Abstract
Distant recurrence of breast cancer results in high lifetime risk and low 5-year survival rate. Early prediction of distant recurrent breast cancer could facilitate intervention and improve patients’ life quality. In this study, we designed an EHR-based predictive model to estimate distant recurrent probability of breast cancer patients. We studied the pathology reports and progress notes of 6,447 patients, who were diagnosed with breast cancer at Northwestern Memorial Hospital between 2001 and 2015. Clinical notes were mapped to Concept unified identifiers (CUI) using natural language processing tools. Bag-of-words and pre-trained embedding were employed to vectorize words and CUI sequences. These features integrated with clinical features from structured data were downstreamed to conventional machine learning classifiers and Knowledge-guided Convolutional Neural Network (K-CNN). The best configuration of our model yielded an AUC of 0.888 and an F-measure of 0.5. Our work provides an automated method to predict breast cancer distant recurrence using natural language processing and deep learning approaches. We expect that through advanced feature engineering, better predictive performance could be achieved.
Keywords: Breast Cancer, Distant Recurrence, Knowledge-guided Convolutional Neural Network, Word Embeddings, Entity Embeddings
1. Background and Related Work
Breast cancer is the most common cancer in women world-wide. In 2012, there are approximately 1.17 million new cases globally which accounts for one fourth of all the new cases of cancer in women [1]. Breast cancer is also one of the most common cancers diagnosed among women in the United States, accounting for nearly one in three cancers. Breast cancer also has the second-highest mortality rate among female cancer patients [2, 3]. It also accounts for approximately 2.6% lifetime risk [4]. Many efforts have already been made to improve treatment quality [5, 6, 7], to identify new diagnostic biomarkers [8, 9, 10] and to learn the genetic patterns [11, 12, 13]. Breast cancer recurrence or recurrent breast cancer is the cancer that comes back after initial treatment, and after a period of time when the cancer couldn’t be detected. Breast cancer might come back to the original site when it first started, or it might spread to new parts of the body. When the cancer recurs in the place other than the original cancer, it is also known as distant recurrence. In the year of 2018, more than 2 million persons were diagnosed as breast cancer [14]. According to the data from the American Cancer Society, the 5-year relative survival rate is 99% for localized breast cancer, whereas it is as low as 27% for distant recurrent breast cancer [4]. In 2005–2011, the 5- and 10- year distant relapse ratio of breast cancer was 5% and 10%, respectively [15]. In another study, the authors observed a approximately one third increase in the number of women living with metastatic breast cancer from 105,354 in 1990 to 138,622 in 2013 [16]. Estimated by their statistical model, by January 1, 2020, there will be 168,292 women living with metastatic breast cancer. Early detection of distant recurrent breast cancer could facilitate the adjustment of treatment and followup plans and decrease the recurrence rate effectively. Meanwhile, patients will benefit from the early awareness of risk so that the life quality will be significantly improved as a result of reduced disease burden.
In the past decade, with the increasing use of Electronic Health Records (EHR), EHR-derived data were widely applied to high-risk patients identification [17], care quality improvement [18], clinical decision support system design [19] and clinical trials monitoring [20]. In recent years, with the vigorous development of computer technology and computing power, more EHR-based phenotype algorithms that use machine learning and natural language processing (NLP) have attracted our attention due to their excellent performance and clinical significance. The clinical notes stored in EHR, which contain a rich description of symptoms, detailed family history, and disease status, are a great source for NLP to develop computational phenotyping research [21]. For example, Huang et al. were able to predict 30-day unplanned intensive care unit readmission with a high area under receiver operating characteristic of 0.768 using discharge summaries [22]; ChexPert, an automated labeling NLP tool, can identify 14 different kinds of thoracic diseases from radiology reports and yielded a high accuracy which defeated human annotator [23]. Therefore, it is natural to employ NLP towards the research of breast cancer recurrence prediction.
Numbers of prior studies have been conducted on breast cancer recurrence with the aid of NLP or machine learning approach. Chen et al. predicted recurrence in triple-negative breast cancer using 35 clinical features. They yielded an AUC of 0.90 and balanced sensitivity and specificity. However, the approach was based on data suitably structured in proper database tables, and it did not consider free text and NLP techniques to analyze and extract variables. Moreover, the authors only collected a small cohort of 114 patients [24]. Kim et al. developed a naive Bayesian model for the prediction of breast cancer recurrence [25]. They achieved a moderate AUC of 0.81 without discriminating local or distant recurrence. However, only structured variables were considered, of which not all of those variables are routinely collected in structured EHR and need manual curation. This is also one of the primary motivations for employing NLP approaches to automate this task from free text. Banerjee et al. applied NLP approaches and neural networks to detect the timeline of metastatic recurrence of breast cancer [26]. They achieved an averaged sensitivity of 0.83 and specificity of 0.73. Their work required intensive clinical domain knowledge to process pathology reports and progress notes. Ling et al. were able to distinguish de novo or recurrent metastatic breast cancer patients using NLP on cancer registry data [27]. Zeng et al. identified distant recurrences in breast cancer using NLP approaches on patients’ progress notes [28]. However, all those three studies using NLP techniques focused on the detection or identification of breast cancer recurrences. When performing detection or identification tasks, the models are allowed to access the data at or after the diagnosis. In this case, the information fed into the model is significantly richer than the development of a predictive model. Despite the technical difficulties of conducting predictive models, it is still desired in that patients can benefit from the prediction of the recurrence through early interventions.
Motivated by the limitations of previous studies that either focus on identification or require intensive feature engineering, we aimed at developing a model to accurately predict distant recurrence of breast cancer. To the best of our knowledge, this is the first study that applies NLP approaches to the prediction of breast cancer recurrences. Such a model should not rely on massive clinical knowledge during the pre-processing stage, which could save the intensive labor consumption on chart review. Furthermore, the model is expected to send the alert of distant recurrence to patients and clinicians so that early intervention could be made. With these aims, we applied conventional machine learning configurations and knowledge-guided convolutional neural networks (K-CNN) [29] on progress notes and pathology reports using various NLP techniques and integrated clinical features to estimate the likelihood of distant recurrence for breast cancer.
