Abstract
Bladder cancer-associated transcript 1 (BLACAT1) is one of the most common cancer-associated long non-coding RNAs (lncRNAs), which has been reported as a tumor promotor in several malignancies. Previously, BLACAT1 was found to be overexpressed in glioma tissues and cell lines. Functional assays determined that BLACAT1 promoted glioma cell proliferation, migration, invasion and epithelial-mesenchymal transition, suggesting that BLACAT1 might serve as an oncogene in glioma. In the present study, we aimed to investigate its clinical significance and prognostic value in glioma patients.
A total of 137 paired glioma tissue samples and adjacent normal brain tissue samples were collected from 137 glioma patients who underwent surgery from May 2014 to February 2019. The Student t test was applied to determine the statistical significance of the observed differences between 2 groups. Survival curves were constructed and differences among groups were calculated using the Kaplan–Meier method.
The relative expression of BLACAT1 in glioma samples was significantly higher than that of matched normal tissues (P < .001). The expression level of tissue BLACAT1 was statistically correlated with tumor size (P = .04), Karnofsky Performance Status (KPS) (P = .006), and WHO grade (P = .017). Kaplan–Meier analysis with the log-rank test revealed that BLACAT1 up-regulation was correlated with shorter overall survival time of patients with glioma (Log Rank test, P = .012). In multivariate Cox analysis, BLACAT1 expression was found to be an independent prognostic factor for overall survival in patients with glioma (HR = 2.739; 95% CI: 1.785–8.229; P = .035). Our study demonstrates that up-regulation of BLACAT1 is able to predict aggressive clinicopathologic characteristics and poor prognosis of glioma patients. These findings may have significant implications for potential treatment options and prognosis for patients with glioma.
Keywords: bladder cancer-associated transcript 1, expression, glioma, prognosis
1. Introduction
Glioma is the most common brain cancer and is the leading cause of brain tumor mortality worldwide.[1] There are 4 different grades (2 classes) of glioma: grade I and II (low8 #1" wglioma) as well as grade III and IV (high low8 glioma).[2] Glioma genesis is a complex evolutionary process involving multiple molecular mechanisms that eventually leads to malignant lesions. Though some progresses in early diagnosis and treatment, majority of the patients with glioma are still detected at advanced stages, and clinical outcomes of them are still disappointing after treatment.[3] Therefore, determining molecular targets for new therapies is imperative.
Long non-coding RNAs (lncRNAs) are RNA genomically transcribed non-coding transcripts longer than 200 nucleotides.[4,5] Dysregulation of lncRNAs expression has been found in various types of human cancers, including glioma.[6,7] Increasing studies have shown that the dysregulation of lncRNAs are closely related to the occurrence and development of glioma.[7] This offers new insight in the diagnosis, targeted therapy and prognosis for glioma.
Bladder cancer-associated transcript 1 (BLACAT1) is one of the most common cancer-associated lncRNAs, which has been reported as tumor promotor in several malignancies, such as non-small-cell lung cancer (NSCLC), hepatocellular carcinoma (HCC), breast cancer, osteosarcoma, cervical cancer, gastric cancer, papillary thyroid cancer (PTC), and colorectal cancer.[8–15] Previously, the role of BLACAT1 has also been investigated in glioma. Li et al found that BLACAT1 expression was significantly increased in glioma tissues compared to adjacent non-tumor tissues (P < .05). BLACAT1 expression was also markedly increased in glioma tumors of high grade (III-IV) compared with low grade (I-II) tumors (P < .05). Functional assays determined that BLACAT1 promoted glioma cell proliferation, migration, invasion and epithelial-mesenchymal transition in vitro. In addition, it was demonstrated that BLACAT1 activated the Wnt/β-catenin signaling pathway.[16] However, its clinical significance and prognostic value in glioma patients have not been investigated.
2. Materials and methods
2.1. Patient information and tissue samples
A total of 137 paired glioma tissue samples and adjacent normal brain tissue samples were collected from 137 patients who underwent surgery at Department of Craniocerebral Surgery, People's Hospital of Lanling County from May 2014 to February 2019. All the glioma patients have not received preoperative chemotherapy or radiotherapy. The inclusion criteria were as follows:
-
1.
a histological diagnosis of glioma,
-
2.
patients with explicit clinical prognostic data.
