Abstract
GINS subunits, a protein complex composed of GINS1, GINS2, GINS3 and GINS4 in the human genome and the expression level of each GINS subunits plays an important role in different human cancers. As one of the most common malignancies after lung cancer in the world, precise biomarkers for early diagnosis and treatment in breast cancer are important. The purpose of our study was to elucidate the expression and prognostic value of GINS subunits in breast cancer.
The purpose of present study was to explore the expression level of GINS subunits in breast cancer patients.
In the present study, we investigated the gene alteration, gene expression and potential prognostic value of GINS subunits by using the Gene Expression Profiling Interactive Analysis (GEPIA), UALCAN, cBioPortal, and bc-GenExMiner databases. Then, the GeneMANIA database was used to show the genes that associated with GINS subunits. Furthermore, gene ontology pathway analysis was conducted by using the Metascape database. Finally, immune infiltration analysis in GINS subunits were evaluated using the Tumor Immune Estimation Resource (TIMER) database.
Our analyses demonstrated that the expression levels of different GINS subunits were different between breast cancer and normal breast tissues. The expression levels of GINS1, GINS2, and GINS4 were significantly higher in breast cancer tissues than in normal tissues. Survival analysis revealed that increased the expression levels of GINS subunits were associated with poor prognoses in all patients with breast cancer. Gene ontology pathway enrichment analysis of the GINS subunits suggested that GINS subunits involved in pathways including the cell cycle checkpoint, DNA replication and other meaningful signaling pathways.
We systemically analyzed the expression, prognostic, clinicopathologic values, and potential functional networks of GINS subunits in breast cancer. Our findings showed that individual GINS subunits could be new potential prognostic biomarkers for breast cancer. However, further verification studies are still needed to prove the clinical value of GINS subunits in breast cancer patients.
Keywords: biomarker, breast cancer, GINS subunits
1. Introduction
Breast cancer is one of the leading causes of cancer-related deaths in women.[1] With the development of disease diagnosis by the detection of gene expression in breast tumor tissues and cells, an increasing number of breast cancer patients can be diagnosed at the early stage. Multigene tests, such as Oncotype Dx, MammaPrint, and PAM50, have also been used to increase the survival rate in breast cancer.[2] However, according to recent research, the morbidity and mortality of breast cancer in females have evidently increased. Approximately 1,671,000 new cases and 522,000 breast cancer-related deaths are expected to occur every year.[3,4] Therefore, exploring some sensitive molecular biomarkers associated with the prognosis of breast cancer has significant clinical value.
GINS subunits, a protein complex composed of GINS1, GINS2, GINS3, and GINS4 in the human genome.[5–7] GINS subunits are also a key component of the CMG complex (Cdc45–MCM–GINS), a eukaryotic replicative helicase that unwinds double-stranded DNA at replication forks.[8,9] As a result of the close link between DNA helicases and human cancers, the expression level of each GINS subunits plays an important role in different human cancers.[10–13] For example, Lian et al reported that individual GINS subunit could be potential prognostic biomarkers for hepatocellular carcinoma patients by using the online data mining websites.[14] However, bioinformatics analyses have not yet been employed to explore the role of GINS subunits in breast cancer.
In the present study, Gene Expression Profiling Interactive Analysis (GEPIA) databases were used to show the difference expression level of GINS subunits between tumor and normal tissues in breast cancer patients. Subsequently, the relationship between GINS subunits and clinical pathological parameters, such as lymph node metastasis, breast cancer subclasses, individual cancer stages and distinct prognostic values of GINS subunits in breast cancer were examined by mining accessible public databases. In addition, genes that associated with GINS subunits and the correlation among each GINS subunit in breast cancer were also analyzed using GeneMANIA and GEPIA database. Besides, the enrichment and Immune infiltration analysis of GINS subunits were also conducted by using the metascape and Tumor Immune Estimation Resource (TIMER) database. Results from these different databases will help us find the value of GINS subunits as survival biomarkers in breast cancer patients.
2. Materials and methods
2.1. Expression and mutation analysis of GINS subunits
The GEPIA database is an online website based on The Cancer Genome Atlas and the Genotype-Tissue Expression projects, which contain 9736 tumors and 8587 normal samples (http://gepia.cancer-pku.cn/).[15] To analyze the expression levels of GINS subunits between breast cancer and normal breast tissues, we used the GEPIA dataset. Furthermore, the associations among GINS subunits in breast cancer were also explored by using the GEPIA dataset. The frequencies of gene alteration of the GINS subunits in breast cancer and pan-cancer were assessed by using the cBioPortal dataset (https://www.cbioportal.org/).[16]
2.2. Survival and clinical pathological analysis of GINS subunits
To analyze the association between the expression of each GINS subunits and lymph node metastasis in breast cancer patients, the open access database UALCAN was used (http://ualcan.path.uab.edu/).[17] Besides, the UALCAN database was also used to identify the relationship between the expression level of GINS subunits and individual cancer stages as well as breast cancer subclasses. Breast cancer Gene–expression miner v4.1 (bc-GenExMiner v4.1) (bcgenex.centregauducheau.fr) is an online database that has gene expression and survival data in breast cancer patients.[18,19] To analyze the prognostic values of the GINS subunits, the bc–GenExMiner v4.1 was used.
