Abstract
Celiac disease (CD) is an autoimmune enteropathy triggered by ingestion of gluten present in wheat, barley, and rye. Gluten along with environmental trigger starts an inflammatory reaction which results in damage to small intestine. Human leukocyte antigen (HLA)-DQA1∗05, -DQB1∗02, and -DQB1∗03:02 are the known risk alleles of CD. The diagnostic method for CD involves serological or intestinal biopsy, but genetic test could be implemented. HLA typing precludes the need for further diagnosis and it has high negative predictive value. The aim of this study was to make aware of HLA molecular typing for celiac disease among local laboratories and healthcare professionals. The prevalence and frequency distribution of HLA-DQ2 and -DQ8 haplotypes in 175 pediatric unrelated healthy controls, celiac patients, and CD with concurrent diabetes mellitus type 1 (DM1) was evaluated. The most common haplotype was DQ2 followed by DQ8. In control group only DQ2 was observed with frequency of 8.5%. In celiac patients 85.7% were DQ2, 11.4% were DQ8, and rest were DQ2/DQ8 (2.8%), and all had CD. In the group of CD with DM1, 31.4% had DQ2, 25% had DQ8, and 34% having both the haplotypes; while only 9 of these patients were suffering from CD. It was concluded that Celiac disease is frequently unrecognized by physicians, in part because of its variable clinical presentation and symptoms. Thus genetic testing for celiac disease could be an additive tool for diagnosis to exclude ambiguity.
Keywords: celiac disease, diabetes mellitus type 1, gluten, human leukocyte antigen genes
1. Introduction
There are wide range of diseases related to human leukocyte antigen (HLA) family of genes. One commonly associated autoimmune disorder is celiac disease (CD); that is the intolerance towards gluten. Gluten is a major protein found in wheat, rye, and barley. The merely treatment for celiac disease is gluten free diet throughout the life span.[1] Gluten when enters in to the gastrointestinal tract is broken down into smaller peptide known as gliadin. Gliadin is noxious for celiac patients. Individuals who are sensitive to gliadin have HLA-DQ2 or HLA-DQ8 heterodimers transcribed from major histocompatibility complex (MHC) class II genes of chromosome number 6. These HLA molecules react to gliadin as soon as it enters in lamina propria of small intestine. Three types of antibodies are produced by B-cell mediated response; anti-gliadin, Immunoglobulin A (IgA) anti-tissue transglutaminase (IgA-tTG), and anti-endomysium (IgA-EMA). Cytokines are also released by T-cell mediated response. These inflammatory reactions cause destruction of enterocytes, which eventually leads to villous atrophy.[2–4] The damage to enterocytes result in malabsorption of the nutrients which cause diarrhea, anemia, growth retardation, lethargy, and in chronic conditions; it could lead to osteoporosis, reproductive disorders, and dermatitis herpetiformis. The standard diagnostic method for celiac disease is the detection of gliadin, endomisial, and more recently, transglutaminase antibodies in the sera of patients through enzyme-linked immunosorbent assay (ELISA). Diagnosis is further confirmed by intestinal biopsy, according to European Society of Pediatric Gastroenterology, Hepatology and Nutrition (ESPGHAN).
The genetic susceptibility of celiac disease involves HLA class II genes encoding MHC II heterodimer molecules; dedicated to presentation of antigenic peptides to CD4+ cells. The MHC II heterodimers namely HLA-DQ2 and HLA-DQ8 comprise of alpha and beta chains, encoded by HLA DQA1∗05–DQB1∗02 (DQ2) and DQA1∗03–DQB1∗03:02 (DQ8) genes respectively.