The exclusion criteria were as follows:
-
1.
patients have history of other tumors,
-
2.
patients received radiotherapy, chemotherapy, targeted treatment and immunotherapy before surgery.
Tissue samples were snap-frozen in liquid nitrogen at the time of surgery and subsequently stored at −80°C. Final histological classifications and findings were made by 2 pathologists. The clinical information of the glioma patients were shown in Table 1. This study was approved by the Ethics Committee of People's Hospital of Lanling County. Informed written consents were obtained from all recruited patients.
Table 1.
Summary of clinicopathologic features of the glioma patients.
| BLACAT1 expression | ||||
| Variables | Cases (n) | High (n = 67) | Low (n = 70) | P value |
| Gender | ||||
| Male | 97 | 51 | 46 | .194 |
| Female | 40 | 16 | 24 | |
| Age (yrs) | ||||
| <60 | 81 | 37 | 44 | .389 |
| ≥60 | 56 | 30 | 26 | |
| Tumor size (cm) | ||||
| <5 | 73 | 42 | 31 | .040 |
| ≥5 | 64 | 25 | 39 | |
| KPS | ||||
| <70 | 45 | 30 | 15 | .006 |
| ≥70 | 92 | 37 | 55 | |
| WHO grade | ||||
| I/II | 68 | 26 | 42 | .017 |
| III/IV | 69 | 41 | 28 | |
2.2. RNA isolation and quantitative real-time PCR
Total RNA from glioma tissues and normal brain tissues was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. RNA was reverse transcribed to cDNA using a SYBR Green PCR kit (TaKaRa, Otsu, Shiga, and Japan). Quantitative real-time polymerase chain reaction reaction parameters were as follows: predegeneration for 15 minutes at 95°C, degeneration at 94°C for 15 second, annealing at 55°C for 30 second, and extension at 72°C for 30 second, for a total of 40 cycles. The BLACAT1 expression levels were normalized to GAPDH. The primers used were as followed: BLACAT1 forward, 5“-CAAGAGGAGCCGGCTTAGCATCTA-3”, reverse 5“-ACGGTTCCAGTCCTCAGTCAG-3”; GAPDH, forward: 55“-CAAGAGGAGCCGGCTTAGCATCTA-3”, reverse 5“-ACGGTTCCAGTCCTCAGTCA”.
2.3. Statistical analysis
The Student t test was applied to determine the statistical significance of the observed differences between groups. Chi-Squared test was used to compare the expression level of BLACAT1 and clinical variables. Survival curves were constructed and differences among groups were calculated using the Kaplan–Meier method. Significant variables in univariate analyses were used in multivariate analyses according to the Cox regression analyses. SPSS software, version 20.0 for Windows (SPSS Inc., Chicago, IL) was used for statistical analysis. P < .05 was considered statistically significant.
3. Results
3.1. The expression level of BLACAT1 in glioma tissues
To investigate the role of BLACAT1 in glioma development, we tested the expression of BLACAT1 in glioma tissues and adjacent normal brain tissues. The relative expression of BLACAT1 in glioma samples was significantly higher than that of matched normal brain tissues (P < .001, shown in Fig. 1). Hence, we considered the up-regulation of BLACAT1 may contribute to glioma tumorigenesis.
Figure 1.
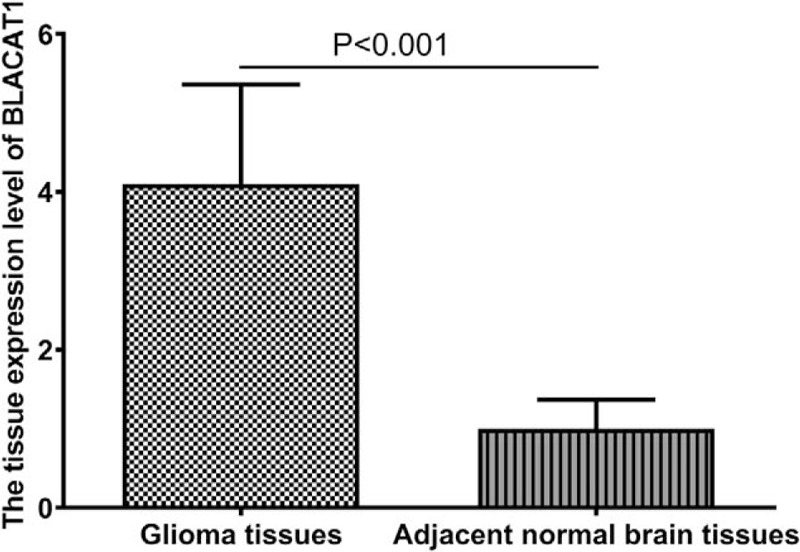
The expression level of BLACAT1 in glioma tissues and adjacent normal brain tissues.
