Abstract
Aims
Angiopoietin-like protein 3 (ANGPTL3) and 4 (ANGPTL4) inhibit lipoprotein lipase (LPL) and represent emerging drug targets to lower circulating triglycerides and reduce cardiovascular risk. To investigate the molecular effects of genetic mimicry of ANGPTL3 and ANGPTL4 inhibition and compare them to the effects of genetic mimicry of LPL enhancement.
Methods and results
Associations of genetic variants in ANGPTL3 (rs11207977-T), ANGPTL4 (rs116843064-A), and LPL (rs115849089-A) with an extensive serum lipid and metabolite profile (208 measures) were characterized in six cohorts of up to 61 240 participants. Genetic associations with anthropometric measures, glucose-insulin metabolism, blood pressure, markers of kidney function, and cardiometabolic endpoints via genome-wide summary data were also explored. ANGPTL4 rs116843064-A and LPL rs115849089-A displayed a strikingly similar pattern of associations across the lipoprotein and lipid measures. However, the corresponding associations with ANGPTL3 rs11207977-T differed, including those for low-density lipoprotein and high-density lipoprotein particle concentrations and compositions. All three genotypes associated with lower concentrations of an inflammatory biomarker glycoprotein acetyls and genetic mimicry of ANGPTL3 inhibition and LPL enhancement were also associated with lower C-reactive protein. Genetic mimicry of ANGPTL4 inhibition and LPL enhancement were associated with a lower waist-to-hip ratio, improved insulin-glucose metabolism, and lower risk of coronary heart disease and type 2 diabetes, whilst genetic mimicry of ANGPTL3 was associated with improved kidney function.
Conclusions
Genetic mimicry of ANGPTL4 inhibition and LPL enhancement have very similar systemic metabolic effects, whereas genetic mimicry of ANGPTL3 inhibition showed differing metabolic effects, suggesting potential involvement of pathways independent of LPL. Genetic mimicry of ANGPTL4 inhibition and LPL enhancement were associated with a lower risk of coronary heart disease and type 2 diabetes. These findings reinforce evidence that enhancing LPL activity (either directly or via upstream effects) through pharmacological approaches is likely to yield benefits to human health.
Keywords: ANGPTL3, ANGPTL4, LPL, Lipoprotein lipids, Lipoprotein subclasses, Amino acids, Glycoprotein acetyls, Mendelian randomization, Drug targets
Graphical Abstract
Introduction
Angiopoietin-like proteins (ANGPTLs) are important regulators of lipoprotein metabolism and have emerged as potential drug targets in managing dyslipidaemia and lowering cardiovascular events.1–3 ANGPTL3 and ANGPTL4 are negative regulators of lipoprotein lipase (LPL), a rate-limiting enzyme in clearing circulating triglycerides. Loss-of-function variants in ANGPTL3 are associated with lower concentrations of triglycerides, low-density lipoprotein (LDL) and high-density lipoprotein (HDL) cholesterol, together with lower risk of coronary heart disease (CHD).1 Loss-of-function variants in ANGPTL4 are associated with lower triglycerides, higher HDL cholesterol, and lower CHD risk.4
ANGPTL3 is exclusively produced in the liver and it primarily acts to inhibit LPL, although it also inhibits endothelial lipase, an enzyme more specific to hydrolysis of lipoprotein phospholipids, particularly in HDL particles.5 In contrast, ANGPTL4 is synthesized in various cells and tissues, including heart, adipose tissue and muscle and serves as an LPL inhibitor under conditions of fasting and exercise.5 Although multiple studies have investigated the effects of ANGPTL3 and ANGPTL4 inhibition on standard lipid measures,1 , 2 , 4 it remains unclear how they affect specific lipoprotein subclasses, particularly as both are LPL inhibitors but with non-identical biological roles. Additionally, inhibition of ANGPTL proteins may have effects on traits beyond lipoprotein metabolism. For example, genetic and pharmacological studies have found that ANGPTL3 inhibition is associated with improved insulin sensitivity and increased circulating ketone bodies.2 Consistent with this, a recent genetic study linked ANGPTL4 inhibition with favourable fat distribution and lower risk of type 2 diabetes.6
In this work, we analysed the metabolic effects of genetic mimicry of ANGPTL3 and ANGPTL4 inhibition in up to 61 240 participants across six population cohorts. Our specific aims were to clarify the detailed effects of genetic mimicry of ANGPTL3 and ANGPTL4 inhibition on lipoprotein metabolism at the subclass level. We also explored the effects on circulating non-lipid biomarkers, including glycolysis-related metabolic measures, amino acids, ketone bodies, and inflammation markers, which are emerging risk factors for cardiometabolic diseases; as well as the effects on fat distribution, and risk of type 2 diabetes and cardiovascular disease (CVD).
Methods
Study design and populations
The study design of the present work is shown in Supplementary material online, Figure S1. The metabolic effects of ANGPTL3, ANGPTL4, and LPL were assessed via Mendelian randomization (MR) analysis.7 ANGPTL3 inhibition, ANGPLT4 inhibition, and LPL enhancement were instrumented by rs11207977-T, rs116843064-A, and rs115849089-A, respectively (details shown Supplementary material online, Supplemental Note, Table S1-2, Figure S2-3). Mendelian randomization analyses were conducted using individual participant data where both genotype and serum nuclear magnetic resonance (NMR) metabolic profiling were measured for 61 240 participants across six cohorts (Table 1 and Supplementary material online, Figure S1). When applicable, pregnant women and participants on lipid-lowering treatment were excluded from the analyses, owing to recognized perturbations in lipids and lipoproteins.8
Table 1.