Other non-HLA genes and environmental triggers like (e.g., viral infections, gut microbiota, and others) have also been reported to play role in onset of disease.[5–7]
The HLA-DQ2 and HLA-DQ8 molecules are also expressed in 40% of Europid population, but only 1% have active CD. But due to high negative predictive value genetic testing could be valuable. More than 90% of celiac patients have HLA-DQ2 or HLA-DQ8 haplotypes, absence of which exclude the further need of diagnosis of celiac.[8–11]
1.1. Association of celiac disease with diabetes mellitus type 1
The individuals with celiac disease have been reported to have other autoimmune disorders due to linkage among HLA genes involved. Almost 90% of celiac patients have HLA-DQ2 or HLA-DQ8 with DR3 haplotype, which is shared by diabetes mellitus type 1 (DM1) as well (60–70%). The occurrence of CD in DM1 is much higher compared with general population. New guidelines by ESPGHAN recommend genotyping for celiac disease as screening method for such high risk patients. Therefore for diabetes mellitus 1 patients, it is advised to undergo regular screening for celiac disease.[12]
Thus HLA typing could be beneficial in early, rapid, and more accurate diagnosis of celiac disease. Also for atypic or latent form of CD, patients with IgA deficiency, familial screening to investigate genetic predisposition to the disease, and discriminating between celiac and other childhood gastrointestinal disorders. Also for patients with other autoimmune and non-autoimmune conditions known to be associated with CD, such as Down syndrome and Turner syndrome. A negative HLA result precludes the need for further celiac testing and reduces the burden from patients and their families.[13,14]
In our knowledge there is no such data published for the genetic testing and allele frequency ratio for celiac disease in Sindh, Pakistan. Thus the aim of this study was to introduce HLA typing for celiac diagnosis and to know the prevalence of celiac disease among local population of Sindh. In this study we have investigated and compared, frequency distribution of CD specific alleles (namely HLA-DQA1∗05, -DQB1∗02, and -DQB1∗02:03) in different groups of pediatric individuals, including healthy control, confirmed patients for CD, individuals with celiac like symptoms, and DM1 patients with/without CD.
2. Materials and methods
2.1. The study design and population
This study was cross-sectional study including 175 pediatric participants (112 males and 63 females; mean age 8.7 ± SD 1.2 years) with non-random purposive sampling technique. The study population was selected as per inclusion and exclusion criteria (Fig. 1). All samples were collected from Liaquat University of medical and health sciences hospital (LUMHS), Isra University hospital, and Asian institute of medical sciences (AIMS) hospital, Hyderabad. The duration of the study was 2 years. The individuals were divided into 5 groups (n = 35); Control (Group A), diagnosed cases of celiac disease (Group B), patients with celiac-like symptoms (Group C), diabetes mellitus type 1 patients with celiac-like symptoms (Group D), and diabetes mellitus type 1 patients (Group E). A detailed questionnaire was developed by the help of Canadian Celiac Society to know the status of disease in each patient.
Figure 1.
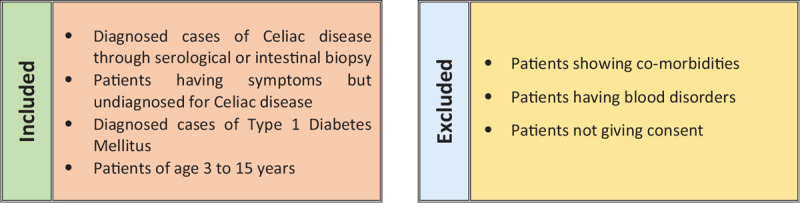
Inclusion criteria of the study population (n = 175).
2.2. Sample collection and genomic DNA extraction
Fresh blood (3 mL) was drawn from all the individuals in Ethylenediamine tetraacetic acid tubes. The blood was stored at 4 °C until the extraction of genome. DNA was extracted from 200 μL of whole blood by Favorgen DNA extraction kit, according to the manufacturer's instructions. Isolated DNA samples were quantified by UV spectrophotometry at 260 nm absorbance and diluted with distilled water to the concentration required for polymerase chain reaction (PCR) (100 ng/μL).
