Abstract
Local invasion and distant metastasis are the key hallmarks in the aggressive progression of malignant tumors, including the ability of cancer cells to detach from the extracellular matrix overcome apoptosis, and disseminate into distant sites. It is generally believed that this malignant behavior is stimulated by epithelial-mesenchymal transition (EMT). Musashi (MSI) RNA-binding proteins, belonging to the evolutionarily conserved RNA-binding proteins (RBP) family, were originally discovered to regulate asymmetric cell division during embryonic development. Recently, Musashi-2 (MSI2), as a key member of MSI family, has been prevalently reported to be tightly associated with the advanced clinical stage of several cancers. Multiple oncogenic signaling pathways mediated by MSI2 play vital roles in EMT. Here, we systematically reviewed the detailed role and signal networks of MSI2 in regulating cancer development, especially in EMT signal transduction, involving EGF, TGF-β, Notch, and Wnt pathways.
Keywords: Musashi-2, EMT, EGF, TGF-β, notch, Wnt, targeted therapy
Background
Over two decades ago, the MSI gene was first identified to encode a neural RNA-binding protein (RBP), which plays an important role in regulating asymmetric cell division of sensory organ precursor (SOP) cells in Drosophila.1 The SOP normally divides to generate a neuronal precursor cell, a socket cell, and a bristle shaft; however, MSI mutated SOP cells produce an alternative phenotype with two shafts in a single bristle. The name of the gene suggests a similarity of this phenotype to the image of the Japanese national hero, Miyamoto Musashi, who fought with two swords.1
Currently, two members of the human MSI family have been identified: MSI1 and MSI2. They are evolutionarily conserved and share ~75% homology of the amino acid sequences.2,3 In early studies, MSI1 was found to be highly expressed in the nervous system of mice,4 especially in the undifferentiated neural stem and precursor cells.5,6 Moreover, the elevated expression of MSI1 was also observed in solid tumors such as malignant glioma, esophageal squamous cell carcinoma and gastric cancer.7–9 MSI2, mainly expressed in the hematopoietic system, is an important regulator of hematopoietic stem cells (HSCs) as well as hematopoietic malignancies.10–13 In 2003, the MSI2 gene was found to be rearranged to form a fusion gene with HOXA9 in chronic myeloid leukemia (CML), first indicating its potential link to cancer.14 Later, MSI2 was identified as a crucial regulator during the sperm and embryo formation.15–17 Recently, numerous studies strongly suggest that MSI2 promotes tumor proliferation,18 migration, invasion,12,19,20 autophagy,21 and acts as an important regulator of EMT in cancer.22
In this review, we will provide an overview of MSI2 protein, MSI2-dependent regulatory mechanisms, and clinical relevance, especially its role in human cancer and EMT. We aim to understand the role and functional mechanisms of MSI2 during the tumorigenesis and disease progression, which may provide new insights into the development of targeted treatment strategies.
Musashi-2 Protein Structure and Its Post-Transcriptional Regulation Mechanism
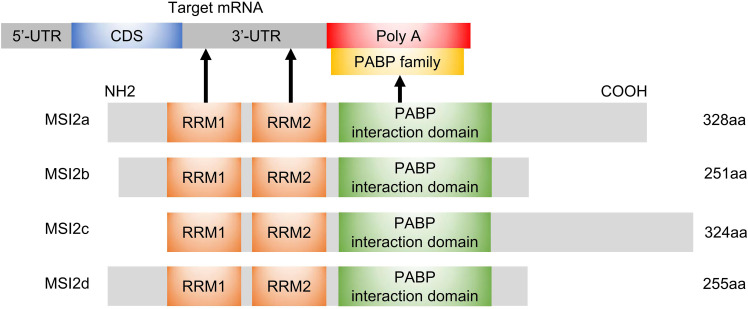
The MSI2 gene, physically located on chromosome 17q22,10 produces four mRNA transcripts, resulting in 4 corresponding protein isoforms (MSI2a, MSI2b, MSI2c, MSI2d).23 All of these four isoforms contain two conserved N-terminal RNA-recognition motifs (RRM1 and RRM2) but differ in the N-terminal or C-terminal amino acids. Biochemical and structural studies showed that the RRM1 contributes to the majority of the binding energy and specificity, while RRM2 has a more supportive role.3 MSI2 usually binds to 3ʹ-end of target RNAs by preferentially interacting with an ACCUUUUUAGAA motif or poly-U sequences,24 UAG motifs,25 and UAG-containing motifs +/‒ additional flanking nucleotides,26 which allows it to both induce and repress protein translation.
However, MSI2 protein itself has no enzymatic activity or inhibitory activity. It has been suggested that additional translation co-factors are required for MSI2 to modulate translation following binding to the mRNA,27 but the molecular mechanism underlying MSI2-dependent translational control and the identity of these co-factors are still not well understood. In the C-terminal half of Musashi1, a poly(A)-binding protein (PABP)-interaction domain is found adjacent to RRM2, which allows the co-factors to induce or repress protein translation. This PABP interaction domain disrupts the ability of PABP to interact with the eukaryotic translation initiation factor 4 gamma (eIF4G), and inhibits the initiation of translation of MSI1 targets.28 In contrast, PABPC1 and ePABP, members of the poly(A)-binding protein family, were found to preferentially interact with MSI1 to promote transcription initiation.29 However, PABPs interact with MSI2 to a lesser extent, mainly due to specific sequence divergence within the C-terminal 190–240 interaction domain.29 In addition, poly-A polymerase Germline Development 2 (GLD2) can interact with the C-terminal domain of MSI1 but not mammalian MSI2, to increase polyadenylation, mRNA stabilization, and higher protein expression in GLD2-rich oocytes.27 Considering that MSI1 has only one isoform, it is relatively complicated to study the additional domains of MSI2 with four isoforms. According to NCBI database (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi), All MSI2 isoforms, like MSI1, may also harbor a PABP interaction domain. This indicates that MSI2 protein rely on RRMs to recognize the 3ʹ-UTR of the target mRNA and might depend on PABP domain to influence translation initiation. The isoforms of MSI2 and its interaction with mRNA/PABP are shown in Figure 1.
Figure 1.
Schematic diagram of MSI2 isoforms, molecular domains and interactions. The N-terminal and C-terminal of MSI2b are different from those of MSI2a. The RRM1 motif of MSI2c is close to the N-terminal, and the C-terminal is 18 aa more than that of MSI2a. MSI2d harbors the same N-terminal as MSI2a and the same C-terminal as MSI2b. All isoforms harbor RRM1 and RRM2 domains, which recognize 3ʹ-UTR of target mRNAs, as well as a PABP interaction domain that may regulate translation initiation through binding to PABP. The arrows represent interactions.
Abbreviations: aa, amino acids; CDS, coding regions; Poly A, polyadenylate; UTR, untranslated region.
In addition, the effect of MSI2 in regulating specific mRNA translation differs depending on MSI2 protein isoforms, phosphorylation status, and cellular context.23,30–33 For example, Li et al demonstrated that overexpression of MSI2a in TNBC cells inhibits EMT, while MSI2b shows no significant effect in TNBC progression.23 Similarly, ectopic expression of MSI2a, but not MSI2b, enhances the self-renewal capacity of embryonic stem cells (ESCs).30 MacNicol et al demonstrated that the canonical MSI2a is subject to site-specific phosphorylation at C-terminus, converting MSI2 from a translational repressor to an activator, while the truncated isoform of human MSI2b that lacks regulatory phosphorylation sites fails to promote translation of target mRNAs.31 These studies strongly suggest that MSI2a may have more important functions than the truncated MSI2b. Other research in myeloid leukemia demonstrated that MSI2 binds Numb mRNAs and inhibits Numb translation;32 however, Numb protein level remained unchanged after the depletion of MSI2 in HSCs.33 In general, all the evidence suggests that the regulation of target mRNA by MSI2 depends on different MSI2 isoforms, phosphorylation status, and cellular context, with mechanisms still to be exactly defined.
