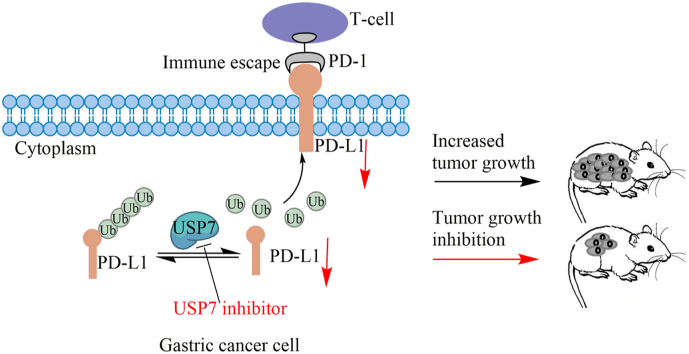
Abstract
Targeting immune checkpoints such as programmed cell death protein 1 (PD-1) and programmed death ligand-1 (PD-L1) have been approved for treating melanoma, gastric cancer (GC) and bladder cancer with clinical benefit. Nevertheless, many patients failed to respond to anti-PD-1/PD-L1 treatment, so it is necessary to seek an alternative strategy for traditional PD-1/PD-L1 targeting immunotherapy. Here with the data from The Cancer Genome Atlas (TCGA) and our in-house tissue library, PD-L1 expression was found to be positively correlated with the expression of ubiquitin-specific processing protease 7 (USP7) in GC. Furthermore, USP7 directly interacted with PD-L1 in order to stabilize it, while abrogation of USP7 attenuated PD-L1/PD-1 interaction and sensitized cancer cells to T cell killing in vitro and in vivo. Besides, USP7 inhibitor suppressed GC cells proliferation by stabilizing P53 in vitro and in vivo. Collectively, our findings indicate that in addition to inhibiting cancer cells proliferation, USP7 inhibitor can also downregulate PD-L1 expression to enhance anti-tumor immune response simultaneously. Hence, these data posit USP7 inhibitor as an anti-proliferation agent as well as a novel therapeutic agent in PD-L1/PD-1 blockade strategy that can promote the immune response of the tumor.
KEY WORDS: USP7, PD-L1, Epigenetics, Immunotherapy, Ubiquitination, Gastric cancer, Immunosuppression, Cancer biology
Abbreviations: BCA, bicinchoninic acid; CHX, cycloheximide; CSN5, COP9 signalosome 5; DUB, deubiquitinating enzymes; EBNA1, Epstein–Barr nuclear antigen 1; FDA, U.S. Food and Drug Administration; FOXO4, forkhead box O4; GC, gastric cancer; GEPIA, Gene-Expression Profiling Interactive Analysis; H2O2, hydrogen peroxidase; HAUSP, herpes virus-associated ubiquitin-specific protease; HDN, well differentiated matched adjacent normal tissues; HDT, well differentiated tumor tissues; ICP0, infected cell protein 0; IL-2, interleukin 2; irAEs, immune-related adverse effects; MDM2, murine double minute-2; PBMC, peripheral blood mononuclear cells; PBS, phosphate buffer saline; PD-1, programmed cell death protein 1; PD-L1, programmed death ligand-1; PDN, poor differentiated matched adjacent normal tissues; PDT, poor differentiated tumor tissues; PTMs, post-translational modifications; qRT-PCR, quantitative real time polymerase chain reaction; RIPA, radioimmunoprecipitation; TCGA, the Cancer Genome Atlas; TCR, T cell receptor; TILs, tumor-infiltrating T cells; USP18, ubiquitin specific peptidase 18; USP22, ubiquitin specific peptidase 22; USP38, ubiquitin specific peptidase 38; USP7, ubiquitin-specific processing protease 7; USP9X, ubiquitin specific peptidase 9 X-linked; WB, Western blotting
Graphical abstract
This study supports that small molecule USP7 inhibitors may be used as novel promoter of the tumor immune response.
1. Introduction
Gastric cancer (GC) is the sixth common malignant tumor all over the world1. The development of GC is a complicated, multistep process including various genetic and epigenetic alterations2. Treatment of GC depends on where the disease initiated and the extent of its metastasis throughout the body, and several types of standard treatment are used clinically, including surgery, chemotherapy and targeted therapy. Nevertheless, the 5-year overall survival of GC is 31.5% in the United States, and for patients with metastasis, 5-year overall survival rate is only 5.3%3. Immunotherapy is a rising therapy choice in a series of solid tumors, and has shown promising effect in patients with GC. As a part of immunotherapy, checkpoint blockades targeting the programmed cell death protein 1/programmed death ligand-1 (PD-1/PD-L1) axis have more superiority in efficiency and fewer side effects4. The interaction between PD-L1 derived from tumor cells with PD-1 derived from tumor-infiltrating T cells (TILs) results in inhibition of the T cell receptor (TCR) pathway and suppression of T cell activity5. Particularly, the use of antibody to block the PD-1/PD-L1 pathway can reactivate the exhausted immune cells in the tumor microenvironment to kill tumor cells. This treatment method adjusts the unbalanced anti-tumor immunity and has acquired 10%–40% response clinically6, and trials that evaluated immune checkpoint blockade in gastrointestinal cancers are ongoing (NCT04152889, NCT04157985, NCT03954756, and NCT03755440). Due to inspiring effects for GC patients, the NCT01848834 trial has demonstrated that treatment by pembrolizumab (antibody against PD-1) in PD-L1-positive recurrent or metastatic GC patients showed prospective antitumor activity and controllable toxicity7. Based on the results of NCT02335411, pembrolizumab has been approved for patients with advanced, PD-L1-positive GC or gastroesophageal cancer by the U.S. Food and Drug Administration (FDA)8,9. Nevertheless, another trial (NCT02370498) showed that pembrolizumab failed to prolong overall survival compared to paclitaxel as second-line therapy for advanced gastric or gastro-oesophageal junction cancer with PD-L1 combined positive score of 1 or higher10. Although anti-PD-1/PD-L1 antibodies have been approved for tumor immunotherapy, they still display several limitations, for example, short half-life, immune-related adverse effects (irAEs), low permeability, immunogenicity and complex production process. As a result, small molecule immune checkpoint inhibitors of PD-1/PD-L1 have been proved to be a meaningful research filed in the drug discovery11, 12, 13, 14. Besides, chemo-immunotherapy15 or combination small molecule inhibitor with anti-PD-1/PD-L116 shows a higher therapeutic efficacy. Patients with expression of PD-L1 on either cancer cells or on TILs prefer to respond to anti-PD-1/PD-L1 therapy17, 18, 19. Meanwhile, the expression of PD-L1 can be regulated by many factors20,21. Hence, it is critical to comprehend the pathways regulating expression and stability of PD-L1.
As a reversible process that can be erased by deubiquitinating enzymes (DUB), ubiquitination plays an important role in multiple cellular processes, for instance involving in embryonic development, advancement of cell cycle, development and progress of tumor and cellular signaling transduction22,23. Among the diverse deubiquitinases, USP7, also known as herpes virus-associated ubiquitin-specific protease (HAUSP), belongs to ubiquitin-specific proteases subfamily24. Thus far, role of USP7 in development and progress of tumor has drew more and more attention25. As USP7 participates in regulating the P53-murine double minute-2 (MDM2) axis26, abrogation of USP7 is considered to inactivate MDM2 and subsequently reactivate P53, leading to cell cycle arrest and apoptosis27. According to these vital discoveries, specific small molecule USP7 inhibitors have been identified for the treatment of diverse human cancers recently24,28,29. Yet, the thorough mechanism by which USP7 modifies antitumor immunity still needs to be illuminated. Especially, whether and how USP7 controls PD-L1-mediated antitumor immunity is still unknown.
In this study, we show that USP7 is overexpressed in GC and correlates with PD-L1 expression. Moreover, USP7 is responsible for PD-L1 protein stabilization in gastric cancer cell lines. Importantly, abrogation of USP7 not only sensitizes GC cells to T cell mediated killing by downregulating cell surface PD-L1 levels and attenuating its interaction with PD-1, but also suppresses the proliferation of GC cells by stabilizing P53 in vitro and in vivo. Our data highlight the therapeutic implications of USP7 inhibitors for treatment of GC.
2. Methods and materials
2.1. Cell lines and compounds
Human gastric cancer cell lines BGC-823, MGC-803, HGC-27 and MKN45 and HEK293 were obtained from Cell Bank of the Chinese Academy of Sciences, Shanghai, China. AGS, NCI-N87, SGC-7901 and MFC were obtained from Cell Bank of the Chinese Academy of Sciences, Beijing, China. Cells were grown in corresponding medium with 10% fetal bovine serum (FBS), at 37 °C with 5% CO2. Almac4 was a gift from Dr. Timothy Harrison. P5091 (P005091) was purchased from Meilun (Dalian, China).
2.2. Bioinformatics analysis
Two public web servers, Gene-Expression Profiling Interactive Analysis (GEPIA, http://gepia.cancer-pku.cn), and Kaplan-Meier plotter (KM-plotter, http://kmplot.com) were used to conduct bioinformatics analysis, respectively. In short, GEPIA was applied to calculate correlation coefficient of genes; KM-plotter was applied to explore cancer patient overall survival.
2.3. Immunohistochemical analysis (IHC)
Gastric carcinoma tissues were acquired from the First Affiliated Hospital of Zhengzhou University. Tissue was collected according to the clinical protocol approved by the ethics review board of the First Affiliated Hospital of Zhengzhou University, Zhengzhou, China. Before taking written consents, patients were informed about the study. Collected specimens were anonymously controlled that supported the ethical and legal standards. Deparaffinization of the slides containing histological sample were performed by xylene and different concentrations of ethanol and washed with phosphate buffer saline (PBS) three times. Then the slides were incubated with 3% hydrogen peroxidase (H2O2) at room temperature for 20 min and 5% goat serum was applied to block the nonspecific binding of antibody. Anti-USP7 antibody was used on the slides at 4 °C overnight. Following day, the slides washing were done with PBS for three times and were probed with secondary antibody for 2 h at 37 °C. Subsequently, the slides were stained with DAB and hematoxylin. Finally, the slides were dehydrated and mounted. The slides were scanned and analyzed on Aperio AT2 (Leica, Wetzlar, Germany).