2.3. Gene interaction, co-expression, immune infiltration, and enrichment analysis of GINS subunits
TIMER (https://cistrome.shinyapps.io/timer/) is a database for analyzing the immune cell infiltration in different cancers.[20,21] By using the TIMER database, we explore the immune cell infiltration in different subtypes of breast cancer. The GeneMANIA database was used to show the genes that associated with GINS subunits (https://genemania.org/).[22,23] Metascape (http://metascape.org/) is an enrichment analysis tool for gene analysis. In this study, the gene ontology (GO) term pathways of GINS subunits and their neighboring genes were enriched on this online tool.[24]
2.4. Statistical analysis
The screening conditions of the expression level of GINS subunits in breast cancer from GEPIA database were set as follows: |Log2FC| cutoff was 1, P value was .01. Student t test was used to explore the expression level of each GINS subunits in the UALCAN database and P < .05 was considered as statistically significant different. To analyze the prognostic values of the GINS subunits, Kaplan–Meier analysis was used in the bc-GenExMiner v4.1 database and P value <.05 was consider as statistically significance. Results from the TIMER database were displayed with cor and P value by using the Spearman correlation analysis, P value smaller than .05 is regarded statistically significant. Terms with P value <.01, minimum count of 3, and enrichment factor of >1.5 were considered as statistically significant in metascape enrichment analysis.
2.5. Ethical statement
All analyses were based on the online open-access database, thus the ethical approval was not required.
3. Results
3.1. Expression and mutation levels of GINS subunits in breast cancer.
The expression level of GINS subunits in breast cancer tissues compared with normal tissues were analyzed using the GEPIA dataset. These data indicated that the expression of GINS1, GINS2, and GINS4 were markedly higher in breast cancer samples than in normal breast tissues (Fig. 1E-H). The expression of GINS subunits in pan-cancer were also shown in Figure 1A-1D. Besides, the frequencies of gene alteration of the GINS subunits in breast cancer and pan-cancer were assessed using the cBioPortal. Among the breast cancer cases with gene alteration of GINS subunits, amplification was the most common alteration type (Fig. 1I). As for the gene alteration of GINS subunits in pan-cancer, amplification was also the most common alteration type (Fig. 1J).
Figure 1.
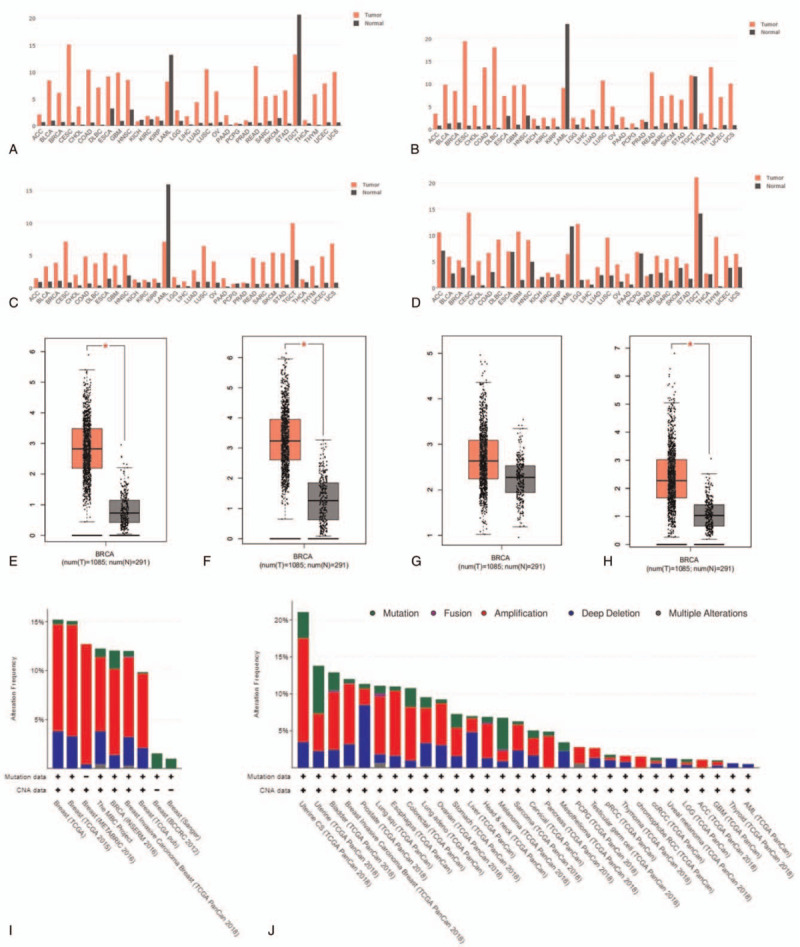
The Expression and alteration of GINS subunits in breast cancer. (A) The expression of GINS1 in pan-cancer. (B) The expression of GINS2 in pan-cancer. (C) The expression of GINS3 in pan-cancer. (D) The expression of GINS4 in pan-cancer. (E) The expression of GINS1 in breast cancer. (F) The expression of GINS2 in breast cancer. (G) The expression of GINS3 in breast cancer. (H) The expression of GINS4 in breast cancer. (I) GINS subunits alteration across breast cancer and (J) across 28 tumor types.