2.3. PCR amplification of celiac disease susceptible alleles
PCR methodology was used for the rapid typing of the HLA-DQA1∗05, DQB1∗02, and DQB∗03:02 alleles, the known celiac specific HLA variants. Sequence specific primers were designed for these 3 alleles (Table 1). Sequences of these primers differed in 2 or 3 nucleotides from those of the other known alleles at the same HLA loci. Part of Beta globin gene was used as positive control (GH20- product size 406 bp). DNA was extracted from 200 μL of blood. All the groups A–E were examined for the presence of 3 alleles. Different PCR mixtures were used for each primer pair. The DNA was amplified following initial denaturation at 95 °C for 5 minutes by 30 cycles at 95 °C for 1 minute, 61 °C for 30 seconds and 72 °C for 30 seconds, and a final extension of 10 minutes at 72 °C. The PCR products were separated by 1.3% agarose gel electrophoresis stained with ethidium bromide and visualized using gel documentation system.
Table 1.
Sequence of primers and product size.
| Target allele | Forward primer | Reverse primer | Product size (bp) |
| HLA-DQA1∗05 | CTGACCACGTCGCCTCTTATGGTGT | ACTGTTCAAGTTATGTTTTAGGACAG | 209 |
| HLA-DQB1∗02 | ACAGAGCGCGTGCGTCTTGTGAGCA | CACCCTGTCCACCGCCGCCCGTTTC | 174 |
| HLA-DQB1∗03:02 | AGCGCGTGCGTCTTGTGACCAGATA | TCACCGCCCGATACACCCCCACGT | 87 |
| GH20 (control) | GAAGAGCCAAGGACAGGTAC | GGAAAATAGACCAATAGGCAG | 408 |
HLA = human leukocyte antigen.
2.4. Data collection and analysis
The data of control and patients were gathered by the help of questionnaire. The data focused on the patients already tested for anti-tTG antibodies (Group B). The patients having celiac like symptoms were tested for anti-tTG (Group C and D). Statistical analysis and data interpretation were carried out by using statistical package for the social sciences (SPSS) version 21.0 by IBM, USA and ANalysis Of VAriance was used for statistical significance with P-value ≤.05.
3. Results
3.1. Clinical data and diagnostic tests
The diagnostic tests for all the groups are summarized in Table 2, along with HLA typing results. Most of the cases were late diagnosis either by anti-tTG or intestinal biopsy. Group B patients were already tested for anti-tTG. For other groups C and D the antibody test was carried out.
Table 2.
Diagnostic tests for the case groups.
| Laboratory tests frequency (%) | |||||||
| Anti-tTG | SBB | HLA result | |||||
| Patient groups | Positive | Negative | Positive | Negative | Positive | Negative | Notes |
| A | – | – | – | – | 3 [8.5%] | 27 [77.1%] | |
| B | 29 [82.8%] | 6 [17.1%] | 20 [57.1%] | 3 [8.5%] | 35 [100%] | 0 | |
| C | 24 [68%] | 11 [31%] | 3 [8.5%] | 6 [17.1%] | 24 [68%] | 11 [31.4%] | 24 had CD |
| D | 9 [25.7%] | 26 [74.2%] | 4 [11.4%] | 0 | 32 [91.4%] | 3 [8.6%] | 9 had CD |
| E | – | – | – | – | 9 [25.7%] | 26 [74.2%] | None had CD |
Group A: control; Group B: diagnosed cases of celiac disease; Group C: patients with celiac-like symptoms; Group D: type 1 diabetes patients with celiac-like symptoms; Group E: type 1 diabetes patients.
HLA-typing: alleles detected: HLA-DQA1∗05, DQB1∗02, and/or DQB1∗03:02.
Anti-tTG = anti tissue transglutaminase; CD = celiac disease; SBB = small bowel biopsy.
