Abstract
The protein tyrosine phosphatase Src homology phosphotyrosyl phosphatase 2 (SHP2) is implicated in various cancers, and targeting SHP2 has become a promising therapeutic approach. We herein described a robust cross-validation high-throughput screening protocol that combined the fluorescence-based enzyme assay and the conformation-dependent thermal shift assay for the discovery of SHP2 inhibitors. The established method can effectively exclude the false positive SHP2 inhibitors with fluorescence interference and was also successfully employed to identify new protein tyrosine phosphatase domain of SHP2 (SHP2-PTP) and allosteric inhibitors. Of note, this protocol showed potential for identifying SHP2 inhibitors against cancer-associated SHP2 mutation SHP2-E76A. After initial screening of our in-house compound library (∼2300 compounds), we identified 4 new SHP2-PTP inhibitors (0.17% hit rate) and 28 novel allosteric SHP2 inhibitors (1.22% hit rate), of which SYK-85 and WS-635 effectively inhibited SHP2-PTP (SYK-85: IC50 = 0.32 μmol/L; WS-635: IC50 = 4.13 μmol/L) and thus represent novel scaffolds for designing new SHP2-PTP inhibitors. TK-147, an allosteric inhibitor, inhibited SHP2 potently (IC50 = 0.25 μmol/L). In structure, TK-147 could be regarded as a bioisostere of the well characterized SHP2 inhibitor SHP-099, highlighting the essential structural elements for allosteric inhibition of SHP2. The principle underlying the cross-validation protocol is potentially feasible to identify allosteric inhibitors or those inactivating mutants of other proteins.
KEY WORDS: SHP2, High-throughput screening, Enzyme assay, Thermal shift assay, Allosteric inhibitors
Abbreviations: AKT, protein kinase B; ALK, anaplastic lymphoma kinase; AML, acute myelogenous leukemia; Bis-tris, bis-(2-hydroxyethyl)amino-tris(hydroxymethyl)methane; BTLA, B and T lymphocyte attenuator; DiFMUP, 6,8-difluoro-4-methylumbelliferyl phosphate; DiFMU, 6,8-difluoro-4-methylumbelliferyl hydroxid; DTT, dithiothreitol; FI, fluorescence intensity; HEPES, 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid; HTS, high-throughput screening; IC50, half maximal inhibitory concentration; JAK, janus kinase; JMML, juvenile myelomonocytic leukaemia; LB, lysogeny broth; LOC, ligand only control; LS, LEOPARD syndrome; MAPK, mitogen-activated protein kinase; MEK, extracellular regulated protein kinase kinases; NPC, no protein control; NS, Noonan syndrome; OD, optical density; p-IRS1, phosphorylated insulin receptor substrate 1; PD-1, programmed death 1; PI3K, phosphatidylinositol 3 kinase; PMSF, phenylmethanesulfonyl fluoride; PTP, protein tyrosine phosphatase; R2, coefficient of determination; RAS, rat sarcoma; S/B, signal over background; SD, standard deviation; SH2, Src homology 2; SHP2, Src homology phosphotyrosyl phosphatase 2; SHP2-WT, wild type Src homology phosphotyrosyl phosphatase 2; SHP2-PTP, protein tyrosine phosphatase domain of Src homology phosphotyrosyl phosphatase 2; SDS-PAGE, sodium dodecyl sulphate polyacyrlamide gel electrophoresis; STAT, signal transducer and activator of transcription; Tm, melting temperature; ΔTm, melting temperature change
Graphical abstract
This work established a cross-validation screening protocol based on fluorescence-based enzyme assay and conformation-dependent thermal shift assay. An in-house compound library consisting of about 2300 compounds was screened based on the protocol, leading to the discovery of 4 new SHP2-PTP inhibitors and 28 novel allosteric SHP2 inhibitors.
1. Introduction
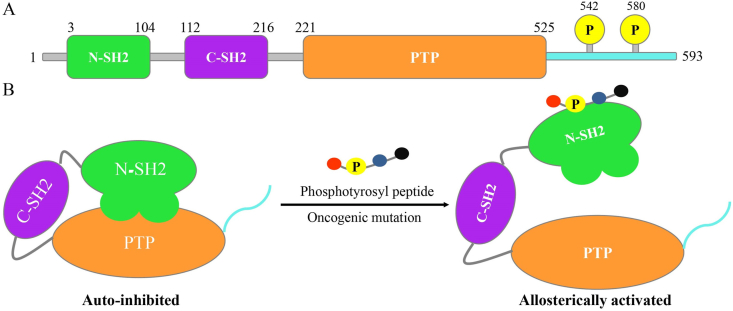
The firstly identified oncogenic protein tyrosine phosphatase, Src homology phosphotyrosyl phosphatase 2 (SHP2), encoded by oncogene PTPN11, is a cytoplasmic non-receptor protein tyrosine phosphatase (PTP) participating in dephosphorylation of phosphoryl-tyrosine1,2. Unlike other phosphatases, SHP2 is a signal-enhancing component that sustains activation of canonical signaling pathways such as rat sarcoma (RAS)-mitogen-activated protein kinase (MAPK), phosphatidylinositol 3 kinase (PI3K)-protein kinase B (AKT), janus kinase (JAK)-signal transducer and activator of transcription (STAT), programmed cell death 1 (PD1), and immune checkpoint (B and T lymphocyte attenuator, BTLA)3, 4, 5. SHP2 contains two tandem Src homology 2 (SH2) domains (N-SH2 and C-SH2), a catalytic PTP, and a poorly ordered C-terminal tail containing two phosphorylation sites (Tyr542 and Tyr580, Fig. 1A). The “molecular switch” allosteric mechanism of SHP2 is unique. Under basal state, SHP2 adopts an inactive and self-blocking conformation with low enzyme activity. X-ray crystal structure of auto-inhibited SHP2 shows that N-SH2 domain packs closely against the PTP domain and blocks the access of substrate to active sites by steric hindrance. Once upon binding to tyrosine-phosphorylated growth factor receptors via its tandem SH2 domains or oncogenic mutation of PTPN11, this specific multi-domain interaction is disrupted accompanied by the dissociation of N-SH2 domain from the PTP domain and the exposure of active sites, then SHP2 is dramatically activated (Fig. 1B)6,7. Germline mutations of PTPN11 can cause developmental disorders such as Noonan syndrome (NS) and LEOPARD syndrome (LS). Somatic mutations within SHP2 are frequently discovered in childhood hematologic malignancies counting the juvenile myelomonocytic leukemia (JMML)2,8,9. Biochemical and functional studies of SHP2 provide insights into the pathogenesis of cancer, suggesting that suppression of SHP2 activity is a promising therapeutic approach for cancer therapy.
Figure 1.
The structural and allosteric regulation mechanism of Src homology phosphotyrosyl phosphatase 2 (SHP2). (A) The secondary structure of SHP2. SHP2 is a protein tyrosine phosphatase with 593 amino acids, containing N-Src homology 2 (SH2) domain (residues 3–104, green), C-SH2 domain (residues 112–216, purple), protein tyrosine phosphatase (PTP) catalytic domain (residues 221–525, orange), and C-terminal tail (residues 526–593, light blue) with two phosphorylation sites at Tyr542 and Tyr580 (yellow). (B) The “molecular switch” allosteric mechanism of SHP2. SHP2 toggles between the auto-inhibited (left) and allosterically activated (right) conformations. Upon binding with a phosphotyrosyl peptide or oncogenic mutations of PTPN11, SHP2 is dramatically activated along with enhanced enzyme activity.