3.2. GINS subunits expression associated with individual breast cancer stages, lymph node metastasis, breast cancer subclasses, and poor prognosis
To further identify the expression level of GINS subunits in different groups of patients, UALCAN database analysis was used. As was shown in Figure 2, it showed that patients with breast cancer had different GINS subunits expression level between subgroups of individual cancer stages, lymph node metastasis and breast cancer subclasses. Expression levels of GINS1, GINS2, GINS3, and GINS4 in breast cancer based on individual cancer stages were presented in Figure 2A, 2B, 2C, and 2D, respectively. Figure 2E, 2F, 2G, and 2H provided the expression levels of GINS1, GINS2, GINS3, and GINS4 in breast cancer based on lymph node metastasis, respectively. Figure 2I, 2J, 2K, and 2L provided the expression levels of GINS1, GINS2, GINS3, and GINS4 in breast cancer based on breast cancer subclasses, respectively. All subgroup comparisons were presented in Tables 1 and 3. The survival was also analyzed for each GINS subunit by using the bc-GenExMiner database. It was demonstrated that high GINS1, GINS2, GINS3, and GINS4 expression level predicted a worse prognosis in all patients with breast cancer (Fig. 3A, HR:1.43, P < .0001. Fig. 3B, HR:1.54, P < .0001, Fig. 3C, HR:1.55, P < .0001. Fig. 3D, HR:1.33, P < .0001.).
Figure 2.
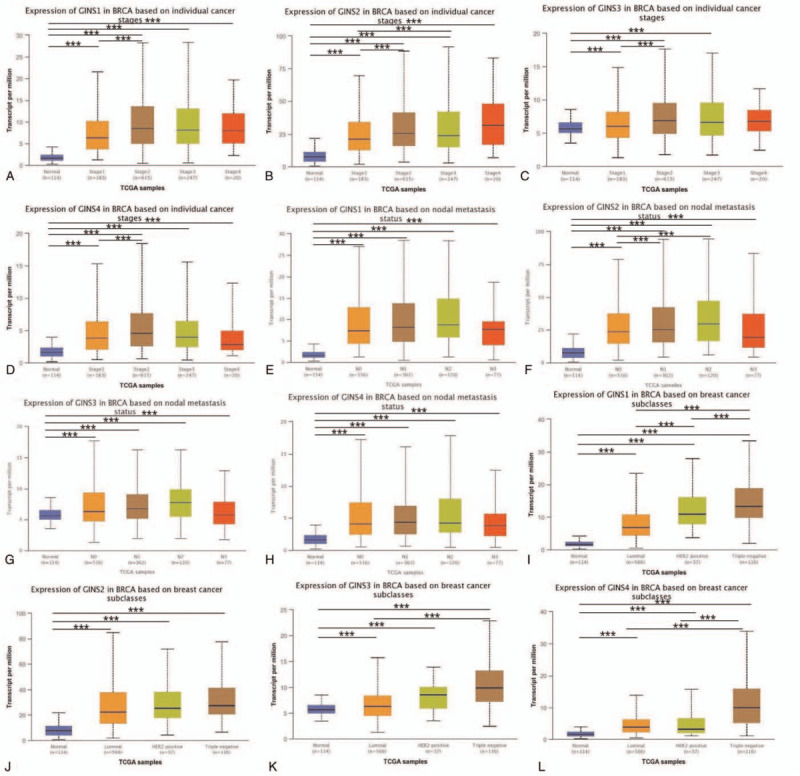
The relationship between the expression level of GINS subunits and individual cancer stages, lymph node metastasis status as well as breast cancer subclasses. (A-D) The expression level of GINS1-4 in breast cancer patients with different individual cancer stages. (E-H) The expression level of GINS1-4 in breast cancer patients with different lymph node metastasis status. (I-L) The expression level of GINS1-4 in breast cancer patients with different breast cancer subclasses. (∗∗∗P < .05).
Table 1.
The Statistical significance between each comparison (Expression level of GINS subunits and individual cancer stages).
| Comparison | GINS1 | GINS2 | GINS3 | GINS4 |
| Normal VS Stage1 | <1E−12 | <1E−12 | 1.17E−04 | 1.97E−12 |
| Normal VS Stage2 | 1.62E−12 | 1.62E−12 | 1.62E−12 | <1E−12 |
| Normal VS Stage3 | 1.62E−12 | 1.62E−12 | 9.07E−10 | <1E−12 |
| Normal VS Stage4 | 1.72E−05 | 3.03E−04 | 5.75E−02 | 8.40E−03 |
| Stage1 VS Stage2 | 1.41E−02 | 7.82E−05 | 1.02E−03 | 6.30E−03 |
| Stage1 VS Stage3 | 6.26E−02 | 1.36E−03 | 1.10E−01 | 2.42E−01 |
| Stage1 VS Stage4 | 8.20E−01 | 9.19E−02 | 9.22E−01 | 9.12E−01 |
| Stage2 VS Stage3 | 9.31E−01 | 8.54E−01 | 1.25E−01 | 1.82E−01 |
| Stage2 VS Stage4 | 4.94E−01 | 3.89E−01 | 1.33E−01 | 4.17E−01 |
| Stage3 VS Stage4 | 2.29E−01 | 4.78E−01 | 5.69E−01 | 7.10E−01 |
Table 3.