3.2. Association between BLACAT1 expression levels and clinicopathologic parameters in glioma patients
We evaluated the correlation of tissue BLACAT1 expression level with clinicopathological parameters of glioma patients. The expression level of tissue BLACAT1 was statistically correlated with tumor size (P = .04), KPS (P = .006), and WHO grade (P = .017); whereas no significant correlation between tissue BLACAT1 expression level and gender, and age (all P > .05). The results suggest that tissue BLACAT1 expression level may be a potential marker for the reflection of disease status. These data are summarized in Table 1.
3.3. Correlation between tissue BLACAT1 expression level and prognosis of glioma patients
To further evaluate the prognostic value of tissue BLACAT1 expression level for glioma patients, we also analyzed the association between BLACAT1 expression and survival duration using Kaplan–Meier analysis with the log-rank test. The results revealed that BLACAT1 up-regulation was correlated with shorter overall survival time of patients with glioma (Log Rank test, P = .012, shown in Fig. 2). In addition, univariate and multivariate Cox analysis were used to examine whether tissue BLACAT1 expression was an independent risk factor for prognosis in glioma patients. The parameters that significantly correlated with survival in the univariate analysis were further assessed by multivariate analysis. In multivariate analysis, BLACAT1 expression was found to be an independent prognostic factor for overall survival in patients with glioma (HR = 2.739; 95% CI: 1.785–8.229; P = .035, shown in Table 2).
Figure 2.
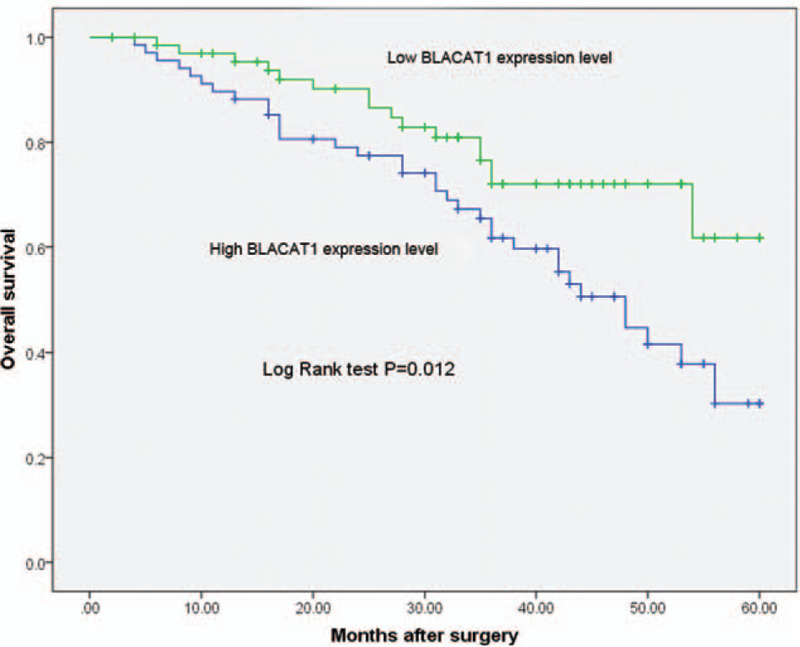
Kaplan–Meier overall survival curve for high and low BLACAT1 expression.
Table 2.