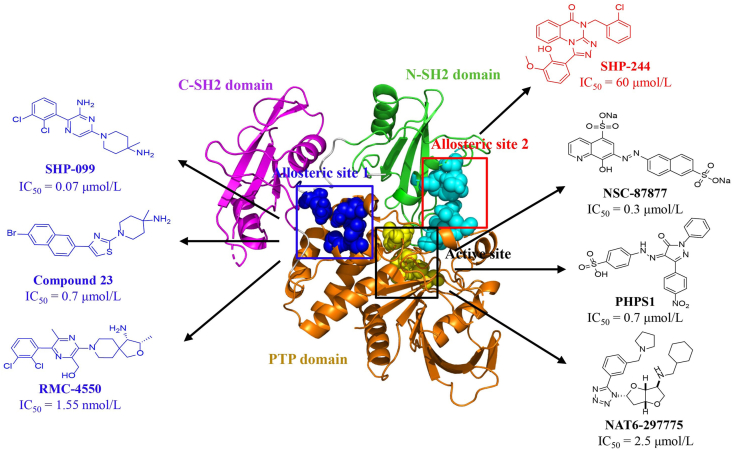
During the past two decades, dozens of SHP2 inhibitors have been identified10, 11, 12, 13, 14, 15 (Fig. 2). However, inhibitors binding to the PTP catalytic site of SHP2 fail to advance into clinical trial due to the poor selectivity and limited bioavailability16,17. Until 2016, discovery of the allosteric inhibitor SHP-099, also known as “molecular glue”, revealed the druggable property of SHP2 by utilizing the auto-regulation allosteric nature of SHP218. SHP-099 serves as a valuable tool for probing the biological functions of SHP2 in normal hematopoiesis and tumorigenesis. SHP-099 in combination with other anti-cancer drugs, such as extracellular regulated protein kinase kinases (MEK), anaplastic lymphoma kinase (ALK) inhibitors, or anti-programmed death 1 (PD-1) antibody, had showed superior therapeutic efficacy than either monotherapy in controlling tumor growth19, 20, 21. According to the allosteric regulation mechanism, the same team also predicted another two allosteric binding pockets and discovered several allosteric compounds. SHP-244, occupying a different allosteric site from SHP-099, could cooperate with SHP-099 to enhance therapeutic effects in cells11. Very recently, Wang et al.22 designed the first potent and effective SHP2 degrader SHP2-D26 based on the proteolysis-targeting chimera (PROTAC) concept, which was 30-fold more potent than SHP099 in inhibition of p-ERK and cell growth. To date, four allosteric SHP2 inhibitors, JAB-3068, JAB-3312, TNO155, and RMC-4630, have progressed into clinical phase1/2 for treating solid tumors4.
Figure 2.
Representative SHP2 (PDB: 2SHP) inhibitors targeting the PTP catalytic active site (black box) and allosteric sites (red and blue boxes).
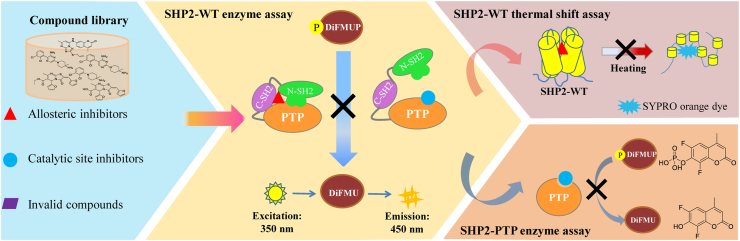
For identifying novel allosteric SHP2 inhibitors, establishment of reliable cross-validation protocols is the key to success. To date, only three research groups have reported the screening protocols10,18,23. Despite of enzyme assay and differential scanning fluorimetry reported by them, several unsolved issues still make them difficult to repeat for other groups. Firstly, the fluorescence interference of substrates or compounds can result in false positive data. Secondly, hit compounds targeting the PTP catalytic active sites should be excluded to identify true allosteric inhibitors of SHP2. Thirdly, there lacks correlation analysis of cross-validation methods to confirm hit compounds. Finally, whether the reported methods are also feasible for multiple cancer-associated SHP2 mutants? Aiming at solving the above problems and accelerating the identification of new SHP2 inhibitors, herein we described a simple, reliable and sensitive cross-validation protocol, which combined a newly designed fluorescence-based enzyme assay and the conformation-dependent thermal shift assay. The established method can effectively exclude the false positive data caused by the fluorescence interference and facilitate the discovery of new SHP2 inhibitors targeting the SHP2-PTP catalytic site and allosteric sites. Based on this cross-validation protocol, we also screened our in-house compound library consisting of over 2300 molecules, leading to the discovery of several types of new allosteric SHP2 inhibitors with the IC50 values up to nanomolar levels (Fig. 3).
Figure 3.
Scheme of the cross-validation protocol for high-throughput screening of new SHP2 inhibitors targeting the PTP active site and allosteric sites. The in-house compound library was firstly screened with wild type SHP2 (SHP2-WT) enzyme assay, which utilized the non-fluorogenic 6,8-difluoro-4-methylumbelliferyl phosphate (DiFMUP) as substrate to characterize the phosphatase activity of SHP2. DiFMUP can be hydrolyzed by SHP2 to remove phosphate group with production of fluorogenic 6,8-difluoro-4-methylumbelliferyl hydroxid (DiFMU), whose fluorescence intensity (FI) could be monitored at 350 nm excitation and 450 nm emission. Then hit compounds would be further cross-validated with the SHP2-WT thermal shift assay and SHP2-PTP enzyme assay respectively to discriminate the new SHP2 inhibitors targeting the PTP active site and allosteric sites. (Red triangle: allosteric inhibitors; Blue circular: catalytic site inhibitors; Purple rhombus: invalid compounds).
2. Materials and methods
2.1. Plasmid construction
The construct of human SHP2-WT lacking of the C-terminal tail was generated by cloning the PTPN11 (accession number NP_002825.3) gene sequence encoding residues Met1–Ser535 into pET-28a (+) expression vector between the Nde I and Xho I restriction sites. The SHP2-PTP construct (residues Ala237–Ser535) was amplified from pET-28a-SHP2-WT construct using forward primer 5ʹ-CGGCAGCCATATGGCTAGCATGGCTGAGACCACAGATAAA-3ʹ and reverse primer 5ʹ-GACTTTATCTGTGGTCTCAGCCATGCTAGCCATATGGCTG-3ʹ by deletion PCR. The cancer-associated SHP2-E76A construct was generated using site-directed mutagenesis with forward primer 5ʹ-AAATTTGCCACTTTGGCTGCGTTGGTCCAGTATTAC-3ʹ and reverse primer 5ʹ-GTAATACTGGACCAACGCAGCCAAAGTGGCAAATTT-3ʹ [synthesized by SunYa (Henan) Ltd., China]. A coding sequence for 6×histidine tag was added to the N-terminus of the SHP2 gene sequence. Constructs identity was verified by DNA sequencing.
2.2. Protein expression and purification
Same procedure was used for the expression and purification of three SHP2 constructs. SHP2 recombinant plasmids were transformed into BL21 Rosetta (DE3) Escherichia coli cells and cultured in lysogeny broth (LB) medium supplemented with 34 mg/L ampicillin and 25 mg/L chloramphenicol. The cells were induced with 0.5 mmol/L isopropyl-β-d-thiogalactoside after growing at 37 °C until the optical density (OD)600 reached 0.6–0.8. After growing overnight at 16 °C, cells were harvested by centrifuging at 7000 rpm (CR22N, HITACHI, Tokyo, Japan) for 8 min.
Cell pellets were resuspended in lysis buffer [20 mmol/L Tris-HCl pH 8.5, 300 mmol/L NaCl, 10 mmol/L β-mercaptoethanol and 2.5 mmol/L phenylmethanesulfonyl fluoride (PMSF)] and lysed by Cell Disrupter (JN-02C, JNBIO) followed by centrifuging at 12,000 rpm (CR22N, HITACHI) for 1 h. Then the supernatant was incubated with Ni2+-NTA agarose resin (QIAGEN, Cat. 1018240, Shanghai, China) for 1 h at 4 °C by gently rocking. After binding, Ni2+-NTA agarose was washed three times with wash buffer (20 mmol/L Tris-HCl pH 8.5, 300 mmol/L NaCl, 5 mmol/L imidazole) and then eluted by elution buffer (20 mmol/L Tris-HCl pH 8.5, 300 mmol/L NaCl, 200 mmol/L imidazole). The protein was subsequently diluted and then purified by Htrap™ Q HP anion exchange column (GE Healthcare) equilibrated with buffer containing 22 mmol/L Tris-HCl pH 8.5 and 2 mmol/L dithiothreitol. The protein was eluted with a 25-column volume of NaCl, linear gradient from 0 to 1 mol/L NaCl. Fractions containing SHP2 protein were pooled and concentrated to 2 mL, and then purified by Hiprep™ 16/60 Sephacryl™ S-200 HR column (GE Healthcare) equilibrated with buffer containing 22 mmol/L Tris-HCl pH 8.5, 220 mmol/L NaCl and 2.2 mmol/L dithiothreitol. Fractions were collected and purity was assessed via sodium dodecyl sulphate polyacyrlamide gel electrophoresis (SDS-PAGE). Pure fractions were concentrated in the buffer [20 mmol/L Tris-HCl pH 7.5, 200 mmol/L NaCl, 2 mmol/L dithiothreitol (DTT) and 5% glycerol], flash cooled in liquid nitrogen, and stored at −80 °C.