The Statistical significance between each comparison (Expression level of GINS subunits and breast cancer subclasses).
| Comparison | GINS1 | GINS2 | GINS3 | GINS4 |
| Normal VS Luminal | <1E−12 | <1E−12 | 6.65E−11 | 1.62E−12 |
| Normal VS HER2 Positive | 2.31E−12 | 4.11E−08 | 1.38E−03 | 1.12E−03 |
| Normal VS TNBC | <1E−12 | 1.62E−12 | 1.11E−16 | <1E−12 |
| Luminal VS HER2 Positive | 1.08E−02 | 8.53E−01 | 7.98E−02 | 6.72E−01 |
| Luminal VS TNBC | 8.86E−10 | 9.10E−02 | 7.32E−10 | 4.74E−10 |
| HER2 Positive VS TNBC | 1.30E−02 | 4.47E−01 | 7.50E−02 | 1.04E−04 |
Figure 3.
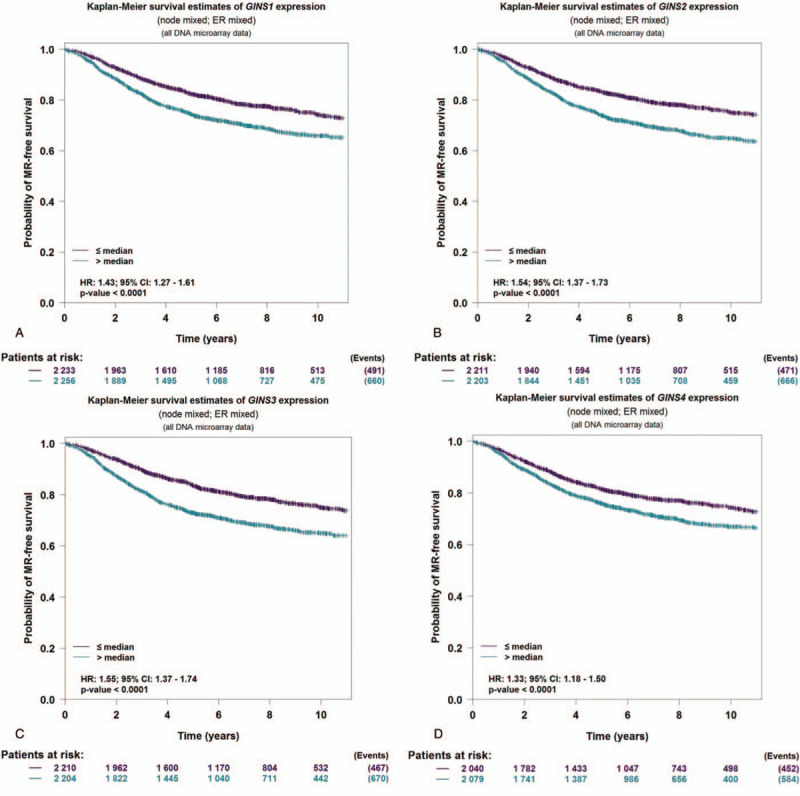
The prognostic values of GINS subunits in all breast cancer patients. (A) GINS1 expression level in all breast cancer patients. (B) GINS2 expression level in all breast cancer patients. (C) GINS3 expression level in all breast cancer patients. (D) GINS4 expression level in all breast cancer patients.
Table 2.
The Statistical significance between each comparison (Expression level of GINS subunits and lymph node metastasis status).
| Comparison | GINS1 | GINS2 | GINS3 | GINS4 |
| Normal VS N0 | <1E−12 | <1E−12 | 1.49E−14 | 1.62E−12 |
| Normal VS N1 | 1.62E−12 | 1.62E−12 | 2.11E−15 | <1E−12 |
| Normal VS N2 | <1E−12 | <1E−12 | 2.33E−09 | 6.00E−12 |
| Normal VS N3 | 8.31E−12 | 5.65E−09 | 3.55E−02 | 1.90E−05 |
| N0 VS N1 | 9.00E−01 | 3.92E−03 | 6.62E−01 | 6.80E−01 |
| N0 VS N2 | 1.42E−01 | 3.72E−02 | 6.17E−01 | 6.83E−01 |
| N0 VS N3 | 2.85E−01 | 9.57E−01 | 1.49E−01 | 7.40E−01 |
| N1 VS N2 | 1.52E−01 | 8.77E−01 | 8.78E−01 | 9.11E−01 |
| N1 VS N3 | 2.82E−01 | 1.65E−01 | 6.72E−01 | 5.75E−01 |
| N2 VS N3 | 6.83E−02 | 1.84E−01 | 9.32E−01 | 5.78E−01 |
3.3. The correction between each GINS subunit and interaction analysis of GINS subunits in breast cancer
We first analyzed the association between GINS1, GINS2, GINS3, and GINS4 in breast cancer by using the GEPIA database and summarized these data into a heatmap (Fig. 4A). We found that GINS1 was positively associated with GINS2 (R = 0.43, P < .05), GINS3 (R = 0.52, P < .05) and GINS4 (R = 0.42, P < .05). GINS2 was positively associated with GINS3 (R = 0.45, P < .05) and GINS4 (R = 0.23, P < .05). GINS3 was associated with GINS4 (R = 0.3, P < .05).
Figure 4.
