Abstract
Osteoarthritis (OA) is a progressive degenerative disease that affects all synovial joints, causing the disability of the main locomotor diarthrodial joints. OA pathogenesis is caused by a complex interplay between a number of genetic and environmental risk factors, involved in the early onset and progression of this chronic inflammatory joint disease. Uncovering the underlying immunological and genetic mechanisms will enable an insight into OA pathophysiology and lead to novel and integrative approaches in the treatment of OA patients, together with a reduction of the disease risk, or a delay of its onset in susceptible patients.
Keywords: Osteoarthritis, immunology, genetic polymorphisms, genetic susceptibility
INTRODUCTION
Osteoarthritis (OA) is the most common chronic inflammatory joint disease that leads to chronic conditions and disabilities as a result of various pathological changes, including synovial inflammation, cartilage degradation, subchondral bone alterations, and impairment of the supporting musculature. It is reported that 10% of men and 18% of women over 60 years are affected by this joint disabling disorder worldwide [1]. The prevalence and incidence of OA increase with age and are higher among women, and the prevalence is significantly rising among postmenopausal women [2]. This degenerative disease predominantly involves weight-bearing joints of lower extremities (the hip, knee, and ankle), exposed to constant and excessive mechanical loading. Approximately 80% of OA patients have a reduced range of joints’ motion, while 25% of patients have impaired quality of life due to limitations in activity, pain, deformity, and swelling [3]. Moreover, this condition has a considerable personal and social impact due to the aging of the global population and the increase in obesity rates [4]. The complex interactions between a number of genetic and environmental risk factors, including developmental disorders, obesity, metabolic factors and prior joint injuries, contribute to OA initiation and progression (Figure 1) [4].
FIGURE 1.
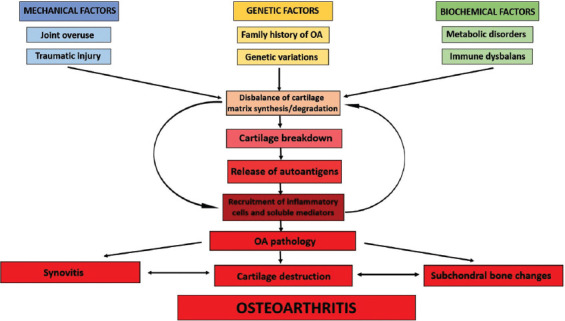
Factors implied in osteoarthritis onset, central events, and disease features. OA: Osteoarthritis.
There is a continuous effort for discovering the predisposing loci in OA genome-wide association studies (GWASs) and their functional relevancy, as well as their interaction with the known risk-conferring etiological factors. Although there is limited information available from family studies, genetic factors are well recognized as contributing factors to OA susceptibility [5]. Genomic studies have shown that genetic polymorphisms, predominantly single nucleotide polymorphisms (SNPs), are linked with OA occurrence, but could also be associated with OA progression or could delay/enhance its onset in susceptible individuals. Previous studies reported SNPs in various genes as genetic risk markers for OA, including genes encoding cartilage and bone metabolism mediators, such as vitamin D receptor (VDR) [6], pro-inflammatory cytokines, different members of the metalloproteinase family [7,8], and related genes, such as growth differentiation factor 5 (GDF5) [9]. However, the genetic background of OA has complex origins and effects, and many studies show conflicting results.
IMMUNOLOGY OF OA
A variety of immune mediators and metabolic intermediates affect the homeostasis of joint structures, causing the imbalance between anabolic and catabolic processes, which leads to the OA development and progression [10].
One of the risk factors for OA degenerative processes is an inflammatory reaction. Elevated levels of inflammatory mediators are detected in almost every OA joint tissue – synovial fluid, synovial membrane, subchondral bone, and cartilage [11]. A critical factor in OA development is the damage of cellular and extracellular matrix (ECM) upon joint injuries, chronic mechanical insults, trauma, or aging. The endogenous damage-associated molecules, released upon cellular stress or tissue injury, lead to the activation of innate immunity through receptor-dependent mechanisms, pattern-recognition receptors, and consequent inflammatory reaction [12]. When triggered, the inflammatory response leads to the upregulation of mediators that further propagate inflammation and cause disbalance of homeostasis and pro-catabolic reprogramming [13].
ROLE OF CYTOKINES IN OA
The initiation and maintenance of the inflammatory processes in OA joints are controlled by different pro- and anti-inflammatory cytokines. Interleukin (IL)-1β and tumor necrosis factor-α (TNF-α), key pro-inflammatory cytokines that exhibit a synergistic effect, are elevated in synovial fluid, synovial membrane, cartilage, and the subchondral bone of OA joints [14].
IL-1β-mediated catabolism is considered as the key pathophysiological process in the course of OA [15]. An important mechanism in the OA pathogenesis is IL-1β-induced upregulation of matrix metalloproteinases (MMPs), the group of enzymes involved in the degradation of collagen and other proteins in the ECM. Interstitial collagenase (MMP-1), stromelysin-1 (MMP-3), and collagenase 3 (MMP-13) appear to degrade the components of the cartilage ECM in arthritis [16]. The A disintegrin and metalloproteinase with thrombospondin motifs (ADAMTS) metalloproteinase family, aggrecan degradation molecules, are also involved in IL-1β-induced cartilage matrix degradation [14]. Production of ADAMTS proteases upon IL-1β stimulation was demonstrated both in human OA chondrocytes [17] and synovial fibroblasts [18]. IL-1β is also associated with the regulation of apoptosis in cultured human OA chondrocytes, through the upregulation of Bcl2 family [19] and the activation and/or production of the oxidative stress intermediates nitric oxide (NO), reactive oxygen species (ROS), and prostaglandin [20]. Also, IL-1β suppresses the anabolic mechanisms in cartilage tissue by repressing the gene expression of ECM constituents (aggrecan and type II collagen) [21].
TNF-α is another important pleiotropic cytokine involved in OA pathogenesis. It plays a dual role in the regulation of immune responses, acting both as a pro-inflammatory mediator, through the initiation of an inflammatory response, and as an immunosuppressive mediator, regulating the duration of the inflammatory response that limits the tissue damage [13]. Similarly to IL-1β, TNF-α induces the expression of matrix-remodeling enzymes in chondrocytes [13,22], and its overproduction is also associated with increased oxidative stress [20].
