Abstract
Tumor-infiltrating lymphocytes (TIL) have potential prognostic value in melanoma and have been considered for inclusion in the American Joint Committee on Cancer (AJCC) staging criteria. However, inter-observer discordance continues to prevent the adoption of TIL into clinical practice. Computational image analysis offers a solution to this obstacle, representing a methodological approach for reproducibly counting TIL. We sought to evaluate the ability of a TIL-quantifying machine learning algorithm to predict survival in primary melanoma. Digitized hematoxylin and eosin (H&E) slides from prospectively-enrolled patients in the NYU melanoma database were scored for % TIL using machine learning and manually graded by pathologists using Clark’s model. We evaluated the association of % TIL with recurrence-free survival (RFS) and overall survival (OS) using Cox proportional hazards modeling and concordance indices. Discordance between algorithmic and manual TIL quantification was assessed with McNemar’s test and visually by an attending dermatopathologist. 453 primary melanoma patients were scored using machine learning. Automated % TIL scoring significantly differentiated survival using an estimated cutoff of 16.6% TIL (Log Rank P<0.001 for RFS; P=0.002 for OS). % TIL was associated with significantly longer RFS (adjusted HR = 0.92 [0.84–1.00] per 10% increase in % TIL) and OS (adjusted HR = 0.90 [0.83–0.99] per 10% increase in % TIL). In comparison, a subset of the cohort (n=240) was graded for TIL by melanoma pathologists. However, TIL did not associate with RFS between groups (P>0.05) when categorized as brisk, non-brisk, or absent. A standardized and automated % TIL scoring algorithm can improve the prognostic impact of TIL. Incorporation of quantitative TIL scoring into the AJCC staging criteria should be considered.
INTRODUCTION
Tumor-infiltrating lymphocytes (TIL), a surrogate for the host immune response against tumor cells, has been proposed as a predictor of immunotherapy response and long-term survival in many cancers, including melanoma [1–8]. Yet, despite its potential, the use of TIL as a prognostic biomarker in the American Joint Committee on Cancer (AJCC) staging criteria is precluded by the inherently subjective nature of extant grading systems such as Clark’s methodology, which leads to poor interrater consistency [9–17]. Thus, despite the release of consensus guidelines for TIL scoring and development of immunohistochemistry-based (IHC) quantification, our group and others have shown that human-reliant assessments of TIL on their own do not reliably predict survival in melanoma [11, 18, 19]. The field of dermatopathology has therefore explored whether machine learning algorithms can be employed as automated, objective, and easily scalable assays for quantifying TIL.
Automated cell segmentation and detection algorithms have recently been investigated as a method to standardize TIL scoring in digitized hematoxylin and eosin (H&E) slides [2, 10, 20–24]. Although the initial training and optimization of computer vision algorithms are cumbersome, implementation typically utilizes few resources and can therefore potentially decrease healthcare costs and physician workload [25]. Machine learning algorithms also derive power from its ability to detect data patterns not discernible to humans, paving the way for novel scientific developments [25, 26]. Despite the growing prominence of machine learning-based techniques in medical literature, however, there are limited reports of successful clinical implementation [25, 27]. Several challenges hindering clinical adoption of machine learning algorithms have been identified [25–28], with ubiquitous concerns over external validation, contextualization, technical difficulties, and propagation of endemic biases. Rigorous testing and refinement is therefore required to ensure accuracy and clinical applicability, especially when considering the ethical implications of improper algorithmic usage [25].
Recent work showed that the novel neural network-based classifier NN192 is capable of generating an automated TIL score from digital H&Es, termed % TIL, that predicts disease-specific overall survival (DSOS) in melanoma [24]. Our objective in this study was to compare the prognostic accuracy of an automated % TIL score using the NN192 algorithm to that of Clark’s grading, the pathologist-based standard for TIL assessment. We then performed an in-depth pathological analysis of discordant cases between the human- and machine-based modalities to facilitate algorithmic refinement. In doing so, our goal was to uncover an optimal integration strategy of % TIL into the AJCC staging criteria, bridging the “AI chasm” that befalls the majority of machine learning methodologies [28].
MATERIALS AND METHODS
Patient cohorts and tissue preparation
457 patients were included in this retrospective analysis, with a median follow-up time of 87.2 months (Interquartile range [IQR]: 56.9–118.7). All patients were accrued to the IRB-approved New York University Interdisciplinary Melanoma Cooperative Group (NYU IMCG) protocol from 2002–2017. The IMCG program has prospectively enrolled more than 4,300 patients since 2002. Developed protocols and Standard Operating Procedure (SOP) guide patient biospecimen and clinicopathological data collection, with no identifying information used in publications [29]. Patients were included on the basis of available, digitized slides at the time of the study, with an enrichment of stage II and III disease, as well as for absent and brisk cases. This was done in order to test the generalizability of an algorithm that was trained using a different distribution of disease severity and to facilitate concordance analyses between the TIL quantification methodologies [24]. An unequivocal diagnosis of melanoma in each case was determined by the presence of prominent cytological atypia, mitotic activity, a patchy asymmetrical host response/inflammatory infiltrate, and an in situ component. BAP1 inactivated tumors were excluded. The reporting guidelines for tumor marker prognostic studies (REMARK) were followed in this study [30]. Representative tumor areas on 457 formalin-fixed paraffin embedded (FFPE) H&E-stained slides were demarcated by an attending pathologist for accurate algorithm implementation solely within these regions. For validation purposes, two pathologists graded a subset of 240 slides out of the 457 using Clark’s model. TIL were graded brisk if they were present throughout or infiltrating across the entire base of the vertical growth phase (VGP), non-brisk if they were in ≥ 1 foci of the VGP, or absent if none were associated with the VGP, as previously described [31].
