Summary
Addressing bioenergetics is key to evaluate the impact of metabolism on the regulation of biological processes and its alteration in disease. Organoids are in vitro grown self-organizing structures derived from healthy and diseased tissue that recapitulate with high fidelity the tissue of origin. Bioenergetics is commonly analyzed by Seahorse XF analysis. However, its application to organoid studies is technically challenging. Here, we share our in-house optimized protocols to examine organoid bioenergetics in response to drugs, gene knockdown, or to characterize the metabolism of specific cell types.
For complete details on the use and execution of this protocol, please refer to Ludikhuize et al. (2020).
Subject areas: Metabolism, Stem cells, Organoids
Graphical abstract
Highlights
-
•
Analysis of bioenergetics in organoids is technically challenging
-
•
We share a detailed protocol to overcome the pitfalls of Seahorse analysis in organoids
-
•
Accurate timing of organoid re-plating improves the robustness of Seahorse analysis
-
•
DNA-based normalization overcomes organoid-related limitations for normalization
Addressing bioenergetics is key to evaluate the impact of metabolism on the regulation of biological processes and its alteration in disease. Organoids are self-organizing structures derived from healthy and diseased tissue that closely recapitulate the tissue of origin. Bioenergetics is commonly analyzed by Seahorse XF analysis. However, its application to organoid studies is technically challenging. Here, we share our in-house optimized protocols to examine organoid bioenergetics in response to drugs, gene knockdown, or to characterize the metabolism of specific cell types.
Before you begin
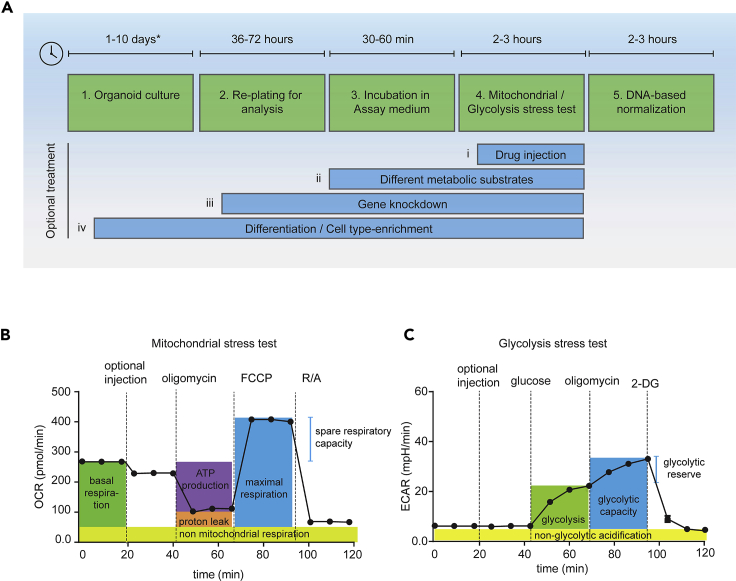
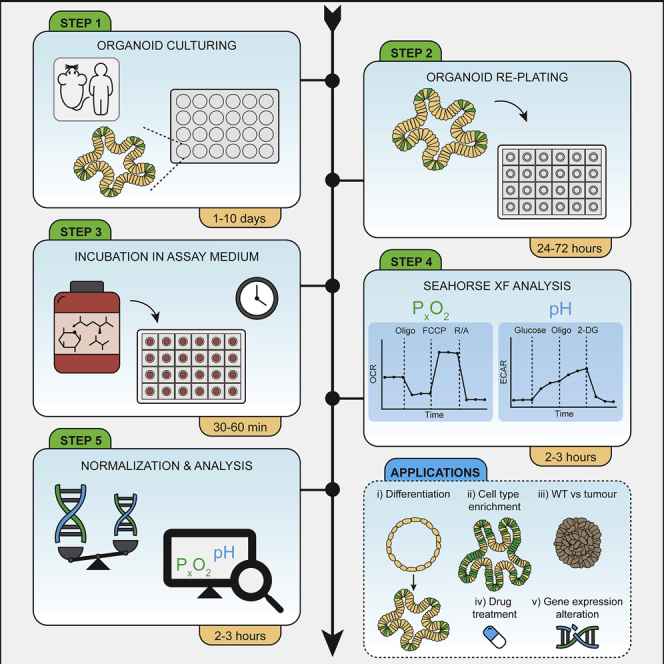
Organoids closely recapitulate the in vivo mechanisms of epithelial homeostasis and harbor all different cell types of the tissue of origin (Schutgens and Clevers, 2020). Importantly, organoids are rapidly gaining ground as research tools and are used in a wide range of scientific disciplines including fundamental biology, hereditary diseases, and cancer (Kim et al., 2020). The analysis of bioenergetics is key to better understand, and potentially target, the supporting role of metabolism in such processes. Glycolysis in the cytosol and oxidative phosphorylation in the mitochondria produce energy in the form of ATP, at different rates and with different requirements and side-products. While in the absence of oxygen cells primarily convert the glycolytic product pyruvate into lactate, under aerobic conditions, pyruvate can fuel the TCA cycle and mitochondrial respiration. Interestingly, most actively proliferating cells produce and secrete lactate independently of oxygen availability, which is known as aerobic glycolysis and defines the “Warburg effect” in cancer cells (Vander Heiden et al., 2009). Medium acidification by lactate secretion and oxygen consumption by mitochondrial respiration can be used to monitor the activity of these metabolic pathways. In brief, Seahorse-based bioenergetics analysis is based on the simultaneous and over time measurements of the oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) as read-outs of mitochondrial respiration and glycolysis, respectively. Changes in OCR and ECAR are measured in response to the presence of metabolic substrates and inhibitors and serve to calculate several bioenergetic parameters. In this manuscript, we share our protocol to measure bioenergetics in organoids (Figure 1A). Here we focus on the most frequently used tests, the mitochondrial and glycolysis stress tests (Figures 1B and 1C). We provide detailed information on how to successfully perform these analyses in organoids derived from mouse small intestine, human colon, and colorectal tumors. Our optimized protocols include how to prepare organoids for analysis, how to adjust the assay medium composition and the concentrations of substrates and inhibitors, the specific settings for the XF Analyzer, and how to normalize the measurements. These protocols have broad applications, such as metabolic profiling of specific cell types, determination of the metabolic changes upon stem cell differentiation or addressing metabolic programs resulting from genetic mutations (Figure 1A). We have listed a number of interesting applications developed by others and us in Table 1.
Figure 1.
Experimental setup and mitochondrial and glycolysis stress test parameters
(A) Experimental setup for profiling bioenergetics in intestine-derived organoids. This setup can be used to analyze organoid bioenergetics upon various treatments. Examples are (i) short treatment with metabolic inhibitors that are injected during the assay, (ii) incubation of organoids in the presence of different metabolic substrates such as glucose vs palmitate in the assay medium, (iii) induction or gene knockdown after re-plating organoids for analysis, or (iv) induction of organoid differentiation or cell-type enrichment which is already started prior to re-plating the organoids for analysis. ∗For other organoid types or specific differentiation protocols, culturing for longer than 10 days might be required (B and C). Setup of and respiratory parameters measured during (B) a mitochondrial stress test or (C) a glycolysis stress test.
Table 1.
