Abstract
Aims
During virally suppressed chronic HIV infection, persistent inflammation contributes to the development of cardiovascular disease (CVD), a major comorbidity in people living with HIV (LWH). Classical blood monocytes (CMs) remain activated during antiretroviral therapy and are a major source of pro-inflammatory and pro-thrombotic factors that contribute to atherosclerotic plaque development and instability.
Methods and results
Here, we identify transcriptomic changes in circulating CMs in peripheral blood mononuclear cell samples from participants of the Women’s Interagency HIV Study, selected by HIV and subclinical CVD (sCVD) status. We flow-sorted CM from participants of the Women’s Interagency HIV Study and deep-sequenced their mRNA (n = 92). CMs of HIV+ participants showed elevated interleukin (IL)-6, IL-1β, and IL-12β, overlapping with many transcripts identified in sCVD+ participants. In sCVD+ participants LWH, those reporting statin use showed reduced pro-inflammatory gene expression to a level comparable with healthy (HIV−sCVD−) participants. Statin non-users maintained an elevated inflammatory profile and increased cytokine production.
Conclusion
Statin therapy has been associated with a lower risk of cardiac events, such as myocardial infarction in the general population, but not in those LWH. Our data suggest that women LWH may benefit from statin therapy even in the absence of overt CVD.
Keywords: Cardiovascular disease, NGS, Statins, Gene signature, Antiretroviral therapy
1. Introduction
Monocytes are key cells in the innate immune response and pathogenesis of HIV infection.1–3 In human blood, surface expression of CD14 and CD16 distinguishes three monocyte subsets,4 each with unique transcriptomic properties.5,6 Classical monocytes (CMs), the majority of circulating monocytes,4 function as the main source of inflammatory cytokines.7 In persistent HIV infection, monocytes have been implicated as important reservoirs for replication-competent virus.8,9 The viral proteins Nef and Tat have also been detected in monocytes in virally suppressed patients,10 which along with the integrated virus11 could influence cell phenotype.
Today almost half of the people living with HIV (LWH) worldwide are on antiretroviral therapy (ART), which can lead to low or undetectable HIV viral loads.12 In the Women’s Interagency HIV Study (WIHS), more than 94% of participants with HIV are on ART, but measurable levels of systemic inflammation remain.13 Chronic inflammation is associated with serious non-AIDS-related events that are major causes of morbidity and mortality.14,15 One study found that surface markers of activation on CM predicted carotid artery intima-media thickness (cIMT) in virally suppressed HIV participants independent of levels of circulating plasma biomarkers, such as C-reactive protein, LPS, and sCD14.2 A fraction of CM remain activated after ART-induced viral suppression and produce proinflammatory cytokines,16 likely contributing to the known increased cardiovascular risk in people LWH.17,18
In atherosclerosis, inflammatory cells accumulate in lipid-rich plaques in large and medium-sized arteries.19 Atherosclerotic plaque rupture or erosion causes most cases of cardiovascular disease (CVD). Atherosclerotic plaques contain numerous immune cells, including many macrophages,19 many of which are monocyte-derived.20 Circulating monocytes, which feed atherosclerotic plaque formation,21–23 track closely with atheroprogression and CVD risk in both mice and humans.24,25 Specifically, blood levels of CM predict cardiovascular events.26 Transcriptome studies in people with HIV suggest that these cells maintain an activated phenotype despite ART and may contribute to dysregulation of lipid metabolism.27 Hyperlipidaemia is a hallmark risk factor for CVD28,29 and induces increased numbers of CM. Elevated low-density lipoprotein (LDL) cholesterol is treated with statins30 or PCSK9 inhibitors.31 Reducing LDL cholesterol greatly reduces the risk of adverse cardiovascular events in the general population.32–34 It is predicted by 2030 that 78% of people LWH will be diagnosed with CVD.35 Thus, it is of the utmost importance to investigate and understand how CM contribute to atheroprogression and persistent inflammation in chronic HIV infection.
Prior work has characterized the monocyte transcriptome in people without HIV infection,5 people LWH,27,36 or people with CVD,37 but the CM transcriptome has not been interrogated for possible interactions between HIV and subclinical CVD (sCVD). Here, we investigated the CM transcriptomes of 92 participants of the WIHS with chronic, treated HIV in the presence and absence of sCVD and identified transcriptome pathways and networks in each condition. Additionally, we reasoned that our study may provide a mechanistic underpinning for the ongoing NIH-funded REPRIEVE clinical trial that explores pitavastatin treatment to prevent future vascular events in people LWH.38 We find that use of statin treatment in participants with HIV and sCVD (H+C+) is associated with a CM gene expression profile resembling that of the control participants, while H+C+ participants not use of statins display inflammatory gene expression. After stimulation with LPS, statin-treated H+C+ participants produce significantly less tumour necrosis factor (TNF), interleukin (IL)-1beta, and IL-6 cytokines compared to H+C+ participants not reporting use of statins. In a comprehensive analysis of all CM transcriptomes, we identified a gene expression signature in CM that is enriched in transcriptomes across several cohorts of patients with coronary artery disease and atherosclerosis and is associated with increased risk for cardiovascular events.
2. Methods
2.1 Study setting, sample selection, and inclusion criteria
The Women’s Interagency HIV Study (WIHS) is an ongoing longitudinal cohort study of over 4000 women with or at risk of HIV infection that was initiated in 1994 at six (now expanded to 10) US sites.39,40 Recruitment in the WIHS occurred in four waves (1994–1995, 2001–2002, 2010–2012, and 2013–2015) from HIV primary care clinics, hospital-based programmes, community outreach, support groups, and other locations. Briefly, the WIHS involves semi-annual follow-up visits, during which participants undergo similar detailed examinations, specimen collection, and structured interviews assessing health behaviours, medical history, and medication use. All participants provided informed consent. Each site’s Institutional Review Board approved the studies and complied with the Declaration of Helsinki.
Participants from the current analysis were part of a vascular substudy nested within the WIHS.41,42 The baseline visit for the vascular substudy occurred between 2004 and 2006, and a follow-up visit occurred on average 7 years later. Participants underwent high-resolution B-mode carotid artery ultrasound to image six locations in the right carotid artery: the near and far walls of the common carotid artery, carotid bifurcation, and internal carotid artery. A standardized protocol was used at all sites,43 and measurements of carotid artery focal plaque, a marker of subclinical atherosclerosis, were obtained at a centralized reading centre (University of Southern California). sCVD was defined based on the presence of one or more carotid artery lesions present.43
Because we were interested in the independent and joint relationships of HIV infection and sCVD with RNA expression by CMs, we used a two-by-two factorial design based on HIV status (H+ or H−) and presence of carotid artery focal plaque at either vascular substudy visit (C+ or C−) to define four groups of participants: H+C+, H+C−, H−C+, and H−C−. HIV infection status was ascertained by enzyme-linked immunosorbent assay (ELISA) and confirmed by western blot. C+ participants either had one or more plaques at each vascular substudy visit, or more than one plaque at a single visit. C− participants with self-reported coronary heart disease or current lipid-lowering therapy use were excluded.
From the initial 1865 participants in the WIHS vascular substudy, we selected 92 participants for RNA-Seq analysis: 23 H+C+, 23 H+C−, 23 H−C+, and 23 H−C−. We created 23 quartets with one participant from each of the four groups, all of whom were matched within the quartet by the following characteristics: race/ethnicity, age at the baseline vascular substudy visit (within 5 years), smoking history, and date of specimen collection (within 1 year). We relaxed the matching criteria (e.g. age within 10 years instead of 5 years) for three additional quartets in order to increase the sample size.
Demographic, clinical, and laboratory variables were assessed from the same study visit using standardized protocols. Supplementary material online, Table S1 shows characteristics of the study population. The median age at the baseline study visit was 45 years [interquartile range (IQR) 40–51], and 96% of participants were either of black race or Hispanic ethnicity. Most (86%) reported a history of smoking. Substance use was highly prevalent, with 43% of HIV+ and 50% of HIV− participants reporting either a history of injection drug use; current use of crack, cocaine, or heroin; or alcohol use (≥14 drinks per week). Among HIV+ participants, over 80% reported use of HAART at the time PBMCs were obtained, and 59% reported an undetectable HIV-1 RNA level. The median CD4+ T-cell count was 585 cells/µL (IQR 382–816) in HIV+ women without sCVD and 535 cells/µL (IQR 265–792) in HIV+ women with sCVD.
2.2 PBMC thawing and monocyte isolation
In the WIHS, blood was taken at each visit, PBMCs isolated by standardized methods, frozen, and stored in liquid nitrogen (lN2) in the WIHS’s specimen repository. This study is based on transcriptomes of CMs isolated from frozen PBMCs obtained from the 92 matched WIHS participants at WIHS Visits 27–36. PBMCs were thawed from cryopreserved specimens collected from the semi-annual WIHS core visit that occurred as close as possible in time to the most recent vascular substudy visit and had adequate volume.