2.4. Protein extraction and Western blotting (WB)
To prepare samples for immunoblotting, protein was extracted from cells with Radioimmunoprecipitation (RIPA) lysis buffer containing protease inhibitor to protect protein from degradation. Cells were washed with PBS, mixed with lysis buffer and then centrifugation was applied at 12,000×g for 10 min at 4 °C. The supernatant was then collected and the protein concentration was measured by a bicinchoninic acid (BCA) protein assay kit. Following denatured at 100 °C for 10 min samples were subjected to SDS-PAGE and then protein was transferred to 0.22 μm blotting membrane (nitrocellulose membrane). Then the membranes blocking was done with 5% nonfat milk prepared in PBS and incubated at room temperature 2 h. Then, the blot was incubated with specific primary antibody diluted in PBST at 4 °C overnight. Followed by, membranes were incubated with horseradish-peroxidase (HRP) conjugated secondary antibody for 2 h at room temperature. After the application of first and second antibody, the membranes were washed for 4 times with 5 min in every time. Finally, the membranes were autographed to X-ray film by chemiluminescence kit from Thermo Fisher Scientific, Waltham, MA, USA. The relative expression ratio for the control and experimental groups were analyzed on the basis of density by Image J software and the GAPDH signal as a reference. Antibodies were used against USP7 (catalog ab10893; Abcam, Cambridge, UK), human PD-L1 (catalog 13684s; Cell Signaling Technology, Danvers, MA, US), mouse PD-L1 (catalog ab213480; Abcam), MDM2 (catalog 86934s; Cell Signaling Technology), P53 (catalog 9282T; Cell Signaling Technology), P21 (catalog 2946T; Cell Signaling Technology), GAPDH (GoodHere No. AB-M-M 001, Hangzhou, China).
2.5. sgRNA-lentivirus and siRNA transfection and plasmids
Cells were seeded in 96-well plate at 20%–30% confluence for 24 h before sgRNA-lentivirus treatment. After transfection, the medium was replaced 12 h later. The following sequence was used: USP7 sgRNA1: GAGTGATGGACACAACACCG; USP7 sgRNA2 sequence: TCTTCAGCACTGCTTGTGCA; mUsp7 sgRNA: AGACCACACCAAAAAAGCGT. The production and packaging of the lentivirus for USP7 knockout was done by Shanghai Genechen Co., Ltd., China. USP7 siRNA1: GCAUAGUGAUAAACCUGUA; USP7 siRNA2: UAAGGACCCUGCAAAUUAU. The production of the siRNA for USP7 knockdown was done by GenePharma, Shanghai, China.
For plasmids, full-length expression cDNA of PD-L1 (Flag-PD-L1, HG10084-NF), HA-USP7 (HG11681-CY) and pCMV3-untagged negative control vector were purchased from Sino Biological Inc. (Beijing, China). pCDNA-HA-His-Ub was generated by GENEWIZ, Suzhou, China.
2.6. Detection of cell surface PD-L1
To analyze the cell surface PD-L1, 100 μL staining buffer containing human PE-PD-L1 antibody (catalog557924; BD Biosciences, Franklin Lakes, NJ, USA) was prepared and cells were suspended in it, and then incubated the mixture at room temperature for 30 min. Same procedure was followed for MFC cells with mouse PE-PD-L1 antibody (catalog 124308; BD Bioscience), after which, the cells were washed by the staining buffer and subjected to FACS analysis using BD FACSCanto flow cytometer (BD Biosciences).
2.7. Immunoprecipitation (IP)
For protein extraction, HEK293 cells were seeded into 60 mm plates and transiently transfected with 2.5 μg Flag-PD-L1 and 0, 0.625, 1.25, and 2.5 μg HA-USP7. At 48 h after transfection, cells were collected for protein extraction and WB analysis. Antibodies against Flag (catalog 14793s; Cell Signaling Technology), and HA (GenScript, Nanjing, China) were used.
For immunoprecipitation, HEK293 cells were transfected with the indicated plasmids and collected at 48 h after transfection. The cells were lysed in IP lysis buffer (26147, Thermo Fisher Scientific) and were incubated overnight with 1 μg of anti-Flag-M2 affinity gel (FLAGIPT1, Sigma–Aldrich, St. Louis, MO, USA) or anti-HA magnetic beads (88836, Thermo Fisher Scientific) at 4 °C. The beads were then washed with lysis buffer and eluted with elution buffer. The immunoprecipitated protein complex was analyzed using SDS-PAGE.
2.8. In vivo ubiquitination assay
HEK293 cells were transfected with 10 μg His-ub, 10 μg Flag-PD-L1, and 10 μg HA-USP7 by H4000 (Engreen Biosystem Co., Ltd., Beijing, China) according to the instruction. The cells were treated with 10 μmol/L MG132 for 6 h at 48 h post-transfection and then lysed for immunoprecipitated using anti-Flag-M2 affinity gel.
2.9. Protein half-life assay
Initially, cell transfection with sgRNA lentivirus or treated with Almac4 (5 μmol/L) were done under indicated parameters. After cycloheximide (CHX, 20 μmol/L) was applied to the medium and at indicated time points, cells were collected and immunoblotting was performed to evaluate the corresponding protein levels.
2.10. Quantitative reverse transcription (qRT) PCR assay
Quantitative real-time polymerase chain reaction (qRT-PCR) assay was performed to analyze the expression level of mRNA. Total RNA extraction was performed with the TaKaRa MiniBEST Universal RNA Extraction Kit (Code No. 9767, TaKaRa, Shiga, Japan). To measure the expression level of mRNA, cDNA was synthesized from 2 μg purified total RNA using Revert Aid First Strand cDNA Synthesis Kit (K1622, Thermo Fisher Scientific) based on the manufacturer's instructions. qPCR was done in a real-time PCR machine Quant Studio 6 Flex (Life Technologies, Rockville, IN, US) using the following primers: human PD-L1, 5′-TCACTTGGTAATTCTGGGAGC-3′ (forward) and 5′-CTTTGAGTTTGTATCTTGGATGCC-3′ (reverse)30; β-actin, 5′-GCAAAGACCTGTACGCCAACA-3′ (forward) and 5′-TGCATCCTGTCGGCAATG-3′ (reverse). All data was analyzed by the comparative Ct method. Internal control β-actin mRNA was used to normalize the results.
2.11. PD-L1/PD-1 binding assay
Cells were treated with 20 μg/mL recombinant human PD-1 Fc protein (Sino Biological) at 25 °C for 2 h. Then cells were washed with staining buffer and stained with anti-human Alexa Fluor 488 dye conjugated antibody (A32731, Thermo Fisher Scientific) at 25 °C for 30 min. Next cells were washed by staining buffer and stained with DAPI (Biosharp, Hefei, China) at 25 °C for 10 min and were examined by Nikon C2 Plus confocal microscope (Nikon, Tokyo, Japan).
2.12. T cell killing assay and IL-2 expression measurement
MGC-803 Con and USP7 KO cells were planted in a 24-well plate. Jurkat T cells were incubated with Dynabeads Human T-Activator CD3/CD28 beads (Gibco, NewYork, NY, USA) for 12 h to activate Jurkat T cells activity. Then MGC-803 Con and USP7 KO cells were incubated with activated Jurkat T cells for 84 h, after which, the cells were fixed. After washing, the cells were stained with (4′,6-diamidino-2-phenylindole) DAPI for 10 min. The cell counts were measured and analyzed by high content screening system (ArrayScan XTI, Thermo Fisher Scientific).
MGC-803 Con and USP7 KO cells were planted in a 96-well plate and put into Xcell Ligence RTCA eSight (Agilent, San Diego, CA, USA). Human peripheral blood mononuclear cells (PBMC, derived from health donor) were incubated with Dynabeads Human T-Activator CD3/CD28 beads and then cocultured with MGC-803 Con and USP7 KO cells at 0.1:1, 1:1, 5:1, 10:1 ratio (PBMC:tumor cell). After 84 h, the cells were fixed. After washing, the cells were stained with DAPI for 10 min. The cell counts were measured and analyzed by high content screening system. The level of IL-2 secreted by the PBMC cells was detected according to the manufacturer instructions (human IL-2 ELISA kits, Thermo Scientific).
2.13. Animals and tumor xenograft model
All animal experiments were performed based on the guidelines of the Institutional Animal Care and Use Committee of Zhengzhou University, Zhengzhou, China. Female BALB/c mice (4–5 week-old, weighing 18–21 g) were obtained from Hunan Slack Scene of Laboratory Animal Company Ltd. (Hunan, China). Optimum food and water were supplied to every animal and the nursing was done under sterilized condition. The right scapular region of the mice was selected and the cancer cells (5 × 106) were injected. The weight of mice and the tumors were measured every 3 days. At the 10th day, the tumors were obtained from mice and weighted. The size of tumor was measured using vernier caliper measurements.
2.14. Tumor-infiltrating lymphocyte profiling analysis
MFC tumors were stripped and the TIL cells were collected after digestion with collagenase type 4 (LS004188, Worthington, Columbus, OH, USA)/hyaluronidase (Sigma–Aldrich). TIL were stained with APC-conjugated anti-mouse CD8a (catalog 100714, BioLegend, San Diego, CA, USA)/FITC-conjugated with anti-mouse CD3 (catalog 100204, BioLegend) and analyzed by FACS.
