Abstract
IL-7 is required for T cell differentiation and mature T cell homeostasis and promotes pro-B cell proliferation and survival. Tyrosine phosphorylation plays a central role in IL-7 signaling. We identified by two-dimensional electrophoresis followed by anti-phosphotyrosine immunoblotting and mass spectrometry sixteen tyrosine phosphorylated proteins from the IL-7-dependent cell line D1. IL-7 stimulation induced the phosphorylation of the proteins STI1, ATIC and hnRNPH, involved in pathways related to survival, proliferation and gene expression, respectively, and increased the phosphorylation of CrkL, a member of a family of adaptors including the highly homologous Crk isoforms CrkII and CrkI, important in multiple signaling pathways. We observed an increased phosphorylation of CrkL in murine pro-B cells and in murine and human T cells. In addition, IL-7 increased the association of CrkL with the transcription factor Stat5, essential for IL-7 pro-survival activity. The selective tyrosine kinase inhibitor Imatinib. counteracted the IL-7 pro-survival effect in D1 cells and decreased CrkL phosphorylation. These data suggested that CrkL could play a pro-survival role in IL-7-mediated signaling. We observed that pro-B cells also expressed, in addition to CrkL, the Crk isoforms CrkII and CrkI and therefore utilized pro-B cells conditionally deficient in all three to evaluate the role of these proteins. The observation that the IL-7 pro-survival effect was reduced in Crk/CrkL conditionally-deficient pro-B cells further pointed to a pro-survival role of these adaptors. To further evaluate the role of these proteins, gene expression studies were performed in Crk/CrkL conditionally-deficient pro-B cells. IL-7 decreased the transcription of the receptor LAIR1, which inhibits B cell proliferation, in a Crk/CrkL-dependent manner, suggesting that the Crk family of proteins may promote pro-B cell proliferation. Our data contribute to the understanding of IL-7 signaling and suggest the involvement of Crk family proteins in pathways promoting survival and proliferation.
Keywords: Interleukin 7, Apoptosis, Tyrosine phosphorylation, Crk-like, Imatinib, LAIR1
1. Introduction
IL-7 is essential for the development, proliferation and survival of immature T cells and promotes pro-B cell proliferation and survival [1–6]. Genetic defects in IL-7 receptor (IL-7R) components are a common cause of human severe combined immunodeficiency disease (SCID), consisting in a reduction of thymic cellularity and severe lymphopenia [1,2]. In addition, mature T cells require IL-7 for survival and homeostatic proliferation to maintain a stable cell number [3–6].
IL-7 pro-survival activities are mainly mediated by the inhibition of the mitochondrial apoptotic pathway regulated by Bcl-2 family members [4,5]. For instance, IL-7 deprivation causes a decline in the synthesis of the anti-apoptotic protein Bcl-2, coupled with the activation of proteins that damage mitochondria, such as Bax and Bad [3,4]. Tyrosine phosphorylation plays a central role in IL-7 signaling. The tyrosine kinases Jak1 and Jak3 are bound to the α and γc chain composing the IL-7R. Following IL-7-IL-7R binding, phosphorylation of the α chain by these kinases creates docking sites for Src family kinases, the p85 regulatory subunit of PI3 kinase, the transcription factor Stat5 and other transducing molecules [3,5]. Importantly, the upregulation of Bcl-2 is mediated by the transcriptional activity of tyrosine phosphorylated Stat-5 [4,7]. Stat-5a/b-deficient mice undergo perinatal mortality, and show atrophic thymus and spleen [7]. In addition, Stat5 is essential for B cell development and for the proliferative response of pro-B cells induced by IL-7 [8,9].
Knowing the molecular endpoints of the survival pathways, we sought to identify the intermediates in signals downstream from IL-7R. Tyrosine phosphorylated proteins from the murine IL-7-dependent thymocyte cell line D1 [10] were analysed by two-dimensional (2D) electrophoresis followed by immunoblotting with an antiphosphotyrosine monoclonal antibody and matrix-assisted laser desorption/ionisation (MALDI) mass spectrometry. We identified 16 proteins and one of these, as we will show, was the adaptor protein Crk-like (CrkL).