The levels of key cytokine receptors (IL-1RI, IL-1RII, and TNF receptor p75) are reported to be significantly higher in both OA cartilage and synovium [23], indicating the higher sensitivity of joint cells to stimulation by those cytokines.
IL-6, one of the highly elevated pro-inflammatory cytokines in OA, has a complex role in OA as a modulator of inflammatory reaction. Binding to a membrane-bound IL-6 receptor (mIL-6Ra) is involved in one of the biological functions of IL-6, which is known as the “classical” pathway. This results in downstream activation of the Janus kinase/signal transduction and activators of transcription (JAK/STAT) signaling pathway, which promotes STAT proteins to act as transcription factors in an inflammatory reaction. Another pathway, based on the interaction of IL-6 with a soluble IL-6 receptor (sIL-6R) and referred to as the “trans-signaling”, is involved in the modulation of the MMP-9 activity, crucial for the cartilage degeneration in OA [24]. A number of studies in OA have shown that IL-6 production is upregulated by IL-1β and MMPs [25]. An increased concentration of circulating IL-6, as well as a high body mass index, was associated with the development of knee OA [26]. Furthermore, higher levels of IL-6 were detected in the synovial fluid of obese OA patients, compared to normal-weight patients [27]. The elevated levels of IL-6 are manifested by abundant infiltration of T-cells in the synovial tissues of OA patients, enhanced production of matrix-degrading enzymes, and inhibition of the ECM constituent synthesis [28].
Several studies also showed a positive association between arthritic disorders and the diverse roles of IL-17 [29]. Patients with OA, especially those with advanced OA stages, are reported to have significantly higher levels of IL-17 in the serum and synovial fluid, compared to healthy controls [30,31]. Besides, IL-17 contributes to cartilage degeneration and synovial infiltration in OA by stimulating chondrocytes and synovial fibroblasts to release IL-8, growth regulated oncogene-α (GRO-α) and monocyte chemoattractant protein-1 (MCP-1) and, to a lesser extent, by inducing the IL-1β synthesis by chondrocytes [32]. Moreover, IL-6 and IL-17 act synergistically with IL-1β and TNF-α in inducing the release of inflammatory mediators in affected joint tissues in OA [33].
Several other cytokines and chemokines, such as IL-8, IL-15, and IL-18, have also been identified as elevated in the constituent cells of the joint [14]. These mediators have a role in OA pathogenesis by favoring the persistent inflammation in joints.
It is known that IL-1β plays a substantial role in the course of OA through transcriptional activation of inflammation-related genes [34]. The polymorphisms in the gene encoding IL1-β, rs16944 (-511, G>A) and rs1143634 (+3594, C>T), were previously associated with an increased risk for OA (Table 1). A study on the Croatian Caucasian population reported an association between rs16944 and rs1143634 in female OA patients, indicating these polymorphisms as possible gender-dependent factors for OA susceptibility [35]. Recently, the positive association of the IL-1 gene cluster (IL-1α [IL1A], IL-1β [IL1B], and interleukin-1 receptor antagonist [IL-1RA, IL1RN] genes) with severe knee OA was reported [36].
TABLE 1.
Single nucleotide polymorphisms in immune-related genes linked with OA susceptibility/severity
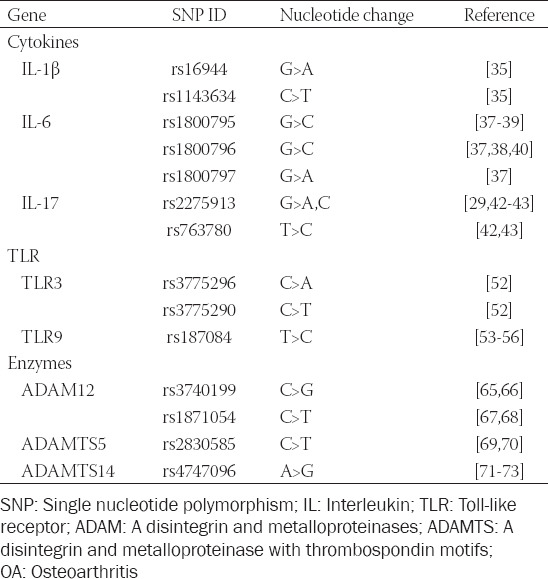
Polymorphisms in the gene coding for the pro-inflammatory cytokine IL-6 are established as one of the key genetic factors for OA susceptibility. The IL-6 promoter variants rs1800797, rs1800796, and rs1800795 (-597, G>A; -572, G>C; and -174, G>C, respectively) were reported to affect both the transcription and secretion of IL-6 in OA [37]. The IL-6 SNP rs1800795 was linked to hip and knee OA susceptibility and severity [38,39], while another IL-6 polymorphism rs1800796 was indicated as a protective factor for the occurrence and severity of hip and knee OA in the elderly [38,40]. In addition, transcriptional regulation of the IL-6 rs1800795 is controlled by two other polymorphisms, rs1800796 and rs1800797, indicating the complex and cooperative regulation of various genetic polymorphisms in the modulation of IL-6 expression [41].
Several studies investigated the role of IL-17 in OA pathology, but data on this cytokine gene polymorphisms and OA susceptibility are still inconsistent. The study by Bai et al. reported the IL-17 polymorphisms rs2275913 (-197, G>A,C) and rs763780 (7488, T>C) as closely linked to knee OA susceptibility [42]. On the other hand, the study by Jiang et al. reported a significant association of knee OA with rs2275913 but not with rs763780 [43]. In addition, conflicting data regarding rs2275913 and OA susceptibility are available, since there is no evidence of an association between this genetic variation and hip/knee OA in a study by Vrgoc et al. [29].