2.15. Statistical analysis
Data were represented as the mean ± standard deviation (SD). The statistical significance of the difference between two groups was measured with Student's t-test. The real-time PCR data was represented by analyzing with the 2–ΔΔCT method. P < 0.05 and P < 0.01 were considered statistically significant.
3. Results
3.1. USP7 and PD-L1 were positively correlated in GC tissues
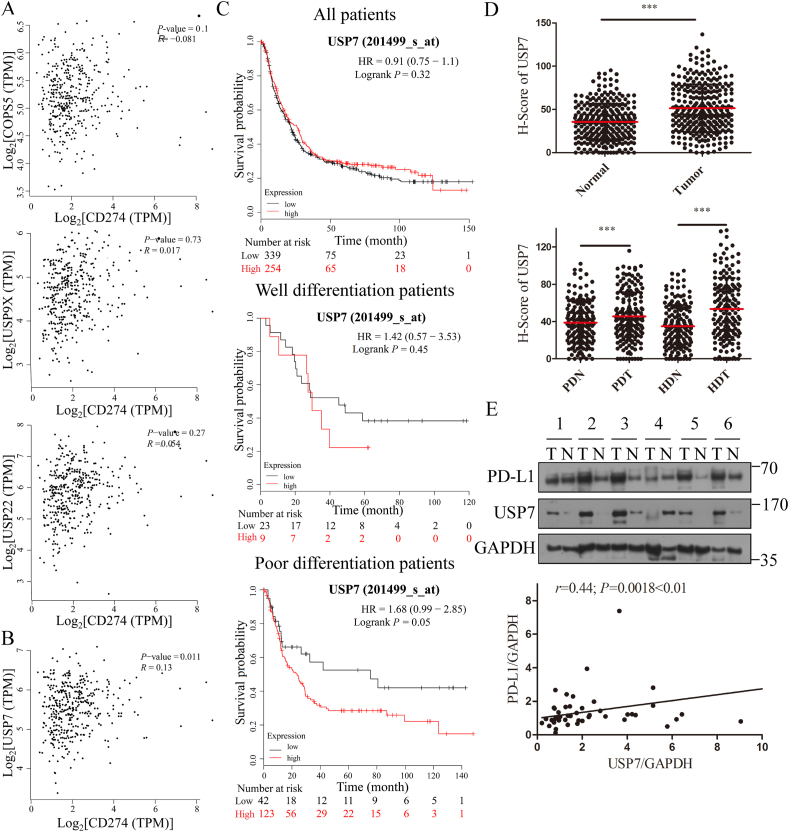
To overcome the clinical response problem, it is crucial to find the upstream regulator that may modulate the amount of PD-L1 in GC31,32. Hence, deubiquitinase attracted our eyes as there have been three deubiquitinases reported to stabilize PD-L1, including COP9 signalosome 5 (CSN5), ubiquitin specific peptidase 9 X-linked (USP9X) and ubiquitin specific peptidase 22 (USP22)33, 34, 35. Nevertheless, correlation analysis using GEPIA (http://gepia.cancer-pku.cn)36 indicated that PD-L1 is not correlated with CSN5, USP9X and USP22 in GC (Fig. 1A), indicating that all these three deubiquitinases may not be the upstream regulator of PD-L1 in GC. Hence, the correlation coefficients between PD-L1 and other different 66 deubiquitinases were analyzed (Supporting Information Table S1), and the results indicate that PD-L1 is correlated with USP7 (Fig. 1B), ubiquitin specific peptidase 18 (USP18, Supporting Information Fig. S1A) and ubiquitin specific peptidase 38 (USP38, Fig. S1A) in GC. In addition, Kaplan-Meier analysis from Kmplot reveals that the poor differentiated patients with high USP7 expression indicated worse overall survival time than those with low levels of USP7 (P = 0.05, Fig. 1C, bottom), but not all patients (P = 0.32, Fig. 1C, top) and well differentiated patients (P = 0.45, Fig. 1C, middle)37. Besides, the expression of USP18 and USP38 had no correlation with overall survival in GC (Fig. S1B). Based on the above data, USP7 was supposed to be the upstream regulator of PD-L1. To explore the expression of USP7 in diverse tumor samples and paired normal tissues, Fig. S1C show that USP7 is overexpressed in breast cancer (BRCA), kidney chromophobe (KICH), kidney renal clear cell carcinoma (KIRC), kidney renal papillary cell carcinoma (KIRP), prostate cancer (PRAD) and GC. In order to further verify this result, expression of USP7 in 286 human GC tissues and paired adjacent normal tissues was examined with our in-house tissue library using IHC. The IHC results also show that USP7 is overexpressed in GC patient tumors compared with adjacent normal tissues significantly (Fig. 1D) and the expression levels of USP7 in tumors are higher than adjacent normal tissues no matter the stage of differentiation (Fig. 1D). Besides, clinical relevance of USP7 in the progression of GC was also investigated. Table 1 shows that USP7 expression is correlated with the stage of differentiation (P = 0.033 < 0.05), but not age, gender and metastasis of GC patients. To further validate this finding in human GC patient samples, the correlation between PD-L1 and USP7 expressions in GC patient samples was analyzed by WB. The WB results indicate that PD-L1 expression levels are highly correlated with USP7 expression (r = 0.44, P < 0.01, Fig. 1E and Supporting Information Table S2). These results suggest that USP7 is correlated with PD-L1 expression in GC.
Figure 1.
USP7 and PD-L1 expressions are positively correlated in GC tissues. (A) Correlation between PD-L1 and CSN5 (top), PD-L1 and USP9X (middle), and PD-L1 and USP22 (bottom) in GC (http://gepia.cancer-pku.cn). (B) Correlation between PD-L1 and USP7 in GC (http://gepia.cancer-pku.cn). (C) Overall survival analysis conducted by Kaplan-Meier method according to USP7 expression. Top, all patients; middle, well differentiated patients; bottom, poor differentiated patients (http://kmplot.com). (D) The expression of USP7 in tumor and adjacent normal tissues of GC patients without classification (top) and under the classification of poor and well differentiation (bottom). (E) Correlation between PD-L1 and USP7 expressions in human GC tissues (T, tumor; N, normal). ∗∗∗P < 0.001. P values were calculated using two-tailed t-test statistical analysis. PDT, poor differentiated tumor tissues; PDN, poor differentiated matched adjacent normal tissues; HDT, well differentiated tumor tissues; HDN, well differentiated matched adjacent normal tissues.
Table 1.
Clinical characteristics of GC patients with different USP7 expressions.
| Variable | No. of patient | USP7 expression |
P value | |
|---|---|---|---|---|
| High | Low | |||
| Gender | ||||
| Male | 216 | 105 | 111 | 0.492 |
| Female | 70 | 38 | 32 | |
| Age | ||||
| ≥51 | 240 | 119 | 121 | 0.872 |
| <51 | 46 | 24 | 22 | |
| Lymph node metastasis | ||||
| Yes | 197 | 101 | 96 | 0.610 |
| No | 89 | 42 | 47 | |
| Differentiation | ||||
| Well/moderate | 135 | 77 | 58 | 0.033 |
| Poor | 151 | 66 | 85 | |
3.2. Inhibition of USP7 decreased PD-L1 expression in GC cells
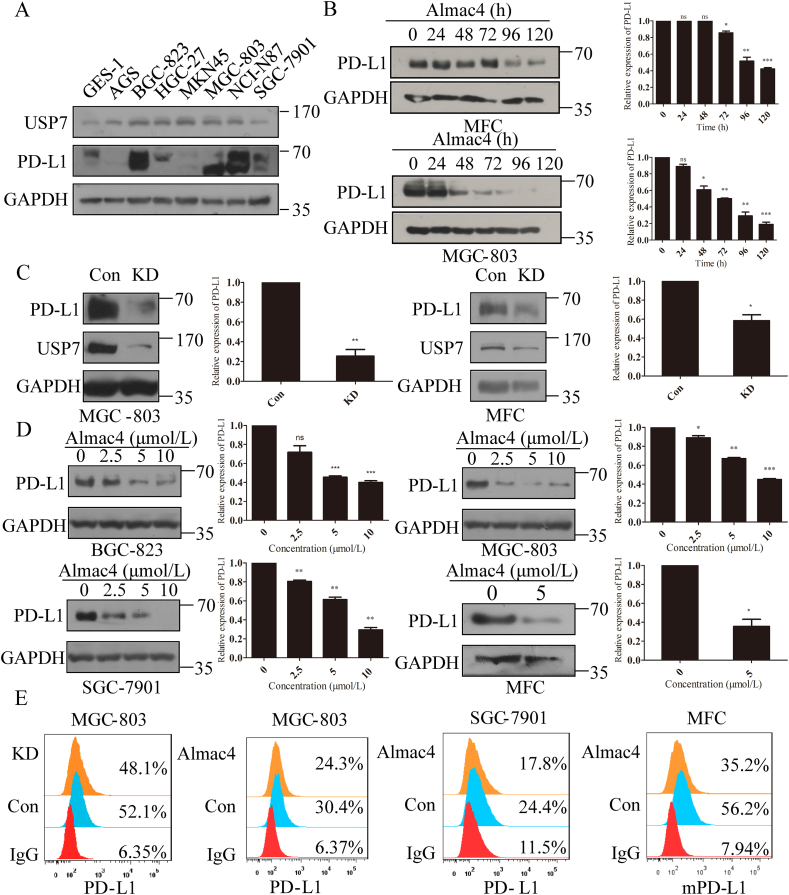
Due to the identification of the positive correlation between USP7 and PD-L1, there may be inter-regulation between USP7 and PD-L1. As PD-L1 is a transmembrane protein and USP7 distributes from nucleus to the cytoplasm as a deubiquitinase, USP7 was supposed as an upstream regulator of PD-L1. To study the regulation of USP7 on PD-L1, expressions of USP7 and PD-L1 were evaluated in eight gastric cell lines. As shown in Fig. 2A, USP7 was ubiquitously expressed in all tested cell lines and six GC cell lines express high amount of USP7 comparing with the cell line of normal gastric epithelium or GES-1. In addition, BGC-823, MGC-803, NCI-N87 and SGC-7901 cells lines expressed higher level of PD-L1 than other GC cell lines, while PD-L1 was absent in MKN45 cells. Among these cell lines, MGC-803 cell line was chosen to explore the effect of endogenous USP7 on regulating PD-L1 expression due to its high expression of PD-L1 and USP7. Firstly, USP7 inhibitor Almac429 was applied to MGC-803 and mouse gastric cancer cell MFC. As indicated in Fig. 2B, when cells were treated with Almac4 to inhibit USP7 activity, expression of PD-L1 was decreased in both of the two cell lines in a time-dependent manner (Fig. 2B). Moreover, to further verify these results, USP7 was knocked down in MGC-803 and MFC cell lines, and PD-L1 expression was significantly reduced compared to control cells (Fig. 2C). Subsequently, BGC-823, MGC-803 and SGC-7901 cells were incubated with Almac4, and results in Fig. 2D suggest that Almac4 treatment decreased the total amount of PD-L1 in all these three cell lines in a dose-dependent way. Moreover, treatment of MFC with Almac4 also dramatically decreased mPD-L1 expression either (Fig. 2D). As PD-L1 on membrane of cancer cells exhibits immunosuppressive effect through binding to PD-1 on activated T cells38, whether USP7 positively regulates membrane PD-L1 remains unclear. Results in Fig. 2E reveal that the amount of membrane PD-L1 was lower than that in parental cells for MGC-803 cells when USP7 was abrogated genetically or pharmacologically in the presence of Almac4, respectively (Fig. 2E). Likewise, in the presence of Almac4, amount of membrane of PD-L1 was decreased in SGC-7901 and MFC cells (Fig. 2E). Meanwhile, in the presence of P5091, another USP7 inhibitor, amount of membrane of PD-L1 also decreased in MGC-803 and BGC-823 cells (Supporting Information Fig. S2). All these results indicate that USP7 abrogation can decrease the expression level of PD-L1 in GC cells.