Regulation of Musashi-2
The signaling pathways that regulate MSI2 expression are currently less known. A list of factors involved in MSI2 expression and function are shown in Table 1, including transcription factors or inhibitors, non-coding RNAs, ubiquitin proteins, and small molecule compounds targeting MSI2.
Table 1.
Factors Influence MSI2 Expression or MSI2 Function
| Gene/Molecule | Change of MSI2 | Cell Type | References |
|---|---|---|---|
| RANKL | ↑ | Osteoclast | [34] |
| ETV4 | ↑ | Lung adenocarcinoma | [35] |
| KLF4 | ↓ | Pancreatic cancer | [36] |
| DBC2 | ↓ | Breast cancer | [37] |
| HMGA2 | ↑ | MPNSTs | [21] |
| USP10 | ↑ | Colon cancer | [38] |
| PLAG1, USF2 | ↑ | HSCs | [39] |
| miR-203 | ↓ | Epidermal cells | [40] |
| miR-149 | ↓ | Bladder cancer | [22] |
| miR-143/miR-107 | ↓ | Cervical cancer | [41] |
| miR-143-3p | ↓ | Thyroid carcinoma | [42] |
| miR-145 | ↓ | Endometriosis stem cells | [43] |
| MSI1 | ↓ | Spermatogonia cells | [44] |
| Ro 08–2750 | ↓ | AML | [46] |
| Gossypolone | ↓ | Colon cancer | [47] |
| Largazole | ↓ | NSCLC, CML | [48] |
Note: “↓” represents factors that inhibit MSI2 expression/function, and “↑” represents the opposite.
As some examples, MSI2 expression is upregulated by RANKL, which is a receptor activator of NF-kB ligand, during osteoclast differentiation.34 Chromatin immunoprecipitation (ChIP) and luciferase reporter assays showed that ETV4 directly binds to the promoter of MSI2 and promotes its transcription in lung adenocarcinoma,35 while KLF4 represses MSI2 transcription by directly binding to its promoter in pancreatic cancer cells.36 In breast cancer, DBC2 directly interacts with MSI2, to promote MSI2 polyubiquitination, suppress MSI2-associated oncogenic functions, and induce apoptosis.37 HMGA2 directly activates the MSI2 promoter in NF1-associated malignant peripheral nerve sheath tumors (MPNSTs).21 In colon cancer, USP10 positively regulates the expression of MSI2 by de-ubiquitination.38 In hematopoietic stem cells, ChIP-seq analyses confirmed a preferential co-occupancy of PLAG1 and USF2 at the promoter of MSI2 that promotes MSI2 transcription.39
Post-transcriptional regulation of MSI2 is mediated mainly by non-coding RNAs and other RBPs. For example, miR-203 directly targets the 3ʹ-UTR of MSI2 mRNA, and co-suppression of individual targets, including MSI2, p63 and Skp2, is required for its function of promoting the cell cycle exit and inhibiting the long-term proliferation.40 Long non-coding RNA DANCR upregulates the expression of MSI2 through neutralizing miR-149.22 miR-143/miR-107 are two p53-targeted tumor suppressive miRNAs that directly bind to MSI2 mRNA and inhibit its expression in cervical cancer cells.41 Similarly, in papillary thyroid carcinoma, upregulation of miR-143-3p suppresses tumor progression by directly down-regulating MSI2.42 Highly expressed miR‐145 inhibits invasiveness and proliferation of endometriosis stem cells partially via targeting MSI2.43 Intriguingly, in mouse spermatogonia cells, MSI2 mRNA appears to be directly targeted by MSI1, leading to downregulation of MSI2 expression.44
Small molecule inhibitors are molecule compounds that interact with proteins and reduce the biological activity of the target proteins. Recently, some small molecule inhibitors for MSI1 and MSI2 have been discovered, which may provide a novel therapeutic strategy.45 In acute myeloid leukemia, a small molecule Ro 08–2750 directly interacts with the MSI2 RRM1 and competes for its RNA binding in biochemical assays.46 Gossypolone can disrupt the binding of MSI2 to Numb RNA mainly by acting on RRM1, thereby inducing apoptosis in colon cancer cells.47 The small compound largazole was shown to reduce the protein and mRNA levels of MSI2 and suppress its downstream mammalian target of rapamycin signaling pathway.48 In short, these studies position MSI2 as an available and valuable therapeutic target, providing a basis for future drug research. However, currently discovered small molecule inhibitors of MSI2 mainly act on RRMs, especially RRM1. However, the small molecule inhibitors of MSI2 currently found mainly act on RRMs, especially RRM1. In order to further detect the specificity of the inhibitors, it would have special significance to test MSI2 closely related RBPs containing RRMs with similar sequence, which may greatly reduce the occurrence of adverse reactions.
The Clinical Significance of Musashi-2 Expression in Malignancies
A critical role for MSI2 over-expression in regulation of malignant tumor progression was first demonstrated in 2010. In this study, Ito and colleagues used mouse models of CML to demonstrate that the upregulation of MSI2 and the downregulation of Numb inhibit the differentiation and promote the progression of chronic CML to blast crisis.32 Over the past 10 years, MSI2 has been found to be over-expressed in a variety of tumors, including acute lymphoblastic leukemia (ALL),49 acute myeloid leukemia (AML),50,51 breast,37 pancreas,19 colon,52 liver53,54 lung,55 ovary,56 cervical,41,57 bladder,58 gastric cancer,59 brain cancer,60 and others. The elevated expression of MSI2 in tumor tissues is often positively associated to tumor size, lymph node metastasis, distant metastasis, degree of differentiation, insensitivity to chemoradiotherapy, and poor prognosis. However, some studies have obtained different results. In gastric cancer, Emadi-Baygi et al suggested that MSI2 expression is related with tumor grade, and there is no difference between tumoral and non-tumoral tissues.61 MSI2a is significantly downregulated in TNBC tumors, which is associated with a higher histological grade and poor prognosis.23 Table 2 lists the clinical significance of MSI2 expression that has been reported.
Table 2.
Association of Elevated MSI2 Expression with Clinical Features in Cancers
| MSI2 Status | Cancer Type | Detection Method | Association |
|---|---|---|---|
| Increased expression | ALL49 | qRT-PCR | Poor prognosis |
| AML50,51 | qRT-PCR | Poor prognosis | |
| Breast cancer37 | Tissue microarray | Poor prognosis | |
| Pancreatic cancer19,36 | qRT-PCR,19 IHC,19,36 WB19,36 | Advanced stages,19,36 Tumor size,19 Poor prognosis,19 Differentiation,36 Lymph node metastasis36 | |
| Colorectal cancer52 | Tissue microarray, IHC | Depth of invasion, Lymph node metastasis, Distant metastasis, Advanced stage | |
| Liver cancer53,54 | qRT-PCR,53 WB,53 IHC54 | Chemoresistance,53 Poor prognosis54 | |
| Lung cancer55 | IHC | Poor prognosis | |
| Ovarian cancer56 | Tissue microarray, IHC | Advanced stages | |
| Cervical cancer41,57 | qRT-PCR,41,57 WB57 | Lymph node metastasis,57 Advanced stages,57 Poor prognosis41,57 | |
| Bladder cancer58 | qRT-PCR, WB | Lymph node metastasis, Poor prognosis | |
| Gastric cancer59 | IHC | Invasion depth, Lymph node metastasis, Degree of differentiation, Tumor size | |
| Glioblastoma60 | IHC, WB | Malignancy | |
| Reduced expression | TNBC23 | IHC, qRT-PCR, RNA sequencing | Differentiation, Distant metastasis, Poor prognosis |
Considering the differential expression of MSI2 in cancer tissues and adjacent tissues, it is necessary to conduct a more systematic analysis of the regulation of MSI2 expression. This will greatly help to better understand how MSI2 is abnormallyactivated in cancer.