INVOLVEMENT OF TOLL-LIKE RECEPTORS (TLRs) IN OA
TLRs represent a group of highly conserved innate immunity receptors, involved in the initiation of protective responses to pathogens. TLRs have a pivotal role in the detection of viral, bacterial, and fungal infection-associated molecular patterns (pathogen-associated molecular patterns, PAMPs). TLRs are engaged in the activation of diverse signaling pathways and transcriptional activation of various adapter molecules, kinases, and transcription factors such as nuclear factor-kB (NF-κB) and interferon (IFN) regulatory factors (IRFs). The downstream TLR signaling, activated by ligand-receptor interactions, results in initiation and perpetuation of an inflammatory immune response, through transcriptional activation of the genes encoding cytokines (IL-1, IL-6, IL-8, IL-12, and TNF-α), chemokines (C-C motif chemokine ligand 5, CCL5), and cellular adhesion molecules (intercellular adhesion molecule-1 [ICAM-1] and its counterpart lymphocyte function-associated antigen-1 [LFA-1]) [44]. Damage-associated molecular patterns (DAMPs) are endogenous TLR ligands that are also generated during inflammation and are associated with joint damage processes such as matrix degradation and bone remodeling [45]. A wide range of DAMPs within the OA joint initiates TLR-dependent response and leads to coordinated recruitment and intercellular communication among immune cells, such as macrophages and neutrophils, and resident cells that create an inflammatory environment [46]. The synthesis of pro-inflammatory cytokines through activation of different TLRs (TLR1, TLR2, TLR3, TLR4, and TLR9) is detected in arthritic synovial fibroblasts and articular chondrocytes [47-49]. Endogenous activators of TLR-mediated immunity such as components of the cartilage ECM (low-molecular-weight hyaluronan, heparan sulfate, biglycan, fibronectin, and tenascin c) and alarmins, such as S100 proteins and high-mobility group protein B1 (HMGB1), were found in OA synovial fluid [50]. Sillat et al. also reported a link between increased expression of TLR1, TLR2, TLR4, and TLR9 with the severity of OA [47].
One feature of OA is excessive activation of TLR and their downstream signaling molecules that contribute to OA pathogenesis through systemic sustaining of low-grade inflammation. Inflammation-related mediators (NO, prostaglandins) and inflammatory cytokines (IL-1β, TNF-α, IL-6, and IL-8), which induce catabolic responses within cartilage, are actively produced by OA chondrocytes [25]. The study on catabolic pathways mediated by TLRs in OA cartilage showed abundant expression of TLR-2 and TLR-4 in OA cartilage lesions [51]. Sillat et al. demonstrated an aberrant TLR profile in osteoarthritic cartilage with upregulated expression of not only TLR2 and TLR4 but also TLR1 and TLR9 in cartilage lesions of advanced OA [47].
The genetic variability of TLRs could lead to the activation of the downstream signaling and subsequent elevation of cytokine levels, such as IL-1β, IL-6 and IL-17, whose roles in OA pathophysiology are thoroughly examined (Table 1). However, there are limited data regarding the genetic polymorphisms of the TLR receptor family in OA. Association between the TLR-3 promoter polymorphisms rs3775296 (-7, C>A) and rs3775290 (1377, C>T) and knee OA susceptibility was found [52]. Several studies reported rs187084 (-1486, T>C) polymorphism of the TLR9 gene as a predisposing genetic marker for knee OA development in Chinese [53,54] and Turkish population [55]. The polymorphism rs187084 was also found to be significantly associated with hip OA risk [56].
REMODELING OF THE ECM IN OA
The dysregulated expression of matrix-degrading enzymes that leads to degradation of the cartilage is considered as a hallmark of OA. The degradation of collagen and aggrecan molecules, induced upon the expression of inflammatory mediators such as IL-1β and TNF-α, is directly associated with the progression of OA [57] (Figure 2). MMPs and the related families that share the metalloproteinase domain, membrane-anchored A disintegrin and metalloproteinases (ADAMs) and the secreted ADAMs with thrombospondin repeats (ADAMTSs), play an essential role in the ECM and the cartilage degeneration during OA progression. Members of ADAMS superfamily have diverse functions in cellular processes related to immune response, such as cellular adhesion, cell signaling, and the cleavage of the extracellular domains of membrane-associated proteins, a process known as “ectodomain shedding” [57,58].
FIGURE 2.
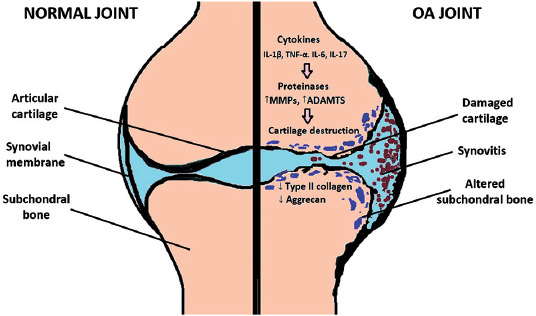
Normal joint and joint in osteoarthritis. OA: Osteoarthritis; MMPs: Matrix metalloproteinases; ADAMTS: A disintegrin and metalloproteinase with thrombospondin motifs; IL: Interleukin; TNF-α: Tumor necrosis factor α.
ADAM10 and ADAM12 are identified as osteoclastic cell differentiation factors [58]. ADAM10 is considered as a progenitor cell fate determination factor during bone development and remodeling [59]. ADAM15 is upregulated in chondrocytes from the early stages of OA and is involved in cartilage remodeling by exerting anti-apoptotic effects [60]. The association between the radiographic features of knee OA and ADAM12 at both gene and protein levels [61] indicated the potential role of ADAM12 in the development of OA.
The ADAMTS group of zinc-dependent proteases is implicated in physiological and pathological tissue remodeling and inflammation [62]. ADAMTS4 and ADAMTS5, also known as aggrecanases 1 and 2, respectively, have been reported as crucial for cartilage aggrecan degradation in OA. ADAMTS4 and ADAMTS5 have differential regulation. While the ADAMTS4 gene expression is found to be upregulated as the result of pro-inflammatory IL-1β, TNF-α, and TGF-β activity, ADAMTS5 does not respond to this pro-inflammatory cytokine stimulation [63]. A study on the expression profiling of ECM degradation mediators in OA demonstrated a dysregulated pattern of ADAMTS genes expression, with upregulated ADAMTS2, -12, -14, and -16 in OA cartilage, compared to normal cartilage samples [64].
Polymorphisms in genes encoding metalloproteinases, ECM-degrading proteases, have also been identified as genetic risk factors in OA (Table 1). Several polymorphisms within the ADAM12 gene have been reported to have a significant impact on OA occurrence. The ADAM12 rs3740199 polymorphism has been associated with knee OA in male patients [65,66], indicating that inflammatory responses in OA differ between genders. The association of the other ADAM12 polymorphism, rs1871054, has also been linked to knee OA susceptibility and severity in the Asian population [67], but not in the Caucasian population [68].