3.2. HLA typing
All the positive celiac patients and patients having clinical presentation for celiac with or without DM1 were tested for HLA typing. The results of HLA typing are summarized in Table 3. The PCR for all 3 alleles namely HLA-DQA1∗05, HLA-DQB1∗02, and HLA-DQB1∗03:02 was carried on all the groups; A to D (Fig. 2). The individuals were termed according to the presence of HLA alleles. Subjects carrying both HLA-DQA1∗05, HLA-DQB1∗02 are: DQ2, Subjects carrying only HLA-DQB1∗03:02 are: DQ8 (the alpha chain allele for DQ8 in most celiac positive cases is DQA1∗03, typing for which was not carried out here. The presence of HLA-DQB1∗03:02 is sufficient to tell that a celiac patient is positive for DQ8 molecule.) Subjects carrying all 3 alleles HLA-DQA1∗05, HLA-DQB1∗02, and HLA-DQB1∗03:02 are: DQ2/DQ8 (heterozygous for beta chain). There were few samples that possessed only HLA-DQA1 are: α5; and those with only HLA-DQB1∗02 are: β2
Table 3.
Distribution of DQ2 and DQ8 haplotypes in patient groups.
| Patient groups | DQ2 | DQ8 | DQ2/DQ8 | α5 | β2 | Negative | Chi square value | P-value |
| A | 3 (8.5%) | – | – | 2 (5.7%) | 3 (8.5%) | 27 | 143.8713 | <.00001 |
| B | 30 (85.7%) | 4 (11.4%) | 1 (2.8%) | – | – | – | ||
| C | 21 (60%) | 3 (8.5%) | – | 2 (5.7%) | 9 (25%) | –– | ||
| D | 11 (31.4%) | 9 (25%) | 12 (34%) | – | – | 3 | ||
| E | 2 (5.7%) | 1 (2.8%) | 6 (17.1%) | – | – | 26 |
Group A: control, Group B: diagnosed cases of celiac disease, Group C: patients with celiac-like symptoms, Group D: diabetes mellitus type 1 patients with celiac-like symptoms, Group E: diabetes mellitus type 1 patients ∗ Subjects carrying; both HLA-DQA1∗05, HLA-DQB1∗02 are: DQ2, only HLA-DQB1∗03:02 are: DQ8, HLA-DQA1∗05, HLA-DQB1∗02 and HLA-DQB1∗03:02 are: DQ2/DQ8, only HLA-DQA1 are: α5, and only HLA-DQB1∗02 are: β2
Figure 2.
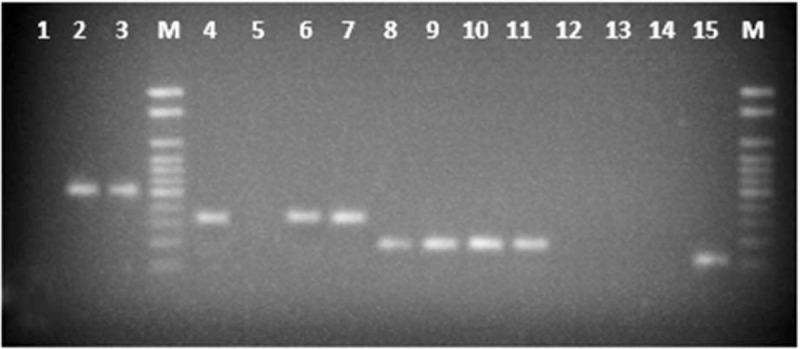
PCR amplification of celiac alleles from pediatric patients. Amplification of DQA1∗05 (209 bp), DQB1∗02 (174 bp), and DQB1∗03:02 (87 bp) alleles was observed in celiac disease samples (lane 4–15). There was no amplification in negative control (lane 1). Lane 2 and 3 are the positive control (408 bp) for healthy and celiac respectively. 1 kb ladder was used as marker (Lane-M). PCR = polymerase chain reaction.