Figure 2.
Abrogation of USP7 decreased PD-L1 expression in GC cells. (A) USP7 and PD-L1 expression in eight gastric cell lines. (B) PD-L1 expression in MFC and MGC-803 cells treated with Almac4 (5 μmol/L) for the indicated times. The values were normalized on PD-L1 expression in control samples. (C) PD-L1 expression in MGC-803 and MFC cells when USP7 was knocked down. The values were normalized on the PD-L1 expression in control samples. (D) PD-L1 expression in BGC-823, MGC-803, SGC-7901 and MFC cells in the presence of different concentrations of Almac4 for 4 days. The values were normalized on the PD-L1 expression in control samples. (E) FACS analysis for cell surface PD-L1 expression in MGC-803 USP7 KD cells or MGC-803, SGC-7901 and MFC cells treated with Almac4 (5 μmol/L). Data are shown as mean ± SD (n = 3), n.s., not significant; ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001. P values were calculated using two-tailed t-test statistical analysis.
3.3. USP7 directly interacted with PD-L1 and promoted its deubiquitination and stabilization
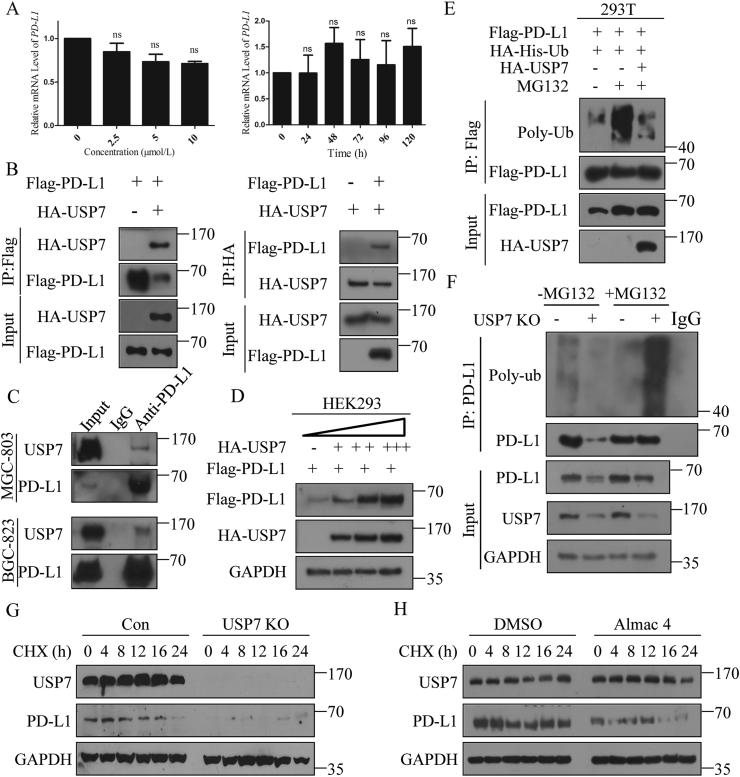
As USP7 expression was positively correlated with PD-L1 expression in human GC tissues and abrogation of USP7 decreased the expression of PD-L1 in different GC cell lines, we next asked how USP7 regulates the PD-L1 expression. First, PD-L1 mRNA levels in MGC-803 cells treated with or without Almac4 were examined. Nevertheless, no significant alteration was discovered for the mRNA of PD-L1 after Almac4 treatment with different concentrations or times (Fig. 3A). As USP7 abrogation can decrease the expression of PD-L1 without impacting on its mRNA level, and PD-L1 was reported to be ubiquitinated and subjected to proteasome mediated degradation33,39, whether USP7 erases the ubiquitination of PD-L1 needs to be answered.
Figure 3.
Abrogation of USP7 downregulated PD-L1 expression by increasing PD-L1 polyubiquitination. (A) mRNA levels of PD-L1 when MGC-803 cells were treated with different concentrations of Almac4 or 5 μmol/L Almac4 for different times; the values were normalized on the PD-L1 expression in control samples. (B) Interaction analysis between Flag-PD-L1 and HA-USP7 using IP. HEK293 cells were transiently transfected with the indicated plasmids Flag-PD-L1 (left) and HA-USP7 (right) was immunoprecipitated and subjected to WB analysis with the HA-tag (left) and Flag-tag antibody (right). (C) Endogenous PD-L1 IP in MGC-803 and BGC-823 cells. (D) WB analysis of Flag-PD-L1 expression in HEK293 cells transfected with Flag-PD-L1 and dose increasing amounts of HA-USP7. (E) Polyubiquitination of PD-L1 in HEK293 cells when Flag-PD-L1 and HA-His-ubiquitin were co-transfected with HA-USP7 or not; cells were treated with MG132 (10 μmol/L) or not 6 h prior to ubiquitination analysis. (F) Polyubiquitination of PD-L1 in MGC-803 cells when USP7 was abrogated genetically, cells were treated with MG132 (10 μmol/L) or not 6 h prior to ubiquitination analysis. Turnover of PD-L1 in MGC-803 cells when USP7 was abrogated genetically (G) or pharmacologically (H) with 20 μmol/L CHX for different times as indicated. Data are shown as mean ± SD (n = 3); ns, not significant; ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001. P values were calculated using two-tailed t-test statistical analysis.
When Flag-PD-L1 and HA-USP7 were co-transfected in HEK293 cells, results in Fig. 3B suggest that they can interact with each other. To further validate these results, endogenous PD-L1 from MGC-803 and BGC-823 cells was enriched with immunoprecipitation and the endogenous USP7 can be detected (Fig. 3C). Moreover, USP7 can promote the amount of PD-L1 in a dose-dependent manner (Fig. 3D). Then, to further verify that USP7 can deubiquitinate PD-L1, Flag-PD-L1 was co-transfected with HA-His-Ub in the presence of HA-USP7 or not in HEK293 cells. Results in Fig. 3E suggest that PD-L1 was strongly ubiquitinated (lane 2, Fig. 3E) in the presence of MG132 but without HA-USP7, while HA-USP7 abolished PD-L1 ubiquitination (lane3, Fig. 3E). Similar results in Fig. 3F show that PD-L1 ubiquitination was significantly increased when USP7 was absent in MGC-803 cells in the presence of MG132, while absence of MG132 led to the sharply decreasing of PD-L1 expression when USP7 was abrogated. To test whether endogenous PD-L1 was also subjected to similar regulation by USP7, USP7 was knocked out genetically (Fig. 3G and Supporting Information Fig. S3A) or pharmacologically (Fig. 3H and Fig. S3B) in MGC-803 cells, and results suggest that endogenous PD-L1 became unstable and degraded rapidly. Similarly, treatment of MFC cells with mUSP7-sgRNA-lentivirus (Fig. S3C) or Almac4 (Fig. S3D) also led to similar results which further confirm the results derived from Fig. 3G and H. All these results indicate that USP7 interacts with PD-L1 and deubiquitinates PD-L1 to promote the stability of PD-L1.