Different applications for profiling bioenergetics in organoids
| Application | Example | Treatment | Reference |
|---|---|---|---|
| Metabolic characterization of specific cell types | Determination of stem cell or enterocyte metabolism of mSIOs | Adjustment of medium composition to enrich organoids with stem cells or with enterocytes | (Lindeboom et al., 2018), (Okkelman et al., 2020) |
| Metabolic profiling during differentiation. | Metabolic profiling of proliferative vs. differentiating mSIOs | Adjustment of medium composition to prevent or enhance differentiation of organoids | (Rodríguez-Colman et al., 2017) |
| Metabolic profiling during maturation of human cardiac organoids | Adjustment of the medium to induce human heart organoid maturation | (Mills et al., 2017) | |
| Determining the effect or effectiveness of metabolic drugs on bioenergetics. | Determination of the contribution of specific metabolic pathways to energy metabolism of mSIOs | Treatment with DCA, Piercidin, Azide, UK5099, Phloretin, PCMB, or Antimycin A | (Rodríguez-Colman et al., 2017) |
| Determination of contribution of Mdivi-1 on energy metabolism in mSIOs | Mdivi-1 treatment | (Ludikhuize et al., 2020) | |
| Determination of effectiveness complex I inhibitor rotenone in mSIOs | Rotenone treatment | (Ludikhuize et al., 2020) | |
| Determination of FA oxidation in human heart organoids | Etomoxir treatment | (Mills et al., 2017) | |
| Determination of FA oxidation mSIOs | Etomoxir treatment | (Schell et al., 2017) | |
| Determination of bioenergetics upon metformin treatment in CRC PDX-derived organoids | Metformin treatment | (Mohamed Suhaimi et al., 2017) | |
| Assessment of substrates for mitochondrial respiration. | Determination of mitochondrial respiration upon lactate supplementation in mSIOs | Glucose vs lactate supplementation | (Rodríguez-Colman et al., 2017) |
| Determination of FA oxidation with palmitate as substrate in mSIOs | Palmitate supplementation | (Schell et al., 2017) | |
| Profiling bioenergetics upon gene knockdown. | Examining the role of FOXO on mSIO bioenergetics | Doxycycline-induced FOXO1/3 KD | (Ludikhuize et al., 2020) |
| Determining the effect of mitochondrial fission on mSIO bioenergetics | Doxycycline-induced Drp1 KD | (Ludikhuize et al., 2020) | |
| Profiling bioenergetics upon gene knockout. | Examining the role of MPC1 on mSIO bioenergetics | Stem cell-specific MPC1 KO | (Schell et al., 2017) |
| Determining activity of O2-producing or consuming enzymes. | Examining the oxygen consumption by the enzymes LbNOX or TpNOX in HeLa cells | Doxycycline-induced expression of LbNOX or TpNOX | (Cracan et al., 2017) |
Abbreviations: mSIO: mouse small intestinal organoid, CRC: colorectal cancer, DCA: dichloroacetate, Drp1: Dynamin-Related Protein 1, FOXO: Forkhead Box O transcription factor, LbNOX: Lactobacillus brevis H2O-forming NADH oxidase, Mdivi-1: mitochondrial division inhibitor 1, MPC1: Mitochondrial pyruvate carrier 1, PCMB: p-chloromercuribenzoate, PDX: patient-derived xenograft, TpNOX: triphosphopyridine nucleotide oxidase.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, peptides, and recombinant proteins | ||
| 2-Deoxyglucose | Sigma-Aldrich | Cat# D8375, CAS 154-17-6 |
| Advanced DMEM/F-12 | Life Technologies | Cat# 12634-010 |
| Antimycin A | Sigma-Aldrich | Cat# A8674, CAS 1397-94-0 |
| Cell recovery solution | Corning | Cat# 354253 |
| DMSO | VWRC | Cat# 23488.294 CAS 67-68-5 |
| FCCP | Sigma-Aldrich | Cat# C2920, CAS 370-86-5 |
| Glucose | Merck Millipore | Cat# 1.08337.1000, CAS 50-99-7 |
| Glutamax | Life Technologies | Cat# 35050087 |
| HEPES | Life Technologies | Cat# 15630-056, CAS 7365-45-9 |
| L-Glutamine | Lonza | Cat# 17-605E |
| Matrigel growth factor matrix | Corning | Cat# 356231 |
| NaOH | Merck Millipore | Cat# 1.06498.1000, CAS #: 1310-73-2 |
| Oligomycin | Sigma-Aldrich | Cat# 75351, CAS 579-13-5 |
| Penicillin-streptomycin | Lonza | Cat# DE17602E |
| Pyruvate | Sigma-Aldrich | Cat# S8636, CAS 113-24-6 |
| Rotenone | Sigma-Aldrich | Cat# R8875, CAS 83-79-4 |
| Seahorse XF base medium | Agilent | Cat# 102353-100 |
| Seahorse XF Calibrant | Agilent | Cat# 102353-100 |
| TrypLE Express Enzyme (1×) phenol red | Sigma-Aldrich | Cat# 12605010 |
| Trypsin solution | Sigma-Aldrich | Cat# T3924 |
| Critical commercial assays | ||
| QIAamp DNA Micro Kit | Qiagen | Cat# 56304 |
| Seahorse XF24 V28 PS cell culture microplates | Agilent | Cat# 100882-004 |
| Seahorse XFe24 analyzer | Agilent | 420017 |
| Seahorse XFe24 FluxPak | Agilent | Cat# 102340-100 |
| Experimental models: cell lines | ||
| shFoxo1/3 mouse small intestinal organoids | (Ludikhuize et al., 2020) | N/A |
| P9 CRC-derived organoids | (van de Wetering et al., 2015) | N/A |
| P16 CRC-derived organoids | (van de Wetering et al., 2015) | N/A |
| P19b CRC-derived organoids | (van de Wetering et al., 2015) | N/A |
| Human WT colon organoids | (Drost et al., 2015) | N/A |
| Software and algorithms | ||
| GraphPad Prism 8 | https://www.graphpad.com/scientific-software/prism/ | N/A |
| Wave Desktop and Controller 2.6 Software | https://www.agilent.com/en/product/cell-analysis/real-time-cell-metabolic-analysis/xf-software/seahorse-wave-desktop-software-740897 | N/A |
| Other | ||
| EVOS M5000 imaging system | Invitrogen | AMF5000 |
| Nanodrop 2000 spectrophotometer | Thermo Scientific | ND-2000 |
| Centrifuge | Thermo Scientific | SL40R |
| Rotor centrifuge | Thermo Scientific | 75003607 |
| Eppendorf centrifuge | Eppendorf | 5417R |
| Rotor Eppendorf centrifuge | Eppendorf | F45-30-11 |
Materials and equipment
Seahorse XF analyzer
Agilent provides multiple Seahorse XF Analyzers (8 well, 24 well, or 96 well systems). Our protocol is optimized for the Seahorse XFe24 system (24 wells). General manufacturer’s instructions for performing a mitochondrial or glycolysis stress test can be respectively found at https://www.agilent.com/cs/library/usermanuals/public/XF_Cell_Mito_Stress_Test_Kit_User_Guide.pdf and https://www.agilent.com/cs/library/usermanuals/public/XF_Glycolysis_Stress_Test_Kit_User_Guide.pdf.
Alternatives: Seahorse XFe96 (96 wells) or Seahorse XF HS mini (8 wells) Analyzers can be alternatively used.
Preparation of assay medium
In this protocol, we prepare assay medium with base medium with phenol red (Agilent, #102353-100) and we buffer it to pH 7.4 with NaOH. This medium can be stored for 24 h at 4°C.
Alternatives: Assay medium can be prepared based on different types of media. For Agilent’s recommendation on alternative media visit: https://www.agilent.com/cs/library/selectionguide/public/5991-7878EN.pdf
Preparation of stocks of substrates and inhibitors
Stocks of substrates and inhibitors can be prepared in advance by dissolving them as depicted below. Glucose can be stored at 4°C. All other stocks should be stored at −20°C. All stocks can be stored for at least six months.
For mitochondrial stress tests, prepare the following stocks:
2.5 mM Oligomycin in DMSO
2.5 mM FCCP in DMSO (prepare in fume hood)
2.5 mM Rotenone in DMSO
2.5 mM Antimycin A in DMSO
For glycolysis stress tests, prepare the following stocks:
2.5 mM Oligomycin in DMSO
1 M glucose in H2O
DNA-based normalization
For normalization, we isolate and quantify organoid DNA by using the QIAamp DNA Micro Kit. Although alternative DNA extraction methods could be applied, the expected amounts of purified DNA are low in concentration, hence the method needs to be able to account for that.