Adaptors interact with at least two proteins, offering flexible mechanisms to regulate signal transduction. The Crk family of adaptors includes CrkI, II, and III, derived from a single gene by alternative splicing, and CrkL, encoded by a distinct highly homologous gene [11,12]. Crk II, also called Crk “isoform a”, is the full length product of the Crk gene, CrkI, or Crk “isoform b”, is shorter and its C terminus ends before tyrosine 207. CrkL and CrkII share 56% overall aminoacid identity, and in human proteins > 70% of the aminoacid sequences in their SRC homology domain are conserved [11–13]. This high level of similarity may explain their overlapping functions. CrkL and Crk-null mice die during embryogenesis, and show evidence of common and distinct developmental defects [14,15]. CrkII and CrkL, expressed by mature T and B lymphocytes [11,12] show similar molecular weights: CrkL = 36–38 kDa and CrkII = 40 kDa [11,12,16]. Functional redundancy is also demonstrated by studies employing mice with floxed alleles indicating that the absence of both adaptors was necessary to reveal structural and functional defects in T cells, fibroblasts, muscle cells and neurons [17].
CrkL contains one SH2, a typical N terminal and an atypical C terminal SH3 domain [18]. The SH2 domain binds tyrosine phosphorylated proteins allowing CrkL to link these substrates to SH3 binding downstream effectors [11,12]. CrkL participates in pathways regulating proliferation, apoptosis, differentiation, cell adhesion and migration [11,12]. It is found in multiprotein complexes induced by triggering T and B cell receptors, and cytokine receptors [11,12] and may be involved in TCR-induced human T cell proliferation [19]. Type I and type II IFN, SCF, IL-2, IL-5, FGF and PDGF induce or increase tyrosine phosphorylation of CrkL [20–26]. In addition, IL-2, IL-5, and type I IFN, induce or increase the formation of a complex between CrkL and Stat5 [25,27,28]. The binding of this complex to Stat5 responsive elements in the DNA has been demonstrated in vitro [24,28–31]. Nevertheless, the functional consequences of this binding remain to be determined.
CrkL overexpression has been reported in numerous solid tumors including glioblastoma, rhabdomyosarcoma, lung, colon, breast, ovarian, thyroid and gastric cancer [32]. In chronic myeloid leukemia (CML) cells from patients, CrkL is constitutively phosphorylated by the p210 BCR-ABL oncoprotein [33]. Noteworthy, Stat5 is constitutively active in haematological malignancies, including CML [34] and both CrkL and Stat5 are among the BCR-ABL substrates required for leukemogenesis [35–37].
In this study we report the identification of proteins with IL-7-induced or increased tyrosine phosphorylation and investigate the role of Crk family proteins in IL-7 signaling.
2. Materials and methods
2.1. Mice
C57BL/6 mice were obtained from the Animal Production Facility of the National Cancer Institute –Frederick Cancer Research and Development Center (Frederick, MD) and Rag1-deficient mice, B6 background, from The Jackson Laboratory (Bar Harbor, ME). The mice carrying the Cre-dependent conditional alleles for Crk and CrkL, generously provided by Dr. A Imamoto, were generated in accordance to animal protocols approved by the IACUC of the University of Chicago. The Crk conditional allele was generated by inserting two loxP sites upstream and downstream of Exon 1 by homologous recombination in a line of ES cells with a B6 inbred background. A mouse strain carrying a CrkL allele, CrkLtm1a(EUCOMM)Hmgu, was generated using ES cells in a B6-inbred background, obtained from EUCOMM/Helmholtz Zentrum Muenchen [38,39]. To make it a Cre-dependent conditional allele, the lacZ and neo cassettes were removed by genetic cross with a FLP strain to derive a conditional allele, CrkLtm1c(EUCOMM)Hmgu, leaving only one FRT and one loxP sites upstream of Exon 2 and one loxP site downstream of Exon 2. For the current study, the Crk and CrkL conditional strains were intercrossed to obtain compound homozygotes. Animal care was provided in accordance with procedures outlined in the “Guide for Care and Use of Laboratory animals” (National Institute of Health, Bethesda, MD, 1996).