2.3. Quantification of basal SHP2 phosphatase activity in the absence of phosphorylated insulin receptor substrate 1 (p-IRS1) peptide
SHP2 phosphatase activity was measured by monitoring the dephosphorylation of the synthetic surrogate substrate 6,8-difluoro-4-methylumbelliferyl phosphate (DiFMUP, Invitrogen, Cat. D22065) to product 6,8-difluoro-4-methylumbelliferyl hydroxid (DiFMU), which has excitation/emission maxima at ∼350/450 nm37. To determine the enzyme concentration linear with velocity, 10 μmol/L DiFMUP was incubated with varying concentration of SHP2 proteins ranging from 1.5 pmol/L to 20 nmol/L to initiate the dephosphorylation reaction. The reaction was proceeded at room temperature in a black, shallow 384-well polystyrene plate (PerkinElmer, Cat. 6008260) for 30 min in the buffer containing 60 mmol/L 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) pH = 7.2, 75 mmol/L KCl, 75 mmol/L NaCl, 1 mmol/L EDTA, 0.05% Tween-20 and 5 mmol/L DTT. The fluorescence signal was continuously monitored using PerkinElmer EnSpire plate reader at 350 nm excitation and 450 nm emission. Raw data were processed with Graphpad Prism 5.0 software using the linear regression to identify the enzyme concentration range linear with velocity. The center of enzyme concentration in linear response was selected and used for the following experiment.
To determine the enzyme kinetics of SHP2 in basal status, SHP2-WT protein was diluted to 0.5 nmol/L in the assay buffer and then added to different concentration of DiFMUP varying from 1 mmol/L to 2.0 μmol/L. DiFMUP dephosphorylation was measured as above. All the reactions were performed in triplicate. Raw data were converted to DiFMU/time and normalized to enzyme concentration to produce relative velocity. Velocity data were fitted by Michaelis–Menton equation following Eq. (1) to extrapolate kinetic parameters:
| (1) |
where V is enzyme velocity, Vmax is maximal enzyme velocity, KM is the Michaelis–Menton constant, [S] is substrate concentration.
2.4. Quantification of SHP2 phosphatase activity in the presence of p-IRS1 peptide
To determine the SHP2 phosphatase activity upon bis-phosphorylated peptide p-IRS1 [sequence: H2N-LN(pY)IDLDLV(dPEG8)LST(pY)ASINFQK-amide] stimulation, pre-incubate 1.65 nmol/L SHP2-WT with varying concentration of p-IRS1 peptide [synthesized by GL Biochem (Shanghai) Ltd., China] ranging from 0.02 to 10 μmol/L. Then DiFMUP dephosphorylation under activated condition was quantified using PerkinElmer EnSpire plate reader at 350 nm excitation and 450 nm emission as above. Raw data were converted to DiFMU/time and then fitted to Eq. (2) following three-parameter dose–response curve to extrapolate EC50 values:
| (2) |
where V is enzyme velocity, basal is the basal response, max is the maximal response, EC50 is the substrates concentration that yields a response representing 50% of the difference between maximal and basal and [S] is substrate concentration.
2.5. Enzyme assay
To quantify the potency of test compounds in inhibiting the enzyme activity of SHP2-WT, we utilized the surrogate substrate DiFMUP to measure the catalytic activity of SHP2 by modified end-point fluorescence enzyme assay to determine IC50 values of test compounds. The dephosphorylation reaction was performed in black, shallow 384-well polystyrene plate at room temperature. The total reaction volume was set as 20 μL per well. To keep the DMSO concentration at a low level, test compounds were firstly serially diluted in DMSO in 96-well clear V-bottom polypropylene plates (Beyotime Cat. FPT019) and then diluted 10-fold into reaction buffer (60 mmol/L HEPES pH 7.2, 75 mmol/L NaCl, 75 mmol/L KCl, 1 mmol/L EDTA, 0.05% Tween-20, and 5 mmol/L DTT), to achieve 10-fold higher than the final DMSO concentration of detection system. 0.5 nmol/L SHP2 was pre-incubated with 1.0 μmol/L p-IRS1 peptide for 5–10 min to activate the enzyme, followed by addition of different concentrations of compounds varying from 0.0003 to 10 μmol/L or DMSO as control. The reaction was incubated for 60 min at room temperature before the substrates 10 μmol/L DiFMUP was added. The enzyme velocity was measured by monitoring fluorescence signal change that reflected accumulation of product DiFMU at room temperature using multi-mode plate reader with excitation and emission wavelengths at 350 and 450 nm, respectively. The relative fluorescence intensity (FI) was obtained by subtracting the initial (0 min) FI value from the ending (30 min) FI value. Then inhibitor dose–response curves were plotted using normalized IC50 regression curve fitting with control-based normalization to extrapolate IC50 values.
This modified end-point fluorescence enzyme assay was also explored to measure the inhibition potency of test compounds against SHP2-PTP. Different from SHP2-WT, p-IRS1 peptide wasn't used because the enzyme activity of SHP2-PTP had no difference in the presence or absence of p-IRS1 peptide. Therefore, 0.14 nmol/L SHP2-PTP was directly incubated with small-molecule inhibitors, varying from 0.1 to 50 μmol/L. The enzyme reaction was initiated by dispensing 100 μmol/L DiFMUP into reaction system and measured as above.
SHP2-E76A enzyme assay was performed as the same protocol. The total reaction volume was set as 20 μL per well containing 8 μL 0.15 nmol/L SHP2-E76A, 2 μL SHP-099 (varying from 0.3 nmol/L to 6 μmol/L) and 50 μmol/L DiFMUP to determine the IC50 value of SHP-099 against SHP2-E76A.
2.6. Thermal shift assay
Purified SHP2-WT and SYPRO orange dye (5000 × concentrated in DMSO, Life Technologies, Cat. S6650) were diluted in the assay buffer containing 100 mmol/L bis-(2-hydroxyethyl)amino-tris(hydroxymethyl)methane (Bis-tris) pH 6.5, 100 mmol/L NaCl, and 1 mmol/L DTT. The reaction system was set as 20 μL containing 2 μL 1.8 μmol/L SHP2-WT, 2 μL 7.5 × SYPRO orange dye and 2 μL 100 μmol/L or 50 μmol/L compounds or 1% DMSO used as control. The PCR plates were sealed after all the reagents were added and then centrifuged at 2000 rpm for 30 s. Thermal scanning was performed using the real-time PCR instrument (Quantstudio™6 Flex System, Thermo Fisher Scientific) with excitation at 465 nm and emission at 580 nm. The temperature was ramped from 25 to 75 °C with 0.05 °C/s increment. The FI was measured every second during the same time melting curves were recorded. Then the raw data were fitted to Boltzmann sigmoidal equation to calculate the melting temperature (Tm). Curve fitting, melting temperature calculation and report generation based on the raw data were performed as the paper reported. Through comparing the melting temperature change (ΔTm) between apo-SHP2 and SHP2 in complex with compound, ΔTm value can be determined. The Tm values are the average of three individual experiments and presents as mean ± standard deviation (SD, n ≥ 3). SHP2-E76A thermal shift assay were performed as the same protocol.
2.7. Cell proliferation assay
Cells (3000 cells/well) of MDA-MB-231/HeLa cell line were seeded in 96-well plates in 200 μL media (RPMI-1640/DMEM containing 10% fetal bovine serum, Israel) and grown overnight. Then, cells were synchronized for 6 h and treated with SHP2 inhibitors at gradient concentrations [0 (control), 0.625, 1.25, 2.5, 5, and 10 μmol/L for WS-635 and TK147, and 0 (control), 62.5, 125, 250, 500, and 1000 nmol/L for SHP-099] for 24, 48, and 72 h. Finally, cell viability was examined using MTT and DMSO. MTT reduction was quantified by measuring the absorbance at 490 nm (OD).
2.8. Other experimental details
The details about the assay robustness evaluation and data analysis and statistics are provided in the Supporting Information.