Musashi-2 in Cancer EMT
It has been well documented that EMT is a complex and dynamic process by which epithelial cells acquire a mesenchymal phenotype.62 Specifically, epithelial cells undergo remarkable morphologic conversion to the elongated fibroblastic phenotype from a cobblestone phenotype.62 During the EMT process, epithelial cells lose their cell-cell junction and initiate actin cytoskeleton reorganization. Moreover, cells lose the expression of epithelial markers such as E-cadherin, ZO-1, Claudins, while cells gain the expression of mesenchymal markers including N-cadherin, Vimentin, fibronectin, α-SMA (α-smooth muscle actin), Twist, Snail/Snail1, Slug/Snail2, ZEB1, and ZEB2.63,64 At the same time, cells have increased activity of matrix metalloproteinases (MMPs) such as MMP-2, MMP-3, and MMP-9.64 After the acquisition of EMT features, cells have increased motility and invasiveness, which promotes tumor progression and metastasis.63
Currently, various studies have shown that MSI2 is associated with increased expression of EMT makers. For example, MSI2 promotes extrahepatic cholangiocarcinoma (eCCA) EMT by down-regulating E-cadherin and up-regulating N-cadherin and vimentin expression. Similarly, in bladder cancer, decreased MSI2 protein caused by knocking down DANCR enhances E-cadherin expression and reduces N-cadherin and vimentin expression, promoting cell migration, invasion, and EMT of bladder cancer cells.22 The expression level of MSI2 is positively related to the expression level of vimentin, a-SMA, and N-cadherin, and negatively correlated with ZO-1 and E-cadherin in HCC.54 In papillary thyroid carcinoma, the silencing of MSI2 exhibited significantly decreased expression of MMP-2 and MMP-9.42 We recently identified MSI2 accelerates the migration and invasion of pancreatic cancer cells through ISYNA1/ZEB-1 pathway.65
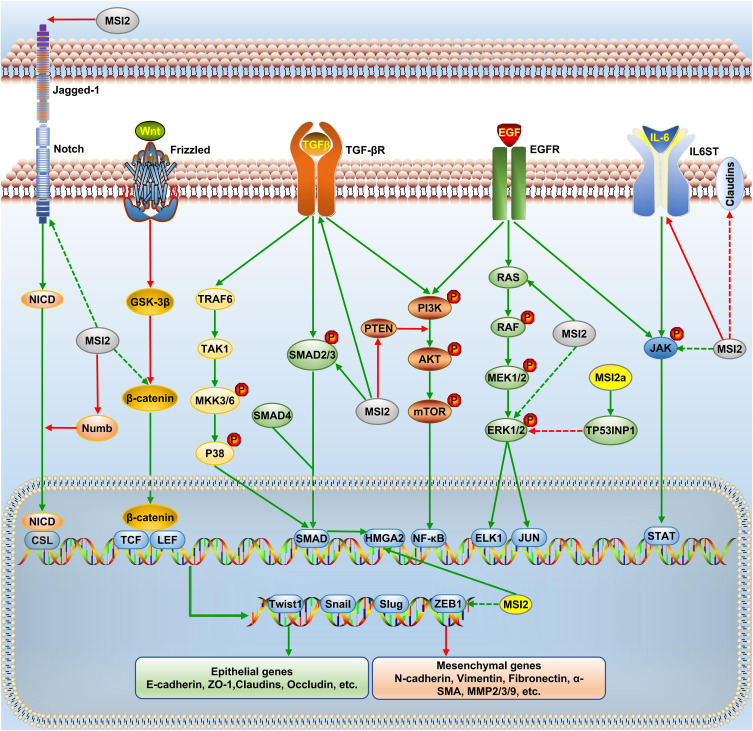
The TGF-β, EGF, Notch, and Wnt signaling are key transduction pathways that are involved in EMT. Later in this review, we will discuss the crosstalk between MSI2 and these EMT-related signal pathways. The regulatory network is shown in Figure 2. In particular, we hope to provide a perspective on the underlying mechanism of MSI2-mediated EMT and provide insights into the intervention of EMT.
Figure 2.
MSI2 and EMT signaling networks. MSI2 affects the expression of EMT transcription factors (Twist1, Snail, Slug and ZEB1) and key proteins (epithelial and mesenchymal genes) through EGF, TGF-β, Notch, and Wnt pathways. Green arrow, positive regulation, and red, negative regulation. Solid arrow, direct regulation, and dashed arrow, indirect regulation.
Crosstalk Between Musashi-2 and EGF Signaling Pathway
The EGFR family includes 4 different receptors: EGFR/ErbB-1, HER2/ErbB-2, HER3/ErbB-3, and HER4/ErbB-4.66,67 A number of different ligands, including EGF-like molecules, transforming growth factor-alpha (TGF-α), and neuregulin, activate the receptors by binding to the extracellular domain, which induces the formation of receptor homodimers or heterodimers.67 After dimerization, autophosphorylation of tyrosine residues in the cytoplasmic region occurs, which provides docking sites for signal transducers, such as Ras, to bind, and therefore, initiating intracellular signaling cascades and gene transcription.66,67 Downstream signaling transduction cascades of EGFR can be roughly divided into the following types: MAPK/ERK, PI3K/Akt/mTOR/NF-κB, protein kinase C (PKC), and JAK/STAT pathways.68 These well-studied signaling cascades are known to control gene expression and cancer-promoting phenotypes including EMT. ERK phosphorylation inhibits the degradation of pro-mesenchymal transcription factors such as ZEB, Snail, Slug, and Twist.69 Several E-cadherin repressors such as Snail, Slug, Twist, Sip1, ZEB1, and ZEB2 are controlled by the NF-κB pathways.70,71 STAT3 is activated and binds to the promoter of the gene encoding Twist upon EGF stimulation.72
Through PAR-CLIP and RNA-Seq analysis, several pathways have been shown to be significantly affected by MSI2 knockdown, including eIF2, eIF4/p70S6K, and EGF and HGF growth factor pathways.73 The KRAS gene is known as one of the most common mutated genes in human tumors and encodes a vital component of the MAPK/ERK pathway; In 1994, Shirayama et al identified MSI2 as a suppressor of the heat shock sensitivity caused by the loss of the IRA1 product, a negative regulator of the RAS protein, suggesting that the MSI2 protein may interfere with the activity of the RAS protein. A recent study has shown that MSI2 positively regulates KRAS expression in bladder cancer cells by directly binding to the KRAS mRNA and promoting its translation.74 In our previous study, the silencing of MSI2 inhibited EGF-mediated EGFR phosphorylation at tyrosine 1068 and reversed EGF-induced expression of the key proteins in EMT (ZEB1, E-cadherin, ZO-1, β-catenin, and c-Myc) through the MAPK/ERK pathway.75 More importantly, MSI2 not only functions an RNA-binding protein, but also binds to ZEB1, c-Myc and p-ERK, which provides an additional mechanism of EMT in pancreatic cancer.75
In another study, knockout of MSI2 reduces phosphorylation of ERK and S6 in LSCs, indicating that MSI2 interacts with MAPK and mTOR signaling pathways. However, in bladder cancer, overexpression of MSI2 promotes migration and invasion by positively regulating the phosphorylation of JAK2 and STAT3.58 In ovarian cancer, miR-149-induced MSI2 silencing led to increased E-cadherin expression but decreased expression of p-PI3K, p-AKT, vimentin, and N-cadherin.76 Over-expression of MSI2 induces a phenotype of intestinal epithelial transformation similar to the activation of WNT pathway; however, this phenotype is mediated by inhibiting PTEN translation and promoting AKT/mTORC1 signaling, independent of WNT activation.24 One study also demonstrated that MSI2 promotes proliferation, migration, and invasion, and inhibits apoptosis by accelerating AKT and STAT3 phosphorylation in bladder cancer.77
However, under the stimulation of IL-6, MSI2 inhibits the phosphorylation of STAT3 and ERK proteins by directly binding and degrading the mRNA of IL6ST, which in turn affects JAK/STAT and MAPK signaling pathways.73 Similarly, a study in triple-negative breast cancer demonstrated that the MSI2a isoform is able to downregulate the expression of p-ERK1/2 and Slug, N-cadherin, and vimentin, but up-regulate ZO-1, β-catenin, and E-cadherin by combining and stabilizing TP53INP1 mRNA.23 Such discrepancies might be attributed to the different cellular environment.
