Abstract
Objective
We examined whether the single-nucleotide polymorphism (SNP) rs13181 in the gene encoding excision repair cross complementation group 2 (ERCC2) is associated with the risk and prognosis of nasopharyngeal carcinoma (NPC).
Methods
SNPs at rs13181 were genotyped in 439 NPC patients (NPC group) and 431 age- and gender-matched cancer-free controls (control group) from a region of China where NPC is endemic, and frequencies of GG, GT and TT genotypes were compared between the two groups in the case–control study. In a subset of 365 NPC cases, SNPs were examined for potential correlation with tumor-free survival time (TFS) and overall survival (OS).
Results
Relative to NPC risk with a TT genotype, NPC risk was similar with GT + GG genotypes (OR 1.052, 95% CI 0.656–1.688), after adjusting for gender, age, smoking history, and immunoglobin A against Epstein-Barr virus capsid antigen (EBV-VCA-IgA) status. Univariate analysis showed that the GG or GT genotype was associated with significantly worse TFS (p<0.001) and OS (p=0.010) than the TT genotype. Prognosis was significantly worse for men than for women (TFS, p=0.045; OS, p=0.031), for T3–T4 classification than for T1–T2 (TFS, p=0.009; OS, p=0.007), for N3 than for N0+N1+N2 (TFS, p<0.001; OS, p<0.001). Based on multivariate analysis, independent risk factors for poor TFS were GG or GT genotype (HR 2.629, 95% CI 1.625–4.254, p<0.001), T3–T4 classification (HR 2.146, 95% CI 1.244–3.701, p=0.006) and N3 (HR 2.527, 95% CI 1.574–4.059, p<0.001). GG or GT genotype (HR 2.217, 95% CI 1.283–3.832, p=0.004), gender (HR 1.989, 95% CI 1.046–3.785, p=0.036), T3–T4 (HR 2.431, 95% CI 1.306–4.526, p=0.005) and N3 (HR 2.693, 95% CI 1.637–4.432, p<0.001) were independent risk factors for poor OS.
Conclusion
The rs13181 SNP in ERCC2 does not appear to be associated with NPC risk, but it may serve as an independent prognostic factor for NPC recurrence and death.
Keywords: single nucleotide polymorphism, rs13181, nasopharyngeal carcinoma, excision repair cross complementation group 2, tumor-free survival, overall survival
Introduction
Nasopharyngeal carcinoma (NPC) is one of the most common malignant tumors of the head and neck in southern China. In this endemic area, the annual incidence of NPC is as high as 25–50 cases per 10,000 people.1 NPC shows a high degree of malignancy and early lymph node metastasis,2 resulting in poor prognosis.3 Although the clinical outcome of NPC has greatly improved due to the rapid development of modern medical imaging, chemotherapy and radiotherapy technology,2,4 distant metastasis and/or local recurrence still occur in 20–30% of patients, especially those with advanced disease.5 This highlights the importance of finding out what causes this malignancy and what factors can predict the risk of recurrence and death.
The detailed mechanisms and risk factors of NPC are not completely understood. Increasing evidence suggests that NPC susceptibility is increased when DNA repair pathways are impaired.6,7 Indeed, NPC risk in a population in Sichuan, China, showed a trend to be linked to several single nucleotide polymorphisms (SNPs) that lead to amino acid changes such as Lys751Gln (rs13181) in the gene encoding excision repair cross complementation group 2 (ERCC2), which is a key player in the nucleotide excision repair (NER) pathway.8 Significant associations between NPC susceptibility and rs13181 SNP were also found in a Malaysian population.9 We are unaware of reports addressing whether this correlation holds in disease-endemic areas of China.
The rs13181 SNP was also investigated regarding its association with the clinical outcome of several malignant tumors. For example, genotype at rs13181 may affect the prognosis of cutaneous melanoma: patients with the GG genotype are at a 2.2-fold higher risk of death than those with the TT genotype.10 In pancreatic cancer, patients with GG genotype at rs13181 are at higher risk of death and progression, compared with those carrying TT or GT genotype.11 In gastric and colorectal cancer, GG and GT genotypes at rs13181 were found to be correlated with poorer progression-free survival and overall survival (OS) than TT genotype.12 These studies indicate a possible role of SNP at rs13181 in prognosis, and it could be used as a reliable factor to evaluate the death and progression risk of cancers. However, we are aware of only two studies that examined whether rs13181 may predict the prognosis of NPC patients.13,14 In one study in Malaysia,13 no significant relationship was found between rs13181 and NPC prognosis. In another study in Guangdong, China, there was also no significant association between rs13181 and clinical outcome of metastatic or recurrent NPC patients.14 The information regarding the role of this variant in NPC prognosis is limited, and whether this variant affects the prognosis of the patients, especially for those without recurrence and distant metastatic disease, needs further inspections.
In the present study, we analyzed the possible associations of genotype at rs13181 with NPC risk, OS and tumor-free survival (TFS) in a population from Guangxi, China, which is an endemic area of NPC. Our aim in this study was to obtain clearer evidence for or against rs13181 SNP for predicting NPC susceptibility and long-term prognosis of NPC patients.