3. Results and discussion
3.1. Purification and biochemical characterization of SHP2 recombinant proteins
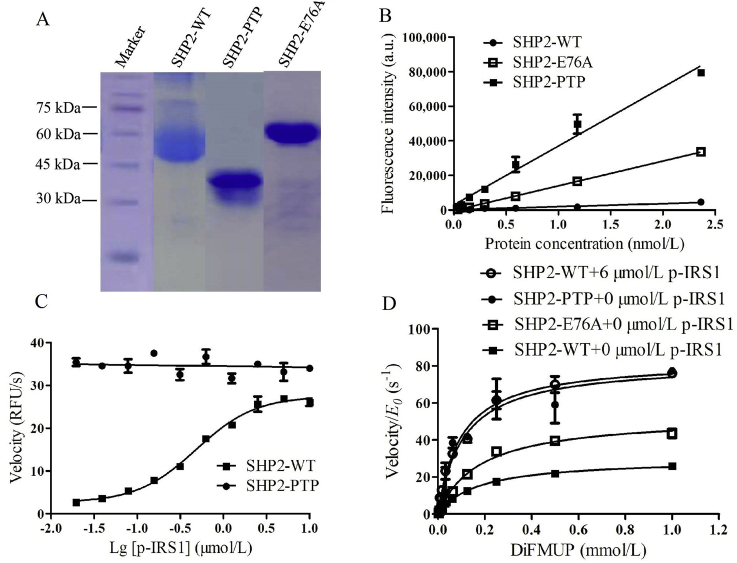
The his-tagged SHP2 recombinant proteins were expressed in E. coli cells. Three of them were purified via affinity purification, ion exchange chromatography and size-exclusion chromatography following previously reported procedures18. Purity and molecular weight were identified by SDS-PAGE (Fig. 4A). To characterize the enzyme activity of these proteins, dephosphorylation level was measured by monitoring fluorescence signal change resulting from the conversion of DiFMUP to DiFMU. As expected, the result of enzyme titration experiment showed the descending trend of dephosphorylation activity as SHP2-PTP > SHP2-E76A > SHP2-WT, in the absence of p-IRS1 peptide (Fig. 4B). While upon p-IRS1 peptide stimulation, the enzyme activity of SHP2-WT was gradually improved with the increasing concentration of peptide until saturation. Two proteins demonstrated similar dephosphorylation activity, and the EC50 value of p-IRS1 peptide for SHP2-WT was 0.448 ± 0.05 μmol/L (previously reported value was 0.25 μmol/L24) even though the PTP domain was unsusceptible to tyrosine-phosphorylated peptide (Fig. 4C). In addition, the steady state analysis of enzyme kinetics was conducted to evaluate the binding affinity of substrate, DiFMUP, toward SHP2-PTP, SHP2-E76A, and SHP2-WT under basal and fully activated state. The Michaelis–Menten kinetic parameters of DiFMUP dephosphorylation were shown in Table 1. Results showed that the KM value for DiFMUP under fully activated SHP2-WT was 0.098 ± 0.01 mmol/L (KM = 0.1 mmol/L in the literature18,24), similar to that of SHP2-PTP (Fig. 4D).
Figure 4.
Biological characterization of his-tagged SHP2 recombinant proteins. (A) Purity and molecular weight analysis of his-tagged SHP2-WT (61.4 kDa), SHP2-PTP (35.0 kDa) and SHP2-E76A (61.4 kDa) proteins by Coomassie Brilliant Blue staining. (B) Basal enzyme activity comparison between SHP2 proteins in the absence of p-IRS1 peptide. 10 μmol/L DiFMUP was titrated with different concentration of protein ranging from 0.037 to 2.36 nmol/L. (C) Activated enzyme activity comparison between SHP2-WT and SHP2-PTP enzymes. 10 μmol/L DiFMUP and 0.5 nmol/L enzyme were titrated with different concentration of p-IRS1 peptide ranging from 0.02 to 10 μmol/L. (D) Enzyme kinetic parameters of DiFMUP dephosphorylation by SHP2-PTP (0.225 nmol/L), SHP2-E76A (0.15 nmol/L), and SHP2-WT (0.5 nmol/L) under basal (0 μmol/L p-IRS1 peptide) and fully activated (6 μmol/L p-IRS1 peptide) condition. Data are shown as mean ± SD; n ≥ 3.
Table 1.
Michaelis–Menten parameters of DiFMUP dephosphorylation by SHP2-E76A, SHP2-PTP, and SHP2-WT under fully activated condition.
| Protein | KM (mmol/L) | Kcat (s−1) | Kcat/KM (L/mol·s) |
|---|---|---|---|
| SHP2-E76A | 0.177 ± 0.02 | 52.90 ± 1.78 | 2.99 × 105 |
| SHP2-PTP | 0.108 ± 0.02 | 81.77 ± 5.84 | 7.57 × 105 |
| SHP2-WT | 0.098 ± 0.01 | 83.08 ± 2.72 | 8.45 × 105 |
Raw data shown as mean ± SD (n ≥ 3) are fitted to the standard Michaelis–Menten equation to extrapolate Kcat and KM values.
Kcat: the first-order rate constant; KM: the Michaelis–Menton constant.
3.2. Enzyme assay of SHP2-WT
Under basal condition, the N-SH2 domain of SHP2 inserts a loop into the conserved PTP catalytic domain. SHP2 stays in closed and auto-inhibited conformation which spatially blocks substrate access to the catalytic domain. While upon stimulation of phosphorylated tyrosine peptide or variant cancer-associated mutations locating in the auto-inhibited interface between the N-SH2 and PTP domains, N-SH2 is released from the PTP domain. The active sites involved in substrate recognition and dephosphorylation are exposed6,7. Then the phosphatase activity of SHP2 will increase as a result of large conformational change, which causes dysregulation of relevant signaling pathways and even promotes tumorigenesis24.
To discover novel inhibitors that block SHP2 in auto-inhibited conformation, fluorescence-based enzyme assay was previously reported using DiFMUP as substrate. However, the fluorescence interference problem of substrates or compounds is still a big challenge. Therefore, newly designed enzyme assay with kinetic readouts was developed based on the principle that kinetic measurement was independent on the initial fluorescence of assay25,26. Two time points within the initial velocity phase of the reaction, rather than one end-point, were chosen to measure the FI in our assay. The relative FI was obtained through subtracting the initial (0 min) FI value from the ultimate (30 min) FI value. Then the raw data were processed to extrapolate IC50 value of the compound. Specially, SHP2-WT (1–535) protein that functioned alike full-length protein but only lacked C-terminal tail and p-IRS1 peptide were used to imitate the enzyme activity of SHP2 under pathologic condition, which was gain-of-function mutation in most cases. Fluorogenic substrate DiFMUP was applied to initiate the dynamic enzyme reaction.
The enzyme reaction was influenced by various factors, including enzyme and substrate concentration, incubation time, buffer condition, and so on26. So we optimized these factors in turn to set up the enzyme assay. As shown in Fig. 5A, the range of linear concentration of SHP2-WT protein was narrowed with the increasing concentration of p-IRS1 peptide. In consideration of linear enzyme concentration and to avoid the ‘assay wall’ phenomenon, 0.5 nmol/L SHP2-WT was primarily set as enzyme concentration used for the following experiments. The substrate DiFMUP concentration and incubation time were further optimized through substrate titration and time course experiments based on the signal over background ratio (S/B). Results showed that the fluorescence signal increased linearly within 30 min under each tested substrate concentration and 10 μmol/L DiFMUP was enough to yield relatively high S/B ratio for high-throughput screening (Fig. 5B). Thus, 30 min incubation time and 10 μmol/L DiFMUP were selected in consideration of efficiency and economy. Further optimization was the concentration of p-IRS1 peptide. In order to achieve approximately 50% activation of SHP2 (a dynamic equilibrium between active and inactive conformation of SHP2 with the ratio of 1:1), 1.0 μmol/L p-IRS1 peptide was selected (Fig. 4C). To further evaluate the stability of SHP2-WT enzyme assay for high-throughput screening (HTS), we also tested the DMSO tolerance up to 5%. Results showed that the DMSO concentration didn't have statistically significant difference on SHP2 enzyme activity (Fig. 5C). To avoid protein denaturation, we chose a relatively low DMSO concentration (1%). Final enzyme assay for SHP2-WT was performed containing 0.5 nmol/L SHP2-WT, 1.0 μmol/L p-IRS1 peptide, 10 μmol/L DiFMUP, and varying concentrations of compounds. The kinetic signal was measured at two time points (0 and 30 min). The data were normalized and then Z factor and S/B were calculated for each plate. In addition, the well-known SHP2 allosteric inhibitor SHP-099 was used as the control compound to verify the feasibility of the enzyme assay. Consistent with the data reported previously, SHP-099 potently inhibited SHP2-WT with an IC50 value of 0.069 ± 0.005 μmol/L (0.07 μmol/L reported in the literature18, Fig. 5D).
Figure 5.
SHP2-WT enzyme assay. (A) Linear titration experiment of SHP2-WT under different concentrations of p-IRS1 peptide. 10 μmol/L DiFMUP was titrated with varying concentrations of SHP2-WT ranging from 0.065 to 16.62 nmol/L under different concentration of p-IRS1 peptide. (B) Substrate DiFMUP titration and incubation time optimization. 0.5 nmol/L SHP2-WT was titrated with different concentrations of DiFMUP ranging from 6.25 to 200 μmol/L. The dephosphorylation reaction signal was continuously monitored for 30 min. (C) DMSO concentration optimization. DMSO concentration ranging from 1% to 5% was tested to evaluate the tolerance of SHP2-WT enzyme assay toward DMSO. (D) SHP-099 verified the reliability of SHP2-WT enzyme assay. (E) Assay robustness evaluation through Z score and the signal over background ratio (S/B) analysis. A series of raw data were derived from experiments conducted at the same time and then fitted to Z score normalization and S/B analysis. Data are shown as mean ± SD; n ≥ 3.