Participant PBMCs were thawed and processed in groups of 6–8 at a time. Cryopreserved samples were transported on dry ice from liquid nitrogen stock. Samples were warmed in 37°C water bath until a small ball of ice remained, then removed and immediately diluted with 1 mL of warm cRPMI and transferred and diluted again in an additional 8 mL of warm cRPMI. Samples were then washed with PBS and stained with viability reagent (Ghost Dye™ Red 710, Tonbo Bioscience) according to manufacturer recommendation. Samples were stained with antibody cocktail containing CD14 (M5E2, Biolegend), CD16 (3G8, Biolegend), and dump channel: CD3 (OKT3, Tonbo Bioscience), CD19 (HIB19, Tonbo Bioscience), CD56 (5.1H11, Biolegend), CD66b (G10F5, Biolegend), gated based on living cells and excluding dump channel-positive cells. CMs were defined as CD14++CD16- and sorted directly into TRIzol® LS Reagent (Life Technologies) and frozen at −80°C. FCS files were exported from FACS Diva (BD Bioscience) and processed using FlowJo v10.2 (FlowJo, LLC).
2.3 Flow cytometry analysis
PBMCs were thawed and resuspended in complete RPMI 1640 at a concentration of 106 cells per well in a 96-well plate. Cells were washed and resuspended in complete media with brefeldin-A (5 µg/mL: Sigma) and stimulated with 100 ng of ultrapure LPS (Sigma) for 6 h at 37°C. Samples were then washed with PBS and stained with viability reagent (FVS-700, BD Biosciences) according to manufacturer recommendation. Samples were washed again and stained with extracellular antibody cocktail containing CD14 (M5E2, Biolegend), CD16 (3G8, Biolegend), CD142 (HTF-1, BD Biosciences), HLA-DR (G46-6, BD Biosciences), CD86 (IT2.2, Biolegend), CD36 (CB38, BD Biosciences), and dump channel: CD3 (OKT3, Tonbo Bioscience), CD19 (HIB19, Tonbo Bioscience), CD20 (2H7, Biolegend), CD2 (RPA-2.10, Biolegend), CD56 (5.1H11, Biolegend), and CD66b (G10F5, Biolegend). Cells were then fixed and permeabilized using the eBioscience™ Intracellular Fixation & Permeabilization Buffer Set and subsequently stained with the following intracellular antibodies: IL-6 (MQ2-13A5, BD Biosciences), IL-1beta (JK1B-1, BD Biosciences), IL-8 (E8N1, BD Biosciences), TNFa (Mab11, BD Biosciences), and CCL2 (2H5, BD Biosciences). Data were acquired on a BD LSRII. FCS files were exported from FACS Diva (BD Bioscience) and processed using FlowJo v10.2 (FlowJo, LLC). Analysis was based on living cells and excluding dump channel-positive cells.
2.4 RNA isolation and sequencing
All sorted samples frozen at −80°C were thawed to room temperature together and processed in one batch. A custom script on a Beckman Coulter Biomek FXP was used to extract total RNA from Trizol using the Direct-zol 96 MagBead RNA kit (Zymo, R2100). Ribosomal RNA was depleted using Ribo-Zero rRNA Removal Kit (Illumina, MRZH11124). A 100 ng aliquot of each sample’s RNA was then prepared into sequencing libraries, according to manufacturer’s instructions, using the Truseq Stranded Total RNA Library Prep Kit (Illumina, RS-122-2203). The resulting libraries were deep sequenced on the Illumina HiSeq 4000, using single-end reads with lengths of 50 nucleotides. The single end, 50 bp RNA-Seq reads that passed Illumina filters, were filtered for reads aligning to tRNA, rRNA, and adapter sequences before RNA metrics were calculated. The reads were then aligned to human genome version hg19 (GRCh37.p13, https://www.gencodegenes.org) using TopHat v1.4.1.44 Post mapping QC was conducted with RSeQC package45 for quality parameters such as read quality, read distribution, junction saturation, junction annotation and gene body coverage. Differential expression analysis was performed on the protein coding genes using edgeR46 with upper quartile normalization.47 The differentially expressed genes with a significance cut-off of FDR <0.05 were considered for the downstream analysis.
The 92 samples were sequenced at a read depth of about 75 million per sample. The read quality score was 40 (highest possible, Supplementary material online, Figure S5A), and the percentage of unmapped reads was below 20% in all groups (Supplementary material online, Figure S5B). The 5′ to 3′ coverage was excellent (Supplementary material online, Figure S5C). Most reads mapped to exons and the 5′ and 3′ untranslated regions, with less than 5% of reads mapping to other sites (Supplementary material online, Figure S5D). The number of detected splice sites showed saturation (Supplementary material online, Figure S5E), and most mapped to known splice sites (Supplementary material online, Figure S5F).
Preliminary validation experiments on fresh and frozen samples showed that RNA quality did not change after ∼6 months of storage in liquid nitrogen (IN2, data not shown). WIHS samples were stored for an average of ∼4.5 years (4.69 ± 0.99 years). There were no correlations between RIN numbers and duration of storage (data not shown).
2.5 Qiagen ingenuity pathway analysis
Differentially expressed gene lists were uploaded to IPA and processed using the Core Analysis function.48 Disease and Biological Functions were used after negative log transformation of the P-value overlap and application of a strict significance cut-off (P < 0.001). The data were further analysed using the comparison analysis function within IPA and subsequently filtered for upstream regulators. P-value overlap and activation z-score were used to show significant activation or inhibition as a prediction of its effect on observed gene expression. P-value overlap was used to explain gene expression changes by measuring whether there was a statistically significant overlap between the dataset genes and the genes that are regulated by a transcriptional regulator, as calculated by Fisher’s exact test. The activation z-score was used to infer the activation of inhibition states of predicted transcriptional regulators based on the published literature.
2.6 Gene signature analysis
In order to identify co-expression patterns in the RNA-seq dataset, PRESTO49 was employed on the RPKM-normalized gene matrix of all participants. Variable genes threshold was used to achieve roughly 3000 input genes for maximal recovery and resolution of subsequent clusters (3300 total genes). All samples were used with additional pre-processing requiring expression of each gene to be greater than 0 in at least 1 participant. tSNE settings: perplexity of 30 and 2000 iterations. Clustering was performed using DBScan and exported for subsequent analyses.
The gene set enrichment analysis (GSEA) tool50 was used as previously described.51 In brief, the tool determines whether a list of genes (signature or cluster genes) is represented in differentially expressed genes of two given conditions (for example, control versus disease). We used the GSEA tool embedded in the GenePattern 2.0 framework52 with standard settings (weighted, 100 iterations).
2.7 Statistical analysis
Participant clinical data were summarized using SAS 9.4 (SAS Institute, Cary, NC, USA). The χ2 tests or Fisher’s exact tests were used for comparing categorical variables among 4 and between 2 groups; Kruskal–Wallis/Wilcoxon tests were used for comparing continuous variables among 4 or between 2 groups. Flow cytometry data and RNA quality control data were further analysed in PRISM v7 and v8 (GraphPad Software). Kaplan–Meier analysis was performed in Prism v8; significance determined by log-rank test (Mantel–Cox). RNA and FACS quality control data normality was determined using the D’Agostino and Pearson normality test and significance was determined by the Kruskal–Wallis test with Dunn’s test used for multiple comparisons or t-test; significance was attributed to P < 0.05. Heatmaps and hierarchical clustering were performed using Morpheus (https://software.broadinstitute.org/morpheus/).
3. Results
3.1 In HIV and sCVD, CMs highly express pro-inflammatory gene programmes
To determine how gene expression in CMs is altered in HIV or sCVD, we utilized cryopreserved PBMC samples from participants of the WIHS, purified CMs by fluorescent activated cell sorting, and subsequently prepared Illumina libraries from these samples for sequencing.
The WIHS enrolled women with and at risk for HIV infection.39 All participants provided informed consent prior to enrolment. Each site’s Institutional Review Board approved the studies and complied with the Declaration of Helsinki (see Section 2). Carotid artery B mode ultrasound was used to define sCVD as the presence of at least one carotid artery plaque as evidenced by focal intima-media thickness greater than 1.5 mm measured at six arterial locations.43Ninety-two participants were matched by age, race/ethnicity, and smoking status into 23 quartets, containing one participant from each of the following groups: HIV−sCVD− (H−C−), HIV+sCVD− (H+C−), HIV−sCVD+ (H−C+), and HIV+sCVD+ (H+C+). Most clinical parameters were similar among the four groups, with the exception of LDL cholesterol, which was higher among sCVD+ participants, and statin use, which was present in 5 of 23 H−C+ participants and 10 of 23 H+C+ participants, but in none of the sCVD− participants (Supplementary material online, Tables S1 and S2). Additionally, there was no apparent difference in the frequency of monocyte subsets between the groups (Supplementary material online, Figure S1).
