Abstract
Plants undergo profound physiological changes when transitioning from vegetative to reproductive growth. These changes affect crop production, as in the case of leafy vegetables. Lettuce is one of the most valuable leafy vegetable crops in the world. Past genetic studies have identified multiple quantitative trait loci (QTLs) that affect the timing of the floral transition in lettuce. Extensive functional molecular studies in the model organism Arabidopsis provide the opportunity to transfer knowledge to lettuce to explore the mechanisms through which genetic variations translate into changes in flowering time. In this review, we integrated results from past genetic and molecular studies for flowering time in lettuce with orthology and functional inference from Arabidopsis. This summarizes the basis for all known genetic variation underlying the phenotypic diversity of flowering time in lettuce and how the genetics of flowering time in lettuce projects onto the established pathways controlling flowering time in plants. This comprehensive overview reveals patterns across experiments as well as areas in need of further study. Our review also represents a resource for developing cultivars with delayed flowering time.
Keywords: flowering time, genetic mapping, quantitative trait loci (QTL) mapping, genome-wide association study, circadian clock, lettuce, crop breeding
Introduction
Flowering plants dominate terrestrial landscapes and play a central role in the human food system. The timing of flowering is critical for the survival and adaptation of species to their environments because of the various ways a plant’s life cycle depends on the correct combination of external factors and internal signals, ranging from temperature, water availability, and pollinator activities, to the plants’ hormonal and carbon accumulation status. From the perspective of agricultural production, the timing of flowering often affects the quality and quantity of harvests. Stable flowering time has been a major objective for breeding endeavors because it provides a foundation for reliable crop production (Leijten et al., 2018). Shifting the flowering time of a crop has allowed breeders to introduce varieties adapted to new or changing growing conditions (Jung and Müller, 2009).
Due to its importance in both basic and translational biology, flowering time has been extensively studied in model plant species, particularly in Arabidopsis thaliana. In Arabidopsis, flowering time is controlled through an intricate network of genes that respond to diverse environmental stimuli and developmental signals. Externally, flowering time is primarily regulated by temperature and day length; internally, flowering time is regulated by gibberellic acid, carbohydrates, and age. Information from different pathways gets synthesized through key floral integrator genes, whose expression initiates the vegetative-to-floral identity shift of the apical meristem (Lee and Lee, 2010; Srikanth and Schmid, 2011). This network has been shown to be conserved across diverse angiosperms with various degrees of modification (Hecht et al., 2005; Abou-Elwafa et al., 2010; Luo et al., 2013; Liu et al., 2018a). This genetic and molecular research in Arabidopsis has provided a framework for understanding flowering time regulation in other plants.
Lettuce (Lactuca sativa) is one of the most popular leafy vegetables in the United States. With an annual domestic farm-gate value of more than $2.9 billion USD, lettuce was the highest-valued fresh-market vegetable in the country in 2019 (USDA-NASS., 2019). On a global scale, China is the world leader in lettuce production, producing four times as much as the United States and contributing to approximately half of the world’s lettuce production (FAO, 2020). Lettuce is harvested while it is in its vegetative growth state as it is consumed solely for its leaves in the US. The initiation of transitioning to reproductive growth is marked by the elongation of the stem, an event referred to as “bolting.” Bolting renders the crop bitter and unmarketable (Ryder, 1996). Therefore, delayed bolting and flowering is preferred in lettuce for maximizing harvestable yield while maintaining culinary quality; however, overly delayed flowering is unfavorable for seed production purposes. Lettuce is a self-pollinating plant whose flowering is accelerated under longer day lengths (Waycott, 1995) and higher ambient temperatures (Ryder, 1996). Wild lettuce (L. serriola), the wild progenitor of cultivated lettuce (Zhang et al., 2017), commonly exhibits a summer annual or winter annual growth habit. As a summer annual, wild lettuce seeds imbibe, germinate and flower rapidly under the long-day condition of spring and summer; as a winter annual, they germinate in the fall, overwinter as a vegetative rosette, and flower in the spring or summer in the following year. Vernalization, a period of cold treatment below a certain temperature threshold, is therefore required for some wild lettuce accessions to transition to reproductive growth (Prince et al., 1978). Understanding the genetic regulation of these biological processes as they related to flowering time in cultivated and wild lettuce is of great interest to biologists and breeders alike.
Over the past decade, numerous genetic and genomic resources have become available for lettuce. In 2013, an ultra-high density genetic map for lettuce was published (Truco et al., 2013); in 2017, a chromosome-scale lettuce reference genome assembly became available (Reyes-Chin-Wo et al., 2017). Multiple mapping populations have been developed and studied across the globe to investigate the genetic regulation of various agronomic traits of this internationally enjoyed vegetable (Silva et al., 1999; Hartman et al., 2013a,b; Truco et al., 2013). Sequencing of diverse accessions of wild and cultivated lettuce has enabled genome-wide association studies (GWAS) of commercially valuable traits (Kwon et al., 2013; Sthapit Kandel et al., 2020). Functional analyses of putative flowering time genes in lettuce have unveiled some of the molecular mechanisms underlying flowering time regulations. The latter approach is mostly enabled by the identification of lettuce orthologs to Arabidopsis flowering time genes through sequence similarity (Abbott, 2010; Fukuda et al., 2011, 2017; Han Y. et al., 2016a; Chen et al., 2018a,b).
The aim of this review is to consolidate current knowledge of the genetic and molecular control of flowering time in lettuce in the context of the current lettuce reference genome. In the Genetic Analysis section of this review, we collate information from all published and several unpublished genetic mapping experiments on bolting and flowering time in lettuce to compose a comprehensive list of the known quantitative trait loci (QTLs) segregating among cultivars and wild accessions. By anchoring flanking markers of these QTL on version 8 of the lettuce reference genome assembly, we provide a common framework for comparing and cross-referencing these QTLs as well as rationalizing their nomenclature. This broad view enables an appreciation of the genetic diversity underlying the phenotypic variation of flowering time in lettuce. Using sequence homology, we located putative orthologs of flowering time genes identified in Arabidopsis and compared their physical coordinates in reference to the bolting and flowering time QTLs in lettuce. In the Integration section of this review, we provide a detailed summary of the experimental evidence relating to the molecular mechanism of flowering time regulation in lettuce and organize these molecular experiments according to the pathways they pertain to. By comparing the assembled knowledge of flowering time regulation in lettuce to its well-studied counterpart in Arabidopsis, we were able to map conservation and divergence of the flowering time regulation pathways between the two species as well as identify underexplored areas and approaches that can potentially benefit the scientific and breeding communities.
Materials and Methods
Physical Location of Published QTLs
Physical coordinates of the markers flanking significant QTLs were reported in seven studies (Niroula, 2017; Mamo et al., 2019; Sandoya et al., 2020; Seki et al., 2020; Sthapit Kandel et al., 2020; Niroula et al., unpublished; You et al., unpublished). DNA variant detector array probe sequences of the lettuce expression sequence tag (EST)-based single nucleotide polymorphism (SNP) markers used in four studies (Hartman et al., 2013b; Jenni et al., 2013; Kwon et al., 2013; Niroula, 2017) were blasted against version 8 of the reference genome (Reyes-Chin-Wo et al., 2017; NCBI: GCA_002870075.2) to determine their physical locations. The coordinates of amplified fragment length polymorphism (AFLP) markers used in two studies could not be determined (Lavelle, 2009; Hartman et al., 2012, 2013b); therefore, the genetic locations of the peaks of the QTLs discovered in these studies were reported instead. QTL intervals of the GWAS were determined by extending the physical coordinates of flanking markers of significant peaks by 4 Mb on each side, according to the results of the linkage disequilibrium analysis in Kwon et al. (2013).
Unpublished Studies
A population consisting of 97 F8 recombinant inbred lines (RILs) had been derived by single-seed descent from a cross between L. sativa cv. Salinas and L. serriola accession US96UC23 (Truco et al., 2013). This RIL population was planted in spring 2009 in an experimental field in Salinas, CA. Each experiment was arranged in a randomized complete block design with two blocks and was surrounded by guard rows. The plants were transplanted in 1 day. All plots were overhead-irrigated immediately after transplanting. Fertilization, pest control, and disease control were carried out according to standard protocols. The developmental stage of the plants was scored once a week. Four plants at the center of each plot were scored. The first scoring was on June 5th and the last scoring was on September 16th.
DNA from parental lines and the RILs were genotyped for 3,696 SNPs using the lettuce GeneChip genotyping protocol (Truco et al., 2013). Polymorphic markers were used to construct a genetic map using procedures described in Truco et al. (2013). A QTL analysis was conducted using composite interval mapping in QTL Cartographer 2.0 (Basten et al., 1994). Significance thresholds at p < 0.05 were calculated for each trait by permutation analysis with 1,000 permutations.
Two (Davis02 and Salinas04) out of the three trials using RILs of the Salinas × US96UC23 population reported by Lavelle (2009) were reanalyzed using the 3,696 lettuce GeneChip SNP markers, with composite interval mapping in QTL Cartographer 2.0 (Basten et al., 1994). Significance thresholds at p < 0.05 were calculated for each trait by permutation analysis with 1,000 permutations.
QTL Name Rationalization
When necessary, QTLs were named or renamed according to the convention established for lettuce with the abbreviation of the associated phenotype and chromosomal linkage group preceded by the letter “q.” “FLT” was used for flowering time and “BLT” for bolting time. The numbering of QTLs on the same chromosome was assigned according to the time of publication. Priority was given to the earliest publication when the same QTL had been identified in more than one study or when the same designation had been used for different QTLs.
Orthology
Orthofinder (Emms and Kelly, 2015) was used for genome-wide prediction of lettuce orthologs of flowering-time-related genes characterized in A. thaliana. Amino acid sequences of seven eudicot genomes, A. thaliana, Solanum lycopersicum, Daucus carota, Helianthus annuus, Cichorium intybus, L. serriola, and L. sativa, were used in the orthology analysis. The list of flowering time genes was obtained from the interactive database of flowering-time gene networks in A. thaliana (Bouché et al., 2015)1.
Expression Analysis
RNA-seq reads from the time course experiment described in Higashi et al. (2016) were quality and adapter trimmed using BBDuk2 and mapped to version 8 of the lettuce reference genome (Reyes-Chin-Wo et al., 2017) using STAR (Dobin et al., 2013). Oscillation of transcription level (in reads per million mapped reads) and period of oscillation were detected using R package DiscoRhythm (Carlucci et al., 2019). Transcripts with p < 0.05 over a 24-h oscillation period are reported.