The prerequisite of acceptable assay performance for a high-throughput screening method is the robustness and reliability. To make sure the enzyme assay developed by us is suitable for high-throughput screening for discovering SHP2 allosteric inhibitors, the raw data were normalized and then Z factor and S/B were calculated for each plate. The assay performance and robustness were identified reliable with the average Z score of 0.75 and S/B ratio of 5.07 (Fig. 5E). All of these results suggested that the enzyme assay of SHP2-WT established by us was reliable and can be used for high-throughput screening.
3.3. Thermal shift assay of SHP2-WT
Thermal shift assay is a general method used for identifying new bioactive compounds, which preferentially reflects the physical property of purified protein based on the specific conformational change caused by the binding of low-molecule-weight ligand during thermal denaturation. An environmentally sensitive dye is used to monitor the transition of protein conformation. The dye favors binding with the exposed hydrophobic region of a protein while it unfolds upon heating27,28. Thus, protein denaturation can be monitored by the fluorescence change induced by protein–dye interaction, which generates a thermal melting curve measured by the real-time PCR machine. Then the Tm value denoted as the midpoint of melting curve can be calculated using the previously reported protocol29. ΔTm values are correlated to the protein conformational stability. Upon binding to a protein of interest, the ligand has the propensity to increase the protein thermal stability. Difference of melting temperature (sensitive to the concentration and binding affinity of ligands) between protein and protein–ligand complex can be calculated to reflect the potency of the compound against protein of interest28. SHP2 is a suitable candidate due to its thermally stable property and unique allosteric regulation mechanism. Therefore, the thermal shift assay is chosen to cross validate the inhibitory activity of the hit compounds in the enzyme assay.
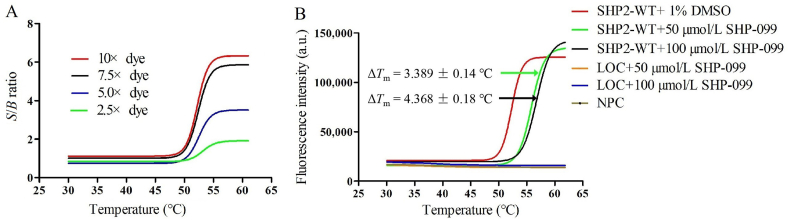
Several assay conditions that influence the fluorescence signal such as buffer condition and protein/dye ratio were optimized. The most important property of the buffer is capable of stabilizing target protein from denaturation or inactivation during the cycle of freezing and thawing. Moreover, buffer should also be chemically inactive and unreactive with the dye or other components in the reaction system. And the temperature-dependent reagent Tris-HCl is not recommended because of its high coefficient of ΔpH/ΔT30. So in the experiment the Bis-Tris buffer was used instead of the Tris-HCl buffer. Subsequently, a panel of dye concentration was titrated with fixed protein concentration to identify the optimal ratio of protein and dye for excellent assay performance with high S/B ratio. Results showed that 7.5 × concentrated dye was sufficient to generate acceptable S/B ratio (Fig. 6A). So the final reaction was performed with 1.8 μmol/L SHP2-WT, 7.5 × SYPRO orange dye and different concentrations of compound (50 and 100 μmol/L) in the buffer condition containing 100 mmol/L Bis-tris pH 6.5, 100 mmol/L NaCl, and 1 mmol/L DTT. For the thermal profile, it is believed that the ramp speed and ramp rate affect protein unfolding and the resulting melting curves30. To obtain well-resolved melt phases, the ramp speed and ramp rate were further optimized. Results showed that 25–75 °C and 0.05 °C/s were suitable for SHP2-WT. The reaction system was set as 20 μL containing 2 μL 1.8 μmol/L SHP2-WT, 2 μL 7.5 × SYPRO orange dye and 2 μL 50 or 100 μmol/L compounds or 1% DMSO used as control. Both ligand only control (LOC) and no protein control (NPC) were used as negative control to exclude the contamination in wells or protein melting reactions and ligand–dye interactions interference. SHP-099 was also used to verify the reliability of the thermal shift assay. The ΔTm value induced by 100 μmol/L of SHP-099 was beyond 4 °C, consistent with that previously reported11, indicating that the established thermal shift assay is reliable (Fig. 6B).
Figure 6.
SHP2-WT thermal shift assay. (A) Protein/dye ratio optimization. Different protein/dye ratios were tested and the best one was chosen based on the principle of cost-saving and acceptable S/B ratio (S/B ≥ 4). (B) Thermal shift assay reliability verification by SHP-099. The unchanged curves of no protein control (NPC) and ligand only control (LOC) suggested there were no contamination in wells or protein melting reactions and ligand–dye interactions interference. Two concentrations (50 and 100 μmol/L) of SHP-099 were tested, whose ΔTm values were 3.389 and 4.368 °C, respectively. Data are shown as mean ± SD; n ≥ 3.
3.4. Enzyme assay of SHP2-PTP
The catalytic domain of SHP2 is conserved among the PTPs family. SHP2 shares more than 75% sequence homology of phosphatase domain with SHP16. Besides, PTP active sites are characterized by its highly solvated and positively charged nature, which easily attracts negatively charged molecules. Thereby, inhibitors targeting PTP active sites suffer from the poor selectivity and limited bioavailability16. In contrast, allosteric inhibitors are believed to possess improved selectivity through targeting the less-conserved binding pocket beyond the active site. Thus, development of allosteric SHP2 inhibitors has been highly pursued for cancer therapy.
To obtain allosteric SHP2 inhibitors, inhibitors targeting the PTP active site should be firstly removed from the hit compounds derived from SHP2-WT enzyme assay through examining whether these inhibitors could block the dephosphorylation activity of SHP2-PTP. The dephosphorylation activity was further measured utilizing the SHP2-PTP protein in the absence of p-IRS1 peptide. The linear enzyme concentration and substrate concentration were also optimized as the way in the SHP2-WT enzyme assay (Figure 4, Figure 7D). 0.125 nmol/L SHP2-PTP, 100 μmol/L DiFMUP, and varying concentrations of compounds were used in the SHP2-PTP enzyme assay. A nonspecific phosphatase inhibitor Na3VO4 was used as control compound to verify the reliability of the assay. The IC50 value was 11.85 ± 0.26 μmol/L, similar to that (10 μmol/L) reported previously31 (Fig. 7B).
Figure 7.
SHP2-PTP enzyme assay. (A) Linear enzyme concentration optimization of SHP2-PTP. 10 μmol/L DiFMUP was titrated with varying concentrations of SHP2-PTP ranging from 0.13 to 16.88 nmol/L. (B) Reliability verification of SHP2-PTP enzyme assay by control compound Na3VO4. Data are shown as mean ± SD; n ≥ 3.
3.5. Assay performance and correlation analysis
The goodness-of-fit coefficient of determination (R2) was used to evaluate the assay performance. Duplicate experiments of 302 compounds in the SHP2-WT enzyme assay showed a high degree of correlation with an R2 of 0.82 (Fig. 8A). The IC50 and ΔTm, obtained from the enzyme assay and thermal shift assay correspondingly, were also processed to determine the correlation of these two cross-validation assays. Interestingly, allosteric site and PTP active site inhibitors of SHP2 showed totally different correlation between two parameters. For allosteric inhibitors (black dots in Fig. 8B), these two parameters showed superior correlation with the R2 of 0.82, suggesting the reliability of two cross-validation protocols. While for PTP active site inhibitors (red dots in Fig. 8B), no significant correlation was observed (Fig. 8B). This result can be explained by the fact that allosteric inhibitors stabilized the basal SHP2-WT conformation during thermal denaturation and inhibited the enzyme activity by blocking SHP2 with an inactive and self-blocking conformation, so compounds showing higher potency in the enzyme assay also had higher positive ΔTm value in thermal shift assay. In contrast, compounds targeting the SHP2 catalytic domain cannot stabilize the basal conformation of SHP2 reflected by no obvious Tm value change. However, it may demand further statistical analysis based on more experimental data of active site inhibitors to verify the above hypothesis. Collectively, we successfully established two highly correlated high-throughput screening protocols (fluorescence-based enzyme assay and thermal shift assay) based on the characteristically physiochemical property of SHP2. These two protocols can be used to exclude false positive compounds with fluorescence interference and then discriminate the active site and allosteric site inhibitors of SHP2, which facilitates the subsequently targeted structural optimization.
Figure 8.