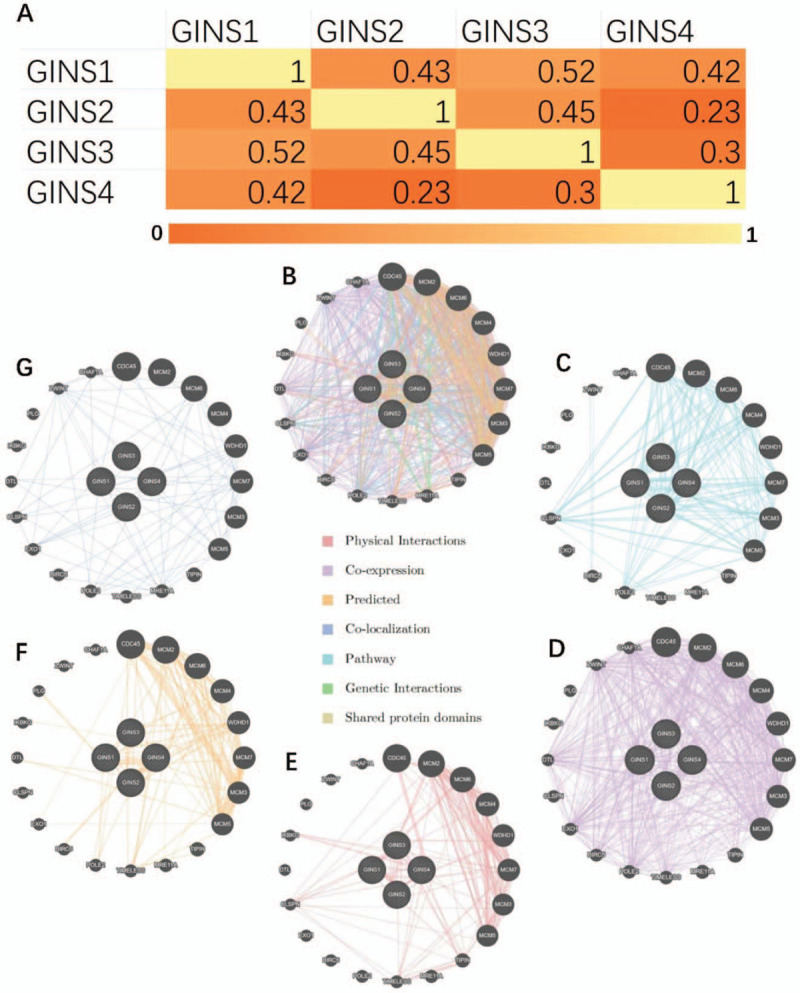
The correction between each GINS subunit and Interaction analysis of GINS subunits in breast cancer. (A) Heatmap of the correction between GINS subunits in breast cancer. (B) Gene–gene integrated interaction network among GINS subunits. (C) Pathway. (D) Co-expression. (E) Physical interaction. (F) Predicted interaction. (G) Co-localization.
To further explore the gene–gene interaction network of GINS subunits in breast carcinoma, we then constructed an interaction network by using the GeneMANIA online database and found that GINS subunits were associated with CDC45, MCM2, MCM6, MCM4, WDHD1, MCM7, MCM3, MCM5, TIPIN, MRE11A, TIMELESS, POLE2, BIRC5, EXO1, CLSPN, DTL, IKBKG, PLG, ZWINT, and CHAF1A (Fig. 4B). Furthermore, the top 5 additional ways that GINS subunits were found to be correlated with other genes were: pathway (Fig. 4C), co-expression (Fig. 4D), physical interaction (Fig. 4E), predicted interaction (Fig. 4F), and co-localization (Fig. 4G).
3.4. The enrichment and immune infiltration analysis of the GINS subunits in breast cancer.
The GO terms of GINS subunits and their neighboring genes (CDC45, MCM2, MCM6, MCM4, WDHD1, MCM7, MCM3, MCM5, TIPIN, MRE11A, TIMELESS, POLE2, BIRC5, EXO1, CLSPN, DTL, IKBKG, PLG, ZWINT, and CHAF1A) were enriched in metascape. DNA replication, double-strand break repair via break-induced replication, DNA strand elongation, replication fork protection complex, cell cycle checkpoint, telomere maintenance, transferase complex, transferring phosphorus-containing groups, chromatin binding, cell division, and regulation of chromosome organization were mainly enriched in these input genes (Fig. 5A-C).
Figure 5.
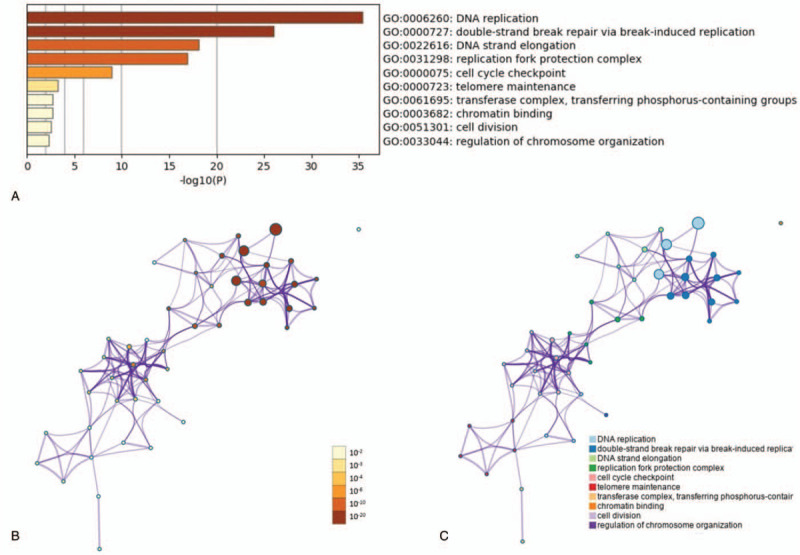
The enrichment analysis of the GINS subunits and neighboring genes in breast cancer. (A) Heatmap of Gene Ontology (GO) enriched terms. B-C, Network of enriched terms: (B) colored by P value (C) colored by enriched terms.