3.3. Dissemination of DQA1∗05, DQB1∗02, and DQB1∗03:02 alleles within control and patients
All the groups were tested for DQ alleles sensitive to celiac. Among celiac disease patients DQ2 (85.7%) was the most common, followed by DQ8 (11.4%) and DQ2/DQ8 (2.8%). While patients with celiac like symptoms had DQ2 (60%) and DQ8 (8.5%) and all were positive for anti-tTG test. The patients with diabetes mellitus type 1 having celiac like symptoms possessed DQ2 (31.4%), DQ8 (25%), and DQ2/DQ8 (34%). On the other hand control had only DQ2 (8.5%) and other alleles were negative (Fig. 3). The difference of frequency among patients of CD and CD-DM1 with control was statistically significant with <.00001 P-value.
Figure 3.

Prevalence of different HLA alleles among studied groups. (Group A: control, Group B: diagnosed cases of celiac disease, Group C: patients with celiac-like symptoms, Group D: diabetes mellitus type 1 patients with celiac-like symptoms, Group E: diabetes mellitus type 1 patients).
4. Discussion
Because of the poor nutritious values and hygiene problems, gastrointestinal disorders are very common among local population of Sindh, Pakistan. On top children suffer the most from gastrointestinal and other related diseases. But among these pediatric patients another common autoimmune disorder also lives that is celiac disease; which is most frequently misdiagnosed by clinicians. The serological test for anti-tTG is the most common way for diagnosis and is further confirmed by intestinal biopsy, here in Pakistan. But according to the new guidelines such as North American Society for Pediatric Gastroenterology, Hepatology, and Nutrition (NASPGHAN), European Society of Pediatric Gastroenterology, Hepatology and Nutrition (ESPGHAN), and British Society of Gastroenterology (BSG),[1,9,15] it is very useful to include HLA typing based diagnosis. If the child is negative for both HLA-DQ2 and HLA-DQ8 haplotypes then it is very unlikely that he has celiac disease. The HLA typing has high accuracy with highest negative predictive value; thus reduces the burden on patients and families.
There were 175 participants in this study with main focus on celiac and celiac like symptomatic patients having comorbidity with diabetes mellitus type 1. Among celiac patients (Group B), all of them were positive for HLA typing. For group C patients the majority had symptoms which seemed like gluten intolerance, few had negative anti-tTG or intestinal biopsy. These patients were tested for HLA celiac alleles and were positive for either DQ2 or DQ8. There were patients who were negative for the 3 alleles tested. These patients may need further investigation for other similar symptomatic disorders like non-celiac gluten susceptibility or wheat sensitivity. Such GIT problems are still in debate whether other components in wheat are causing the problems or is it really gluten.[16,17] Few cases as shown in Table 2 had negative serological tests or biopsy, but were positive for celiac and they were also suffering from mild symptoms of celiac disorder. Patients with IgA antibodies could generate false negative results. Therefore along with serological tests genetic diagnosis should be implemented for more accurate diagnosis. Few studies also report for the incorrect diagnosis where intestinal biopsy for celiac was confirmed without the presence of DQ2/DQ8 alleles.[18,19] A large population study in Spain observed that biopsy could be avoided in high proportion of cases with celiac symptoms without the risk of misdiagnosis.[20] Thus current reviews regarding CD diagnosis have drawn attention to the inclusion of HLA-DQ genotyping in addition to serological tests especially having high negative predictive value.[21,22,9] The HLA typing result shows that group B has 85.7% HLA-DQ2 and 11.4% HLA-DQ8 in Sindh population. In control (Group A) only 8.5% participants had DQ2 allele, rest were negative or had other alleles. The allele frequency observed is similar to the ranges in European population, where HLA-DQ2 is 90% and HLA-DQ8 is 10%.[23,24]
There is considerable amount of genetic overlap among celiac disease and diabetes mellitus type 1, which explains the increase in prevalence of CD among DM1 patients compared with healthy individuals. Polymorphisms related to HLA-II-DQ and DR alleles are responsible for the risk of developing DM1 and CD together.[25,26] In this survey for Group D 37% DM1 patients were positive for CD (via anti-tTG or intestinal biopsy screening), whereas 91% patients were positive for HLA DQ2 or DQ8, and only 25% of these patients were suffering from celiac disease. Thus for such cases patients must routinely be screened for trigger of CD at later stages. Though the prevalence of celiac disorder among DM1 children was found to be 25.7%, the occurrence of CD in concurrent disorders is higher than general population (∼5%).[25,27] The worldwide prevalence of celiac in comorbidity to diabetes mellitus type 1 is ∼1% to 10% and it has elevated in recent years: the United Kingdom (4.4%), Israel (3.7%), Greece (4.8%), and Germany (6.4%); followed by Brazil (10.5%) and India (11.1%).[28–34] Thus such patients need regular screening to have proper glycemic control along with gluten free diet to prevent long term complications.