Genes that encode ADAMTS family members are also considered as novel OA candidate genes, but with inconsistent findings in different populations. A positive association between the ADAMTS5 rs2830585 genetic polymorphism and OA development was found in the Chinese population [69], while in the Turkish population no association of rs2830585 with OA susceptibility was observed [70]. The involvement of ADAMTS14 genetic polymorphisms in OA development has been also noted recently [71]. A positive association between the ADAMTS14 gene polymorphism rs4747096 and an increased risk of OA was reported [72], with a possible gender-dependent association, especially for knee and hand OA susceptibility [71,73].
FUTURE PERSPECTIVES
Multifactorial diseases, such as OA, are challenging to study and treat due to the involvement of polymorphisms in multiple genes and association with various environmental causes. Elucidation of the diverse immunoregulatory network along with the genetic architecture of OA would provide an insight into mechanisms underlying this complex disease as well as gene-environment interactions.
There is a continuous effort for discovering novel predisposing loci in OA GWAS studies and functional relevancy of genes previously implicated in OA. However, the genetic background of OA is complex, and inheritance patterns that could be responsible for triggering disease onset and/or progression, are generally unknown. Moreover, gene-environment interactions most likely play an important role in the development of OA and significant clinical variations, including joint pain, stiffness, and locomotor restriction.
Establishing the genetic network of OA in the future will provide guidance for precision medicine and identify the predisposing genetic risk factors. Also, revealing possible therapeutic targets in the treatment of OA and further development of immunotherapies will provide an optimal strategy for personalized approaches in OA therapy, based on the patient’s genotype [74]. Further genetic studies identifying the underlying mechanisms of OA could also give recommendations for individualized nutrition that may slow down the progression, improve symptoms, and prevent or delay surgical treatment in patients with advanced disease. Poor diet and adverse lifestyle factors such as low physical activity, obesity, high alcohol intake, and smoking are major risk factors for developing certain complex diseases, including OA. What is more, emerging evidence indicates that various bioactive dietary components, as well as caloric intake, could play a protective role in the immunomodulation through their anti-inflammatory and antioxidative effects [75]. Classification of relevant genetic changes may enable early identification of potential therapeutic targets in susceptible individuals and personalized approaches to achieve optimal clinical care. There are growing efforts to develop relevant scores to achieve an optimal diagnosis and clinical care of patients with OA [76]. Several protein-based markers from serum and urine have been assessed as indicators of OA burdens [77,78]. Moreover, a study of knee OA [26] assessed a serum IL-6 assay and clinical features such as patient’s age and BMI in the stratification of the patient’s risk.
Although there are a number of recent studies on genetic polymorphisms in OA [79], the actual SNPs and genes are still not included in clinical protocols of OA patients’ treatment. Integration of biochemical and genetic biomarkers, as well as clinical data, will improve clinical care of OA patients, beyond reporting associations of biomarkers with OA.
Footnotes
Conflict of interest statement: The authors declare no conflict of interests
REFERENCES
- 1.Roseti L, Desando G, Cavallo C, Petretta M, Grigolo B. Articular cartilage regeneration in osteoarthritis. Cells. 2019;8(11):1305. doi: 10.3390/cells8111305. https://doi.org/10.3390/cells8111305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mahajan A, Patni R. Menopause and osteoarthritis:Any association? J Midlife Health. 2018;9(4):171–2. doi: 10.4103/jmh.JMH_157_18. https://doi.org/10.4103/jmh.JMH_157_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ma VY, Chan L, Carruthers KJ. Incidence, prevalence, costs, and impact on disability of common conditions requiring rehabilitation in the United States:Stroke, spinal cord injury, traumatic brain injury, multiple sclerosis, osteoarthritis, rheumatoid arthritis, limb loss, and back pain. Arch Phys Med Rehabil. 2014;95(5):986–95. doi: 10.1016/j.apmr.2013.10.032. https://doi.org/10.1016/j.apmr.2013.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Egloff C, Hügle T, Valderrabano V. Biomechanics and pathomechanisms of osteoarthritis. Swiss Med Wkly. 2012;142:w13583. doi: 10.4414/smw.2012.13583. https://doi.org/10.4414/smw.2012.13583. [DOI] [PubMed] [Google Scholar]
- 5.Zeggini E, Panoutsopoulou K, Southam L, Rayner NW, Day-Williams AG, Lopes MC, et al. Identification of new susceptibility loci for osteoarthritis (arcOGEN):A genome-wide association study. Lancet. 2012;380(9844):815–23. doi: 10.1016/S0140-6736(12)60681-3. https://doi.org/10.1016/s0140-6736(12)60681-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Colombini A, Cauci S, Lombardi G, Lanteri P, Croiset S, Brayda-Bruno M, et al. Relationship between Vitamin D receptor gene (VDR) polymorphisms, Vitamin D status, osteoarthritis and intervertebral disc degeneration. J Steroid Biochem Mol Biol. 2013;138:24–40. doi: 10.1016/j.jsbmb.2013.03.001. https://doi.org/10.1016/j.jsbmb.2013.03.001. [DOI] [PubMed] [Google Scholar]