Assay performance and correlation analysis. (A) Assay performance and correlation analysis of 302 compounds tested at 10 μmol/L in SHP2-WT enzyme assay. Data represented by inhibition rate were obtained from two independent experiments. (B) Correlation analysis of two cross-validation high-throughput screening assays through a panel of hit compounds derived from subsequent screening of our in-house compound library. Red dots represent active site inhibitors identified from SHP2-PTP enzyme assay. Black dots represent allosteric site inhibitors cross-validated through SHP2-WT enzyme assay and thermal shift assay.
3.6. Expanding cross-validation screening protocol toward SHP2 mutants
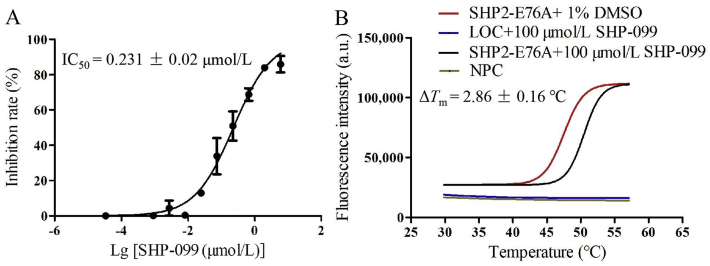
Oncogenic mutations of PTPN11 are frequently observed in residues mainly locating in the interface between N-SH2 and PTP domains of SHP2. These mutations weaken the interaction of multi-domain interface and enhance the enzyme activity of SHP2. So activating mutations of SHP2 are the leading cause of serious diseases, such as D61Y-associated acute myelogenous leukemia (AML), E76A-associated juvenile myelomonocytic leukemia (JMML), and E76D-associated NS9. Among these mutations, residue E76 of the N-SH2 domain is the most frequently occurred and highly activated mutation. Crystallographic studies showed that E76 mutation yielded a weaker and less negative multi-domain interaction, leading to N-SH2 domain shifting away from PTP domain and improving the enzyme activity of SHP2 mutants24. Biochemical studies indicated that compared with SHP2-WT, the inhibitory potency of SHP-099 against gain-of-function SHP2 mutants declined. These findings suggest that development of effective SHP2 inhibitors targeting the oncogenic SHP2 mutants in cancer is in high demand7,32. However, it is of great challenge to establish enzyme assay for identifying effective inhibitors against SHP2 mutants because of the enhanced reaction velocity and contracted initial velocity phase. Besides, the structures of GOF SHP2 mutants have propensity to adopt open conformation, making it difficult to establish thermal shift assay for the stability of apo-protein. In order to verify whether the cross-validation screening protocol originally established for SHP2-WT is feasible to SHP2-E76A mutation, we examined the inhibition potency of SHP-099. As shown in Fig. 9, the IC50 and ΔTm values of SHP-099 against SHP2-E76A were 0.231 ± 0.02 μmol/L and 2.86 ± 0.16 °C, respectively, similar to that (IC50 = 0.124 μmol/L) previously reported10. Therefore, this established cross-validation protocol was also suitable for SHP2-E76A despite of the enhanced enzyme activity. It could be potentially used to identify new SHP2 inhibitors against cancer-associated SHP2 mutations including SHP2-E76A.
Figure 9.
The feasibility of the cross-validation screening protocol against SHP2-E76A characterized by SHP-099. The IC50 and ΔTm value were 0.231 μmol/L (A) and 2.86 °C (B), respectively. Data were shown as mean ± SD; n ≥ 3.
3.7. Discovery of new SHP2 inhibitors targeting the SHP2-PTP catalytic and allosteric sites based on the cross-validation high-throughput screening protocol
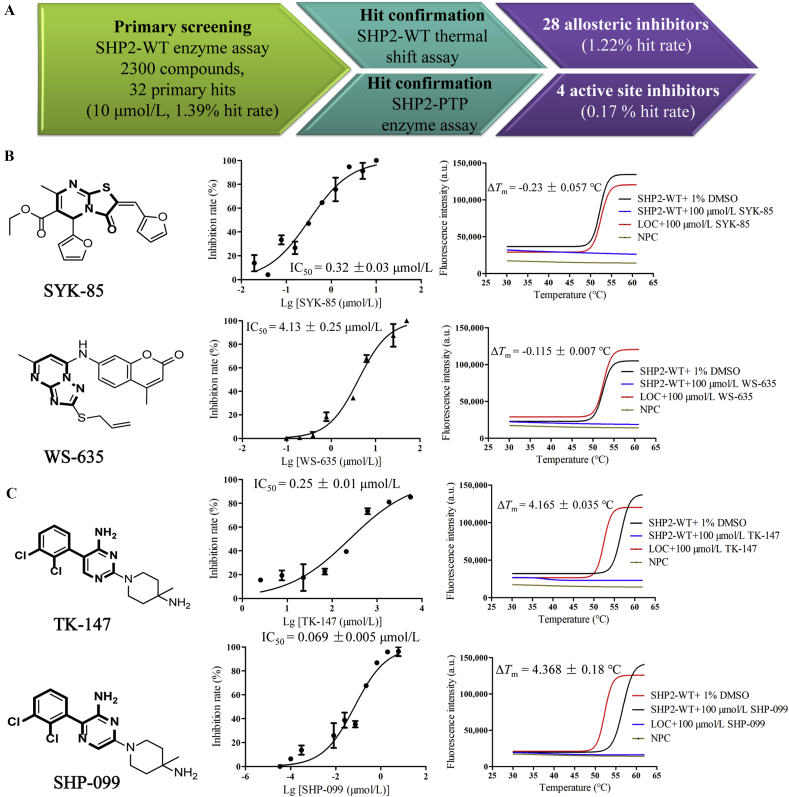
Having the protocol established, we first screened our in-house compound library containing approximately 2300 compounds using the SHP2-WT enzyme assay at the concentration of 10 μmol/L, 70 hit compounds with the inhibition rate over 30% were identified (∼3% of hit rate). Interestingly, 32 hit compounds showed over 50% of inhibition rate against SHP2-WT and were further cross-validated by the thermal shift assay and the SHP2-PTP enzyme assay. Compounds with negative ΔTm values in thermal shift assay also potently inhibited SHP2-PTP and were considered to be SHP2-PTP inhibitors, while compounds with positive ΔTm values didn't inhibit the enzymatic activity of SHP2-PTP and were identified as new allosteric SHP2 inhibitors, the corresponding hit rate was ∼1.22% (Fig. 10A). To our delight, four compounds showed acceptable potency against SHP2-PTP (IC50 < 5.0 μmol/L), among which SYK-85 showed the best inhibitory activity against SHP2-PTP with an IC50 value of 0.32 ± 0.03 μmol/L and the ΔTm value of −0.23 ± 0.057 °C (Fig. 10B), WS-635 bearing the privileged coumarin and [1,2,4]triazolo[1,5-a]pyrimidine scaffold also inhibited SHP2-PTP moderately with an IC50 value of 4.13 ± 0.25 μmol/L and the ΔTm value of −0.115 ± 0.007 °C (Fig. 10B). These two compounds represent novel structural skeletons for designing new SHP2-PTP inhibitors. In contrast, 28 compounds were found to be allosteric SHP2 inhibitors with superior potency (IC50 < 1.0 μmol/L, 0.74% hit rate). Among these allosteric inhibitors, TK-147 displayed good potency (IC50 = 0.25 ± 0.01 μmol/L), but was about three-fold less potent than SHP-099 (IC50 = 0.069 ± 0.005 μmol/L). In the thermal shift assay, the ΔTm value for TK-147 was 4.165 ± 0.035 °C, comparable to that (4.368 ± 0.18 °C) of SHP-099 (Fig. 10C). In structure, TK-147 could be considered as a bioisostere of SHP-099, which may be responsible for its allosteric regulation potency. The structural features of TK-147 and SHP-099 may suggest that the biologically relevant biaryl scaffold may be a promising substructure for designing new allosteric SHP2 inhibitors33, 34, 35, 36. Collectively, based on the established cross-validation protocol, we identified 4 SHP2-PTP inhibitors and 28 allosteric SHP2 inhibitors with good potency from our in-house compound library. We believe that SYK-85, WS-635, and TK-147 could be used as templates for designing new SHP2-PTP and allosteric SHP2 inhibitors, respectively. Further structural modifications are undergoing in our lab and will be reported in due course. Other bioactive compounds against SHP2 identified in this work are not shown here.
Figure 10.