Crosstalk Between Musashi-2 and TGF-β Signaling Pathway
In human cells, TGF-β-induced activation of the receptor complex (two copies of TGF-βR1, two copies of TGF-βR2) leads to phosphorylation and activation of SMAD2 and SMAD3. Phosphorylated SMAD2 and SMAD3 then form trimers with SMAD4, and translocate into the nucleus, where they associate and cooperate with DNA binding transcription factors to activate or repress target gene transcription.78 In the canonical TGF-β signaling pathway, TGF-β-induced SMAD complexes activate the transcription of the Snail, Slug, ZEB1, Twist, and HMGA2.79 HMGA2, as a group of transcription factors, induces the expression of Snail, Slug, and Twist1.80 Other than the canonical SMAD-dependent pathway, TGF-β receptors also relay the signaling by some additional signal transduction pathways, such as PI3K/AKT, ERK1/2, and JNK/P38, which also contribute in various ways to the EMT process.81,82 Phosphorylated P38 cooperates with Smad3/4 in TGF-β associated EMT through the transcription factor ATF2.83
In canonical SMAD-dependent TGF-β signaling pathway, downregulation of MSI2 significantly inhibits the expression of TGF-βR1, pSMAD3, Snail, Slug, and vimentin protein, but increased E-cadherin protein expression; interestingly, TGF-β also increases MSI2 protein expression, which suggests a positive feedback loop of MSI2-TGF-β/TGF-βR1/SMAD3 signaling.60 In non-small cell lung cancer, MSI2 depletion not only downregulates pro-EMT factors vimentin, Snail, Slug, and anti-EMT factors E-cadherin, but also up-regulates partial pro-EMT factors ZEB1, ZEB2, and FOXC2 through supporting TGF-βR1/SMAD3 expression, reflecting a mixed effect on EMT.55 It has also been demonstrated that loss of MSI2 can increase claudin-3/claudin-5/claudin-7 expression independently of the TGF-βR1/SMAD3 pathway.55 Similarly, in the hematopoietic system, MSI2 was identified as a stem cell regulator, by directly regulating the expression of SMAD3.33 In addition, analysis of 3ʹUTRs binding-sites and RIP-PCR identified BRD4, c-MET, and HMGA2 as direct targets of MSI2 in pancreatic cancer,84 suggesting its role in regulating EMT.
MSI2 silencing enhances the chemical sensitivity of acute lymphoblastic leukemia cells to daunorubicin, induces cell cycle arrest by downregulating CyclinD1 and upregulating p21 in the G0/G1 phase, and induces apoptosis by upregulating Bax expression and inhibiting p-AKT, p-ERK1/2, p-p38, and Bcl-2 expression.85 The same result is also seen in breast cancer.37 In addition, MSI2 knockdown inhibits proliferation and promotes apoptosis by deducting the phosphorylation of ERK and p38, and the expression of downstream targets, c-Myc, c-Fos, and MAPKAPK2, independent of changes in p-AKT expression.86
Crosstalk Between Musashi-2 and Notch Signaling Pathway
The activation of the Notch signaling pathway is initiated after the interaction of Notch ligands and its receptor, presented on the surface of neighboring cells. So far, four receptors (Notch1-4) and two families of ligands (Delta-like and Jagged) have been identified.87 Subsequently, Notch is cleaved and released to the Notch intracellular domain (NICD) through two proteolytic cleavage events by tumor necrosis factor-α-converting enzyme (TACE) and γ-secretase complex.87 NICD translocates to the nucleus and binds to CSL (C protein binding factor 1/Suppressor of Hairless/Lag-1). The formation of a complex of NICD and CSL converts CSL from a transcriptional repressor to a transcriptional activator, leading to expression of target genes (Hey1, Hes1, etc.).88 Currently, Notch is considered to activate EMT through transcriptional upregulation of several EMT-associated TFs, such as Snail and Slug.89,90
Bennett et al demonstrated Dll1, Jagged-1 and Notch2 as the high-confidence MSI2 targets through high-throughput studies of MSI2-binding targets.26 Troschel et al demonstrated that knockdown of MSI1 and MSI2 suppresses the Notch pathway and EGFR protein expression in triple-negative breast cancer.91 LFNG is known to be a Notch1 receptor glycosyltransferase that regulates the expression of Notch1 receptor. Recently, MSI2 was identified as a molecule that maintains the stemness properties of CD44v6+ LCSCs through directly bounding to Lunatic fringe (LFNG) mRNA and protein, which in turn activates the Notch1 signaling pathway.92 In osteoclast, MSI2 is required for the optimal differentiation and survival by activating Notch2 and Hes1, which leads to the activation of NF-κB signaling.34 In contrast, MSI1/2 inhibits EMT in vitro by repressing the translation of Jagged-1 ligand and the notch signaling.93
Numb, which negatively regulates Notch pathway by ubiquitylation and degradation of NICD, can antagonize Notch-mediated EMT in cancers.94 The inhibitory effect of MSI2 on Numb protein expression and activity has been widely confirmed (citations?). MSI2 can unlock the differentiation potential of blast crisis CML and impair its growth by binding Numb mRNAs in 3ʹ-UTR.32 Similarly, knockdown of MSI2 reduces the growth of glioblastoma cells and medulloblastoma cells by upregulating Numb.18 Subsequently, inhibition of the MSI2/Numb/c-Myc signaling pathway is also demonstrated to induce apoptosis and arrest cell cycle at the G0/G1 phase in B-cell lymphoma (BCL) cells.95
In addition, the alternate estrogen receptor has been shown to induce EMT through activation of the Notch pathway.96 High MSI2 expression accelerates the growth of cells by binding estrogen receptor 1 (ESR1) mRNA and increasing the stability of ESR1 protein in breast cancer.97 Thus, MSI2/ESR may be a novel mechanism by which MSI2 regulates Notch signaling and promotes the occurrence of EMT in breast cancer.
Crosstalk Between Musashi-2 and Wnt Signaling Pathway
The canonical WNT signaling pathway is initiated by soluble Wnt ligands that bind to the Frizzled receptors, and subsequently, trigger a series of signaling events that culminate in the nuclear translocation of β-catenin.98 In the nucleus, β-Catenin directly binds to other transcription factors associated with the promoters of Slug, ZEB1, and Twist and induces their expression.99,100
In AML, one study strongly demonstrated that MSI2 knockdown significantly decreases expression of the downstream genes of Wnt, Ras-MAPK, and Myc pathways.11 MSI2 knockdown repressed β-catenin and LEF-1/TCF-4 protein and mRNA levels, inhibited tumor cell migration and invasion in HCC.101 MSI2 knockdown suppresses pro-EMT factors MMP-2, MMP-9, N-cadherin, and c-Myc, and increases anti-EMT E-cadherin expression through up-regulating β-catenin, Gli-1 and Patch, which suggested that the Wnt/β-catenin and Hedgehog signaling pathways are involved in the regulation of EMT and metastasis in esophageal squamous cell carcinoma.102 A list of EMT-related genes targeted by MSI2 is summarized in Table 3.
Table 3.
Summary of EMT-Associated Genes Directly Targeted by MSI2
| Target Gene | Target Change | Cancer Type | Effect of MSI2 in EMT-Related Signals | Reference |
|---|---|---|---|---|
| KRAS | ↑ | Bladder cancer | Activation of MAPK/ERK signal | [74] |
| PTEN | ↓ | Colorectal cancer | Activation of PI3K/AKT/mTOR signal | [24] |
| TGF-βR1 | ↑ | NSCLC | Activation of PI3K/AKT, ERK1/2 and JNK/P38 signals | [55] |
| SMAD3 | ↑ | NSCLC, HSCs | Transcriptional activation of Snail, Slug, ZEB1, Twist and HMGA2 | [33,55] |
| HMGA2 | ↑ | Pancreatic cancer | Transcriptional activation of Snail, Slug and Twist1 | [84] |
| Jagged-1 | ↓ | Epithelial-luminal cell | Down-regulation of Notch signal | [93] |
| Numb | ↓ | CML | Up-regulation of Notch signal | [32] |
| IL6ST | ↓ | Embryonic kidney cells | Suppression of JAK/STAT signal | [73] |
| TP53INP1 | ↑ | TNBC | Activates P53 to suppress MAPK/ERK signal | [23] |
Notes: “↓” means the target gene is down-regulated by MSI2, and “↑” means up-regulation under the action of MSI2.