Methods
Study Participants
Participants in Analysis of NPC Risk
Unrelated patients who were diagnosed with NPC at the Cancer Hospital of Guangxi Medical University (Guangxi, China) in the period from July 2012 to June 2015 were eligible for enrollment, and their clinical data were retrospectively reviewed in the study. These eligible patients also met the following inclusion criteria: (1) were newly diagnosed by pathology; (2) were above 18 years of age; and (3) were with no history of other primary malignant tumors. Controls were chosen among subjects undergoing routine physical examinations at the Cancer Hospital and the First Affiliated Hospital of Guangxi Medical University. To be enrolled as a control, subjects had to be local residents of Guangxi province with no history of any malignant tumors who were receiving a health examination during the same period of enrollment as the NPC cases. A total of 440 NPC patients and 431 age- and gender-matched cancer-free controls were enrolled. One NPC case was later excluded from the study because the SNP at rs13181 could not be genotyped, leaving a final number of 439 NPC patients. The Ethics Committee of Guangxi Medical University approved this study, and all study participants provided written informed consent prior to enrollment. This study was conducted in accordance with the Declaration of Helsinki.
Patients in Analysis of Prognosis
For this analysis, a subset of NPC patients was chosen from the patients (439 cases) included in NPC risk analysis according to the following criteria: (1) completed radiotherapy or radiochemotherapy after NPC diagnosis, (2) lacked any signs of distant metastasis at initial diagnosis, (3) showed no other severe chronic diseases, and (4) were followed up for more than 6 months. In accordance with the criteria, 74 NPC patients were excluded from this study including 49 cases with signs of distant metastasis, 18 cases who did not receive a complete course of treatment, 6 cases who were followed up for less than 6 months, and 1 case with severe chronic disease. In the end, 365 eligible patients were included in the analysis.
Blood Collection
Peripheral venous blood (3 mL) was collected into Ethylene Diamine Tetraacetic Acid (EDTA)-containing tubes from all participants. Using the TGuide Blood Genomic DNA Kit (Tiangen, Beijing, China), genomic DNA was isolated following the manufacturer’s instructions. DNA quality was assessed by agarose gel electrophoresis. Extracted DNA was stored at –20°C for future investigation or processed for genotyping immediately.
Mass Spectrometry-Based Genotyping
The rs13181 SNP in the ERCC2 gene was genotyped by BGI (Beijing, China) as described15 using allele-specific, matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Primers designed using MassARRAY Assay Design 3.1 software (Sequenom, San Diego, CA, USA) were used to amplify fragments spanning the rs13181SNP in a 384-well ABI Veriti PCR System (Applied Biosystems, Foster City, CA, USA) in accordance with the manufacturer’s instructions. Amplification reactions (5 μL) consisted of 1 μL DNA (20 ng/μL) and 4 μL Master Mix, and reaction conditions were as follows: 94°C for 5 min, then 45 cycles of 94°C for 20 sec and 56°C for 30 sec, followed by 72°C for 1 min. Genotypes at rs13181 were analyzed using MassARRAY TYPER 4.0 software (Sequenom). Rates of successful genotyping were 99.8% (439/440) among the NPC patients and 100% (431/431) in controls.
Patient Treatment
The 365 patients chosen for prognostic analysis underwent intensity-modulated radiation therapy (358, 98.1%) or conventional radiotherapy. Most (347, 95.1%) including 26 with stage II and 321 with stage III+IV also received combination chemotherapy consisting of concurrent platinum-based chemotherapy and platinum- or platinum plus taxane (docetaxel)-based induction chemotherapy. The remaining 18 patients including 10 with stage II and 8 with stage III+IV did not receive chemotherapy.
Follow-Up
After treatment, follow-up was carried out every 3–6 months for three years and then every 3–12 months thereafter. The results of follow-up were obtained by telephone interview or from medical records.
Variables and Definitions
The following variables were considered: age, gender, history of drinking alcohol and smoking, rs13181 SNP genotype, presence of immunoglobulin A against Epstein-Barr virus capsid antigen (EBV-VCA-IgA), use of any chemotherapy, NPC clinical stage (I through IV), tumor (T) classification, lymph node (N) classification, and TNM stage based on the American joint Committee on Cancer (AJCC) (2010) clinical staging system. The TNM stage was determined based on nasopharyngeal and neck magnetic resonance imaging, as well as pathology results from nasopharyngoscopy. OS was defined as the time from the first visit to our hospital until the patient’s death or, if the patient was still alive, until the patient’s last visit. TFS was defined as the time from the first visit to our hospital until death, first relapse, metastasis, or last visit (if the patient was still alive).16
Statistical Analysis
Statistical analysis was performed using SPSS 22.0 (IBM, Chicago, IL, USA) and GraphPad Prism 7 software (GraphPad Software, Inc., La Jolla, USA). An unpaired t-test was used to compare the ages of patients and controls. Genotype distribution of SNPs at rs13181 was compared against the expected distribution as determined from Hardy–Weinberg equilibrium using a chi-squared test. Comparisons of SNP genotype distributions among various groups were also performed using chi-squared tests. Logistic regression was used to assess associations between rs13181 genotypes and NPC risk with adjustment for age, gender, smoking history and EBV-VCA-IgA status.