Step-by-step method details
This step-by-step protocol consists of the following sections: culturing organoids, re-plating organoids for analysis, cartridge hydration, set up of mitochondrial and glycolysis stress tests and normalization by DNA content (Figure 1A). All steps have been optimized for examining bioenergetics in organoids derived from mouse intestine, human colon, and from colorectal tumors.
Culturing organoids
Timing: 1–2 weeks
Organoid growth rates vary depending on the tissue of origin, differentiation rates, and genetic mutations. To achieve an accurate and reliable assay, the number of organoids per well needs to be comparable within and across conditions. To better control for differences in growth rates, we first expand our organoids in normal culture plates and re-plate the organoids in Seahorse Cell Culture microplates plates 24–72 h prior to analysis. This re-plating step is meant to better control for the differences in number and size of the organoids between the conditions to be compared on the day of the Seahorse analysis. Thus, this timing should be adjusted depending on the growth rate of the organoid type. For mouse small intestine-, human colon- or colorectal tumor-derived organoids, 36–48 h prior to analysis is an adequate re-plating time.
When long treatments are needed, for instance for organoid differentiation protocols, the treatments can be started during normal culture and continued after re-plating into Seahorse microplates. Here, we introduce our culturing methods for organoids derived from mouse small intestine, human colon, and colorectal tumors.
For mouse intestinal organoids:
-
1.Disrupt organoids by mechanical shearing and plate them in a 24-well plate.
-
a.Collect organoids in 1 mL of ice cold DMEM F12 advanced medium supplemented with penicillin/streptomycin, 10 mM HEPES, and 20 mM glutamax (+++ medium).Note: Collecting 100 μL of organoids in Matrigel at the density depicted in Figure 2Ci is sufficient.
-
b.Disrupt organoids by pipetting up and down with a 1000 μL pipet.
-
i.First pipet up and down with a single 1,000 μL tip (Figure 2A).
-
ii.When organoids are not disrupted yet, pipet up and down with a 200 μL pipet tip on top of a 1,000 μL pipet tip (Figure 2B) until organoids are broken into small crypts. The grade of mechanical shearing of the organoids should be monitored and visualized in between steps using a microscope.Note: To prevent organoids from sticking to the pipet tips, pre-wet the pipet tips by pipetting op and down with cold +++ medium before starting the mechanical shearing.
-
iii.Add 5 mL of ice cold +++ medium to the organoids.
-
i.
-
c.Centrifuge at 140–220 × g at 4°C for 5 min (Thermo Scientific SL40R centrifuge with Thermo Scientific 75003607 rotor, 800–1,000 rpm).
-
d.Re-suspend the pellet in 200–300 μL of Matrigel to reach the following density (Figure 2Cii)
-
i.200 μL of organoids in Matrigel is sufficient for the later re-plating step in the Seahorse cell culture microplate.
-
i.
-
e.Plate organoids in a 24-well plate and culture for 36–48 h.
-
i.Plate 50 μL Matrigel per well divided in 3–4 droplets.
-
i.
-
f.Incubate the plate upside down at 37°C for 30 min.
-
g.Add culture medium.
-
a.
-
2.
Culture organoids for 36–48 h prior to re-plating them in a Seahorse Cell Culture microplate (Figure 2Ciii).
Note: The time of culturing before re-plating organoids in Seahorse microplate largely relies on the growth rate of the organoids. As an example of this, medium composition favoring organoid expansion by preventing differentiation, such as Wnt addition to mouse small intestinal organoids (Rodríguez-Colman et al., 2017), requires a shorter culture time from splitting to re-plating. In addition to that, the selection of the culture timing should be adjusted to properly address the research question.
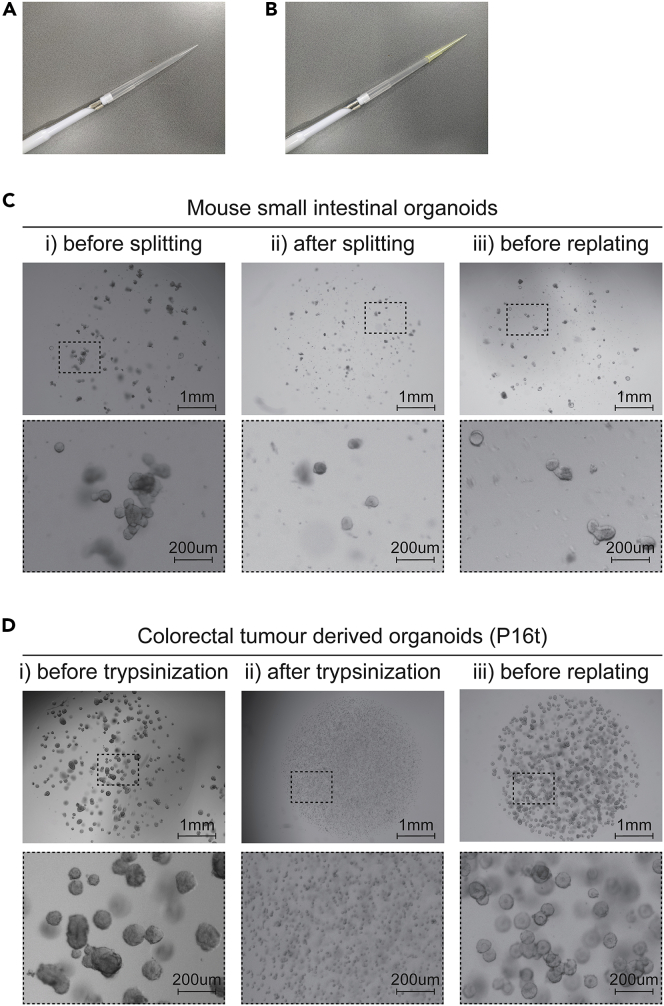
Figure 2.
Organoid culturing
(A) 1,000 μL tip on a 1,000 μL pipet.
(B) 200 μL tip on top of a 1,000 μL tip.
(C) Mouse small intestinal organoids before splitting, after splitting, and before re-plating.
(D) P16t colorectal tumor-derived organoids before trypsinization, after trypsinization, and before re-plating.
For colorectal cancer or colon organoids:
-
3.Disrupt organoids by trypsinization and plate in a 24-well plate.
-
a.Collect organoids in 10 mL of ice cold +++ medium.Note: Collecting 50 μL of organoids in Matrigel in the following density (Figure 2Di) will be sufficient.
-
b.Centrifuge at 140–220 × g at 4°C for 5 min (Thermo Scientific SL40R centrifuge with Thermo Scientific 75003607 rotor, 800–1,000 rpm).
-
c.Trypsinize the organoids in 2 mL of trypsin or trypLE solution until organoids are disrupted (single cells or small clumps of cells depending on the organoid line).
-
d.Add 10 mL of ice cold +++ medium.
-
e.Centrifuge at 220–320 × g at 4°C for 5 min (1,000–1,200 rpm).
-
f.Remove supernatant and add 10 mL of ice cold +++ medium.
-
g.Centrifuge at 220–320 × g at 4°C for 5 min (1,000–1,200 rpm).
-
h.Re-suspend the pellet in 150–300 μL Matrigel to reach the following density (Figure 2Dii).
-
i.200 μL of organoids in Matrigel is sufficient for the later re-plating step in the Seahorse cell culture microplate.
-
i.
-
i.Plate organoids in a 24-well plate
-
i.Plate 50 μL Matrigel per well, divided in 3–4 droplets.
-
i.
-
j.Incubate the plate upside down at 37°C for 30 min.
-
k.Add culture medium.
-
a.
-
4.
Culture organoids until the desired size prior to re-plating them in a Seahorse Cell Culture microplate (Figure 2Diii). Check Note in step 2 for a comment on this.
Plate organoids for analysis
Timing: 1–3 days
Seahorse analysis has to be performed by plating organoids in Seahorse XF24 V28 PS cell culture microplates (Agilent, #100882-004) (Figure 3A). Below we describe step-by-step how to accurately re-plate organoids to guarantee reliable measurements.
-
5.