Conclusions
Mesenchymal-to-epithelial transition (MET) is the reverse process of EMT. The mutual transformation of tumor cells between an epithelial phenotype and a mesenchymal phenotype illustrates the plastic nature of this transition. The process of EMT is regulated by multiple signaling in cancer, including but not limited to EGF, TGF-β, Notch, and the Wnt pathway. Numerous crosstalk sites exist between MSI2 and these signal transduction pathways, which increase our knowledge of the regulatory mechanisms underlying EMT. Moreover, all these findings further highlight the importance of post-transcriptional control in EMT. However, the mechanisms by which MSI2 regulates the Wnt/β-catenin, JAK/STAT, and MAPK/p38 pathways remain to be fully understood, and further experiments are needed to explore. Additionally, different MSI2 isoforms, especially classical isoform a may play significant regulatory roles in cancer, which may become a research hotspot. Given the expression characteristics of MSI2 in malignant tumors, MSI2-targeted therapy may become a new strategy to reverse EMT-related cancer progression. Recently, small molecule inhibitors of MSI2 have been shown to be effective in vivo and in vitro. However, attempts to develop MSI2 inhibitors are still in the early stages. In the future, the research and development of MSI2-specific inhibitors, and even MSI2 isoform-specific inhibitors, will contribute to the precise treatment of tumors. It is believed that MSI2-targeted drugs will be available in the clinic in the near future.
Acknowledgments
We thank the General Laboratory of the First Hospital of China Medical University for technical supports.
Funding Statement
This work was supported by Chinese National Science Foundation (NO. 81672835 to Ming Dong).
Abbreviations
ALL, acute lymphoid leukemia; AML, acute myeloid leukemia; BCL, B-cell lymphoma; ChIP-seq, chromatin immunoprecipitation sequencing; CML, chronic myeloid leukemia; eIF4G, eukaryotic translation initiation factor 4 gamma; ePABP, embryonic poly(A)-binding protein; EGFR, epidermal growth factor receptor; EMT, epithelial-mesenchymal transition; ESCs, embryonic stem cells; HCC, hepatocellular carcinoma; HSCs, hematopoietic stem cells; LCSCs, liver cancer stem cells; LSCs, leukemia stem cells; MET, mesenchymal-to-epithelial transition; MLL, mixed-lineage leukemia; MMPs, matrix metalloproteinases; MPNSTs, Malignant peripheral nerve sheath tumors; MSI, Musashi; NICD, Notch intracellular domain; PABP, poly(A)-binding protein; PABPC1, poly(A)-binding protein cytoplasmic 1; PKC, protein kinase C; RBP, RNA-binding proteins; RRM, RNA recognition motifs; SOP, sensory organ precursor; TGF-α, transforming growth factor-alpha; TGF-β, transforming growth factor-beta; TFs, transcription factors; TNBC, triple-negative breast cancer.
Disclosure
The authors declare that they have no competing interest.
References
- 1.Nakamura M, Okano H, Blendy JA, Montell C. Musashi, a neural RNA-binding protein required for Drosophila adult external sensory organ development. Neuron. 1994;13:67–81. doi: 10.1016/0896-6273(94)90460-X [DOI] [PubMed] [Google Scholar]
- 2.Good P, Yoda A, Sakakibara S, et al. The human Musashi homolog 1 (MSI1) gene encoding the homologue of Musashi/Nrp-1, a neural RNA-binding protein putatively expressed in CNS stem cells and neural progenitor cells. Genomics. 1998;52:382–384. doi: 10.1006/geno.1998.5456 [DOI] [PubMed] [Google Scholar]
- 3.Sakakibara S, Nakamura Y, Satoh H, Okano H. Rna-binding protein Musashi2: developmentally regulated expression in neural precursor cells and subpopulations of neurons in mammalian CNS. J Neurosci. 2001;21:8091–8107. doi: 10.1523/JNEUROSCI.21-20-08091.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sakakibara S, Nakamura Y, Yoshida T, et al. RNA-binding protein Musashi family: roles for CNS stem cells and a subpopulation of ependymal cells revealed by targeted disruption and antisense ablation. Proc Natl Acad Sci U S A. 2002;99:15194–15199. doi: 10.1073/pnas.232087499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaneko Y, Sakakibara S, Imai T, et al. Musashi1: an evolutionally conserved marker for CNS progenitor cells including neural stem cells. Dev Neurosci. 2000;22:139–153. doi: 10.1159/000017435 [DOI] [PubMed] [Google Scholar]
- 6.Sakakibara S, Okano H. Expression of neural RNA-binding proteins in the postnatal CNS: implications of their roles in neuronal and glial cell development. J Neurosci. 1997;17:8300–8312. doi: 10.1523/JNEUROSCI.17-21-08300.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin JC, Tsai JT, Chao TY, Ma HI, Chien CS, Liu WH. MSI1 associates glioblastoma radioresistance via homologous recombination repair, tumor invasion and cancer stem-like cell properties. Radiother Oncol. 2018;129:352–363. doi: 10.1016/j.radonc.2018.09.014 [DOI] [PubMed] [Google Scholar]
- 8.Moghbeli M, Forghanifard MM, Sadrizadeh A, Mozaffari HM, Golmakani E, Abbaszadegan MR. Role of Msi1 and MAML1 in regulation of notch signaling pathway in patients with esophageal squamous cell carcinoma. J Gastrointest Cancer. 2015;46:365–369. doi: 10.1007/s12029-015-9753-9 [DOI] [PubMed] [Google Scholar]
- 9.Nikpour P, Emadi-Baygi M, Mohhamad-Hashem F, Maracy MR, Haghjooy-Javanmard S. MSI1 overexpression in diffuse type of gastric cancer. Pathol Res Pract. 2013;209:10–13. doi: 10.1016/j.prp.2012.09.008 [DOI] [PubMed] [Google Scholar]
- 10.De Weer A, Speleman F, Cauwelier B, et al. EVI1 overexpression in t(3;17) positive myeloid malignancies results from juxtaposition of EVI1 to the MSI2 locus at 17q22. Haematologica. 2008;93:1903–1907. doi: 10.3324/haematol.13192 [DOI] [PubMed] [Google Scholar]
- 11.Kharas MG, Lengner CJ, Al-Shahrour F, et al. Musashi-2 regulates normal hematopoiesis and promotes aggressive myeloid leukemia. Nat Med. 2010;16:903–908. doi: 10.1038/nm.2187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sheng W, Dong M, Chen C, et al. Cooperation of Musashi-2, Numb, MDM2, and P53 in drug resistance and malignant biology of pancreatic cancer. FASEB J. 2017;31:2429–2438. doi: 10.1096/fj.201601240R [DOI] [PubMed] [Google Scholar]
- 13.Park SM, Gonen M, Vu L, et al. Musashi2 sustains the mixed-lineage leukemia-driven stem cell regulatory program. J Clin Invest. 2015;125:1286–1298. doi: 10.1172/JCI78440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barbouti A, Hoglund M, Johansson B, et al. A novel gene, MSI2, encoding a putative RNA-binding protein is recurrently rearranged at disease progression of chronic myeloid leukemia and forms a fusion gene with HOXA9 as a result of the cryptic t(7;17)(p15;q23). Cancer Res. 2003;63:1202–1206. [PubMed] [Google Scholar]