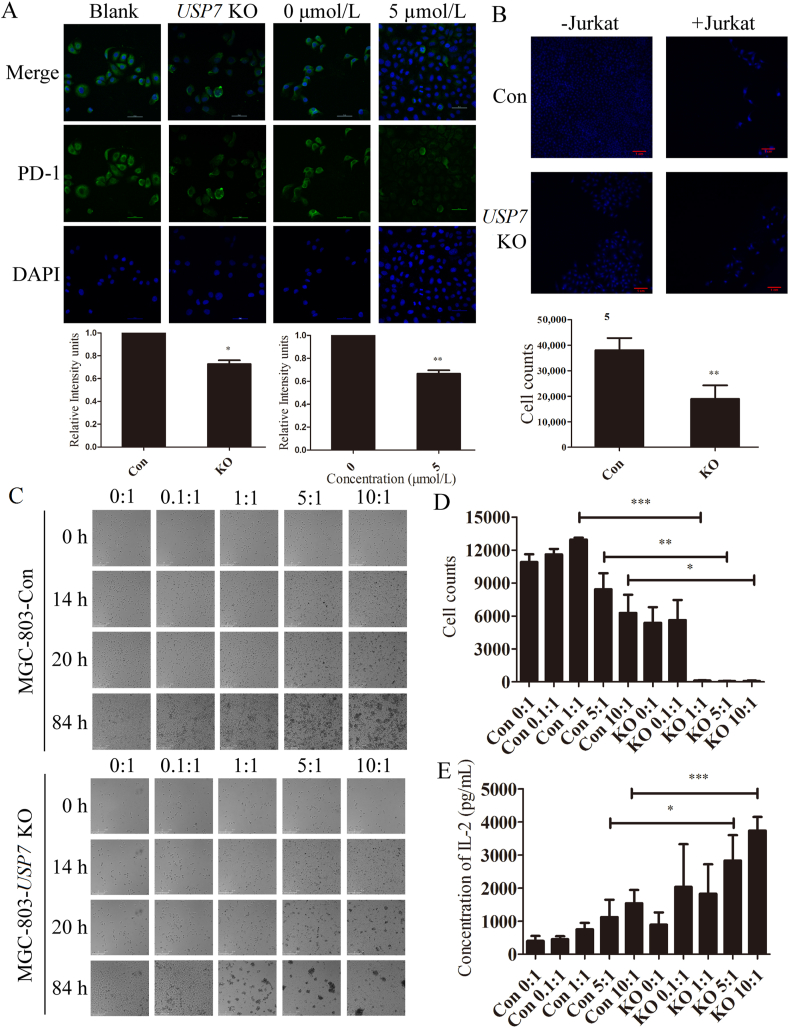
3.4. Blockade of USP7 sensitized cancer cells to T cell killing
Based on the above results, whether USP7 abrogation induced decreasing expression of PD-L1 may affect the binding between PD-L1 and PD-1 and sensitivity of tumor cells to T cell-mediated killing remains unclear. Hence, MGC-803 cells were treated with USP7 sgRNA-lentivirus or Almac4, respectively, followed by incubation with recombinant human PD-1 Fc chimaera protein that enabled fluorescent labeling and visualization. As expected, the binding of PD-1 on the cell surface decreased as a result of abrogation of USP7 both genetically and pharmacologically (Fig. 4A). Moreover, the question regarding whether USP7 KO-mediated PD-L1 downregulation may affect T-cell function remains to be answered. Hence, T cell killing assay was performed by co-culturing activated Jurkat cells with MGC-803 cells in the presence of USP7 or not. This mechanistic assay reveals that knockout of USP7 uninterruptedly activates the cytotoxic T cell towards tumor cells (Fig. 4B). Moreover, another T cell killing assay using activated human PBMCs also confirmed that abrogation of USP7 sensitized MGC-803 cells to T cell in a time- and T cell number-dependent manner (Fig. 4C D, and Supporting Information Videos S1–S10). Interleukin 2 (IL-2) was one of the first cytokines to be discovered and produced by activated T cells and promoted CD8+ T cell cytotoxicity activity40. USP7 knockout enhances IL-2 expression in PBMC cells (Fig. 4E). These results demonstrate that abrogation of USP7 can attenuate the PD-L1/PD-1 interaction and sensitize gastric cancer cells to T cell killing.
Figure 4.
Abrogation of USP7 sensitizes gastric cancer cells to T cell killing. (A) Binding of PD-1 with cell surface PD-L1 of MGC-803 cells when USP7 was abrogated genetically using sgRNA lentivirus or pharmacologically using Almac4 (5 μmol/L) for 4 days. (B) MGC-803 Con and USP7 KO cells were co-cultured with or without activated Jurkat cells for 4 days. Upper, the cells were stained with DAPI; down, the cell counts of MGC-803 Con and USP7 KO cells co-cultured with activated Jurkat cells. (C) MGC-803 Con (upper) and USP7 KO cells (down) were co-cultured with activated PBMC cells for 84 h. (D) The cell counts of (C). (E) Soluble IL-2 levels in MGC-803 Con and USP7 KO cells co-cultured with activated PBMC cells for 84 h. Data are shown as mean ± SD (n = 3); ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001. P values were calculated using two-tailed t-test statistical analysis.
The following is/are the supplementary data related to this article:
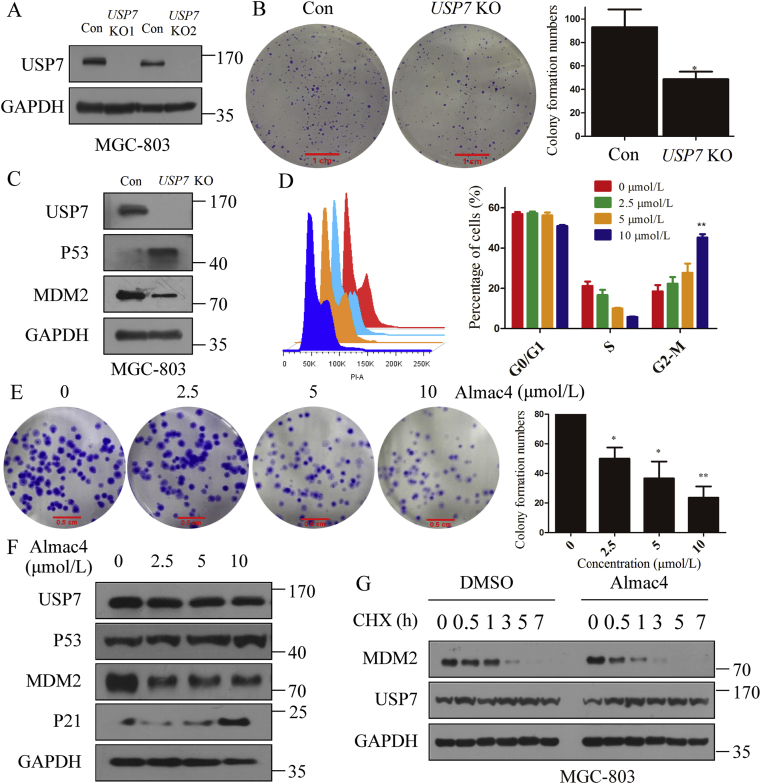
3.5. Abrogation of USP7 suppressed GC cell proliferation and cell cycle progression
As the above results confirmed that USP7 knockout enhanced the sensibility to T cell killing, it was also reported that abrogation of USP7 can suppress tumor growth24,29,41. Next, whether the same function exits in GC needs to be elucidated. To explore whether abrogation of USP7 can suppress GC cell proliferation, knockout of USP7 in MGC-803 was applied. As indicated in Fig. 5A, when USP7 was genetically inactivated in MGC-803 cells by two different sgRNA-lentivirus, the colony formation efficiency of MGC-803 was inhibited (Fig. 5B). Because MDM2 and P53 are the substrates of USP741,42, expression of MDM2 and P53 in MGC-803 cells were investigated, and the results show that MDM2 was reduced and P53 was accumulated upon USP7 knockout (Fig. 5C). As P53 controls the G2/M checkpoint of cell cycle and regulates growth arrest43, the cell cycle analysis was performed when cells were exposed to Almac4 and results in Fig. 5D indicate that Almac4 can induce G2/M phase arrest (42.22%). Besides, Almace4 can also reduce the colony formation activity in MGC-803 cells in a dose-dependently manner (Fig. 5E). Moreover, Almac4 induced the expression of P53 and P21 in MGC-803 cells in a dose-dependent manner (Fig. 5F) and accelerated endogenous MDM2 turnover with CHX (20 μmol/L, Fig. 5G). Hence, abrogation of USP7 inhibits proliferation of GC cells through promoting the stability of P53.
Figure 5.
Abrogation of USP7 suppresses GC cell proliferation and arrested cell cycle. (A) Knockout of USP7 in MGC-803 cells using two different sgRNA lentivirus. (B) Colony formation analysis when USP7 was knocked out in MGC-803 cells. (C) Expression of MDM2 and P53 when USP7 was knocked out in MGC-803 cells. (D) Cell cycle analysis when MGC-803 cells were treated with different concentrations of Almac4 for 48 h. (E) Colony formation when MGC-803 cells were treated with different concentrations of Almac4 for 10 days. (F) Expression of P53 and P21 in MGC-803 treated with different concentrations of Almac4 for 48 h. (G) Turnover of MDM2 in MGC-803 treated with Almac4 (5 μmol/L) for different times as indicated. Data are shown as mean ± SD (n = 3); ∗P < 0.05; ∗∗P < 0.01. P-values were calculated using two-tailed t-test statistical analysis.