- 7.Chow YY, Chin KY. The role of inflammation in the pathogenesis of osteoarthritis. Mediators Inflamm. 2020;2020:8293921. doi: 10.1155/2020/8293921. https://doi.org/10.1155/2020/8293921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pérez-García S, Carrión M, Gutiérrez-Cañas I, Villanueva-Romero R, Castro D, Martínez C, et al. Profile of matrix-remodeling proteinases in osteoarthritis:Impact of fibronectin. Cells. 2019;9(1):40. doi: 10.3390/cells9010040. https://doi.org/10.3390/cells9010040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miyamoto Y, Mabuchi A, Shi D, Kubo T, Takatori Y, Saito S, et al. A functional polymorphism in the 50 UTR of GDF5 is associated with susceptibility to osteoarthritis. Nat Genet. 2007;39:529–33. doi: 10.1038/2005. https://doi.org/10.1038/2005. [DOI] [PubMed] [Google Scholar]
- 10.Hunter DJ, Bierma-Zeinstra S. Osteoarthritis. Lancet. 2019;393(10182):1745–59. doi: 10.1016/S0140-6736(19)30417-9. https://doi.org/10.1016/s0140-6736(19)30417-9. [DOI] [PubMed] [Google Scholar]
- 11.Orlowsky EW, Kraus VB. The role of innate immunity in osteoarthritis:When our first line of defense goes on the offensive. J Rheum. 2015;42(3):363–71. doi: 10.3899/jrheum.140382. https://doi.org/10.3899/jrheum.140382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gómez R, Villalvilla A, Largo R, Gualillo O, Herrero-Beaumont G. TLR4 signaling in osteoarthritis finding targets for candidate DMOADs. Nat Rev Rheumatol. 2014;11:159–70. doi: 10.1038/nrrheum.2014.209. https://doi.org/10.1038/nrrheum.2014.209. [DOI] [PubMed] [Google Scholar]
- 13.Kapoor M, Martel-Pelletier J, Lajeunesse D, Pelletier JP, Fahmi H. Role of proinflammatory cytokines in the pathophysiology of osteoarthritis. Nat Rev Rheumatol. 2010;7:33–42. doi: 10.1038/nrrheum.2010.196. https://doi.org/10.1038/nrrheum.2010.196. [DOI] [PubMed] [Google Scholar]
- 14.Wojdasiewicz P, Poniatowski ŁA, Szukiewicz D. The role of inflammatory and anti-inflammatory cytokines in the pathogenesis of osteoarthritis. Mediators Inflamm. 2014;2014:561459. doi: 10.1155/2014/561459. https://doi.org/10.1155/2014/561459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pierzchala AW. Expression of IL-1β receptor antagonist in patients with severe osteoarthritis of the knee the mechanism limiting the inflammation in osteoarthritis. Rheumatology (Sunnyvale) 2017;7:2. https://doi.org/10.4172/2329-8731.1000217. [Google Scholar]
- 16.Vincenti MP, Brinckerhoff CE. Transcriptional regulation of collagenase (MMP-1, MMP-13) genes in arthritis:Integration of complex signaling pathways for the recruitment of gene-specific transcription factors. Arthritis Res. 2002;4(3):157–64. doi: 10.1186/ar401. https://doi.org/10.1186/ar401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fan Z, Bau B, Yang H, Soeder S, Aigner T. Freshly isolated osteoarthritic chondrocytes are catabolically more active than normal chondrocytes, but less responsive to catabolic stimulation with interleukin-1b. Arthritis Rheum. 2005;52(1):136–43. doi: 10.1002/art.20725. https://doi.org/10.1002/art.20725. [DOI] [PubMed] [Google Scholar]
- 18.Pérez-García S, Gutiérrez-Cañas I, Seoane IV, Fernandez J, Mellado M, Leceta J, et al. Healthy and osteoarthritic synovial fibroblasts produce a disintegrin and metalloproteinase with thrombospondin motifs 4, 5, 7, and 12. Am J Pathol. 2016;186(9):2449–61. doi: 10.1016/j.ajpath.2016.05.017. https://doi.org/10.1016/j.ajpath.2016.05.017. [DOI] [PubMed] [Google Scholar]
- 19.Lopez-Armada MJ, Carames B, Lires-Dean M, Cillero-Pastor B, Ruiz-Romero C, Galdo F, et al. Cytokines, tumor necrosis factor-a and interleukin-1b, differentially regulate apoptosis in osteoarthritis cultured human chondrocytes. Osteoarthritis Cart. 2006;14(7):660–9. doi: 10.1016/j.joca.2006.01.005. https://doi.org/10.1016/j.joca.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 20.Afonso V, Champy R, Mitrovic D, Collin P, Lomri A. Reactive oxygen species and superoxide dismutases:Role in joint diseases. Joint Bone Spine. 2007;74(4):324–9. doi: 10.1016/j.jbspin.2007.02.002. https://doi.org/10.1016/j.jbspin.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 21.Goldring MB, Birkhead JR, Sandell LJ, Kimura T, Krane SM. Interleukin 1 suppresses expression of cartilage-specific Types II and IX collagens and increases Types I and III collagens in human chondrocytes. J Clin Invest. 1988;82(6):2026–37. doi: 10.1172/JCI113823. https://doi.org/10.1172/jci113823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Séguin CA, Bernier SM. TNFa suppresses link protein and Type II collagen expression in chondrocytes: Role of MEK1/2 and NF-κB signaling pathways. J Cell Physiol. 2003;197(3):356–69. doi: 10.1002/jcp.10371. https://doi.org/10.1002/jcp.10371. [DOI] [PubMed] [Google Scholar]
- 23.Silvestri T, Pulsatelli L, Dolzani P, Frizziero L, Facchini A, Meliconi R. In vivo expression of inflammatory cytokine receptors in the joint compartments of patients with arthritis. Rheum Int. 2005;26(4):360–8. doi: 10.1007/s00296-005-0586-x. https://doi.org/10.1007/s00296-005-0586-x. [DOI] [PubMed] [Google Scholar]