Discovery of new SHP2 inhibitors targeting the SHP2-PTP catalytic and allosteric sites. (A) High-throughput screening workflow for the identification of new SHP2 inhibitors. (B) Cross-validation of new SHP2-PTP inhibitors SYK-85 and WS-635 through the SHP2-PTP enzyme assay and thermal shift assay. (C) Cross-validation of the allosteric inhibitor TK-147 and SHP-099 through SHP2-WT enzyme assay and thermal shift assay. The core scaffolds are highlighted in bold. Data are shown as mean ± SD; n ≥ 3.
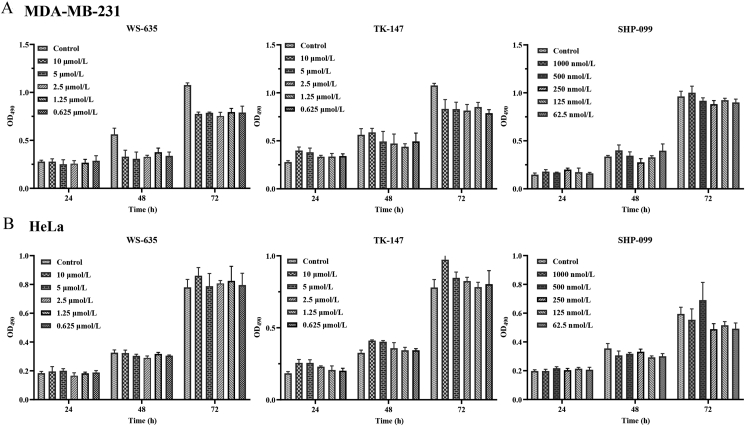
To determine whether hit compounds have effects on cell growth, SHP2-dependent MDA-MB-231 and HeLa cells were treated with gradient concentrations of test compounds38,39. Results shown in Fig. 11 suggested that three compounds WS-635, TK-147, and SHP-099 had no significant effects on the proliferation of MDA-MB-231 and HeLa cells in each test concentration. These results are consistent with previous reports that SHP2 inhibitors do not function by affecting the proliferation of tumor cells22.
Figure 11.
Compounds do not inhibit cancer cell proliferation in vitro. (A) MDA-MB-231 and (B) HeLa cells were treated with compounds at gradient concentrations [0 (control), 0.625, 1.25, 2.5, 5, 10 μmol/L for WS-635 and TK147, and 0 (control), 62.5, 125, 250, 500, 1000 nmol/L for SHP-099] for 24, 48, and 72 h. Cell viability was examined by MTT. MTT reduction was quantified by measuring the absorbance at 490 nm (optical density, OD). Data are shown as mean ± SD; n = 3.
4. Conclusions
SHP2 is the firstly identified oncoprotein of protein tyrosine phosphatases family whose dysregulation is the leading cause of several cancers. Suppression of SHP2 activity is a promising therapeutic approach for cancer therapy. In this work, we established the cross-validation screening protocol based on the fluorescence-based enzyme assay and conformation-dependent thermal shift assay. The cross-validation protocol could exclude the false positive compounds with fluorescence interference and can also be used to identify new SHP2 inhibitors targeting the SHP2-PTP catalytic active site and allosteric site. Particularly, this protocol was also viable for identifying SHP2 inhibitors against cancer-associated SHP2 mutations including SHP2-E76A. SHP-099 potently inhibited SHP2-E76A with an IC50 value of 0.231 ± 0.02 μmol/L and the ΔTm of 2.86 ± 0.16 °C. Based on the protocol, we screened our in-house compound library consisting of about 2300 compounds, leading to the discovery of 4 new SHP2-PTP inhibitors (IC50 < 5.0 μmol/L, 0.17% hit rate) and 28 novel allosteric SHP2 inhibitors (up to nanomolar levels, 1.22% hit rate). SYK-85 and WS-635 potently inhibited SHP2-PTP (SYK-85: IC50 = 0.32 ± 0.03 μmol/L, ΔTm = −0.23 ± 0.057 °C; WS-635: IC50 = 4.13 ± 0.25 μmol/L, ΔTm = −0.115 ± 0.007 °C) and thus represent novel scaffolds for designing new SHP2-PTP inhibitors. Among these allosteric inhibitors, TK-147 demonstrated good inhibition against SHP2 (IC50 = 0.25 ± 0.01 μmol/L, ΔTm = 4.165 ± 0.035 °C), comparable to those of the well characterized SHP2 inhibitor SHP-099. Like SHP-099, TK-147 features a biaryl scaffold, which may be responsible for the allosteric regulation activity against SHP2. The general principle underlying the cross-validation protocol could be potentially employed to identify new inhibitors binding to allosteric sites or inhibiting mutant forms of proteins of interest.
Acknowledgments
We sincerely acknowledge the financial support from the National Natural Science Foundation of China (Nos. 31900875, 81773562, 81973177, and 81703326), Program for Science & Technology Innovation Talents in Universities of Henan Province (No. 21HASTIT045, China), China Postdoctoral Science Foundation (Nos. 2019M662518, 2018M630840, and 2019T120641, China), and Postdoctoral Starting Foundation of Henan Province (No. 201903007, China). We thank Prof. Stephen C. Blacklow at Department of Biological Chemistry & Molecular Pharmacology of Harvard Medical School (Boston, MA, USA) for providing useful suggestions in designing the enzyme assay, and Kai Tang, Yunkai Shi, and Shuai Wang (Zhengzhou University, China) for providing the compound library for screening.
Author contributions
Yihui Song designed the experiments. Min Zhao and Yahong Wu performed the experiment and analyzed the data. Yihui Song and Min Zhao wrote the draft. Bin Yu and Yihui Song revised and submitted the manuscript on behalf of other authors. Hong-Min Liu provided the financial support for this project.
Conflicts of interest
The authors have no conflicts of interest to declare.
Footnotes
Peer review under responsibility of Institute of Materia Medica, Chinese Academy of Medical Sciences and Chinese Pharmaceutical Association.
Supporting data to this article can be found online at https://doi.org/10.1016/j.apsb.2020.10.021.
Contributor Information
Bin Yu, Email: yubin@zzu.edu.cn.
Hong-Min Liu, Email: liuhm@zzu.edu.cn.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Chan R.J., Feng G.S. PTPN11 is the first identified proto-oncogene that encodes a tyrosine phosphatase. Blood. 2007;109:862–867. doi: 10.1182/blood-2006-07-028829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ostman A., Hellberg C., Bohmer F.D. Protein-tyrosine phosphatases and cancer. Nat Rev Canc. 2006;6:307–320. doi: 10.1038/nrc1837. [DOI] [PubMed] [Google Scholar]
- 3.Yang W., Klaman L.D., Chen B., Araki T., Harada H., Thomas S.M. An Shp2/SFK/Ras/Erk signaling pathway controls trophoblast stem cell survival. Dev Cell. 2006;10:317–327. doi: 10.1016/j.devcel.2006.01.002. [DOI] [PubMed] [Google Scholar]
- 4.Liu Q., Qu J., Zhao M., Xu Q., Sun Y. Targeting SHP2 as a promising strategy for cancer immunotherapy. Pharmacol Res. 2020;152:104595. doi: 10.1016/j.phrs.2019.104595. [DOI] [PubMed] [Google Scholar]
- 5.Marasco M., Berteotti A., Weyershaeuser J., Thorausch N., Sikorska J., Krausze J. Molecular mechanism of SHP2 activation by PD-1 stimulation. Sci Adv. 2020;6:4458. doi: 10.1126/sciadv.aay4458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hof P., Pluskey S., Dhe-Paganon S., Eck M.J., Shoelson S.E. Crystal structure of the tyrosine phosphatase SHP-2. Cell. 1998;92:441–450. doi: 10.1016/s0092-8674(00)80938-1. [DOI] [PubMed] [Google Scholar]
- 7.Pádua R.A.P., Sun Y., Marko I., Pitsawong W., Stiller J.B., Otten R. Mechanism of activating mutations and allosteric drug inhibition of the phosphatase SHP2. Nat Commun. 2018;9:4507. doi: 10.1038/s41467-018-06814-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grossmann K.S., Rosário M., Birchmeier C., Birchmeier W. The tyrosine phosphatase Shp2 in development and cancer. Adv Canc Res. 2010;106:53–89. doi: 10.1016/S0065-230X(10)06002-1. [DOI] [PubMed] [Google Scholar]
- 9.Bentires-Alj M., Paez J.G., David F.S., Keilhack H., Halmos B., Naoki K. Activating mutations of the noonan syndrome-associated SHP2/PTPN11 gene in human solid tumors and adult acute myelogenous leukemia. Canc Res. 2004;64:8816–8820. doi: 10.1158/0008-5472.CAN-04-1923. [DOI] [PubMed] [Google Scholar]
