakpoint cluster region is the main transcript in this patient, and the other breakpoints stem from alternative splicing events. The copy number of the internal reference gene, ABL1, was 157000. The relative expression of PML/RARα transcripts was 19%. However, the cytogenetic study showed an aberrant karyotype of a structural aberration in the short arm of chromosome 11 and translocation of chromosome 13 and chromosome 20, described as 46,XY,add(11)(p15), and ?t(13;20)(q12;q11.2)[20]/ (Figure 3) according to ISCN 1995 [9]. Furthermore, the next-generation sequencing (NGS) detected FLT3, WT1, and KRAS mutations (Table 1).
FIGURE 2.
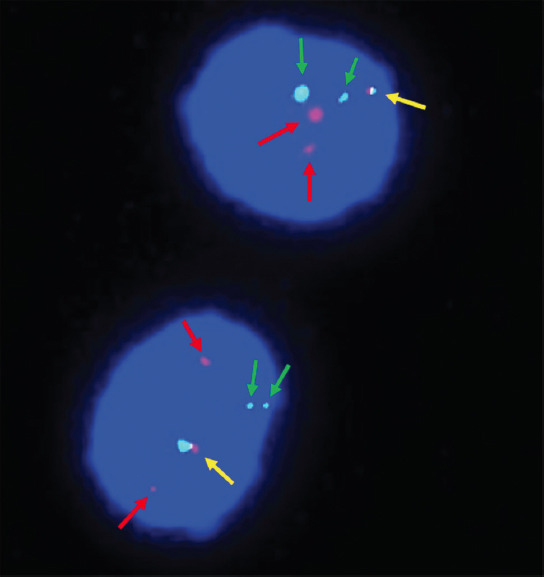
The fluorescence in situ hybridization analysis with painting probes for chromosomes 15 (green arrows) and 17 (red arrows). The fusion signals were marked by yellow arrows.
FIGURE 3.
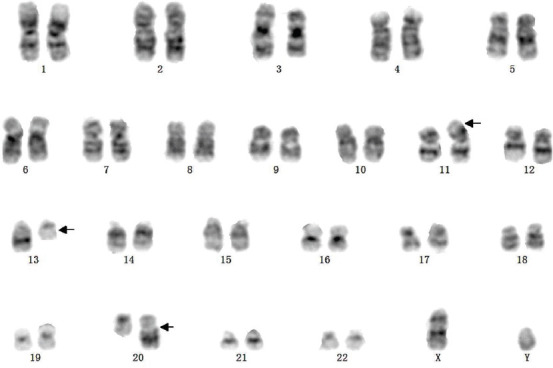
G-banded karyotype of the patient: 46,XY,add(11)(p15),?t(13;20)(q12;q11.2)[20]. The abnormal chromosomes are indicated by arrows.
TABLE 1.
The Mutation types are detected by next-generation sequence
After 21 days, the patient obtained morphology complete remission (CR) with a significant reduction of myeloid promyelocytes from 95.2% to 0.4%. The blasts (CD34+ CD117+ CD33+ HLA-DR + CD38+) were accounted for 0.012% detected by flow cytometry, while there was no significant improvement in leucocytes or hemoglobin. FISH still can detect 80% atypical 2R1G1Y/2R2G1Y fusion fluorescence signal in interphase cells, and 23.1% PML/RARα transcripts were detected by RT-qPCR. Subsequently, the patient received the second cycle induction treatment with ATRA and ATO.
After the second treatment cycle, cytogenetic remission was still achieved. About 1% (2/200) 3R3G atypical fluorescence signal but no fusion signal was detected by FISH, 0.053% PML/RARα transcripts were detected by RT-qPCR, and 0.24% blasts (CD34-CD117+ CD56+ HLA-DR+CD38+) were detected by flow cytometry. The results of RT-qPCR and flow cytometry suggested minimal residual disease (MRD). NGS and CC analyses showed that the mutations and the karyotypes presented normal. After a recovery phase, a comprehensive review showed that there were no blasts or PML/RARα fusion genes in BM, which indicated a stable CR status. The patient then received post-remission therapy, including ATO, Ara-C, and DNR. The treatment process is shown in Figure 4, where the patient was still under the CR status.