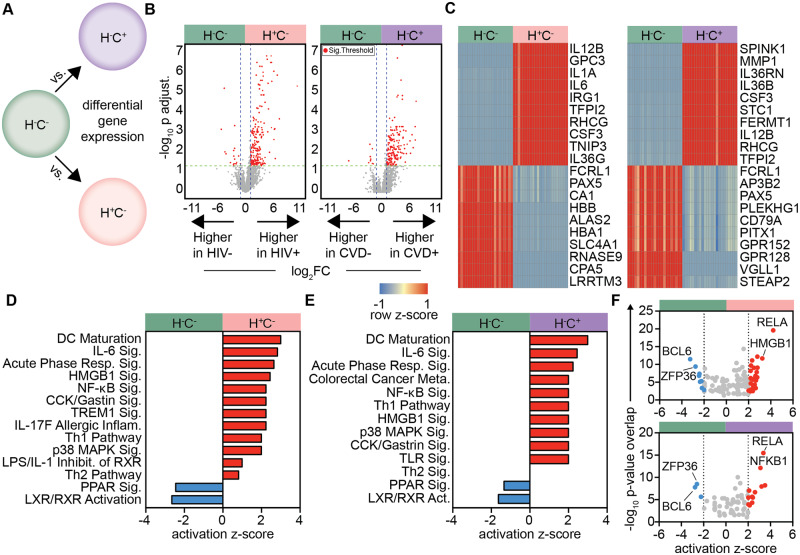
In order to determine gene regulation differences in HIV and sCVD participants, we first performed differential gene expression (DE) analysis,46 using H−C− as a healthy reference group (Figure 1A). CMs from H+C− and H−C+ participants significantly overexpressed 204 and 179 genes, respectively, compared to the healthy reference (Figure 1B, Supplementary material online, Tables S3–S6). The top over- and under-expressed genes in HIV or sCVD were uniformly expressed in monocytes of all participants within each group. The gene sets shared several notable similarities between HIV and sCVD, including IL12B, encoding for the p40 subunit of IL-12 and IL-23, TFPI2, encoding for tissue factor pathway inhibitor-2, and CSF3, encoding for G-CSF (Figure 1C). Forty-two overexpressed genes (∼20%) were commonly regulated in HIV and sCVD (data not shown). The overlap was even more striking when looking at canonical pathway analysis of DE genes (Figure1D andE). In both H+C− and H−C+ participants, we saw activation and significant enrichment for inflammatory pathways such as DC maturation, IL-6 signalling, Th1 pathway and NFκB signalling compared with H−C− participants (Figure1D andE). Likewise, inhibition of PPAR signalling and liver X and retinoic acid receptor (LXR/RXR) pathways was common between the two groups (Figure1D andE). PPAR, LXR and RXR pathways are known down-regulators of pro-inflammatory gene expression53 and inhibitors of inflammatory cytokine secretion in monocytes and macrophages.54 The common highly-expressed genes in HIV and sCVD CMs included both subunits of IL-23, p19 (IL23a) and p40 (IL12b), IL-1α and IL-6, a classical pro-inflammatory cytokine and one of the best-established blood biomarkers for CVD and atherosclerosis.55–57
Figure 1.
Inflammatory transcriptome signatures and pathways in CM associated with HIV and sCVD. Differential gene expression analysis was performed, using edgeR, between (A) H+C− vs. H−C− (HIV signature), H−C+ vs. H−C− (sCVD signature); n = 23 each group. (B) Volcano plots for these comparisons; red genes indicate significance by FDR < 0.05 and |log2FC|>1. (C) Heatmaps of top 10 up- and down-regulated genes by log2FC for each comparison, hierarchically clustered on genes (rows) and participants (columns); gene expression was normalized by row z-score. Full gene lists for each group shown in Supplementary material online, Tables S3–S6. (D) DE gene lists were uploaded to Qiagen IPA and used in individual core analyses. Log fold change between (E) H+C− vs. healthy or (F) H−C+ vs. healthy was used to predict significantly enriched activated and inhibited pathways, and upstream transcription regulators; significance calculated by right-tailed Fisher’s exact test, P < 0.05, all transcription regulators in Supplementary material online, Table S7.
Next, we investigated the relatedness of upstream control of gene expression as revealed by potential transcriptional regulators in each condition (Figure 1F). The NFκB subunits RELA and NFKB1 were highly activated and enriched in both H+C− and H−C+ groups, as well as the known inflammatory macrophage-driving transcription factor IRF5.58 H+C− participants showed additional activation for IRF1 and 8, which have been shown to play a crucial role in macrophage host inflammatory and defense responses59 and are specifically induced in monocyte-derived DCs infected by HIV.60 Additional activation of STAT1 suggest CMs from these participants may be further along in their maturation to macrophages.61 A comprehensive list of transcription regulators and their activation scores is shown in Supplementary material online, Table S7.
3.2 Statin use in HIV correlates with a strong anti-inflammatory phenotype in CM
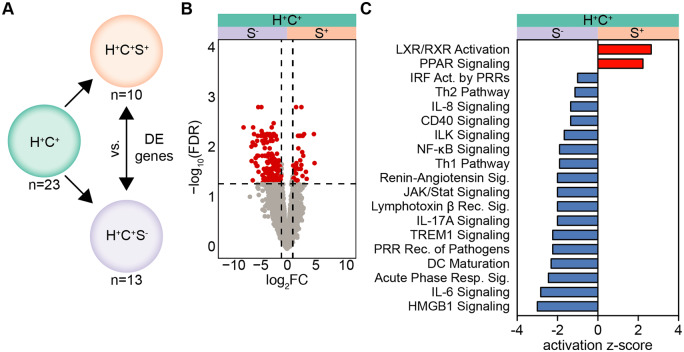
Gene expression analysis of CMs from H+C+ participants yielded only 9 DE genes when compared to the H−C− reference group. We reasoned that this lack of DE genes might be caused by a confounder. Indeed, almost half of the H+C+ participants (10 of 23) reported taking LDL cholesterol-lowering statins at the time of blood draw (Supplementary material online, Table S2). To examine whether statin treatment might influence gene expression in CM of these participants, we performed differential gene expression analysis between participants reporting statin use (H+C+S+) and not reporting statin use (H+C+S−) participants within the H+C+ group (Figure 2A). Statin use (S+) was associated with significantly lower expression of most differentially expressed genes (Figure 2B, Supplementary material online, Tables S8 and S9), but there were minimal clinical differences between these groups (Supplementary material online, Table S10). Next, we asked how statins may influence known canonical pathways by using the DE gene list as input to IPA. Analysis of statin-users (H+C+S+), predicted activation of the anti-inflammatory LXR/RXR and PPAR signalling pathways and inhibition of many of the pro-inflammatory and monocyte activation pathways that were indicative of HIV and sCVD CMs (Figure 2C). These data suggest that CMs from H+C+ participants without statin treatment may be enriched for pro-inflammatory gene programmes.
Figure 2.
Anti-inflammatory transcriptome modification in CM of statin-using H+C+ participants. (A) Differentially expressed genes (FDR < 0.05, |log2FC|>1) within H+C+ participants taking statins (S+, n = 10) or not (S−, n = 13) at the time of PBMC visit, determined using edgeR. (B) Hierarchically clustered heatmap of DE genes; heatmap normalized by row z-score. Gene lists in Supplementary material online, Tables S8 and S9. DE gene list with log2FC was used as input into Qiagen IPA core analyses. (C) Predicted regulation of canonical pathways between S+ and S−; red bar indicates activation, blue bar indicates inhibition in H+C+S+. All pathways shown were significantly enriched; significance calculated by right-tailed Fisher’s exact test, P < 0.05.
3.3 Statin use is linked with reduced inflammatory-associated gene expression in CM
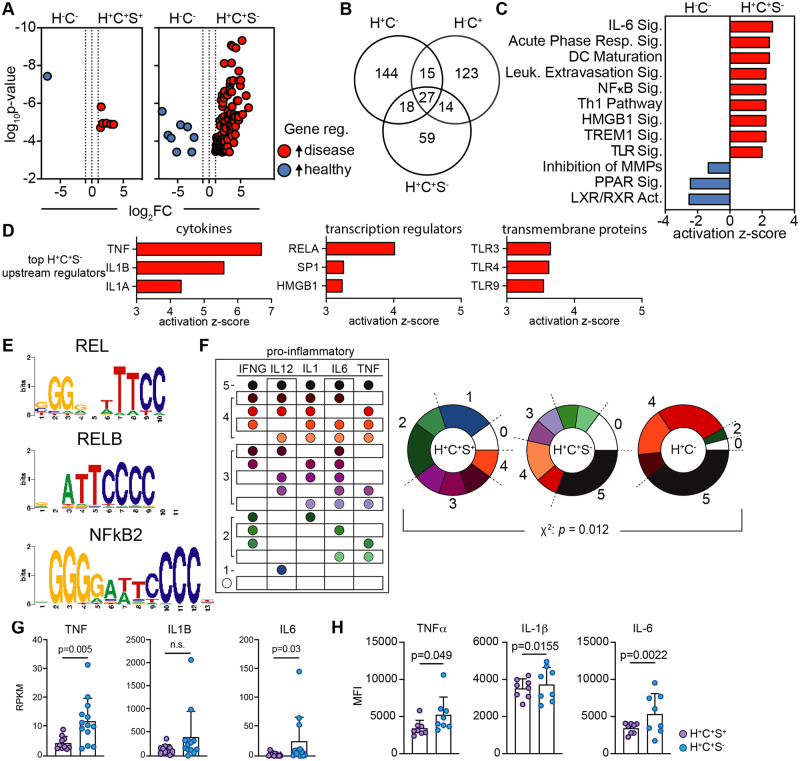
Next, we contrasted the CM transcriptomes of H+C+S+ and H+C+S− participants with those of healthy (H−C−) participants. We found that the CM transcriptomes of H+C+S+ participants were quite similar to healthy (H−C−) participants (Figure 3A, left, Supplementary material online, Tables S11 and S12). However, H+C+S− participants displayed a large number of highly expressed transcripts (Figure 3A, right, Supplementary material online, Tables S13 and S14) compared to healthy. There were both unique and shared genes regulated in CM of H+C−, H−C+, and H+C+S− participants (Figure 3B, additional comparisons in Supplementary material online, Tables S15–S18). The expression of 27 genes was significantly increased in all three diseased participant groups (H+C−, H−C+, H+C+S−), suggesting there may be a core programme of inflammatory genes involved in CMs that is shared between HIV and sCVD. Half of the genes up-regulated in H+C+S− participants (59 of 108) were uniquely expressed. To further investigate the role of these genes in CMs, we looked at enriched canonical pathways (Figure 3C). Here, we saw pro-inflammatory pathways such as IL-6 signalling, DC maturation, and leucocyte extravasation signalling, suggesting that CM of H+C+ participants had an enhanced potential to extravasate and differentiate into mo-DCs or inflammatory macrophages. Furthermore, we identified key upstream regulating cytokines, transcriptional regulators, and transmembrane receptors likely to be influencing monocyte gene expression (Figure 3D). The cytokines TNF, IL1B, and IL1A were highly enriched in H+C+S−, as were the transcriptional regulators RELA, SP1, and HMGB1, which are associated with M1-macrophage polarization.51,62 TLR3, 4, and 9 pathways were also activated. TLR3 increases with the transition from monocytes to mo-DCs63 and TLR4 is involved in the NFκB-mediated activation of IL6 and NOS2,64 which are both potent inflammatory molecules. NFκB mediated gene activation was further implicated by identifying common DNA-binding motifs in the up-regulated genes by PScan65 (Figure 3E), where several NFκB/REL motifs were highly enriched (Figure 3E).