In the prognostic study, differences of TFS and OS between patients with TT and those with GG+GT were assessed using the Kaplan-Meier method (log-rank test). And univariate Cox regression analyses were carried out to assess the effects of the following categorical variables on OS and TFS: gender (male or female), age (>46 or ≤46 yr), history of smoking (yes or no), history of drinking (yes or no), EBV-VCA-IgA (positive or negative), chemotherapy (yes or no), rs13181 genotype (TT or GT+GG), T classification (T1–T2 or T3–T4), N classification (N0+N1+N2 or N3), and clinical stage (stage I–II or III–IV). Any variables showing a correlation of P<0.05 in the univariate analyses were included in subsequent Cox multivariate analysis of OS and TFS. Hazard ratios (HRs) and 95% confidence intervals (95% CIs) were calculated as appropriate. A two-sided P value of <0.05 was considered statistically significant.
Results
Patient Characteristics
Altogether, 439 NPC patients and 431 cancer-free controls were involved in the present study. The two groups significantly differed in smoking history and EBV-VCA-IgA status, while they were similar in age and gender proportion (Table 1). Frequencies of SNP genotypes among the controls were in agreement with Hardy–Weinberg equilibrium (Table 2).
Table 1.
Clinical Characteristics of Patients with Nasopharyngeal Carcinoma and Cancer-Free Controls
| Characteristics | Cases | Controls | χ2/t | P |
|---|---|---|---|---|
| n | 439 | 431 | ||
| Age, yr | 46.78±11.43 | 47.29±13.50 | 0.597 | 0.550a |
| Gender, n | 0.153 | 0.696b | ||
| Male | 334 | 323 | ||
| Female | 105 | 108 | ||
| EBV-VCA-IgA status, n | 372.604 | <0.001b | ||
| Positive | 283 | 11 | ||
| Negative | 156 | 420 | ||
| Smoking history, n | 4.397 | 0.036b | ||
| Yes | 150 | 119 | ||
| No | 289 | 312 |
Notes: aUnpaired, two-tailed Student’s t-test. bChi-squared test. Age is presented as average±standard deviation.
Abbreviation: EBV-VCA-IgA, immunoglobin A against Epstein–Barr virus capsid antigen.
Table 2.
Frequencies of Genotypes at ERCC2 rs13181 in Patients with Nasopharyngeal Carcinoma and Controls
| Genotype | Cases | Controls | Unadjusted OR (95% CI) | P | Adjusted ORa (95% CI) | Adjusted P | PHWEb |
|---|---|---|---|---|---|---|---|
| TT | 369 | 361 | 1.000 | – | 1.000 | – | 0.998 |
| GT | 67 | 67 | 0.964 (0.668–1.392) | 0.845 | 1.008 (0.621–1.635) | 0.976 | – |
| GG | 3 | 3 | – | – | – | – | – |
| GG+GT | 70 | 70 | 0.978 (0.681–1.405) | 0.905 | 1.052 (0.656–1.688) | 0.832 | – |
Notes: aCalculated using multiple logistic regression analysis after controlling for age, gender, smoking history, and EBV infection factors; bP value of Hardy–Weinberg equilibrium for frequencies of SNP genotypes among the controls.
Abbreviations: 95% CI, 95% confidence interval; HWE, Hardy–Weinberg equilibrium; OR, odds ratio.
A total of 365 NPC patients were chosen for prognostic analysis, including 270 males (74.0%) and 95 females (26.0%). Patients were divided into two age groups based on the median age of 46 years: 184 (50.4%) patients were ≤46 years, while 181 (49.6%) were >46 years. Similarly, patients were divided into two groups based on rs13181 genotype: 309 (84.7%) patients had the TT genotype, and 56 (15.3%) had the GT or GG genotype (Table 3). More of these patients were classified as T3–T4 (257, 70.4%) than as T1–T2 (108, 29.6%), and more were classified as N0+N1+N2 (297, 81.4%) than as N3 (68, 18.6%). The difference was even greater between patients in clinical stages III–IV (329, 90.1%) than in stages I–II (36, 9.9%). Follow-up lasted a median of 52 months (range, 7 to 81) and ended on March 1, 2019. By the end of follow-up, 85 (23.3%) patients had experienced local recurrence or distant metastasis, and 72 (19.7%) had died.
Table 3.
Characteristics of the 365 Patients for Prognostic Study
| Characteristics | No. (%) | Genotype (n) | P | |
|---|---|---|---|---|
| TT | GT+GG | |||
| Age (y) | ||||
| ≤46 | 184 (50.4) | 156 | 28 | 0.947 |
| >46 | 181 (49.6) | 153 | 28 | |
| Gender | ||||
| Male | 270 (74.0) | 228 | 42 | 0.849 |
| Female | 95 (26.0) | 81 | 14 | |
| Tumor classification | ||||
| T1+2 | 108 (29.6) | 88 | 20 | 0.275 |
| T3+4 | 257 (70.4) | 221 | 36 | |
| Lymph node classification | ||||
| N0+1+2 | 297 (81.4) | 250 | 47 | 0.593 |
| N3 | 68 (18.6) | 59 | 9 | |
| TNM stage (AJCC) | ||||
| I + II | 36 (9.9) | 28 | 8 | 0.228 |
| III + IV | 329 (90.1) | 281 | 48 | |
| Smoking history | ||||
| Yes | 112 (30.7) | 98 | 14 | 0.316 |
| No | 253 (69.3) | 211 | 42 | |
| Drinking history | ||||
| Yes | 74 (20.3) | 69 | 5 | 0.022 |
| No | 291 (79.7) | 240 | 51 | |
| Chemotherapy | ||||
| Yes | 347 (95.1) | 294 | 53 | 0.746 |
| No | 18 (4.9) | 15 | 3 | |
| EBV-VCA-IgA | ||||
| Negative | 126 (34.5) | 106 | 20 | 0.838 |
| Positive | 239 (65.5) | 203 | 36 | |
Abbreviations: AJCC, American Joint Committee on Cancer; EBV-VCA-IgA, immunoglobulin A against Epstein–Barr virus capsid antigen.