Univariate and multivariate analysis of overall survival in 137 patients with glioma.
| Univariate analysis | Multivariate analysis | |||||
| Variable | Hazard ratio | 95% CI | P value | Hazard ratio | 95% CI | P value |
| Gender | 0.692 | 0.478–2.738 | .736 | |||
| Age | 1.784 | 0.815–3.664 | .163 | |||
| Tumor size | 2.183 | 0.892–6.284 | .075 | |||
| KPS | 4.281 | 2.284–12.283 | .009 | 3.928 | 2.775–11.923 | .007 |
| WHO grade | 3.957 | 1.559–10.034 | 0.013 | 4.012 | 2.192–10.273 | .009 |
| BLACAT1 expression | 2.885 | 1.628–8.285 | 0.047 | 2.739 | 1.785–8.229 | .035 |
4. Discussion
Glioma is one of the most aggressive and common primary malignant brain tumors, and it accounts for approximately 80% of all primary malignant brain tumor cases.[2] In spite of the progress that researchers and medical workers made in the treatment of patients with glioma over the last decades, the prognosis and survival rate are still unpleasant.[17–19] Therefore, it is critical to develop accurate risk evaluation methods with the aim of preventing the recurrence of glioma.
LncRNAs play important roles in some crucial biological processes, such as cellular proliferation, differentiation, apoptosis, and tumorigenesis.[4,20,21] Recently, accumulating evidence indicates that lncRNAs expression level in tissues or serum can be served as novel diagnostic or prognostic biomarkers in many types of cancers.[22,23] BLACAT1 is one of the most common cancer-associated lncRNAs, which has been reported as tumor promotor in several malignancies, such as hepatocellular carcinoma, colorectal cancer, osteosarcoma, esophageal cancer, gallbladder carcinoma, endometrial cancer, and bladder cancer. There are a lot of studies investigating its mechanism and clinical significance. For example, Xu et al found that non-small-cell lung cancer (NSCLC) patients with high BLACAT1 expression had shorter overall survival and progression-free survival than those with low BLACAT1 expression. Multivariate analyses showed that BLACAT1 was an independent prognostic factor of survival in NSCLC patients. In vitro assays showed that the downregulation of BLACAT1 significantly suppressed cell progression, migration, and invasion. The epithelial-mesenchymal transition was also inhibited when BLACAT1 was silenced, indicated by an increase in E-cadherin expression and a decrease in vimentin expression by mediating Wnt/β-catenin signaling pathway.[8] Peng et al found that BLACAT1 was aberrantly upregulated in HCC and its inhibition had tumor suppressing effects in human HCC, possibly through endogenously competing against has-miR-485–5p. The BLACAT1/ has-miR-485-5p regulatory axis may be a molecular target for future HCC therapy.[9] Hu et al found that BLACAT1 took part in breast cancer cell aggressive phenotype. The real-time PCR result showed that BLACAT1 was up-regulated in tumor tissues compared to adjacent normal tissues. The molecular mechanism experiments demonstrated that BLACAT1 down-regulation suppressed the proliferation and metastasis of human breast cancer cells by regulating miR-150-5p targeting CCR2. The clinical studies indicated that lack of BLACAT1 was related to tumor size, and metastasis.[10] Dong et al found that BLACAT1 was upregulated in osteosarcoma tissues and cells. Upregulation of BLACAT1 predicted unfavorable prognosis for patients with osteosarcoma. Downregulation of BLACAT1 inhibited osteosarcoma cell proliferation and invasion, whereas upregulation of BLACAT1 accelerated cell proliferation and invasion. More importantly, BLACAT1 could interact with STAT3 and regulate the phosphorylation of STAT3.[11] Su et al found that increased BLACAT1 was an independent unfavorable prognostic indicator for colorectal cancer, and revealed that BLACAT1 knockdown significantly repressed proliferation, both in vitro and in vivo. Mechanistic investigations demonstrated that BLACAT1 had a key role in G1/G0 arrest, and showed that BLACAT1 can repress p15 expression by binding to EZH2, thus contributing to the regulation of colorectal cancer cell cycle and proliferation. These results suggest that BLACAT1, as a cell cycle regulator, may serve as a potential target for cancer prevention and treatment in human colorectal cancer.[15]