Figure 3.
Statin use associated with reduced pro-inflammatory cytokine and gene expression in H+C+ participants. (A) Differentially expressed genes were determined, by edgeR, for H+C+S+ (n = 10) and H+C+S− (n = 13) participants vs. H−C− (n = 23, Supplementary material online, Tables S11–S14), respectively; significance defined by FDR < 0.05, |log2FC|>1, red genes higher in H+C+, blue higher in H−C−. (B) Venn diagram of highly expressed genes from DE analyses between each disease group vs. H−C−. DE genes and log2FC from H+C+S− vs. H−C− were used for a core pathway analysis in Qiagen IPA. (C) Significantly enriched, activated and inhibited canonical pathways and (D) the predicted top upstream cytokines, transcriptional regulators, and transmembrane receptors influencing gene expression. (E) Up-regulated gene list (H+C+S−) was used as input into PScan to identify significantly enriched common DNA-binding motifs; P < 0.05. RPKM gene expression of top 5 pro-inflammatory cytokines, INFG, IL12 (IL12B), IL1 (IL1B), IL6, and TNF, for each participant within the H+C−, H+C+S−, and H+C+S+ groups compared to the median RPKM expression of the H−C− participants. Participants with higher expression than the median (greater than healthy) were considered positive expressers of the gene. (F) All combinations of gene expression patterns of top 5 inflammatory genes. Fraction of participants overexpressing 0–5 inflammatory genes. The χ2 analysis was used to determine significance in pattern differences. (G) RPKM normalized gene expression for H+C+S− (n = 13) and H+C+S+ (n = 10) participants for inflammatory cytokines TNF, IL1B, and IL6; significance by Mann–Whitney test. Cryopreserved PBMCs were stimulated with LPS for 6 h and CM subsequently analysed by flow cytometry for intracellular cytokine expression (median fluorescence intensity) for (H) TNFα, IL-1β, and IL-6; H+C+S− (n = 8) and H+C+S+ (n = 8), significance determined by unpaired t-test.
NFκB is an integral transcription factor in the function and activation of macrophages and is required for the induction of macrophage-associated pro-inflammatory genes, including IFNG, IL12B, TNF, IL1B, and IL6.66 To determine the expression patterns of these genes across participants, we counted the number of individuals whose CMs expressed IFNG, IL12B, TNF, IL1B, or IL6 at a level higher than CM of healthy (H−C−) participants. We observed significant differences in the number of expressed pro-inflammatory cytokines (Figure 3F). About one-third of H+C− and H+C+S− participants expressed all five pro-inflammatory cytokines. H+C+S+ participants, however, showed expression of only 1, 2, or 3 of these top inflammatory cytokines, suggesting a lower inflammatory burden. In order to determine the actual cytokine production of CMs in H+C+S− and H+C+S+ at the protein level, we stimulated participant PBMCs with LPS and measured cytokine expression by flow cytometry. CMs from H+C+S− participants produced more inflammatory cytokines at the protein level (Supplementary material online, Figure S2) than H+C+S+, as was apparent in the gene expression data (Figure 3F). TNF and IL-6, cytokines downstream of NFκB, were increased in H+C+S− CMs at the mRNA (Figure 3G) and protein level (Figure 3H) compared to H+C+S+ participant CMs. However, only IL-1β protein differed, not mRNA in this comparison (Figure 3H).
3.4 CVD risk assessment by co-expression gene networks in CM
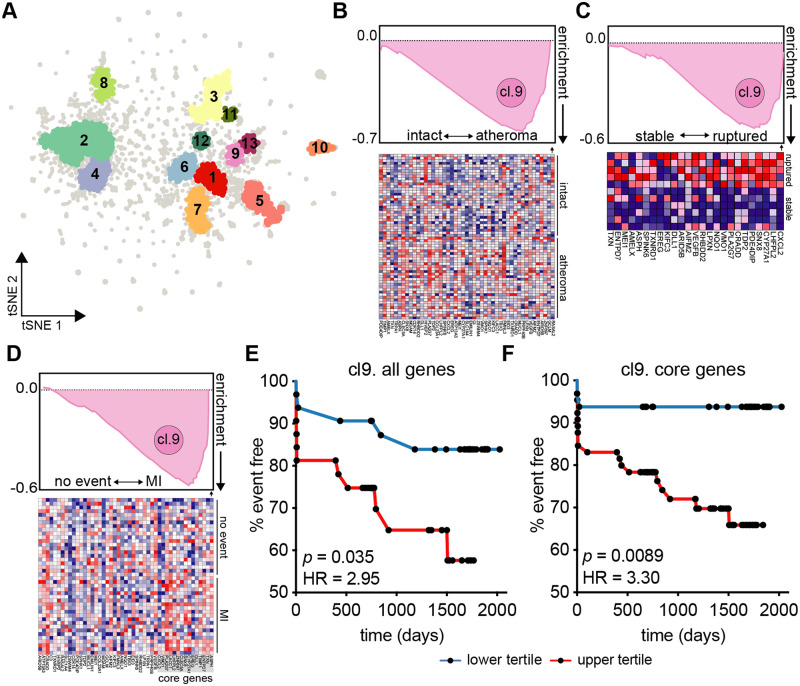
The present study lacks participant outcome data from the WIHS, and therefore cannot be used directly to determine an association between monocyte gene expression and cardiac event risk. However, by using the RPKM-normalized gene expression data and PRESTO,49 an algorithm that identifies co-expression networks, we were able to identify 13 clusters of co-regulated genes (Figure 4A). We applied the gene lists from each cluster to external datasets using Gene Set Enrichment Analysis (GSEA)50 to suggest an association of cluster gene expression and disease phenotypes. Cluster 9 genes were evaluated in non-WIHS patients without HIV and with CVD, including blood transcriptomes of patients with or without acute myocardial infarction67 (Supplementary material online, Figure S3A), and patients with stable coronary artery disease or ST-segment elevation myocardial infarction68 (Supplementary material online, Figure S3B). In both instances, cluster 9 genes were enriched in patients that presented with myocardial infarction. When applied to tissue biopsy transcriptomes from patients who underwent carotid endarterectomy,69 cluster 9 genes were found to be highly enriched in atheroma plaque compared to macroscopically intact adjacent tissue (Figure 4B). Cluster 9 genes were also more enriched in samples taken from ruptured plaque compared to stable plaque70 (Figure 4C).
Figure 4.
Higher cardiovascular risk associated with CM gene cluster in H+C+ participants. (A) To identify gene co-expression networks, the entire RPKM gene expression matrix (n = 92) was used as input into PRESTO. The top ∼3300 variable genes segregated into 13 clusters identified using DBScan. (B) Cluster 9 gene list was applied to tissue biopsy transcriptomes of hypertensive patient-matched carotid endarterectomy macroscopically intact tissue vs. atheroma plaque (n = 32, GSE43292); all cluster 9 genes across all patients shown below. (C) Transcriptomes of laser micro-dissection macrophage-rich (CD68+) carotid plaque regions of stable (n = 5) or ruptured (n = 6) plaques (GSE41571) using gene set enrichment analysis (GSEA). Top 25 genes and their expression across patient plaques shown below. (D) GSEA applied to PBMC transcriptomes (n = 97) of the BiKE cohort (GSE21545) and identified core enrichment genes within cluster 9. (E) Patients were divided into top and bottom tertiles based on their PBMC mean expression of all cluster 9 gene [HR = 2.95, 95% CI (1.08–8.04)] or (F) core enrichment genes [15 genes, HR = 3.30, 95% CI (1.35–8.06)] and analysed for gene expression-associated ischaemic event risk. Kaplan Meier curves show time (days) until ischaemic event; significance by log-rank test, hazard ratio (HR) by Mantel–Haenszel analysis.