Digital Image Analysis (DIA)-generated % TIL scores
457 FFPE H&E-stained slides were digitized using an Aperio ScanScope AT2 (Leica Biosystems, Wetzlar, Germany) at 20x magnification to generate whole slide images (WSI), with a resolution of 0.503 microns per pixel. For accurate segmentation of cells regardless of the temporal differences in staining, pixel values in each image were first optimized to ensure white balancing and to prevent color oversaturation, while refinement of H&E stain estimates was achieved using the “estimated stain vectors” command in the open source software QuPath [32]. Cell segmentation, which explicitly labels the pixels of a given cell, was then performed on a manually selected region of interest (ROI) using watershed cell detection with the following settings: Detection image: hematoxylin OD; requested pixel size: 0.5 μm; background radius: 8 μm; median filter radius: 0 μm; sigma: 1.5 μm; minimum cell area: 10 μm2; maximum cell area: 400 μm2; threshold 0.1; maximum background intensity [24, 32]. All ROIs were chosen within the tumor areas demarcated by attending pathologists and were re-verified after digital selection. Only 1 ROI was necessary to encompass the entirety of tumors ≤ 1 mm2 in histologic area (Fig. 1). In order to circumvent limitations in computational processing, several ROIs were selected and summed to fully incorporate tumors that were 1 to 5 mm2. The largest tumors (> 5 mm2) could not be completely selected even when using multiple ROIs. A row of up to 10 ROIs, ranging from the basal to apical layer of the tumor, was used instead as a representative area. After detection, smoothed object features at 25 and 50 μm radius were applied to aid in the classification of cells into tumor cells, TIL, stromal cells, or others (i.e. false detections, background). Automated TIL classification and quantification were performed using the neural network-based classifier “NN192,” currently available at GitHub (Fig. 2) [24]. The NN192 algorithm calculated the percentage of machine defined TIL (% TIL) using the formula: (TIL/[TIL + Tumor cells]) × 100. We then compared the concordance between Clark’s grading and automated % TIL scoring. Cases graded as absent and scored as high % TIL by the algorithm, as well as cases graded brisk and scored as low % TIL, were deemed discordant and examined in detail by an IMCG dermatopathologist to determine the reason for the discrepancy.
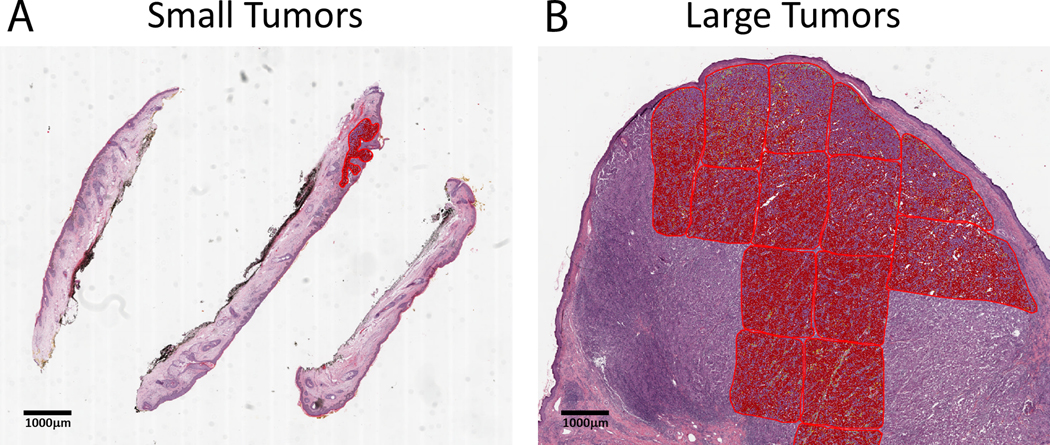
Fig. 1. Digital selection of regions of interest (ROIs) in QuPath relies on the size of the tumor.
(a) Melanoma tumors that are ≤1 mm2 in histologic area can be assessed using a single fully encompassing ROI. (b) Larger tumors (>1 mm2) require the selection and averaging of multiple, smaller ROIs for cell segmentation and % TIL quantification due to computational limitations.
Fig. 2. The NN192 classifier assists in the visualization and quantification of multiple cell types.
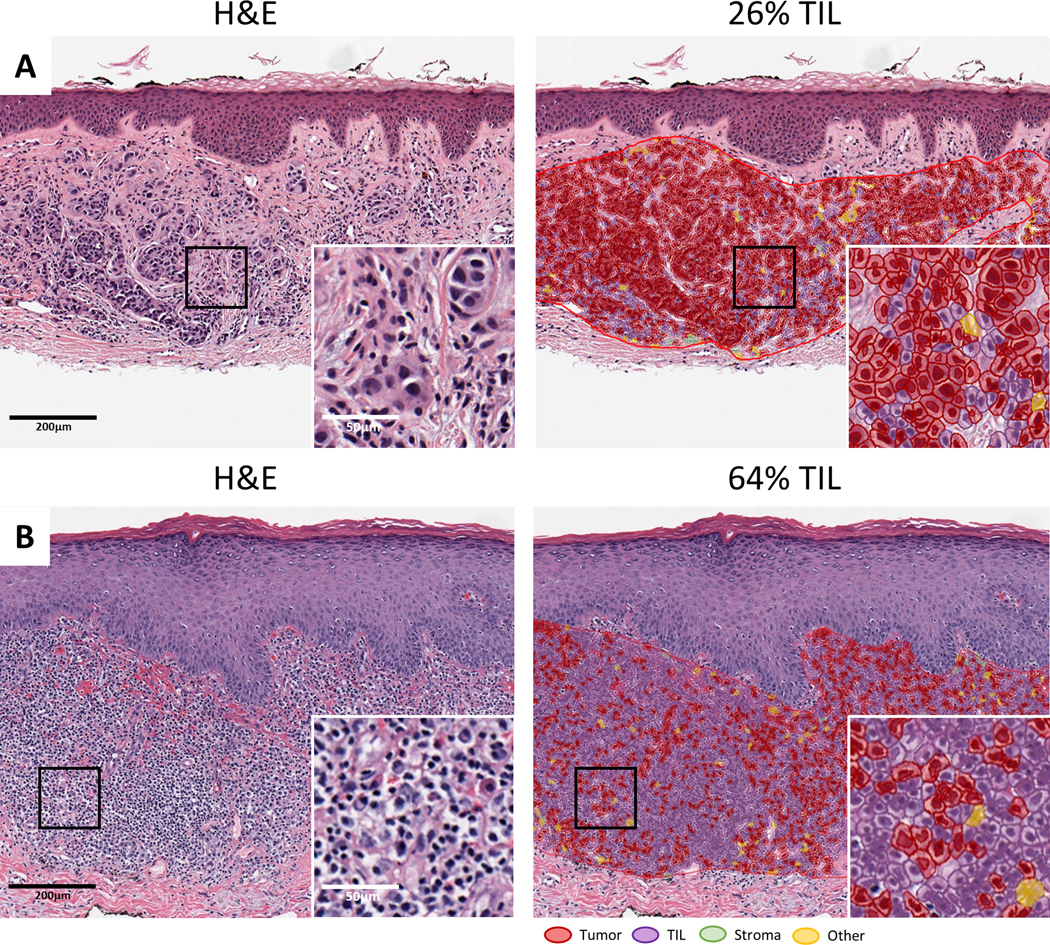
(a-b) Two representative melanoma cases are shown, with the 20x H&E digitized slide on the left and the same image with the NN192 classifier overlay applied on the right. Classification of cells are as follows: red denotes tumor cells, purple indicates TIL, green is stroma, and yellow signifies other components, such as false detections or background.