Previously, the role of BLACAT1 has also been investigated in glioma. Li et al found that BLACAT1 expression was significantly increased in glioma tissues compared to adjacent non-tumor tissues (P < .05). BLACAT1 expression was also markedly increased in glioma tumors of high grade (III-IV) compared with low grade (I-II) tumors (P < .05). Functional assays determined that BLACAT1 promoted glioma cell proliferation, migration, invasion and epithelial-mesenchymal transition in vitro. In addition, it was demonstrated that BLACAT1 activated the Wnt/β-catenin signaling pathway.[16] However, its clinical significance and prognostic value in glioma patients have not been investigated. In the present study, to investigate the role of BLACAT1 in glioma development, we tested the expression of BLACAT1 in glioma tissues and adjacent normal brain tissues. The relative expression of BLACAT1 in glioma samples was significantly higher than that of matched normal brain tissues. Hence, we considered the up-regulation of BLACAT1 may contribute to glioma tumorigenesis. We then evaluated the correlation of tissue BLACAT1 expression level with clinicopathological parameters of glioma patients. The expression level of tissue BLACAT1 was statistically correlated with tumor size, KPS, and WHO grade; whereas no significant correlation between tissue BLACAT1 expression level and gender, and age. The results suggest that tissue BLACAT1 expression level may be a potential marker for the reflection of disease status. To further evaluate the prognostic value of tissue BLACAT1 expression level for glioma patients, we also analyzed the association between BLACAT1 expression and survival duration using Kaplan–Meier analysis with the log-rank test. The results revealed that BLACAT1 up-regulation was correlated with shorter overall survival time of patients with glioma. In addition, univariate and multivariate Cox analysis were used to examine whether tissue BLACAT1 expression was an independent risk factor for prognosis in glioma patients. The parameters that significantly correlated with survival in the univariate analysis were further assessed by multivariate analysis. In multivariate analysis, BLACAT1 expression was found to be an independent prognostic factor for overall survival in patients with glioma. In the present study, we have not investigated the expression level of BLACAT1 in serum. This is the limitation of our study, and we will assess the BLACAT1 level in paired serum samples from glioma patients and healthy controls, and also investigate the prognostic value of serum BLACAT1 level in the future.
In conclusion, up-regulation of BLACAT1 is associated with aggressive clinicopathologic characteristics and poor prognosis in glioma patients. These findings may have significant implications for potential treatment options and prognosis for patients with glioma. Further study with larger sample size is warranted to confirm our results.
Author contributions
Conceptualization: Xiaojue Zhang.
Data curation: Peng Jin.
Investigation: Xiaojue Zhang, Xiuchuan Wei, Jie Liu.
Methodology: Xiaojue Zhang, Xiuchuan Wei.
Project administration: Xiaojue Zhang.
Software: Xiaojue Zhang, Jie Liu, Jiaying Yang.
Visualization: Xiaojue Zhang.
Writing – original draft: Xiaojue Zhang, Xiuchuan Wei.
Writing – review & editing: Xiaojue Zhang, Jiaying Yang, Peng Jin.
Footnotes
Abbreviations: BLACAT1 = bladder cancer-associated transcript 1, KPS = Karnofsky Performance Status, lncRNAs = long non-coding RNAs.
How to cite this article: Zhang X, Wei X, Liu J, Yang J, Jin P. Up-regulation of long non-coding RNA BLACAT1 predicts aggressive clinicopathologic characteristics and poor prognosis of glioma. Medicine. 2021;100:11(e20722).
The authors have no conflicts of interests to disclose.