Because cluster 9 genes were associated with CVD events (MI), atheroma, and ruptured plaque phenotypes, we sought to determine if cluster 9 genes could independently predict cardiovascular events. We utilized patient PBMC transcriptomes from the Biobank of Karolinska Endarterectomy (BiKE), which prospectively collected atherosclerotic plaque tissue and PBMCs from patients undergoing carotid endarterectomy for high-grade carotid artery stenosis.71 First, we confirmed that cluster 9 genes were associated with myocardial infarction events in patient PBMCs (Figure 4D). Next, we ranked patients by their mean expression of all cluster 9 genes and divided high and low expressing patients into two groups. We found that high expression of cluster 9 genes was significantly associated with myocardial infarction [hazard ratio (HR) = 2.95, 95% confidence interval (CI) (1.08–8.04), Figure 4E]. Using a set of core genes (15 genes) that were enriched in BiKE patient PBMCs isolated from patients who had an ischaemic event (Figure 4D), we were additionally able to identify that high expression of these core genes was associated with myocardial infarction [HR = 3.30, 95% CI (1.35–8.06), Figure 4F]. Expression of cluster 9 genes was also elevated in H+C+S− participants compared to H+C+S+ in our study (Supplementary material online, Figures S4 and S5), suggesting a higher CVD event risk in these individuals compared to statin-using participants.
4. Discussion
ART is of vital importance for controlling the viral burden in HIV, but the ramifications of persistent inflammation remain to be addressed. In people LWH, non-AIDS-related comorbidities, like CVD, present at an earlier age than in the general population.72 Cardiovascular events are a major cause of death.14,73 Therefore, it remains a priority to understand how persistent inflammation contributes to accelerated comorbid disease in higher-risk people LWH.17 Previously, we found that circulating serum biomarkers of monocyte-associated inflammation, soluble CD14 and soluble CD163, were associated with increased atherosclerotic plaque in CVD and HIV.13 We also found that women with both CVD and HIV showed a loss of CXCR4 cell surface expression on non-CMs.74 It is well-known that monocytes, particularly CM, are major contributors to innate inflammatory responses75,76 and are prominently involved in CVD progression24 and event prediction.26 Here, we implemented the first study investigating the CM transcriptome and co-expression pathways likely responsible for enhanced progression and risk of CVD in people LWH.
Differential gene expression analysis revealed that both HIV and sCVD are associated with high expression of hundreds of pro-inflammatory genes, many of which are connected with known inflammatory pathways or recognized disease biomarkers.1,57 Many pro-inflammatory pathways induced by chronic HIV infection or sCVD are overlapping. These CM switch on the transcription of several pro-inflammatory programmes. Two pathways of anti-inflammatory genes, the PPAR and LXR pathways, were inhibited in H+C− and H−C+ participant groups.53 These results support the hypothesis of a chronic inflammatory state in people LWH, where CM remain activated and contribute to systemic inflammation. Under these conditions, CMs have been found to possess higher surface expression of integrins,77 therefore, likely possessing a higher potential for tissue recruitment and extravasation.
Unexpectedly, the CM transcriptomes of participants with both HIV and CVD did not show any synergy and not even an additive effect compared to H+C− or H−C+. We suspected a confounding parameter that we had not considered. Indeed, about half of the H+C+ participants reported taking cholesterol-lowering statins, which are known to wield several additional pleiotropic benefits beyond lipid lowering.78 We speculated that statin therapy may alter the systemic inflammatory environment and influence the CM transcriptome. Interestingly, we found a large number of pro-inflammatory transcripts to be suppressed in H+C+ participants taking statins, including pro-inflammatory pathways, IL-6 and TREM1 signalling. Integrin signalling and the M1-associated transcription factor HMGB1 signalling were also suppressed by statin treatment. Upstream analysis showed suppression of the inflammatory cytokines TNF, IL1α, IL1β, and IFNγ. The M1-associated transcription factors RELA, HMGB1, and NFKB151 were also suppressed in statin-users. This suggests that monocytes of statin-using H+C+ participants may have a lower propensity to extravasate, and a reduced potential to mature into inflammatory DCs and M1-macrophages. In these participants, statin treatment was also associated with activation of the anti-inflammatory LXR and PPAR signalling pathways, which are known to play a major role in lipid homeostasis and metabolic disease.79 The PPAR pathway was also activated in monocytes of statin-using individuals, which inhibits their ability to produce some pro-inflammatory cytokines.54
Monocytes are the major source for several cytokines, both systemically and locally, in response to inflammation.80 Statin treatment appears to modulate the expression of combinations of central pro-inflammatory cytokines compared to both non-statin-treated participants and H+C− participants. These findings are consistent with so-called pleiotropic anti-inflammatory effects of statins.78 Statin pleiotropic effects have not yet been studied in HIV. An ongoing prospective clinical trial (REPRIEVE) aims to examine the ability of statin therapy to prevent vascular events in people with HIV without known CVD.38 Our data provides monocyte transcriptomic evidence supporting the REPRIEVE hypothesis, which holds that the use of statins (pitavastatin) will reduce the incidence of future major adverse cardiovascular events in people LWH on any ART programme, even if they do not have evidence of CVD and are low-to-moderate CVD risk. We think statin treatment should be combined with ART in people LWH, even those with no or only early signs of CVD to limit pro-atherogenic immune phenotypes.
By comparing statin-using and -non-using H+C+ groups to healthy controls, we establish the combinatorial inflammatory phenotype of HIV and CVD. Here, statin-using H+C+ participant CM transcriptomes approach those seen in healthy (H−C−) participants, with only a handful of genes being significantly different. In contrast, statin-untreated H+C+ participants not reporting statin use show many highly expressed inflammatory transcripts. These transcripts overlap with transcripts from both HIV and sCVD, but many are unique to H+C+ participants. Further investigation of the CM machinery of these individuals aligns with common M1-macrophage and mo-DC genes and pathways. This is most clear in upstream analysis where the top 3 transcriptional regulators are M1-associated factors,51 and in a canonical pathway comparison with both HIV and sCVD associated pathways. Several pathways highest in H+C+ participants suggest an increased capacity for these monocytes to adhere, extravasate, and secrete cytokines/chemokines and pro-thrombotic factors. These data suggest CM in H+C+ participants possess an even more pro-atherogenic20 phenotype than in participants with single disease (H+C− or H−C+).
The subclinical vascular inflammation that is reflected in the CM transcriptomes of HIV+ individuals is of great concern. Addressing and treating this inflammation is expected to alleviate the development and burden of CVD in chronic HIV. Here, we were able to identify major co-expression networks associated with each participant group and pinpoint a specific group of CM genes that highly correlate with cardiovascular risk and plaque instability. These genes were most highly expressed in the H+C− and H+C+S− groups, and lowest in H+C+S+. Cluster 9 showed enrichment for many TNF-related cytokines and receptors, as well as Trx pathway signalling. The 15 core genes were applied as a biomarker signature to patient blood. Expression of this signature significantly predicted ischaemic events such as myocardial infarction. This signature suggests a new and robust method for measuring ischaemic risk in patients. An advantage of our signature is that it was derived through hypothesis-free co-expression analysis. Thus, the presence and stability of atherosclerotic plaques can be determined by a simple multiplex qPCR of patient blood to determine the expression of these 15 core signature genes. The monotonic nature of the transcriptome signature then provides a robust predictor of CVD risk. It is also important to note that this type of analysis is robust to biological patient-to-patient variability, which means this signature provides a better risk assessment across a population of patients than other traditional biomarkers.
As with any study, the present work has some limitations. It uses a surrogate marker (carotid lesions) instead of overt clinical disease to define CVD. cIMT has been shown to be highly predictive of clinical coronary events.81,82 There is value of studying preclinical forms of CVD, because it suggests strategies to avert adverse events later. We focused only on CM in this study; non-classical or intermediate monocyte gene expression may provide further insights. Our collection of statin use is from participant self-report and dichotomized here simply as reported use versus no use without regard to dose and adherence. In addition to the possibility of misclassification, the lack of randomization of statins opens the possibility for other confounding factors to influence our results. We also are unable to explicitly rule out the lipid-lowering effect of statins as a contributor to reduced inflammation, due to the cross-sectional nature of our study. Finally, our results entirely within a cohort of women may not be generalizable to men.
Despite these limitations, our study provides (i) the first comprehensive examination of the transcriptomes of CM associated with CVD and HIV in a well-established and well-characterized cohort; (ii) evidence that statin therapy may alleviate expression of pro-inflammatory genes and cytokines in CM; and (iii) evidence for gene networks in CM that are associated with cardiovascular risk, plaque destabilization, and ischaemic events.
Data availability
The data underlying this article will be shared on reasonable request to the corresponding author.
Supplementary material
Supplementary material is available at Cardiovascular Research online.
Authors’ contributions
K.L. and R.C.K. designed the study. E.E., K.M., and L.B. performed experiments. E.E., P.A.B., Y. G., J.L., D.B.H., K.M., T.W., and K.B. analysed data and performed bioinformatic analysis. E.E., K.L., R.C.K., D.B.H. wrote the manuscript. K.L., R.C.K., Q.Q., K.A., J.M.L., W.J.M., P.C.T., M.H.C., I.O., S.G., S.L.H., H.N.H., R.P.T., Y.G., C.P.D., and A.L.L. provided critical insight on the manuscript, data analysis, and study design.