To explore the immune cell infiltration in different subtypes breast cancer (basal-like, HER-2, and luminal) we used the TIMER database. By using this database, the correlation of the expression level of GINS subunits with various immune cells (B cells, CD4+ T cells, CD8+ T cells, Neutrophils, Macrophages, and Dendritic cells) and tumor purity in breast cancer were explored. The expression level of GINS1 had a positive correlation with tumor purity (r = 0.205, P = 6.51e−11), besides, it also had positive correlations with levels of infiltrating B cell (r = 0.187, P = 3.95e−09), CD8 + T cell (r = 0.127, P = 6.87e−05), Neutrophil (r = 0.165, P = 2.97e−07) and Dendritic cell (r = 0.162, P = 5.35e−07) (Fig. 6A). Similarly, the GINS2 expression level had a positive correlation with tumor purity (r = 0.264, P = 2.25e−17), a positive correlation with level of infiltrating B cell (r = 0.116, P = 2.81e−04) and a negative correlation with level of infiltrating Macrophage (r = −0.122 P = 1.30e−04) (Fig. 6B). As for GINS3, the GINS3 expression level had a positive correlation with tumor purity (r = 0.123, P = 9.80e−05) and had positive correlation with levels of infiltrating B cell (r = 0.205, P = 1.09e−10), CD8 + T cell (r = 0.186, P = 4.87e−09), neutrophil (r = 0.184, P = 1.02e−08) and dendritic cell (r = 0.193, P = 1.81e−09) (Figure 6C). For GINS4, the GINS4 expression level had a positive correlation with tumor purity (r = 0.174, P = 3.44e−08) and had positive correlation with levels of infiltrating B cell (r = 0.136, P = 1.89e−05), Neutrophil (r = 0.16, P = 7.31e−07), and Dendritic cell (r = 0.131, P = 4.88e−05) (Fig. 6D). These results showed that some GINS subunits expression is correlated with the level of different immune infiltration in breast cancer.
Figure 6.
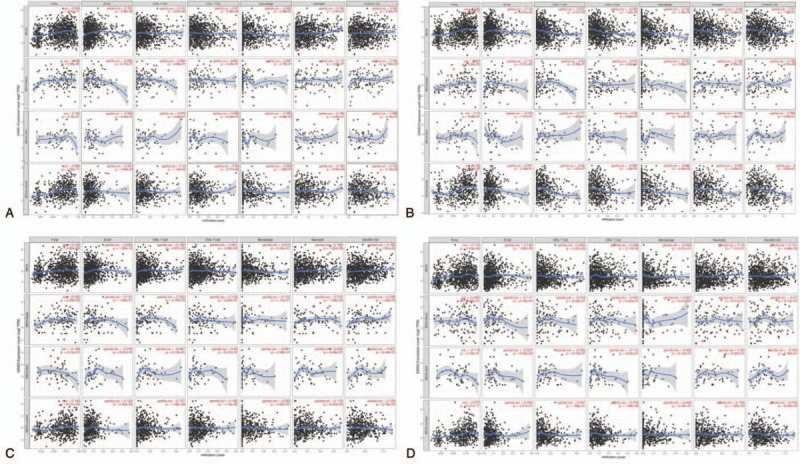
Correlation between GINS subunits expression and immune infiltration levels in breast cancer. Scatter plots showed the correlations between (A) GINS1, (B) GINS2, (C) GINS3, (D) GINS4 expression and immune infiltration levels in different subtypes of breast cancer.
4. Discussions
After diagnosis, the determination of prognosis and identification of appropriate treatments have become the most immediate challenge in patients with breast cancer. In recent years, emerging evidence has continued to identify the effect of novel multigene prognostic biomarkers, such as Oncotype Dx and MammaPrint in breast cancer. These novel multigene prognostic biomarkers have significantly contributed toward the understanding of the pathogenesis and development of breast cancer.[25,26] Therefore, biomarkers play an essential role in breast cancer.
The GINS complex, a circular nucleic acid replication factor, was first discovered by Takayama et al.[27] In recent years, researchers have found that the GINS complex is highly expressed in many tumors, such as breast cancer, melanoma, and colon cancer.[28–30] Additionally, the GINS subunits show an important effect in cell cycle regulation and apoptosis.[31–33] However, the role of each GINS subunits in breast cancer remains unknown. According to GEPIA databases, the expression level of GINS1, GINS2 and GINS4 was significantly higher in breast cancer tissues than that in normal tissues. In addition, we analyzed the association among GINS1, GINS2, GINS3, and GINS4 in breast cancer using the GEPIA database and found that these GINS subunits are positively correlated in terms of expression. Besides, the results from UALCAN database suggested that the expression level of GINS subunits in breast cancer patients were associated with some clinical pathological parameters, such as lymph node metastasis, individual cancer stages and breast cancer subclasses.