- 15.Siddall NA, McLaughlin EA, Marriner NL, Hime GR. The RNA-binding protein Musashi is required intrinsically to maintain stem cell identity. Proc Natl Acad Sci U S A. 2006;103:8402–8407. doi: 10.1073/pnas.0600906103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sutherland JM, Sobinoff AP, Gunter KM, et al. Knockout of RNA binding protein MSI2 impairs follicle development in the mouse ovary: characterization of MSI1 and MSI2 during folliculogenesis. Biomolecules. 2015;5:1228–1244. doi: 10.3390/biom5031228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sutherland JM, Fraser BA, Sobinoff AP, et al. Developmental expression of Musashi-1 and Musashi-2 RNA-binding proteins during spermatogenesis: analysis of the deleterious effects of dysregulated expression. Biol Reprod. 2014;90:92. doi: 10.1095/biolreprod.113.115261 [DOI] [PubMed] [Google Scholar]
- 18.Cox JL, Wilder PJ, Gilmore JM, Wuebben EL, Washburn MP, Rizzino A. The SOX2-interactome in brain cancer cells identifies the requirement of MSI2 and USP9X for the growth of brain tumor cells. PLoS One. 2013;8:e62857. doi: 10.1371/journal.pone.0062857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sheng W, Dong M, Chen C, Li Y, Liu Q, Dong Q. Musashi2 promotes the development and progression of pancreatic cancer by down-regulating Numb protein. Oncotarget. 2017;8:14359–14373. doi: 10.18632/oncotarget.8736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu C, Wyatt AW, Lapuk AV, et al. Integrated genome and transcriptome sequencing identifies a novel form of hybrid and aggressive prostate cancer. J Pathol. 2012;227:53–61. doi: 10.1002/path.3987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang K, Guo W, Ren T, et al. Knockdown of HMGA2 regulates the level of autophagy via interactions between MSI2 and Beclin1 to inhibit NF1-associated malignant peripheral nerve sheath tumour growth. J Exp Clin Cancer Res. 2019;38:185. doi: 10.1186/s13046-019-1183-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhan Y, Chen Z, Li Y, et al. Long non-coding RNA DANCR promotes malignant phenotypes of bladder cancer cells by modulating the miR-149/MSI2 axis as a ceRNA. J Exp Clin Cancer Res. 2018;37:273. doi: 10.1186/s13046-018-0921-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li M, Li A-Q, Zhou S-L, Lv H, Wei P, Yang W-T. RNA-binding protein MSI2 isoforms expression and regulation in progression of triple-negative breast cancer. J Exp Clin Cancer Res. 2020;39:92. doi: 10.1186/s13046-020-01587-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang S, Li N, Yousefi M, et al. Transformation of the intestinal epithelium by the MSI2 RNA-binding protein. Nat Commun. 2015;6:6517. doi: 10.1038/ncomms7517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zearfoss NR, Deveau LM, Clingman CC, et al. A conserved three-nucleotide core motif defines Musashi RNA binding specificity. J Biol Chem. 2014;289:35530–35541. doi: 10.1074/jbc.M114.597112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bennett CG, Riemondy K, Chapnick DA, et al. Genome-wide analysis of Musashi-2 targets reveals novel functions in governing epithelial cell migration. Nucleic Acids Res. 2016;44:3788–3800. doi: 10.1093/nar/gkw207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cragle C, MacNicol AM. Musashi protein-directed translational activation of target mRNAs is mediated by the poly(A) polymerase, germ line development defective-2. J Biol Chem. 2014;289:14239–14251. doi: 10.1074/jbc.M114.548271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kawahara H, Imai T, Imataka H, Tsujimoto M, Matsumoto K, Okano H. Neural RNA-binding protein Musashi1 inhibits translation initiation by competing with eIF4G for PABP. J Cell Biol. 2008;181:639–653. doi: 10.1083/jcb.200708004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cragle CE, MacNicol MC, Byrum SD, et al. Musashi interaction with poly(A)-binding protein is required for activation of target mRNA translation. J Biol Chem. 2019;294:10969–10986. doi: 10.1074/jbc.RA119.007220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wuebben EL, Mallanna SK, Cox JL, Rizzino A, Zhou Z. Musashi2 is required for the self-renewal and pluripotency of embryonic stem cells. PLoS One. 2012;7:e34827. doi: 10.1371/journal.pone.0034827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.MacNicol MC, Cragle CE, McDaniel FK, et al. Evasion of regulatory phosphorylation by an alternatively spliced isoform of Musashi2. Sci Rep. 2017;7:11503. doi: 10.1038/s41598-017-11917-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ito T, Kwon HY, Zimdahl B, et al. Regulation of myeloid leukaemia by the cell-fate determinant Musashi. Nature. 2010;466:765–768. doi: 10.1038/nature09171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Park S-M, Deering RP, Lu Y, et al. Musashi-2 controls cell fate, lineage bias, and TGF-β signaling in HSCs. J Exp Med. 2014;211:71–87. doi: 10.1084/jem.20130736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fujiwara T, Zhou J, Ye S, Zhao H. RNA-binding protein Musashi2 induced by RANKL is critical for osteoclast survival. Cell Death Dis. 2016;7:e2300. doi: 10.1038/cddis.2016.213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cheng T, Zhang Z, Cheng Y, et al. ETV4 promotes proliferation and invasion of lung adenocarcinoma by transcriptionally upregulating MSI2. Biochem Biophys Res Commun. 2019;516:278–284. doi: 10.1016/j.bbrc.2019.06.115 [DOI] [PubMed] [Google Scholar]
- 36.Guo K, Cui J, Quan M, et al. The novel KLF4/MSI2 signaling pathway regulates growth and metastasis of pancreatic cancer. Clin Cancer Res. 2017;23:687–696. doi: 10.1158/1078-0432.CCR-16-1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Choi YM, Kim KB, Lee JH, et al. DBC2/RhoBTB2 functions as a tumor suppressor protein via Musashi-2 ubiquitination in breast cancer. Oncogene. 2017;36:2802–2812. doi: 10.1038/onc.2016.441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ouyang SW, Liu TT, Liu XS, et al. USP10 regulates Musashi-2 stability via deubiquitination and promotes tumour proliferation in colon cancer. FEBS Lett. 2019;593:406–413. doi: 10.1002/1873-3468.13323 [DOI] [PubMed] [Google Scholar]
- 39.Belew MS, Bhatia S, Keyvani Chahi A, Rentas S, Draper JS, Hope KJ. PLAG1 and USF2 co-regulate expression of Musashi-2 in human hematopoietic stem and progenitor cells. Stem Cell Rep. 2018;10:1384–1397. doi: 10.1016/j.stemcr.2018.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jackson SJ, Zhang Z, Feng D, et al. Rapid and widespread suppression of self-renewal by microRNA-203 during epidermal differentiation. Development. 2013;140:1882–1891. doi: 10.1242/dev.089649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dong P, Xiong Y, Hanley SJB, Yue J, Watari H. Musashi-2, a novel oncoprotein promoting cervical cancer cell growth and invasion, is negatively regulated by p53-induced miR-143 and miR-107 activation. J Exp Clin Cancer Res. 2017;36:150. doi: 10.1186/s13046-017-0617-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang Z-L, Wang C, Liu W, Ai Z-L. Upregulation of microRNA-143-3p induces apoptosis and suppresses proliferation, invasion, and migration of papillary thyroid carcinoma cells by targeting MSI2. Exp Mol Pathol. 2020;112:104342. doi: 10.1016/j.yexmp.2019.104342 [DOI] [PubMed] [Google Scholar]
- 43.Wang L, Xing Q, Feng T, He M, Yu W, Chen H. SNP rs710886 A>G in long noncoding RNA PCAT1 is associated with the risk of endometriosis by modulating expression of multiple stemness-related genes via microRNA-145 signaling pathway. J Cell Biochem. 2020;121:1703–1715. doi: 10.1002/jcb.29406 [DOI] [PubMed] [Google Scholar]