Correlation of rs13181 SNP with NPC Risk
The frequency of GG + GT genotypes at rs13181 was not significantly different between NPC patients (15.9%) and controls (16.2%). The risk of NPC for these genotypes relative to the TT genotype was OR of 0.978 (95% CI 0.681–1.405) without adjustment and OR of 1.052 (95% CI 0.656–1.688) with adjustment (Table 2).
Correlation of rs13181 SNP and Clinicodemographic Factors with TFS
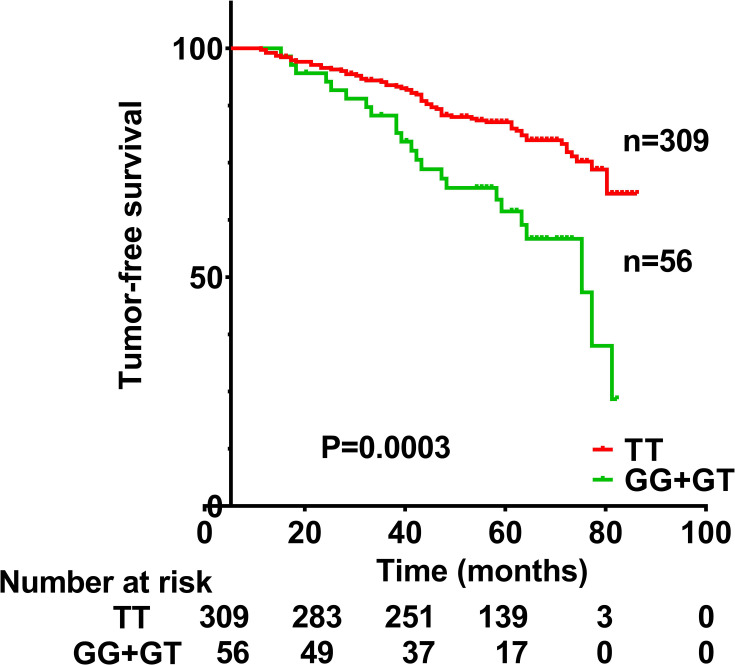
TFS was significantly lower among patients with the GG or GT genotype at rs13181 than among those with the TT genotype (Figure 1). TFS was significantly worse for men than women (HR 1.799, 95% CI 1.014–3.191, p=0.045), for patients with T3–T4 classification than for those with T1–T2 classification (HR 2.062, 95% CI 1.197–3.551, p=0.009), for those with N3 classification than for those with N0+N1+N2 (HR 2.315, 95% CI 1.446–3.708, p<0.001). TFS did not correlate significantly with clinical stage (HR 3.158, 95% CI 0.998–9.997, p=0.051), age (HR 1.006, 95% CI 0.640–1.579, p=0.981), smoking history (HR 1.474, 95% CI 0.953–2.277, p=0.081), or drinking history (HR 0.986, 95% CI 0.580–1.676, p=0.958), or chemotherapy (HR 1.348, 95% CI 0.426–4.271, p=0.612), or EBV-VCA-IgA status (HR 1.422, 95% CI 0.893–2.266, p=0.138).
Figure 1.
Tumor-free survival of nasopharyngeal carcinoma patients with a TT genotype or GG+GT genotypes at rs13181 in the ERCC2 gene. Survival differed significantly between the two groups (P=0.0003).
After univariate analysis, multivariate analysis was carried out to further assess the relationship between TFS and these various factors (Table 4). TFS remained significantly associated with genotype (HR 2.629, 95% CI 1.625–4.254, p<0.001), T classification (HR 2.146, 95% CI 1.244–3.701, p=0.006) and N classification (HR 2.527, 95% CI 1.574–4.059, p<0.001), but not with gender (HR 1.713, 95% CI 0.963–3.045, p=0.067).
Table 4.
Cox Multivariate Model to Analyze Prognostic Factors for Tumor-Free Survival in 368 NPC Patients
| Prognostic Factor | P | HR | 95% CI for HR |
|---|---|---|---|
| Genotype of rs13181 | <0.001 | 2.629 | 1.625–4.254 |
| Gender | 0.067 | 1.713 | 0.963–3.045 |
| T classification | 0.006 | 2.146 | 1.244–3.701 |
| N classification | <0.001 | 2.527 | 1.574–4.059 |
Abbreviations: HR, hazard ratio; NPC, nasopharyngeal carcinoma; 95% CI, 95% confidence interval.