- 10.Xie J., Si X., Gu S., Wang M., Shen J., Li H. Allosteric inhibitors of SHP2 with therapeutic potential for cancer treatment. J Med Chem. 2017;60:10205–10219. doi: 10.1021/acs.jmedchem.7b01520. [DOI] [PubMed] [Google Scholar]
- 11.Fodor M., Price E., Wang P., Lu H., Argintaru A., Chen Z. Dual allosteric inhibition of SHP2 phosphatase. ACS Chem Biol. 2018;13:647–656. doi: 10.1021/acschembio.7b00980. [DOI] [PubMed] [Google Scholar]
- 12.Tang K., Jia Y.N., Yu B., Liu H.M. Medicinal chemistry strategies for the development of protein tyrosine phosphatase SHP2 inhibitors and PROTACs degraders. Eur J Med Chem. 2020;204:112657. doi: 10.1016/j.ejmech.2020.112657. [DOI] [PubMed] [Google Scholar]
- 13.Song Z.D., Wang M.J., Ge Y., Chen X.P., Xu Z.Y., Sun Y. Tyrosine phosphatase SHP2 inhibitors in tumor-targeted therapies. Acta Pharm Sin B. 2021;11:13–29. doi: 10.1016/j.apsb.2020.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bagdanoff J.T., Chen Z., Acker M., Chen Y.N., Chan H., Dore M. Optimization of fused bicyclic allosteric SHP2 inhibitors. J Med Chem. 2019;62:1781–1792. doi: 10.1021/acs.jmedchem.8b01725. [DOI] [PubMed] [Google Scholar]
- 15.Wu X., Xu G., Li X., Xu W., Li Q., Liu W. Small molecule inhibitor that stabilizes the autoinhibited conformation of the oncogenic tyrosine phosphatase SHP2. J Med Chem. 2019;62:1125–1137. doi: 10.1021/acs.jmedchem.8b00513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stanford S.M., Bottini N. Targeting tyrosine phosphatases: time to end the stigma. Trends Pharmacol Sci. 2017;38:524–540. doi: 10.1016/j.tips.2017.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen Y.F., Fu L.W. Mechanisms of acquired resistance to tyrosine kinase inhibitors. Acta Pharm Sin B. 2011;1:197–207. doi: 10.1016/j.apsb.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen Y.N., LaMarche M.J., Chan H.M., Fekkes P., Garcia-Fortanet J., Acker M.G. Allosteric inhibition of SHP2 phosphatase inhibits cancers driven by receptor tyrosine kinases. Nature. 2016;535:148–152. doi: 10.1038/nature18621. [DOI] [PubMed] [Google Scholar]
- 19.Ruess D.A., Heynen G.J., Ciecielski K.J., Ai J., Berninger A., Kabacaoglu D. Mutant KRAS-driven cancers depend on PTPN11/SHP2 phosphatase. Nat Med. 2018;24:954–960. doi: 10.1038/s41591-018-0024-8. [DOI] [PubMed] [Google Scholar]
- 20.Dardaei L., Wang H.Q., Singh M., Fordjour P., Shaw K.X., Yoda S. SHP2 inhibition restores sensitivity in ALK-rearranged non-small-cell lung cancer resistant to ALK inhibitors. Nat Med. 2018;24:512–517. doi: 10.1038/nm.4497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao M., Guo W., Wu Y., Yang C., Zhong L., Deng G. SHP2 inhibition triggers anti-tumor immunity and synergizes with PD-1 blockade. Acta Pharm Sin B. 2019;9:304–315. doi: 10.1016/j.apsb.2018.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang M., Lu J., Wang M., Yang C.Y., Wang S. Discovery of SHP2-D26 as a first, potent, and effective PROTAC degrader of SHP2 protein. J Med Chem. 2020;63:7510–7528. doi: 10.1021/acs.jmedchem.0c00471. [DOI] [PubMed] [Google Scholar]
- 23.Nichols R.J., Haderk F., Stahlhut C., Schulze C.J., Hemmati G., Wildes D. RAS nucleotide cycling underlies the SHP2 phosphatase dependence of mutant BRAF-, NF1- and RAS-driven cancers. Nat Cell Biol. 2018;20:1064–1073. doi: 10.1038/s41556-018-0169-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.LaRochelle J.R., Fodor M., Xu X., Durzynska I., Fan L., Stams T. Structural and functional consequences of three cancer-associated mutations of the oncogenic phosphatase SHP2. Biochemistry. 2016;55:2269–2277. doi: 10.1021/acs.biochem.5b01287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Walters W.P., Namchuk M. Designing screens: how to make your hits a hit. Nat Rev Drug Discov. 2003;2:259–266. doi: 10.1038/nrd1063. [DOI] [PubMed] [Google Scholar]
- 26.Copeland R.A. 2nd ed. John Wiley & Sons, Inc.; New Jersey: 2013. Evaluation of enzyme inhibitors in drug discovery: a guide for medicinal chemists and pharmacologists. [PubMed] [Google Scholar]
- 27.Renaud J.P., Chung C.W., Danielson U.H., Egner U., Hennig M., Hubbard R.E. Biophysics in drug discovery: impact, challenges and opportunities. Nat Rev Drug Discov. 2016;15:679–698. doi: 10.1038/nrd.2016.123. [DOI] [PubMed] [Google Scholar]
- 28.Lo M.C., Aulabaugh A., Jin G., Cowling R., Bard J., Malamas M. Evaluation of fluorescence-based thermal shift assays for hit identification in drug discovery. Anal Biochem. 2004;332:153–159. doi: 10.1016/j.ab.2004.04.031. [DOI] [PubMed] [Google Scholar]
- 29.Lucet I.S., Hildebrand J.M., Czabotar P.E., Zhang J.G., Nicola N.A., Silke J. Determination of pseudokinase-ligand interaction by a fluorescence-based thermal shift assay. Bio-protocol. 2014;4:1135. [Google Scholar]
- 30.Niesen F.H., Berglund H., Vedadi M. The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nat Protoc. 2007;2:2212–2221. doi: 10.1038/nprot.2007.321. [DOI] [PubMed] [Google Scholar]
- 31.Irving E., Stoker A.W. Vanadium compounds as PTP inhibitors. Molecules. 2017;22:2269. doi: 10.3390/molecules22122269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.LaRochelle J.R., Fodor M., Vemulapalli V., Mohseni M., Wang P., Stams T. Structural reorganization of SHP2 by oncogenic mutations and implications for oncoprotein resistance to allosteric inhibition. Nat Commun. 2018;9:4508. doi: 10.1038/s41467-018-06823-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yuan S., Yu B., Liu H.M. Palladium-catalyzed ‘on-water’ tandem cyclization reactions for the synthesis of biologically important 4-arylquinazolines. Chem Eur J. 2019;25:13109–13113. doi: 10.1002/chem.201903464. [DOI] [PubMed] [Google Scholar]
- 34.Yuan S., Chang J., Yu B. Construction of biologically important biaryl scaffolds through direct C–H bond activation: advances and prospects. Top Curr Chem. 2020;378:23. doi: 10.1007/s41061-020-0285-9. [DOI] [PubMed] [Google Scholar]
- 35.Yuan S., Wang S., Zhao M., Zhang D., Chen J., Li J.X. Brønsted acid-promoted ‘on-water’ C(sp3)-H functionalization for the synthesis of isoindolinone/[1,2,4]triazolo[1,5-a]pyrimidine derivatives targeting the SKP2–CKS1 interaction. Chin Chem Lett. 2020;31:349–352. [Google Scholar]
- 36.Yuan S., Yu B., Liu H.M. Brønsted acid-catalyzed direct C(sp2)-H heteroarylation enabling the synthesis of structurally diverse biaryl derivatives. Adv Synth Catal. 2019;361:59–66. [Google Scholar]
- 37.Welte S., Baringhaus K.H., Schmider W., Muller G., Petry S., Tennagels N. 6,8-Difluoro-4-methylumbiliferyl phosphate: a fluorogenic substrate for protein tyrosine phosphatases. Anal Biochem. 2005;338:32–38. doi: 10.1016/j.ab.2004.11.047. [DOI] [PubMed] [Google Scholar]
- 38.Vazhappilly C.G., Saleh E., Ramadan W., Menon V., Al-Azawi A.M., Tarazi H. Inhibition of SHP2 by new compounds induces differential effects on RAS/RAF/ERK and PI3K/AKT pathways in different cancer cell types. Invest New Drugs. 2019;37:252–261. doi: 10.1007/s10637-018-0626-5. [DOI] [PubMed] [Google Scholar]
- 39.Chen C., Xue T., Fan P., Meng L., Wei J., Luo D. Cytotoxic activity of Shp2 inhibitor fumosorinone in human cancer cells. Oncol Lett. 2018;15:10055–10062. doi: 10.3892/ol.2018.8593. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.