At least 24 h before plating, incubate the Seahorse XF24 V28 PS cell culture microplate to be used and a flask filled with water (Figure 3B) at 37°C. This step enhances the adhesion of the Matrigel drop to the bottom of the plate.
-
6.
Collect the previously cultured organoids in 1 mL of ice cold +++ medium.
Note: It is important not to disrupt the integrity of the organoids while collecting. For mouse small intestinal organoids cultured for 36 h on medium supplemented with EGF, Noggin, and R-spondin, it is recommended to collect 4 wells of a 24-well plate (50 μL Matrigel each) to fill 20 wells of a Seahorse microplate. For colon- or colorectal tumor-derived organoids cultured for 7 days after trypsinization, it is recommended to collect 1 well of a 24-well plate to fill 20 wells of a Seahorse microplate.
-
7.
Centrifuge at 140–220 × g at 4°C for 5 min (800–1,000 rpm).
Note: Depending on the organoid type, centrifugation can affect organoid integrity. Choose the gravitational force that efficiently pulls down the organoids, but does not cause organoid disruption.
-
8.Re-suspend the organoid pellet in a volume of Matrigel that leads to the appropriate organoid density (steps b–d).
-
a.It is important to reduce the variability in organoid density across and within experiments.
-
b.For mouse small intestine- and colon- or colorectal tumor-derived organoids, 20–40 organoids per well should be plated to measure OCR and ECAR within the range of sensitivity of the method (Figures 4A and 4B). Organoid density can be further adjusted to the specific requirements of the organoid type, under the consideration that the obtained measurements are within the range of sensitivity of the method (see CRITICAL).
-
c.As a guideline for mouse small intestinal organoids, collect 4 wells of a 24-plate, start by re-suspending the organoids in 100 μL of Matrigel and dilute further when necessary.
-
d.As a guideline for colon- or colorectal tumor-derived organoids, collect 1 well of a 24-plate, start by re-suspending the organoids in 150 μL of Matrigel and dilute further when necessary.
-
a.
CRITICAL: Small volumes of Matrigel rapidly polymerize at room temperature. Prevent this by keeping the organoid-containing tubes on ice at all possible times. The density of organoids in Matrigel droplet needs attention: if the density of the organoids is too low or too high, the sensitivity of the method may limit the reliability of the measurements. ECAR can be reliably measured between 20 and 200 mpH/min. OCR can be reliably measured between 100 and 1,000 pmol O2/min (higher OCR measurements could be considered upon validation).
-
9.Plate organoids in a Seahorse Cell Culture Microplate [Troubleshooting 1] [Troubleshooting 2].
-
a.Place the Seahorse microplate on top of the pre-warmed water-filled flask (Figure 3B).Note: Placing the microplate on top of a warm surface facilitates plating 3 μL Matrigel droplets in the middle of Seahorse Cell Culture as the warmer temperature enhances the stability of the droplet, as such, and accelerates its polymerization.
-
b.Plate 3 μL of Matrigel (without organoids) in 4 control wells (Figures 4A and 4B).Note: OCR and ECAR measurements are sensitive to the presence of Matrigel. Therefore, in control wells Matrigel should also be plated in the same volume.
- c.
-
d.After plating, place the Seahorse microplate plate back in the incubator for 10–15 min at 37°C.
 CRITICAL: Normal plating of organoids involves turning the plate upside down. Here that step should be avoided to prevent disruption of the Matrigel droplets. As small volumes of Matrigel polymerize fast, do not leave the plate for longer than the stated time to prevent drying out of the Matrigel droplet.
CRITICAL: Normal plating of organoids involves turning the plate upside down. Here that step should be avoided to prevent disruption of the Matrigel droplets. As small volumes of Matrigel polymerize fast, do not leave the plate for longer than the stated time to prevent drying out of the Matrigel droplet. -
e.Add 200 μL of culture medium in each well.
 CRITICAL: Due to the limited plating area, organoids should be plated in the middle of the well and in between the three nodes (Figure 3C). Inaccurate plating leads to its adherence to the walls of the well and consequently to inaccurate measurements.
CRITICAL: Due to the limited plating area, organoids should be plated in the middle of the well and in between the three nodes (Figure 3C). Inaccurate plating leads to its adherence to the walls of the well and consequently to inaccurate measurements.
-
a.
-
10.
When required add the treatment of interest to the selected wells.
-
11.
Culture organoids in the Seahorse microplate for 1–3 days until a mitochondrial or glycolysis stress test is performed.
Note: As described in the “culturing organoids” section, this time frame is dependent on the growth rates of the organoids and the desired effect of the optional treatment.
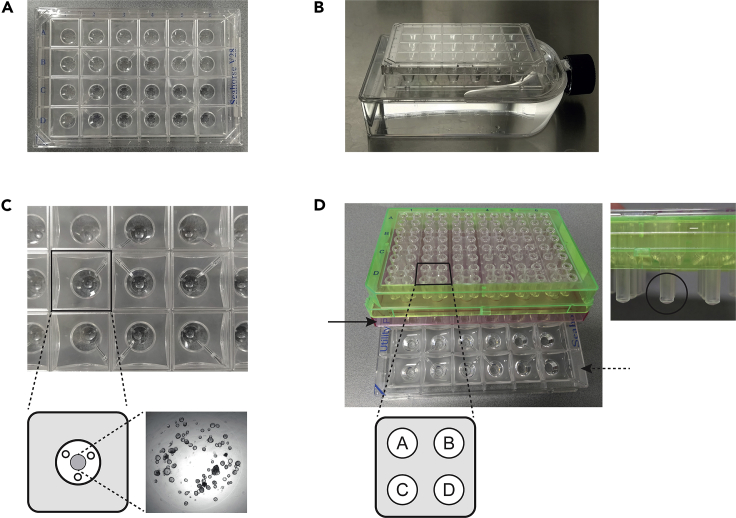
Figure 3.
Organoid plating in Seahorse cell culture microplate and preparation of sensor cartridge
(A) Seahorse XF24 V28 PS cell culture microplate.
(B and C) Plating organoids in a Seahorse cell culture microplate placed on top of a pre-warmed water-filled flask.
(D) Seahorse Sensor Cartridge on top of a Hydro Booster (black arrow) and Utility plate (dashed arrow) with injection ports A–D and O2- and pH-sensitive probe tips (black circle).
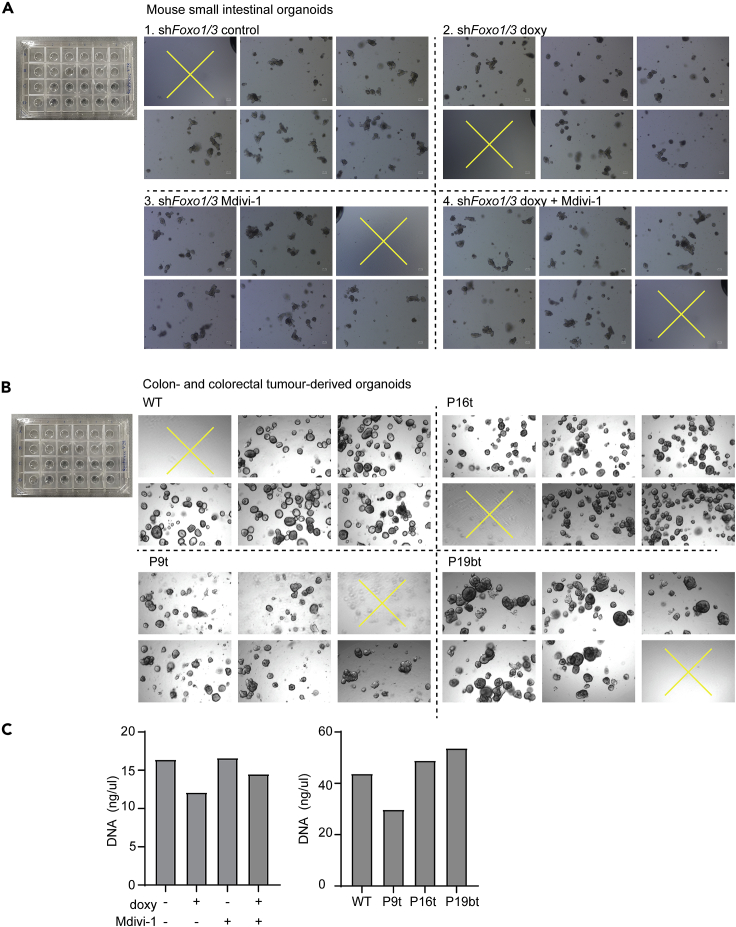
Figure 4.