Correlation of rs13181 SNP and Clinicodemographic Factors with OS
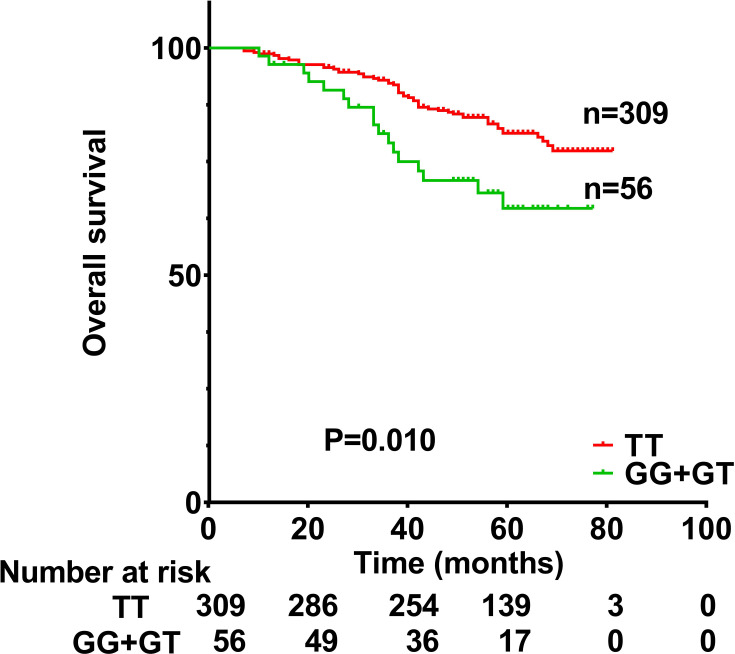
OS was significantly lower among patients with the GT or GG genotype at rs13181 than among those patients with the TT genotype (Figure 2). OS was significantly worse for men than for women (HR 2.027, 95% CI 1.067–3.853, p=0.031), for patients with T3–T4 classification than for those with T1–T2 classification (HR 2.333, 95% CI 1.255–4.338, p=0.007), for patients with N3 classification than for those with N0+1+2 (HR 2.487, 95% CI 1.514–4.085, p<0.001). OS did not correlate significantly with clinical stage (HR 2.703, 95% CI 0.850–8.594, p=0.092), or EB-VCA-IgA status (HR 1.636, 95% CI 0.969–2.765, p=0.066), or age (HR 1.155, 95% CI 0.714–1.869, p=0.556), or drinking history (HR 1.050, 95% CI 0.594–1.854, p=0.867), or chemotherapy (HR 1.141, 95% CI 0.359–3.631, p=0.823) or smoking history (HR 1.556, 95% CI 0.969–2.499, p=0.068).
Figure 2.
Overall survival of nasopharyngeal carcinoma patients with a TT genotype or GG+GT genotypes at rs13181 in the ERCC2 gene. Survival differed significantly between the two groups (P=0.010).
Multivariate analysis (Table 5) showed that OS remained significantly correlated with genotype (HR 2.217, 95% CI 1.283–3.832, p=0.004), gender (HR 1.989, 95% CI 1.046–3.785, p=0.036), T classification (HR 2.431, 95% CI 1.306–4.526, p=0.005) and N classification (HR 2.693, 95% CI 1.637–4.432, p<0.001).
Table 5.
Cox Multivariate Model to Analyze Prognostic Factors for Overall Survival in 368 NPC Patients
| Prognostic Factor | P | HR | 95% CI for HR |
|---|---|---|---|
| Genotype of rs13181 | 0.004 | 2.217 | 1.283–3.832 |
| Gender | 0.036 | 1.989 | 1.046–3.785 |
| N classification | <0.001 | 2.693 | 1.637–4.432 |
| T classification | 0.005 | 2.431 | 1.306–4.526 |
Abbreviations: EBV-VCA-IgA, immunoglobulin A against Epstein–Barr virus capsid antigen; HR, hazard ratio; NPC, nasopharyngeal carcinoma; 95% CI, 95% confidence interval.
Discussion
Genomic DNA damage and the consequent genomic instability are some of the early carcinogenic events and risk factors for the occurrence of malignant tumors. NER is an intrinsic defense mechanism to remove various helix-distorting DNA lesions, such as large chemical adducts, pyrimidine dimers, and DNA crosslinks, in order to repair genomic instability.10 ERCC2, an indispensable component in the NER pathway, is responsible for detecting genomic lesions and maintaining the accuracy of repair, and it contains ATPase and helicase functions that mediate DNA unwinding around lesion sites. At the rs13181 SNP, a nucleotide change from T to G at codon 751 of ERCC2, which replaces a lysine with a glutamine and affects its function of DNA repair,17 is associated with increased genome stability, potentially decreasing the risk of various cancers.18–20 Nevertheless, the rs13181 SNP may be also associated with increased DNA-repair function of ERCC2 in cancer cells, leading to their poor response to therapy, and thus resulting in a poorer prognosis.11,17,19
Data linking rs13181 to NPC are very limited. A study in a population from Sichuan province of China showed a non-significant tendency for TT genotype to confer increased NPC risk,8 while a Malaysian study9 found this association to be significant, even after adjusting for cigarette smoking, alcohol drinking and salted fish consumption. In contrast, the results in our investigation showed no significant difference in TT genotype frequency between NPC cases and controls, who were from Guangxi province, a region in southern China where NPC is endemic. These results suggest that SNPs at rs13181 may not be linked to NPC susceptibility, which is inconsistent with the previous studies.8,9 These conflicting results may be attributed to geographical and ethnic factors and reflect different roles of rs13181 SNP in the carcinogenesis process of NPC among populations with different genetic backgrounds.