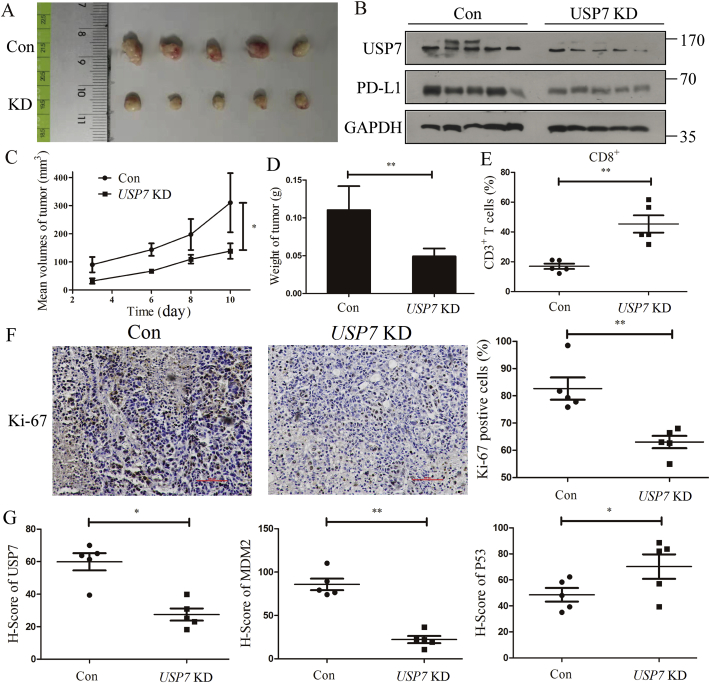
3.6. Inhibition of USP7 decreased PD-L1 expression in vivo
The results from in vitro analysis prompted us to make a hypothesis that knockdown of USP7 can affect mPD-L1 expression in vivo, we assessed the ability of USP7-depleted cells to impair tumor growth in BALB/c mice bearing MFC Con and USP7 KD cells. Results in Fig. 6A show that knockdown of USP7 in MFC reduces tumor size compared to control group, and mice bearing MFC USP7 KD cells had lower mPD-L1 expression in tumors compared to control mice (Fig. 6B). Moreover, abrogation of USP7 decreased the tumor volume and weight in USP7 KD group compared with the control group (Fig. 6C and D). As cytotoxic CD8+ T cells play a critical role in meditating the antitumor effect of anti-PD-L1 treatment44 and the tumor-infiltrated activated CD8+ T-cells were quantified, result in Fig. 6E suggests that the tumor-infiltrated activated CD8+ T-cell population was significantly increased in mice bearing MFC USP7 KD cells (Fig. 6E). Because the expression of Ki-67, which is widely used in routine pathological investigation as a proliferation marker, has strong association with tumor cell proliferation and growth45, Ki-67 of each tumor was examined. Result in Fig. 6F shows that Ki-67 positive tumor cells in MFC USP7 KD significantly decreased compared with MFC Con group. Moreover, it is commonly known that P53 is a powerful tumor suppressor and can be functionally activated to eradicate tumors. However, its function was effectively inhibited through direct interaction with MDM2. Since both P53 and MDM2 are substrates of USP7, expression of P53 and MDM2 in each tumor was tested, and result in Fig. 6G reveals that MDM2 expression decreased in MFC USP7 KD compared with MFC Con group. At the same time, expression of P53 accumulated in MFC USP7 KD compared with MFC Con group. Together, abrogation of USP7 decreased the expression of mPD-L1 and suppressed the tumor growth.
Figure 6.
Abrogation of USP7 decreases mPD-L1 expression and reduces mouse gastric cancer tumorigenesis in vivo. (A) Representative tumors formed in BALB/c mice bearing MFC Con and USP7 KD cells. (B) Expression of USP7 and mPD-L1 in tumors stripped from BALB/c mice bearing MFC Con and USP7 KD cells. Tumor volumes (C) and weight (D) in BALB/c mice bearing MFC Con and USP7 KD cells. (E) Ratio of CD8+ cells in CD3+ T cell populations from the isolated tumor-infiltrating lymphocytes. (F) Ki-67 staining of tumor tissues stripped from BALB/c mice bearing MFC Con and USP7 KD cells, scale bar = 500 μm. (G) USP7, MDM2 and P53 staining of tumors stripped from BALB/c mice bearing MFC Con and USP7 KD cells. Data are shown as mean ± SD (n = 3); ns, not significant; ∗P < 0.05; ∗∗P < 0.01. P values were calculated using two-tailed t-test statistical analysis.
4. Discussion
USP7 was primarily discovered to be associated with viral proteins such as infected cell protein (ICP0)46 and Epstein-Barr nuclear antigen 1 (EBNA1)47. Besides, a lot of research work reported that USP7 promotes the stability of several other proteins, including MDM2 and forkhead box O4 (FOXO4)48, 49, 50, 51, indicating that USP7 plays as an oncogene in tumorigenesis. Here, we reported that USP7 is upregulated in clinical gastric cancer tissues, and poor differentiation patients with high USP7 expression show worse overall survival rate compared to the patients with low levels of USP7. Further, we found that USP7 inhibitor inhibited tumor proliferation via promoting the stability of P53 in GC and arrested the cell cycle at G2–M phase. Hence, we demonstrate that abrogation of USP7 can suppress GC cells proliferation via stabilizing P53 in vitro and in vivo.
Recently, immunotherapies, such as PD-1/PD-L1 targeting therapy and chimeric antigen receptor (CAR) T-cell therapy, have been proved as powerful treatments in clinical oncology. However, PD-L1 inhibitor pembrolizumab only achieved 22% response clinically52. To crack this problem, multifactorial biomarkers, including tumor mutational load, infiltrating CD8+ T cell intensity, and PD-L1 expression levels, have been proposed as bona fide biomarkers of therapeutic response to anti-PD-L1 therapies53. However, the overall response rate was 30% in gastric and gastroesophageal junction (GEJ) cancers irrespective of PD-L1 status54. Moreover, changes in the expression of PD-L1 and tumor mutational burden are independent biomarkers in most cancers55. As PD-L1 expression status is insufficient in determining which patient may be benefit from PD-1/PD-L1 targeting therapy56, findings of new drug or combinational therapy approaches that modulate the tumor microenvironment to promote antitumor immunity are in urgent6,57. In this study, abrogation of USP7 reduced the expression of PD-L1 in a dose- and time-dependent manner in GC cells and results in Fig. 2B show that it took 48 h to decrease expression of PD-L1. We supposed that USP7 may stabilize PD-L1 in an indirect manner although USP7 can interact with PD-L1 directly. These results demonstrate that abrogation of USP7 increased PD-L1 polyubiquitinaion, decreased the PD-1/PD-L1 interaction and sensitized gastric cancer cells to T cell-mediated killing. All these data suggest that USP7 acted as an upstream deubiquitinase of PD-L1 in GC and abrogation of USP7 suppressed tumor growth via downregulating PD-L1-associated immunosuppression, and thus stabilizing P53 and subsequent cell cycle arrest simultaneously.
As PD-L1 can be regulated at the transcriptional level, post-transcriptional, post-translational and extracellular levels58, several post-translational modifications (PTMs) of PD-L1, including phosphorylation 30,59, 60, 61, 62, ubiquitination33,39,63,64, glycosylation65, 66, 67, 68 and palmitoylation69,70, have been reported to regulate PD-L1 stability. In the current study, we show that USP7 deubiquitinates PD-L1 to cause cancer immune resistance to promote cancer growth.
5. Conclusions
In summary, stabilization of PD-L1 by USP7 supports a role for USP7 in antitumor immunity and provides insight into the mechanism how USP7 inhibitor elicits its immunomodulatory effect. Our data support that small molecule USP7 inhibitors may be used as novel promoter of the tumor immune response. The application of USP7 inhibitors to pharmacologically promote antitumor immunity gives a novel insight for novel drug combinations with checkpoint inhibitor agents that can broaden the population of patients that respond to PD-L1/PD-1-targeted therapies.
Acknowledgments
The authors thank Dr. Timothy Harrison for USP7 inhibitor Almac4. This work was supported by National Natural Science Foundation of China (Project No. 81602961 for Yichao Zheng; Nos. 81430085, 81773562, and 82020108030 for Hongmin Liu); National Key Research Program of Proteins (Nos. 2016YFA0501800 and 2018YFE0195100 for Hongmin Liu, China); Key Research Program of Henan Province (No. 161100310100, for Hongmin Liu, China). Science and Technology Innovation Talents of Henan Provincial Education Department (19IRTSTHN001, China).
Footnotes
Peer review under responsibility of Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences.
Supporting data to this article can be found online at https://doi.org/10.1016/j.apsb.2020.11.005.
Contributor Information
Yichao Zheng, Email: yichaozheng@zzu.edu.cn.
Hong-Min Liu, Email: liuhm@zzu.edu.cn.
Author contributions
Zhiru Wang and Wenting Kang designed this study and finalized the manuscript. Wenting Kang and Fengyu Qi performed the USP7 immunohistochemistry and analyzed the data. Zhiru Wang and Ouwen Li conducted most of experiments and collected and analyzed the data. Junwei Wang and Yinghua You performed the mice experiment. Pingxing He did the vector construction. Zhenhe Suo critically revised the manuscript for important intellectual content. Yichao Zheng supervised the whole study. Hongmin Liu sincerely provided his time to obtain funding and gave valuable suggestions to improve the manuscript. All authors read and approved the final version of the manuscript.
Conflicts of interest
The authors of this manuscript clearly declare no conflicts of interest.