Statistical Analysis
Continuous and categorical variables are presented as means with standard deviations and as frequencies with proportions, respectively. We used Youden’s index to calculate the optimal threshold of high versus low % TIL groups for differentiating patient survival outcomes in the NYU patient cohort, and compared it to the recommended cutoff (16.6% TIL) generated from an independent cohort [24, 33]. Recurrence-free survival (RFS) was calculated as the length of time between initial diagnosis and first recurrence or death, while overall survival (OS) was defined as time from diagnosis until death from any cause. Kaplan-Meier curves were generated and compared by log-rank test between the high and low % TIL groups. We used Cox proportional hazards models to assess the correlation between % TIL, as a continuous covariate, with RFS and OS outcomes. Multivariable Cox regression was used to assess the prognostic significance of % TIL independent of other covariates. % TIL, age, gender, and stage were included as candidate covariates. We did not include Breslow thickness or ulceration as they are included in AJCC staging criteria. Backward stepwise model selection was used to derive the final model with covariates of p-values less than 0.05. The concordance index (C-index), which is similar to area under the curve for binary outcomes, was used to indicate discriminatory ability to predict RFS and OS, respectively. A value of 0.5 indicates that the model has no discriminatory ability, and a value of 1.0 indicates that the model has perfect discrimination ability. A nonparametric test was used to compare C-indices from a Cox regression with stage only to a model with both stage and % TIL score [34]. McNemar’s test was used to assess concordance between the TIL quantification methodologies, juxtaposing pathologist-based Clark’s grading with that of % TIL algorithmic scoring. Concordance was defined as absent-low % TIL (<16.6%) and brisk-high % TIL (≥16.6%). 32 slides deemed as non-brisk were excluded from the concordance analysis as they could not be directly compared to either the high or low % TIL cohorts, leading to a total of 208 out of 240 cases. All tests were two-sided and the level of significance was set at P<0.05. All data were analyzed using R version 3.6.2 (https://www.r-project.org/).
RESULTS
Patient characteristics
453 out of 457 digitized H&E slides were successfully analyzed for % TIL by the NN192 algorithm. The remaining 4 slides were unable to be analyzed for % TIL due to high concentrations of melanin in the tumor image. A summary of clinicopathological features for the 453 patients with primary melanoma included in this study is shown in Table 1. Of the 453 primary melanoma patients scored using automated TIL assessment, 17.9% (n=81) were stage I, 42.4% (n=192) were stage II, and 39.7% (n=180) stage III. The stage distribution of our cohort differs from that seen in the study by Acs et al., in which the proportion of stage III patients was 19.8% (59/76) at its highest [24]. The threshold of % TIL most robust at separating patient survival in our cohort was identified at 16.24% TIL by Youden’s Index. This is similar to and provides external data validation for the recommended 16.6% TIL threshold generated by Acs et al., with only a 6-patient difference between the two cutoffs [24]. Therefore, 16.6% TIL is the threshold used in the rest of the paper. 201 (44.4%) patients were classified as low % TIL, with the remaining 252 (55.6%) classified as high % TIL. The distribution of sex, age, sentinel lymph node status, and frequency of adjuvant therapy between the low (<16.6) and high (≥16.6) % TIL patients is comparable. However, high % TIL patients had thinner melanomas than low % TIL patients (3.2 vs 4.7 mm, P<0.001), as well as a lower frequency of ulceration (48.4 vs 58.2%, P=0.038). High % TIL patients were also more skewed towards stage I compared to low % TIL patients (24.6 vs 9.5%, P<0.001). As expected, the majority of high and low % TIL patients were graded by pathologists as brisk and absent, respectively (58.4% and 60.5%). Median follow-up time was 91.3 months for high % TIL patients and 80.0 months for the low % TIL patients.