2.2. Preparation of cells
The culture medium for murine cells was RPMI 1640 supplemented with 10% fetal bovine serum (Hyclone, Logan, UT), 2 mM glutamine, 100 U/ml penicillin, 100 μg/ml streptomycin and 50 μM β-mercaptoethanol (Invitrogen, CA, USA). For human cells β-mercaptoethanol was omitted. The murine IL-7-dependent cell line D1 [10] was maintained in medium supplemented with 50 ng/ml of murine IL-7 (Peprotech Inc., Rocky Hill, NJ). Cells, washed twice with phosphate buffered saline (PBS) were deprived of IL-7 for the indicated times (5 × 105/ml) prior stimulation with IL-7 (50 ng/ml). T cells from lymph nodes of C57BL/6 mice and splenocytes from Rag-1-deficient mice were obtained as previously described [40]. Pro-B cells were negatively selected by cell sorting using the following moAb: Fitc-anti-Gr-1 and anti-TER-119 moAb, and PE-anti-Mac-1 moAb (Pharmingen) [40]. To obtain pro-B cells from C57BL/6 mice Fitc-anti-Ig-M and anti-CD3 Moab (Pharmingen) were added. Cells, separated using a MoFlo high speed cell sorter (Dako Cytomation, Fort Collins, Co) yielded 90% B220+ CD19+ IgM− cells. Cells were cultured without IL-7 for at least 48 h prior IL-7 stimulation. Human T lymphocytes were isolated from leukopacks (Transfusion Medicine Department, NIH Clinical Center, Bethesda, MD) as previously described [41], and were > 80% CD3+. Human recombinant IL-7 was generously provided by Dr. S. Giardina (NCI). Freshly isolated cells or cells cultured in medium alone exhibited variable basal levels of CrkL phosphorylation, possibly attributable to exposure to IL-7 or other stimuli in vivo, or to the presence of IL-7 in murine and human serum [5,42]. Therefore, as previously described by others [43], T cells were cultured for 5 days in medium containing murine or human IL-7, deprived of IL-7 for at least 48 h and stimulated with IL-7 (50 ng/ml) for 15 min or the indicated times. Viability, evaluated by trypan blue staining before and after IL-7 deprivation, was higher than 90%.
2.3. Two-dimensional (2D) gel electrophoresis preparation and detection of tyrosine phosphorylated proteins
Preparation of soluble proteins: D1 cells (6–8 × 107) were washed twice in cold PBS. Cell pellets were resuspended in 1 ml of cold hypotonic buffer 1 (Biorad) containing protease inhibitors (Roche, Mannheim, Germany) and 1 mM activated sodium orthovanadate, sonicated with a microultrasonic cell disrupter (Kontes, Vineland, NJ) on ice and spun down twice (20,000× g at 4 °C for 20 min). Supernatants, supplemented with 1 mM MgCl2, were treated with benzonase (250 units) (Sigma, St Louis, MO) at 37 °C for 30 min, centrifuged (20,000× g at 4 °C for 20 min) and stored at −70 °C.
First-dimension electrophoresis: protein samples (150 μg/20 μl) in 320 μl IPG (immobilized pH gradient) gel rehydration buffer containing 8 M urea, 2% (wt/vol) 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS), 2% (vol/vol) IPG (pH 4–7) buffer, 40 mM DTT, and 0.01% (wt/vol) bromophenol blue were loaded onto 18-cm long Immobiline DryStrips (pH 4–7) allowing proteins to enter the gel during the overnight rehydration and equilibration process. After equilibration proteins were separated by isoelectric focusing using an Ettan IPGphor II electro-phoresis unit (GE Healthcare). The strips were run for a total of 35 kVh. After the isoelectric focusing strips were equilibrated for 15 min with SDS equilibration buffer containing 50 mM Tris-HCl (pH 8.8), 6 M urea, 30% glycerol, 2% SDS, and a trace of bromophenol blue and used in second-dimension electrophoresis.