Characteristics of study populations
| Cohorts | FINRISK2007 | FINRISK1997 | NFBC1966 | NFBC1986 | YFS2001 | INTERVAL |
|---|---|---|---|---|---|---|
| Number of participants | 3875 | 6642 | 4923 | 2819 | 2009 | 40 972 |
| Male (%) | 45 | 48 | 50 | 49 | 46 | 49 |
| Age (years) | 50 (13) | 48 (13) | 31 (0) | 16 (0) | 32 (5) | 44 (14) |
| BMI (kg/m2) | 26.9 (4.8) | 26.6 (4.5) | 24.6 (4.1) | 21.3 (3.5) | 25.1 (4.5) | 26.0 (5.0) |
| Systolic blood pressure (mmHg) | 136 (20) | 136 (20) | 125 (13) | 116 (13) | 117 (13) | Not measured |
| Diastolic blood pressure (mmHg) | 79 (11) | 82 (11) | 78 (11) | 68 (8) | 71 (11) | Not measured |
| LDL cholesterol (mmol/L) | 1.6 (0.5) | 1.9 (0.6) | 2.0 (0.7) | 1.5 (0.5) | 1.9 (0.6) | 1.4 (0.5) |
| HDL cholesterol (mmol/L) | 1.5 (0.3) | 1.6 (0.4) | 1.6 (0.5) | 1.4 (0.3) | 1.6 (0.4) | 1.4 (0.4) |
| Triglycerides (mmol/L) | 1.1 (0.5) | 1.3 (0.7) | 1.2 (0.6) | 1.0 (0.4) | 1.3 (0.7) | 1.2 (0.5) |
| Apolipoprotein B (g/L) | 0.8 (0.2) | 1.0 (0.2) | 1.0 (0.3) | 0.8 (0.2) | 1.0 (0.2) | 0.8 (0.2) |
| Apolipoprotein A-1 (g/L) | 1.5 (0.2) | 1.6 (0.2) | 1.7 (0.3) | 1.5 (0.2) | 1.7 (0.3) | 1.5 (0.3) |
Values presented are means with standard deviations in parentheses.
Outcomes: lipoprotein, lipid, and metabolite quantification (individual participant data)
High-throughput NMR spectroscopy-based metabolic profiling was used to quantify over 200 metabolic measures from fasting and semi-fasting serum samples in the six cohorts. The metabolic profiles include routine lipids and individual lipids and their composition in 14 lipoprotein subclasses, fatty acids, amino acids, ketone bodies, glycolysis-related metabolites, and various other measures.9
Outcomes: risk factors and disease endpoints (summary data)
We performed look-ups of the associations of ANGPTL3 rs11207977-T, ANGPTL4 rs116843064-A, and LPL rs115849089-A with multiple risk factors and disease endpoints using publicly available summary statistics. Details of these summary data are listed in Supplementary material online, Table S3. Furthermore, we used PhenoScanner to examine the potential pleiotropic effects of the genotypes by conducting PheWAS across any disease or trait in the catalogue.10
Statistical analysis
Linear regression models were fitted to assess the associations of the triglyceride-lowering alleles in ANGPTL3 (rs11207977-T), ANGPTL4 (rs116843064-A), and LPL (rs115849089-A) with metabolic profiles, using each metabolic measure as an outcome and each genotype as an explanatory variable. All analyses assumed an additive effect and were adjusted for age, sex, and the first four genetic principal components. The metabolic measures were scaled to SD units to allow easier comparison of the results. Effect sizes from each cohort were combined using inverse variance-weighted fixed-effect meta-analysis. To ease the comparison of the association pattern across the three genotypes, the metabolic associations for ANGPTL3 rs11207977-T, ANGPTL4 rs116843064-A, and LPL rs115849089-A were scaled such that each genotype was associated with the same amount of triglyceride lowering (1-SD, being equivalent to about 0.55 mmol/L or 48.7 mg/dL). The scaled metabolic associations with ANGPTL3 and ANGPTL4 were compared to those of LPL via z-tests. In sensitivity analyses, the metabolic associations of the genotypes were compared with the results of the second genetic proxy and were stratified by fasting status and country origin. Also, we applied 2*2 factorial MR analysis11 to investigate for potential interactions of ANGPTL inhibitors and statins via assessing the interactions of LPL, ANGPTL3, and ANGPTL4 genotypes with HMGCR genotype (rs12916) (Supplementary material online, Figure S4).
Bonferroni correction was applied to correct for multiple testing. Here our primary analyses were to assess the effects of genetic mimicry of ANGPTL3 inhibition, ANGPTL4 inhibition, and LPL enhancement on 208 metabolite measures and also explore the differences between these three drug targets. Thus, in total, there were 5*208 tests. However, the metabolic measures are highly correlated, e.g. over 99% of the variation in the 208 biomarkers could be explained by 44 principal components. Thus, in total, there were 5*44 independent tests and accordingly, we used P < 0.0002 (0.05/(5*44)) to guide the interpretation of the results. All analyses were undertaken in the statistical software package R (version 3.4.3).
Results
Associations with lipoprotein-related measures
The associations of triglyceride-lowering alleles in LPL, ANGPTL4, and ANGPTL3 with total lipoprotein lipids and fatty acid measures are presented in Figures 1 and 2. Results for other measures are given in Supplementary material online, Figures S5 and S6 and Table S5.
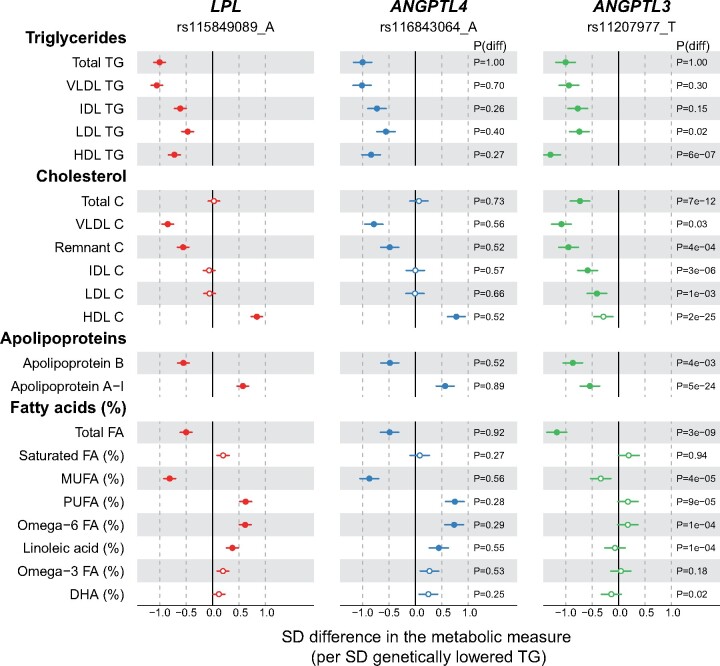
Figure 1.
Associations of LPL (red), ANGPTL4 (blue), and ANGPTL3 (green) genotype with lipoprotein lipids and fatty acids. The associations were scaled so that each genotype is associated with 1-SD (∼0.55 mmol/L) lower concentration of triglycerides. The associations were meta-analysed across six population cohorts, up to 61 240 participants. P(diff) denotes the P-value for the comparison of the metabolic associations of ANGPTL3 (or ANGPTL4) with those of the LPL genotype. Closed symbols: P < 0.0002; open symbols: P ≥ 0.0002. C, cholesterol; DHA, Docosahexaenoic acid; FA, fatty acids; HDL, high-density lipoprotein; IDL, intermediate-density lipoprotein; LDL, low-density lipoprotein; MUFA, monounsaturated fatty acids; PUFA, polyunsaturated fatty acids; TG, triglycerides; VLDL, very-low-density lipoprotein.