The relationship between the rs13181 SNP and NPC prognosis is also controversial. We found this SNP to be significantly associated with both TFS and OS of Chinese NPC patients, in contrast to a Malaysian study13 and a study in Guangdong, China14 that found no such association. In our study, compared to patients with a TT genotype, the risk of recurrence or death was, respectively, 2.629-fold and 2.217-fold higher in those with a GG or GT genotype. The disparity between our results and those of the prior studies could be explained by the studied populations with different genetic background or patients with heterogeneity of disease progression. In the Malaysian study, nearly 30% of the patients belonged to ethnicity of Malay,13 whose genetic background may be different from that of Chinese patients in our study. We speculate that rs13181 SNP may play a different role in therapeutic sensitivity of NPC among different ethnic populations. In the Guangdong study,14 the involved patients were with recurrent or metastatic NPC, unlike those with newly diagnosed and non-metastatic disease in our study. Among recurrent or metastatic patients, the objective response rate to cisplatin-based chemotherapy was reported to decrease by a previous study,21 suggesting resistance of recurrent or metastatic disease to cisplatin. We propose that rs13181 SNP, which affects the function of ERCC2 to repair DNA damages induced by cisplatin, may play little role in cisplatin response of recurrent or metastatic NPC, and thus may not be associated with its outcome. Our data, which was based on newly diagnosed Chinese patients without distant metastasis, suggested that SNP at rs13181 may be a reliable marker to independently predict NPC prognosis, especially for those with non-relapsed or non-metastatic disease.
How SNPs at rs13181 influence NPC prognosis is unclear, though they are likely to exert an influence by affecting DNA repair, which in turn would affect the repair of double- and single-stranded breaks caused by radiotherapy,22,23 as well as the repair of platinum-DNA-adducts induced by platinum-based chemotherapy, such as the first-line NPC drug cisplatin.24–26 Indeed, radiotherapy and platinum-based chemotherapy are the main therapies to treat NPC. Our results are consistent with the idea that rs13181 SNP alters the activity of the ERCC2 protein and thereby efficacy of radiotherapy and platinum-based chemotherapy, as suggested in previous studies.12,27 In addition, nearly a third of our patients (100/368) also received platinum plus taxane-based chemotherapy, our results further suggest that the SNPs could also alter the sensitivity of cancer cells to taxanes, consistent with previous studies.28–30 We hypothesize that NPC patients with GT or GG genotypes at rs13181 have a greater ability to repair DNA damage in NPC cells induced by radiotherapy or platinum- or platinum plus taxane-based chemotherapy than patients with the TT genotype, leading to poorer response of tumor cells to treatments, and thus significantly shortening TFS and OS. This hypothesis needs to be further validated. If it is confirmed, rs13181 SNP could be used as a reliable indicator identifying those NPC patients who are sensitive or resistant to radiotherapy or platinum-based chemotherapy, so that the doses of radiation or platinum drugs could be optimized to achieve maximal tumor control and minimal side effects. Another possibility of how SNPs at rs13181 affect NPC prognosis is that rs13181 lies within the last exon, SNPs may affect poly(A) adenylation of the transcript.9,31 In this way, SNPs at rs13181 may regulate transcription and translation of ERCC2, ultimately altering DNA repair ability.9 A third possibility is that rs13181 SNPs affect other ERCC2 SNPs with which it is in linkage disequilibrium,10 such as rs238406 and rs1799793. These two SNPs have been linked to outcomes of several cancers,32–34 such as esophageal cancer and gastric cancer. The same may be true for genetic interactions between rs13181 in ERCC2 and SNPs in other genes, such as the excision repair cross complementing 1 (ERCC1) gene in NER.35
Some limitations in our study should be mentioned. A relatively small sample was included in our single-center study, which may cause selection bias. Studies from multiple institutions involving larger cohorts should be undertaken. Although we focused on only one SNP, it shows linkage disequilibrium with other SNPs in ERCC2 that modulate treatment sensitivity and with genes involved in DNA damage repair. Future investigations should explore ERCC2 and other genes for other SNPs possibly associated with NPC prognosis. No related functional assays were carried out in the present study. Detailed mechanistic studies are needed to explain the relationship between rs13181 SNP and NPC prognosis.
Despite these limitations, our study is the first to provide evidence suggesting that rs13181 variants in ERCC2 may be a reliable marker predicting NPC prognosis, even if the genotype at this locus does not appear to be associated with risk of NPC. Functional studies should assess the effects of these nucleotide variations on ERCC2 expression and activity.
Acknowledgments
This study was supported by the Guangxi Natural Science Foundation of China (2017GXNSFAA198064), the Key Laboratory Foundation (GKE2019-18), and the National Natural Science Foundation of China (81760361, 81360406).