There is not sufficient literature found to report the overall prevalence rate of celiac disease in Pakistani population. But it seems to be a common disorder and it is over presented among diabetes mellitus type 1 patients. Thus celiac screening of children with long term gastrointestinal complaints should be tested for anti-tTG or intestinal biopsy, along with HLA genotyping for accuracy and for CD patients with DM1 screening for antibodies must be done at regular intervals.
5. Conclusions
The HLA-DQ2 and HLA-DQ8 genotyping can be beneficial for accurate diagnosis, particularly in areas with poor awareness and diagnostic aptitude. In interior Sindh population, celiac patients had DQ2 (85.7%), DQ8 (11.4%), and DQ2/DQ8 (2.8%). While in CD with DM1, 31.4% had DQ2, 25% had DQ8, and 34% having both the haplotypes; while only 9 patients were having CD. We suggest that this procedure could represent an adjunctive tool in the diagnostic approach to celiac disease; it is suitable for family screening, epidemiological studies, and for diagnosis particularly when histological and immunological patterns are ambiguous.
Acknowledgments
The authors wish to thank the Canadian Celiac society for providing help in questionnaire and to LUMHS, Isra University, and AIMS hospitals Hyderabad, Pakistan for the help in sample collection.
Author contributions
Conceptualization: Komal Siddiqui.
Data curation: Komal Siddiqui, Arsalan Ahmed Uqaili.
Funding acquisition: Komal Siddiqui.
Investigation: Komal Siddiqui.
Methodology: Komal Siddiqui.
Project administration: Muhammad Rafiq, Muhammad Aqeel Bhutto.
Resources: Muhammad Rafiq, Arsalan Ahmed Uqaili, Komal Siddiqui, Muhammad Aqeel Bhutto.
Software: Arsalan Ahmed Uqaili.
Supervision: Muhammad Rafiq, Muhammad Aqeel Bhutto.
Writing – original draft: Komal Siddiqui.
Writing – review & editing: Komal Siddiqui, Arsalan Ahmed Uqaili.
Footnotes
Abbreviations: ANOVA = ANalysis Of VAriance, BSG = British Society of Gastroenterology, CD = celiac disease, DNA = deoxyribonucleic acid, EDTA = Ethylenediamine tetraacetic acid, ELISA = enzyme-linked immunosorbent assay, EMA = endomysial antibodies, ESPGHAN = European Society of Pediatric Gastroenterology, Hepatology and Nutrition, HLA = human leukocyte antigen, IgA = Immunoglobulin A, MHC = major histocompatibility complex, NASPGHAN = North American Society for Pediatric Gastroenterology, Hepatology, and Nutrition, PCR = polymerase chain reaction, SPSS = statistical package for the social sciences, tTG = tissue transglutaminase.
How to cite this article: Siddiqui K, Uqaili AA, Rafiq M, Bhutto MA. Human leukocyte antigen (HLA)-DQ2 and -DQ8 haplotypes in celiac, celiac with type 1 diabetic, and celiac suspected pediatric cases. Medicine. 2021;100:11(e24954).
Compliance with ethical standards: All procedures performed in study involving human participants were in accordance with the ethical standards. This article does not contain any studies with animals performed by any of the authors. Blood samples and data were collected after informed consent of the participants or their parents in this study.