Research has shown that expression of GINS1 is high in bladder cancer and that overexpression of GINS1 may lead to the abnormal proliferation of cancer cells.[34] However, the association between GINS1 expression and breast cancer is not yet clear. Therefore, analysis via the bc-GenExMiner database laid out that increased the expression level of GINS1 indicated poor survival in all breast cancer patients. Overexpression of GINS2 has been found in breast cancer, and interfering with the expression level of GINS2 could promote the proliferation of breast cancer cells.[35] However, the clinical significance of GINS2 in breast cancer remains unclear. The results from the bc-GenExMiner database revealed that higher expression of GINS2 levels result in poor survival in breast cancer patients. Unlike other GINS subunits, GINS3 was not significantly highly expressed in breast cancer tissue. Nevertheless, similar to GINS1 and GINS2, GINS3 overexpression could result in poorer survival in breast cancer patients. Recent studies have mainly focused on the role of GINS4 in human lung adenocarcinoma, and a study found that lung adenocarcinoma patients with higher expression level of GINS4 had a lower survival rate.[36,37] Nevertheless, the role of GINS4 has not been discussed in breast cancer. According to our database analysis, the increased expression level of GINS4 showed poorer survival in breast cancer patients.
As far as we know, this study is the first to analyze the role of GINS subunits as a whole in breast cancer with their prognostic values, expression pattern, and potential regulatory mechanisms. However, there are still some limitations in our study. This is a bioinformatics analysis based on different online databases, and thus caution should be practiced regarding the background heterogeneity in the overall estimations provided in this study.
5. Conclusions
In summary, based on bioinformatics analysis, we systemically analyzed the expression, prognostic, and clinicopathologic values of GINS subunits in breast cancer. The present study showed that increased expression level of GINS subunits might play a significant role in breast cancer patients. In summary, GINS subunits might act as new survival biomarkers or potential therapeutic target for breast cancer patients, and we hope that our study will enhance the accuracy of prognostic determination in breast cancer patients.
Acknowledgments
The authors thank the Science and Technology Branch Project of Xinjiang Uygur Autonomous Region, People's Republic of China for funding to do this research.
Author contributions
Conceptualization: Hongtao Li, Jing Ma, Binlin Ma.
Data curation: Hongtao Li.
Formal analysis: Jing Ma.
Funding acquisition: Binlin Ma.
Investigation: Jing Ma.
Methodology: Jing Ma, Lin Luo.
Project administration: Lin Luo, Binlin Ma.
Resources: Binlin Ma.
Software: Hongtao Li, Yanzhen Cao, Lin Luo.
Supervision: Hongtao Li, Binlin Ma.
Validation: Lin Luo.
Visualization: Binlin Ma.
Writing – original draft: Hongtao Li, Yanzhen Cao.
Writing – review & editing: Hongtao Li.
Footnotes
Abbreviations: GEPIA = Gene Expression Profiling Interactive Analysis, GO = gene ontology, TIMER = Tumor Immune Estimation Resource.
How to cite this article: Li H, Cao Y, Ma J, Luo L, Ma B. Expression and prognosis analysis of GINS subunits in human breast cancer. Medicine. 2021;100:11(e24827).
This study was supported by Science and Technology Branch Project of Xinjiang Uygur Autonomous Region, People's Republic of China (2017E0262).
The authors have no conflicts of interests to disclose.
The datasets generated during and/or analyzed during the current study are publicly available.