FIGURE 4.
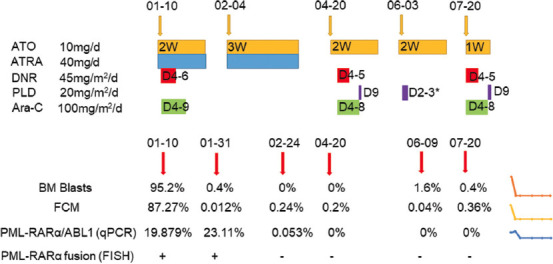
The treatment process of the patient. *D3 used half the drug dose. Aberration: ATO, arsenic trioxide; ATRA, all-trans retinoic acid; DNR, daunorubicin; PLD: pegylated liposomal doxorubicin; Ara-C: Cytarabine; FCM: Flow cytometer; qPCR: Quantitative polymerase chain reaction; FISH: Fluorescence in situ hybridization.
DISCUSSION
The clinical features of APL patients can be similar to those of other AML subtypes, and disseminated intravascular coagulation (DIC) and hyperfibrinolysis are the dominating complications directly affecting the clinical outcomes [10]. With ATRA and/or ATO, more than 90% of patients achieve long-term remission, and BM transplantation is limited to patients who relapse for the first time [11,12]. The classic t(15;17) leads to PML and RARα rearrangement at the molecular level [4]. About 9% of APL patients don’t have t(15;17) translocation, although some are still expressed PML/RARα fusion gene. These cases are believed to be caused by sub-microscopically inserted PML/RARα or more complex rearrangements, with hidden or masked transcripts, which could not be detected by the CC test. When karyotype fails to provide effective information, either due to a lack of metaphases or poor-quality metaphases or due to complex or cryptic rearrangements, the FISH analysis may help to verify the PML/RARα fusion and immediately initiate or maintain specific APL treatment. However, in some cases of APL, FISH does not detect the t(15;17) due to technical limitations, such as the probe size [5,13]. In these rare cases, the cryptic transcript is usually detected by RT-qPCR, and these patients can still benefit from ATRA and ATO since both are targeted therapies against the PML/RARa protein activity, although the patients are lacking the classic translocation.
Here, we reported a masked t(15;17) APL patient with a complex karyotype add(11)(p15) and t(13;20)(q12;q11.2). To the best of our knowledge, this is the first time this karyotype is reported in APL. Typical APL with PML/RARα and 1R1G2F signals are commonly detected by FISH, which indicates the classical t(15;17). In our patient, 182 out of 200 cells (91%) exhibited 2R2G1F or 2R1G1F signals which have also been reported in some cases with complex translocations such as three-way or four-way translocation [7,8]. Another indirect evidence was the 1% 3R3G atypical fluorescence signal that appeared at the third assessment. Although the signal did not reach the cutoff value of over 1%, considering the timepoint of CR, it still provides some information that the extra one red and one green signal represent chromosome 15 and 17, and that they may participate in translocation with other chromosomes. Normally, a three- or four-way translocation can be detected by CC, but in our case, we only detected a t(13;20)(q12;q11.2) and add(11)(p15) without t(15;17) which implied the submicroscopic insert. Kouichi Haraguchi et al. reported a case of masked t(15;17) with a PML/RARα insertion into chromosome 4 detected by spectral karyotyping (SKT) and FISH analyses [14]. Gudrun Göhring et al. reported a masked t(15;17) with a complex karyotype involving chromosomes 4, 6, 8, 10, 17, 18, and 21, a translocation between chromosomes 5 and 17 was detected by multi-color FISH (mFISH) analysis [15]. SKT and mFISH are both effectively applied to cases with complex karyotypes [16,17].