Appendix A. Supplementary data
The following is/are the supplementary data to this article:
References
- 1.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Nagini S. Carcinoma of the stomach: A review of epidemiology, pathogenesis, molecular genetics and chemoprevention. World J Gastrointest Oncol. 2012;4:156–169. doi: 10.4251/wjgo.v4.i7.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Howlader N.N.A., Krapcho M., Miller D., Brest A., Yu M. National Cancer Institute; Bethesda, MD, USA: 1975–2016. SEER cancer statistics review.https://seer.cancer.gov/csr/1975_2016/ Based on November 2018 SEER data submission, posted to the SEER web site, April 2019. Available from: [Google Scholar]
- 4.Robert C., Schachter J., Long G.V., Arance A., Grob J.J., Mortier L. Pembrolizumab versus ipilimumab in advanced melanoma. N Engl J Med. 2015;372:2521–2532. doi: 10.1056/NEJMoa1503093. [DOI] [PubMed] [Google Scholar]
- 5.Schildberg F.A., Klein S.R., Freeman G.J., Sharpe A.H. Coinhibitory pathways in the B7-CD28 ligand-receptor family. Immunity. 2016;44:955–972. doi: 10.1016/j.immuni.2016.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zou W., Wolchok J.D., Chen L. PD-L1 (B7-H1) and PD-1 pathway blockade for cancer therapy: Mechanisms, response biomarkers, and combinations. Sci Transl Med. 2016;8:328rv4. doi: 10.1126/scitranslmed.aad7118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Muro K., Chung H.C., Shankaran V., Geva R., Catenacci D., Gupta S. Pembrolizumab for patients with PD-L1-positive advanced gastric cancer (KEYNOTE-012): A multicentre, open-label, phase 1b trial. Lancet Oncol. 2016;17:717–726. doi: 10.1016/S1470-2045(16)00175-3. [DOI] [PubMed] [Google Scholar]
- 8.Fuchs C.S., Doi T., Jang R.W., Muro K., Satoh T., Machado M. Safety and efficacy of pembrolizumab monotherapy in patients with previously treated advanced gastric and gastroesophageal junction cancer: Phase 2 clinical KEYNOTE-059 trial. JAMA Oncol. 2018;4 doi: 10.1001/jamaoncol.2018.0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bang Y.J., Kang Y.K., Catenacci D.V., Muro K., Fuchs C.S., Geva R. Pembrolizumab alone or in combination with chemotherapy as first-line therapy for patients with advanced gastric or gastroesophageal junction adenocarcinoma: Results from the phase II nonrandomized KEYNOTE-059 study. Gastric Cancer. 2019;22:828–837. doi: 10.1007/s10120-018-00909-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shitara K., Ozguroglu M., Bang Y.J., Di Bartolomeo M., Mandala M., Ryu M.H. Pembrolizumab versus paclitaxel for previously treated, advanced gastric or gastro-oesophageal junction cancer (KEYNOTE-061): A randomised, open-label, controlled, phase 3 trial. Lancet. 2018;392:123–133. doi: 10.1016/S0140-6736(18)31257-1. [DOI] [PubMed] [Google Scholar]
- 11.Guzik K., Tomala M., Muszak D., Konieczny M., Hec A., Blaszkiewicz U. Development of the inhibitors that target the PD-1/PD-L1 interaction-a brief look at progress on small molecules, peptides and macrocycles. Molecules. 2019;24:2071. doi: 10.3390/molecules24112071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kopalli S.R., Kang T.B., Lee K.H., Koppula S. Novel small molecule inhibitors of programmed cell death (PD)-1, and its ligand, PD-L1 in cancer immunotherapy: A review update of patent literature. Recent Pat Anti-Cancer Drug Discov. 2019;14:100–112. doi: 10.2174/1574892813666181029142812. [DOI] [PubMed] [Google Scholar]
- 13.Smith W.M., Purvis I.J., Bomstad C.N., Labak C.M., Velpula K.K., Tsung A.J. Therapeutic targeting of immune checkpoints with small molecule inhibitors. Am J Transl Res. 2019;11:529–541. [PMC free article] [PubMed] [Google Scholar]
- 14.Shaabani S., Huizinga H.P.S., Butera R., Kouchi A., Guzik K., Magiera-Mularz K. A patent review on PD-1/PD-L1 antagonists: Small molecules, peptides, and macrocycles (2015–2018) Expert Opin Ther Pat. 2018;28:665–678. doi: 10.1080/13543776.2018.1512706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wei J., Long Y., Guo R., Liu X., Tang X., Rao J. Multifunctional polymeric micelle-based chemo-immunotherapy with immune checkpoint blockade for efficient treatment of orthotopic and metastatic breast cancer. Acta Pharm Sin B. 2019;9:819–831. doi: 10.1016/j.apsb.2019.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao M., Guo W., Wu Y., Yang C., Zhong L., Deng G. SHP2 inhibition triggers anti-tumor immunity and synergizes with PD-1 blockade. Acta Pharm Sin B. 2019;9:304–315. doi: 10.1016/j.apsb.2018.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Herbst R.S., Soria J.C., Kowanetz M., Fine G.D., Hamid O., Gordon M.S. Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature. 2014;515:563–567. doi: 10.1038/nature14011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ribas A., Hu-Lieskovan S. What does PD-L1 positive or negative mean?. J Exp Med. 2016;213:2835–2840. doi: 10.1084/jem.20161462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sznol M., Chen L. Antagonist antibodies to PD-1 and B7-H1 (PD-L1) in the treatment of advanced human cancer. Clin Canc Res. 2013;19:1021–1034. doi: 10.1158/1078-0432.CCR-12-2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deng H., Tan S., Gao X., Zou C., Xu C., Tu K. Cdk5 knocking out mediated by CRISPR-Cas9 genome editing for PD-L1 attenuation and enhanced antitumor immunity. Acta Pharm Sin B. 2020;10:358–373. doi: 10.1016/j.apsb.2019.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu Y., Liu X., Zhang N., Yin M., Dong J., Zeng Q. Berberine diminishes cancer cell PD-L1 expression and facilitates antitumor immunity via inhibiting the deubiquitination activity of CSN5. Acta Pharm Sin B. 2020;10:2299–2312. doi: 10.1016/j.apsb.2020.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Craney A., Rape M. Dynamic regulation of ubiquitin-dependent cell cycle control. Curr Opin Cell Biol. 2013;25:704–710. doi: 10.1016/j.ceb.2013.07.004. [DOI] [PubMed] [Google Scholar]
- 23.Hochstrasser M. Ubiquitin, proteasomes, and the regulation of intracellular protein degradation. Curr Opin Cell Biol. 1995;7:215–223. doi: 10.1016/0955-0674(95)80031-x. [DOI] [PubMed] [Google Scholar]
- 24.McFarlane C., Kelvin A.A., de la Vega M., Govender U., Scott C.J., Burrows J.F. The deubiquitinating enzyme USP17 is highly expressed in tumor biopsies, is cell cycle regulated, and is required for G1–S progression. Canc Res. 2010;70:3329–3339. doi: 10.1158/0008-5472.CAN-09-4152. [DOI] [PubMed] [Google Scholar]
- 25.Wang Z., Kang W., You Y., Pang J., Ren H., Suo Z. USP7: Novel drug target in cancer therapy. Front Pharmacol. 2019;10:427. doi: 10.3389/fphar.2019.00427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mungamuri S.K., Qiao R.F., Yao S., Manfredi J.J., Gu W., Aaronson S.A. USP7 enforces heterochromatinization of p53 target promoters by protecting SUV39H1 from MDM2-mediated degradation. Cell Rep. 2016;14:2528–2537. doi: 10.1016/j.celrep.2016.02.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li M., Brooks C.L., Kon N., Gu W. A dynamic role of HAUSP in the p53–Mdm2 pathway. Mol Cell. 2004;13:879–886. doi: 10.1016/s1097-2765(04)00157-1. [DOI] [PubMed] [Google Scholar]
- 28.Kategaya L., Di Lello P., Rouge L., Pastor R., Clark K.R., Drummond J. USP7 small-molecule inhibitors interfere with ubiquitin binding. Nature. 2017;550:534–538. doi: 10.1038/nature24006. [DOI] [PubMed] [Google Scholar]
- 29.Gavory G., O'Dowd C.R., Helm M.D., Flasz J., Arkoudis E., Dossang A. Discovery and characterization of highly potent and selective allosteric USP7 inhibitors. Nat Chem Biol. 2018;14:118–125. doi: 10.1038/nchembio.2528. [DOI] [PubMed] [Google Scholar]
- 30.Li C.W., Lim S.O., Xia W., Lee H.H., Chan L.C., Kuo C.W. Glycosylation and stabilization of programmed death ligand-1 suppresses T-cell activity. Nat Commun. 2016;7:12632. doi: 10.1038/ncomms12632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Teng F., Meng X., Kong L., Yu J. Progress and challenges of predictive biomarkers of anti PD-1/PD-L1 immunotherapy: A systematic review. Canc Lett. 2018;414:166–173. doi: 10.1016/j.canlet.2017.11.014. [DOI] [PubMed] [Google Scholar]
- 32.Koemans W.J., Chalabi M., van Sandick J.W., van Dieren J.M., Kodach L.L. Beyond the PD-L1 horizon: In search for a good biomarker to predict success of immunotherapy in gastric and esophageal adenocarcinoma. Canc Lett. 2019;442:279–286. doi: 10.1016/j.canlet.2018.11.001. [DOI] [PubMed] [Google Scholar]
- 33.Lim S.O., Li C.W., Xia W., Cha J.H., Chan L.C., Wu Y. Deubiquitination and stabilization of PD-L1 by CSN5. Canc Cell. 2016;30:925–939. doi: 10.1016/j.ccell.2016.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jingjing W., Wenzheng G., Donghua W., Guangyu H., Aiping Z., Wenjuan W. Deubiquitination and stabilization of programmed cell death ligand 1 by ubiquitin-specific peptidase 9, X-linked in oral squamous cell carcinoma. Canc Med. 2018;7:4004–4011. doi: 10.1002/cam4.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang X., Zhang Q., Lou Y., Wang J., Zhao X., Wang L. USP22 deubiquitinates CD274 to suppress anticancer immunity. Canc Immunol Res. 2019;7:1580–1590. doi: 10.1158/2326-6066.CIR-18-0910. [DOI] [PubMed] [Google Scholar]