- 44.Sutherland JM, Sobinoff AP, Fraser BA, et al. RNA binding protein Musashi-1 directly targets Msi2 and Erh during early testis germ cell development and interacts with IPO5 upon translocation to the nucleus. FASEB j. 2015;29:2759–2768. doi: 10.1096/fj.14-265868 [DOI] [PubMed] [Google Scholar]
- 45.Minuesa G, Antczak C, Shum D, et al. A 1536-well fluorescence polarization assay to screen for modulators of the MUSASHI family of RNA-binding proteins. Comb Chem High Throughput Screen. 2014;17:596–609. doi: 10.2174/1386207317666140609122714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Minuesa G, Albanese SK, Xie W, et al. Small-molecule targeting of MUSASHI RNA-binding activity in acute myeloid leukemia. Nat Commun. 2019;10:2691. doi: 10.1038/s41467-019-10523-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lan L, Liu H, Smith AR, et al. Natural product derivative Gossypolone inhibits Musashi family of RNA-binding proteins. BMC Cancer. 2018;18:809. doi: 10.1186/s12885-018-4704-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang M, Sun X-Y, Zhou Y-C, et al. Suppression of Musashi‑2 by the small compound largazole exerts inhibitory effects on malignant cells. Int J Oncol. 2020;56:1274–1283. doi: 10.3892/ijo.2020.4993 [DOI] [PubMed] [Google Scholar]
- 49.Aly RM, Ghazy HF. Prognostic significance of MSI2 predicts unfavorable outcome in adult B-acute lymphoblastic leukemia. Int J Lab Hematol. 2015;37:272–278. doi: 10.1111/ijlh.12284 [DOI] [PubMed] [Google Scholar]
- 50.Ye A-F, Han Y-X, Zhang S-H, Li H-Y, Chen C-Q, Wu J-B. [Expression of musashi-2 gene in leukemia stem cells from acute myeloid leukemia patients]. Zhongguo Shi Yan Xue ye Xue Za Zhi. 2014;22:1235–1238. doi: 10.7534/j.issn.1009-2137.2014.05.009.Chinese. [DOI] [PubMed] [Google Scholar]
- 51.Lu Y, Yu M, Mu Q, et al. [Expression of Musashi2 gene in de novo acute myeloid leukemia and its clinical implications]. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2014;31:713–718. doi: 10.3760/cma.j.issn.1003-9406.2014.06.007.Chinese. [DOI] [PubMed] [Google Scholar]
- 52.Zong Z, Zhou T, Rao L, et al. Musashi2 as a novel predictive biomarker for liver metastasis and poor prognosis in colorectal cancer. Cancer Med. 2016;5:623–630. doi: 10.1002/cam4.624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fang T, Lv H, Wu F, et al. Musashi 2 contributes to the stemness and chemoresistance of liver cancer stem cells via LIN28A activation. Cancer Lett. 2017;384:50–59. doi: 10.1016/j.canlet.2016.10.007 [DOI] [PubMed] [Google Scholar]
- 54.He L, Zhou X, Qu C, et al. Musashi2 predicts poor prognosis and invasion in hepatocellular carcinoma by driving epithelial-mesenchymal transition. J Cell Mol Med. 2014;18:49–58. doi: 10.1111/jcmm.12158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kudinov AE, Deneka A, Nikonova AS, et al. Musashi-2 (MSI2) supports TGF-beta signaling and inhibits claudins to promote non-small cell lung cancer (NSCLC) metastasis. Proc Natl Acad Sci U S A. 2016;113:6955–6960. doi: 10.1073/pnas.1513616113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee J, An S, Choi YM, et al. Musashi-2 is a novel regulator of paclitaxel sensitivity in ovarian cancer cells. Int J Oncol. 2016;49:1945–1952. doi: 10.3892/ijo.2016.3683 [DOI] [PubMed] [Google Scholar]
- 57.Liu Y, Fan Y, Wang X, Huang Z, Shi K, Zhou B. Musashi-2 is a prognostic marker for the survival of patients with cervical cancer. Oncol Lett. 2018;15:5425–5432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yang C, Zhang W, Wang L, et al. Musashi-2 promotes migration and invasion in bladder cancer via activation of the JAK2/STAT3 pathway. Lab Investig. 2016;96:950–958. doi: 10.1038/labinvest.2016.71 [DOI] [PubMed] [Google Scholar]
- 59.Yang Z, Li J, Shi Y, Li L, Guo X. Increased musashi 2 expression indicates a poor prognosis and promotes malignant phenotypes in gastric cancer. Oncol Lett. 2019;17:2599–2606. doi: 10.3892/ol.2019.9889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jiang X, Tan J, Wen Y, et al. MSI2-TGF-beta/TGF-beta R1/SMAD3 positive feedback regulation in glioblastoma. Cancer Chemother Pharmacol. 2019;84:415–425. doi: 10.1007/s00280-019-03892-5 [DOI] [PubMed] [Google Scholar]
- 61.Emadi-Baygi M, Nikpour P, Mohammad-Hashem F, Maracy MR, Haghjooy-Javanmard S. MSI2 expression is decreased in grade II of gastric carcinoma. Pathol Res Pract. 2013;209:689–691. doi: 10.1016/j.prp.2013.07.008 [DOI] [PubMed] [Google Scholar]
- 62.De Craene B, Berx G. Regulatory networks defining EMT during cancer initiation and progression. Nat Rev Cancer. 2013;13. [DOI] [PubMed] [Google Scholar]
- 63.Thiery JP, Acloque H, Huang RYJ, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007 [DOI] [PubMed] [Google Scholar]
- 64.Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15:178–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhou L, Sheng W, Jia C, et al. Musashi2 promotes the progression of pancreatic cancer through a novel ISYNA1-p21/ZEB-1 pathway. J Cell Mol Med. 2020;24:10560–10572. doi: 10.1111/jcmm.15676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Herbst RS. Review of epidermal growth factor receptor biology. Int J Radiat Oncol Biol Phys. 2004;59:21–26. doi: 10.1016/j.ijrobp.2003.11.041 [DOI] [PubMed] [Google Scholar]
- 67.Yewale C, Baradia D, Vhora I, Patil S, Misra A. Epidermal growth factor receptor targeting in cancer: a review of trends and strategies. Biomaterials. 2013;34:8690–8707. [DOI] [PubMed] [Google Scholar]
- 68.Chong CR, Jänne PA. The quest to overcome resistance to EGFR-targeted therapies in cancer. Nat Med. 2013;19:1389–1400. doi: 10.1038/nm.3388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.McCubrey JA, Steelman LS, Chappell WH, et al. Mutations and deregulation of Ras/Raf/MEK/ERK and PI3K/PTEN/Akt/mTOR cascades which alter therapy response. Oncotarget. 2012;3:954–987. doi: 10.18632/oncotarget.652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chua HL, Bhat-Nakshatri P, Clare SE, Morimiya A, Badve S, Nakshatri H. NF-kappaB represses E-cadherin expression and enhances epithelial to mesenchymal transition of mammary epithelial cells: potential involvement of ZEB-1 and ZEB-2. Oncogene. 2007;26:711–724. [DOI] [PubMed] [Google Scholar]
- 71.Criswell TL, Arteaga CL. Modulation of NFkappaB activity and E-cadherin by the type III transforming growth factor beta receptor regulates cell growth and motility. J Biol Chem. 2007;282:32491–32500. doi: 10.1074/jbc.M704434200 [DOI] [PubMed] [Google Scholar]