Genetic Analysis
A systematic literature review and consultation with the research community identified nine published studies (Hartman et al., 2012, 2013a,b; Jenni et al., 2013; Kwon et al., 2013; Mamo et al., 2019; Sandoya et al., 2020; Seki et al., 2020; Sthapit Kandel et al., 2020), two dissertations (Lavelle, 2009; Niroula, 2017), and four unpublished datasets (Niroula et al., unpublished; You et al., unpublished; Han et al., unpublished; M.-J. Truco, unpublished) pertinent to this study. These data represent a comprehensive collection of genetic mapping studies of flowering time and bolting time in lettuce. A total of 56 field and greenhouse experiments have been conducted between 2002 and 2019, testing 11 mapping populations (for QTL mapping) and 2 diversity panels (for GWAS). Data including the parental lines and generation of the mapping populations, the number of lines, the type and number of markers, mapping software, and parameters used in the analysis, as well as the time and location of each experiment are summarized in Supplementary Table 1. Most experiments tracked either the flowering time (23) or bolting time (13) phenotype, while 20 studies tracked both. Over three quarters (41) of the experiments were conducted in the field. The geographic locations of experiments included Chile, Japan, the Netherlands, southeastern Canada, and West Coast of the United States. These growing areas differ substantially in temperature and humidity. Notably, however, nearly all experiments (50) were conducted under long-day (LD) conditions with photoperiods longer than 12 h per day. Only three mapping populations have been tested in a total of six experiments under both LD and short-day (SD) conditions.
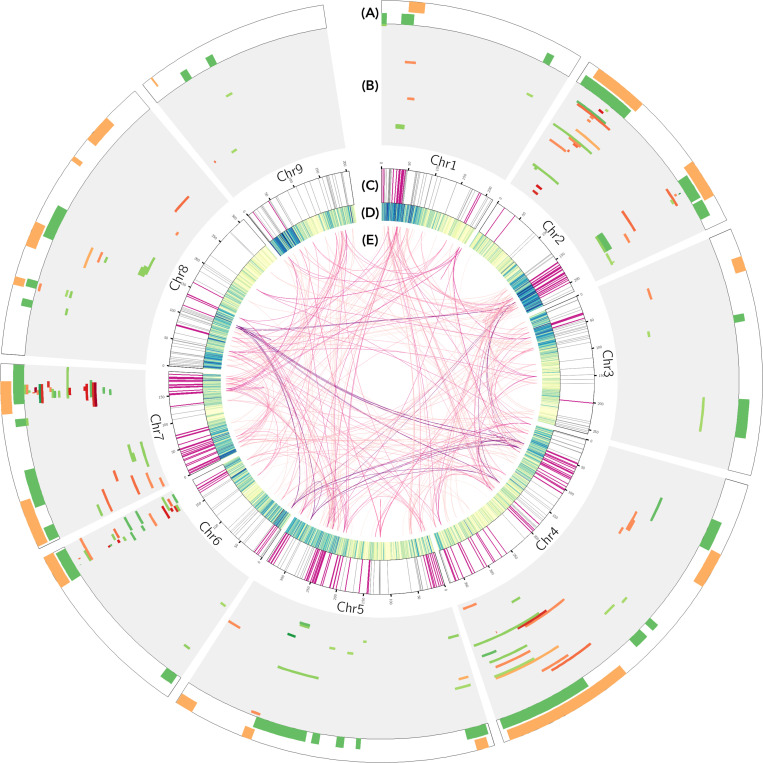
A total of 167 QTLs have been reported for bolting and flowering time in lettuce. Merging QTLs with extensively overlapping intervals reduces this number to 67. Bolting and flowering time QTLs are located on all nine lettuce chromosomes (Figure 1). Chromosomes 2 and 7 have the highest counts of QTLs. Specifications of each QTL including physical and genetic locations as well as effect sizes in each experiment are provided in Supplementary Table 2. Seven major QTLs each explained more than 30% of phenotypic variance in their respective experiments. The effect of any one QTL can be highly variable across trials. For instance, qFLT7.1 explained 11.2, 30.23, 39.6, and 51.7% of phenotypic variance in separate experiments that used the same mapping population, PI251246 × Salinas. The vast majority of the reported QTLs are environmentally sensitive; their effects on the phenotype only manifested in a subset of the experiments that used a specific mapping population. This is in line with the current understanding of the mechanism of flowering time regulation as one that integrates both internal and external signals. It is possible that the environment-sensitive QTLs represent genetic variations in the upstream signaling of the flowering time pathway, where environmental cues are perceived and transduced into molecular signals, while QTLs that are consistently detected across all environments represent variation in downstream components that affect flowering time regardless of external cues. Some QTLs identified under SD conditions overlapped with QTLs identified under LD conditions in the same mapping populations (qBLT6.1, 6.5 and qFLT2.1, 4.3, and 6.1), while other SD QTLs were novel (qFLT1.2, 4.5, and 9.5); all novel SD QTLs were detected in the Armenian × PI251246 population (Han et al., unpublished). The detection of novel SD QTL suggests this population segregates for photoperiod sensitivity; and photoperiod sensitivity is regulated separately from daylength-independent flowering time. Fifteen of the 67 QTLs were discovered in multiple populations that do not share parents; nevertheless, the parental lines could share regions of identity by descent. All other QTLs were identified in only one mapping population. These results suggest that modification of flowering time phenotype is achieved through a variety of strategies in different lettuce cultivars and wild lettuce accessions as in other species. The lack of overlapping results in past studies also suggests that there are likely to be additional polymorphic loci regulating flowering time that remain to be discovered.
FIGURE 1.
(A) Physical location of 35 bolting time (green) and 32 flowering time (yellow) consensus quantitative trait loci (QTLs) in lettuce. (B) Physical location of 167 QTLs reported in a total of 56 field and greenhouse experiments. Each track represents one experiment. Earlier reported experiments are in the outer tracks. Bolting time QTLs are indicated by green color blocks and flowering time QTLs by orange blocks. Saturation of the color blocks indicates percent phenotypic variance explained by the QTLs. The physical locations of 12 QTLs from three experiments (Summer 2003 from Lavelle, 2009 and both experiments from Hartman et al., 2013b) could not be located on the genome because amplified fragment length polymorphism (AFLP) markers were used. (C) Location of lettuce orthologs of genes with flowering time function in Arabidopsis. Flowering time orthologs within known QTLs are highlighted in fuchsia. (D) Gene density of the lettuce genome. (E) Flowering time orthologs within the same orthogroup are connected. Darker shade connections indicate larger orthogroups.
About half (35) of the reported QTLs are for the bolting time phenotype, despite the over-representation of flowering time being the primary phenotype of interest. Nine QTLs have a pleiotropic effect on both bolting time and flowering time as shown by their colocation on the lettuce genome (Figure 1). The low frequency of collocation between bolting and flowering time QTLs can be partly explained because most studies only tracked one of the two phenotypes. Nevertheless, this might also reflect genetic differences where the pathways controlling bolting and flowering are distinct, thus providing opportunities for understanding the transition between stem elongation and floral initiation.
Integration of Functional Information From Model Systems
Adjustment of flowering time based on environmental and internal cues is realized via the intricate interplay of seven major genetic pathways in Arabidopsis. These include the vernalization pathway, the autonomous pathway, the ambient temperature pathway, the photoperiod pathway, the hormone pathway, the aging pathway, and the sugar pathway (Srikanth and Schmid, 2011). Extensive research in A. thaliana has revealed the identity and function of pivotal genes in these pathways. We review these pathways and their key genic components in A. thaliana, followed by a summary of functional analyses that have been performed on their orthologs in lettuce. All pathways contain genes that collocate with known QTLs. The number of QTLs each pathway intersects with is relatively proportional to the number of genes present in the pathway. By contrasting our knowledge of flowering time regulation in lettuce to its counterpart in Arabidopsis, we aim to identify key differences between the two systems and provide data-informed suggestions for future breeding and scientific endeavors.
An analysis of orthology placed 306 Arabidopsis flowering time genes into 222 orthogroups; 237 of these genes have orthologs in lettuce (Supplementary Table 3). Four hundred and five lettuce genes were identified as flowering time orthologs representing 217 out of the 222 orthogroups (Table 1).
TABLE 1.
Summary statistics of lettuce and Arabidopsis genes within each flowering time regulatory pathway.
| Pathways | Genes in lettuce | Genes in Arabidopsis |
| Vernalization and autonomous | 30 | 27 |
| Ambient temperature | 5 | 7 |
| Photoperiodism and circadian clock | 171 | 110 |
| Gibberellic acid | 37 | 28 |
| Aging | 18 | 22 |
| Sugar | 24 | 9 |
| Floral integrator | 17 | 8 |
| Flower development and meristem identity | 25 | 9 |
| General | 168 | 117 |
| Two pathways | 48 | 73 |
| Three pathways | 17 | 18 |
| Four pathways and above | 2 | 3 |
The majority of the 405 orthologs have at least two paralogs within the lettuce genome. Only 110 Arabidopsis flowering time genes have single lettuce orthologs. The largest flowering time orthogroups, which contains Arabidopsis FT-INTERACTING PROTEIN 1 (FTIP1), has eight orthologs in the lettuce genome. The lettuce genome has undergone duplication and triplication events (Reyes-Chin-Wo et al., 2017); however, locations of paralogs within flowering time orthogroups is not obviously consistent with the triplication signature basal to the Compositae family (Figure 1; Reyes-Chin-Wo et al., 2017). The lack of correspondence to the triplication event may reflect gene expansion in earlier duplication events. Nearly half of the orthologs (190/405) lie within the known QTLs, while the rest do not. This supports the idea that there is undiscovered natural genetic variation for bolting and flowering time phenotypes. Conversely, four out of the 64 QTLs do not have putative orthologs within their intervals. Under 12 h light/12 h dark conditions and in 3-week-old vegetative plants, 43 orthologs show oscillating expression following 24-h periods (Supplementary Table 3). Twenty-five out of the forty-three oscillating transcripts are putative members of the photoperiod pathway and/or the circadian clock. Their oscillation provides support for the function of these genes in pathways that involve light sensing and time tracking.