2. Methods
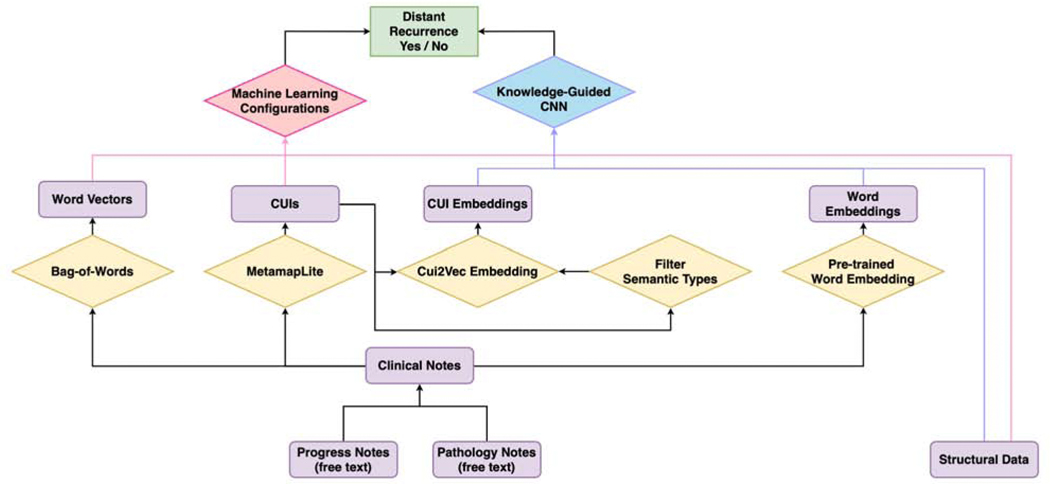
The summarised workflow employed in this study is illustrated in Fig. 1 We first extract progress notes and pathology reports before the cancer recurrence and aggregate them together for each patient. We process these clinical notes to obtain word vectors and UMLS Concept Unique Identifier (CUI) [30]. Based on previous experiences, another subset of CUIs, which is more closely related to diseases, is generated based on the semantic types [29]. Different combinations of word vectors, CUIs, subset of CUIs and structured clinical data are fed into various machine learning configurations to predict distant recurrence of breast cancer. Furthermore, we obtained word embedding and CUI embedding from pre-trained embedding dictionaries. Those embeddings, coupled with structured clinical data, are trained and evaluated using knowledge-guided convolutional neural network (K-CNN).
Figure 1:
Diagram of the workflow. Processing steps are in the diamond boxes; narratives, concepts, and features are in the rectangular boxes. Two major types of configurations are employed in this study, conventional machine learning classifiers and knowledge-guided convolutional neural network (K-CNN). Features are built from free-texted progress notes and pathology reports, as well as structured clinical data. Word vectors and Unified Medical Language System (UMLS) Concept Unique Identifier (CUI) are generated from clinical notes using natural language processing (NLP) techniques. Based on the previous knowledge, a subset of disease-re I a ted CUIs is extracted. Different combinations of word vectors, CUIs, a subset of CUIs, and structured clinical data are fed into various machine learning classifiers for distant recurrence prediction. On the other hand, we generate word embedding and CUI embedding using pre-trained embedding dictionaries. The embedding integrated with structured clinical data is utilized for training and evaluating the K-CNN configuration on breast cancer distant recurrence prediction.
2.1. Dataset
Data are extracted from Northwestern Medicine Enterprise Warehouse (NMEDW). NMEDW is designed as a comprehensive and integrated repository of clinical and research data across Northwestern University Feinberg School of Medicine and Northwestern Memorial Healthcare. Patients diagnosed with breast cancer ICD9 codes at Northwestern Memorial Hospital between 2001 and 2015 are included in this study. In total, 6,899 patients are identified. 452 of these patients are excluded since they have no available progress note or pathology note in the database before the distant recurrence to fulfill the need of our prediction task. In total, 6,447 subjects were included in the final analysis, among which 446 patients had distant recurrence of breast cancer. The distant recurrence of the patients happened between 11/14/2005 and 02/19/2017. The status and date of distant recurrence are annotated by an expert group formed by breast surgery fellow, medical student, and medical informatics scholars using patients’ progress notes. The process of annotation and validation were detailed in [28].
In order to predict the distant recurrence, we extracted the progress notes and pathology notes prior to the date of distant recurrence. While for patients without distant recurrence diagnosed, all the available progress notes and pathology notes were included. The average time from the patients’ first available progress notes to the recurrence is 1409.92 days (~3.86 years), while 1455.08 days for the pathology notes (~3.99 years).
2.2. Feature Generation
2.2.1. Bag-of-Words
Bag-of-Words [31] is utilized in this study for information retrieval from the free-form clinical notes. We use Bag-of-Words with uni-gram. Only single words are collected and counted, whereas phrases (the combinations of words) of any length are ignored. Meanwhile, we exclude the words in the English Stopwords list according to the National Center for Biotechnology Information (NCBI) Stopwords guide. More rules are applied to the vocabulary construction: terms contain special characters or numbers are excluded; terms that have more than 0.99 or less than 0.01 document frequency are ignored. To retrieve the more meaningful information, we employ term frequency-inverse document frequency (tf-idf) to apply weights to tokens of various diversities.