We also reviewed APL case reports that included the aberrant karyotypes of chromosome 13, 20, and 11, listed in Table 2. A complex four-way translocation has been reported, including chromosome 13 and chromosome 20, but the breakpoints were located at 13q22 and 20p13. This patient received ATRA treatment but soon died a week after admission [3]. As for the chromosome 13 band, 13q14 [18], 13q31 [19], and 13q12 [20] also have been reported as unusual karyotypes in APL patients. Paola Temperani et al. reported a case of APL patient whose t(12;13)(p13.2;q14) karyotype was detected not at the time of diagnosis but the second relapse[18]. Yuting Tang et al. described a case of a young APL patient with a masked del13(q31) who obtained CR 3 times but died soon after the third relapse due to sepsis [19]. Giorgina Specchia et al. described a secondary APL case following a renal transplant with complex karyotypes of +8,t(13;22)(q12;q13). The patient also achieved CR without extra hematologic toxicity during induction and consolidation treatment [20]. As for the chromosome 20 band, besides, 20q11.2[7], 20p13[3,22] and 20q12[21] have also been reported in APL patients. Zaida Garcı´a-Casado et al. reported a patient with t(17;20)(q21;q12) whose survival time was more than 5 years [21]. Another case with 20p13 was described that included a complex three-way translocation [22], but the outcome of the patient was favorable which was contrary to the patient with four-way translocation [3]. Jun Yamanouchi et al. described a case that also had a complex four-way translocation, including chromosome 20, the breakpoint was q11.2 which was the same as in our patient, but the clinical character of the karyotype remains to be explored [7]. As for the chromosome 11 band, 11q23 usually induces PLZF/RARa fusion protein [23], Trisomy 11 [24], and 11q13 [25-27] have also been reported. The role of 11p15 has been well described in non-M3 AML, usually participating in the karyotype of t(11;12)(p15;q13), which leads to the NUP98-HOXC11[28,29] or NUP98-HOXC13[30,31] fusion genes. Esperanza et al. reported an APL patient with t(11;12)(p15;q13) karyotype and unusual NUP98/RARG fusion genes, who achieved a favorable outcome with standard chemotherapy [32]. However, the role of add(11)(p15) in APL has not been explored. The three breakpoints have been reported before but the combination is a novel finding, which implies non-random events. As far as we know, the presence of complex karyotypes of our patient does not affect the therapeutic effect of chemotherapy combined with ATRA and ATO.
TABLE 2.
Cases of APL with aberrant chromosome 13, 20, and 11

In addition, FLT3, KRAS, and WT1 mutations were detected by NGS in the present case. Based on the classical molecular landscape of 1540 AML patients, 4% (60/1540) of patients were classified with typical t(15;17)(q22;q12), and the most common mutants were FLT3-ITD and WT1 that account for 35% and 17%, respectively, in APL cases [2]. FLT3 encodes FMS-like tyrosine kinase 3, and the internal tandem duplication (ITD) is the most common aberration in APL [33]. FLT3-ITD induces constitutive activation of the tyrosine kinase receptor which may contribute to the pathogenesis and outcome of APL patients. About 8% of APL patients harbor another point mutation of FLT3 in the codon for aspartic acid 835 (D835). Researches have reported that both two types of mutated FLT3 are related to worse outcomes and high leukocyte counts [34,35]. The Wilms tumor 1 (WT1) gene encodes a protein that contains zinc-finger motifs, which plays an important role in hematopoiesis and elevated expression in AML as well as APL. The role of WT1 in APL still needs to be explored, but a piece of indirect evidence showed that the WT1 gene was the only mutated gene in relapsed patients and may influence the clinical outcomes [36]. KRAS is one of the three small GTP proteins of the RAS family which plays a crucial role in AML oncogenesis. However, the knowledge about the prevalence of KRAS mutation and the significance in APL is limited [37].