Data Sharing Statement
The raw data of this study are available from the corresponding author on reasonable request.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Kamran SC, Riaz N, Lee N. Nasopharyngeal carcinoma. Surg Oncol Clin N Am. 2015;24(3):547–561. doi: 10.1016/j.soc.2015.03.008 [DOI] [PubMed] [Google Scholar]
- 2.Tang L, Li L, Mao Y, et al. Retropharyngeal lymph node metastasis in nasopharyngeal carcinoma detected by magnetic resonance imaging: prognostic value and staging categories. Cancer. 2008;113(2):347–354. doi: 10.1002/cncr.23555 [DOI] [PubMed] [Google Scholar]
- 3.Li J, Jiang R, Liu WS, et al. A large cohort study reveals the association of elevated peripheral blood lymphocyte-to-monocyte ratio with favorable prognosis in nasopharyngeal carcinoma. PLoS One. 2013;8(12):e83069. doi: 10.1371/journal.pone.0083069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wei WI, Mok VW. The management of neck metastases in nasopharyngeal cancer. Curr Opin Otolaryngol Head Neck Surg. 2007;15(2):99–102. doi: 10.1097/MOO.0b013e3280148a06 [DOI] [PubMed] [Google Scholar]
- 5.Lee AW, Lau WH, Tung SY, et al. Preliminary results of a randomized study on therapeutic gain by concurrent chemotherapy for regionally-advanced nasopharyngeal carcinoma: NPC-9901 trial by the Hong Kong Nasopharyngeal Cancer Study Group. J Clin Oncol. 2005;23(28):6966–6975. doi: 10.1200/JCO.2004.00.7542 [DOI] [PubMed] [Google Scholar]
- 6.Cho EY, Hildesheim A, Chen CJ, et al. Nasopharyngeal carcinoma and genetic polymorphisms of DNA repair enzymes XRCC1 and hOGG1. Cancer Epidemiol Biomarkers Prev. 2003;12(10):1100–1104. [PubMed] [Google Scholar]
- 7.Cao Y, Miao XP, Huang MY, et al. Polymorphisms of XRCC1 genes and risk of nasopharyngeal carcinoma in the Cantonese population. BMC Cancer. 2006;6(1):167. doi: 10.1186/1471-2407-6-167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang ZH, Du B, Wei YS, et al. Genetic polymorphisms of the DNA repair gene and risk of nasopharyngeal carcinoma. DNA Cell Biol. 2007;26(7):491–496. doi: 10.1089/dna.2006.0537 [DOI] [PubMed] [Google Scholar]
- 9.Lye MS, Visuvanathan S, Chong PP, Yap YY, Lim CC, Ban EZ. Homozygous wildtype of XPD K751Q polymorphism is associated with increased risk of nasopharyngeal carcinoma in Malaysian population. PLoS One. 2015;10(6):e0130530. doi: 10.1371/journal.pone.0130530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schrama D, Scherer D, Schneider M, et al. ERCC5 p.Asp1104His and ERCC2 p.Lys751Gln polymorphisms are independent prognostic factors for the clinical course of melanoma. J Invest Dermatol. 2011;131(6):1280–1290. doi: 10.1038/jid.2011.35 [DOI] [PubMed] [Google Scholar]
- 11.Avan A, Pacetti P, Reni M, et al. Prognostic factors in gemcitabine-cisplatin polychemotherapy regimens in pancreatic cancer: XPD-Lys751Gln polymorphism strikes back. Int J Cancer. 2013;133(4):1016–1022. doi: 10.1002/ijc.28078 [DOI] [PubMed] [Google Scholar]
- 12.Yin M, Yan J, Martinez-Balibrea E, et al. ERCC1 and ERCC2 polymorphisms predict clinical outcomes of oxaliplatin-based chemotherapies in gastric and colorectal cancer: a systemic review and meta-analysis. Clin Cancer Res. 2011;17(6):1632–1640. doi: 10.1158/1078-0432.CCR-10-2169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ban EZ, Lye MS, Chong PP, Yap YY, Lim SYC, Abdul Rahman H. Association of hOGG1 Ser326Cys, ITGA2 C807T, TNF-A −308G>A and XPD Lys751Gln polymorphisms with the survival of Malaysian NPC patients. PLoS One. 2018;13(6):e0198332. doi: 10.1371/journal.pone.0198332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen C, Wang F, Wang Z, et al. Polymorphisms in ERCC1 C8092A predict progression-free survival in metastatic/recurrent nasopharyngeal carcinoma treated with cisplatin-based chemotherapy. Cancer Chemother Pharmacol. 2013;72(2):315–322. doi: 10.1007/s00280-013-2196-8 [DOI] [PubMed] [Google Scholar]
- 15.Schaeffeler E, Zanger UM, Eichelbaum M, Asante-Poku S, Shin JG, Schwab M. Highly multiplexed genotyping of thiopurine s-methyltransferase variants using MALD-TOF mass spectrometry: reliable genotyping in different ethnic groups. Clin Chem. 2008;54(10):1637–1647. doi: 10.1373/clinchem.2008.103457 [DOI] [PubMed] [Google Scholar]
- 16.Li Y, Liang L, Dai W, et al. Prognostic impact of programmed cell death-1 (PD-1) and PD-ligand 1 (PD-L1) expression in cancer cells and tumor infiltrating lymphocytes in colorectal cancer. Mol Cancer. 2016;15(1):55. doi: 10.1186/s12943-016-0539-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lunn RM, Helzlsouer KJ, Parshad R, et al. XPD polymorphisms: effects on DNA repair proficiency. Carcinogenesis. 2000;21(4):551–555. doi: 10.1093/carcin/21.4.551 [DOI] [PubMed] [Google Scholar]