Second-dimension electrophoresis: electrophoresis was performed using a Multiphor II (GE Healthcare) flatbed system with an ExcelGel SDS 12–14% precast polyacrylamide gradient gel (GE Healthcare). Each sample was simultaneously run on two gels under the same conditions. One gel was stained with Commassie Simply Blue (Life Technologies) and used as a control. The proteins from the second gel were electroblotted at a constant current of 250 mA by a semidry horizontal transfer technique using a Multiphor II NovaBlot unit, onto polyvinylidene difluoride Hybond-P membranes (GE Healthcare) in 25 mM Tris base, 192 mM glycine, and 20% methanol.
Tyrosine phosphorylated proteins were detected by immunoblotting using biotinylated 4G10 moAb (Upstate, Millipore) and HRP conjugated streptavidin (Millipore).
2.4. Protein identification
Corresponding images from Coomassie stained precast gels and developed Western blots were superimposed and analysed using PDQuest 2-D Analysis Software (Bio-Rad). Coomassie stained spots were excised and analysed by MALDI mass spectrometry using an EX-Quest spot cutter (Bio-Rad), processed and analysed as previously described [44]. Briefly, proteins were in-gel digested overnight at 37 °C with trypsin (Promega) at a concentration of 10 ng/μl in a solution of 25 mM ammonium bicarbonate and 10% acetonitrile. Peptide mass fingerprinting was done on a Bruker Reflex III MALDI-TOF mass spectrometer equipped with a SCOUT 384 multiprobe inlet and a 337 nm nitrogen laser using α-cyano-4-hydroxycinnamic acid as the matrix. Tryptic digests were concentrated and desalted using ZipTip pipette tips with C18 resin and a 0.2-μl bed volume (Millipore). The mass spectra were internally calibrated with trypsin autolysis fragments (842.51, 1045.56, 2211.10, and 2283.18 m/z). Data searches were done using the MASCOT software package available from Matrix Science, allowing up to one missed trypsin cleavage, carbamidomethylation of Cys and oxidation of Met as variable modifications, and a mass tolerance of ± 0.1 Da. In the MASCOT program, probability-based molecular weight search (MOWSE) scores > 55 were considered significant (P < 0.05) searching Mus musculus sequences deposited in the Swiss-Prot database [45].
2.5. Western blot analysis and co-immunoprecipitation
Western blot analysis was performed as previously described [46]. For immunoprecipitation cells were lysed in a 0.5% Tryton X-100, 50 mM Tris HCL, 150 mM NaCL, 2 mM EDTA buffer supplemented with protease inhibitors (Roche) and 1 mM activated sodium orthovanadate for 15 min on ice. Equal amounts of proteins were incubated overnight with the appropriate antibodies at 4 °C, and then loaded onto protein G-Agarose beads (Invitrogen). After 1 h beads were washed 5 times with 0.1% Tryton X-100, 50 mM Tris HCL, 150 mM NaCL, 2 mM EDTA washing buffer and boiled in SDS sample buffer. For immunoblotting the following antibodies were utilized: polyclonal rabbit anti-phospho-CrkL tyrosine 207/CrkII tyrosine 221, anti-phospho-Jak1, (Cell Signaling, Beverly, MA), anti-CrkL, anti-Stat5 (Santa Cruz, Ca, USA), murine anti-CrkL specific moAb (clone 5–6, Millipore), anti Crk moAb (clone 22/Crk (BD, Transduction Laboratory)) recognizing both CrkII and CrkL [16], polyclonal anti-rabbit and anti-murine horseradish peroxidase conjugated antibodies and rabbit anti-β/γ actin horseradish peroxidase conjugated moAb (Cell Signaling).
2.6. DNA content analysis
Imatinib, kindly provided by Novartis was solubilized in dymethylsulfoxide (DMSO) (Sigma) and stored in aliquots at −20° C. D1 cells were deprived of IL-7 for 6 h in the presence of Imatinib or of DMSO alone, at the indicated concentrations, then were cultured in the presence of IL-7 for 24 h. Nuclear hypodiploidy and condensation were detected by propidium iodide staining. Cells were washed twice with PBS and fixed in 75% cold ethanol for 1 h. After fixation cells were washed twice with PBS and stained with 50 μg/ml propidium iodide (Sigma) in the presence of 25 μg/ml ribonuclease A (Sigma) overnight at 4 °C and analysed by flow cytometry.