Table 1.
Clinicopathologic characteristics of 453 primary melanoma patients separated into high and low % TIL cohortsa
| Low % TIL (n=201) | High % TIL (n=252) | P-value | |
|---|---|---|---|
| Male sex (%) | 121 (60.2) | 146 (57.9) | 0.627 |
| Age (Mean ± SD) | 62.7 ± 16.3 | 60.8 ± 16.9 | 0.228 |
| Thickness (Mean mm ± SD) | 4.7 ± 4.6 | 3.2 ± 3.3 | <0.001 |
| Ulceration present (%) | 117 (58.2) | 122 (48.4) | 0.038 |
| SLN biopsy performed (%) | 178 (88.6) | 184 (73.3) | <0.001 |
| Positive SLN status (%) | 77 (38.5) | 83 (32.9) | 0.219 |
| AJCC 8th Edition staging (%) | <0.001 | ||
| I | 19 (9.5) | 62 (24.6) | |
| II | 93 (46.3) | 99 (39.3) | |
| III | 89 (44.3) | 91 (36.1) | |
| Clark’s grade | <0.001 | ||
| Absent | 52 (60.5) | 46 (29.9) | |
| Non-brisk | 14 (16.3) | 18 (11.7) | |
| Brisk | 20 (23.3) | 90 (58.4) | |
| Adjuvant therapy received (%) | 69 (34.3) | 71 (28.2) | 0.159 |
| Follow-up (Months, median [IQR]) | 80.0 [54.5 – 112.9] | 91.3 [58.9 – 119.6] |
Using a threshold of 16.6% TIL
% TIL thresholds significantly improve prediction of survival outcomes compared to Clark’s grading
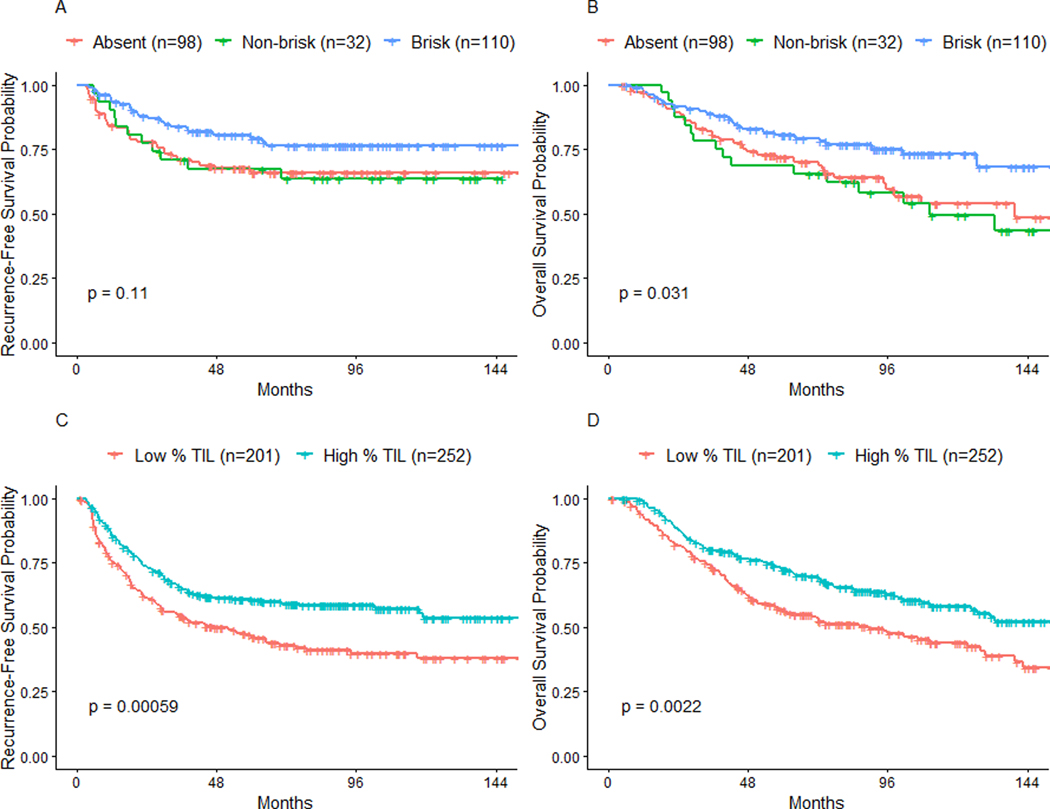
Assessment of TIL using semi-quantitative Clark’s grading (absent, non-brisk, brisk) did not significantly differentiate RFS (Log rank P=0.110; Fig. 3a). Brisk-graded melanoma patients had improved OS compared to absent and non-brisk patients (Log rank P=0.03). However, no differences in OS were observed between absent and non-brisk patients (Fig. 3b). In contrast, the automated 16.6% TIL threshold significantly differentiated patient survival outcomes. High % TIL patients had a more favorable prognosis, with significantly longer RFS (Log rank P< 0.001) and OS (Log rank P=0.002) compared to patients categorized as low % TIL (Fig. 3c–d). Median RFS and OS for high % TIL patients was consequently longer at 155.0 and 155.0 months, versus 48.0 and 89.0 months, when compared to the low % TIL cohort.
Fig. 3. A 16.6% TIL threshold more significantly differentiates patient survival than Clark’s TIL grades.
(a) When using Clark’s grades, Kaplan-Meier curves of RFS do not significantly separate. (b) Brisk-graded patients perform significantly better in terms of OS; however, non-brisk and absent patients are unable to be significantly differentiated. (c-d) When applying a threshold of 16.6% TIL, patients above the threshold have significantly better RFS and OS than those below.
% TIL is an independent prognostic variable and improves the prognostic capability of AJCC 8th Edition staging
When analyzed as a continuous covariate by Cox regression, % TIL score was associated with better RFS (unadjusted HR = 0.85 [95% CI: 0.78–0.92] per 10% increase in % TIL, P<0.001) and OS (unadjusted HR=0.86 [0.79–0.94] per 10% increase in % TIL, P=0.001) in univariate analyses. In the selected multivariable Cox regression models, only % TIL and stage remained significant. % TIL remained a significant prognostic factor for better survival outcomes in multivariate Cox proportional hazards models adjusting for stage (adjusted HR = 0.92 [0.84–1.00] per 10% increase in % TIL, P=0.05 for RFS; adjusted HR = 0.90 [0.83–0.99] per 10% increase in % TIL, P=0.026 for OS). Compared to a Cox regression with stage only, the addition of % TIL significantly increased prognostic discrimination ability for both RFS (C-index improved from 0.68 to 0.70, P=0.02) and OS (C-index improved from 0.62 to 0.64, P=0.01).
Discordance between Clark’s TIL grading and % TIL scoring
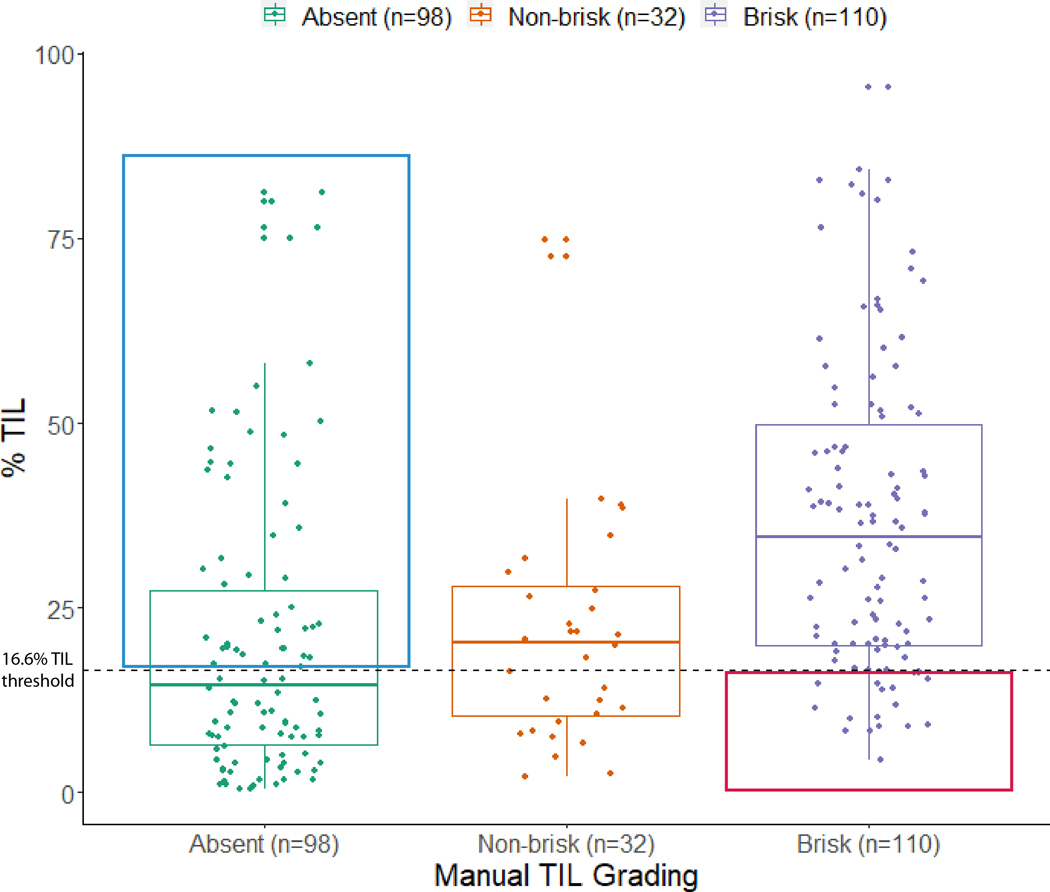