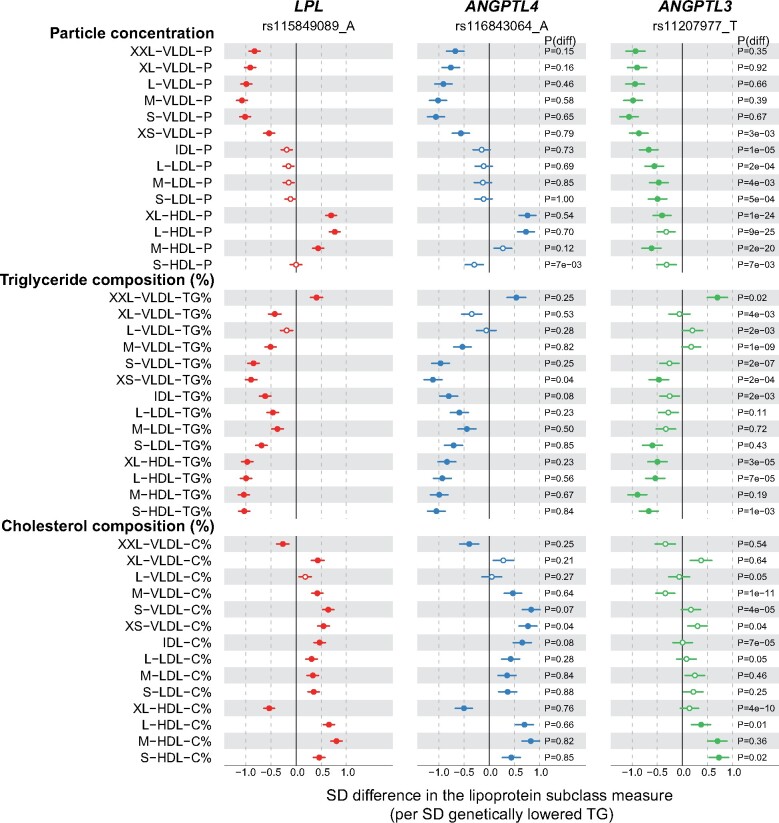
Figure 2.
Associations of LPL (red), ANGPTL4 (blue), and ANGPTL3 (green) genotypes with the lipoprotein lipid compositions. Analyses details are as in Figure 1. TG% refers to the triglyceride concentration relative to total lipids within a single lipoprotein particle. Similar definition applied to C%. Closed symbols: P < 0.0002; open symbols: P ≥ 0.0002.
LPL rs115849089-A was associated with lower triglycerides, lower cholesterol in very-low-density lipoprotein (VLDL) and remnant particles, and higher HDL cholesterol (Figure 1). Apolipoprotein B was lower and apolipoprotein A-I higher. The detailed lipoprotein profiling further revealed that particle concentrations of all VLDL subclasses were lower (between 0.5 and 1 SD per 1-SD lowered triglycerides), whilst those of HDL subclasses were higher by ∼0.5 SDs except for the smallest HDL subclass (Figure 2). Evidence for lower intermediate-density lipoprotein (IDL) and LDL particle concentrations was weaker. For the lipoprotein particle compositions, the triglyceride proportion within each lipoprotein particle across the subclasses was lower except for the large VLDL subclass (Figure 2). Concomitantly, the cholesterol composition of the particles was mostly larger, except for the large VLDL subclass.
ANGPTL4 rs116843064-A displayed a near-identical pattern to that of the LPL allele across all lipoprotein and lipid measures. While ANGPTL3 rs11207977-T showed a similar association profile to ANGPTL4 rs116843064-A in relation to triglyceride concentrations in VLDL, IDL, and LDL lipoprotein fractions and apolipoprotein B (Figure 1), ANGPTL3 rs11207977-T gave a distinct association profile for many other traits. For example, the magnitudes of associations with HDL triglycerides and almost all the non-HDL cholesterol measures were larger than the equivalent values seen for ANGPTL4 and LPL. ANGPTL3 was associated with lower concentrations of IDL and LDL cholesterols but there were no associations of these traits with ANGPTL4 or LPL. Furthermore, while both ANGPTL4 and LPL associated with higher apolipoprotein A-I, the direction of association was reversed for ANGPTL3.
Associations with non-lipid metabolic measures
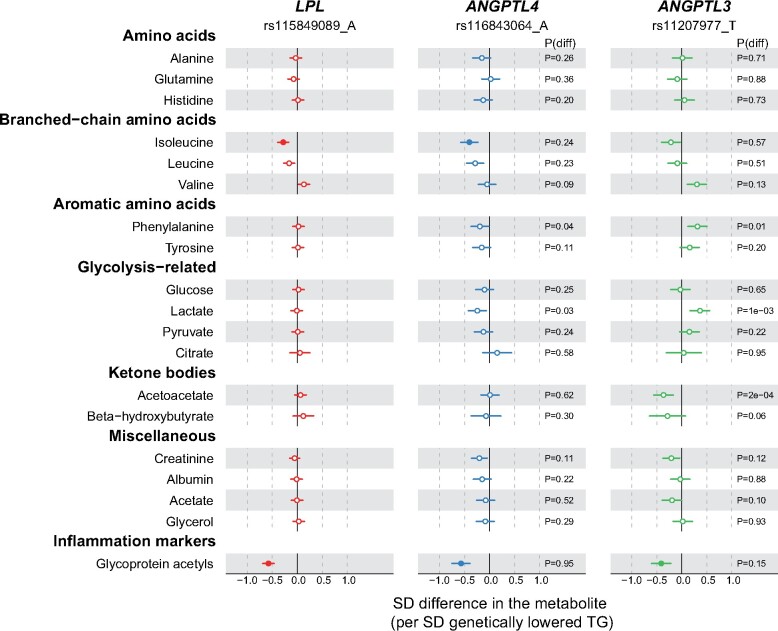
Figure 3 illustrates the associations of LPL rs115849089-A, ANGPTL4 rs116843064-A, and ANGPTL3 rs11207977-T with 19 circulating non-lipid measures. The associations of the LPL allele with these measures were generally null or very weak, except for lower concentrations of isoleucine (P = 5 × 10−7) and glycoprotein acetyls (GlycA; P = 4 × 10−23). The ANGPTL4 allele displayed a very similar association pattern as the LPL allele.
Figure 3.
Associations of LPL (red), ANGPTL4 (blue), and ANGPTL3 (green) genotypes with non-lipid measures. Analyses details are as in Figure 1. Closed symbols: P < 0.0002; open symbols: P ≥ 0.0002.