- 24.Akeson G, Malemud C. A role for soluble IL-6 receptor in osteoarthritis. J Funct Morphol Kinesiol. 2017;2(3):27. doi: 10.3390/jfmk2030027. https://doi.org/10.3390/jfmk2030027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pelletier JP, Martel-Pelletier J, Abramson SB. Osteoarthritis, an inflammatory disease: Potential implication for the selection of new therapeutic targets. Arthritis Rheum. 2001;44(6):1237–47. doi: 10.1002/1529-0131(200106)44:6<1237::AID-ART214>3.0.CO;2-F. https://doi.org/10.1002/1529-0131(200106)44:6<1237::aid-art214>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 26.Livshits G, Zhai G, Hart DJ, Kato BS, Wang H, Williams FM, et al. Interleukin-6 is a significant predictor of radiographic knee osteoarthritis: The Chingford study. Arthritis Rheumatol. 2009;60(7):2037–45. doi: 10.1002/art.24598. https://doi.org/10.1002/art.24598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pearson MJ, Herndler-Brandstetter D, Tariq MA, Nicholson TA, Philp AM, Smith HL, et al. IL-6 secretion in osteoarthritis patients is mediated by chondrocyte-synovial fibroblast cross-talk and is enhanced by obesity. Sci Rep. 2017;7:3451. doi: 10.1038/s41598-017-03759-w. https://doi.org/10.1038/s41598-017-03759-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hashizume M, Mihara M. The roles of interleukin-6 in the pathogenesis of rheumatoid arthritis. Arthritis. 2011;2011:765624. doi: 10.1155/2011/765624. https://doi.org/10.1155/2011/765624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vrgoc G, Vrbanec J, Eftedal RK, Dembic PL, Balen S, Dembic Z, et al. Interleukin-17 and toll-like receptor 10 genetic polymorphisms and susceptibility to large joint osteoarthritis. J Orthop Res. 2018;36(6):1684–93. doi: 10.1002/jor.23823. https://doi.org/10.1002/jor.23823. [DOI] [PubMed] [Google Scholar]
- 30.Chen B, Deng Y, Tan Y, Qin J, Chen LB. Association between severity of knee osteoarthritis and serum and synovial fluid interleukin 17 concentrations. J Int Med Res. 2013;42(1):138–44. doi: 10.1177/0300060513501751. https://doi.org/10.1177/0300060513501751. [DOI] [PubMed] [Google Scholar]
- 31.Liu Y, Peng H, Meng Z, Wei M. Correlation of IL-17 level in synovia and severity of knee osteoarthritis. Med Sci Monit. 2015;21:1732–6. doi: 10.12659/MSM.893771. https://doi.org/10.12659/msm.893771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Honorati MC, Bovara M, Cattini L, Piacentini A, Facchini A. Contribution of interleukin 17 to human cartilage degradation and synovial inflammation in osteoarthritis. Osteoarthritis Cart. 2002;10(10):799–807. doi: 10.1053/joca.2002.0829. https://doi.org/10.1053/joca.2002.0829. [DOI] [PubMed] [Google Scholar]
- 33.Gabr MA, Jing L, Helbling AR, Sinclair SM, Allen KD, Shamji MF, et al. Interleukin-17 synergizes with IFNg or TNFa to promote inflammatory mediator release and intercellular adhesion molecule-1 (ICAM-1) expression in human intervertebral disc cells. J Orthop Res. 2010;29(1):1–7. doi: 10.1002/jor.21206. https://doi.org/10.1002/jor.21206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Attur M, Wang HY, Kraus VB, Bukowski JF, Aziz N, Krasnoutsky S, et al. Radiographic severity of knee osteoarthritis is conditional on interleukin 1 receptor antagonist gene variations. Ann Rheum Dis. 2009;69(5):856–61. doi: 10.1136/ard.2009.113043. https://doi.org/10.1136/ard.2009.113043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kaarvatn MH, Jotanovic Z, Mihelic R, Etokebe GE, Mulac-Jericevic B, Tijanic T, et al. Associations of the interleukin-1 gene locus polymorphisms with risk to hip and knee osteoarthritis:Gender and subpopulation differences. Scand J Immunol. 2013;77(2):151–61. doi: 10.1111/sji.12016. https://doi.org/10.1111/sji.12016. [DOI] [PubMed] [Google Scholar]
- 36.Jotanovic Z, Etokebe GE, Mihelic R, Kaarvatn MH, Mulac-Jericevic B, Tijanic T, et al. IL1B-511(G>A) and IL1RN (VNTR) allelic polymorphisms and susceptibility to knee osteoarthritis in Croatian population. Rheumatol Int. 2012;32(7):2135–41. doi: 10.1007/s00296-011-1946-3. https://doi.org/10.1007/s00296-011-1946-3. [DOI] [PubMed] [Google Scholar]
- 37.Kämäräinen OP, Solovieva S, Vehmas T, Luoma K, Riihimaki H, Ala-Kokko L, et al. Common interleukin-6 promoter variants associate with the more severe forms of distal interphalangeal osteoarthritis. Arthritis Res Ther. 2008;10(1):R21. doi: 10.1186/ar2374. https://doi.org/10.1186/ar2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pola E, Papaleo P, Pola R, Gaetani E, Tamburelli FC, Aulisa L, et al. Interleukin-6 gene polymorphism and risk of osteoarthritis of the hip:A case control study. Osteoarthritis Cart. 2005;13(11):1025–8. doi: 10.1016/j.joca.2005.07.011. https://doi.org/10.1016/j.joca.2005.07.011. [DOI] [PubMed] [Google Scholar]
- 39.Honsawek S, Deepaisarnsakul B, Tanavalee A, Yuktanandana P, Bumrungpanichthaworn P, Malila S, et al. Association of the IL-6-174G/C gene polymorphism with knee osteoarthritis in a Thai population. Genet Mol Res. 2011;10(3):1674–80. doi: 10.4238/vol10-3gmr1161. https://doi.org/10.4238/vol10-3gmr1161. [DOI] [PubMed] [Google Scholar]
- 40.Fernandes MT, Fernandes KB, Marquez AS, Colus IM, Souza MF, Santos JP, et al. Association of interleukin-6 gene polymorphism (rs1800796) with severity and functional status of osteoarthritis in elderly individuals. Cytokine. 2015;75(2):316–20. doi: 10.1016/j.cyto.2015.07.020. https://doi.org/10.1016/j.cyto.2015.07.020. [DOI] [PubMed] [Google Scholar]
- 41.Terry F, Loukaci V, Green FR. Cooperative influence of genetic polymorphisms on interleukin 6 transcriptional regulation. J Biol Chem. 2000;275:18138–44. doi: 10.1074/jbc.M000379200. https://doi.org/10.1074/jbc.m000379200. [DOI] [PubMed] [Google Scholar]