Supplementary Material
Acknowledgements
We thank Cheryl Kim, Lara Boggeman, and Kurt Van Gunst for their help with flow sorting, and Jeremy Day and Kristen Jepsen for help with RNA sequencing. Data in this article were collected by the MACS/WIHS Combined Cohort Study (MWCCS). The contents of this publication are solely the responsibility of the authors and do not represent the official views of the National Institutes of Health (NIH). MWCCS (Principal Investigators): Bronx CRS (Kathryn Anastos and Anjali Sharma), U01-HL146204-01; Brooklyn CRS (Deborah Gustafson and Tracey Wilson), U01-HL146202-01; Data Analysis and Coordination Center (Gypsyamber D’Souza, Stephen Gange, and Elizabeth Golub), U01-HL146193-01; Chicago-Cook County CRS (Mardge Cohen and Audrey French); Connie Wofsy Women’s HIV Study, Northern California CRS (Bradley Aouizerat and Phyllis Tien), U01-HL146242-01; Los Angeles CRS (Roger Detels), U01-HL146333-01; Metropolitan Washington CRS (Seble Kassaye and Daniel Merenstein), U01-HL146205-01. The MWCCS is funded primarily by the National Heart, Lung, and Blood Institute (NHLBI), with additional co-funding from the Eunice Kennedy Shriver National Institute Of Child Health & Human Development (NICHD), National Human Genome Research Institute (NHGRI), National Institute On Aging (NIA), National Institute Of Dental & Craniofacial Research (NIDCR), National Institute Of Allergy And Infectious Diseases (NIAID), National Institute Of Neurological Disorders And Stroke (NINDS), National Institute Of Mental Health (NIMH), National Institute On Drug Abuse (NIDA), National Institute of Nursing Research (NINR), National Cancer Institute (NCI), National Institute on Alcohol Abuse and Alcoholism (NIAAA), National Institute on Deafness and Other Communication Disorders (NIDCD), National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK). MWCCS data collection is also supported by UL1-TR000004 (UCSF CTSA).
Conflict of interest: none declared.
Funding
E.E. was supported in part by the UCSD Graduate Training Program in Cellular and Molecular Pharmacology through an institutional training grant from the National Institute of General Medical Sciences (T32 GM007752). Q.Q. is supported by a Research Scientist Development Award (K01-HL-129892) from the National Heart, Lung, and Blood Institute (NHLBI). D.B.H. is supported by a Research Scientist Development Award (K01-HL-137557) from NHLBI. This work was supported by NIH grants R01 HL126543 and R01 HL148094-01 to R.C.K. and K.L., R35 HL145241 to K.L., and R01 HL126543, R01 HL132794, R01 HL083760, and R01 HL095140 to R.C.K., and HL146193 to S.G.
Translational perspective
Monocytes from women living with HIV express many more pro-inflammatory genes than uninfected controls. An overlapping list of genes is expressed in samples from women with ultrasound evidence of carotid plaque. The inflammatory burden is enhanced in women with both HIV and carotid plaque, and this is mitigated by statin treatment, almost to the level of healthy participants. Thus, the present monocyte transcriptome data from 92 women support the idea that participants with HIV may specifically benefit from statin treatment, perhaps more so than seronegative subjects.
References
- 1. Wilson EM, Singh A, Hullsiek KH, Gibson D, Henry WK, Lichtenstein K, Onen NF, Kojic E, Patel P, Brooks JT, Sereti I, Baker JV; for the Study to Understand the Natural History of HIV/AIDS in the Era of Effective Therapy (SUN Study) Investigators. Monocyte-activation phenotypes are associated with biomarkers of inflammation and coagulation in chronic HIV infection. J Infect Dis 2014;210:1396–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Westhorpe CLV, Maisa A, Spelman T, Hoy JF, Dewar EM, Karapanagiotidis S, Hearps AC, Cheng W-J, Trevillyan J, Lewin SR, Sviridov D, Elliott JH, Jaworowski A, Dart AM, Crowe SM.. Associations between surface markers on blood monocytes and carotid atherosclerosis in HIV-positive individuals. Immunol Cell Biol 2014;92:133–138. [DOI] [PubMed] [Google Scholar]
- 3. Brenchley JM, Price DA, Schacker TW, Asher TE, Silvestri G, Rao S, Kazzaz Z, Bornstein E, Lambotte O, Altmann D, Blazar BR, Rodriguez B, Teixeira-Johnson L, Landay A, Martin JN, Hecht FM, Picker LJ, Lederman MM, Deeks SG, Douek DC.. Microbial translocation is a cause of systemic immune activation in chronic HIV infection. Nat Med 2006;12:1365–1371. [DOI] [PubMed] [Google Scholar]
- 4. Passlick B, Flieger D, Ziegler-Heitbrock H.. Identification and characterization of a novel monocyte subpopulation in human peripheral blood. Blood 1989;74:2527–2534. [PubMed] [Google Scholar]
- 5. Anbazhagan K, Duroux-Richard I, Jorgensen C, Apparailly F.. Transcriptomic network support distinct roles of classical and non-classical monocytes in human. Int Rev Immunol 2014;33:470–489. [DOI] [PubMed] [Google Scholar]
- 6. Ingersoll MA, Spanbroek R, Lottaz C, Gautier EL, Frankenberger M, Hoffmann R, Lang R, Haniffa M, Collin M, Tacke F, Habenicht AJR, Ziegler-Heitbrock L, Randolph GJ.. Comparison of gene expression profiles between human and mouse monocyte subsets. Blood 2010;115:e10–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Boyette LB, Macedo C, Hadi K, Elinoff BD, Walters JT, Ramaswami B, Chalasani G, Taboas JM, Lakkis FG, Metes DM.. Phenotype, function, and differentiation potential of human monocyte subsets. PLoS One 2017;12:e0176460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sonza S, Mutimer HP, Oelrichs R, Jardine D, Harvey K, Dunne A, Purcell DF, Birch C, Crowe SM.. Monocytes harbour replication-competent, non-latent HIV-1 in patients on highly active antiretroviral therapy. AIDS 2001;15:17–22. [DOI] [PubMed] [Google Scholar]
- 9. Honeycutt JB, Wahl A, Baker C, Spagnuolo RA, Foster J, Zakharova O, Wietgrefe S, Caro-Vegas C, Madden V, Sharpe G, Haase AT, Eron JJ, Garcia JV.. Macrophages sustain HIV replication in vivo independently of T cells. J Clin Invest 2016;126:1353–1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wang T, Green LA, Gupta SK, Amet T, Byrd DJ, Yu Q, Twigg HL, Clauss M.. Intracellular Nef detected in peripheral blood mononuclear cells from HIV patients. AIDS Res Hum Retrov 2015;31:217–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wong ME, Jaworowski A, Hearps AC.. The HIV reservoir in monocytes and macrophages. Front Immunol 2019;10:1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Montaner JSG, Lima VD, Harrigan PR, Lourenço L, Yip B, Nosyk B, Wood E, Kerr T, Shannon K, Moore D, Hogg RS, Barrios R, Gilbert M, Krajden M, Gustafson R, Daly P, Kendall P.. Expansion of HAART coverage is associated with sustained decreases in HIV/AIDS morbidity, mortality and HIV transmission: the ‘HIV Treatment as Prevention’ experience in a Canadian setting. PLoS One 2014;9:e87872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hanna DB, Lin J, Post WS, Hodis HN, Xue X, Anastos K, Cohen MH, Gange SJ, Haberlen SA, Heath SL, Lazar JM, Liu C, Mack WJ, Ofotokun I, Palella FJ, Tien PC, Witt MD, Landay AL, Kingsley LA, Tracy RP, Kaplan RC.. Association of macrophage inflammation biomarkers with progression of subclinical carotid artery atherosclerosis in HIV-infected women and men. J Infect Dis 2017;215:1352–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mocroft A, Reiss P, Gasiorowski J, Ledergerber B, Kowalska J, Chiesi A, Gatell J, Rakhmanova A, Johnson M, Kirk O, Lundgren J.. Serious fatal and nonfatal non-AIDS-defining illnesses in Europe. J Acquir Immune Defic Syndr 2010;55:262–270. [DOI] [PubMed] [Google Scholar]
- 15. Neuhaus J, Angus B, Kowalska JD, Rosa AL, Sampson J, Wentworth D, Mocroft A; INSIGHT SMART and ESPRIT study groups. Risk of all-cause mortality associated with nonfatal AIDS and serious non-AIDS events among adults infected with HIV. AIDS 2010;24:697–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Schechter ME, Andrade BB, He T, Richter GH, Tosh KW, Policicchio BB, Singh A, Raehtz KD, Sheikh V, Ma D, Brocca-Cofano E, Apetrei C, Tracy R, Ribeiro RM, Sher A, Francischetti IMB, Pandrea I, Sereti I.. Inflammatory monocytes expressing tissue factor drive SIV and HIV coagulopathy. Sci Transl Med 2017;9:eaam5441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Saves M, Chene G, Ducimetiere P, Leport C, Moal GL, Amouyel P, Arveiler D, Ruidavets J-B, Reynes J, Bingham A, Raffi F; French WHO MONICA Project and the APROCO (ANRS EP11) Study Group. Risk factors for coronary heart disease in patients treated for human immunodeficiency virus infection compared with the general population. Clin Infect Dis 2003;37:292–298. [DOI] [PubMed] [Google Scholar]