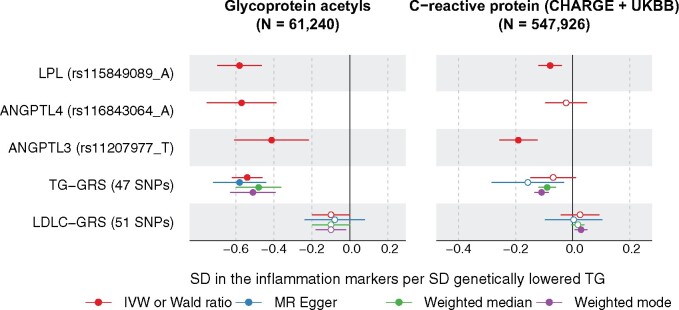
All three genetic instruments associated with lower GlycA, an emerging inflammation marker. Using the genetic summary data from the CHARGE consortium, we looked up the associations with C-reactive protein (CRP), a common inflammation biomarker used in clinical trials. Inverse associations were found for ANGPTL3 inhibition (−0.18 SD CRP per SD genetically lowered triglycerides, P = 0.003) and LPL activation (−0.11, P = 0.003), but the estimate for ANGPTL4 (Beta = 0.03, P = 0.69) was imprecise (Supplementary material online, Figure S9). Results were consistent when we further looked up the associations of the variants with CRP in UK Biobank (Figure 4 and Supplementary material online, Figure S9). To elucidate whether the associations with inflammation markers were specific to LPL pathways or as a general result of lowering triglycerides, we assessed the relationship of a triglycerides polygenic instrument on CRP and GlycA using two-sample MR analysis.12 Four methods (including inverse variance-weighted, MR-Egger, weighted median as well as weighted mode) using 47 triglyceride-associated single nucleotide polymorphisms as the instrument provided consistent results of lower triglycerides being causal for lower CRP and lower GlycA (Figure 4 and Supplementary material online, Figures S7 and S8 and Table S4). Also, we assessed the causal role of LDL cholesterol on CRP and GlycA using 52 SNPs associated with LDL cholesterol as the instrument; the same four methods showed null or weak associations of LDL cholesterol with CRP and GlyA, indicating probable lack of a causal effect (Figure 4 and Supplementary material online, Figures S7 and S8 and Table S4). The consistency of the associations for LPL, ANGPTL4, and ANGPTL3 as compared with the genetic risk score of triglycerides with the inflammation markers for a given lowering of triglycerides (Figure 4), suggests that these effects are a common feature of triglyceride lowering, rather than an effect that is unique to LPL-related pathways. In addition, we also looked up the associations of the three triglyceride-lowering variants with various cytokines; however, no strong associations were found (Supplementary material online, Figure S10).
Figure 4.
The effects of LPL, ANGTPL4, ANGPLT3 genotypes on GlycA (left) and CPR (right) compared with the effects of triglycerides on these markers. Associations of the variants and GRSs with GlycA were analysed in the present study, whilst associations with CRP were meta-analysed from CHARGE and UKBB. Details on the triglyceride and LDL cholesterol GRS are given in Supplementary material online, Figures S7 and S8. Closed symbols: P < 0.01 (corrected for multiple testing 0.05/5). Open symbols: P ≥ 0.01.
Associations with risk factors and disease endpoints
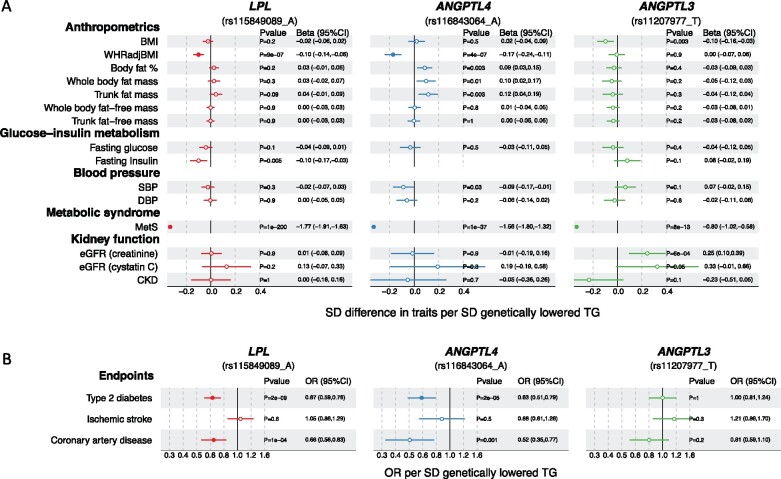
Figure 5 shows the associations of LPL rs115849089-A, ANGPTL4 rs116843064-A, and ANGPTL3 rs11207977-T with multiple risk factors and disease endpoints using publicly available genome-wide summary data (Supplementary material online, Table S3). Across the risk factors, all three variants were associated with lower risk of metabolic syndrome, yet LPL and ANGPTL4 variants displayed specific associations with lower waist-to-hip ratio (P < 0.0002) whilst ANGPTL3 variant showed a unique association with higher levels of estimated glomerular filtration rate assessed by creatinine (eGFR, P = 6 × 10-4). None of the variants were, however, associated with blood pressure traits.
Figure 5.
Associations of LPL (red), ANGPTL4 (blue), and ANGPTL3 (green) genotypes with (A) risk factors and (B) cardiometabolic endpoints. The detailed information on the source data (consortia or UK Biobank) used here is summarized in Supplementary material online, Table S3. For fasting insulin, as the instrument SNP was not available, proxy SNPs were used (rs3850634 for ANGPTL3 and rs11991231 for LPL, R 2 > 0.85 with the instrument SNP). However, no proxy with R 2 > 0.85 was found for ANGPTL4 rs116843064 and thus the result is missing from the figure. BMI, body mass index; WHRadjBMI, Waist-to-hip ratio adjusted for BMI. Closed symbols: P < 0.0002; open symbols: P ≥ 0.0002.
With the cardiometabolic endpoints, LPL and ANGPTL4 variants were associated with lower risk of type 2 diabetes and coronary artery disease. The directions of association of the ANGPTL3 allele in the case of CAD were consistent with the other two genotypes though the 95% CI included the null. Similar findings were identified when scaled to a 1-SD lower apolipoprotein B (Supplementary material online, Figure S11): while the effect estimates for the odds ratio of CAD ranged from 0.26 (for ANGPTL4) to 0.78 (for ANGPLT3), the 95% CI were wide meaning there was no heterogeneity between the estimates. Also, the associations of the three genotypes scaled to apolipoprotein B showed comparable estimates to the causal effects of apolipoprotein B to CAD (Supplementary material online, Figure S11).