2.2.2. Bag-of-CUIs
Clinical texts are mapped to UMLS concepts unique identifiers (CUI) using MetaMapLite [32]. With the CUIs, we construct Bag-of-CUIs similarly to how we construct Bag-of-Words. According to previous experiences, we further create a subset of CUIs selected by 13 semantic types that are closely related with diseases (T023: Body Part, Organ, or Organ Component / T033: Finding / T034: Laboratory or Test Result / T047: Disease or Syndrome / T048: Mental or Behavioral Dysfunction / T049: Cell or Molecular Dysfunctions / T059: Laboratory Procedure / T060: Diagnostic Procedure / T061: Therapeutic or Preventive Procedure / T121: Pharmacologic Substance / T122 / Biomedical or Dental Material / T123: Biologically Active Substance / T184: Sign or Symptom) [29].
2.2.3. Structured Data
Seven clinical features in the structured data are selected to improve the performance of prediction, which include: race, ethnicity, marital status, smoking status, alcohol usage, family history of cancer, and age at diagnosis of breast cancer. We ensure that all the information we use is collected prior to the distance recurrence to maintain the essence of prediction.
2.2.4. Word Embedding
We use 100, 200, 300, 400, 500 and 600 dimensional word embeddings pre-trained from MIMIC-III clinical notes [33]. We select the best performed word embedding for each configuration based on the performance on validation data.
2.2.5. CUI Embedding
cui2vec, which is a pre-trained 500 dimensional CUI embedding, is employed [34].
2.3. Classifiers
2.3.1. Machine Learning Classifiers
Several machine learning classifiers are experimented on features generated from the Bag-of-Words model. Clinical features and CUIs are added on with an intention to improve the performance. Classifiers we used include random forest [35], support vector machine with linear kernel (linearSVC) [36], logistic regression [37], stochastic gradient descent classifier (SGDC) [38] and naïve Bayes [39]. Some parameters are further specified. In LinearSVC, both L1- and L2-regularization are tried. In SGDC, L1-, L2- and elastic net regularization are used. Multinomial and Bernoulli models are applied to naïve Bayes classifier. In all classifiers except naïve Bayes, the parameter class_weight is set to be balanced, which can reduce the impact of data imbalance. Furthermore, we use Youden’s J statistic to search for the most suitable cut-off value for positive and negative labels.
2.3.2. Knowledge-guided CNN
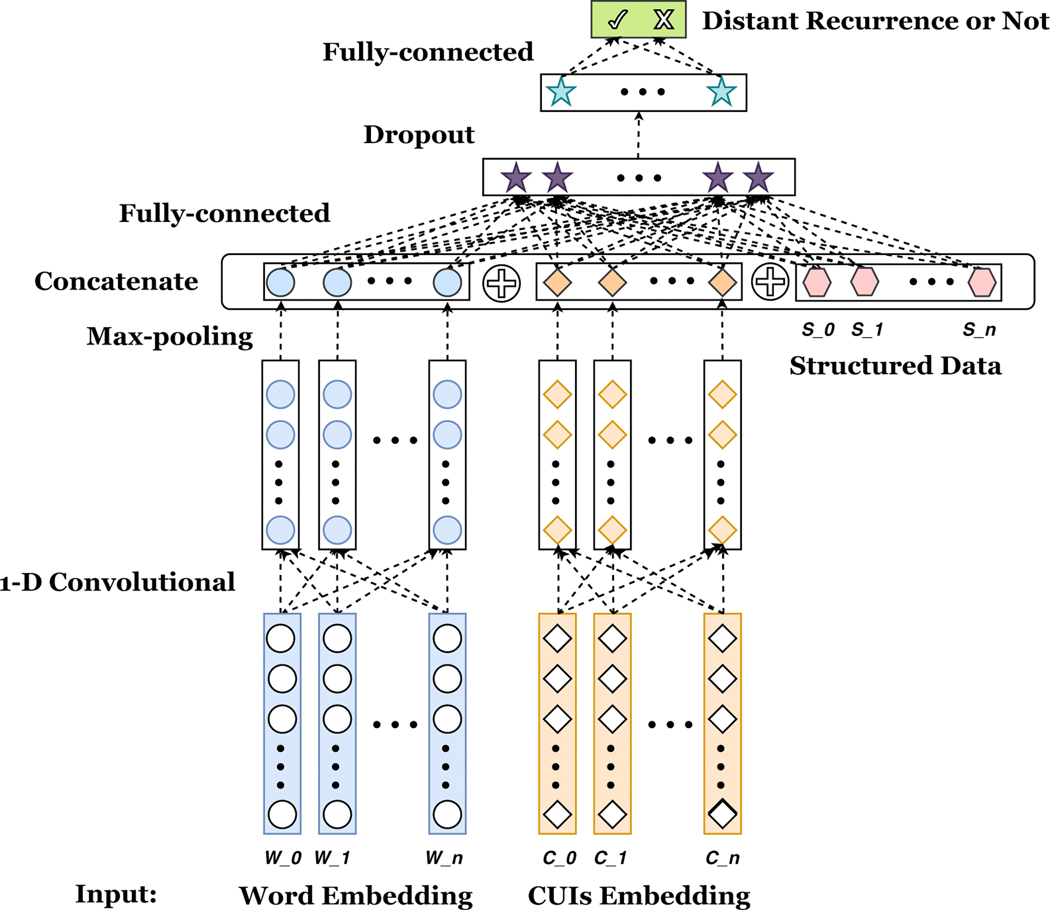
We adopt a deep learning framework similar to knowledge-guided CNN (K-CNN) designed by Yao et al. [29] shown in Fig. 2. The pre-trained word and CUI embedding are first downstreamed to a 1-dimensional convolutional layer, respectively. Then, a max pooling is added to select the most important feature with the highest value in the convolutional feature map. After that, the two parts of selected features, as well as structured data are concatenated together. Next, the concatenated hidden layer is fed into a fully connected hidden layer, followed by a dropout layer and ReLU activation layer. Finally, a fully connected layer with softmax activation is used to derive the probability result of the distant recurrence outcome. We implement the K-CNN configuration described above on different combinations of features, which include: using the word embedding only (word), the combination of word embedding and structured clinical features (word+str), the combination of word embedding and CUI embedding (word+CUI), the combination of word embedding and semantic selected CUI embedding (word+CUIsem) and the combination of word embedding, CUI embedding and structured data (word+CUI+str). Similarly as conventional machine learning configurations, we also utilize Youden’s J statistics to find the best threshold for positive and negative labels.