- 18. Kaplan RC, Kingsley LA, Sharrett AR, Li X, Lazar J, Tien PC, Mack WJ, Cohen MH, Jacobson L, Gange SJ.. Ten-year predicted coronary heart disease risk in HIV-infected men and women. Clin Infect Dis 2007;45:1074–1081. [DOI] [PubMed] [Google Scholar]
- 19. Winkels H, Ehinger E, Vassallo M, Buscher K, Dinh HQ, Kobiyama K, Hamers AAJ, Cochain C, Vafadarnejad E, Saliba A-E, Zernecke A, Pramod AB, Ghosh AK, Michel NA, Hoppe N, Hilgendorf I, Zirlik A, Hedrick CC, Ley K, Wolf D.. Atlas of the immune cell repertoire in mouse atherosclerosis defined by single-cell RNA-sequencing and mass cytometry. Circ Res 2018;122:1675–1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Woollard KJ, Geissmann F.. Monocytes in atherosclerosis: subsets and functions. Nat Rev Cardiol 2010;7:77–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ley K, Miller YI, Hedrick CC.. Monocyte and macrophage dynamics during atherogenesis. Arterioscler Thromb Vasc Biol 2011;31:1506–1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Swirski FK, Nahrendorf M.. Leukocyte behavior in atherosclerosis, myocardial infarction, and heart failure. Science 2013;339:161–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Soehnlein O, Drechsler M, Hristov M, Weber C.. Functional alterations of myeloid cell subsets in hyperlipidaemia: relevance for atherosclerosis. J Cell Mol Med 2009;13:4293–4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Swirski FK, Pittet MJ, Kircher MF, Aikawa E, Jaffer FA, Libby P, Weissleder R.. Monocyte accumulation in mouse atherogenesis is progressive and proportional to extent of disease. Proc National Acad Sci 2006;103:10340–10345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Waterhouse DF, Cahill RA, Sheehan F, McCreery C.. Prediction of calculated future cardiovascular disease by monocyte count in an asymptomatic population. Vasc Heal Risk Management 2008;4:177–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Berg KE, Ljungcrantz I, Andersson L, Bryngelsson C, Hedblad B, Fredrikson GN, Nilsson J, Björkbacka H.. Elevated CD14 ++ CD16− monocytes predict cardiovascular events. Circ Cardiovasc Genet 2012;5:122–131. [DOI] [PubMed] [Google Scholar]
- 27. den Bergh RV, Florence E, Vlieghe E, Boonefaes T, Grooten J, Houthuys E, Tran HTT, Gali Y, Baetselier PD, Vanham G, Raes G.. Transcriptome analysis of monocyte-HIV interactions. Retrovirology 2010;7:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hunninghake DB, Stein EA, Dujovne CA, Harris WS, Feldman EB, Miller VT, Tobert JA, Laskarzewski PM, Quiter E, Held J, Taylor AM, Hopper S, Leonard SB, Brewer BK.. The efficacy of intensive dietary therapy alone or combined with lovastatin in outpatients with hypercholesterolemia. N Engl J Med 1993;328:1213–1219. [DOI] [PubMed] [Google Scholar]
- 29. Fontes JD, Yamamoto JF, Larson MG, Wang N, Dallmeier D, Rienstra M, Schnabel RB, Vasan RS, Keaney JF, Benjamin EJ.. Clinical correlates of change in inflammatory biomarkers: the Framingham Heart Study. Atherosclerosis 2013;228:217–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pedersen TR, Kjekshus J, Berg K, Haghfelt T, Faergeman O, Faergeman G, Pyörälä K, Miettinen T, Wilhelmsen L, Olsson AG, Wedel H; Scandinavian Simvastatin Survival Study Group. Randomised trial of cholesterol lowering in 4444 patients with coronary heart disease: the Scandinavian Simvastatin Survival Study (4S). Atheroscler Suppl 2004;5:81–87. [DOI] [PubMed] [Google Scholar]
- 31. Stein EA, Mellis S, Yancopoulos GD, Stahl N, Logan D, Smith WB, Lisbon E, Gutierrez M, Webb C, Wu R, Du Y, Kranz T, Gasparino E, Swergold GD.. Effect of a monoclonal antibody to PCSK9 on LDL cholesterol. N Engl J Med 2012;366:1108–1118. [DOI] [PubMed] [Google Scholar]
- 32. Ridker PM, Danielson E, Fonseca FAH, Genest J, Gotto AM, Kastelein JJP, Koenig W, Libby P, Lorenzatti AJ, MacFadyen JG, Nordestgaard BG, Shepherd J, Willerson JT, Glynn RJ; JUPITER Study Group. Rosuvastatin to prevent vascular events in men and women with elevated C-reactive protein. N Engl J Med 2008;359:2195–2207. [DOI] [PubMed] [Google Scholar]
- 33. Sabatine MS, Giugliano RP, Wiviott SD, Raal FJ, Blom DJ, Robinson J, Ballantyne CM, Somaratne R, Legg J, Wasserman SM, Scott R, Koren MJ, Stein EA; Open-Label Study of Long-Term Evaluation against LDL Cholesterol (OSLER) Investigators. Efficacy and safety of evolocumab in reducing lipids and cardiovascular events. N Engl J Med 2015;372:1500–1509. [DOI] [PubMed] [Google Scholar]
- 34. Robinson JG, Farnier M, Krempf M, Bergeron J, Luc G, Averna M, Stroes ES, Langslet G, Raal FJ, Shahawy ME, Koren MJ, Lepor NE, Lorenzato C, Pordy R, Chaudhari U, Kastelein JJP; ODYSSEY LONG TERM Investigators. Efficacy and safety of alirocumab in reducing lipids and cardiovascular events. N Engl J Med 2015;372:1489–1499. [DOI] [PubMed] [Google Scholar]
- 35. Twigg HL, Crystal R, Currier J, Ridker P, Berliner N, Kiem H-P, Rutherford G, Zou S, Glynn S, Wong R, Peprah E, Engelgau M, Creazzo T, Colombini-Hatch S, Caler E.. Refining current scientific priorities and identifying new scientific gaps in HIV-related Heart, Lung, Blood, and Sleep Research. AIDS Res Hum Retrov 2017;33:889–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Levine AJ, Horvath S, Miller EN, Singer EJ, Shapshak P, Baldwin GC, Martínez-Maza O, Witt MD, Langfelder P.. Transcriptome analysis of HIV-infected peripheral blood monocytes: gene transcripts and networks associated with neurocognitive functioning. J Neuroimmunol 2013;265:96–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zeller T, Wild P, Szymczak S, Rotival M, Schillert A, Castagne R, Maouche S, Germain M, Lackner K, Rossmann H, Eleftheriadis M, Sinning CR, Schnabel RB, Lubos E, Mennerich D, Rust W, Perret C, Proust C, Nicaud V, Loscalzo J, Hübner N, Tregouet D, Münzel T, Ziegler A, Tiret L, Blankenberg S, Cambien F.. Genetics and beyond—the transcriptome of human monocytes and disease susceptibility. PLoS One 2010;5:e10693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gilbert JM, Fitch KV, Grinspoon SK.. HIV-related cardiovascular disease, statins, and the REPRIEVE trial. Top Antivir Med 2015;23:146–149. [PMC free article] [PubMed] [Google Scholar]
- 39. Bacon MC, von Wyl V, Alden C, Sharp G, Robison E, Hessol N, Gange S, Barranday Y, Holman S, Weber K, Young MA.. The Women’s Interagency HIV study: an observational cohort brings clinical sciences to the bench. Clin Diagn Lab Immunol 2005;12:1013–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Barkan SE, Melnick SL, Preston-Martin S, Weber K, Kalish LA, Miotti P, Young M, Greenblatt R, Sacks H, Feldman J.. The Women’s interagency HIV study. Epidemiology 1998;9:117–125. [PubMed] [Google Scholar]
- 41. Hanna DB, Post WS, Deal JA, Hodis HN, Jacobson LP, Mack WJ, Anastos K, Gange SJ, Landay AL, Lazar JM, Palella FJ, Tien PC, Witt MD, Xue X, Young MA, Kaplan RC, Kingsley LA.. HIV infection is associated with progression of subclinical carotid atherosclerosis. Clin Infect Dis 2015;61:640–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hodis HN, Mack WJ, Lobo RA, Shoupe D, Sevanian A, Mahrer PR, Selzer RH, Liu C-R, Liu C-h, Azen SP; for the Estrogen in the Prevention of Atherosclerosis Trial Research Group. Estrogen in the prevention of atherosclerosis. A randomized, double-blind, placebo-controlled trial. Ann Intern Med 2001;135:939–953. [DOI] [PubMed] [Google Scholar]
- 43. Kaplan RC, Kingsley LA, Gange SJ, Benning L, Jacobson LP, Lazar J, Anastos K, Tien PC, Sharrett AR, Hodis HN.. Low CD4+ T-cell count as a major atherosclerosis risk factor in HIV-infected women and men. Aids Lond Engl 2008;22:1615–1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Trapnell C, Pachter L, Salzberg SL.. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 2009;25:1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wang L, Wang S, Li W.. RSeQC: quality control of RNA-seq experiments. Bioinformatics 2012;28:2184–2185. [DOI] [PubMed] [Google Scholar]
- 46. Robinson MD, McCarthy DJ, Smyth GK.. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010;26:139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Robinson MD, Oshlack A.. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol 2010;11:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Krämer A, Green J, Pollard J, Tugendreich S.. Causal analysis approaches in ingenuity pathway analysis. Bioinformatics 2014;30:523–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. McArdle S, Buscher K, Ehinger E, Pramod AB, Riley N, Ley K. PRESTO, a new tool for integrating large-scale -omics data and discovering disease-specific signatures. Biorxiv2018. 10.1101/302604. [DOI]