There have been two genome-wide expression experiments that capture the transition between vegetative and reproductive growth in lettuce (Chen et al., 2018a,b; Liu et al., 2018b). One experiment compared heat-treated bolting lettuce with non-heat-treated rosette samples and identified 2,149 differentially expressed genes (DEGs; Liu et al., 2018b). The other experiment was performed in laser capture micro-dissected apical meristematic tissues (Chen et al., 2018a,b). This study reported 21 DEGs at the initiation of bolting (formation of dome-shaped apical meristem) and 365 DEGs after the apical meristem committed to reproductive growth (elongation of apical meristem; Chen et al., 2018a). The same group later used the same dataset to analyze the expression profile of 15 putative flowering-time orthologs in lettuce (Chen et al., 2018b); 4 showed upregulation at the initiation of bolting while 7 showed upregulation after the apical meristem committed to reproductive growth. Several of these key regulatory flowering-time orthologs are discussed in detail below according to the regulatory pathway they belong to.
Vernalization and Autonomous Pathway
The vernalization and autonomous pathways are often discussed together because of the central role of FLC in both of them. Vernalization refers to the requirement of exposure of imbibed seeds or vegetative seedlings to a prolonged period of cold temperature to induce flowering. In Arabidopsis, vernalization is realized through FRIGIDA (FRI) and FLOWERNG LOCUS C (FLC). Naturally occurring mutations in FRI have been associated with the loss of the vernalization requirement in summer annual Arabidopsis accessions (Johanson et al., 2000). FRI is an up-regulator of FLC (Geraldo et al., 2009), which encodes a MADS box protein that directly represses flowering time genes FD, FLOWERING LOCUS T (FT), and SUPPRESSOR OF OVEREXPERSSION OF CONSTANS 1 (SOC1; Searle et al., 2006). The expression of FLC is suppressed by gene VERNALIZATION INSENSITIVE 3 (VIN3) and two antisense long RNAs of FLC itself, COOLAIR and COLDAIR (Sung and Amasino, 2004; Heo and Sung, 2011). The maintenance of the silencing of FLC is carried out through VERNALIZATION 1 and 2 (VRN1 and VRN2; Gendall et al., 2001; Levy et al., 2002). In contrast to vernalization pathway mutants, those in the autonomous pathway are characterized by delayed flowering regardless of environmental conditions. Genes on the autonomous pathways, including FLOWERING CONTROL LOCUS A (FCA), FLOWERING LOCUS D and K (FLD and FLK), FPA, FY, LUMINIDEPENDENS (LD), FVE, and RELATIVE OF EARLY FLOWERING 6 (REF6), modulate FLC expression by either changing the chromatin configuration or participating in mRNA modification (Simpson, 2004).
Many accessions of wild lettuce (L. serriola) require vernalization to transition to reproductive growth. Early studies identified 10°C to be the upper limit for vernalizing wild lettuce (Warne, 1947). The response is quantitative, with longer cold-treatment periods associated with more accelerated flowering after the treatment. Imbibed seeds and older vegetative seedlings were both more responsive to vernalization treatment than cotyledon-stage seedlings (Warne, 1947; Prince et al., 1978). Vernalization can accelerate flowering in some cultivars of lettuce (L. sativa), such as the crisphead cultivar “Great Lake” (Rappaport et al., 1956), but not others such as the leafy cultivars studied by Zhang et al. (2016).
No lettuce ortholog of AtFLC has been identified, despite numerous attempts using multiple bioinformatic and molecular approaches (Reeves et al., 2007; Lavelle, 2009; Abbott, 2010), suggesting a potentially different molecular mechanism underlying vernalization in lettuce. FLC belongs to a distinct clade within the MLKC-type MADS box gene family (Hileman et al., 2006). The presence and function of genes in the FLC clade have been challenging to predict across eudicot lineages regardless of the evolutionary relationship between the species of interest (Becker and Theißen, 2003; Hecht et al., 2005; Hileman et al., 2006). Searches for FLC homologs have been made in legumes (Fabaceae; rosid), sunflower (Helianthus annuus; asterid), and tomato (Solanum lycopersicum; asterid), but with negative results (Hecht et al., 2005; Hileman et al., 2006; Blackman et al., 2011b). Nevertheless, an FLC homolog with conserved functions was identified in sugar beets (Beta vulgaris; Reeves et al., 2007), a member of the Caryophyllales more closely related to lettuce (asterid) than Arabidopsis (rosid). An FLC-like gene, CiFL1, has also been cloned in chicory (Cichorium indybus), a biennial Compositae species closely related to lettuce; expression analysis suggested it functions as a floral suppressor during the vernalization process in chicory (Périlleux et al., 2013). Orthologs of many other genes that function upstream of FLC in Arabidopsis have been identified and cloned in lettuce including FCA, FLD, FLK, FPA, FY, LD, and FVE. The identity between the amino acid sequence of the lettuce genes and their respective Arabidopsis orthologs ranges from 43 to 79% (Abbott, 2010). Among these genes, LsFVE exhibits peak expression before the vegetative–reproductive transition; maximum expression of LsFLD and LsLD overlapped temporally with the vegetative–reproductive transition period (Fukuda et al., 2017), suggesting the important role of the autonomous pathway in flowering time regulation of cultivated lettuce. Our orthology analysis identified a potential lettuce ortholog for FRI on Chromosome 5, Lsat_1_v5_gn_5_9321 (Supplementary Table 3); no molecular analyses have been conducted to study its function. The elusive evolutionary history of the FLC gene clade and the unresolved regulatory mechanism of vernalization in lettuce creates an interesting opportunity for future genetic and molecular studies.
Ambient Temperature Pathway
High ambient temperature accelerates flowering in Arabidopsis (Balasubramanian et al., 2006). Although the regulatory mechanisms of ambient temperature influences on flowering time is not as fully elucidated as some of the other pathways, several genes that play key roles in this thermo-sensory response have been identified. The loss of SHORT VEGETATIVE PHASE (SVP) function resulted in insensitivity to ambient temperature changes and plants deficient in ACTIN RELATED PROTEIN 6 (ARP6) display constitutive warm temperature responses. SVP is a MADS-box protein that binds to FT and SOC1 promoters and acts as a repressor. It also mediates the temperature-dependent functions of FCA and FVE in the autonomous pathway (Lee et al., 2007). Chromatin immunoprecipitation (ChIP) analysis suggests that SVP may function in an FLC-dependent manner (Li et al., 2008). ARP6 functions by introducing into nucleosomes, rather than H2A, a special histone H2A.Z, whose DNA wrapping capacities exhibits temperature dependency (Kumar and Wigge, 2010). There is extensive crosstalk between the ambient temperature and the vernalization/autonomous pathways. FLC is a potent suppressor of thermo-induced flowering, while FLOWERING LOCUS M (FLM), a MADS-box protein with extensive sequence similarity to FLC, co-locates with a QTL modulating thermosensitivity (Balasubramanian et al., 2006).
Heat-accelerated bolting and flowering is a common phenomenon that impacts agricultural production of diverse lettuce cultivars. Cultivars exhibit broad variation in their bolting behavior in response to high ambient temperature, making this trait an important subject of genotype × environment studies (Hwang et al., 2007; Jenni and Yan, 2009; Jenni et al., 2013; Han Y.Y. et al., 2016b; Lafta et al., 2017; Holmes et al., 2019). Intersecting QTLs with physical coordinates of lettuce flowering time orthologs suggests Lsat_1_v5_gn_2_93321, a MADS-box gene, might function in heat-induced bolting (Jenni et al., 2013). No definitive lettuce ortholog of SVP has been identified; this is at least partly due to the sequence-level similarity between SVP and AGAMOUS-LIKE 24 (AGL24), a floral meristem identity gene that primarily functions downstream of the floral initiation pathway (Gregis et al., 2008). Four putative orthologs of SVP and AGL24 are present in the lettuce genome; one of them, Lsat_1_v5_gn_3_62800, has been considered either an ortholog of SVP or AGL24 (Supplementary Table 3; Huo et al., 2016; Chen et al., 2018b). Another ortholog of SVP and AGL24, Lsat_1_v5_gn_6_105061 (Supplementary Table 3), is located within qFLT6.2 but has yet to be studied at the molecular level. The lack of clear lettuce orthologs of FLC and SVP has made it difficult to form testable hypotheses regarding the regulatory mechanism through which ambient temperatures accelerate bolting in lettuce. In addition, two studies have reported binding motifs of heat shock transcription factors in the promoter regions of LsSOC1 and LsMADS55, putative lettuce orthologs to Arabidopsis SOC1 and APETALA1 (AP1), both known to function downstream of FLC in the floral induction pathway (Chen et al., 2018b; Ning et al., 2019). These reports postulate the possibility that the thermal control of flowering time in lettuce might have a different and more concise regulatory architecture than its counterpart in Arabidopsis. Exploratory studies using differential expression and protein accumulation analyses have suggested genetic elements actively involved in this process. Expression levels of lettuce HEAT SHOCK PROTEIN 70 (HSP70), an output of the ambient temperature sensing pathway in Arabidopsis (Kumar and Wigge, 2010), differ between heat sensitive and heat tolerant lettuce lines, suggesting its conserved function in temperature sensing in lettuce (Li et al., 2017). A differential expression analysis comparing the gene expression profiles of heat-treated (bolted) and control (non-bolted) lettuce plants on the seventh day after the initiation of heat treatment revealed significant changes in C2H2 zinc finger, Ap2-EREBP, and WRKY transcription factor families, indicating their potential functions in heat-induced bolting (Liu et al., 2018b). The same study also identified increased gibberellic acid (GA) and indole-3-acetic acid (IAA) prior to heat induced bolting. Two comparative proteomic studies have investigated differential protein accumulation in heat-treated lettuce plants. One study compared the heat response of an early bolting lettuce genotype with that of a late bolting genotype (Han Y. et al., 2016a) and reported elevated accumulation of metabolism-related proteins in the early bolting genotype and protein synthesis-related proteins up regulated in the late bolting genotype. The other compared heat-treated, bolted lettuce plants with non-treated, unbolted plants of the same genotype (Hao et al., 2018); this reported enrichment of proteins is associated with photosynthesis, tryptophan metabolism, and IAA biosynthesis. Additional studies are needed to disentangle the molecular signal of the bolting process from that of the thermo-sensory pathway that precedes bolting.