2.7. Retroviral transduction of Cre recombinase in bone marrow pro-B cells
Bone marrow cells from C57BL/6 male mice and from age-and sex-matched C57BL/6 mice bearing loxP-flanked CrkL alleles or Crk/CrkL alleles were depleted of mature T and B cells and enriched in cycling B220+ pro-B cells by negative selection using anti-Pe microbeads recognizing Pe-anti-Gr-1, -TER-119, -Mac-1,-IgM, -CD4 -CD8 moAbs (Pharmingen) and LS MACS columns (Miltenyi). Cells bearing loxP-flanked CrkL or Crk/CrkL alleles were transduced with the retroviral vector pMSCV-Cre recombinase-IRES-GFP or with the pMSCV-IRES-GFP empty vector using Lipofectamine 2000 (Invitrogen) in the presence IL-7 for 67 h. At 67 h Crk protein expression was markedly reduced, but the cells were ≥90% viable. To exclude toxicity due to non-specific Cre-recombinase activity cells from wild type C57BL/6 mice were transduced with pMSCV-Cre recombinase-IRES-GFP vector or with the empty vector and they were ≥ 90% viable (data not shown). Cells stained with APC anti-B220 moAb (Pharmingen) were purified using a MoFlo high speed cell sorter (Dako) yielding > 90% B220+ pro-B cells from wild type mice, B220 + GFP+ and B220 + and B220 + GFP− pro-B cells from mice bearing loxP-flanked CrkL or Crk/CrkL alleles transduced with pMSCV-Cre recombinase-IRES-GFP or with the empty vector as indicated. Viability was evaluated by trypan blue staining.
2.8. RNA extraction and microarray analysis
Total RNA was extracted from purified pro-B cells and residual DNA was digested using the RNA easy kit (Qiagen, Valencia, CA, USA). Quality of independently prepared triplicate RNA samples was evaluated using the RNA Integrity Number (RIN) on the Agilent Bioanalyzer. The microarrays were performed using the low-input Affymetrix Gene Chip Mouse Genome 430 2.0 at the Laboratory of Molecular Technology, Leidos Biomedical Research Inc., Frederick, MD. Two sets of samples were compared: wild type purified pro-B cells after culture with or without IL-7 for 67 h (IL-7 vs No IL7) and purified pro-B cells bearing lox-P flanked Crk and CrkL alleles transduced with pMSCV-Cre recombinase-IRES-GFP or with the control vector pMSCV-IRES-GFP, cultured with IL-7 for 67 h (pMSCV-Cre recombinase vs empty PMCSV).
The Affymetrix CEL files, representing triplicates for each of the experimental groups, were uploaded onto Partek Genomics Suite v6.6. The Gene Expression module workflow was executed for analysis of the raw data that passed all initial quality checks, including principal component analysis (PCA). To assess for differential expression of genes, Analysis of Variance (ANOVA) test was implemented to compare between the respective sample groups (IL-7 vs No IL-7 and pMSCV-Cre recombinase vs empty pMSCV) of the microarray experiments. To compare the IL-7 vs No IL-7 sample groups the default statistical cutoffs of adjusted P-value (False Discovery Rate, FDR) < 0.05 and fold-change of > 4 or < −4 were applied to generate a list of significantly differentially expressed genes. To compare pMSCV-Cre recombinase vs empty pMSCV sample groups the default statistical cutoffs of adjusted P-value (False Discovery Rate, FDR) < 0.05 and fold-change of > 2 or < −2 were applied. The same differential expression test (ANOVA) was applied between IL-7 treated wild type pro-B cells and IL-7 treated pro-B cells transduced with the empty vector (empty pMSCV). Zero genes failed to pass the nominal P-val < 0.05.