Intestine-derived organoids in a Seahorse cell culture microplate and DNA quantification
(A and B) Organoids plated in a Seahorse cell culture plate. Organoid densities have been adjusted to fit the requirements of the organoid type. Mouse intestinal organoids undergo crypt formation-based morphological changes and are plated at a lower density than wild-type colon and colorectal tumor organoids. The depicted densities result in OCR and ECAR measurements within the sensitive range. (A) Mouse small intestinal shFoxo1/3 organoids treated with doxycycline or Mdivi-1 for 36 h. (B) Wild-type colon and colorectal tumor-derived organoids P9t, P16t, and P19bt. In (A) and (B), yellow crosses indicate control wells.
(C) DNA quantification of mouse small intestine- and human colon- and colorectal tumor-derived organoids in (A) and (B).
Cartridge hydration
Timing: 4–72 h
Each probe tip of the sensor cartridge contains sensor material that detects changes in O2 concentration and pH (Figure 3D). For correct functioning, the probe tips of the sensor cartridge have to be hydrated in XF Calibrant for a minimum of 4 h and a maximum of 72 h. For practical reasons we recommend to hydrate the cartridge for 12 to 24 h.
-
12.
Lift the sensor cartridge from the Utility plate, and pipet 1 mL of XF Calibrant into each well of the Utility plate (Figure 3D).
-
13.
Place the sensor cartridge back on the Utility plate with the Hydro Booster in between, in such a way that the probe tips of the Sensor Cartridge are submerged in XF Calibrant.
-
14.
Place the Sensor Cartridge in a non-CO2 37°C humidified incubator for at least 4 h, but at maximum for 72 h.
XF mitochondrial stress test or glycolysis stress test
Timing: 3 h
Agilent offers multiple assays to analyze different bioenergetic parameters. We have optimized the assay medium composition, the concentrations of substrates and inhibitors and the measurement settings for the XF Analyzer for performing mitochondrial and glycolysis stress tests in mouse small intestine, colon, and colorectal tumor-derived organoids.
-
15.
Take pictures of the organoids in all wells of the Seahorse microplate (Figures 3C, 4A, and 4B) by a bright-field microscope.
Note: Imaging the organoids helps to control for viability and to compare differences in organoid growth and morphology across conditions and among experiments.
-
16.Prepare 50 mL assay medium.
Note: Following Agilent’s recommendations, substrates should be provided in saturating concentrations to ensure that they will not be limiting OCR and ECAR measurements. In Tables 2 and 3 we have listed the saturating substrate concentrations adjusted for both, mouse small intestine- and human colon- or colorectal tumor-derived organoid analysis. Note that these concentrations are organoid type-specific and need to be determined empirically when applied to other organoid types.
-
17.Prepare organoids for the XF analysis.
-
a.Remove the culture medium from the organoids.
-
b.Carefully wash organoids twice with 500 μL assay medium.
-
c.Add 525 μL assay medium into each well.
-
d.Incubate organoids in an incubator without CO2 control at 37°C for at least 30 min, but at maximum 1 h.
-
a.
Optional: When analyzing the effect of drugs/treatment on bioenergetics, it could be necessary to maintain the treatment during the assay. This needs to be decided depending on the mechanism of action of the drug and the research question. Note that the assay medium is not buffered. Therefore, depending on the compound type, it can be required to readjust the pH of the compound’s stock.
-
18.During the incubation time of organoids in assay medium, prepare the injection solutions. Injections should be prepared in assay medium and 75 μL of these injections should be pipetted in the injection ports (Figure 3D). The device will inject the drugs in the order of port A to port D.
- a.
- b.
-
c.Upon completion of preparation of the injection ports, place the sensor cartridge hold in the Utility plate back into the incubator.
Note: When other organoid types are used, optimal substrate and inhibitor concentrations have to be determined. We recommend to start with titrating Oligomycin, followed by respectively FCCP and Rotenone and Antimycin A. The lowest concentration that causes maximal inhibition of every drug should be selected. For an indication of inhibitor concentrations used for cell lines, check the Seahorse XF Analyzer guides: https://www.agilent.com/en/support/cell-analysis/seahorse-assay-guides-templates
Optional: For analysis of the immediate metabolic response to a certain drug, drugs can be injected during the mitochondrial or glycolysis stress test via the injection ports (Figures 1B and 1C). For such a bioenergetics assay, use injection port A for the drug of interest and move the injections corresponding to the mitochondrial or glycolysis stress test to port B, C and D respectively (Figure 3D). An example of such an application in a mitochondrial stress test, can be found in our recent study (Ludikhuize et al., 2020). Note that the concentrations of the inhibitors in injection ports need to be adjusted according to the change in the volume resulting from adding preceding injection step in A. Based on the research question, the order of the substrates and inhibitors can also be re-arranged. Then, the experimental settings can be updated in the Wave software accordingly.
-
19.Prepare settings of the XF Analyzer.
-
a.Open the Wave software.
-
b.Open a mitochondrial or glycolysis stress test template, and fill in the assay details. A detailed explanation on how to create and customize assay template files using the Wave software can be found in section 2.4 in: https://www.agilent.com/en/product/cell-analysis/how-to-run-an-assay.
-
c.Make sure you adjust the XF analysis settings in the “Instrument Protocol” section.
-
d.Save your template.
-
a.
CRITICAL: compared to cell lines, organoids show a slower response to Oligomycin, possibly due to the presence of Matrigel. Therefore, we adjusted the settings by applying a 10-min waiting step right after the injection of the inhibitor in both, the mitochondrial stress test and the glycolysis stress test (Tables 6 and 7).
-
20.Run the mitochondrial or glycolysis stress test [Troubleshooting 3].
-
a.Upon clicking on “Run,” the Wave software will ask to insert the sensor cartridge in the XF Analyzer.
-
b.Remove the Hydro Booster and the lid (Figure 3D), and place the sensor cartridge hold in the Utility plate in the XF Analyzer.
-
c.After the calibration step which takes approximately 15 min, the Wave software will request to replace the Utility plate by the Seahorse Cell Culture microplate containing the organoids.
-
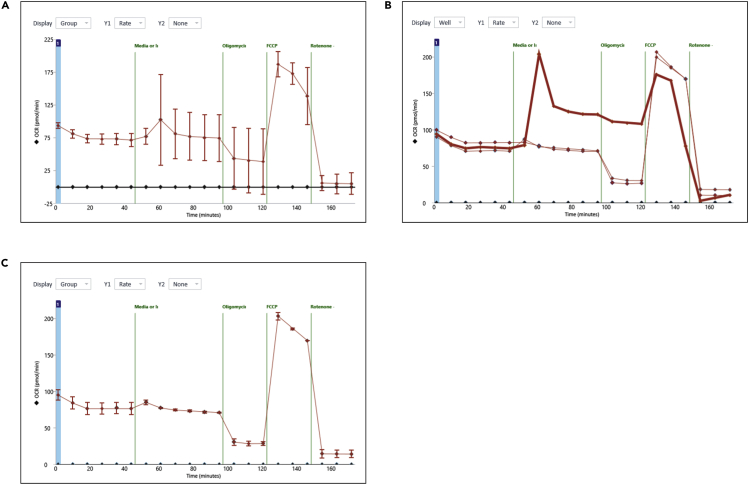
d.The XF analysis will automatically begin.Note: Once the experiment has finished, the OCR and ECAR measurements will be displayed on the XF Analyzer screen. A first inspection of the results can be done at this point. When high standard deviations within one condition are observed, switching from “group” view to “well” view, allows to detect possible outliers (Figure 5), which can result, for instance, from inaccurate plating or differential organoid viability. Pictures taken at step 15 can be used to judge the cause of large standard deviations. Excluding outliers from the analysis can be considered (Figures 5A–5C) [Troubleshooting 4].