- 42.Bai Y, Gao S, Liu Y, Jin S, Zhang H, Su K. Correlation between interleukin-17 gene polymorphism and osteoarthritis susceptibility in Han Chinese population. BMC Medi Genet. 2019;1:20. doi: 10.1186/s12881-018-0736-0. https://doi.org/10.1186/s12881-018-0736-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jiang L, Zhou X, Xiong Y, Bao J, Xu K, Wu L. Association between interleukin-17A/F single nucleotide polymorphisms and susceptibility to osteoarthritis in a Chinese population. Medicine (Baltimore) 2019;98:e14944. doi: 10.1097/MD.0000000000014944. https://doi.org/10.1097/md.0000000000014944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Barreto G, Manninen M, Eklund K. Osteoarthritis and toll-like receptors:When innate immunity meets chondrocyte apoptosis. Biology. 2020;9(4):65. doi: 10.3390/biology9040065. https://doi.org/10.3390/biology9040065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Berenbaum F. Osteoarthritis as an inflammatory disease (osteoarthritis is not osteoarthrosis!) Osteoarthritis Cart. 2013;21(1):16–21. doi: 10.1016/j.joca.2012.11.012. https://doi.org/10.1016/j.joca.2012.11.012. [DOI] [PubMed] [Google Scholar]
- 46.Haraden CA, Huebner JL, Hsueh M, Li YJ, Kraus VB. Synovial fluid biomarkers associated with osteoarthritis severity reflect macrophage and neutrophil related inflammation. Arthritis Res Ther. 2019;21(1):146. doi: 10.1186/s13075-019-1923-x. https://doi.org/10.1186/s13075-019-1923-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sillat T, Barreto G, Clarijs P, Soininen A, Ainola M, Pajarinen J, et al. Toll-like receptors in human chondrocytes and osteoarthritic cartilage. Acta Orthop. 2013;84(6):585–92. doi: 10.3109/17453674.2013.854666. https://doi.org/10.3109/17453674.2013.854666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hu F, Li Y, Zheng L, Shi L, Liu H, Zhang X, et al. Toll-like receptors expressed by synovial fibroblasts perpetuate Th1 and Th17 cell responses in rheumatoid arthritis. PLoS One. 2014;9(6):e100266. doi: 10.1371/journal.pone.0100266. https://doi.org/10.1371/journal.pone.0100266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bobacz K, Sunk IG, Hofstaetter JG, Amoyo L, Toma CD, Akira S, et al. Toll-like receptors and chondrocytes:The lipopolysaccharide-induced decrease in cartilage matrix synthesis is dependent on the presence of toll-like receptor 4 and antagonized by bone morphogenetic protein 7. Arthritis Rheum. 2007;56(6):1880–93. doi: 10.1002/art.22637. https://doi.org/10.1002/art.22637. [DOI] [PubMed] [Google Scholar]
- 50.Van den Bosch MH. Inflammation in osteoarthritis:Is it time to dampen the alarm-in this debilitating disease? Clin Exp Immunol. 2019;195(2):153–66. doi: 10.1111/cei.13237. https://doi.org/10.1111/cei.13237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim HA, Cho ML, Choi HY, Yoon CS, Jhun JY, Oh HJ, et al. The catabolic pathway mediated by Toll-like receptors in human osteoarthritic chondrocytes. Arthritis Rheum. 2006;54(7):2152–63. doi: 10.1002/art.21951. https://doi.org/10.1002/art.21951. [DOI] [PubMed] [Google Scholar]
- 52.Yang HY, Lee HS, Lee CH, Fang WH, Chen HC, Salter DM, et al. Association of a functional polymorphism in the promoter region of TLR-3 with osteoarthritis:A two-stage case-control study. J Orthop Res. 2013;31(5):680–5. doi: 10.1002/jor.22291. https://doi.org/10.1002/jor.22291. [DOI] [PubMed] [Google Scholar]
- 53.Su SL, Yang HY, Lee CH, Huang GS, Salter DM, Lee HS. The 235 (-1486T/C) promoter polymorphism of the TLR-9 gene is associated with end-stage knee osteoarthritis in a Chinese population. J Orthop Res. 2011;30(1):9–14. doi: 10.1002/jor.21494. https://doi.org/10.1002/jor.21494. [DOI] [PubMed] [Google Scholar]
- 54.Zheng M, Shi S, Zheng Q, Wang Y, Ying X, Jin Y. Association between TLR -9 gene rs1∄4 polymorphism and knee osteoarthritis in a Chinese population. Biosci Rep. 2017;37(5):BSR20170844. doi: 10.1042/BSR20170844. https://doi.org/10.1042/bsr20170844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Balbaloglu O, Ozcan OS, Korkmaz M, Yilmaz N. Promoter 238 polymorphism (T-1486C) of TLR-9 gene is associated with knee osteoarthritis in a 239 Turkish population. J Orthop Res. 2017;35(11):2484–9. doi: 10.1002/jor.23559. https://doi.org/10.1002/jor.23559. [DOI] [PubMed] [Google Scholar]
- 56.Yi X, Xu E, Xiao Y, Cai X. Evaluation of relationship between common variants in TLR-9 gene and hip osteoarthritis susceptibility. Genet Test Mol Biomarkers. 2019;23(6):373–9. doi: 10.1089/gtmb.2019.0010. https://doi.org/10.1089/gtmb.2019.0010. [DOI] [PubMed] [Google Scholar]
- 57.Yang CY, Chanalaris A, Troeberg L. ADAMTS and ADAM metalloproteinases in osteoarthritis - looking beyond the 'usual suspects'. Osteoarthritis Cart. 2017;25(7):1000–9. doi: 10.1016/j.joca.2017.02.791. https://doi.org/10.1016/j.joca.2017.02.791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Verrier S, Hogan A, McKie N, Horton M. ADAM gene expression and regulation during human osteoclast formation. Bone. 2004;35(1):34–46. doi: 10.1016/j.bone.2003.12.029. https://doi.org/10.1016/j.bone.2003.12.029. [DOI] [PubMed] [Google Scholar]
- 59.Dallas D, Genever P, Patton A, Millichip MI, McKie N, Skerry TM. Localization of ADAM10 and Notch receptors in bone. Bone. 1999;25(1):9–15. doi: 10.1016/s8756-3282(99)00099-x. https://doi.org/10.1016/s∴-3282(99)00099-x. [DOI] [PubMed] [Google Scholar]
- 60.Böhm BB, Aigner T, Roy B, Brodie TA, Blobel CP, Burkhardt H. Homeostatic effects of the metalloproteinase disintegrin ADAM15 in degenerative cartilage remodeling. Arthritis Rheum. 2005;52(4):1100–9. doi: 10.1002/art.20974. https://doi.org/10.1002/art.20974. [DOI] [PubMed] [Google Scholar]
- 61.Kerna I, Kisand K, Laitinen P, Tamm AE, Kumm J, Lintrop M, et al. Association of ADAM12-S protein with radiographic features of knee osteoarthritis and bone and cartilage markers. Rheum Int. 2011;32(2):519–23. doi: 10.1007/s00296-010-1717-6. https://doi.org/10.1007/s00296-010-1717-6. [DOI] [PubMed] [Google Scholar]