- 36.Tang Z., Li C., Kang B., Gao G., Li C., Zhang Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98–W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Szasz A.M., Lanczky A., Nagy A., Forster S., Hark K., Green J.E. Cross-validation of survival associated biomarkers in gastric cancer using transcriptomic data of 1,065 patients. Oncotarget. 2016;7:49322–49333. doi: 10.18632/oncotarget.10337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Topalian S.L., Drake C.G., Pardoll D.M. Targeting the PD-1/B7-H1(PD-L1) pathway to activate anti-tumor immunity. Curr Opin Immunol. 2012;24:207–212. doi: 10.1016/j.coi.2011.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang J., Bu X., Wang H., Zhu Y., Geng Y., Nihira N.T. Cyclin D-CDK4 kinase destabilizes PD-L1 via cullin 3-SPOP to control cancer immune surveillance. Nature. 2018;553:91–95. doi: 10.1038/nature25015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bachmann M.F., Oxenius A. Interleukin 2: From immunostimulation to immunoregulation and back again. EMBO Rep. 2007;8:1142–1148. doi: 10.1038/sj.embor.7401099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kon N., Kobayashi Y., Li M., Brooks C.L., Ludwig T., Gu W. Inactivation of HAUSP in vivo modulates p53 function. Oncogene. 2010;29:1270–1279. doi: 10.1038/onc.2009.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ma J., Martin J.D., Xue Y., Lor L.A., Kennedy-Wilson K.M., Sinnamon R.H. C-terminal region of USP7/HAUSP is critical for deubiquitination activity and contains a second mdm2/p53 binding site. Arch Biochem Biophys. 2010;503:207–212. doi: 10.1016/j.abb.2010.08.020. [DOI] [PubMed] [Google Scholar]
- 43.Agarwal M.L., Agarwal A., Taylor W.R., Stark G.R. p53 controls both the G2/M and the G1 cell cycle checkpoints and mediates reversible growth arrest in human fibroblasts. Proc Natl Acad Sci U S A. 1995;92:8493–8497. doi: 10.1073/pnas.92.18.8493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Binnewies M., Roberts E.W., Kersten K., Chan V. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat Med. 2018;24:541–550. doi: 10.1038/s41591-018-0014-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li L.T., Jiang G., Chen Q., Zheng J.N. Ki67 is a promising molecular target in the diagnosis of cancer. Mol Med Rep. 2015;11:1566–1572. doi: 10.3892/mmr.2014.2914. (review) [DOI] [PubMed] [Google Scholar]
- 46.Daubeuf S., Singh D., Tan Y., Liu H., Federoff H.J., Bowers W.J. HSV ICP0 recruits USP7 to modulate TLR-mediated innate response. Blood. 2009;113:3264–3275. doi: 10.1182/blood-2008-07-168203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Holowaty M.N., Zeghouf M., Wu H., Tellam J., Athanasopoulos V., Greenblatt J. Protein profiling with Epstein-Barr nuclear antigen-1 reveals an interaction with the herpesvirus-associated ubiquitin-specific protease HAUSP/USP7. J Biol Chem. 2003;278:29987–29994. doi: 10.1074/jbc.M303977200. [DOI] [PubMed] [Google Scholar]
- 48.Cummins J.M., Rago C., Kohli M., Kinzler K.W., Lengauer C., Vogelstein B. Tumour suppression: Disruption of HAUSP gene stabilizes p53. Nature. 2004;428:1–2. doi: 10.1038/nature02501. [DOI] [PubMed] [Google Scholar]
- 49.Saridakis V., Sheng Y., Sarkari F., Holowaty M.N., Shire K., Nguyen T. Structure of the p53 binding domain of HAUSP/USP7 bound to Epstein-Barr nuclear antigen 1 implications for EBV-mediated immortalization. Mol Cell. 2005;18:25–36. doi: 10.1016/j.molcel.2005.02.029. [DOI] [PubMed] [Google Scholar]
- 50.Sheng Y., Saridakis V., Sarkari F., Duan S., Wu T., Arrowsmith C.H. Molecular recognition of p53 and MDM2 by USP7/HAUSP. Nat Struct Mol Biol. 2006;13:285–291. doi: 10.1038/nsmb1067. [DOI] [PubMed] [Google Scholar]
- 51.van der Horst A., de Vries-Smits A.M., Brenkman A.B., van Triest M.H., van den Broek N., Colland F. FOXO4 transcriptional activity is regulated by monoubiquitination and USP7/HAUSP. Nat Cell Biol. 2006;8:1064–1073. doi: 10.1038/ncb1469. [DOI] [PubMed] [Google Scholar]
- 52.Brar G., Shah M.A. The role of pembrolizumab in the treatment of PD-L1 expressing gastric and gastroesophageal junction adenocarcinoma. Therap Adv Gastroenterol. 2019;12 doi: 10.1177/1756284819869767. 1756284819869767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Topalian S.L., Taube J.M., Anders R.A., Pardoll D.M. Mechanism-driven biomarkers to guide immune checkpoint blockade in cancer therapy. Nat Rev Canc. 2016;16:275–287. doi: 10.1038/nrc.2016.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kang Y.K., Boku N., Satoh T., Ryu M.H., Chao Y., Kato K. Nivolumab in patients with advanced gastric or gastro-oesophageal junction cancer refractory to, or intolerant of, at least two previous chemotherapy regimens (ONO-4538-12, ATTRACTION-2): A randomised, double-blind, placebo-controlled, phase 3 trial. Lancet. 2017;390:2461–2471. doi: 10.1016/S0140-6736(17)31827-5. [DOI] [PubMed] [Google Scholar]
- 55.Yarchoan M., Albacker L.A., Hopkins A.C., Montesion M., Murugesan K., Vithayathil T.T. PD-L1 expression and tumor mutational burden are independent biomarkers in most cancers. JCI Insight. 2019;4:1286–1295. doi: 10.1172/jci.insight.126908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shen X., Zhao B. Efficacy of PD-1 or PD-L1 inhibitors and PD-L1 expression status in cancer: Meta-analysis. BMJ. 2018;362:k3529. doi: 10.1136/bmj.k3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sznol M. Blockade of the B7-H1/PD-1 pathway as a basis for combination anticancer therapy. Canc J. 2014;20:290–295. doi: 10.1097/PPO.0000000000000056. [DOI] [PubMed] [Google Scholar]
- 58.Cha J.H., Chan L.C., Li C.W., Hsu J.L., Hung M.C. Mechanisms controlling PD-L1 expression in cancer. Mol Cell. 2019;76:359–370. doi: 10.1016/j.molcel.2019.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jiao S., Xia W., Yamaguchi H., Wei Y., Chen M.K., Hsu J.M. PARP inhibitor upregulates PD-L1 expression and enhances cancer-associated immunosuppression. Clin Canc Res. 2017;23:3711–3720. doi: 10.1158/1078-0432.CCR-16-3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cha J.H., Yang W.H., Xia W., Wei Y., Chan L.C., Lim S.O. Metformin promotes antitumor immunity via endoplasmic-reticulum-associated degradation of PD-L1. Mol Cell. 2018;71:606–620. doi: 10.1016/j.molcel.2018.07.030. e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li H., Li C.W., Li X., Ding Q., Guo L., Liu S. MET inhibitors promote liver tumor evasion of the immune response by stabilizing PDL1. Gastroenterology. 2019;156:1849–1861 e13. doi: 10.1053/j.gastro.2019.01.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chan L.C., Li C.W., Xia W., Hsu J.M., Lee H.H., Cha J.H. IL-6/JAK1 pathway drives PD-L1 Y112 phosphorylation to promote cancer immune evasion. J Clin Invest. 2019;129:3324–3338. doi: 10.1172/JCI126022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mezzadra R., Sun C., Jae L.T., Gomez-Eerland R., de Vries E., Wu W. Identification of CMTM6 and CMTM4 as PD-L1 protein regulators. Nature. 2017;549:106–110. doi: 10.1038/nature23669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Burr M.L., Sparbier C.E., Chan Y.C., Williamson J.C., Woods K., Beavis P.A. CMTM6 maintains the expression of PD-L1 and regulates anti-tumour immunity. Nature. 2017;549:101–105. doi: 10.1038/nature23643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li C.W., Lim S.O., Chung E.M., Kim Y.S., Park A.H., Yao J. Eradication of triple-negative breast cancer cells by targeting glycosylated PD-L1. Canc Cell. 2018;33:187–201. doi: 10.1016/j.ccell.2018.01.009. e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hsu J.M., Xia W., Hsu Y.H., Chan L.C., Yu W.H., Cha J.H. STT3-dependent PD-L1 accumulation on cancer stem cells promotes immune evasion. Nat Commun. 2018;9:1908. doi: 10.1038/s41467-018-04313-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.D'Arrigo P., Russo M., Rea A., Tufano M., Guadagno E., Del Basso De Caro M.L. A regulatory role for the co-chaperone FKBP51s in PD-L1 expression in glioma. Oncotarget. 2017;8:68291–68304. doi: 10.18632/oncotarget.19309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Maher C.M., Thomas J.D., Haas D.A., Longen C.G., Oyer H.M., Tong J.Y. Small-molecule sigma1 modulator induces autophagic degradation of PD-L1. Mol Canc Res. 2018;16:243–255. doi: 10.1158/1541-7786.MCR-17-0166. [DOI] [PubMed] [Google Scholar]
- 69.Yang Y., Hsu J.M., Sun L., Chan L.C., Li C.W., Hsu J.L. Palmitoylation stabilizes PD-L1 to promote breast tumor growth. Cell Res. 2019;29:83–86. doi: 10.1038/s41422-018-0124-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yao H., Lan J., Li C., Shi H., Brosseau J.P., Wang H. Inhibiting PD-L1 palmitoylation enhances T-cell immune responses against tumours. Nat Biomed Eng. 2019;3:306–317. doi: 10.1038/s41551-019-0375-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.