Overall concordance of the associations across the three genotypes
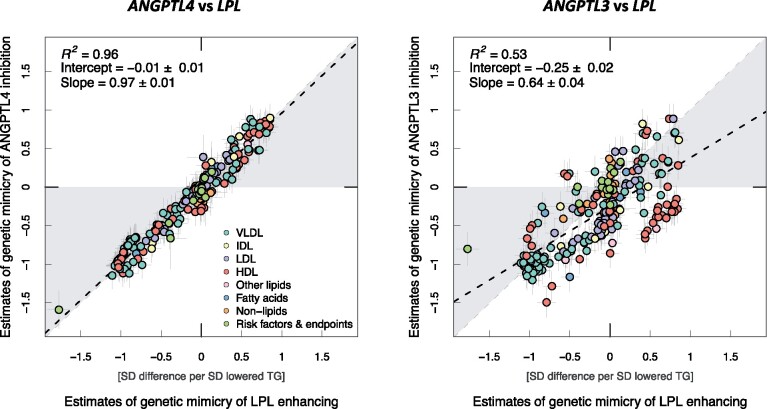
The graphical abstract summarizes the correspondence of the metabolic associations across the 208 metabolic measures and the 18 risk factors and disease endpoints for the ANGPTL4 and LPL genotype (left panel) and for the ANGPTL3 and LPL genotype (right panel). The metabolic associations of genetic mimicry of LPL enhancement were highly similar to those of ANGPTL4 inhibition, the effect sizes falling close to a straight line (slope = 0.97, R 2 = 0.96). The correspondence between the associations for the genetic mimicry of ANGPTL3 inhibition and LPL enhancement was, however, modest (slope = 0.64, R 2 = 0.54).
Results of various sensitivity analyses are summarized in Supplementary material online, Supplemental Note.
Discussion
In this study, we sought to compare and contrast the metabolic associations of genetic variants in ANGPTL3 and ANGPTL4 with LPL. This is timely owing to ongoing phase 2 clinical trials of ANGPTL3 inhibitors, and major investment by pharmaceutical companies in this area.13
Lipoprotein lipase-related genotypes and lipoprotein lipids
Genetic studies have suggested that lowering circulating triglycerides might be beneficial for CHD prevention, but triglyceride-lowering trials have so far provided mixed results.14 Recently, genetic studies have suggested that circulating levels of apolipoprotein B might account for the associations of triglycerides or LDL cholesterol with risk of CHD.11 The fundamental fact that the apolipoprotein B protein molecule does not appear in circulation without lipids, leads to a natural corollary that LDL cholesterol, remnant cholesterol, lipoprotein(a), and triglycerides will all appear on the causal pathway to CHD and that the number of apolipoprotein B-containing lipoprotein particles can explain them all in an elementary way. Genetic mimicry of ANGPTL3 and ANGPTL4 inhibition as well as LPL enhancing were all associated with lower concentrations of triglycerides and apolipoprotein B. Here, the factorial MR analyses showed similar apolipoprotein B-lowering effects for all the three genetic variants with or without stratification by the HMGCR genotype. This genetic evidence supports the view that pharmacologically enhancing LPL-mediated lipolysis is likely to provide cardiovascular benefits in addition to existing LDL cholesterol lowering by statins.6
Lipoprotein lipase-related genotypes and inflammation
The current results demonstrate a novel association that genetic mimicry of ANGPTL3 and ANGPTL4 inhibition as well as LPL enhancement were associated with lower concentration of circulating GlycA. We extended these findings by further looking up the associations with CRP, a well-known marker of systemic inflammation, in the CHARGE consortium and UK Biobank data, and confirmed consistent associations with lower CRP concentrations arising from genetic mimicry of ANGPTL3 inhibition and LPL enhancement. Our two-sample MR analyses assessing the role of triglycerides and these inflammation markers indicated that higher triglycerides are causally associated with higher circulating concentrations of both GlycA and CRP. These findings support an interpretation that the lowering of the inflammation markers derived from these three genotypes (ANGTPL3, ANGPLT4, and LPL, each orientated to lower triglycerides) would most likely be due to lowered circulating triglycerides, and not via attributes that are specific to LPL-related molecular pathways. Recent genetic studies and randomized controlled trials indicate a causal role for inflammation in CVDs, including findings from genetic studies of the interleukin-6 receptor and phase III clinical trials of anti-inflammatory drugs for the treatment of CHD, such as CANTOS trial of canakinumab,15 a monoclonal antibody to interleukin-1 beta, and the recent trial of colchicine.16 Although genetic studies do not support a causal role of CRP for CHD and causal evidence for GlycA is lacking, these markers can be used to assess overall systemic inflammation that is likely to play a role in cardiometabolic diseases.
Lipoprotein lipase-related genotypes and metabolic risk factors and diseases
Metabolic syndrome is a constellation of multiple risk factors, including dyslipidaemia, hypertension, insulin resistance, and adiposity. Our results suggest that the genetic mimicry of ANGPTL3 and ANGPTL4 inhibition and LPL enhancement are all associated with decreased risk of metabolic syndrome. These associations are largely driven by dyslipidaemia, and possibly also by glucose and adiposity traits, but not related to blood pressure traits. Also, we found that the genetic mimicry of ANGPTL3 inhibition is associated with higher levels of eGFR (a directional concordant association with lower risk of chronic kidney disease, though P = 0.1), suggesting ANGPTL3 inhibition may improve kidney function. This is consistent with previous animal experiments in which the Angptl3 knockout was associated with improved renal structure and function and also a delayed disease progression.17 It might thus be worth to assess the role of ANGPTL3 inhibitors in possible prevention and treatment of kidney diseases in forthcoming trials.
The results presented here also suggest that the triglyceride-lowering LPL variant is, in addition to CVD risk reduction, associated with improved glucose-insulin metabolism and a lower risk of type 2 diabetes. This finding is in line with previous studies.2 , 6 In addition, recent MR analyses provided evidence that the lowering of type 2 diabetes risk would be specific to molecular pathways related to LPL.6 In this study, we also revealed that the LPL and ANGPTL4 variants are associated with lower levels of isoleucine—a branched-chain amino acid that have been implicated as potentially lying on the causal pathway to type 2 diabetes.18 The LPL and ANGPTL4 variants were also associated with an improved body fat distribution, a marker reflecting the capacity of peripheral adipose tissue to store surplus energy and a key element in contributing to the development of type 2 diabetes.