References
- [1].Torre LA, Siegel RL, Ward EM, et al. Global cancer incidence and mortality rates and trends-an update. Cancer Epidemiol Biomarkers Prev 2016;25:16–27. [DOI] [PubMed] [Google Scholar]
- [2].Nicolini A, Ferrari P, Duffy MJ. Prognostic and predictive biomarkers in breast cancer: past, present and future. Semin Cancer Biol 2018;52:56–73. [DOI] [PubMed] [Google Scholar]
- [3].DeSantis C, Ma J, Bryan L, et al. Breast cancer statistics, 2013. CA Cancer J Clin 2014;64:52–62. [DOI] [PubMed] [Google Scholar]
- [4].DeSantis C, Siegel R, Bandi P, et al. Breast cancer statistics, 2011. CA Cancer J Clin 2011;61:409–18. [DOI] [PubMed] [Google Scholar]
- [5].Kimura T, Cui D, Kawano H, et al. Induced expression of GINS complex is an essential step for reactivation of quiescent stem-like tumor cells within the peri-necrotic niche in human glioblastoma. J Cancer Res Clin Oncol 2019;145:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].NM, ERB, Makarova KS, et al. GINS, a central nexus in the archaeal DNA replication fork. Embo J 2006;7:539–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Labib K. Agnieszka Gambus. A key role for the GINS complex at DNA replication forks. Trends Cell Biol 2007;17:271–8. [DOI] [PubMed] [Google Scholar]
- [8].Onesti S, Macneill SA. Structure and evolutionary origins of the CMG complex. Chromosoma (Berlin) 2013;122:47–53. [DOI] [PubMed] [Google Scholar]
- [9].Kamada K. The GINS complex: structure and function. Subcell Biochem 2012;62:135–56. [DOI] [PubMed] [Google Scholar]
- [10].Karow JK, Wu L, Hickson ID. RecQ family helicases: roles in cancer and aging. Curr Opin Genet Dev 2000;10:32–8. [DOI] [PubMed] [Google Scholar]
- [11].Pyle AM. Translocation and Unwinding Mechanisms of RNA and DNA Helicases. Annu Rev Biophys 2008;37:317–36. [DOI] [PubMed] [Google Scholar]
- [12].Abdelhaleem M. Do human RNA helicases have a role in cancer? Biochimica et Biophysica Acta 2004;1704:37–46. [DOI] [PubMed] [Google Scholar]
- [13].B.R.. DNA helicases involved in DNA repair and their roles in cancer. Nat Rev Cancer 2013;13:542–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Lian Yi-Fan, Li Shan-Shan, Huang Yan-Lin, et al. Up-regulated and interrelated expressions of GINS subunits predict poor prognosis in hepatocellular carcinoma. Biosci Rep 2018;38:1–0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Tang Z, Li C, Kang B, et al. GEPIA: a web server for cancer and normal gene expression profifiling and interactive analyses. Nucleic Acids Res 2017;45:98–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Cerami Ethan, Gao Jianjiong, Dogrusoz Ugur, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012;2:401–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Chandrashekar DS, Bashel B, Balasubramanya SAH, et al. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia 2017;19:649–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Jézéquel P, Campone M, Gouraud W, et al. bc-GenExMiner: An easy-to-use online platform for gene prognostic analyses in breast cancer. Breast Cancer Res Treat 2012;131:765–75. [DOI] [PubMed] [Google Scholar]
- [19].Jézéquel P, Frénel JS, Campion L, et al. bc-GenExMiner 3.0: new mining module computes breast cancer gene expression correlation analyses. Database (Oxford) 2013;60:bas060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Bo Li, Eric Severson, Jean-Christophe Pignon, et al. Comprehensive analyses of tumor immunity: implications for cancer immunotherapy. Genome Biol 2016;17:174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Taiwen Li, Jingyu Fan, Binbin Wang, et al. TIMER: a web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res 2017;77:108–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Warde-Farley David, Donaldson Sylva L, Comes Ovi, et al. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res 2010;38:214–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Franz Max, Rodriguez Harold, Lopes Christian, et al. GeneMANIA update 2018. Nucleic Acids Res 2018;46:60–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Zhou Yingyao, Zhou Bin, Pache Lars, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun 2019;10:1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Bedard PL, Mook S, Piccartgebhart MJ, et al. MammaPrint 70-gene profile quantifies the likelihood of recurrence for early breast cancer. Expert Opin Med Diagn 2009;3:193–205. [DOI] [PubMed] [Google Scholar]
- [26].Fabian CJ, Kimler BF. Use of biomarkers for breast cancer risk assessment and prevention. J Steroid Biochem Molec Biol 2007;106:0–39. [DOI] [PubMed] [Google Scholar]
- [27].Takayama Y. GINS, a novel multiprotein complex required for chromosomal DNA replication in budding yeast. Genes Dev 2003;17:1153–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Juha K, Rantala, Henrik Edgren, et al. Integrative functional genomics analysis of sustained polyploidy phenotypes in breast cancer cells identifies an oncogenic profile for GINS2. Neoplasia 2010;12:877–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Ryu B, Kim DS, Deluca AM, et al. Comprehensive expression profiling of tumor cell lines identifies molecular signatures of melanoma progression. PLoS One 2007;7:e594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Nagahama Y, Ueno M, Haraguchi N, et al. PSF3 marks malignant colon cancer and has a role in cancer cell proliferation. Biochem Biophys Res Commun 2010;392:144–54. [DOI] [PubMed] [Google Scholar]
- [31].Gouge CA, Christensen TW. Drosophila Sld5 is essential for normal cell cycle progression and maintenance of genomic integrity. Biochem Biophys Res Commun 2010;400:145–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Bermudez VP, Farina A, Raghavan V, et al. Studies on human DNA polymerase and GINS complex and their role in DNA replication. J Biol Chem 2011;286:28963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Ueno M, Itoh M, Kong L, et al. PSF1 is essential for early embryogenesis in mice. Mol Cell Biol 2005;25:105–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Yamane K, Naito H, Wakabayashi T, et al. Regulation of SLD5 gene expression by miR-370 during acute growth of cancer cells. Sci Rep 2016;6:1–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Nakahara I, Miyamoto M, Shibata T, et al. Up-regulation of PSF1 promotes the growth of breast cancer cells. Genes Cells 2010;15:1015–24. [DOI] [PubMed] [Google Scholar]
- [36].Tane S, Sakai Y, Hokka D, et al. Significant role of Psf3 expression in non-small-cell lung cancer. Cancer Sci 2015;106:1625–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Hokka D, Maniwa Y, Tane S, et al. Psf3 is a prognostic biomarker in lung adenocarcinoma. Lung Cancer 2013;79:77–82. [DOI] [PubMed] [Google Scholar]