Photoperiod Pathway
Arabidopsis is a facultative long-day plant, for which floral transition is promoted by LD conditions and is delayed, but not completely inhibited, by SD conditions (Mouradov et al., 2002; Fornara et al., 2010; Srikanth and Schmid, 2011). The photoperiodic control of flowering in Arabidopsis is realized through the integration of transcriptional and post-transcriptional regulations of a zinc finger transcription factor, CONSTANS (CO). The baseline expression level of CO is circadianly entrained, resulting in a 24-h-phase oscillation of its transcription (Suárez-López et al., 2001). This phasing is further modified by the GIGANTEA–FLAVIN-BINDING, KELCH REPEAT, F-BOX 1–CYCLING DOF FACTOR 1 (GI-FKF1-CDF1) protein triad, resulting in a second peak of CO expression toward the end of the day under LD conditions (Imaizumi et al., 2005; Sawa et al., 2007; Fornara et al., 2009). At the post-transcriptional level, the protein stability of CO is regulated by the E3 ubiquitin ligase CONSTITUTIVELY PHOTOMORPHOGENIC 1 (COP1) and members of the SUPPRESSOR OF PHYA-105 (SPA) protein family in a daylength-dependent fashion (Hoecker and Quail, 2001; Laubinger et al., 2006; Liu et al., 2008b). Other systemic regulators of CO include PHYTOCHROME B (PHYB), PHYC, and DAY NEUTRAL FLOWERING (DNF) (Monte et al., 2003; Morris et al., 2010; Golembeski and Imaizumi, 2015). The expression of CO regulates flowering by promoting the production of a floral induction signal in leaves (Ayre and Turgeon, 2004). The signaling protein and mRNA encoded by FT are both transported to the shoot apical meristem via phloem companion cells to initiate flowering (Kardailsky et al., 1999; Weigel et al., 2000; Corbesier et al., 2007; Li et al., 2011). During vegetative growth, the expression of FT is repressed by histone trimethylation mediated by polycomb repressive complexes (Jiang et al., 2008).
Like Arabidopsis, lettuce is also a facultative long-day plant (Sukprakarn, 1985; Waycott, 1995). Lettuce exhibits an array of responses to changes in photoperiod, varying from the less responsive summer cultivars, such as the North American crispheads “Empire” and “Salinas,” to the highly sensitive winter cultivars, such as the European butterheads “May King” and “Saffier,” whose flowering time is significantly delayed as daylength shortens. However, the genetic determinants underlying the photoperiodic response have not been well-characterized. Three populations have been grown under both LD and SD conditions; the SD experiments detected nine QTLs; three of these SD QTLs were unique to the SD condition (qFLT1.2, 4.5, and 9.5; Supplementary Table 2). The causal genes underlying these QTLs are candidates for key elements of the photoperiod sensing pathway in lettuce.
Six CO-like genes have been identified in the lettuce genome (Supplementary Table 3). Two of these genes, Lsat_1_v5_gn_2_86121 and Lsat_1_v5_gn_5_122401, show oscillating expression in 24-hour periods. Lsat_1_v5_gn_5_122401 is within qFLT5.3 (Seki et al., 2020). No CO-like gene has been analyzed functionally in lettuce. Consequently, the mechanism of the photoperiod control of flowering time in lettuce remains unresolved. Multiple lettuce genes may jointly fulfill the functions of AtCO; complementary functions have been reported for CO-like genes in sugar beet (B. vulgaris; Dally et al., 2018). A functional CO ortholog, HaCOL2, has been identified in sunflower (H. annuus), a close relative to lettuce; overexpression of HaCOL2 complemented the Arabidopsis co mutation (Blackman et al., 2011a).
Gibberellic Acid Pathway
In Arabidopsis, gibberellic acid (GA) acts as a flower promoting agent in parallel to CO activities; deficiency in GA biosynthesis has an additive effect on the late flowering phenotype of co mutants in LD (Putterill et al., 1995). GA functions by targeting DELLA proteins, which function as repressors of plant growth and development (Harberd et al., 2009; Sun, 2010), for ubiquitination, thereby promoting the expression of the floral integrators SOC1 (Liu et al., 2008a) and LEAFY (LFY; Gocal et al., 2001; Achard et al., 2004).
Foliar application of exogenous GA induces bolting in lettuce (Kato, 1964b; Han Y. et al., 2016a) and endogenous GA may play a role in the ambient temperature signaling pathway in lettuce (Kato, 1964a). Under natural growing conditions, GA is barely detectable in lettuce apical meristems during vegetative growth; however, its concentration increases rapidly after floral induction (Kato, 1964a). After heat treatment, GA begins to accumulate shortly after treatment, resulting in a sharp increase similar to the GA accumulation observed during the natural bolting process (Kato, 1964a). This result has been confirmed using mass spectrometry to measure GA levels; in addition, the expression of LsGA3ox1, a GA 3-oxidase gene that metabolizes GA20 to the bioactive form GA1, is significantly upregulated by high temperature (Fukuda et al., 2009). Coupling of GA to the expression of major flower induction genes, such as LsSOC1 and LsLFY, has yet to be demonstrated.
Aging Pathway
Before transitioning to reproductive growth, the vegetative growth of a plant can be temporally divided into a juvenile phase and an adult phase (Huijser and Schmid, 2011). Vegetative phase change is controlled primarily by the interplay of two microRNAs (miRNAs), miR156 and miR172 (Wang et al., 2011). In Arabidopsis, miR156 is highly abundant in juvenile seedlings; it represses flower induction by negatively regulating SQUAMOSA PROMOTER BINDING PROTEIN LIKE (SPL) genes. Both in concert and parallel with FT, SPL genes promote the expression of floral integrator genes SOC1 and LFY, as well as the floral identity genes FRUITFUL (FUL) and AP1 (Wang et al., 2011). MiR172 is a direct target of SPL9 in Arabidopsis (Wu et al., 2009); in contrast to miR156, its expression level gradually increases as the plant ages and the activity of the SPL genes rise. MiR172 targets flower suppressors, such as AP2, SCHLAFMÜTZE (SMZ), SCHNARCHZAPFEN (SNZ), TARGET OF EAT1 (TOE1), TOE2, and TOE3, for silencing (Schmid et al., 2003; Mathieu et al., 2009). This pathway is conserved across divergent monocot and dicot lineages (Lauter et al., 2005).
LsMIR156 and LsMIR172, genes that encode miR156 and miR172 in lettuce, have been cloned and ectopically expressed in Arabidopsis. Consistent with the function of their respective orthologs in Arabidopsis, LsMIR156 delayed flowering and LsMIR172 accelerated flowering when overexpressed. In addition, the mRNA levels of LsSPL3 and 4 were inversely correlated with that of miR156 and positively with miR172, consistent with conservation of the aging pathway between Arabidopsis and lettuce. Interestingly, unlike the case in Arabidopsis, the function of MIR156 in lettuce depends on the gene DELAY OF GERMINATION1 (DOG1). In lettuce, DOG1 has a pleiotropic effect on both seed germination and flowering time, while the Arabidopsis dog1 loss-of-function mutant exhibits deficient seed dormancy but no flowering time phenotype (Huo et al., 2016).
Sugar Pathway
The integration of carbohydrate status into the flowering time pathway is still poorly understood and the effect of sugars on flowering time varies across plant species (Srikanth and Schmid, 2011). In Arabidopsis, extremely high or low concentrations of sucrose delay flowering, while a 1% concentration of sucrose promotes earlier flowering (Ohto et al., 2001). Homeostasis of another sugar, trehalose, is also essential for the normal transition toward reproductive growth (Dijken et al., 2004; Paul et al., 2008).
The effects of sugar on flowering time have not been studied in lettuce. Orthologs of ADG1 (Eimert et al., 1995), HXK1 (Moore et al., 2003), SUS4 (Seo et al., 2011), and TPS1 (Dijken et al., 2004) on the sugar pathway in Arabidopsis are found within lettuce bolting and/or flowering time QTLs; however, orthologs of other flowering time genes were also present within these QTLs, making it unclear whether variations in the sugar pathway were involved in the phenotypic variations (Supplementary Table 3).
Floral Integrators
FT, SOC1, and LFY are often referred to as floral integrators because of their pivotal role in connecting various floral induction pathways to the floral development pathway (Lee and Lee, 2010; Srikanth and Schmid, 2011). Experimental evidence puts the three genes along a signaling pathway from upstream to downstream in the order of FT, SOC1, and LFY (Yoo et al., 2005; Lee et al., 2008), each promoting the expression of its target. FT is directly targeted by FLC, SVP, and CO, thereby integrating signals from the vernalization/autonomous pathway, temperature pathway, and photoperiod pathway. SOC1 is regulated by FLC, SVP, and GA signaling. LFY is regulated by GA and miRNAs (Yamaguchi et al., 2009; Wang et al., 2011). Through this network of interactions, the floral integrator genes are expressed under conducive environmental conditions, resulting in the upregulation of flower homeotic genes such as AP1, APETALA3 (AP3), and AGAMOUS (AG), triggering the transition from vegetative to reproductive growth (Liu et al., 2009).
Lettuce orthologs of FT (Lsat_1_v5_gn_2_17881), SOC1 (Lsat_1_v5_gn_7_6780), and LFY (Lsat_1_v5_gn_4_84380) have been identified and cloned (Abbott, 2010; Fukuda et al., 2011, 2017; Chen et al., 2018a,b). LsFT is located within qFLT2.4 and qFLT2.6, while LsSOC1 is located within qFLT7.3 (Supplementary Table 3). As expected, none of these genes are expressed in vegetative lettuce plants.
LsFT plays an important role in the flowering time pathway in lettuce. Lettuce has only one identifiable FT ortholog, located on Chromosome 2, unlike in sunflower, where three FT orthologs have been identified (Oda et al., 2004; Blackman et al., 2011a; Fukuda et al., 2011; Reyes-Chin-Wo et al., 2017). The amino acid sequence of LsFT is 76% identical to AtFT (Abbott, 2010; Fukuda et al., 2011). Ectopic expression of LsFT using the 35S promoter induces early flowering in wild type Arabidopsis and partially rescues the late flowering phenotype of ft-2 mutant Arabidopsis (Fukuda et al., 2011; Chen et al., 2018a). The expression level of LsFT increases during the transition from vegetative to reproductive growth; it is also up-regulated during heat-induced flowering (Fukuda et al., 2017). RNAi knockdown of LsFT delays bolting and diminishes bolting in response to heat treatment. Knockdown lines also show reduced expression of LsAP1, LsAP3, and LsLFY, suggesting that similar to their orthologs in Arabidopsis, these genes function downstream of FT. Early bolting lettuce cultivars exhibit higher endogenous LsFT expression levels than late bolting lines (Chen et al., 2018a).