-
a.
-
21.
After the experiment, carefully remove medium from the wells.
CRITICAL: The plate should be removed immediately after the experiment has finished in order to prevent loosing organoid viability as a result of the presence of the inhibitors used in the assay.
Pause point: When required, after medium aspiration, the Seahorse Cell Culture Microplate can be stored at −20°C for at least 2 weeks to later proceed with the DNA quantification step.
Table 2.
Assay medium mitochondrial stress test
| Reagent | Default assay medium compositiona | Optimized assay medium composition for intestine-derived organoidsb | Volumes to prepare 50 mL optimized assay medium |
|---|---|---|---|
| Glucose (1 M) | 10 mM | 10 mM | 0.5 mL |
| Pyruvate (100 mM) | 1 mM | 5 mM | 2.5 mL |
| Glutamine (200 mM) | 2 mM | 2 mM | 0.5 mL |
| NaOH (1 M) | Adjust to pH 7.4 | Adjust to pH 7.4 | 25 μL |
| Base medium | n/a | n/a | 46.5 mL |
| Total | n/a | n/a | 50 mL |
Default assay medium composition suggested by Agilent.
Assay medium composition optimized for mouse small intestine- and human colon- or colorectal tumor-derived organoids.
Table 3.
Assay medium glycolysis stress test
| Reagent | Default assay medium compositiona | Optimized assay medium composition for intestine-derived organoidsb | Volumes to prepare 50 mL optimized assay medium |
|---|---|---|---|
| Glutamine (200 mM) | 2 mM | 2 mM | 0.5 mL |
| NaOH (1 M) | Adjust to pH 7.4 | Adjust to pH 7.4 | 25 μL |
| Base medium | n/a | n/a | 49.5 mL |
| Total | n/a | n/a | 50 mL |
Default assay medium composition suggested by Agilent.
Assay medium composition optimized for mouse small intestine- and human colon- or colorectal tumor-derived organoids.
Table 4.
Mitochondrial stress test injections optimized for mouse small intestine-, human colon- and colorectal tumor-derived organoids
| Injection port | Drug | Final concentration in Seahorse plate | Injection concentration | To add from 2.5 mM stock (μL) | XF assay medium (μL) |
|---|---|---|---|---|---|
| Port A | Oligomycin | 5 μM | 40 μM | 32 | 1,968 |
| Port B | FCCP | 2 μM | 18 μM | 14.4 | 1,985.6 |
| Port C | Rotenone and Antimycin A | 2 μM + 2 μM | 20 μM | 16 (each) | 1.68 |
| Port D |
Table 5.
Glycolysis stress test injections optimized for mouse small intestine-, human colon- and colorectal tumor-derived organoids
| Injection port | Drug | Final concentration in Seahorse plate | Injection concentration | To add from stock | XF assay medium (μL) |
|---|---|---|---|---|---|
| Port A | Glucose | 10 mM | 80 mM | 160 μL | 1,840 |
| Port B | Oligomycin | 5 μM | 45 μM | 36 μL | 1,964 |
| Port C | 2-DGa | 100 mM | 1 M | 333 mg | 2,000 |
| Port D |
2-Deoxyglucose solutions are not stable over time and should be dissolved on the day of performing the glycolysis stress test
Table 6.
Settings mitochondrial stress test
| Settings | Defaulta | Intestine-derived organoidsb | Your organoids? | ||
|---|---|---|---|---|---|
| Basal | 3 cycles | mix | 3 min | 4 min | |
| wait | 2 min | 0 min | |||
| measure | 3 min | 2 min | |||
| Oligomycin | 1 cycle | mix | 3 min | 5 min | |
| wait | 2 min | 10 min | |||
| measure | 3 min | 2 min | |||
| 2 cycles | mix | 3 min | 4 min | ||
| wait | 2 min | 0 min | |||
| measure | 3 min | 2 min | |||
| FCCP | 3 cycles | mix | 3 min | 4 min | |
| wait | 2 min | 0 min | |||
| measure | 3 min | 2 min | |||
| Rotenone + Antimycin A | 3 cycles | mix | 3 min | 4 min | |
| wait | 2 min | 0 min | |||
| measure | 3 min | 2 min | |||
Suggested by Agilent.
These settings can be applied to mouse small intestine- and human colon- or colorectal tumor-derived organoids.
Table 7.
Settings glycolysis stress test
| Settings | Defaulta | Intestine-derived organoidsb | Your organoids? | ||
|---|---|---|---|---|---|
| Basal | 3 cycles | Mix | 3 min | 4 min | |
| Wait | 2 min | 0 min | |||
| Measure | 3 min | 2 min | |||
| Glucose | 3 cycles | Mix | 3 min | 4 min | |
| Wait | 2 min | 1 min | |||
| Measure | 3 min | 2 min | |||
| Oligomycin | 1 cycles | mix | 3 min | 5 min | |
| wait | 2 min | 10 min | |||
| measure | 3 min | 2 min | |||
| 2 cycles | mix | 3 min | 4 min | ||
| wait | 2 min | 0 min | |||
| measure | 3 min | 2 min | |||
| 2-DG | 3 cycles | mix | 3 min | 4 min | |
| wait | 2 min | 0 min | |||
| measure | 3 min | 2 min |
Suggested by Agilent.
These settings can be applied to mouse small intestine- and human colon- or colorectal tumor-derived organoids.
Figure 5.
Detection of possible outliers in Seahorse XF Wave software
OCR measurements of an exemplary mitochondrial stress test. In (A), the measurements are displayed as “group view” and in (B) as “well view.” In (B), the line in bold indicates the outlier measurement. (C) “Group view” of the OCR measurements displayed in (A) upon exclusion of the outlier depicted in (B).
DNA quantification
Timing: 2–3 h
Calculation of bioenergetic parameters requires normalization to correct for differences in numbers of cells, organoids, or amount of biological material. Standard normalization methods are based on the determination of cell numbers or protein content. Due to the presence of Matrigel, normalization by protein content is not adequate when using organoids. Quantification of cell numbers in organoids is technically challenging due to their 3D structure and the limited number of cells. A reliable alternative for normalization of OCR and ECAR measurements from organoids is normalization by the DNA content. Below we describe how to collect and isolate DNA from organoids cultured on Seahorse Cell Culture microplates.
-
22.
Add 100 μL ice cold Cell Recovery Solution (Corning, #354253) to each well of the Seahorse Cell Culture microplate.
Note: Upon decision to remove a certain outlier from analysis in step 20, the organoids from this well should not be collected for DNA quantification.
-
23.
Incubate the plate at 4°C for 30–60 min.
Note: Incubation in Cell Recovery Solution facilitates organoid collection and removal of the Matrigel.
-
24.
Collect the organoids and pool the organoids belonging to one condition in an Eppendorf tube.
-
25.
Centrifuge at 10,620 × g at 4°C for 5 min (Eppendorf 5417R centrifuge with an Eppendorf F45-30-11 rotor, 10 000 rpm).
-
26.
Remove supernatant while leaving the pellet undisrupted.
-
27.Isolate genomic DNA by using the QIAamp DNA micro kit according to the manufacturer’s instructions.
-
a.In order to maximize the DNA yield, add 30 μL elution buffer let it stand for 2–3 min to maximize DNA elution.
-
a.
-
28.
Measure genomic DNA by Nanodrop.
CRITICAL: Aiming for similar organoid densities across conditions improves the robustness of the analysis. To be noticed, normalization by pooled DNA corrects for variability across conditions but does not correct for differences within conditions. Therefore, the variation in organoid density within conditions should be kept low.