Figure 2:
Knowledge-guided Convolutional Neural Network. Pre-trained word embeddings and CUI embeddings are first downstreamed to a 1-dimensional (1-D) convolutional layer, followed by a max-pooling layer to select the highest value of each word or CUI embedding. Then, those selected values are concatenated with seven structured clinical features. A fully connected hidden layer is further used, followed by a dropout and ReLU activation layer. Finally, another fully-connected layer with softmax function is used to yield the probability of distant recurrence. 5 different combinations of features are implemented
The technical details of K-CNN are revealed as below: word sequence of each note is truncated to 10,000 words, and CUI sequence is truncated to 1,000 CUIs; 256 1-dimensio al kernels are used; kernel size is 5; the size of fully connected layer hidden neurons is 128; dropout keep probability: 0.8; learning rate: 0.001; batch size is 32; batch normalization is applied. The most exhaustive network has less than 72K trainable parameters, which requires the training time less than 2 hours.
2.4. Evaluation
Training and testing sets are split to approximately 7:3. For conventional machine learning configurations, we employ 5-fold cross validation to avoid overfitting and to select the best parameters. A separate validation set constructed from approximately 10% of the training data is utilized for K-CNN. The area under the receiver operating characteristic curve (AUC), specificity (true negative rate), Youden’s J statistic, binary precision for the positive class as distant recurrent, binary recall for the positive class as distant recurrent, and binary f1 score for the positive class as distant recurrent are recorded as an evaluation of the configurations.
2.5. Implementation
The entire pipeline is built with Python V3.6.3. Bag-of-Words model, machine learning classifiers and model evaluation are implemented with scikit learn V0.19.1 package [40]. Tensorflow V1.9 is used to construct K-CNN architecture [41]. CUDA V8.0 is used to execute deep learning computation on Tesla K40c GPUs. Metamap Lite V3.6.2 is employed to identify clinical entities from free text.
3. Results
3.1. Machine Learning Classifiers
Shown in Table 1 are the results of conventional machine learning configurations. Precision, recall, f1 score, specificity, and Youden’s J statistic are only showing the value for positive class. The results for the nine configurations are listed in the descending order of AUC. We highlight the best f1 scores, as well. The configuration using only the word vectors derived from Bag-of-Word yields the baseline with the F1 score = 0.415. The experiments show that adding structured clinical data, CUIs, CUIs filtered with semantic types, and the combination of them will all improve the performance of prediction. Especially by integrating word vectors, CUIs, and structured data, the configuration achieves the best performance of f1 score = 0.451. Meanwhile, AUCs yielded from most of the configurations are higher than 0.8, with the highest of 0.88l. Furthermore, we achieve a nearly perfect specificity of greater than 0.95.
Table 1.
Results for Machine Learning Configurations
| Features | Classifiers | AUC | Precision | Recall | F1 Score | Specificity | Youden |
|---|---|---|---|---|---|---|---|
| BoW | Random Forest | 0.866 | 0.516 | 0.348 | 0.415 | 0.974 | 0.322 |
| SGDCIassifier_ENP | 0.845 | 0.363 | 0.468 | 0.409 | 0.935 | 0.403 | |
| SGDCIassifier_L2 | 0.842 | 0.378 | 0.461 | 0.415 | 0.940 | 0.401 | |
| SGDCIassifier_L1 | 0.840 | 0.317 | 0.560 | 0.405 | 0.905 | 0.466 | |
| BernoulliNB | 0.836 | 0.275 | 0.631 | 0.383 | 0.869 | 0.500 | |
| Logistic Regression | 0.823 | 0.439 | 0.333 | 0.379 | 0.967 | 0.300 | |
| LinearSVC_L2 | 0.822 | 0.532 | 0.177 | 0.266 | 0.988 | 0.165 | |
| MultinomialNB | 0.815 | 0.247 | 0.638 | 0.356 | 0.847 | 0.486 | |
| LinearSVC_L1 | 0.808 | 0.515 | 0.241 | 0.329 | 0.982 | 0.223 | |
| BoW+str | Random Forest | 0.874 | 0.515 | 0.362 | 0.425 | 0.973 | 0.335 |
| BernoulliNB | 0.836 | 0.276 | 0.631 | 0.384 | 0.870 | 0.501 | |
| SGDCIassifier_L2 | 0.829 | 0.335 | 0.433 | 0.378 | 0.933 | 0.365 | |
| SGDCIassifier_ENP | 0.820 | 0.252 | 0.589 | 0.352 | 0.862 | 0.451 | |
| Logistic Regression | 0.815 | 0.423 | 0.312 | 0.359 | 0.967 | 0.279 | |
| LinearSVC_L1 | 0.800 | 0.550 | 0.156 | 0.243 | 0.990 | 0.146 | |
| LinearSVC_L2 | 0.793 | 0.680 | 0.121 | 0.205 | 0.996 | 0.116 | |
| MultinomialNB | 0.791 | 0.232 | 0.617 | 0.337 | 0.839 | 0.456 | |
| SGDCIassifier_L1 | 0.763 | 0.714 | 0.035 | 0.068 | 0.999 | 0.034 | |
| BoW+CUI | Random Forest | 0.881 | 0.444 | 0.454 | 0.449 | 0.955 | 0.409 |
| SGDCIassifier_ENP | 0.868 | 0.686 | 0.170 | 0.273 | 0.994 | 0.164 | |