LsSOC1, which encodes a MADS box transcription factor, is located on Chromosome 7. There are two other paralogs of LsSOC1 on Chromosomes 3 and 4; however, genetic mapping (Hartman et al., 2013b) and molecular analysis (Chen et al., 2018b) both indicate that Lsat_1_v5_gn_7_6780 is the functional ortholog. LsSOC1 expression increased in both heat-treated and non-heat-treated bolting lettuce plants. RNAi knockdown of LsSOC1 resulted in delayed flowering and insensitivity to heat treatment in terms of bolting. Binding motifs of heat shock transcription factors have been identified in the promoter of LsSOC1, suggesting that LsSOC1 might function in the ambient temperature pathway in lettuce (Chen et al., 2018b). However, the lack of a heat response in RNAi-LsFT lines suggests that LsFT might function as the end point of the ambient temperature pathway upstream of LsSOC1. Further molecular analyses are needed to clarify the regulatory relationship between LsFT and LsSOC1.
Unlike LsFT and LsSOC1, LsLFY is not located within any known QTLs (Supplementary Table 2). LsLFY encodes a transcription factor and is located on Chromosome 4. Under flowering-promoting conditions, its expression is upregulated in a synchronized fashion with the progression of floral development in field-grown lettuce, consistent with a role in flower bud formation (Fukuda et al., 2017).
Conclusion and Future Prospects
This review of genetic mapping experiments of bolting and flowering time in lettuce has documented the diversity of genetic scenarios that underlie the variation in timing of phase transitions in response to environmental and developmental cues. There are several underexplored areas that warrant future research.
Major Findings
-
1.
There are many QTLs that affect bolting and flowering time in lettuce (Figure 1 and Supplementary Table 2). The phenotypic diversity in flowering time in lettuce results from a variety of genetic differences, which collocate with genes encoding different flowering time pathways. Numerous genetic mapping experiments have provided a sample of the potential genetic diversity (Supplementary Table 1). There is likely additional genetic variation affecting lettuce flowering time through other loci.
-
2.
Despite the inevitable association of bolting and flowering time due to the developmental sequence of the two phenotypes, the genetics of these two traits was more distinct from each other than expected. Future studies should record both phenotypes to provide additional data on the genetic and molecular determinants of each.
-
3.
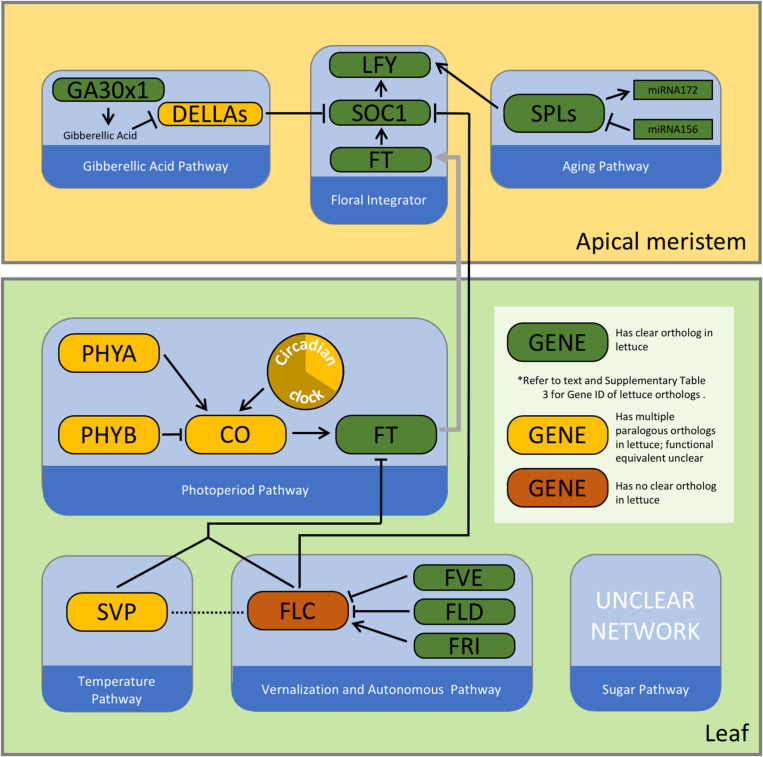
The lettuce genome contains orthologs for many but not all flowering time genes identified in Arabidopsis (Figure 2 and Supplementary Table 3). Results of forward genetic, reverse genetic, and genomic analyses revealed general similarity as well as some differences in the regulation of the flowering time in lettuce compared to Arabidopsis.
FIGURE 2.
A simplified schematic of the core flowering time regulatory network in Arabidopsis. Colors of the nodes represent current information on orthology of these genes in the lettuce genome. Please refer to Srikanth and Schmid (2011); Bouché et al. (2015), and Wils and Kaufmann (2017) for examples of detailed depictions of these complex pathways. (CO: Constans; FLD: Flowering locus D; FRI: Frigida; FT: Flowering locus T; GA3OX1: Gibberellin 3-beta-dioxygenase 1; LFY: Leafy; PHYA: Phytochrome A; PHYB: Phytochrome B; SOC1: Suppressor of CONSTANS overexpression 1; SPL: squamosa promoter binding protein like; SVP: Short vegetative phase.)
-
4.
No clear ortholog of FLC has been found in the lettuce genome. In contrast, there are multiple paralogs with sequence similarity to SVP and CO. Uncovering the identities of the functional equivalents of FLC, SVP, and CO would significantly advance our understanding of the network architecture of flowering time regulation in lettuce. Lsat_1_v5_gn_6_105061 is a potential ortholog of SVP due to its sequence similarity with AtSVP and its collocation with an intermediate-effect QTL, qFTL6.2. Similarly, Lsat_1_v5_gn_5_122401 could be a subject of molecular studies because of its sequence similarity to CO, its oscillating expression pattern that follows a 24-h period and peaks after dusk (Supplementary Table 1), and its collocation with qBLT5.4.
-
5.
Ambient temperature has a major effect of both bolting and flowering. However, additional studies are needed to dissect the molecular link between heat detection and the acceleration in flowering time.
-
6.
Less is known about the molecular basis of day-length sensitivity in lettuce because most experiments have been conducted under LD conditions. More experiments under SD conditions are needed to study the photoperiodic response of flowering time in lettuce. Such studies will facilitate the discovery of functional equivalents of CO in lettuce.
-
7.
Little is currently known regarding the regulation of flowering time by sugar and starch homeostasis in lettuce. This area holds great potential because sugar and starch content could also affect the market quality of lettuce. Genetic variation in the sugar pathway could have positive or negative pleiotropic effects on quality and flowering that are important to understand.
Systems biology approaches are needed to investigate the integration of signaling pathways on genetic, transcriptional, translational, and physiological levels that result in the flowering phenotype. Previous studies have not investigated the crosstalk between pathways controlling flowering time in lettuce. Understanding the interactions between multiple environmental factors in the context of developmental regulation will be informative for guiding agricultural practices.
High throughout phenotyping and genotyping of more mapping populations and diversity panels will allow more extensive analyses on the phenotypic diversity of flowering time in lettuce. Resequencing of diverse cultivars and wild accessions combined with functional studies including genome editing will deliver a deeper understanding of flowering time regulation that can be used to address flowering-time-related breeding goals in this important vegetable crop.
Data Availability Statement
The original contributions presented in the study are publicly available. This data can be found here: accession number DRA004561 on DNA Data Bank of Japan (DDBJ, trace.ddbj.nig.ac.jp/DRASearch/).
Author Contributions
RM and RH conceived the study. RH conducted the literature review, performed the comparative analysis, and drafted the manuscript. MT and DL developed the mapping populations, conducted genotyping by sequencing, and performed the QTL mapping in a previously unpublished study. All authors contributed to writing the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank D. Still, Y. You, M. Niroula, and K. Bradford for providing data and E. Georgian for assistance in editing this manuscript.
Funding. This research was funded by an NSF Graduate Research Fellowship to RH and a USDA NIFA Specialty Crop Research Initiative (SCRI) Grant # 2015-51181-24283 to RM.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.632708/full#supplementary-material
Parental lines and generation of 11 mapping populations, the number of lines, the type and number of markers, mapping software, and parameters used in the analysis, and time and location of 52 experiments reported in the review.
All bolting and flowering time QTLs, their physical and genetic locations, effect sizes in each experiment, flowering time orthologs within them, as well as the populations and experiments they were identified in.
Lettuce orthologs of genes with flowering time function in Arabidopsis, their physical locations, presence within known QTLs, the pathways they belong to, whether they are circadianly expressed in vegetative lettuce, and molecular studies that investigated their functions.