The overall discordance rate between Clark’s grading and automated % TIL scoring is 31.7% (66/208; P=0.002 from McNemar’s test assessing concordance of the two systems; Fig. 4). 32 out of 240 slides were non-brisk and excluded from the analysis. We examined the differences in survival outcomes of the discordant cases. Absent-graded, high % TIL patients (discordant) showed better RFS (Log rank P=0.004) and OS (Log rank P=0.095) than absent-graded, low % TIL patients (concordant; Sup. Fig. 1). In contrast, there were no differences in RFS (Log rank P=0.840) or OS (Log rank P=0.590) between low % TIL (discordant) and high % TIL (concordant) brisk-graded patients. We further investigated the reasons for discordance seen in the 46 absent-graded, high % TIL-scored tumors through detailed pathologic assessment (Fig. 4). Overcalling of inflammatory cells by the NN192 algorithm accounted for 50% (23/46) of the discordance, with most of these cases secondary to the categorization of tumor cells as TIL (Fig. 5a). Undercalling of tumor cells by the algorithm was a subsequent cause for discordance in 19.6% (9/46) slides, which was predominantly due to the classification of tumor cells as stromal cells or due to pigmentation (Fig. 5b–c). Slides containing small, thin melanomas (≤1 mm) with a limited invasive component led to a high % TIL designation in 15.2% (7/46) of absent-graded cases, despite having few absolute TIL (Fig. 5d). The remaining 15.2% (7/46) of discordant slides were re-graded as non-brisk after a second review.
Fig. 4. Juxtaposing Clark’s TIL grades with their respective % TIL scores reveals discordant samples.
46.9% (46/98) of absent-graded slides are scored as high % TIL (blue box), while 18.2% (20/110) of brisk-graded images are categorized as low % TIL (red box).
Fig. 5. The discordance seen in absent-graded slides is mainly due to misclassification of cells by the NN192 algorithm.
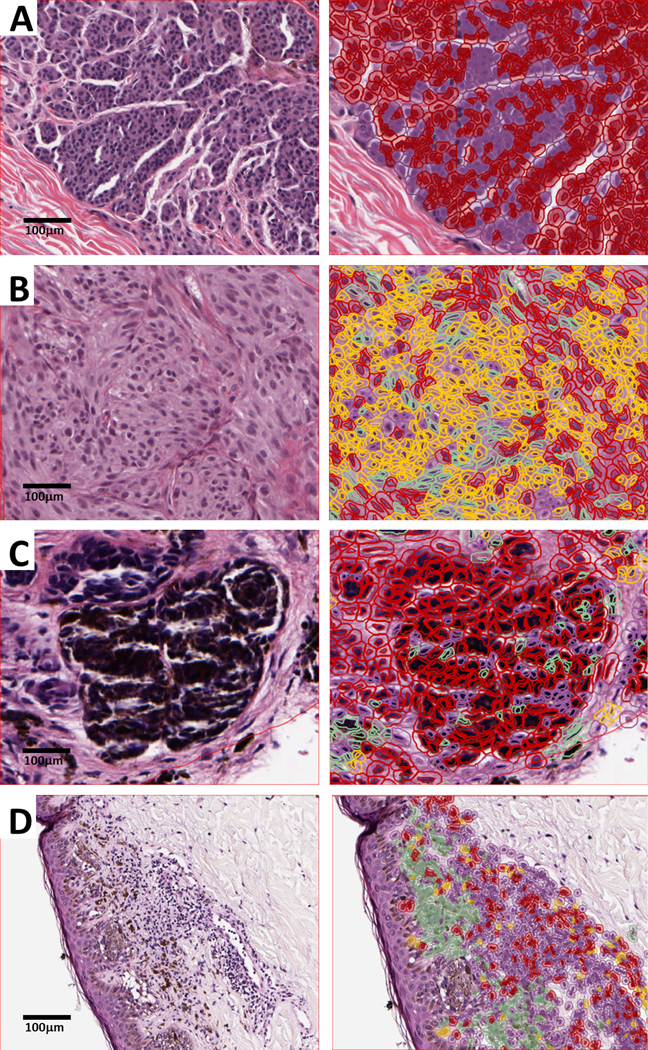
Representative cases are shown, with the 20x H&E digitized slide on the left and the same image with the NN192 classifier overlay applied on the right. Classification of cells are as follows: red denotes tumor cells, purple indicates TIL, green is stroma, and yellow signifies other components, such as false detections or background. (a) Tumor cells are labeled as “TIL” in this example of a nevoid melanoma. (b) Most tumor cells are labeled as “stromal” or “other” cells in this digitized image of a spindle cell melanoma. (c) Coarse pigmentation in macrophages can be interpreted as tumor cells, interfering with TIL counting by the algorithm, while (d) thin melanomas with a limited invasive component can impact the % TIL calculation. Notice the relative abundance of inflammatory cells and the scarcity of tumor cells, leading to the increased % TIL calculation.
DISCUSSION
The utility of TIL as a prognostic biomarker is equivocal due to its subjective grading system and ensuing inter-observer variability [17, 36, 37]. We have previously shown, for instance, that both Clark’s grading and IHC-based TIL quantitation were unable to significantly differentiate survival, particularly between non-brisk and absent-graded melanoma patients [11]. Machine learning modalities hold promise for alleviating this variability and can facilitate the inclusion of TIL into prognostic criteria, such as AJCC staging [2, 10, 17, 24, 36]. Notably, Acs et al. developed a TIL-quantifying neural network capable of predicting DSOS in melanoma with a % TIL threshold of 16.6%. In this study, we first sought to evaluate the clinical significance of % TIL using the NN192 algorithm within an independent cohort, particularly in the context of current methodologies. We found a consistent % TIL threshold using our cohort (Fig. 3; 16.24 vs 16.6% TIL), despite a higher staged study population, and showed improved survival differentiation when compared to Clark’s grading [24]. Furthermore, we confirmed the validity of % TIL as an independent prognostic marker when adjusting for stage, which accounts for the significant differences in thickness and ulceration seen between the high and low % TIL groups in univariate analyses. Our work validating % TIL thereby facilitates a major step towards clinical application unseen by most machine learning modalities [25].