In general, almost all APL patients with complex translocations harboring PML/RARα fusion, including our case, have a good response to ATRA and ATO, similar to the typical patients with t(15;17). Our case may indicate that the translocation involving 13q12 and 20q11.2 and add(11)(p15) has no detrimental effect on the response to the ATRA and ATO. The patient achieved CR, and consolidation treatment will be followed up. However, considering the FLT3 mutation and MRD which indicate poor clinical outcomes and relapse, medical observation must continue closely.
CONCLUSION
We report a cryptic t(15;17) APL patient with a complex karyotype, including add(11)(p15) and t(13;20)(q12;q11.2), who achieved CR with polychemotherapy combined with ATRA and ATO targeted therapy. This is the first report of a cryptic APL contained complex karyotype involving chromosomes 11, 13, and 20, which seems not to alter the effectiveness of chemotherapy combined with ATRA and ATO.
Footnotes
Conflict of interest: The authors declare no conflict of interests
Funding: This work is supported in part by the National Natural Science Foundation of China (81770172), Jiangsu Provincial Special Program of Medical Science (BE2017747), Jiangsu Province “333” project (BRA2019103), The Fundamental Research Funds for the Central Universities (2242019K3DZ02), Milstein Medical Asian American Partnership (MMAAP) Foundation Research Project Award in Hematology (2017), Key Medical of Jiangsu Province (ZDXKB2016020).
REFERENCES
- 1.Ryan MM. Acute promyelocytic leukemia:A summary. J Adv Pract Oncol. 2018;9(2):178–87. [PMC free article] [PubMed] [Google Scholar]
- 2.Papaemmanuil E, Gerstung M, Bullinger L, Gaidzik VI, Paschka P, Roberts ND, et al. Genomic classification and prognosis in acute myeloid leukemia. N Engl J Med. 2016;374(23):2209–21. doi: 10.1056/NEJMoa1516192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zaccaria A, Testoni N, Martinelli G, Pelliconi S, Buzzi M, Farabegoli P, et al. Four-chromosomes complex translocations in acute promyelocytic leukemia:Description of two cases. Eur J Haematol. 1994;52(3):129–33. doi: 10.1111/j.1600-0609.1994.tb01302.x. https://doi.org/10.1111/j.1600-0609.1994.tb01302.x. [DOI] [PubMed] [Google Scholar]
- 4.Grimwade D, Enver T. Acute promyelocytic leukemia:Where does it stem from? Leukemia. 2004;18(3):375–84. doi: 10.1038/sj.leu.2403234. https://doi.org/10.1038/sj.leu.2403234. [DOI] [PubMed] [Google Scholar]
- 5.Kim MJ, Cho SY, Kim MH, Lee JJ, Kang SY, Cho EH, et al. FISH-negative cryptic PML-RARA rearrangement detected by long-distance polymerase chain reaction and sequencing analyses:A case study and review of the literature. Cancer Genet Cytogenet. 2010;203(2):278–83. doi: 10.1016/j.cancergencyto.2010.08.026. https://doi.org/10.1016/j.cancergencyto.2010.08.026. [DOI] [PubMed] [Google Scholar]
- 6.Burns TF, Loo EY, Bengtson EM, Bao L. Cytogenetically cryptic insertion of PML segment into RARA on chromosome 17q resulting PML-RARA fusion in acute promyelocytic leukemia. Ann Hematol. 2019;98(1):211–3. doi: 10.1007/s00277-018-3399-1. https://doi.org/10.1007/s00277-018-3399-1. [DOI] [PubMed] [Google Scholar]