References
- Abbott A. A. (2010). The Isolation of Flowering Time Genes from Lettuce to Enable the Manipulation of Bolting Time. Available online at: http://wrap.warwick.ac.uk/35099/1/WRAP_THESIS_Abbott_2010.pdf (accessed January 26, 2020). [Google Scholar]
- Abou-Elwafa S. F., Büttner B., Chia T., Schulze-Buxloh G., Hohmann U., Mutasa-Göttgens E., et al. (2010). Conservation and divergence of autonomous pathway genes in the flowering regulatory network of Beta vulgaris. J. Exp. Bot. 62 3359–3374. 10.1093/jxb/erq321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achard P., Herr A., Baulcombe D. C., Harberd N. P. (2004). Modulation of floral development by a gibberellin-regulated microRNA. Development 131 3357–3365. 10.1242/dev.01206 [DOI] [PubMed] [Google Scholar]
- Ayre B. G., Turgeon R. (2004). Graft Transmission of a Floral Stimulant Derived from CONSTANS. Plant Physiol. 135 2271–2278. 10.1104/pp.104.040592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian S., Sureshkumar S., Lempe J., Weigel D. (2006). Potent Induction of Arabidopsis thaliana Flowering by Elevated Growth Temperature. PLoS Genet. 2:e106. 10.1371/journal.pgen.0020106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basten C. J., Weir B. S., Zeng Z.-B. (1994). “Zmap-a QTL cartographer,” in Proceedings of the 5th World Congress on Genetics Applied to Livestock Production: Computing Strategies and Software, eds Smith C., Gavora J. S., Benkel B., Chesnais J., Fairfull W., Gibson J. P., et al. (Guelph, ON: ), 65–66. [Google Scholar]
- Becker A., Theißen G. (2003). The major clades of MADS-box genes and their role in the development and evolution of flowering plants. Mol. Phylogenet. Evol. 29 464–489. 10.1016/S1055-7903(03)00207-0 [DOI] [PubMed] [Google Scholar]
- Blackman B. K., Michaels S. D., Rieseberg L. H. (2011a). Connecting the sun to flowering in sunflower adaptation. Mol. Ecol. 20 3503–3512. 10.1111/j.1365-294X.2011.05166.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackman B. K., Rasmussen D. A., Strasburg J. L., Raduski A. R., Burke J. M., Knapp S. J., et al. (2011b). Contributions of flowering time genes to sunflower domestication and improvement. Genetics 187 271–287. 10.1534/genetics.110.121327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouché F., Lobet G., Tocquin P., Périlleux C. (2015). FLOR-ID: an interactive database of flowering-time gene networks in Arabidopsis thaliana. Nucleic Acids Res. 44 D1167–D1171. 10.1093/nar/gkv1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlucci M., Kriščiūnas A., Li H., Gibas P., Koncevičius K., Petronis A., et al. (2019). DiscoRhythm: an easy-to-use web application and R package for discovering rhythmicity. Bioinformatics 36 1952–1954. 10.1093/bioinformatics/btz834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Han Y., Ning K., Ding Y., Zhao W., Yan S., et al. (2018a). Inflorescence development and the role of LsFT in regulating bolting in lettuce (Lactuca sativa L.). Front. Plant Sci 8:2248. 10.3389/fpls.2017.02248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Zhao W., Ge D., Han Y., Ning K., Luo C., et al. (2018b). LCM-seq reveals the crucial role of LsSOC1 in heat-promoted bolting of lettuce (Lactuca sativa L.). Plant J. 95 516–528. 10.1111/tpj.13968 [DOI] [PubMed] [Google Scholar]
- Corbesier L., Vincent C., Jang S., Fornara F., Fan Q., Searle I., et al. (2007). FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316 1030–1033. 10.1126/science.1141752 [DOI] [PubMed] [Google Scholar]
- Dally N., Eckel M., Batschauer A., Höft N., Jung C. (2018). Two CONSTANS-LIKE genes jointly control flowering time in beet. Sci. Rep. 8:16120. 10.1038/s41598-018-34328-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dijken A. J. H., van, Schluepmann H., Smeekens S. C. M. (2004). Arabidopsis Trehalose-6-phosphate synthase 1 is essential for normal vegetative growth and transition to flowering. Plant Physiol. 135 969–977. 10.1104/pp.104.039743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A., Davis C. A., Schlesinger F., Drenkow J., Zaleski C., Jha S., et al. (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29 15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eimert K., Wang S. M., Lue W. I., Chen J. (1995). Monogenic recessive mutations causing both late floral initiation and excess starch accumulation in Arabidopsis. Plant Cell 7 1703–1712. 10.1105/tpc.7.10.1703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emms D. M., Kelly S. (2015). OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 16:157. 10.1186/s13059-015-0721-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- FAO (2020). FAOSTAT: Crops. Available online at: http://www.fao.org/faostat/en/#data/QC (accessed July 20, 2020). [Google Scholar]
- Fornara F., de Montaigu A., Coupland G. (2010). SnapShot: control of flowering in Arabidopsis. Cell 141 550.e1–550.e2. 10.1016/j.cell.2010.04.024 [DOI] [PubMed] [Google Scholar]
- Fornara F., Panigrahi K. C. S., Gissot L., Sauerbrunn N., Rühl M., Jarillo J. A., et al. (2009). Arabidopsis DOF transcription factors act redundantly to reduce CONSTANS expression and are essential for a photoperiodic flowering response. Dev. Cell 17 75–86. 10.1016/j.devcel.2009.06.015 [DOI] [PubMed] [Google Scholar]
- Fukuda M., Matsuo S., Kikuchi K., Kawazu Y., Fujiyama R., Honda I. (2011). Isolation and functional characterization of the FLOWERING LOCUS T homolog, the LsFT gene, in lettuce. J. Plant Physiol. 168 1602–1607. 10.1016/j.jplph.2011.02.004 [DOI] [PubMed] [Google Scholar]
- Fukuda M., Matsuo S., Kikuchi K., Mitsuhashi W., Toyomasu T., Honda I. (2009). The endogenous level of GA1 is upregulated by high temperature during stem elongation in lettuce through LsGA3ox1 expression. J. Plant Physiol. 166 2077–2084. 10.1016/j.jplph.2009.06.003 [DOI] [PubMed] [Google Scholar]
- Fukuda M., Yanai Y., Nakano Y., Sasaki H., Uragami A., Okada K. (2017). Isolation and gene expression analysis of flowering-related genes in Lettuce (Lactuca sativa L.). Hortic. J. 86 283–290. 10.2503/hortj.OKD-036 [DOI] [Google Scholar]
- Gendall A. R., Levy Y. Y., Wilson A., Dean C. (2001). The VERNALIZATION 2 gene mediates the epigenetic regulation of vernalization in Arabidopsis. Cell 107 525–535. [DOI] [PubMed] [Google Scholar]
- Geraldo N., Bäurle I., Kidou S., Hu X., Dean C. (2009). FRIGIDA delays flowering in Arabidopsis via a cotranscriptional mechanism involving direct interaction with the nuclear cap-binding complex. Plant Physiol. 150 1611–1618. 10.1104/pp.109.137448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gocal G. F., Sheldon C. C., Gubler F., Moritz T., Bagnall D. J., MacMillan C. P., et al. (2001). GAMYB-like genes, flowering, and gibberellin signaling in Arabidopsis. Plant Physiol. 127 1682–1693. [PMC free article] [PubMed] [Google Scholar]
- Golembeski G. S., Imaizumi T. (2015). Photoperiodic regulation of Florigen function in Arabidopsis thaliana. Arabidopsis Book 13:e0178. 10.1199/tab.0178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregis V., Sessa A., Colombo L., Kater M. M. (2008). AGAMOUS-LIKE24 and SHORT VEGETATIVE PHASE determine floral meristem identity in Arabidopsis. Plant J. 56 891–902. 10.1111/j.1365-313X.2008.03648.x [DOI] [PubMed] [Google Scholar]
- Han Y., Chen Z., Lv S., Ning K., Ji X., Liu X., et al. (2016a). MADS-box genes and gibberellins regulate bolting in lettuce (Lactuca sativa L.). Front. Plant Sci. 7:1889. 10.3389/fpls.2016.01889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y. Y., Li Y. B., Fan S. X., Liu C. J., Hao J. H., Chen Q. J., et al. (2016b). “Screening and identification of lettuce germplasm for tolerance to high and low temperature,” in Proceedings of the Acta Horticulturae (International Society for Horticultural Science (ISHS), Leuven, 381–388. 10.17660/ActaHortic.2016.1127.59 [DOI] [Google Scholar]
- Hao J.-H., Zhang L.-L., Li P.-P., Sun Y.-C., Li J.-K., Qin X.-X., et al. (2018). Quantitative proteomics analysis of lettuce (Lactuca sativa L.) reveals molecular basis-associated auxin and photosynthesis with bolting induced by high temperature. Int. J. Mol. Sci. 19:2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harberd N. P., Belfield E., Yasumura Y. (2009). The angiosperm gibberellin-GID1-DELLA growth regulatory mechanism: how an “inhibitor of an inhibitor” enables flexible response to fluctuating environments. Plant Cell 21 1328–1339. 10.1105/tpc.109.066969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman Y., Hooftman D. A. P., Eric Schranz M., van Tienderen P. H. (2013a). QTL analysis reveals the genetic architecture of domestication traits in Crisphead lettuce. Genet. Resour. Crop Evol. 60 1487–1500. 10.1007/s10722-012-9937-0 [DOI] [Google Scholar]
- Hartman Y., Hooftman D. A. P., Uwimana B., van de Wiel C. C. M., Smulders M. J. M., Visser R. G. F., et al. (2012). Genomic regions in crop–wild hybrids of lettuce are affected differently in different environments: implications for crop breeding. Evol. Appl. 5 629–640. 10.1111/j.1752-4571.2012.00240.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman Y., Uwimana B., Hooftman D. A. P., Schranz M. E., van de Wiel C. C. M., Smulders M. J. M., et al. (2013b). Genomic and environmental selection patterns in two distinct lettuce crop–wild hybrid crosses. Evol. Appl. 6 569–584. 10.1111/eva.12043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht V., Foucher F., Ferrándiz C., Macknight R., Navarro C., Morin J., et al. (2005). Conservation of Arabidopsis flowering genes in model legumes. Plant Physiol. 137 1420–1434. 10.1104/pp.104.057018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo J. B., Sung S. (2011). Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science 331 76–79. 10.1126/science.1197349 [DOI] [PubMed] [Google Scholar]