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
- [1].Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA 2018;68:7–30. [DOI] [PubMed] [Google Scholar]
- [2].Schwartzbaum JA, Fisher JL, Aldape KD, et al. Epidemiology and molecular pathology of glioma. Nat Clin Pract Neurol 2006;2:494–503. quiz 1 p following 16. [DOI] [PubMed] [Google Scholar]
- [3].Sathornsumetee S, Rich JN. New treatment strategies for malignant gliomas. Expert Review Anticancer Therapy 2006;6:1087–104. [DOI] [PubMed] [Google Scholar]
- [4].Iyer MK, Niknafs YS, Malik R, et al. The landscape of long noncoding RNAs in the human transcriptome. Nat Genetics 2015;47:199–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Development 2009;23:1494–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Wang J, Zhang X, Chen W, et al. Regulatory roles of long noncoding RNAs implicated in cancer hallmarks. Int J Cancer 2019. [DOI] [PubMed] [Google Scholar]
- [7].Zhang X, Sun S, Pu JK, et al. Long non-coding RNA expression profiles predict clinical phenotypes in glioma. Neurobiol Dis 2012;48:1–8. [DOI] [PubMed] [Google Scholar]
- [8].Xu R, Cao XR, Zhang BQ, et al. BLACAT1 is negatively associated with prognosis in patients with NSCLC and inhibits cell progression, metastasis and epithelial-mesenchymal transition through down-regulating Wnt/beta-catenin signaling pathway. Eur Rev Med Pharmacological Sci 2019;23:6217–25. [DOI] [PubMed] [Google Scholar]
- [9].Peng Y, Leng W, Duan S, et al. Long noncoding RNA BLACAT1 is overexpressed in hepatocellular carcinoma and its downregulation suppressed cancer cell development through endogenously competing against hsa-miR-485-5p. Biomed Pharmacothera 2019;116:109027. [DOI] [PubMed] [Google Scholar]
- [10].Hu X, Liu Y, Du Y, et al. Long non-coding RNA BLACAT1 promotes breast cancer cell proliferation and metastasis by miR-150-5p/CCR2. Cell Biosci 2019;9:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Dong Z, Wang Y. LncRNA BLACAT1 accelerates the proliferation and migration of osteosarcoma cells through regulating STAT3. Pathol Res Pract 2019;215:571–9. [DOI] [PubMed] [Google Scholar]
- [12].Wang CH, Li YH, Tian HL, et al. Long non-coding RNA BLACAT1 promotes cell proliferation, migration and invasion in cervical cancer through activation of Wnt/beta-catenin signaling pathway. Eur Rev Med Pharmacological Sci 2018;22:3002–9. [DOI] [PubMed] [Google Scholar]
- [13].Wu X, Zheng Y, Han B, et al. Long noncoding RNA BLACAT1 modulates ABCB1 to promote oxaliplatin resistance of gastric cancer via sponging miR-361. Biomed Pharmacothera 2018;99:832–8. [DOI] [PubMed] [Google Scholar]
- [14].Liao D, Lv G, Wang T, et al. Prognostic value of long non-coding RNA BLACAT1 in patients with papillary thyroid carcinoma. Cancer Cell Int 2018;18:47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Su J, Zhang E, Han L, et al. Long noncoding RNA BLACAT1 indicates a poor prognosis of colorectal cancer and affects cell proliferation by epigenetically silencing of p15. Cell Death Dis 2017;8:e2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Li X, Qi S, Ma D, et al. Long non-coding RNA BLACAT1 promotes the proliferation and invasion of glioma cells via Wnt/beta-catenin signaling. Experimental Therapeutic Med 2019;17:4703–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Ho VK, Reijneveld JC, Enting RH, et al. Changing incidence and improved survival of gliomas. Eur J Cancer 2014;50:2309–18. [DOI] [PubMed] [Google Scholar]
- [18].Rees JH. Diagnosis and treatment in neuro-oncology: an oncological perspective. Br J Radiol 2011;84((Spec No 2)):S82–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Spinelli GP, Miele E, Lo Russo G, et al. Chemotherapy and target therapy in the management of adult high- grade gliomas. Current Cancer Drug Targets 2012;12:1016–31. [DOI] [PubMed] [Google Scholar]
- [20].Noh JH, Kim KM, McClusky WG, et al. Cytoplasmic functions of long noncoding RNAs. Wiley Interdisciplinary Rev RNA 2018;9:e1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Rafiee A, Riazi-Rad F, Havaskary M, et al. Long noncoding RNAs: regulation, function and cancer. Biotechnol Genetic Eng Rev 2018;34:153–80. [DOI] [PubMed] [Google Scholar]
- [22].Prensner JR, Chinnaiyan AM. The emergence of lncRNAs in cancer biology. Cancer Discovery 2011;1:391–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Zhang H, Chen Z, Wang X, et al. Long non-coding RNA: a new player in cancer. J Hematol Oncol 2013;6:37. [DOI] [PMC free article] [PubMed] [Google Scholar]