2.9. Quantitative real-time PCR
For gene detection RNA (1 μg) was used for reverse transcription using TaqMan Reverse Transcription Reagents (Applied Biosystem, Life Technologies) and Real-time PCR was performed by using the SsoAdvanced Universal SYBR Green Supermix with an amplification program as follows: 30 s at 95 °C, 15 s at 95 °C and 30 s at 60 °C. The reaction was followed by a melting curve protocol according to the specification of the CFX96 Real Time PCR System (Biorad). Expression of murine LAIR1 was normalized to murine GAPDH expression. Primers used were as follows: LAIR1 Fw 5ʹ GCTCTGACCAGACCTGGTAAGG-3ʹ, LAIR1 Rev. 5ʹ-CCATGTGTGTCTCCAGGTGTGC-3ʹ, GAPDH, Fw 5ʹ-ATC AACGACCCCTTCAT-3ʹ, GAPDH Rev. 5ʹ-CACACCCATCACAAACAT-3ʹ [47]. Relative quantification of gene expression was calculated according to the comparative method of 2−ΔΔCT.
3. Results
3.1. Identification of tyrosine phosphorylated proteins in IL-7 stimulated D1 cells
IL-7-dependent D1 cells deprived of IL-7 for 12 h were stimulated with IL-7 (50 ng/ml) for 15 min. The soluble protein fractions from unstimulated and IL-7 stimulated cells were separated by 2D gel electrophoresis, subjected to immunoblotting and probed with the anti-phosphotyrosine antibody 4G10. Equal amounts of proteins were separated in parallel and stained with Coomassie blue staining (Fig. 1). Gel spots correspondent to 4G10 immunoreactivity were excised from Coomassie stained gels and analysed by MALDI mass spectrometry.
Fig. 1. 2-D Western blot analysis with anti-phosphotyrosine antibody of lysates from IL-7-stimulated D1 cells.
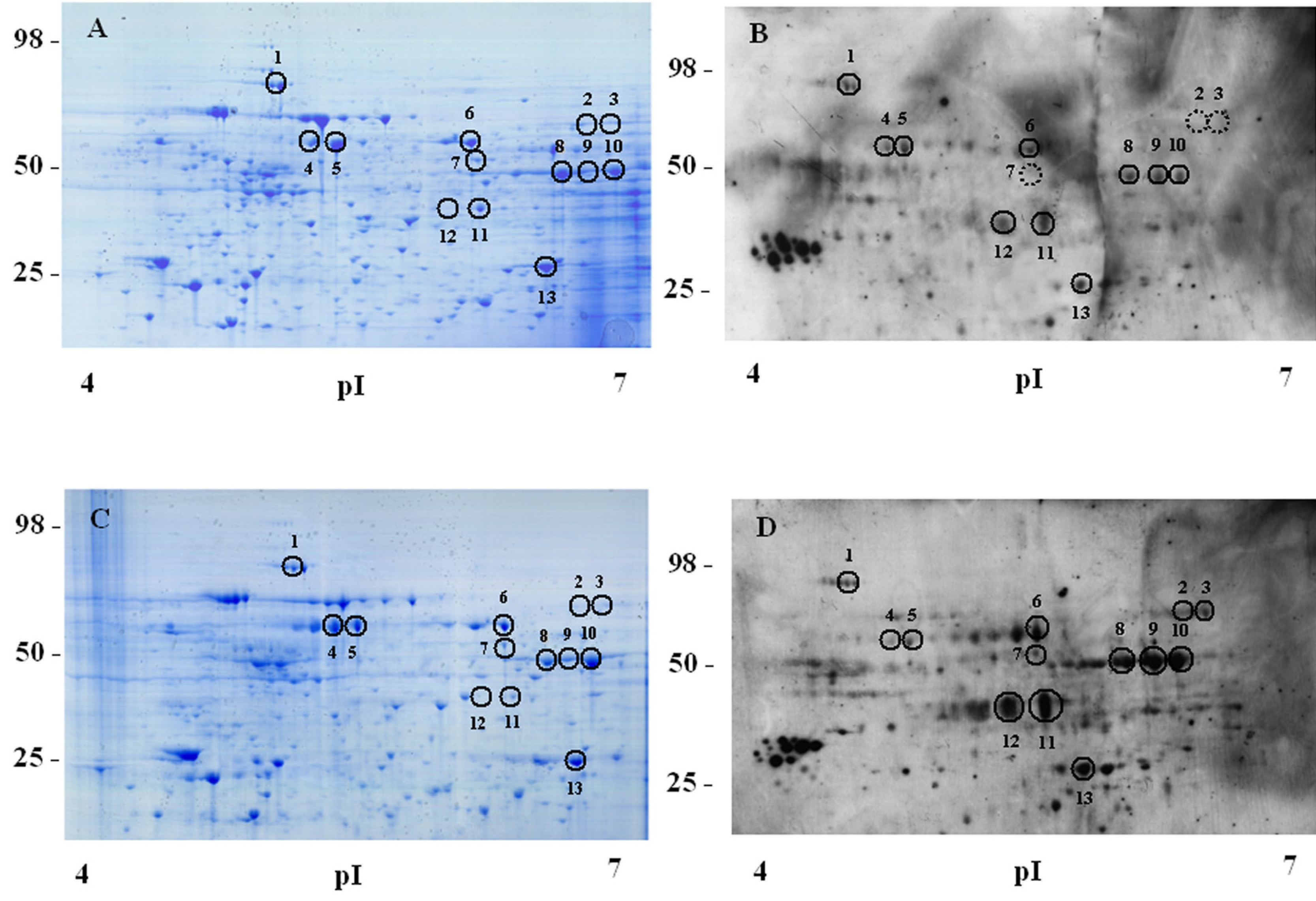
Soluble protein fractions from lysates of unstimulated or IL-7 stimulated D1 cells were analysed by 2D Western blot analysis and Coomassie blue gel staining as described in Materials and Methods. A, Unstimulated cells, Coomassie blue staining; B, Unstimulated cells, immunoblot with anti-phosphotyrosine MoAb; C, IL-7-stimulated cells, Coomassie blue staining; D, IL-7-stimulated cells, immunoblot with anti-phosphotyrosine MoAb. Spots are numbered as reported in Table 1, showing protein identification. Representative of 3 experiments.