Note: In these protocols, we apply DNA normalization; this does not exclude the application of other normalization methods. Importantly, RNA quantification could be considered when differences in DNA amount are expected across compared conditions. Importantly, some organoid types could have a high rate of shedding dead cells. This requires attention, as damaged cells can be metabolically low/inactive but they still contribute to total DNA quantification.
Expected outcomes
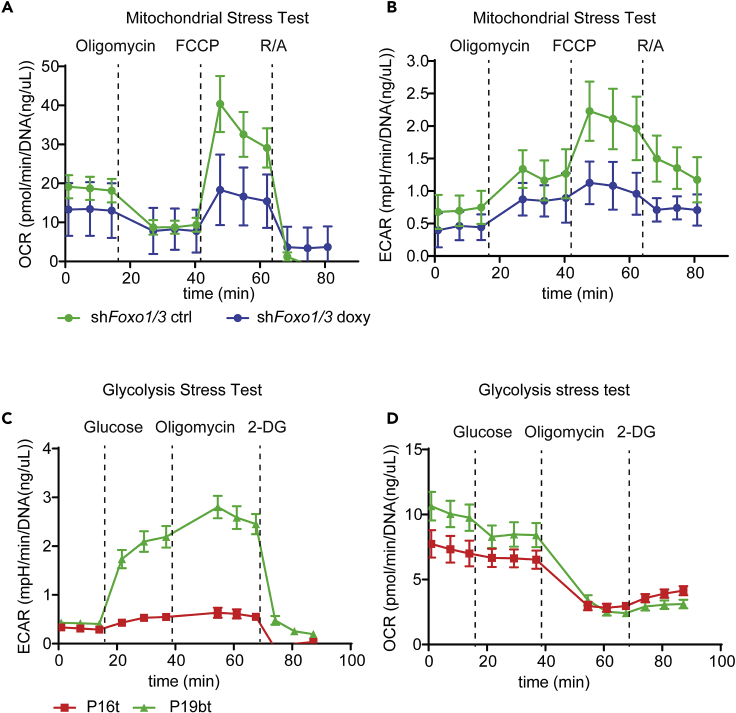
Our protocol describes how to perform mitochondrial stress tests and glycolysis stress tests with mouse intestine-, human colon-, or colorectal tumor-derived organoids. Here we provide an example of a mitochondrial stress test performed with doxycycline-inducible shFoxo1/3 mouse small intestinal organoids in order to examine the respiratory state upon Foxo1/3 knockdown in the small intestine (Ludikhuize et al., 2020). This mitochondrial stress test shows that Foxo1/3 knockdown results in decreased basal and maximal respiration (Figures 1B and 6A). Although a mitochondrial stress test does not enable the analysis of specific glycolytic parameters, ECAR measurements during a mitochondrial stress test do provide information about the glycolytic rates. For instance, the first three time points show that Foxo1/3 knockdown also reduces glycolysis (Figure 6B). For a detailed analysis of bioenergetics upon Foxo1/3 knockdown in mouse small intestinal organoids, please refer to (Ludikhuize et al., 2020). We also provide an example of a glycolysis stress test performed with the colorectal tumor-derived organoids P16t and P19bt (van de Wetering et al., 2015). This glycolysis stress test shows that P16t organoids have a lower glycolytic rate and glycolytic capacity compared to P19bt organoids (Figures 1C and 6C). We also provide examples of quantification of DNA isolated from mouse small intestinal organoids and human colon or colorectal tumor-derived organoids. DNA isolation from 5 Seahorse microplate wells mouse small intestinal organoids results in 10–30 ng DNA/μL in 30 μL elution buffer (Figures 4A and 4C). DNA isolation from 5 Seahorse microplate wells colon- or colorectal tumor-derived organoids results in 20–60 ng DNA/μL in 30 μL elution buffer (Figures 4B and 4C).
Figure 6.
Profiling bioenergetics of intestine-derived organoids
(A and B) OCR (A) and ECAR (B) measurements of a mitochondrial stress test of doxycycline-inducible shFoxo1/3 mouse small intestinal organoids treated with or without doxycycline for 36 h (mean of 5 technical replicates ± SEM). Figure reprinted with permission from Ludikhuize et al. (2020).
(C and D) ECAR (A) and OCR (B) measurements of a glycolysis stress test of two different patient-derived colorectal cancer organoids (P16t and P19bt) (mean of 5 technical replicates ± SEM).
Quantification and statistical analysis
After normalization, the mitochondrial stress tests and glycolysis stress tests can be analyzed by using the Seahorse Wave Desktop software (free to download from the Agilent website: https://www.agilent.com/en/product/cell-analysis/real-time-cell-metabolic-analysis/xf-software/seahorse-wave-desktop-software-740897).
-
1.
Open your experiment in the software.
-
2.
Normalize your data for the DNA content, measured during step 28.
Note: Adjust the DNA concentration to the number of the wells you have pooled for DNA isolation.
-
3.Exporting data.
-
a.For a mitochondrial stress test, export your data as Seahorse XF Cell Mito Stress Test Report Generator.
-
b.For a glycolysis stress test, export your data as Seahorse XF Glycolysis Stress Test Report Generator.
-
c.Raw data can also be exported to Excel or GraphPad Prism.
-
a.
-
4.
Analyze parameters of interest, such as basal respiration, maximal respiration, glycolysis, and glycolytic capacity (Figures 1B and 1C). The values used for each Seahorse XF Report Generator calculation are outlined in the specific Seahorse XF Report Generator assay user guides.
-
5.
Statistical analysis can be applied to the technical replicates, which reflects the technical accuracy of the experiment. To address whether the observed differences are reproducible and biologically relevant, repeat the experiments at least three times [Troubleshooting 5]. After experiments are performed multiple times, statistics can be performed on independent experiments. Statistical analysis needs to be adjusted to the experimental set up and conditions in the assay. For applied examples see (Ludikhuize et al., 2020).
Limitations
The bioenergetic parameters measured by Seahorse analysis are tightly coupled to the in vitro conditions of the analysis. The setup of the assay conditions include the use of saturating concentrations of substrates and drug concentrations. This should be taken into consideration when aiming at drawing conclusions referring to the in vitro culture conditions or the in vivo situation, where substrates can be present at different concentrations.
Due to the low amount of organoid material per well, we indicate to pool the technical replicates of each condition prior DNA extraction and quantification. This strategy increases the accuracy of the DNA measurements and serves to correct for the variability across conditions but not within conditions. Nevertheless, other steps in this protocol such as the re-plating step and guidelines to improve the accuracy of plating are meant to minimize the variability across the wells of the same condition.
Organoids are heterogeneous as they consist of multiple cell types. Thus, bioenergetics analysis of organoids will always reflect the average metabolic phenotype of the different cell types that comprise the organoid. Hence, when analyzing the metabolic consequence of for instance the knockdown of a gene, the effect of this knockdown on differentiation should be taken into consideration. When the cellular heterogeneity is not of interest for the research question, organoid cultures could be modified to prevent differentiation and increase the cellular homogeneity. Examples of such treatments can be found in Table 1. Importantly, organoid cultures can also be modified to enrich organoid cultures for a specific cell type to enable its metabolic characterization (see Table 1). Combining differentiation protocols and Seahorse technology therefore provides a great opportunity to determine the metabolic phenotype of specific cell types, and even the metabolic changes associated to the differentiation process.
Troubleshooting
Problem 1
The Matrigel droplets do not stay in the middle of the Seahorse microplate well after re-plating (step 9).
Potential solution
Incubate the Seahorse microplate at 37°C for at least 24 h and put the plate on top of a pre-warmed water-filled flask. When this is not sufficient, reduce the amount of medium that is mixed with Matrigel. Diluted Matrigel will hardly form droplets. Alternatively, reducing the volume of the Matrigel droplets from 3 to 2.5 μL can help.
Problem 2
The organoids are disrupted or broken or their appearance indicates low fitness or viability in the Seahorse cell culture microplate (step 9).