ANGPTL3 inhibitors and their potential clinical impact
A recent phase 2 proof-of-concept study (NCT02265952) showed that evinacumab, a fully human monoclonal antibody to ANGPTL3, reduced LDL cholesterol levels in patients with homozygous familial hypercholesterolaemia via a mechanism independent of low-density lipoprotein receptors. An antisense therapy has also been developed to reduce the production of ANGPTL3 protein in the liver and is currently being evaluated in a phase 2 study (NCT03371355) in patients with type 2 diabetes, hypertriglyceridaemia, and non-alcoholic fatty liver disease, a patient segment that might benefit from additional lipid-lowering medication. It is estimated that around 20% of adults with diabetes have residual hypertriglyceridaemia despite statin use, making them at moderate or increased risk of future CVD.19 This motivates the exploration of new therapies that lower triglycerides to reduce cardiometabolic risk. Based on the current genetic results on the effects of ANGPTL3 inhibition, it would be expected that these therapies would lower circulating concentrations of cholesterol and triglycerides in all apolipoprotein B-containing lipoproteins, with a commensurate reduction in the risk of cardiovascular outcomes.
Study limitations
Some discrepancies were seen in the systemic effects of genetic mimicry of ANGPTL3 inhibition vs. genetic mimicry of ANGPTL4 inhibition and LPL enhancement. These findings likely reflect downstream consequences and do not allow elucidation of detailed mechanisms. However, the known function of ANGPTL3 also as an inhibitor of endothelial lipase would provide a plausible explanation for differences observed in various HDL measures and apolipoprotein A-I.5 In the genetic analyses confounding by linkage disequilibrium could potentially drive the observed effects. However, the concordance between the estimates of genetic mimicry of ANGPTL4 inhibition and LPL enhancement together with the co-localization analyses suggest this to be highly unlikely. In addition, we acknowledge the potential limitation of statistical power in the factorial MR analyses (comparing small genetic effects across multiple subgroups), and that we cannot preclude the possibility of interaction effects between these emerging triglyceride-lowering therapies and statins. In addition, it would be clinically relevant to address whether the triglyceride modifying therapeutic targets would have the same effects across different baseline triglyceride concentrations. This may be approached by the development of non-linear methods for drug-target MR analyses.20 , 21
Conclusions
Our results demonstrate that genetic mimicry of LPL pathways is associated with lower circulating triglycerides, all VLDL subclasses and apolipoprotein B, and systemic inflammation biomarkers (CRP and GlyA). Also, genetic mimicry of ANGPTL4 inhibition and LPL enhancement were related to lower waist-to-hip ratio as well as a lower risk of type 2 diabetes and CHD, whilst genetic mimicry of ANGPTL3 inhibition was associated with markers of improved kidney function. Genetic mimicry of ANGPTL4 inhibition and LPL enhancement had near-identical systemic metabolic effects, whereas genetic mimicry of ANGPTL3 inhibition appeared to have differing metabolic consequences including beneficial associations with LDL-related measures and kidney function. Ongoing and future randomized controlled trials should take into account that ANGPTL3 inhibitors may have additional effects beyond LPL inhibition.
Supplementary material
Supplementary material is available at European Heart Journal online.
Data availability
The datasets used in the current study are available from the cohortsthrough application process for researchers who meet the criteria for access to confidential data. For the NFBCs please contact the project center (NFBCprojectcenter@oulu.fi) and visit the website (www.oulu.fi/nfbc) for more information. At the time of publication the NFBC66 genome-wide data is unable to be transferred outside of the University of Oulu. Please contact NFBCprojectcenter@oulu.fi for more information on availability. Aggregated statistical YFS data may be accessed through the data controller on case by case basis for scientific research. Please contact the project center (yfs@utu.fi) and visit the website (http://youngfinnsstudy.utu.fi) for more information. Regarding the FINRISK cohorts requests for data availability should be addressed to the THL Biobank as instructed inhttps://thl.fi/en/web/thl-biobank/for-researchers. For the INTERVAL study please contact the project center (helpdesk@intervalstudy.org.uk) and visit the website (http://www.intervalstudy.org.uk) for more information. Results and summary data related to this study can be found in the supplement.
Funding
The Academy of Finland (grant no. 297338, 307247, 322098, 286284, 134309, 126925, 121584, 124282, 129378, 117787, and 41071), Novo Nordisk foundation (NNF17OC0027034 and NNF17OC0026062), Oulu Health and Wellfare Center, Social Insurance Institution of Finland; Competitive State Research Financing of the Expert Responsibility area of Kuopio, ERDF European Regional Development Fund (grant no. 539/2010: A31592), EU Horizon 2020 (grant no. 633595 and 755320), EU Research Council (grant no. 742927) and following foundations: Sigrid Juselius, Finnish Cardiovascular Research, Juho Vainio, Paavo Nurmi, Finnish Cultural, Tampere Tuberculosis, Emil Aaltonen, Yrjö Jahnsson, Signe and Ane Gyllenberg, and Finnish Diabetes Research. Dr M.V.H. is supported by a British Heart Foundation Intermediate Clinical Research Fellowship (FS/18/23/33512) and the National Institute for Health Research Oxford Biomedical Research Centre.
Conflict of interest: V.S. has consulted for Novo Nordisk and Sanofi and received modest honoraria from these companies. He also has ongoing research collaboration with Bayer Ltd (all unrelated to the present study). J.D. reports as member of advisory board for Novartis, Astrazeneca, and Nightingale Health. A.S.B. has received grants unrelated to the present study from AstraZeneca, Biogen, Bioverativ, Merck, Novartis, Pfizer, and Sanofi. M.V.H. has collaborated with Boehringer Ingelheim in research, and in accordance with the policy of the Clinical Trial Service Unit and Epidemiological Studies Unit (University of Oxford), did not accept any personal payment. Q.W., C.O.-W., O.T.L., J.V., T.L., M.K., M.-R.J., M.P., J.K., and M.A.-K. declare no competing interests.
Supplementary Material
Contributor Information
Qin Wang, Systems Epidemiology, Baker Heart and Diabetes Institute, Melbourne, VIC, Australia; Computational Medicine, Faculty of Medicine, University of Oulu and Biocenter Oulu, Oulu, Finland; Center for Life Course Health Research, University of Oulu, Oulu, Finland.
Clare Oliver-Williams, MRC/BHF Cardiovascular Epidemiology Unit, Department of Public Health and Primary Care, University of Cambridge, Cambridge, UK; Homerton College, University of Cambridge, Cambridge, UK.