Secondly, we aimed to enhance the accuracy of the algorithm and to generate a digital pathology workflow for optimal clinical integration. A major obstacle surrounding clinical application of machine learning algorithms revolves around the notion that it is a “black box” in which clinicians are not privy to the various factors considered by the algorithm [38]. This is of particular importance considering that melanoma is a notoriously morphologically heterogeneous tumor that exhibits diverse cytomorphologic and architectural patterns, therefore posing a great technical challenge not only for dermatopathologists, but for image analysis-based machine learning methodologies [20, 39]. To efficiently identify algorithmic inaccuracies in cell classification, we isolated the cases where TIL quantification was discrepant between the human- and machine-based methodologies. Our results indicate that absent-graded slides (46.9%) were more discordantly assessed by the algorithm than brisk-graded patients (18.2%; P=0.002; Fig. 4). We believe that the higher discordance rate seen in the absent patients is due to the lower absolute TIL count, making the % TIL calculation more susceptible to any changes in cell classification. Therefore, we propose that an absolute quantification of TIL be incorporated in future outputs of the algorithm to provide contextualization of the final % TIL measurement.
In the context of our findings, we then performed a focused pathologic assessment of absent-graded, high % TIL cases. Most of the discordant cases were due to overcalling of TIL in nevoid melanomas, a rare melanoma variant [40]. As nevoid melanomas have smaller nuclei than conventional melanomas, which can simulate the appearance of inflammatory cells, the algorithmic misclassifications are understandable (Fig. 5a) [41]. Spindle cell melanomas, another rare variant, similarly led to the misidentification of tumor cells by the algorithm. As its name suggests, these melanomas have nuclear features that can resemble those of stromal fibroblasts, resulting in the misclassification of tumor cells as stromal cells instead (Fig. 5b) [42]. It should be noted, however, that these unique and rare morphologies only account for 6 to 10% of all cases, highlighting the utility of the algorithm for the majority of melanomas. Furthermore, 15.2% of the discordant absent cases were deemed to have the potential of being graded as non-brisk, highlighting the inter-observer variability inherent to human assessment and the functionality of this algorithm in standardizing pathologic assessment [40, 42]. For a minor subset of small, thin melanomas, the limited dermal invasive component present led to a disproportionately high percentage of TIL to tumor cells (Fig. 5d). This finding suggests that the usage of this algorithm may need to be further explored in focused studies on thin melanomas (≤1 mm2). Pigmentation also interfered with the identification of tumor cells, leading to the inability to analyze some cases.
Of note, we examined the impact of discordance on survival and found that the absent-graded, high % TIL discordant patients had better RFS than those who were concordant (Log Rank P=0.004; Sup. Fig. 1a). This result suggests that % TIL scoring may be superior at differentiating survival outcomes for absent-graded patients. This superiority may be secondary to the ability of machine learning-based methodologies to perceive data patterns not readily visible to humans, intimating that the benefit of this algorithm is not solely bound to the accurate calling of individual cells. However, the low discordance rate within the brisk-graded samples prevented us from making definitive conclusions about this subpopulation and this topic should be further studied in the future.
Other limitations and technical challenges must also be acknowledged in this study. A % TIL threshold does not universally discriminate survival outcomes within stages (Sup. Fig. 2). For stage II and III, patients with high % TIL trended towards better survival than patients with low % TIL, although it was not significant due to sample size constraints upon division of our cohort. Survival outcomes for stage I patients with high % TIL, on the other hand, did not differ from those with low % TIL. Furthermore, fewer stage I patients were categorized as low % TIL (19/81) compared to stage II (93/192) and stage III (89/180) patients. These results suggest a less discriminative power for % TIL in stage I patients. Detailed guided training was also required to calibrate the digital image analysis software (QuPath) before usage to prevent inaccurate cell segmentation and classification. Lastly, computational limitations prevented the analysis of high-resolution digitized images of the largest tumors (>5 mm2) in a singular ROI [20]. Interobserver discordance in ROI selection for these cases may lead to variation in % TIL calculation.
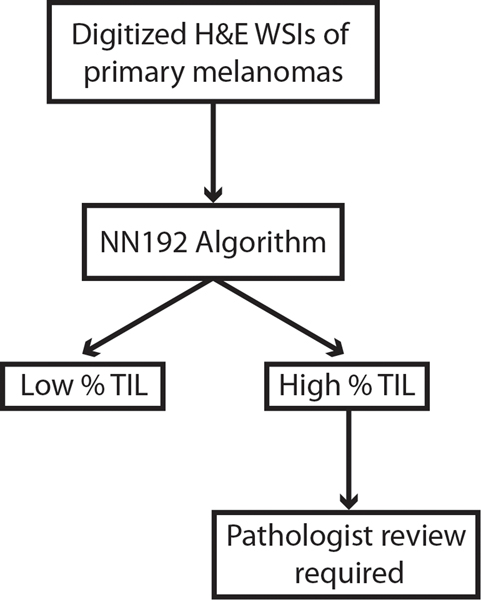
With all results considered, we believe that the survival predictions for cases deemed as low % TIL by the NN192 algorithm appear to be trustworthy, while pathologist supervision and further training will likely be required for the high % TIL cases (Fig. 6). By incorporating human supervision into the workflow, this can increase reliability and efficiency of TIL quantification, while also accounting for sensitivity towards rare variants of melanoma. This approach, termed “human-in-the-loop AI,” integrates the best of human intelligence and machine learning algorithms to collectively outperform either modality individually, a finding reported in prior machine learning studies in the fields of radiology and pathology [43–46]. We believe that our work will help bring the NN192 algorithm closer to clinical application by facilitating the incorporation of % TIL into the AJCC staging criteria. This is of particular significance considering the few examples of peer-reviewed and externally validated machine learning algorithms in use today [28, 47]. The next step following additional training, based on our observations, will be to validate this algorithm prospectively in order to further optimize its clinical applications and improve melanoma prognostication for our patients.
Fig. 6. A digital workflow for the optimal clinical usage of the NN192 algorithm.