Potential solution
Some organoid lines can be particularly sensitive to mechanical shearing compromising their fitness and viability. To minimize this, try to reduce the pipetting movements while re-plating the organoids. Alternatively, try to adjust the timing of the re-plating to when the organoids have a size that is less sensitive to mechanical shearing. Depending of the organoid type, this size can be either larger or smaller. For intestinal organoids, re-plating the organoids when they are rather small (less than 120 μm diameter, Figure 2C), protects them from mechanical disruption.
Problem 3
The organoids do not or only mildly respond to the injections (step 20).
Potential solution
First of all, it is essential to control the stocks of the inhibitors and replace by new fresh stocks when the organoid response to the drugs decreases over time.
When the low response occurs with fresh and correctly prepared drug stocks, it should be checked whether the unresponsiveness of the organoids is a technical artifact or the actual phenotype of the organoids. For instance, when Oligomycin does not increase or only mildly increases the ECAR (Figure 6C), it should be analyzed whether Oligomycin was functional. In this experiment, Oligomycin did reduce the OCR (Figure 6D), indicating that the amount of Oligomycin that was used was correct. Furthermore, it should be examined whether the setup of the experiment was optimal. For instance, it should be checked whether increasing glucose concentrations will enhance the glycolytic capacity, in order to check that glucose availability was not limited in the experiment.
When the ineffectiveness of the drugs cannot be attributed to technical problems, the density of the organoids should be checked. When the density of the organoids is high, the drugs can possibly be less effective. In that case, try to reduce the number of organoids plated in the Seahorse Cell Culture microplate wells. When the organoids are plated in the proper density, titrate the concentrations of the injection reagents. When this does not work, this can be repeated in presence of more or less substrates in the XF assay medium.
Problem 4
There is high variability in OCR and ECAR measurement within technical replicates of an experiment (step 20).
Potential solution
This observation can result from uneven organoid numbers in the different wells. To avoid that, carefully re-suspend the organoids in the Matrigel and plate them immediately in the Seahorse cell culture microplate. Alternatively, inaccurate plating can lead to Matrigel drops fusing to the wall of the plate instead of being centered in the well (Figure 3C). This also leads to variable measurements of OCR and ECAR. Follow the steps concerning microplate incubation at 37°C and the use of a warm water-filled flask (step 9). Plating accuracy can be checked by microscope inspection after plating. Consider to exclude “failed wells” from the analysis. When doing so, restrain to collect that well for DNA extraction.
Problem 5
There is high variability in OCR and ECAR measurement between different experiments (Section Quantification & statistical analysis, step 5).
Potential solution
Make sure that the timing of the experiments is always similar. Reduce variability in culture time after splitting or trypsinizing the organoids and plating organoids in the Seahorse cell culture microplate. Furthermore, always plate the same number of organoids per well in every experiment.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Maria J. Rodríguez Colman (M.J.RodriguezColman@umcutrecht.nl).
Materials availability
This study did not generate new unique reagents, plasmids, or organoid lines.
Data and code availability
This study did not generate new datasets or codes.
Acknowledgments
This work was financially supported by Dutch Cancer Society (KWF 2016-I 10471, KWF 2017-II 11315).
Author contributions
Methodology and Investigation, M.J.R.C., M.C.L., and M.M. Writing – Original Draft, M.J.R.C. and M.C.L. Writing – Review & Editing, M.J.R.C.; Funding Acquisition, M.J.R.C. and B.M.T.B.
Declaration of interests
The authors declare no competing interests.
Contributor Information
Marlies C. Ludikhuize, Email: m.c.ludikhuize-2@umcutrecht.nl.
Maria J. Rodríguez Colman, Email: m.j.rodriguezcolman@umcutrecht.nl.
References
- Cracan V., Titov D.V., Shen H., Grabarek Z., Mootha V.K. A genetically encoded tool for manipulation of NADP+/NADPH in living cells. Nat. Chem. Biol. 2017;13:1088–1095. doi: 10.1038/nchembio.2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drost J., Jaarsveld R.H., van, Ponsioen B., an Jaarsveld, R.H., Ponsioen, B., Zimberlin, C., van Boxtel, R., Buijs, A., Sachs, N., Overmeer, R.M., Offerhaus, G.J., Begthel, H., et al. Sequential cancer mutations in cultured human intestinal stem cells. Nature. 2015;521:43–47. doi: 10.1038/nature14415. [DOI] [PubMed] [Google Scholar]
- Kim J., Koo B.K., Knoblich J.A. Human organoids: model systems for human biology and medicine. Nat Rev Mol Cell Biol. 2020;21:571–584. doi: 10.1038/s41580-020-0259-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindeboom R.G., van Voorthuijsen L., Oost K.C., Rodríguez-Colman M.J., Luna-Velez M.V., Furlan C., Baraille F., Jansen P.W., Ribeiro A., Burgering B.M. Integrative multi-omics analysis of intestinal organoid differentiation. Mol. Syst. Biol. 2018;14:e8227. doi: 10.15252/msb.20188227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludikhuize M.C., Meerlo M., Gallego M.P., Xanthakis D., Burgaya Julià M., Nguyen N.T.B., Brombacher E.C., Liv N., Maurice M.M., Paik J.-H. Mitochondria define intestinal stem cell differentiation downstream of a foxo/notch axis. Cell Metab. 2020;32:889–900.e7. doi: 10.1016/j.cmet.2020.10.005. [DOI] [PubMed] [Google Scholar]
- Mills R.J., Titmarsh D.M., Koenig X., Parker B.L., Ryall J.G., Quaife-Ryan G.A., Voges H.K., Hodson M.P., Ferguson C., Drowley L. Functional screening in human cardiac organoids reveals a metabolic mechanism for cardiomyocyte cell cycle arrest. Proc. Natl. Acad. Sci. U S A. 2017;114:E8372–E8381. doi: 10.1073/pnas.1707316114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohamed Suhaimi N.-A., Phyo W.M., Yap H.Y., Choy S.H.Y., Wei X., Choudhury Y., Tan W.J., Tan L.A.P.Y., Foo R.S.Y., Tan S.H.S. Metformin inhibits cellular proliferation and bioenergetics in colorectal cancer patient-derived xenografts. Mol. Cancer Ther. 2017;16:2035–2044. doi: 10.1158/1535-7163.MCT-16-0793. [DOI] [PubMed] [Google Scholar]
- Okkelman I.A., Neto N., Papkovsky D.B., Monaghan M.G., Dmitriev R.I. A deeper understanding of intestinal organoid metabolism revealed by combining fluorescence lifetime imaging microscopy (FLIM) and extracellular flux analyses. Redox Biol. 2020;30:101420. doi: 10.1016/j.redox.2019.101420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Colman M.J., Schewe M., Meerlo M., Stigter E., Gerrits J., Pras-Raves M., Sacchetti A., Hornsveld M., Oost K.C., Snippert H.J. Interplay between metabolic identities in the intestinal crypt supports stem cell function. Nature. 2017;543:424–427. doi: 10.1038/nature21673. [DOI] [PubMed] [Google Scholar]
- Schell J.C., Wisidagama D.R., Bensard C., Zhao H., Wei P., Tanner J., Flores A., Mohlman J., Sorensen L.K., Earl C.S. Control of intestinal stem cell function and proliferation by mitochondrial pyruvate metabolism. Nat. Cell Biol. 2017;19:1027–1036. doi: 10.1038/ncb3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schutgens F., Clevers H. Human Organoids: Tools for Understanding Biology and Treating Diseases. Annual Review of Pathology: Mechanisms of Disease 2020. 2020;15:1:211–234. doi: 10.1146/annurev-pathmechdis-012419-032611. [DOI] [PubMed] [Google Scholar]
- van de Wetering M., Francies H.E., Francis J.M., Bounova G., Iorio F., Pronk A., van Houdt W., van Gorp J., Taylor-Weiner A., Kester L. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell. 2015;161:933–945. doi: 10.1016/j.cell.2015.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Heiden M.G., Cantley L.C., Thompson C.B. Understanding the Warburg Effect: The Metabolic Requirements of Cell Proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This study did not generate new datasets or codes.