- 18.Dybdahl M, Vogel U, Frentz G, Wallin H, Nexo BA. Polymorphisms in the DNA repair gene XPD: correlations with risk and age at onset of basal cell carcinoma. Cancer Epidemiol Biomarkers Prev. 1999;8(1):77–81. [PubMed] [Google Scholar]
- 19.Banerjee M, Sarkar J, Das JK, et al. Polymorphism in the ERCC2 codon 751 is associated with arsenic-induced premalignant hyperkeratosis and significant chromosome aberrations. Carcinogenesis. 2007;28(3):672–676. doi: 10.1093/carcin/bgl181 [DOI] [PubMed] [Google Scholar]
- 20.Jin D, Zhang M, Hua H. Impact of polymorphisms in DNA repair genes XPD, hOGG1 and XRCC4 on colorectal cancer risk in a Chinese Han population. Biosci Rep. 2019;39(1). doi: 10.1042/BSR20181074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Y, Chen L, Hu GQ, et al. Gemcitabine and cisplatin induction chemotherapy in nasopharyngeal carcinoma. N Engl J Med. 2019;381(12):1124–1135. doi: 10.1056/NEJMoa1905287 [DOI] [PubMed] [Google Scholar]
- 22.Zhang Y, Rohde LH, Wu H. Involvement of nucleotide excision and mismatch repair mechanisms in double strand break repair. Curr Genomics. 2009;10(4):250–258. doi: 10.2174/138920209788488544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lomax ME, Folkes LK, O’Neill P. Biological consequences of radiation-induced DNA damage: relevance to radiotherapy. Clin Oncol (R Coll Radiol). 2013;25(10):578–585. doi: 10.1016/j.clon.2013.06.007 [DOI] [PubMed] [Google Scholar]
- 24.Kong XY, Lu JX, Yu XW, et al. Gemcitabine plus cisplatin versus fluorouracil plus cisplatin as a first-line concurrent chemotherapy regimen in nasopharyngeal carcinoma: a prospective, multi-institution, randomized controlled Phase II study. Cancer Chemother Pharmacol. 2019;84(1):155–161. doi: 10.1007/s00280-019-03858-7 [DOI] [PubMed] [Google Scholar]
- 25.Kusakabe M, Onishi Y, Tada H, et al. Mechanism and regulation of DNA damage recognition in nucleotide excision repair. Genes Environ. 2019;41(1):2. doi: 10.1186/s41021-019-0119-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tezuka S, Ueno M, Kobayashi S, et al. Predictive value of ERCC1, ERCC2, ERCC4, and glutathione S-transferase Pi expression for the efficacy and safety of FOLFIRINOX in patients with unresectable pancreatic cancer. Am J Cancer Res. 2018;8(10):2096–2105. [PMC free article] [PubMed] [Google Scholar]
- 27.Mahimkar MB, Samant TA, Kannan S, Tulsulkar J, Pai PS, Anantharaman D. Polymorphisms in GSTM1 and XPD genes predict clinical outcome in advanced oral cancer patients treated with postoperative radiotherapy. Mol Carcinog. 2012;51(Suppl 1):E94–103. doi: 10.1002/mc.21868 [DOI] [PubMed] [Google Scholar]
- 28.Moisan F, Laroche-Clary A, Auzanneau C, et al. Deciphering the role of the ERCC2 gene polymorphism on anticancer drug sensitivity. Carcinogenesis. 2012;33(5):962–968. doi: 10.1093/carcin/bgs107 [DOI] [PubMed] [Google Scholar]
- 29.Zhou R, Zhu J, Chen X, Liu Y, Wang Y, Zhang T. The efficacy and safety of docetaxel, cisplatin and fluorouracil (TPF)-based induction chemotherapy followed by concurrent chemoradiotherapy for locoregionally advanced nasopharyngeal carcinoma: a meta-analysis. Clin Transl Oncol. 2020;22(3):429–439. doi: 10.1007/s12094-019-02142-7 [DOI] [PubMed] [Google Scholar]
- 30.Petty WJ, Knight SN, Mosley L, et al. A pharmacogenomic study of docetaxel and gemcitabine for the initial treatment of advanced non-small cell lung cancer. J Thorac Oncol. 2007;2(3):197–202. doi: 10.1097/JTO.0b013e318031cd89 [DOI] [PubMed] [Google Scholar]
- 31.Sturgis EM, Zheng R, Li L, et al. XPD/ERCC2 polymorphisms and risk of head and neck cancer: a case-control analysis. Carcinogenesis. 2000;21(12):2219–2223. doi: 10.1093/carcin/21.12.2219 [DOI] [PubMed] [Google Scholar]
- 32.Lu X, Liu Y, Yu T, et al. ERCC1 and ERCC2 haplotype modulates induced BPDE-DNA adducts in primary cultured lymphocytes. PLoS One. 2013;8(4):e60006. doi: 10.1371/journal.pone.0060006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou F, Zhu M, Wang M, et al. Genetic variants of DNA repair genes predict the survival of patients with esophageal squamous cell cancer receiving platinum-based adjuvant chemotherapy. J Transl Med. 2016;14(1):154. doi: 10.1186/s12967-016-0903-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li M, Zhao Y, Zhao E, Wang K, Lu W, Yuan L. Predictive value of two polymorphisms of ERCC2, rs13181 and rs1799793, in clinical outcomes of chemotherapy in gastric cancer patients: a meta-analysis. Dis Markers. 2018;2018:3947626. doi: 10.1155/2018/3947626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yin J, Vogel U, Gerdes LU, Dybdahl M, Bolund L, Nexo BA. Twelve single nucleotide polymorphisms on chromosome 19q13.2-13.3: linkage disequilibria and associations with basal cell carcinoma in Danish psoriatic patients. Biochem Genet. 2003;41(1–2):27–37. doi: 10.1023/A:1020970428370 [DOI] [PubMed] [Google Scholar]