- 7.Yamanouchi J, Hato T, Niiya T, Miyoshi K, Azuma T, Sakai I, et al. A new four-way variant t(5;17;15;20)(q33;q12;q22;q11.2) in acute promyelocytic leukemia. Int J Hematol. 2011;94(4):395–8. doi: 10.1007/s12185-011-0929-1. https://doi.org/10.1007/s12185-011-0929-1. [DOI] [PubMed] [Google Scholar]
- 8.Lv L, Yang L, Cui H, Ma T. A complex translocation (1;17;15) with spliced short-type PML-RARA fusion transcripts in acute promyelocytic leukemia:A case report. Exp Ther Med. 2019;17(2):1360–6. doi: 10.3892/etm.2018.7091. https://doi.org/10.3892/etm.2018.7091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mitelman F. Memphis, Tennessee, USA: Karger Medical and Scientific Publishers; 1995. ISCN 1995:An international system for human cytogenetic nomenclature 1995:Recommendations of the International Standing Committee on Human Cytogenetic Nomenclature. [Google Scholar]
- 10.Park JH, Qiao B, Panageas KS, Schymura MJ, Jurcic JG, Rosenblat TL, et al. Early death rate in acute promyelocytic leukemia remains high despite all-trans retinoic acid. Blood. 2011;118(5):1248–54. doi: 10.1182/blood-2011-04-346437. https://doi.org/10.1182/blood-2011-04-346437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang ZY, Chen Z. Acute promyelocytic leukemia:From highly fatal to highly curable. Blood. 2008;111(5):2505–15. doi: 10.1182/blood-2007-07-102798. https://doi.org/10.1182/blood-2007-07-102798. [DOI] [PubMed] [Google Scholar]
- 12.Lo-Coco F, Avvisati G, Vignetti M, Thiede C, Orlando SM, Iacobelli S, et al. Retinoic acid and arsenic trioxide for acute promyelocytic leukemia. N Engl J Med. 2013;369:111–21. doi: 10.1056/NEJMoa1300874. [DOI] [PubMed] [Google Scholar]
- 13.Campbell LJ, Oei P, Brookwell R, Shortt J, Eaddy N, Ng A, et al. FISH detection of PML-RARA fusion in ins (15;17) acute promyelocytic leukaemia depends on probe size. Biomed Res Int. 2013;2013:164501. doi: 10.1155/2013/164501. https://doi.org/10.1155/2013/164501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haraguchi K, Ohno N, Tokunaga M, Tokunaga M, Itoyama T, Gotoh M, et al. Masked t (15;17) APL with the insertion of PML-RARa fusion gene in 4q21. Leuk Res. 2009;33(11):1552–5. doi: 10.1016/j.leukres.2009.04.033. https://doi.org/10.1016/j.leukres.2009.04.033. [DOI] [PubMed] [Google Scholar]
- 15.Göhring G, Lange K, Atta J, Krauter J, Hölzer D, Schlegelberger B. Cryptic t (15;17) in a patient with AML M3 and a complex karyotype. Cancer Genet Cytogenet. 2007;175(1):77–80. doi: 10.1016/j.cancergencyto.2007.01.004. https://doi.org/10.1016/j.cancergencyto.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 16.Schröck E, Du Manoir S, Veldman T, Schoell B, Wienberg J, Ferguson-Smith M, et al. Multicolor spectral karyotyping of human chromosomes. Science. 1996;273(5274):494–7. doi: 10.1126/science.273.5274.494. https://doi.org/10.1126/science.273.5274.494. [DOI] [PubMed] [Google Scholar]
- 17.Liehr T, Othman MA, Rittscher K, Alhourani E. The current state of molecular cytogenetics in cancer diagnosis. Expert Rev Mol Diagn. 2015;15(4):517–26. doi: 10.1586/14737159.2015.1013032. https://doi.org/10.1586/14737159.2015.1013032. [DOI] [PubMed] [Google Scholar]