- 50. Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP.. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc National Acad Sci USA 2005;102:15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Buscher K, Ehinger E, Gupta P, Pramod AB, Wolf D, Tweet G, Pan C, Mills CD, Lusis AJ, Ley K.. Natural variation of macrophage activation as disease-relevant phenotype predictive of inflammation and cancer survival. Nat Commun 2017;8:16041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Reich M, Liefeld T, Gould J, Lerner J, Tamayo P, Mesirov JP.. GenePattern 2.0. Nat Genet 2006;38:500–501. [DOI] [PubMed] [Google Scholar]
- 53. A-González N, Castrillo A.. Liver X receptors as regulators of macrophage inflammatory and metabolic pathways. Biochim Biophys Acta 2011;1812:982–994. [DOI] [PubMed] [Google Scholar]
- 54. Jiang C, Ting AT, Seed B.. PPAR-gamma agonists inhibit production of monocyte inflammatory cytokines. Nature 1998;391:82–86. [DOI] [PubMed] [Google Scholar]
- 55. Luc G, Bard J-M, Juhan-Vague I, Ferrieres J, Evans A, Amouyel P, Arveiler D, Fruchart J-C, Ducimetiere P, Group PS.. C-reactive protein, interleukin-6, and fibrinogen as predictors of coronary heart disease. Arterioscler Thromb Vasc Biol 2003;23:1255–1261. [DOI] [PubMed] [Google Scholar]
- 56. Ridker PM, Rifai N, Stampfer MJ, Hennekens CH.. Plasma concentration of interleukin-6 and the risk of future myocardial infarction among apparently healthy men. Circulation 2000;101:1767–1772. [DOI] [PubMed] [Google Scholar]
- 57. Volpato S, Guralnik JM, Ferrucci L, Balfour J, Chaves P, Fried LP, Harris TB.. Cardiovascular disease, interleukin-6, and risk of mortality in older women. Circulation 2001;103:947–953. [DOI] [PubMed] [Google Scholar]
- 58. Krausgruber T, Blazek K, Smallie T, Alzabin S, Lockstone H, Sahgal N, Hussell T, Feldmann M, Udalova IA.. IRF5 promotes inflammatory macrophage polarization and TH1-TH17 responses. Nat Immunol 2011;12:231–238. [DOI] [PubMed] [Google Scholar]
- 59. Langlais D, Barreiro LB, Gros P.. The macrophage IRF8/IRF1 regulome is required for protection against infections and is associated with chronic inflammation. J Exp Med 2016;213:585–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Harman AN, Lai J, Turville S, Samarajiwa S, Gray L, Marsden V, Mercier SK, Mercier S, Jones K, Nasr N, Rustagi A, Cumming H, Donaghy H, Mak J, Gale M, Churchill M, Hertzog P, Cunningham AL.. HIV infection of dendritic cells subverts the IFN induction pathway via IRF-1 and inhibits type 1 IFN production. Blood 2011;118:298–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Coccia EM, Russo ND, Stellacci E, Testa U, Marziali G, Battistini A.. STAT1 activation during monocyte to macrophage maturation: role of adhesion molecules. Int Immunol 1999;11:1075–1083. [DOI] [PubMed] [Google Scholar]
- 62. Zhang Y-H, He M, Wang Y, Liao A-H.. Modulators of the balance between M1 and M2 macrophages during pregnancy. Front Immunol 2017;8:120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Visintin A, Mazzoni A, Spitzer JH, Wyllie DH, Dower SK, Segal DM.. Regulation of Toll-like receptors in human monocytes and dendritic cells. J Immunol 2001;166:249–255. [DOI] [PubMed] [Google Scholar]
- 64. Deng S, Yu K, Zhang B, Yao Y, Wang Z, Zhang J, Zhang X, Liu G, Li N, Liu Y, Lian Z.. Toll-like receptor 4 promotes NO synthesis by upregulating GCHI expression under oxidative stress conditions in sheep monocytes/macrophages. Oxid Med Cell Longev 2015;2015:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Zambelli F, Pesole G, Pavesi G.. Pscan: finding over-represented transcription factor binding site motifs in sequences from co-regulated or co-expressed genes. Nucleic Acids Res 2009;37:W247–W252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Wang N, Liang H, Zen K.. Molecular mechanisms that influence the macrophage m1-m2 polarization balance. Front Immunol 2014;5:614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Suresh R, Li X, Chiriac A, Goel K, Terzic A, Perez-Terzic C, Nelson TJ.. Transcriptome from circulating cells suggests dysregulated pathways associated with long-term recurrent events following first-time myocardial infarction. J Mol Cell Cardiol 2014;74:13–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Kiliszek M, Burzynska B, Michalak M, Gora M, Winkler A, Maciejak A, Leszczynska A, Gajda E, Kochanowski J, Opolski G.. Altered gene expression pattern in peripheral blood mononuclear cells in patients with acute myocardial infarction. PLoS One 2012;7:e50054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Ayari H, Bricca G.. Identification of two genes potentially associated in iron-heme homeostasis in human carotid plaque using microarray analysis. J Biosci 2013;38:311–315. [DOI] [PubMed] [Google Scholar]
- 70. Lee K, Santibanez-Koref M, Polvikoski T, Birchall D, Mendelow AD, Keavney B.. Increased expression of fatty acid binding protein 4 and leptin in resident macrophages characterises atherosclerotic plaque rupture. Atherosclerosis 2013;226:74–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Folkersen L, Persson J, Ekstrand J, Agardh HE, Hansson GK, Gabrielsen A, Hedin U, Paulsson-Berne G.. Prediction of ischemic events on the basis of transcriptomic and genomic profiling in patients undergoing carotid endarterectomy. Mol Med 2012;18:669–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Triant VA. Cardiovascular disease and HIV infection. Curr HIV/AIDS Rep 2013;10:199–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Hanna DB, Ramaswamy C, Kaplan RC, Kizer JR, Daskalakis D, Anastos K, Braunstein SL.. Sex- and poverty-specific patterns in HIV-associated cardiovascular disease mortality in New York City, 2007-2017. Clin Infect Dis 2019. 10.1093/cid/ciz852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Mueller KAL, Hanna DB, Ehinger E, Xue X, Baas L, Gawaz MP, Geisler T, Anastos K, Cohen MH, Gange SJ, Heath SL, Lazar JM, Liu C, Mack WJ, Ofotokun I, Tien PC, Hodis HN, Landay AL, Kaplan RC, Ley K.. Loss of CXCR4 on non-classical monocytes in participants of the Women’s Interagency HIV Study (WIHS) with subclinical atherosclerosis. Cardiovasc Res 2019;115:1029–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Shi C, Pamer EG.. Monocyte recruitment during infection and inflammation. Nat Rev Immunol 2011;11:762–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Ingersoll MA, Platt AM, Potteaux S, Randolph GJ.. Monocyte trafficking in acute and chronic inflammation. Trends Immunol 2011;32:470–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Kulkarni M, Bowman E, Gabriel J, Amburgy T, Mayne E, Zidar DA, Maierhofer C, Turner AN, Bazan JA, Koletar SL, Lederman MM, Sieg SF, Funderburg NT.. Altered monocyte and endothelial cell adhesion molecule expression is linked to vascular inflammation in human immunodeficiency virus infection. Open Forum Infect Dis 2016;3:ofw224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Mihos CG, Pineda AM, Santana O.. Cardiovascular effects of statins, beyond lipid-lowering properties. Pharmacol Res 2014;88:12–19. [DOI] [PubMed] [Google Scholar]
- 79. Kidani Y, Bensinger SJ.. Liver X receptor and peroxisome proliferator-activated receptor as integrators of lipid homeostasis and immunity. Immunol Rev 2012;249:72–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Duque GA, Descoteaux A.. Macrophage cytokines: involvement in immunity and infectious diseases. Front Immunol 2014;5:491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Hodis HN, Mack WJ, LaBree L, Selzer RH, Liu CR, Liu CH, Azen SP.. The role of carotid arterial intima-media thickness in predicting clinical coronary events. Ann Intern Med 1998;128:262–269. [DOI] [PubMed] [Google Scholar]
- 82. Hanna DB, Moon J-Y, Haberlen SA, French AL, Palella FJ, Gange SJ, Witt MD, Kassaye S, Lazar JM, Tien PC, Feinstein MJ, Kingsley LA, Post WS, Kaplan RC, Hodis HN, Anastos K.. Carotid artery atherosclerosis is associated with mortality in HIV-positive women and men. AIDS 2018;32:2393–2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article will be shared on reasonable request to the corresponding author.