- Higashi T., Aoki K., Nagano A. J., Honjo M. N., Fukuda H. (2016). Circadian oscillation of the lettuce transcriptome under constant light and light-dark conditions. Front. Plant Sci. 7:1114. 10.3389/fpls.2016.01114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hileman L. C., Sundstrom J. F., Litt A., Chen M., Shumba T., Irish V. F. (2006). Molecular and phylogenetic analyses of the MADS-box gene family in tomato. Mol. Biol. Evol. 23 2245–2258. 10.1093/molbev/msl095 [DOI] [PubMed] [Google Scholar]
- Hoecker U., Quail P. H. (2001). The phytochrome A-specific signaling intermediate SPA1 interacts directly with COP1, a constitutive repressor of light signaling in Arabidopsis. J. Biol. Chem. 276 38173–38178. 10.1074/jbc.M103140200 [DOI] [PubMed] [Google Scholar]
- Holmes S. C., Wells D. E., Pickens J. M., Kemble J. M. (2019). Selection of heat tolerant lettuce (Lactuca sativa L.) cultivars grown in deep water culture and their marketability. Horticulturae 5:50. [Google Scholar]
- Huijser P., Schmid M. (2011). The control of developmental phase transitions in plants. Development 138 4117–4129. 10.1242/dev.063511 [DOI] [PubMed] [Google Scholar]
- Huo H., Wei S., Bradford K. J. (2016). DELAY OF GERMINATION1 (DOG1) regulates both seed dormancy and flowering time through microRNA pathways. Proc. Natl. Acad. Sci. U.S.A. 113 E2199L–E2206. 10.1073/pnas.1600558113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang H.-J., Lee J.-M., An J.-M., Kim S.-Y., Choi G.-W. (2007). Bolting response of various lettuce cultivars affected by seed treatments. Korean J. Environ. Agric. 26 325–331. [Google Scholar]
- Imaizumi T., Schultz T. F., Harmon F. G., Ho L. A., Kay S. A. (2005). FKF1 F-box protein mediates cyclic degradation of a repressor of CONSTANS in Arabidopsis. Science 309 293–297. 10.1126/science.1110586 [DOI] [PubMed] [Google Scholar]
- Jenni S., Truco M. J., Michelmore R. W. (2013). Quantitative trait loci associated with tipburn, heat stress-induced physiological disorders, and maturity traits in crisphead lettuce. Theor. Appl. Genet. 126 3065–3079. 10.1007/s00122-013-2193-7 [DOI] [PubMed] [Google Scholar]
- Jenni S., Yan W. (2009). Genotype by environment interactions of heat stress disorder resistance in crisphead lettuce. Plant Breed. 128 374–380. 10.1111/j.1439-0523.2009.01657.x [DOI] [Google Scholar]
- Jiang D., Wang Y., Wang Y., He Y. (2008). Repression of FLOWERING LOCUS C and FLOWERING LOCUS T by the Arabidopsis Polycomb Repressive Complex 2 Components. PLoS One 3:e3404. 10.1371/journal.pone.0003404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johanson U., West J., Lister C., Michaels S., Amasino R., Dean C. (2000). Molecular analysis ofFRIGIDA, a major determinant of natural variation in arabidopsis flowering time. Science 290 344–347. 10.1126/science.290.5490.344 [DOI] [PubMed] [Google Scholar]
- Jung C., Müller A. E. (2009). Flowering time control and applications in plant breeding. Trends Plant Sci. 14 563–573. 10.1016/j.tplants.2009.07.005 [DOI] [PubMed] [Google Scholar]
- Kardailsky I., Shukla V. K., Ahn J. H., Dagenais N., Christensen S. K., Nguyen J. T., et al. (1999). Activation Tagging of the floral inducer FT. Science 286 1962–1965. 10.1126/science.286.5446.1962 [DOI] [PubMed] [Google Scholar]
- Kato T. (1964a). Physiological studies on the bolting of lettuce plants. I. Relationships between various constituents and flower bud formation. J. Japanese Soc. Hortic. Sci. 33 125–132. 10.2503/jjshs.33.125 [DOI] [Google Scholar]
- Kato T. (1964b). Physiological studies ono the bolting of lettuce plants. II. Relation between the stem elongation and auxin metabolism. J. Japanese Soc. Hortic. Sci. 33 243–250. 10.2503/jjshs.33.243 [DOI] [Google Scholar]
- Kumar S. V., Wigge P. A. (2010). H2A.Z-containing nucleosomes mediate the thermosensory response in Arabidopsis. Cell 140 136–147. 10.1016/j.cell.2009.11.006 [DOI] [PubMed] [Google Scholar]
- Kwon S., Simko I., Hellier B., Mou B., Hu J. (2013). Genome-wide association of 10 horticultural traits with expressed sequence tag-derived SNP markers in a collection of lettuce lines. Crop J. 1 25–33. 10.1016/j.cj.2013.07.014 [DOI] [Google Scholar]
- Lafta A., Turini T., Sandoya G. V., Mou B. (2017). Field evaluation of green and red leaf lettuce genotypes in the imperial, san joaquin, and salinas valleys of california for heat tolerance and extension of the growing seasons. Hortsci. Horts 52 40–48. 10.21273/HORTSCI10835-16 [DOI] [Google Scholar]
- Laubinger S., Marchal V., Gentilhomme J., Wenkel S., Adrian J., Jang S., et al. (2006). Arabidopsis SPA proteins regulate photoperiodic flowering and interact with the floral inducer CONSTANS to regulate its stability. Development 133 3213–3222. 10.1242/dev.02481 [DOI] [PubMed] [Google Scholar]
- Lauter N., Kampani A., Carlson S., Goebel M., Moose S. P. (2005). microRNA172 down-regulates glossy15 to promote vegetative phase change in maize. Proc. Natl. Acad. Sci. U.S.A. 102 9412–9417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavelle D. O. (2009). Genetics of Candidate Genes for Developmental and Domestication-Related Traits in Lettuce. Ph.D. dissertation, University of California, Davis: [Google Scholar]
- Lee J., Lee I. (2010). Regulation and function of SOC1, a flowering pathway integrator. J. Exp. Bot. 61 2247–2254. 10.1093/jxb/erq098 [DOI] [PubMed] [Google Scholar]
- Lee J., Oh M., Park H., Lee I. (2008). SOC1 translocated to the nucleus by interaction with AGL24 directly regulates LEAFY. Plant J. 55 832–843. 10.1111/j.1365-313X.2008.03552.x [DOI] [PubMed] [Google Scholar]
- Lee J. H., Yoo S. J., Park S. H., Hwang I., Lee J. S., Ahn J. H. (2007). Role of SVP in the control of flowering time by ambient temperature in Arabidopsis. Genes Dev. 21 397–402. 10.1101/gad.1518407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leijten W., Koes R., Roobeek I., Frugis G. (2018). Translating flowering time from Arabidopsis thaliana to Brassicaceae and Asteraceae crop species. Plants 7 111. 10.3390/plants7040111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy Y. Y., Mesnage S., Mylne J. S., Gendall A. R., Dean C. (2002). Multiple roles of Arabidopsis VRN1 in vernalization and flowering time control. Science 297 243–246. [DOI] [PubMed] [Google Scholar]
- Li C., Gu M., Shi N., Zhang H., Yang X., Osman T., et al. (2011). Mobile FT mRNA contributes to the systemic florigen signalling in floral induction. Sci. Rep. 1:73. 10.1038/srep00073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D., Liu C., Shen L., Wu Y., Chen H., Robertson M., et al. (2008). A repressor complex governs the integration of flowering signals in Arabidopsis. Dev. Cell 15 110–120. 10.1016/j.devcel.2008.05.002 [DOI] [PubMed] [Google Scholar]
- Li Y., Li T., Han Y., Fan S. (2017). Cloning and function analysis of heat-shock-protein LsHsp70-2711 gene under high temperature stress in leaf lettuce (Lactuca sativa L.). Sci. Agric. Sin. 50 1486–1494. 10.3864/j.issn.0578-1752.2017.08.012 [DOI] [Google Scholar]
- Liu C., Chen H., Er H. L., Soo H. M., Kumar P. P., Han J.-H., et al. (2008a). Direct interaction of AGL24 and SOC1 integrates flowering signals in Arabidopsis. Development 135 1481–1491. 10.1242/dev.020255 [DOI] [PubMed] [Google Scholar]
- Liu C., Xi W., Shen L., Tan C., Yu H. (2009). Regulation of floral patterning by flowering time genes. Dev. Cell 16 711–722. 10.1016/j.devcel.2009.03.011 [DOI] [PubMed] [Google Scholar]
- Liu L., Wu Y., Liao Z., Xiong J., Wu F., Xu J., et al. (2018a). Evolutionary conservation and functional divergence of the LFK gene family play important roles in the photoperiodic flowering pathway of land plants. Heredity 120 310–328. 10.1038/s41437-017-0006-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L.-J., Zhang Y.-C., Li Q.-H., Sang Y., Mao J., Lian H.-L., et al. (2008b). COP1-mediated ubiquitination of CONSTANS is implicated in cryptochrome regulation of flowering in Arabidopsis. Plant Cell 20 292–306. 10.1105/tpc.107.057281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Lv S., Liu R., Fan S., Liu C., Liu R., et al. (2018b). Transcriptomic analysis reveals the roles of gibberellin-regulated genes and transcription factors in regulating bolting in lettuce (Lactuca sativa L.). PLoS One 13:e0191518. 10.1371/journal.pone.0191518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y., Guo Z., Li L. (2013). Evolutionary conservation of microRNA regulatory programs in plant flower development. Dev. Biol. 380 133–144. 10.1016/j.ydbio.2013.05.009 [DOI] [PubMed] [Google Scholar]
- Mamo B. E., Hayes R. J., Truco M. J., Puri K. D., Michelmore R. W., Subbarao K. V., et al. (2019). The genetics of resistance to lettuce drop (Sclerotinia spp.) in lettuce in a recombinant inbred line population from Reine des Glaces × Eruption. Theor. Appl. Genet. 132 2439–2460. 10.1007/s00122-019-03365-6 [DOI] [PubMed] [Google Scholar]
- Mathieu J., Yant L. J., Mürdter F., Küttner F., Schmid M. (2009). Repression of flowering by the miR172 target SMZ. PLoS Biol. 7:e1000148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monte E., Alonso J. M., Ecker J. R., Zhang Y., Li X., Young J., et al. (2003). Isolation and characterization of phyC mutants in Arabidopsis reveals complex crosstalk between phytochrome signaling pathways. Plant Cell 15 1962–1980. 10.1105/tpc.012971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore B., Zhou L., Rolland F., Hall Q., Cheng W.-H., Liu Y.-X., et al. (2003). Role of the Arabidopsis glucose sensor HXK1 in nutrient, light, and hormonal signaling. Science 300 332–336. 10.1126/science.1080585 [DOI] [PubMed] [Google Scholar]
- Morris K., Thornber S., Codrai L., Richardson C., Craig A., Sadanandom A., et al. (2010). DAY NEUTRAL FLOWERING represses CONSTANS to prevent Arabidopsis flowering early in short days. Plant Cell 22 1118–1128. 10.1105/tpc.109.066605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouradov A., Cremer F., Coupland G. (2002). Control of flowering time: interacting pathways as a basis for diversity. Plant Cell 14(Suppl.), S111–S130. 10.1105/tpc.001362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning K., Han Y., Chen Z., Luo C., Wang S., Zhang W., et al. (2019). Genome-wide analysis of MADS-box family genes during flower development in lettuce. Plant Cell Environ. 42 1868–1881. 10.1111/pce.13523 [DOI] [PubMed] [Google Scholar]