The authors have no conflicts of interest to disclose.
The datasets generated during and/or analyzed during the current study are not publicly available, but are available from the corresponding author on reasonable request.
References
- [1].Ludvigsson JF, Leffler DA, Bai JC, et al. The Oslo definitions for coeliac disease and related terms. Gut 2013;62:43–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Rubio-Tapia A, Murray JA. Celiac disease. Curr Opin Gastroenterol 2010;26:116–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Arentz–Hansen H, Mcadam SN, Molberg Ø, et al. Celiac lesion T cells recognize epitopes that cluster in regions of gliadins rich in proline residues. Gastroenterology 2002;123:803–9. [DOI] [PubMed] [Google Scholar]
- [4].Sollid LM, Jabri B. Triggers and drivers of autoimmunity: lessons from coeliac disease. Nat Rev Immunol 2013;13:294–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Lionetti E, Castellaneta S, Francavilla R, et al. Introduction of gluten, HLA status, and the risk of celiac disease in children. N Engl J Med 2014;371:1295–303. [DOI] [PubMed] [Google Scholar]
- [6].Hardy MY, Tye-Din JA. Coeliac disease: a unique model for investigating broken tolerance in autoimmunity. Clin Transl Immunology 2016;5:1–0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Lebwohl B, Ludvigsson JF, Green PH. The unfolding story of celiac disease risk factors. Clin Gastroenterol Hepatol 2014;12:632–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Di Sabatino A, Corazza GR. Coeliac disease. Lancet 2009;373:1480–93. [DOI] [PubMed] [Google Scholar]
- [9].Husby S, Koletzko S, Korponay-Szabo I, et al. European Society for Pediatric Gastroenterology, Hepatology, and Nutrition guidelines for the diagnosis of coeliac disease. J Pediatr Gastroenterol Nutr 2012;54:136–60. [DOI] [PubMed] [Google Scholar]
- [10].Tjon JM-L, van Bergen J, Koning F. Celiac disease: how complicated can it get? Immunogenetics 2010;62:641–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Abadie V, Sollid LM, Barreiro LB, et al. Integration of genetic and immunological insights into a model of celiac disease pathogenesis. Annu Rev Immunol 2011;29:493–525. [DOI] [PubMed] [Google Scholar]
- [12].Matysiak-Budnik T, Malamut G, de Serre NP-M, et al. Long-term follow-up of 61 coeliac patients diagnosed in childhood: evolution toward latency is possible on a normal diet. Gut 2007;56:1379–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Husby S, Murray JA. New aspects of the diagnosis of celiac disease in children, adolescents, and adults. Mayo Clin Proc 2013;88:540–3. [DOI] [PubMed] [Google Scholar]
- [14].Mărginean CO, Meliţ LE, Săsăran VS, et al. Diagnostic challenges of celiac disease in a young child: a case report and a review of the literature. Medicine (Baltimore) 2018;97:1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Hill ID, Dirks MH, Liptak GS, et al. Guideline for the diagnosis and treatment of celiac disease in children: recommendations of the North American Society for Pediatric Gastroenterology, Hepatology and Nutrition. J Pediatr Gastroenterol Nutr 2005;40:1–9. [DOI] [PubMed] [Google Scholar]
- [16].Collaboration C, Higgins JPT, Green S. Cochrane Handbook for Systematic Reviews of Interventions Version 5.1.0.2011; 2015. [Google Scholar]
- [17].Alarida K, Harown J, Di Pierro MR, et al. HLA-DQ2 and-DQ8 genotypes in celiac and healthy Libyan children. Dig Liver Dis 2010;42:425–7. [DOI] [PubMed] [Google Scholar]