- 62.Kelwick R, Desanlis I, Wheeler GN, Edwards DR. The ADAMTS (a disintegrin and metalloproteinase with thrombospondin motifs) family. Genome Biol. 2015;16(1):113. doi: 10.1186/s13059-015-0676-3. https://doi.org/10.1186/s13059-015-0676-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bondeson J, Wainwright S, Hughes C, Caterson B. The regulation of the ADAMTS4 and ADAMTS5 aggrecanases in osteoarthritis:A review. sClin Exp Rheumatol. 2008;26:139–45. https://doi.org/10.1016/s1063-4584(10)60506-7. [PubMed] [Google Scholar]
- 64.Kevorkian L, Young DA, Darrah C, Donell ST, Shepstone L, Porter S, et al. Expression profiling of metalloproteinases and their inhibitors in cartilage. Arthritis Rheum. 2004;50(1):131–41. doi: 10.1002/art.11433. https://doi.org/10.1002/art.11433. [DOI] [PubMed] [Google Scholar]
- 65.Wu Z, Xu XW, Zhang XW. The association of ADAM12 polymorphism with osteoarthritis susceptibility:A meta-analysis. Ther Clin Risk Manag. 2017;13:821–30. doi: 10.2147/TCRM.S134581. https://doi.org/10.2147/tcrm.s134581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Poonpet T, Tammachote R, Tammachote N, Kanitnate S, Honsawek S. Association between ADAM12 polymorphism and knee osteoarthritis in Thai population. The Knee. 2016;23(3):357–61. doi: 10.1016/j.knee.2016.01.007. https://doi.org/10.1016/j.knee.2016.01.007. [DOI] [PubMed] [Google Scholar]
- 67.Wang L, Guo L, Tian F, Hao R, Yang T. Analysis of single nucleotide polymorphisms within ADAM12 and risk of knee osteoarthritis in a Chinese Han Population. Biomed Res Int. 2015;2015:518643. doi: 10.1155/2015/518643. https://doi.org/10.1155/2015/518643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lv ZT, Liang S, Huang XJ, Cheng P, Zhu WT, Chen AM. Association between ADAM12 single-nucleotide polymorphisms and knee osteoarthritis:A meta-analysis. BioMed Res Int. 2017;2017:5398181. doi: 10.1155/2017/5398181. https://doi.org/10.1155/2017/5398181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhou X, Jiang L, Zhang Y, Zhang J, Zhou D, Wu L, et al. Genetic variation of aggrecanase-2 (ADAMTS5) in susceptibility to osteoarthritis. Braz J Med Biol Res. 2019;52(2):e8109. doi: 10.1590/1414-431X20188109. https://doi.org/10.1590/1414-431x20188109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Canbek U, Imerci A, Kara M, Akgun U, Canbek TD, Aydogan NH. Polymorphisms in ADAMTS4 and ADAMTS5 are not linked to susceptibility to knee osteoarthritis in the Turkish population. Genet Mol Res. 2016;15(3):4238. doi: 10.4238/gmr.15038264. https://doi.org/10.4238/gmr.15038264. [DOI] [PubMed] [Google Scholar]
- 71.Poonpet T, Honsawek S, Tammachote N, Kanitnate R, Tammachote R. ADAMTS14 gene polymorphism associated with knee osteoarthritis in Thai women. Genet Mol Res. 2013;12(4):5301–9. doi: 10.4238/2013.November.7.5. https://doi.org/10.4238/2013.november.7.5. [DOI] [PubMed] [Google Scholar]
- 72.Ma S, Ouyang C, Ren S. Relationship between ADAMTS14 rs4747096 gene polymorphism and knee osteoarthritis in Chinese Population. Biosci Rep. 2018;38(5):BSR20181413. doi: 10.1042/BSR20181413. https://doi.org/10.1042/bsr20181413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rodriguez-Lopez J, Pombo-Suarez M, Loughlin J, Tsezou A, Blanco FJ, Meulenbelt I, et al. Association of a nsSNP in ADAMTS14 to some osteoarthritis phenotypes. Osteoarthritis Cart. 2009;17(3):321–7. doi: 10.1016/j.joca.2008.07.012. https://doi.org/10.1016/j.joca.2008.07.012. [DOI] [PubMed] [Google Scholar]
- 74.Knevel R, le Cessie S, Terao CC, Slowikowski K, Cui J, Huizinga TW, et al. Using genetics to prioritize diagnoses for rheumatology outpatients with inflammatory arthritis. Sci Transl Med. 2020;12(545):eaay1548. doi: 10.1126/scitranslmed.aay1548. https://doi.org/10.1126/scitranslmed.aay1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wu D, Lewis ED, Pae M, Meydani SN. Nutritional modulation of immune function: Analysis of evidence, mechanisms, and clinical relevance. Front Immunol. 2019;9:3160. doi: 10.3389/fimmu.2018.03160. https://doi.org/10.3389/fimmu.2018.03160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sawitzke AD. Personalized medicine for osteoarthritis:Where are we now? Ther Adv Musculoskelet Dis. 2013;5(2):67–75. doi: 10.1177/1759720X12470752. https://doi.org/10.1177/1759720X12470752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kraus VB, Kepler TB, Stabler T, Renner J, Jordan J. First qualification study of serum biomarkers as indicators of total body burden of osteoarthritis. PLoS One. 2010;5(3):e9739. doi: 10.1371/journal.pone.0009739. https://doi.org/10.1371/journal.pone.0009739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Karsdal M, Byrjalsen I, Bay-Jensen A, Henriksen K, Riis B, Christiansen C. Biochemical markers identify influences on bone and cartilage degradation in osteoarthritis the effect of sex, Kellgren-Lawrence (KL) score, body mass index (BMI), oral salmon calcitonin (sCT) treatment and diurnal variation. BMC Musculoskelet Disord. 2010;11:12. doi: 10.1186/1471-2474-11-125. https://doi.org/10.1186/1471-2474-11-125. https://doi.org/10.1002/art.24598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang T, Liang Y, Li H, Li H, He Q, Xue Y, et al. Single nucleotide polymorphisms and osteoarthritis:An overview and a meta-analysis. Medicine (Baltimore) 2016;95(7):e2811. doi: 10.1097/MD.0000000000002811. https://doi.org/10.1097/md.0000000000002811. [DOI] [PMC free article] [PubMed] [Google Scholar]