Digitized H&E slides should first be analyzed by the NN192 classifier in order to increase efficiency and reliability of TIL quantification. Following categorization as either high or low % TIL, high % TIL slides should be evaluated in detail by pathologists.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the patients and their families who participated in this study. This work was supported by the NYU Melanoma SPORE grant (P50CA225450), the Perlmutter Cancer Center Support grant (P30CA016087), and the Melanoma Research Alliance.
Footnotes
CONFLICT OF INTEREST
The authors declare that they have no conflicts of interest.
REFERENCES
- 1.Clemente CG, Mihm MC Jr., Bufalino R, Zurrida S, Collini P, Cascinelli N. Prognostic value of tumor infiltrating lymphocytes in the vertical growth phase of primary cutaneous melanoma. Cancer. 1996;77:1303–10. [DOI] [PubMed] [Google Scholar]
- 2.Wong PF, Wei W, Smithy JW, Acs B, Toki MI, Blenman KRM, et al. Multiplex Quantitative Analysis of Tumor-Infiltrating Lymphocytes and Immunotherapy Outcome in Metastatic Melanoma. Clin Cancer Res. 2019;25:2442–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thomas NE, Busam KJ, From L, Kricker A, Armstrong BK, Anton-Culver H, et al. Tumor-infiltrating lymphocyte grade in primary melanomas is independently associated with melanoma-specific survival in the population-based genes, environment and melanoma study. J Clin Oncol. 2013;31:4252–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barroso-Sousa R, Keenan TE, Pernas S, Exman P, Jain E, Garrido-Castro AC, et al. Tumor mutational burden and PTEN alterations as molecular correlates of response to PD-1/L1 blockade in metastatic triple-negative breast cancer. Clin Cancer Res. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Antohe M, Nedelcu RI, Nichita L, Popp CG, Cioplea M, Brinzea A, et al. Tumor infiltrating lymphocytes: The regulator of melanoma evolution. Oncol Lett. 2019;17:4155–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee N, Zakka LR, Mihm MC Jr., Schatton T. Tumour-infiltrating lymphocytes in melanoma prognosis and cancer immunotherapy. Pathology. 2016;48:177–87. [DOI] [PubMed] [Google Scholar]
- 7.Schatton T, Scolyer RA, Thompson JF, Mihm MC Jr. Tumor-infiltrating lymphocytes and their significance in melanoma prognosis. Methods Mol Biol. 2014;1102:287–324. [DOI] [PubMed] [Google Scholar]
- 8.Uryvaev A, Passhak M, Hershkovits D, Sabo E, Bar-Sela G. The role of tumor-infiltrating lymphocytes (TILs) as a predictive biomarker of response to anti-PD1 therapy in patients with metastatic non-small cell lung cancer or metastatic melanoma. Med Oncol. 2018;35:25. [DOI] [PubMed] [Google Scholar]
- 9.Larsen TE, Grude TH. A retrospective histological study of 669 cases of primary cutaneous malignant melanoma in clinical stage I. 3. The relation between the tumour-associated lymphocyte infiltration and age and sex, tumour cell type, pigmentation, cellular atypia, mitotic count, depth of invasion, ulceration, tumour type and prognosis. Acta Pathol Microbiol Scand A. 1978;86A:523–30. [PubMed] [Google Scholar]
- 10.Klauschen F, Muller KR, Binder A, Bockmayr M, Hagele M, Seegerer P, et al. Scoring of tumor-infiltrating lymphocytes: From visual estimation to machine learning. Semin Cancer Biol. 2018;52:151–7. [DOI] [PubMed] [Google Scholar]
- 11.Weiss SA, Han SW, Lui K, Tchack J, Shapiro R, Berman R, et al. Immunologic heterogeneity of tumor-infiltrating lymphocyte composition in primary melanoma. Hum Pathol. 2016;57:116–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barnhill RL, Fine JA, Roush GC, Berwick M. Predicting five-year outcome for patients with cutaneous melanoma in a population-based study. Cancer. 1996;78:427–32. [DOI] [PubMed] [Google Scholar]
- 13.Taylor RC, Patel A, Panageas KS, Busam KJ, Brady MS. Tumor-infiltrating lymphocytes predict sentinel lymph node positivity in patients with cutaneous melanoma. J Clin Oncol. 2007;25:869–75. [DOI] [PubMed] [Google Scholar]
- 14.Mandala M, Imberti GL, Piazzalunga D, Belfiglio M, Labianca R, Barberis M, et al. Clinical and histopathological risk factors to predict sentinel lymph node positivity, disease-free and overall survival in clinical stages I-II AJCC skin melanoma: outcome analysis from a single-institution prospectively collected database. Eur J Cancer. 2009;45:2537–45. [DOI] [PubMed] [Google Scholar]
- 15.Rao UN, Lee SJ, Luo W, Mihm MC Jr., Kirkwood JM. Presence of tumor-infiltrating lymphocytes and a dominant nodule within primary melanoma are prognostic factors for relapse-free survival of patients with thick (t4) primary melanoma: pathologic analysis of the e1690 and e1694 intergroup trials. Am J Clin Pathol. 2010;133:646–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eriksson H, Frohm-Nilsson M, Jaras J, Kanter-Lewensohn L, Kjellman P, Mansson-Brahme E, et al. Prognostic factors in localized invasive primary cutaneous malignant melanoma: results of a large population-based study. Br J Dermatol. 2015;172:175–86. [DOI] [PubMed] [Google Scholar]
- 17.Brambilla E, Le Teuff G, Marguet S, Lantuejoul S, Dunant A, Graziano S, et al. Prognostic Effect of Tumor Lymphocytic Infiltration in Resectable Non-Small-Cell Lung Cancer. J Clin Oncol. 2016;34:1223–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Denkert C, Wienert S, Poterie A, Loibl S, Budczies J, Badve S, et al. Standardized evaluation of tumor-infiltrating lymphocytes in breast cancer: results of the ring studies of the international immuno-oncology biomarker working group. Mod Pathol. 2016;29:1155–64. [DOI] [PubMed] [Google Scholar]