| LinearSVC_L2 | 0.866 | 0.707 | 0.206 | 0.319 | 0.993 | 0.199 | |
| SGDCIassifier_L2 | 0.866 | 0.700 | 0.199 | 0.309 | 0.993 | 0.192 | |
| Logistic Regression | 0.862 | 0.652 | 0.319 | 0.429 | 0.987 | 0.306 | |
| SGDCIassifier_L1 | 0.858 | 0.850 | 0.121 | 0.211 | 0.998 | 0.119 | |
| LinearSVC_L1 | 0.853 | 0.875 | 0.099 | 0.178 | 0.999 | 0.098 | |
| BernoulliNB | 0.834 | 0.348 | 0.574 | 0.433 | 0.915 | 0.490 | |
| MultinomialNB | 0.827 | 0.417 | 0.426 | 0.421 | 0.953 | 0.379 | |
| BoW+CUIsem | Random Forest | 0.875 | 0.398 | 0.468 | 0.430 | 0.957 | 0.412 |
| Logistic Regression | 0.859 | 0.418 | 0.418 | 0.418 | 0.967 | 0.373 | |
| SGDCIassifier_ENP | 0.851 | 0.767 | 0.163 | 0.269 | 0.996 | 0.159 | |
| LinearSVC_L2 | 0.849 | 0.765 | 0.184 | 0.297 | 0.967 | 0.180 | |
| SGDCIassifier_L2 | 0.846 | 0.774 | 0.170 | 0.279 | 0.996 | 0.166 | |
| SGDCIassifier_L1 | 0.845 | 0.714 | 0.142 | 0.237 | 0.996 | 0.137 | |
| BernoulliNB | 0.833 | 0.324 | 0.567 | 0.412 | 0.907 | 0.474 | |
| LinearSVC_L1 | 0.832 | 0.714 | 0.106 | 0.185 | 1.000 | 0.103 | |
| MultinomialNB | 0.727 | 0.319 | 0.475 | 0.382 | 0.964 | 0.395 | |
| BoW+CUI+str | Random Forest | 0.877 | 0.442 | 0.461 | 0.451 | 0.954 | 0.415 |
| LinearSVC_L1 | 0.855 | 0.808 | 0.149 | 0.251 | 0.997 | 0.146 | |
| LinearSVC_L2 | 0.849 | 0.732 | 0.213 | 0.330 | 0.994 | 0.207 | |
| Logistic Regression | 0.838 | 0.258 | 0.617 | 0.364 | 0.861 | 0.478 | |
| BernoulliNB | 0.834 | 0.348 | 0.574 | 0.433 | 0.915 | 0.490 | |
| MultinomialNB | 0.830 | 0.397 | 0.383 | 0.390 | 0.954 | 0.337 | |
| SGDCIassifier_L1 | 0.816 | 0.696 | 0.113 | 0.195 | 0.996 | 0.110 | |
| SGDCIassifier_ENP | 0.797 | 0.684 | 0.092 | 0.163 | 0.997 | 0.089 | |
| SGDCIassifier_L2 | 0.774 | 0.750 | 0.064 | 0.118 | 0.998 | 0.062 | |
Results within one feature combination are ranked by descending AUC; the best f1 score across all feature combinations is highlighted in bold. Precision, recall, f1 score, sensitivity, specificity, and Youden’s J statistic index are only showing the values for the positive class as distant recurrence.
Abbreviation: BoW: Bag-of-Words model; +str: is incorporated with structured data; +CUI: is incorporated with Bag-of-CUIs; +CUIsem: is incorporated with Bag-of-CUIs by semantic selection; BernoulliNB: naïve Bayes using Bernoulli model; MultinomialNB: naïve Bayes using multinomial model; LinearSVC: support vector machine using linear kernel; SGDC: Stochastic Gradient Descent Classifier; L1, L2, ENP: L1, L2, elastic net penalty regularization; AUC: area under the receiver operating characteristic curve.
3.2. Knowledge-guided Convolutional Neural Network
Selected results of knowledge-guided convolutional neural network (K-CNN) are shown in Table 2. Precision, recall, f1 score, specificity, and Youden’s J statistic are showing the values for the positive class as distant recurrence. We selected the best-performed dimension of word embedding based on the performance on validation sets. The configuration with only the word embedding yields a baseline f1 score. Improvements are observed when integrating structured data, CUIs, and the combination of structured data and CUIs. The configuration implemented on the combination of word embedding and structured data out-performs other combinations and achieves an f1 score of 0.5. All the AUCs are higher than 0.8 and even better than machine learning classifiers.
Table 2.
Results for K-CNN
| Features | AUC | Precision | Recall | F1 Score | Specificity | Youden | Best ED |
|---|---|---|---|---|---|---|---|
| word | 0.882 | 0.484 | 0.312 | 0.408 | 0.974 | 0.286 | 300 |
| word + str | 0.888 | 0.537 | 0.468 | 0.500 | 0.968 | 0.436 | 500 |
| word + CUI | 0.869 | 0.440 | 0.468 | 0.454 | 0.953 | 0.421 | 300 |
| word + CUIsem | 0.835 | 0.553 | 0.298 | 0.387 | 0.981 | 0.279 | 300 |
| word + CUI + str | 0.838 | 0.477 | 0.369 | 0.416 | 0.968 | 0.337 | 200 |
The best f1 score is highlighted in bold. Abbreviation: word: word embedding; str: Structured Data; CUI: Clinical Unified Indentifiers embedding; CUIsem: semantic selected CUI; ED: embedding dimension; AUC: area under the receiver operating characteristic curve. Precision, recall, f1 score, sensitivity, specificity and Youden’s J statistic are showing the value for positive class as distant recurrence.
4. Discussion
In this study, we demonstrated the feasibility of performing breast cancer distant recurrence prediction using machine learning as well as knowledge-guided convolutional neural network (K-CNN) with the aid of natural language processing (NLP) techniques. Comparing with previous approaches, we illustrated an implementation that required less clinical expertise and data curation. By using progress notes and pathology reports, our algorithms were able to predict the distant recurrence of breast cancer efficiently.