- 18.Temperani P, Luppi M, Giacobbi F, Medici V, Morselli M, Barozzi P, et al. Late-appearing PML/RARalpha fusion transcript with coincidental t(12;13)(p13.2;q14) in acute promyelocytic leukemia lacking the t(15;17) cytogenetic anomaly. Cancer Genet Cytogenet. 2000;119(2):121–6. doi: 10.1016/s0165-4608(99)00233-2. https://doi.org/10.1016/s0165-4608(99)00233-2. [DOI] [PubMed] [Google Scholar]
- 19.Tang Y, Wang Y, Hu L, Meng F, Xu D, Wan K, et al. Acute promyelocytic leukemia with cryptic t(15;17) on isochromosome 17:A case report and review of literature. Int J Clin Exp Pathol. 2015;8(11):15294–300. [PMC free article] [PubMed] [Google Scholar]
- 20.Specchia G, Storlazzi CT, Cuneo A, Surace C, Mestice A, Pannunzio A, et al. Acute promyelocytic leukemia with additional chromosome abnormalities in a renal transplant case. Ann Hematol. 2001;80(4):246–50. doi: 10.1007/s002770000273. https://doi.org/10.1007/s002770000273. [DOI] [PubMed] [Google Scholar]
- 21.Garcia-Casado Z, Cervera J, Valencia A, Pajuelo JC, Mena-Duran AV, Barragan E, et al. A t(17;20)(q21;q12) masking a variant t(15;17)(q22;q21) in a patient with acute promyelocytic leukemia. Cancer Genet Cytogenet. 2006;168(1):73–6. doi: 10.1016/j.cancergencyto.2005.12.014. https://doi.org/10.1016/j.cancergencyto.2005.12.014. [DOI] [PubMed] [Google Scholar]
- 22.Yamamoto K, Hamaguchi H, Nagata K, Kobayashi M, Takashima T, Taniwaki M. A new complex translocation (15;20;17)(q22;p13;q21) in acute promyelocytic leukemia. Cancer Genet Cytogenet. 1998;101(2):89–94. doi: 10.1016/s0165-4608(97)00251-3. https://doi.org/10.1016/s0165-4608(97)00251-3. [DOI] [PubMed] [Google Scholar]
- 23.McConnell MJ, Licht JD. The PLZF gene of t (11;17)-associated APL. Curr Top Microbiol Immunol. 2007;313:31–48. doi: 10.1007/978-3-540-34594-7_3. https://doi.org/10.1007/978-3-540-34594-7_3. [DOI] [PubMed] [Google Scholar]
- 24.Bastos EF, Silva LA, Ramos MC, Pimenta G, Cortez PI, de Lucena SB, et al. Trisomy 11 as an additional chromosome alteration in a child with acute promyelocytic leukemia with poor prognosis. Case Rep Genet. 2012;2012:659016. doi: 10.1155/2012/659016. https://doi.org/10.1155/2012/659016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wan TS, Ma SK, Yip SF, Yeung YM, Chan LC. Molecular characterization of der(15)t(11;15) as a secondary cytogenetic abnormality in acute promyelocytic leukemia with cryptic PML-RAR alpha fusion on 17q. Cancer Genet Cytogenet. 2000;121(1):90–3. doi: 10.1016/s0165-4608(00)00234-x. https://doi.org/10.1016/s0165-4608(00)00234-x. [DOI] [PubMed] [Google Scholar]
- 26.Temperani P, Luppi M, Giacobbi F, Vaccari P, Longo G, Donelli A, et al. Occurrence of a novel t(11;19)(q13;q13.3) in complete remission of acute promyelocytic leukemia. Cancer Genet Cytogenet. 1998;101(1):35–8. doi: 10.1016/s0165-4608(97)00223-9. https://doi.org/10.1016/s0165-4608(97)00223-9. [DOI] [PubMed] [Google Scholar]
- 27.Najfeld V, Scalise A, Troy K. A new variant translocation 11;17 in a patient with acute promyelocytic leukemia together with t(7;12) Cancer Genet Cytogenet. 1989;43(1):103–8. doi: 10.1016/0165-4608(89)90133-7. https://doi.org/10.1016/0165-4608(89)90133-7. [DOI] [PubMed] [Google Scholar]