- 19.Hendry S, Salgado R, Gevaert T, Russell PA, John T, Thapa B, et al. Assessing Tumor-infiltrating Lymphocytes in Solid Tumors: A Practical Review for Pathologists and Proposal for a Standardized Method From the International Immunooncology Biomarkers Working Group: Part 1: Assessing the Host Immune Response, TILs in Invasive Breast Carcinoma and Ductal Carcinoma In Situ, Metastatic Tumor Deposits and Areas for Further Research. Adv Anat Pathol. 2017;24:235–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vu QD, Graham S, Kurc T, To MNN, Shaban M, Qaiser T, et al. Methods for Segmentation and Classification of Digital Microscopy Tissue Images. Front Bioeng Biotechnol. 2019;7:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saltz J, Gupta R, Hou L, Kurc T, Singh P, Nguyen V, et al. Spatial Organization and Molecular Correlation of Tumor-Infiltrating Lymphocytes Using Deep Learning on Pathology Images. Cell Rep. 2018;23:181–93 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holten-Rossing H, Talman MM, Jylling AMB, Laenkholm AV, Kristensson M, Vainer B. Application of automated image analysis reduces the workload of manual screening of sentinel lymph node biopsies in breast cancer. Histopathology. 2017;71:866–73. [DOI] [PubMed] [Google Scholar]
- 23.Corredor G, Wang X, Zhou Y, Lu C, Fu P, Syrigos K, et al. Spatial Architecture and Arrangement of Tumor-Infiltrating Lymphocytes for Predicting Likelihood of Recurrence in Early-Stage Non-Small Cell Lung Cancer. Clin Cancer Res. 2019;25:1526–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Acs B, Ahmed FS, Gupta S, Wong PF, Gartrell RD, Sarin Pradhan J, et al. An open source automated tumor infiltrating lymphocyte algorithm for prognosis in melanoma. Nat Commun. 2019;10:5440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kelly CJ, Karthikesalingam A, Suleyman M, Corrado G, King D. Key challenges for delivering clinical impact with artificial intelligence. BMC Med. 2019;17:195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shaw J, Rudzicz F, Jamieson T, Goldfarb A. Artificial Intelligence and the Implementation Challenge. J Med Internet Res. 2019;21:e13659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sendak M, Gao M, Nichols M, Lin A, Balu S. Machine Learning in Health Care: A Critical Appraisal of Challenges and Opportunities. EGEMS (Wash DC). 2019;7:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Topol EJ. High-performance medicine: the convergence of human and artificial intelligence. Nat Med. 2019;25:44–56. [DOI] [PubMed] [Google Scholar]
- 29.Wich LG, Hamilton HK, Shapiro RL, Pavlick A, Berman RS, Polsky D, et al. Developing a multidisciplinary prospective melanoma biospecimen repository to advance translational research. Am J Transl Res. 2009;1:35–43. [PMC free article] [PubMed] [Google Scholar]
- 30.McShane LM, Altman DG, Sauerbrei W, Taube SE, Gion M, Clark GM, et al. REporting recommendations for tumour MARKer prognostic studies (REMARK). Br J Cancer. 2005;93:387–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Clark WH Jr., Elder DE, Guerry Dt, Braitman, Trock BJ, Schultz D, et al. Model predicting survival in stage I melanoma based on tumor progression. J Natl Cancer Inst. 1989;81:1893–904. [DOI] [PubMed] [Google Scholar]
- 32.Bankhead P, Loughrey MB, Fernandez JA, Dombrowski Y, McArt DG, Dunne PD, et al. QuPath: Open source software for digital pathology image analysis. Sci Rep. 2017;7:16878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Youden WJ. Index for rating diagnostic tests. Cancer. 1950;3:32–5. [DOI] [PubMed] [Google Scholar]
- 34.Kang L, Chen W, Petrick NA, Gallas BD. Comparing two correlated C indices with right-censored survival outcome: a one-shot nonparametric approach. Stat Med. 2015;34:685–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Note QM on the sampling error of the difference between correlated proportions or percentages. Psychometrika. 1947;12:153–7. [DOI] [PubMed] [Google Scholar]
- 36.Fu Q, Chen N, Ge C, Li R, Li Z, Zeng B, et al. Prognostic value of tumor-infiltrating lymphocytes in melanoma: a systematic review and meta-analysis. Oncoimmunology. 2019;8:1593806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tramm T, Di Caterino T, Jylling AB, Lelkaitis G, Laenkholm AV, Rago P, et al. Standardized assessment of tumor-infiltrating lymphocytes in breast cancer: an evaluation of inter-observer agreement between pathologists. Acta Oncol. 2018;57:90–4. [DOI] [PubMed] [Google Scholar]
- 38.Wang F, Kaushal R, Khullar D. Should Health Care Demand Interpretable Artificial Intelligence or Accept “Black Box” Medicine? Ann Intern Med. 2019. [DOI] [PubMed] [Google Scholar]
- 39.Idriss MH, Rizwan L, Sferuzza A, Wasserman E, Kazlouskaya V, Elston DM. Nevoid melanoma: A study of 43 cases with emphasis on growth pattern. J Am Acad Dermatol. 2015;73:836–42. [DOI] [PubMed] [Google Scholar]
- 40.Blessing K, Evans AT, al-Nafussi A. Verrucous naevoid and keratotic malignant melanoma: a clinico-pathological study of 20 cases. Histopathology. 1993;23:453–8. [DOI] [PubMed] [Google Scholar]
- 41.Magro CM, Crowson AN, Mihm MC. Unusual variants of malignant melanoma. Mod Pathol. 2006;19 Suppl 2:S41–70. [DOI] [PubMed] [Google Scholar]
- 42.Walia R, Jain D, Mathur SR, Iyer VK. Spindle cell melanoma: a comparison of the cytomorphological features with the epithelioid variant. Acta Cytol. 2013;57:557–61. [DOI] [PubMed] [Google Scholar]
- 43.Bien N, Rajpurkar P, Ball RL, Irvin J, Park A, Jones E, et al. Deep-learning-assisted diagnosis for knee magnetic resonance imaging: Development and retrospective validation of MRNet. PLoS Med. 2018;15:e1002699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lakhani P, Sundaram B. Deep Learning at Chest Radiography: Automated Classification of Pulmonary Tuberculosis by Using Convolutional Neural Networks. Radiology. 2017;284:574–82. [DOI] [PubMed] [Google Scholar]
- 45.Patel BN, Rosenberg L, Willcox G, Baltaxe D, Lyons M, Irvin J, et al. Human-machine partnership with artificial intelligence for chest radiograph diagnosis. NPJ Digit Med. 2019;2:111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Steiner DF, MacDonald R, Liu Y, Truszkowski P, Hipp JD, Gammage C, et al. Impact of Deep Learning Assistance on the Histopathologic Review of Lymph Nodes for Metastatic Breast Cancer. Am J Surg Pathol. 2018;42:1636–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Haenssle HA, Fink C, Schneiderbauer R, Toberer F, Buhl T, Blum A, et al. Man against machine: diagnostic performance of a deep learning convolutional neural network for dermoscopic melanoma recognition in comparison to 58 dermatologists. Ann Oncol. 2018;29:1836–42. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.