- [18].Björck S, Brundin C, Lörinc E, et al. Screening detects a high proportion of celiac disease in young HLA-genotyped children. J Pediatr Gastroenterol Nutr 2010;50:49–53. [DOI] [PubMed] [Google Scholar]
- [19].Perez-Bravo F, Araya M, Mondragon A, et al. Genetic differences in HLA-DQA1∗ and DQB1∗ allelic distributions between celiac and control children in Santiago, Chile. Hum Immunol 1999;60:262–7. [DOI] [PubMed] [Google Scholar]
- [20].Donat E, Ramos JM, Sánchez-Valverde F, et al. ESPGHAN 2012 guidelines for coeliac disease diagnosis: validation through a retrospective Spanish multicentric study. J Pediatr Gastroenterol Nutr 2016;62:284–91. [DOI] [PubMed] [Google Scholar]
- [21].Hadithi M, von Blomberg BM, Crusius JB, et al. Accuracy of serologic tests and HLA-DQ typing for diagnosing celiac disease. Ann Intern Med 2007;147:294–302. [DOI] [PubMed] [Google Scholar]
- [22].Kurppa K, Salminiemi J, Ukkola A, et al. Utility of the new ESPGHAN criteria for the diagnosis of celiac disease in at-risk groups. J Pediatr Gastroenterol Nutr 2012;54:387–91. [DOI] [PubMed] [Google Scholar]
- [23].Kaur G, Sarkar N, Bhatnagar S, et al. Pediatric celiac disease in India is associated with multiple DR3-DQ2 haplotypes. Hum Immunol 2002;63:677–82. [DOI] [PubMed] [Google Scholar]
- [24].Krini M, Chouliaras G, Kanariou M, et al. HLA class II high-resolution genotyping in Greek children with celiac disease and impact on disease susceptibility. Pediatr Res 2012;72:625–30. [DOI] [PubMed] [Google Scholar]
- [25].Lundin KE, Wijmenga C. Coeliac disease and autoimmune disease—genetic overlap and screening. Nat Rev Gastroenterol Hepatol 2015;12:507–15. [DOI] [PubMed] [Google Scholar]
- [26].Moheb-Alian A, Forouzesh F, Sadeghi A, et al. Contribution of HLA-DQ2/DQ8 haplotypes in type one diabetes patients with/without celiac disease. J Diabetes Complications 2019;33:59–62. [DOI] [PubMed] [Google Scholar]
- [27].Doolan A, Donaghue K, Fairchild J, et al. Use of HLA typing in diagnosing celiac disease in patients with type 1 diabetes. Diabet Care 2005;28:806–9. [DOI] [PubMed] [Google Scholar]
- [28].Walker-Smith J, Vines R, Grigor W. Coeliac disease and diabetes. Lancet 1969;294:650. [DOI] [PubMed] [Google Scholar]
- [29].Goh C, Banerjee K. Prevalence of coeliac disease in children and adolescents with type 1 diabetes mellitus in a clinic based population. Postgrad Med J 2007;83:132–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Karavanaki K, Kakleas K, Paschali E, et al. Screening for associated autoimmunity in children and adolescents with type 1 diabetes mellitus (T1DM). Horm Res Paediatr 2009;71:201–6. [DOI] [PubMed] [Google Scholar]
- [31].Akirov A, Pinhas-Hamiel O. Co-occurrence of type 1 diabetes mellitus and celiac disease. World J Diabet 2015;6:707–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Sanchez-Albisua I, Wolf J, Neu A, et al. Coeliac disease in children with type 1 diabetes mellitus: the effect of the gluten-free diet. Diabet Med 2005;22:1079–82. [DOI] [PubMed] [Google Scholar]
- [33].Araújo J, da Silva GAP, Melo FMd. Serum prevalence of celiac disease in children and adolescents with type 1 diabetes mellitus. J Pediatr (Rio J) 2006;82:210–4. [DOI] [PubMed] [Google Scholar]
- [34].Bhadada SK, Kochhar R, Bhansali A, et al. Prevalence and clinical profile of celiac disease in type 1 diabetes mellitus in north India. J Gastroenterol Hepatol 2011;26:378–81. [DOI] [PubMed] [Google Scholar]