- 72.Lo H-W, Hsu S-C, Xia W, et al. Epidermal growth factor receptor cooperates with signal transducer and activator of transcription 3 to induce epithelial-mesenchymal transition in cancer cells via up-regulation of TWIST gene expression. Cancer Res. 2007;67:9066–9076. doi: 10.1158/0008-5472.CAN-07-0575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Duggimpudi S, Kloetgen A, Maney SK, et al. Transcriptome-wide analysis uncovers the targets of the RNA-binding protein MSI2 and effects of MSI2’s RNA-binding activity on IL-6 signaling. J Biol Chem. 2018;293:15359–15369. doi: 10.1074/jbc.RA118.002243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tsujino T, Sugito N, Taniguchi K, et al. MicroRNA-143/Musashi-2/KRAS cascade contributes positively to carcinogenesis in human bladder cancer. Cancer Sci. 2019;110:2189–2199. doi: 10.1111/cas.14035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sheng W, Shi X, Lin Y, et al. Musashi2 promotes EGF-induced EMT in pancreatic cancer via ZEB1-ERK/MAPK signaling. J Exp Clin Cancer Res. 2020;39:16. doi: 10.1186/s13046-020-1521-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhao LW, Yu AJ, Zhang YJ, Wang XC, Han B, Wang XH. MicroRNA-149 suppresses the malignant phenotypes of ovarian cancer via downregulation of MSI2 and inhibition of PI3K/AKT pathway. Eur Rev Med Pharmacol Sci. 2020;24:55–64. doi: 10.26355/eurrev_202001_19895 [DOI] [PubMed] [Google Scholar]
- 77.Wang Z-L, Wang C, Liu W, Ai Z-L. Emerging roles of the long non-coding RNA 01296/microRNA-143-3p/MSI2 axis in development of thyroid cancer. Biosci Rep. 2019;39. doi: 10.1042/BSR20182376 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 78.Shi Y, Massagué J. Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell. 2003;113:685–700. doi: 10.1016/S0092-8674(03)00432-X [DOI] [PubMed] [Google Scholar]
- 79.Dongre A, Weinberg RA. New insights into the mechanisms of epithelial-mesenchymal transition and implications for cancer. Nat Rev Mol Cell Biol. 2019;20:69–84. doi: 10.1038/s41580-018-0080-4 [DOI] [PubMed] [Google Scholar]
- 80.Thuault S, Valcourt U, Petersen M, Manfioletti G, Heldin C-H, Moustakas A. Transforming growth factor-beta employs HMGA2 to elicit epithelial-mesenchymal transition. J Cell Biol. 2006;174:175–183. doi: 10.1083/jcb.200512110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chen X-F, Zhang H-J, Wang H-B, et al. Transforming growth factor-β1 induces epithelial-to-mesenchymal transition in human lung cancer cells via PI3K/Akt and MEK/Erk1/2 signaling pathways. Mol Biol Rep. 2012;39:3549–3556. doi: 10.1007/s11033-011-1128-0 [DOI] [PubMed] [Google Scholar]
- 82.Zhang YE. Non-Smad pathways in TGF-beta signaling. Cell Res. 2009;19:128–139. doi: 10.1038/cr.2008.328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sano Y, Harada J, Tashiro S, Gotoh-Mandeville R, Maekawa T, Ishii S. ATF-2 is a common nuclear target of Smad and TAK1 pathways in transforming growth factor-beta signaling. J Biol Chem. 1999;274:8949–8957. doi: 10.1074/jbc.274.13.8949 [DOI] [PubMed] [Google Scholar]
- 84.Fox RG, Lytle NK, Jaquish DV, et al. Image-based detection and targeting of therapy resistance in pancreatic adenocarcinoma. Nature. 2016;534:407–411. doi: 10.1038/nature17988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Han Y, Ye A, Zhang Y, et al. Musashi-2 silencing exerts potent activity against acute myeloid leukemia and enhances chemosensitivity to daunorubicin. PLoS One. 2015;10:e0136484. doi: 10.1371/journal.pone.0136484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhang H, Tan S, Wang J, et al. Musashi2 modulates K562 leukemic cell proliferation and apoptosis involving the MAPK pathway. Exp Cell Res. 2014;320:119–127. doi: 10.1016/j.yexcr.2013.09.009 [DOI] [PubMed] [Google Scholar]
- 87.Kopan R, Ilagan MXG. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137:216–233. doi: 10.1016/j.cell.2009.03.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Miele L. Notch signaling. Clinical Cancer Research. 2006;12:1074–1079. doi: 10.1158/1078-0432.CCR-05-2570 [DOI] [PubMed] [Google Scholar]
- 89.Saad S, Stanners SR, Yong R, Tang O, Pollock CA. Notch mediated epithelial to mesenchymal transformation is associated with increased expression of the Snail transcription factor. Int J Biochem Cell Biol. 2010;42:1115–1122. doi: 10.1016/j.biocel.2010.03.016 [DOI] [PubMed] [Google Scholar]
- 90.Fukusumi T, Guo TW, Sakai A, et al. The pathway induces epithelial-mesenchymal transition in head and neck squamous cell carcinoma. Clinical Cancer Research. 2018;24:619–633. doi: 10.1158/1078-0432.CCR-17-1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Troschel FM, Minte A, Ismail YM, et al. Knockdown of Musashi RNA binding proteins decreases radioresistance but enhances cell motility and invasion in triple-negative breast cancer. Int J Mol Sci. 2020;21:21. doi: 10.3390/ijms21062169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wang X, Wang R, Bai S, et al. Musashi2 contributes to the maintenance of CD44v6+ liver cancer stem cells via notch1 signaling pathway. J Exp Clin Cancer Res. 2019;38:505. doi: 10.1186/s13046-019-1508-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Katz Y, Li F, Lambert NJ, et al. Musashi proteins are post-transcriptional regulators of the epithelial-luminal cell state. eLife. 2014;3:e03915. doi: 10.7554/eLife.03915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhang J, Shao X, Sun H, et al. NUMB negatively regulates the epithelial-mesenchymal transition of triple-negative breast cancer by antagonizing Notch signaling. Oncotarget. 2016;7:61036–61053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yang S, Sheng L, Xu K, et al. Anticancer effect of quinacrine on diffuse large B-cell lymphoma via inhibition of MSI2-NUMB signaling pathway. Mol Med Rep. 2018;17:522–530. doi: 10.3892/mmr.2017.7892 [DOI] [PubMed] [Google Scholar]
- 96.Pupo M, Pisano A, Abonante S, Maggiolini M, Musti AM. GPER activates Notch signaling in breast cancer cells and cancer-associated fibroblasts (CAFs). Int J Biochem Cell Biol. 2014;46:56–67. doi: 10.1016/j.biocel.2013.11.011 [DOI] [PubMed] [Google Scholar]
- 97.Kang MH, Jeong KJ, Kim WY, et al. Musashi RNA-binding protein 2 regulates estrogen receptor 1 function in breast cancer. Oncogene. 2017;36:1745–1752. doi: 10.1038/onc.2016.327 [DOI] [PubMed] [Google Scholar]
- 98.Nusse R, Clevers H. Wnt/β-catenin signaling, disease, and emerging therapeutic modalities. Cell. 2017;169:985–999. doi: 10.1016/j.cell.2017.05.016 [DOI] [PubMed] [Google Scholar]
- 99.Daniels DL, Weis WI. Beta-catenin directly displaces Groucho/TLE repressors from Tcf/Lef in Wnt-mediated transcription activation. Nat Struct Mol Biol. 2005;12:364–371. doi: 10.1038/nsmb912 [DOI] [PubMed] [Google Scholar]
- 100.Sánchez-Tilló E, Lázaro A, Torrent R, et al. ZEB1 represses E-cadherin and induces an EMT by recruiting the SWI/SNF chromatin-remodeling protein BRG1. Oncogene. 2010;29:3490–3500. doi: 10.1038/onc.2010.102 [DOI] [PubMed] [Google Scholar]
- 101.Wang MH, Qin SY, Zhang SG, et al. Musashi-2 promotes hepatitis Bvirus related hepatocellular carcinoma progression via the Wnt/beta-catenin pathway. Am J Cancer Res. 2015;5:1089–1100. [PMC free article] [PubMed] [Google Scholar]
- 102.Li Z, Jin H, Mao G, Wu L, Guo Q. Msi2 plays a carcinogenic role in esophageal squamous cell carcinoma via regulation of the Wnt/β-catenin and Hedgehog signaling pathways. Exp Cell Res. 2017;361:170–177. doi: 10.1016/j.yexcr.2017.10.016 [DOI] [PubMed] [Google Scholar]