In order to improve the performance, we employed MetaMap Lite to identify terms and phrases in the free-text notes and mapped them to the Unified Medical Language System (UMLS) concept unique identifiers (CUI). The configurations benefit from the mapped CUIs because of the increased semantic interoperability. We saw improvements when including CUIs as features in both machine learning configuration and knowledge-guided convolutional neural network (K-CNN). The integration of structured data extracted from the electronic health records (EHR) also elevated the f1 score for the prediction of positive classes. Remarkably, we were able to see further improvement when integrating both structured data and CUIs. The f1 score (0.451) for the 3-way combination configuration was the highest among all the machine learning configurations. The combination of features introduced more predictive powers to the configurations.
We also observed improvement when the configuration was trained using the subset of CUIs filtered by semantic types. The disease-related semantic types list was adopted from the previous experiment that Yao et al. mentioned in their published study [29]. However, the improvement of using the subset of CUIs was not as significant as using all the CUIs. This, in turn, indicated that the distant recurrence of breast cancer is a multi-determined event that needs a more comprehensive vocabulary to make the precise prediction.
In addition to the uni-gram Bag-of-Words model, we also did experiments on multi-gram conditions. However, the results are even worse compared with uni-gram Bag-of-Words, so we decided not to report them in the result section. The best AUC obtained from configurations of multi-gram Bag-of-Words was 0.704, with an f1 score of 0.371. We outlined the reasons why a more sophisticated version of the model did not outperform the simpler one as follows: First of all, our notes are relatively long. We used the multi-gram range of 1 to 3, which resulted in a 90 times larger size of vectorized features. The increment in the number of features is likely to cause overfitting. Secondly, the majority of additional word combinations added by multi-gram Bag-of-Words may not be clinically meaningful, so that it will lead to less optimal performance.
Comparing with the conventional machine learning classifiers, we observed an overall improvement when utilizing knowledge-guided convolutional neural network (K-CNN) for the prediction task. Improvements were detected when structured data and CUIs were integrated into the feature constructions. We experimented on word embedding of different dimensions. Unlike other studies [29] that used fixed dimension word embedding for all models, we observed that various feature constructions benefit from different dimensions of embeddings, and we did not found a perfect dimension that fit all configurations.
The out-of-vocabulary rate for word embedding and entity embedding is approximately 0.23 and 0.51. However, the higher out-of-vocabulary rate might not necessarily be a drawback. Since the pre-trained word and entity embedding were trained on clinical notes extracted from comprehensive and high-quality databases that we could assume them as good representations of meaningful words and entities. The words and entities not found in the embedding dictionaries might be very specific to one disease that was only prevalent in the training database and also possibly to be unrelated. However, the currently available algorithms and pre-trained word and CUI embeddings are still far from perfect and expect further investigations.
The limited pre-trained entity embedding dictionary that we used and the relatively high out-of-vocabulary rate for CUIs also accounted for the performance of K-CNN. The best performance that we observed from the K-CNN configurations was from the combination of word embedding and structured clinical data, which yielded an f1 score of 0.500. The models integrated with CUIs out-performed the configuration with only word embedding, but still less optimal comparing with the configuration with the combination of word embedding and structured clinical data. Furthermore, we restricted the CUI sequence length for computational efficiency. This may also affect the performance of the models using CUI embedding as input features.
In terms of clinical usefulnesses, the model provides clinicians a reference for the development of personalized treatment plans. For a breast cancer patient with a higher risk of distant recurrence yielded by the model, more aggressive operations may be considered during the surgery to lower the chance of metastatic diseases. The prediction of breast cancer distant recurrence can also help to adjust the current medication plans for a better prognosis.
Error analysis.
We investigated the clinical notes of 20 patients in the test set that were inaccurately classified by Random Forest on the combined features of Bag-of-Words and structured clinical data, among which ten were for false-positive cases, and ten were for false-negative cases. We dug into the notes with high predicted probability and tried to find common reasons that caused the misclassification. One of the patients with metastatic breast cancer was classified as no distant recurrence with a 0.902 probability by our model. We reviewed the clinical notes as well as the corresponding clinical features in detail. We found no evidence of metastatic disease in the clinical notes before the recurrence; meanwhile, the physical condition of this patient was much better compared with other metastatic cases. For example, when we detected in her notes, there were sentences like “...These revealed enhancing mass of the left upper inner quadrant with satellite nodules and rounded nodes in the axillae. No evidence of distant metastatic disease was seen...”; “...There is sclerosis at the margins of the sacroiliac joint, most prominent on the right side. This is degenerative in appearance and not the pattern of a metastatic process...”. Furthermore, this patient is a white female with no family history of breast cancer. Therefore, the actual probability for this patient to develop distant recurrence cancer should be lower. The lack and ambiguous information made our model prone to classify this uncommon case wrongly. In another example, the patient was predicted to experience distant recurrence with a probability of 0.732 while actually, they did not have any distant recurrence. This is a very complicated case with a long history of breast cancer. Through the review of the corresponding notes, we found this patient is a “56 year-old female with history of breast carcinoma since 2004 with known bone and lung metastases; bone scan for restaging”. Several narrations of the “metastases” were made in both the progress notes and pathology reports. And also, the reason for readmission was “Indication: 55 year-old female with suspected metastatic breast carcinoma, presenting with recent onset of chest pain and shortness of breath, with focal skeletal lesions demonstrated on CT”. The complexity of the medical history and particular purpose for the readmission makes this case not typical for our configurations. Nevertheless, identifying the causes of inaccurate predictions will help us better understand the shortcomings of the model and urge us to improve performance more specifically.