- Niroula M. (2017). Environmental Sensitivity of Quantitative Trait Loci for Seed Germination and Flowering Time in Lettuce (Lactuca sativa L.). Ph.D. thesis, University of California, Davis, CA. [Google Scholar]
- Oda A., Fujiwara S., Kamada H., Coupland G., Mizoguchi T. (2004). Antisense suppression of the Arabidopsis PIF3 gene does not affect circadian rhythms but causes early flowering and increases FT expression. FEBS Lett. 557 259–264. 10.1016/S0014-5793(03)01470-4 [DOI] [PubMed] [Google Scholar]
- Ohto M., Onai K., Furukawa Y., Aoki E., Araki T., Nakamura K. (2001). Effects of sugar on vegetative development and floral transition in Arabidopsis. Plant Physiol. 127 252–261. 10.1104/pp.127.1.252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul M. J., Primavesi L. F., Jhurreea D., Zhang Y. (2008). Trehalose metabolism and signaling. Annu. Rev. Plant Biol. 59 417–441. 10.1146/annurev.arplant.59.032607.092945 [DOI] [PubMed] [Google Scholar]
- Périlleux C., Pieltain A., Jacquemin G., Bouché F., Detry N., D’Aloia M., et al. (2013). A root chicory MADS box sequence and the Arabidopsis flowering repressor FLC share common features that suggest conserved function in vernalization and de-vernalization responses. Plant J. 75 390–402. 10.1111/tpj.12208 [DOI] [PubMed] [Google Scholar]
- Prince S. D., Marks M. K., Carter R. N. (1978). Induction of flowering in wild lettuce (Lactuca serriola L.). New Phytol. 81 265–277. 10.1111/j.1469-8137.1978.tb02632.x [DOI] [Google Scholar]
- Putterill J., Robson F., Lee K., Simon R., Coupland G. (1995). The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell 80 847–857. [DOI] [PubMed] [Google Scholar]
- Rappaport L., Wittwer S. H., Tukey H. B. (1956). Seed vernalization and flowering in lettuce (Lactuca sativa). Nature 178:51. 10.1038/178051a0 [DOI] [Google Scholar]
- Reeves P. A., He Y., Schmitz R. J., Amasino R. M., Panella L. W., Richards C. M. (2007). Evolutionary conservation of the FLOWERING LOCUS C-mediated vernalization response: evidence from the sugar beet (Beta vulgaris). Genetics 176 295–307. 10.1534/genetics.106.069336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Chin-Wo S., Wang Z., Yang X., Kozik A., Arikit S., Song C., et al. (2017). Genome assembly with in vitro proximity ligation data and whole-genome triplication in lettuce. Nat. Commun. 8:14953. 10.1038/ncomms14953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryder E. J. (1996). Ten lettuce genetic stocks with early flowering genes Ef-1ef-1 and Ef-2ef-2. Hortscience 31 473–475. 10.21273/HORTSCI.31.3.473 [DOI] [Google Scholar]
- Sandoya G., Truco M.-J., Bertier L. D., Subbarao K. V., Simko I., Hayes R. J., et al. (2020). Genetics of partial resistance against Verticillium dahliae Race 2 in wild and cultivated lettuce. Phytopathology. 10.1094/PHYTO-09-20-0396-R [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Sawa M., Nusinow D. A., Kay S. A., Imaizumi T. (2007). FKF1 and GIGANTEA complex formation is required for day-length measurement in Arabidopsis. Science 318 261–265. 10.1126/science.1146994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M., Uhlenhaut N. H., Godard F., Demar M., Bressan R., Weigel D., et al. (2003). Dissection of floral induction pathways using global expression analysis. Development 130 6001–6012. [DOI] [PubMed] [Google Scholar]
- Searle I., He Y., Turck F., Vincent C., Fornara F., Kröber S., et al. (2006). The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis. Genes Dev. 20 898–912. 10.1101/gad.373506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki K., Komatsu K., Tanaka K., Hiraga M., Kajiya-Kanegae H., Matsumura H., et al. (2020). A CIN-like TCP transcription factor (LsTCP4) having retrotransposon insertion associates with a shift from Salinas type to Empire type in crisphead lettuce (Lactuca sativa L.). Hortic. Res. 7:15. 10.1038/s41438-020-0241-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo P. J., Ryu J., Kang S. K., Park C.-M. (2011). Modulation of sugar metabolism by an INDETERMINATE DOMAIN transcription factor contributes to photoperiodic flowering in Arabidopsis. Plant J. 65 418–429. 10.1111/j.1365-313X.2010.04432.x [DOI] [PubMed] [Google Scholar]
- Silva E. C., Maluf W. R., Leal N. R., Gomes L. A. A. (1999). Inheritance of bolting tendency in lettuce Lactuca sativa L. Euphytica 109 1–7. 10.1023/A:1003698117689 [DOI] [Google Scholar]
- Simpson G. G. (2004). The autonomous pathway: epigenetic and post-transcriptional gene regulation in the control of Arabidopsis flowering time. Curr. Opin. Plant Biol. 7 570–574. 10.1016/j.pbi.2004.07.002 [DOI] [PubMed] [Google Scholar]
- Srikanth A., Schmid M. (2011). Regulation of flowering time: all roads lead to Rome. Cell. Mol. Life Sci. 68 2013–2037. 10.1007/s00018-011-0673-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sthapit Kandel J., Peng H., Hayes R. J., Mou B., Simko I. (2020). Genome-wide association mapping reveals loci for shelf life and developmental rate of lettuce. Theor. Appl. Genet. 133 1947–1966. 10.1007/s00122-020-03568-2 [DOI] [PubMed] [Google Scholar]
- Suárez-López P., Wheatley K., Robson F., Onouchi H., Valverde F., Coupland G. (2001). CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature 410 1116–1120. 10.1038/35074138 [DOI] [PubMed] [Google Scholar]
- Sukprakarn S. (1985). A Study of the Effects of Temperature and Photoperiod on Vegetative Growth and Seed Production of Leaf Lettuce (Lactuca sativa L.) : A thesis Presented in Partial Fulfilment of the Requirement for the Degree of Doctor of Philosophy in Seed Technology. Available online at: http://hdl.handle.net/10179/3339 (accessed July 30, 2020). [Google Scholar]
- Sun T. (2010). Gibberellin-GID1-DELLA: a pivotal regulatory module for plant growth and development. Plant Physiol. 154 567–570. 10.1104/pp.110.161554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung S., Amasino R. M. (2004). Vernalization in Arabidopsis thaliana is mediated by the PHD finger protein VIN3. Nature 427 159–164. 10.1038/nature02195 [DOI] [PubMed] [Google Scholar]
- Truco M. J., Ashrafi H., Kozik A., van Leeuwen H., Bowers J., Wo S. R. C., et al. (2013). An ultra-high-density, transcript-based, genetic map of lettuce. G3 3 617–631. 10.1534/g3.112.004929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- USDA-NASS. (2019). CHAPTER IV: STATISTICS OF VEGETABLES AND MELONS. Available online at: https://www.nass.usda.gov/Publications/Ag_Statistics/2019/chapter04.pdf (accessed July 7, 2020). [Google Scholar]
- Wang J.-W., Park M. Y., Wang L.-J., Koo Y., Chen X.-Y., Weigel D., et al. (2011). MiRNA control of vegetative phase change in trees. PLoS Genet. 7:e1002012. 10.1371/journal.pgen.1002012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warne L. G. G. (1947). Vernalization of lettuce. Nature 159 31–32. 10.1038/159031a0 [DOI] [PubMed] [Google Scholar]
- Waycott W. (1995). Photoperiodic response of genetically diverse lettuce accessions. J. Am. Soc. Hortic. Sci. 120 460–467. 10.21273/JASHS.120.3.460 [DOI] [Google Scholar]
- Weigel D., Ahn J. H., Blázquez M. A., Borevitz J. O., Christensen S. K., Fankhauser C., et al. (2000). Activation tagging in Arabidopsis. Plant Physiol. 122 1003–1013. 10.1104/pp.122.4.1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wils C. R., Kaufmann K. (2017). Gene-regulatory networks controlling inflorescence and flower development in Arabidopsis thaliana. Biochim. Biophys. Acta 1860 95–105. 10.1016/j.bbagrm.2016.07.014 [DOI] [PubMed] [Google Scholar]
- Wu G., Park M. Y., Conway S. R., Wang J.-W., Weigel D., Poethig R. S. (2009). The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell 138 750–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi A., Wu M.-F., Yang L., Wu G., Poethig R. S., Wagner D. (2009). The microRNA-regulated SBP-Box transcription factor SPL3 is a direct upstream activator of LEAFY, FRUITFULL, and APETALA1. Dev. Cell 17 268–278. 10.1016/j.devcel.2009.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S. K., Chung K. S., Kim J., Lee J. H., Hong S. M., Yoo S. J., et al. (2005). CONSTANS activates SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 through FLOWERING LOCUS T to promote flowering in Arabidopsis. Plant Physiol. 139 770–778. 10.1104/pp.105.066928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Su W., Tao R., Zhang W., Chen J., Wu P., et al. (2017). RNA sequencing provides insights into the evolution of lettuce and the regulation of flavonoid biosynthesis. Nat. Commun. 8:2264. 10.1038/s41467-017-02445-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L. L., Hao J. H., Han Y. Y., Liu C. J., Su H. N., Li P. P., et al. (2016). Effects of temperature on leaf lettuce vernalization. J. Appl. Ecol. 27 3600–3606. 10.13287/j.1001-9332.201611.014 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Parental lines and generation of 11 mapping populations, the number of lines, the type and number of markers, mapping software, and parameters used in the analysis, and time and location of 52 experiments reported in the review.
All bolting and flowering time QTLs, their physical and genetic locations, effect sizes in each experiment, flowering time orthologs within them, as well as the populations and experiments they were identified in.
Lettuce orthologs of genes with flowering time function in Arabidopsis, their physical locations, presence within known QTLs, the pathways they belong to, whether they are circadianly expressed in vegetative lettuce, and molecular studies that investigated their functions.
Data Availability Statement
The original contributions presented in the study are publicly available. This data can be found here: accession number DRA004561 on DNA Data Bank of Japan (DDBJ, trace.ddbj.nig.ac.jp/DRASearch/).