Olli T Raitakari, Research Centre of Applied and Preventive Cardiovascular Medicine, University of Turku, Turku, Finland; Department of Clinical Physiology and Nuclear Medicine, Turku University Hospital, Turku, Finland; Centre for Population Health Research, University of Turku, Turku, Finland; Turku University Hospital, Turku, Finland.
Jorma Viikari, Department of Medicine, University of Turku, Turku, Finland; Division of Medicine, Turku University Hospital, Turku, Finland.
Terho Lehtimäki, Department of Clinical Chemistry, Fimlab Laboratories, and Finnish Cardiovascular Research Center Tampere, Faculty of Medicine and Health Technology, Tampere University, Tampere, Finland.
Mika Kähönen, Department of Clinical Physiology, Tampere University Hospital, and Finnish Cardiovascular Research Center Tampere, Faculty of Medicine and Health Technology, Tampere University, Tampere, Finland.
Marjo-Riitta Järvelin, Center for Life Course Health Research, University of Oulu, Oulu, Finland; Unit of Primary Health Care, Oulu University Hospital, OYS, Oulu, Finland; Department of Epidemiology and Biostatistics, MRC-PHE Centre for Environment and Health, School of Public Health, Imperial College London, London, UK; Department of Life Sciences, College of Health and Life Sciences, Brunel University London, UK.
Veikko Salomaa, National Institute for Health and Welfare, Helsinki, Finland.
Markus Perola, National Institute for Health and Welfare, Helsinki, Finland; Institute for Molecular Medicine (FIMM), University of Helsinki, Helsinki, Finland; Estonian Genome Center, University of Tartu, Tartu, Estonia.
John Danesh, MRC/BHF Cardiovascular Epidemiology Unit, Department of Public Health and Primary Care, University of Cambridge, Cambridge, UK; National Institute for Health Research Blood and Transplant Research Unit in Donor Health and Genomics, University of Cambridge, Cambridge, UK; Wellcome Trust Sanger Institute, Hinxton, UK; British Heart Foundation Cambridge Centre of Excellence, Department of Medicine, University of Cambridge, Cambridge, UK.
Johannes Kettunen, Computational Medicine, Faculty of Medicine, University of Oulu and Biocenter Oulu, Oulu, Finland; Center for Life Course Health Research, University of Oulu, Oulu, Finland; National Institute for Health and Welfare, Helsinki, Finland.
Adam S Butterworth, MRC/BHF Cardiovascular Epidemiology Unit, Department of Public Health and Primary Care, University of Cambridge, Cambridge, UK; National Institute for Health Research Blood and Transplant Research Unit in Donor Health and Genomics, University of Cambridge, Cambridge, UK.
Michael V Holmes, Medical Research Council Population Health Research Unit, University of Oxford, Oxford, UK; Clinical Trial Service Unit & Epidemiological Studies Unit, Nuffield Department of Population Health, University of Oxford, Big Data Institute Building, Old Road Campus, Roosevelt Drive, Oxford OX3 7LF, UK; National Institute for Health Research, Oxford Biomedical Research Centre, Oxford University Hospital, Oxford, UK; Medical Research Council Integrative Epidemiology Unit, University of Bristol, Bristol, UK.
Mika Ala-Korpela, Computational Medicine, Faculty of Medicine, University of Oulu and Biocenter Oulu, Oulu, Finland; Center for Life Course Health Research, University of Oulu, Oulu, Finland; NMR Metabolomics Laboratory, School of Pharmacy, University of Eastern Finland, Kuopio, Finland.
References
- 1. Dewey FE, Gusarova V, Dunbar RL, O’Dushlaine C, Schurmann C, Gottesman O, McCarthy S, Van Hout CV, Bruse S, Dansky HM, Leader JB, Murray MF, Ritchie MD, Kirchner HL, Habegger L, Lopez A, Penn J, Zhao A, Shao W, Stahl N, Murphy AJ, Hamon S, Bouzelmat A, Zhang R, Shumel B, Pordy R, Gipe D, Herman GA, Sheu WHH, Lee I-T, Liang K-W, Guo X, Rotter JI, Chen Y-DI, Kraus WE, Shah SH, Damrauer S, Small A, Rader DJ, Wulff AB, Nordestgaard BG, Tybjærg-Hansen A, van den Hoek AM, Princen HMG, Ledbetter DH, Carey DJ, Overton JD, Reid JG, Sasiela WJ, Banerjee P, Shuldiner AR, Borecki IB, Teslovich TM, Yancopoulos GD, Mellis SJ, Gromada J, Baras A. Genetic and pharmacologic inactivation of ANGPTL3 and cardiovascular disease. N Engl J Med 2017;377:211–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Graham MJ, Lee RG, Brandt TA, Tai L-J, Fu W, Peralta R, Yu R, Hurh E, Paz E, McEvoy BW, Baker BF, Pham NC, Digenio A, Hughes SG, Geary RS, Witztum JL, Crooke RM, Tsimikas S. Cardiovascular and metabolic effects of ANGPTL3 antisense oligonucleotides. N Engl J Med 2017;377:222–232. [DOI] [PubMed] [Google Scholar]
- 3. Mach F, Baigent C, Catapano AL, Koskinas KC, Casula M, Badimon L, Chapman MJ, De Backer GG, Delgado V, Ference BA, Graham IM, Halliday A, Landmesser U, Mihaylova B, Pedersen TR, Riccardi G, Richter DJ, Sabatine MS, Taskinen M-R, Tokgozoglu L, Wiklund O, ESC Scientific Document Group. 2019 ESC/EAS Guidelines for the management of dyslipidaemias: lipid modification to reduce cardiovascular risk. Eur Heart J 2020;41:111–188. [DOI] [PubMed] [Google Scholar]
- 4. Dewey FE, Gusarova V, O’Dushlaine C, Gottesman O, Trejos J, Hunt C, Van Hout CV, Habegger L, Buckler D, Lai K-MV, Leader JB, Murray MF, Ritchie MD, Kirchner HL, Ledbetter DH, Penn J, Lopez A, Borecki IB, Overton JD, Reid JG, Carey DJ, Murphy AJ, Yancopoulos GD, Baras A, Gromada J, Shuldiner AR. Inactivating variants in ANGPTL4 and risk of coronary artery disease. N Engl J Med 2016;374:1123–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kersten S. Angiopoietin-like 3 in lipoprotein metabolism. Nat Rev Endocrinol 2017;13:731–739. [DOI] [PubMed] [Google Scholar]
- 6. Lotta LA, Stewart ID, Sharp SJ, Day FR, Burgess S, Luan J, Bowker N, Cai L, Li C, Wittemans LBL, Kerrison ND, Khaw K-T, McCarthy MI, O’Rahilly S, Scott RA, Savage DB, Perry JRB, Langenberg C, Wareham NJ. Association of genetically enhanced lipoprotein lipase-mediated lipolysis and low-density lipoprotein cholesterol-lowering alleles with risk of coronary disease and type 2 diabetes. JAMA Cardiol 2018;3:957–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Smith GD, Ebrahim S. ‘ Mendelian randomization’: can genetic epidemiology contribute to understanding environmental determinants of disease? Int J Epidemiol 2003;32:1–22. [DOI] [PubMed] [Google Scholar]