There are still some limitations to our study. Although adding CUIs to the configuration improved the performance of the prediction task, MetaMap Lite is still a heavily rule-based algorithm. It has limitations in sentence boundary recognition, term identification, medical semantic similarity detection, etc. Meanwhile, the dataset used to obtain pre-trained word and entity embedding might not be a perfect fit for our data since neither of these embedding dictionaries was pre-trained based on breast cancer related or even cancer-associated databases.
For future work, we plan to perform more detailed feature engineering, which includes novel algorithms for sentence boundary recognition before feeding into any named entities recognition algorithm. We will also try to develop more advanced word and entity embedding systems to customize for specific tasks and place more emphasis on clinical semantic similarity. The assertion types of clinical concepts will also be incorporated into future study. Furthermore, we have the plan to extend our implementations to the prediction of local recurrence of breast cancer.
5. Conclusion
In conclusion, we employ conventional machine learning classifiers and deep learning frameworks to develop an automated breast cancer distant recurrence predictive model using pathology reports and progress notes, within which we applied NLP techniques to generate features. Our approach has tremendous potential to predict distant recurrence of breast cancer patients. Further development of this model with advanced NLP and deep learning techniques will allow a better predictive performance. More investigations are also called for to validate the clinical utility of our model.
Integration of clinical notes, concept identifiers and structural clinical features improves the performance of distant breast cancer recurrence prediction using Machine Learning and yields high AUC of over 0.88.
Knowledge-guided Convolutonal Neural Network outperforms conventional Machine Learning configurations on the task of distant breast cancer recurrence prediction and yields high f1 score of 0.50.
Natural Language Processing techniques, including Bag-of-Word, Metamap, word and entity embedding are employed to represent progress notes and pathology reports.
Detailed report review and error analysis detect common caveats of using clinical notes for prediction of cancer recurrence which could inspire future investments.
6. Acknowledgement
We thank Ying Liu (ying.liu@utdallas.edu) for providing helpful discussions.
This study was supported in part by grants R21LM012618 and 1R01LM013337 from the National Institutes of Health, Breast Cancer Research Foundation, and the Lynn Sage Cancer Research Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].W. W.C. R. F. I. for Cancer Research), Diet, nutrition, physical activity and cancer: a global perspective. continuous update project expert report (2018). [Google Scholar]
- [2].DeSantis C, Ma J, Bryan L, Jemal A, Breast cancer statistics, 2013, CA: a cancer journal for clinicians 64 (2014) 52–62. [DOI] [PubMed] [Google Scholar]
- [3].DeSantis C, Siegel R, Bandi P, Jemal A, Breast cancer statistics, 2011, CA: a cancer journal for clinicians 61 (2011) 408–418. [DOI] [PubMed] [Google Scholar]
- [4].Siegel RL, Miller KD, Jemal A, Cancer statistics, 2019, CA: a cancer journal for clinicians 69 (2019) 7–34. [DOI] [PubMed] [Google Scholar]
- [5].Turner J, Hayes S, Reul-Hirche H, Improving the physical status and quality of life of women treated for breast cancer: a pilot study of a structured exercise intervention, Journal of surgical oncology 86 (2004) 141–146. [DOI] [PubMed] [Google Scholar]
- [6].Vicini FA, Sharpe M, Kestin L, Martinez A, Mitchell CK, Wallace MF, Matter R, Wong J, Optimizing breast cancer treatment efficacy with intensity-modulated radiotherapy, International Journal of Radiation Oncology* Biology* Physics 54 (2002) 1336–1344. [DOI] [PubMed] [Google Scholar]
- [7].Shulman LN, Willett W, Sievers A, Knaul FM, Breast cancer in developing countries: opportunities for improved survival, Journal of oncology 2010 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Rui Z, Jian-Guo J, Yuan-Peng T, Hai P, Bing-Gen R, Use of serological proteomic methods to find biomarkers associated with breast cancer, Proteomics 3 (2003) 433–439. [DOI] [PubMed] [Google Scholar]
- [9].Sauter ER, Zhu W, Fan X, Wassell R, Chervoneva I, Du Bois GC, Proteomic analysis of nipple aspirate fluid to detect biologic markers of breast cancer, British journal of cancer 86 (2002) 1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Brooks M, Breast cancer screening and biomarkers, in: Cancer Epidemiology, Springer, 2009, pp. 307–321. [DOI] [PubMed] [Google Scholar]
- [11].Ali HR, Chlon L, Pharoah PD, Markowetz F, Caldas C, Patterns of immune infiltration in breast cancer and their clinical implications: a gene-expression-based retrospective study, PLoS medicine 13 (2016) e1002194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Fasching P, Hu C, Hart S, Polley E, Lee K, Gnanolivu R, Lilyquist J, Hartkopf A, Taran F, Janni W, et al. , Abstract pd1–02: Cancer predisposition genes in metastatic breast cancer-association with metastatic pattern, prognosis, patient and tumor characteristics, 2018. [Google Scholar]
- [13].Nakshatri H, Kumar B, Burney HN, Cox ML, Jacobsen M, Sandusky GE, D’Souza-Schorey C, Storniolo AMV, Genetic ancestry-dependent differences in breast cancer-induced field defects in the tumor-adjacent normal breast, Clinical Cancer Research 25 (2019) 2848–2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A, Global cancer statistics 2018: Globocan estimates of incidence and mortality worldwide for 36 cancers in 185 countries, CA: a cancer journal for clinicians 68 (2018) 394–424. [DOI] [PubMed] [Google Scholar]