- 28.Taketani T, Taki T, Shibuya N, Kikuchi A, Hanada R, Hayashi Y. Novel NUP98-HOXC11 fusion gene resulted from a chromosomal break within exon 1 of HOXC11 in acute myeloid leukemia with t(11;12)(p15;q13) Cancer Res. 2002;62(16):4571–4. https://doi.org/10.4267/2042/70186. [PubMed] [Google Scholar]
- 29.Gu BW, Wang Q, Wang JM, Xue YQ, Fang J, Wong KF, et al. Major form of NUP98/HOXC11 fusion in adult AML with t(11;12)(p15;q13) translocation exhibits aberrant trans-regulatory activity. Leukemia. 2003;17(9):1858–64. doi: 10.1038/sj.leu.2403036. https://doi.org/10.1038/sj.leu.2403036. [DOI] [PubMed] [Google Scholar]
- 30.La Starza R, Trubia M, Crescenzi B, Matteucci C, Negrini M, Martelli MF, et al. Human homeobox gene HOXC13 is the partner of NUP98 in adult acute myeloid leukemia with t(11;12)(p15;q13) Genes Chromosomes Cancer. 2003;36(4):420–3. doi: 10.1002/gcc.10182. https://doi.org/10.1002/gcc.10182. [DOI] [PubMed] [Google Scholar]
- 31.Panagopoulos I, Isaksson M, Billström R, Strömbeck B, Mitelman F, Johansson B. Fusion of the NUP98 gene and the homeobox gene HOXC13 in acute myeloid leukemia with t(11;12)(p15;q13) Genes Chromosomes Cancer. 2003;36(1):107–12. doi: 10.1002/gcc.10139. https://doi.org/10.1002/gcc.10139. [DOI] [PubMed] [Google Scholar]
- 32.Such E, Cervera J, Valencia A, Barragán E, Ibañez M, Luna I, et al. A novel NUP98/RARG gene fusion in acute myeloid leukemia resembling acute promyelocytic leukemia. Blood. 2011;117(1):242–5. doi: 10.1182/blood-2010-06-291658. https://doi.org/10.1182/blood-2010-06-291658. [DOI] [PubMed] [Google Scholar]
- 33.Cervera J, Sanz MA. Sheung Wan: AME Publishing Company; 2017. Revealing the Mutational Landscape of Acute Promyelocytic Leukemia. [Google Scholar]
- 34.Beitinjaneh A, Jang S, Roukoz H, Majhail NS. Prognostic significance of FLT3 internal tandem duplication and tyrosine kinase domain mutations in acute promyelocytic leukemia:A systematic review. Leuk Res. 2010;34(7):831–6. doi: 10.1016/j.leukres.2010.01.001. https://doi.org/10.1016/j.leukres.2010.01.001. [DOI] [PubMed] [Google Scholar]
- 35.Picharski GL, Andrade DP, Fabro A, Lenzi L, Tonin FS, Ribeiro RC, et al. The impact of Flt3 gene mutations in acute promyelocytic leukemia:A meta-analysis. Cancers. 2019;11(9):1311. doi: 10.3390/cancers11091311. https://doi.org/10.3390/cancers11091311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao J, Liang JW, Xue HL, Shen SH, Chen J, Tang YJ, et al. The genetics and clinical characteristics of children morphologically diagnosed as acute promyelocytic leukemia. Leukemia. 2019;33(6):1387–99. doi: 10.1038/s41375-018-0338-z. https://doi.org/10.1038/s41375-018-0338-z. [DOI] [PubMed] [Google Scholar]
- 37.Liquori A, Ibañez M, Sargas C, Sanz MÁ, Barragán E, Cervera J. Acute Promyelocytic leukemia:A constellation of molecular events around a single PML-RARA fusion gene. Cancers. 2020;12(3):624. doi: 10.3390/cancers12030624. https://doi.org/10.3390/cancers12030624. [DOI] [PMC free article] [PubMed] [Google Scholar]