- 8. Wang Q, Würtz P, Auro K, Mäkinen V-P, Kangas AJ, Soininen P, Tiainen M, Tynkkynen T, Jokelainen J, Santalahti K, Salmi M, Blankenberg S, Zeller T, Viikari J, Kähönen M, Lehtimäki T, Salomaa V, Perola M, Jalkanen S, Järvelin M-R, Raitakari OT, Kettunen J, Lawlor DA, Ala-Korpela M. Metabolic profiling of pregnancy: cross-sectional and longitudinal evidence. BMC Med 2016;14:205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Würtz P, Kangas AJ, Soininen P, Lawlor DA, Davey Smith G, Ala-Korpela M. Quantitative serum nuclear magnetic resonance metabolomics in large-scale epidemiology: a primer on -Omic Technologies. Am J Epidemiol 2017;186:1084–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Staley JR, Blackshaw J, Kamat MA, Ellis S, Surendran P, Sun BB, Paul DS, Freitag D, Burgess S, Danesh J, Young R, Butterworth AS. PhenoScanner: a database of human genotype-phenotype associations. Bioinformatics 2016;32:3207–3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ference BA, Kastelein JJP, Ray KK, Ginsberg HN, Chapman MJ, Packard CJ, Laufs U, Oliver-Williams C, Wood AM, Butterworth AS, Di Angelantonio E, Danesh J, Nicholls SJ, Bhatt DL, Sabatine MS, Catapano AL. Association of triglyceride-lowering LPL variants and LDL-C–lowering LDLR variants with risk of coronary heart disease. JAMA 2019;321:364–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Davey Smith G, Hemani G. Mendelian randomization: genetic anchors for causal inference in epidemiological studies. Hum Mol Genet 2014;23:R89–R98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Banerjee P, Chan K-C, Tarabocchia M, Benito-Vicente A, Alves AC, Uribe KB, Bourbon M, Skiba PJ, Pordy R, Gipe DA, Gaudet D, Martin C. Functional analysis of LDLR (low-density lipoprotein receptor) variants in patient lymphocytes to assess the effect of evinacumab in homozygous familial hypercholesterolemia patients with a spectrum of LDLR activity. Arterioscler Thromb Vasc Biol 2019;35:2146–2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Navar AM. The evolving story of triglycerides and coronary heart disease risk. JAMA 2019;321:347–349. [DOI] [PubMed] [Google Scholar]
- 15. Ridker PM, Everett BM, Thuren T, MacFadyen JG, Chang WH, Ballantyne C, Fonseca F, Nicolau J, Koenig W, Anker SD, Kastelein JJP, Cornel JH, Pais P, Pella D, Genest J, Cifkova R, Lorenzatti A, Forster T, Kobalava Z, Vida-Simiti L, Flather M, Shimokawa H, Ogawa H, Dellborg M, Rossi PRF, Troquay RPT, Libby P, Glynn RJ, Trial Group C. Antiinflammatory therapy with canakinumab for atherosclerotic disease. N Engl J Med 2017;377:1119–1131. [DOI] [PubMed] [Google Scholar]
- 16. Tardif J-C, Kouz S, Waters DD, Bertrand OF, Diaz R, Maggioni AP, Pinto FJ, Ibrahim R, Gamra H, Kiwan GS, Berry C, López-Sendón J, Ostadal P, Koenig W, Angoulvant D, Grégoire JC, Lavoie M-A, Dubé M-P, Rhainds D, Provencher M, Blondeau L, Orfanos A, L’Allier PL, Guertin M-C, Roubille F. Efficacy and safety of low-dose colchicine after myocardial infarction. N Engl J Med 2019;381:2497–2505. [DOI] [PubMed] [Google Scholar]
- 17. Dai R, Liu H, Han X, Liu J, Zhai Y, Rao J, Shen Q, Xu H. Angiopoietin-like-3 knockout protects against glomerulosclerosis in murine adriamycin-induced nephropathy by attenuating podocyte loss. BMC Nephrol 2019;20:185–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Holmes MV, Ala-Korpela M, Smith GD. Mendelian randomization in cardiometabolic disease: challenges in evaluating causality. Nat Rev Cardiol 2017;14:577–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fan W, Philip S, Granowitz C, Toth PP, Wong ND. Residual hypertriglyceridemia and estimated atherosclerotic cardiovascular disease risk by statin use in U.S. adults with diabetes: National Health and Nutrition Examination Survey 2007-2014. Diabetes Care 2019;42:2307–2314. [DOI] [PubMed] [Google Scholar]
- 20. Sun Y-Q, Burgess S, Staley JR, Wood AM, Bell S, Kaptoge SK, Guo Q, Bolton TR, Mason AM, Butterworth AS, Di Angelantonio E, Vie GÅ, Bjørngaard JH, Kinge JM, Chen Y, Mai X-M. Body mass index and all cause mortality in HUNT and UK Biobank studies: linear and non-linear Mendelian randomisation analyses. BMJ 2019;364:l1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Staley JR, Burgess S. Semiparametric methods for estimation of a nonlinear exposure-outcome relationship using instrumental variables with application to Mendelian randomization. Genet Epidemiol 2017;41:341–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used in the current study are available from the cohortsthrough application process for researchers who meet the criteria for access to confidential data. For the NFBCs please contact the project center (NFBCprojectcenter@oulu.fi) and visit the website (www.oulu.fi/nfbc) for more information. At the time of publication the NFBC66 genome-wide data is unable to be transferred outside of the University of Oulu. Please contact NFBCprojectcenter@oulu.fi for more information on availability. Aggregated statistical YFS data may be accessed through the data controller on case by case basis for scientific research. Please contact the project center (yfs@utu.fi) and visit the website (http://youngfinnsstudy.utu.fi) for more information. Regarding the FINRISK cohorts requests for data availability should be addressed to the THL Biobank as instructed inhttps://thl.fi/en/web/thl-biobank/for-researchers. For the INTERVAL study please contact the project center (helpdesk@intervalstudy.org.uk) and visit the website (http://www.intervalstudy.org.uk) for more information. Results and summary data related to this study can be found in the supplement.