- [15].Malmgren J, Hurlbert M, Atwood M, Kaplan HG, Examination of a paradox: recurrent metastatic breast cancer incidence decline without improved distant disease survival: 1990–2011, Breast cancer research and treatment 174 (2019) 505–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Mariotto AB, Etzioni R, Hurlbert M, Penberthy L, Mayer M, Estimation of the number of women living with metastatic breast cancer in the united states, Cancer Epidemiology and Prevention Biomarkers (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Baillie CA, VanZandbergen C, Tait G, Hanish A, Leas B, French B, William Hanson C, Behta M, Umscheid CA , The readmission risk flag: Using the electronic health record to automatically identify patients at risk for 30-day readmission, Journal of hospital medicine 8 (2013) 689–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Cebul RD, Love TE, Jain AK, Hebert CJ, Electronic health records and quality of diabetes care, New England Journal of Medicine 365 (2011) 825–833. [DOI] [PubMed] [Google Scholar]
- [19].Bell LM, Grundmeier R, Localio R, Zorc J, Fiks AG, Zhang X, Stephens TB, Swietlik M, Guevara JP, Electronic health record-based decision support to improve asthma care: a cluster-randomized trial, Pediatrics 125 (2010) e770–e777. [DOI] [PubMed] [Google Scholar]
- [20].Yi SS, Tabaei BP, Angell SY, Rapin A, Buck MD, Pagano WG, Maselli FJ, Simmons A, Chamany S, Self-blood pressure monitoring in an urban, ethnically diverse population: a randomized clinical trial utilizing the electronic health record, Circulation: Cardiovascular Quality and Outcomes 8 (2015) 138–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Luo Y, Thompson WK, Herr TM, Zeng Z, Berendsen MA, Jonnalagadda SR, Carson MB, Starren J, Natural language processing for ehr-based pharmacovigilance: a structured review, Drug safety 40 (2017) 1075–1089. [DOI] [PubMed] [Google Scholar]
- [22].Huang K, Altosaar J, Ranganath R, Clinicalbert: Modeling clinical notes and predicting hospital readmission, arXiv preprint arXiv:1904.05342 (2019). [Google Scholar]
- [23].Irvin J, Rajpurkar P, Ko M, Yu Y, Ciurea-Ilcus S, Chute C, Marklund H, Haghgoo B, Ball R, Shpanskaya K, et al. , Chexpert: A large chest radiograph dataset with uncertainty labels and expert comparison, arXivpreprintarXiv:1901.07031 (2019). [Google Scholar]
- [24].Chen X, Zhou Z, Thomas K, Folkert M, Kim N, Rahimi A, Wang J, A reliable multi-classifier multi-objective model for predicting recurrence in triple negative breast cancer, in: 2019 41st Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), IEEE, pp. 2182–2185. [DOI] [PubMed] [Google Scholar]
- [25].Kim W, Kim KS, Park RW, Nomogram of naive bayesian model for recurrence prediction of breast cancer, Healthcare informatics research 22 (2016) 89–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Banerjee I, Bozkurt S, Caswell-Jin JL, Kurian AW, Rubin DL, Natural language processing approaches to detect the timeline of metastatic recurrence of breast cancer, JCO clinical cancer informatics 3 (2019) 1–12. [DOI] [PubMed] [Google Scholar]
- [27].Ling AY, Kurian AW, Caswell-Jin JL, Sledge GW, Shah NH, Tamang SR, Using natural language processing to construct a metastatic breast cancer cohort from linked cancer registry and electronic medical records data, JAMIA Open (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Zeng Z, Yao L, Roy A, Li X, Espino S, Clare SE, Khan SA, Luo Y, Identifying breast cancer distant recurrences from electronic health records using machine learning, Journal of Healthcare Informatics Research (2019) 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Yao L, Mao C, Luo Y, Clinical text classification with rule-based features and knowledge-guided convolutional neural networks, BMC medical informatics and decision making 19 (2019) 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Bodenreider O, The unified medical language system (umls): integrating biomedical terminology, Nucleic acids research 32 (2004) D267–D270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Harris ZS, Distributional structure, Word 10 (1954) 146–162. [Google Scholar]
- [32].Demner-Fushman D, Rogers WJ, Aronson AR, Metamap lite: an evaluation of a new java implementation of metamap, Journal of the American Medical Informatics Association 24 (2017) 841–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Johnson AE, Pollard TJ, Shen L, Li-wei HL, Feng M, Ghassemi M, Moody B, Szolovits P, Celi LA, Mark RG, Mimic-iii, a freely accessible critical care database, Scientific data 3 (2016) 160035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Beam AL, Kompa B, Fried I, Palmer NP, Shi X, Cai T, Kohane IS, Clinical concept embeddings learned from massive sources of multimodal medical data, arXiv preprintarXiv:1804.01486 (2018). [PMC free article] [PubMed] [Google Scholar]
- [35].Ho TK, Random decision forests, in: Proceedings of 3rd international conference on document analysis and recognition, volume 1, IEEE, pp. 278–282. [Google Scholar]
- [36].Suykens JA, Vandewalle J, Least squares support vector machine classifiers, Neural processing letters 9 (1999) 293–300. [Google Scholar]
- [37].Kleinbaum DG, Dietz K, Gail M, Klein M, Klein M, Logistic regression, Springer, 2002. [Google Scholar]
- [38].Mei S, Montanari A, Nguyen P-M, A mean field view of the landscape of two-layer neural networks, Proceedings of the National Academy of Sciences 115 (2018) E7665–E7671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].McCallum A, Nigam K, et al. , A comparison of event models for naive bayes text classification, in: AAAI-98 workshop on learning for text categorization, volume 752, Citeseer, pp. 41–48. [Google Scholar]
- [40].Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, Blondel M, Prettenhofer P, Weiss R, Dubourg V, et al. , Scikit-learn: Machine learning in python, Journal of machine learning research 12 (2011) 2825–2830. [Google Scholar]
- [41].Abadi M, Barham P, Chen J, Chen Z, Davis A, Dean J, Devin M, Ghemawat S, Irving G, Isard M, et al. , Tensor flow: A system for large-scale machine learning, in: 12th {USENIX} Symposium on Operating Systems Design and Implementation ({OSDI} 16), pp. 265–283. [Google Scholar]