Our initial analysis (pI 4–7) concerned 13 spots that corresponded to 10 proteins (Fig. 1 A–D, Table 1). The immunoreactivity of spots 2, 3 and 7, was detected only after IL-7 stimulation, these spots were identified as Stress-induced phosphoprotein 1 (STI1), 5-aminoimida-zole-4carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase (ATIC), and Heterogeneous Nuclear Ribonuclease H (hnRNPH). The immunoreactivity of 8 spots (1, 6, 8–13) was increased upon IL-7 stimulation. Six proteins (STI1, ATIC, hnRNPH, CrkL, alpha-enolase and transitional andoplasmic reticulum ATPase) (Table 1) were previously known to be tyrosine phosphorylated. Interestingly, spot 11 was identified as the adaptor protein CrkL and was selected for further study as will be described.
Table 1.
Identification of 4G10 immunoreactive proteins from IL-7 stimulated D1 cells.
| Spot | Proteina | ACb | Mrc(cal) | pId (cal) | Mre(obs) | pIf (obs) | Score | Covg | Peph |
|---|---|---|---|---|---|---|---|---|---|
| 1 | Transitional endopl. retic. ATPasei | Q01853 | 89.3 | 5.1 | 92 | 5.0 | 157 | 30 | 26 |
| 2l | PURH (ATIC)m | Q9CWJ9 | 64.2 | 6.3 | 65 | 6.6 | 70 | 42 | 16 |
| Stress-induced phosphoprotein 1 | Q60864 | 62.5 | 6.4 | 65 | 6.6 | 66 | 39 | 22 | |
| 3 | Stress-induced phosphoprotein 1 | Q9CWJ9 | 62.5 | 6.3 | 65 | 6.8 | 112 | 51 | 26 |
| 4 | Heat shock protein 60 | P63038 | 57.9 | 5.3 | 58 | 5.6 | 156 | 55 | 20 |
| 5 | Heat shock protein 60 | P63038 | 57.9 | 5.3 | 58 | 5.7 | 145 | 57 | 24 |
| 6 | Protein disulfide-isomerase A3 (ERp57) | P27773 | 54.2 | 5.3 | 60 | 6.2 | 71 | 33 | 18 |
| 7 | Heterogeneous nuclear ribonucl. Hn | O35737 | 49.1 | 5.9 | 52 | 6.2 | 86 | 37 | 16 |
| 8 | Alpha-enolase | P17182 | 47.1 | 6.3 | 50 | 6.3 | 72 | 42 | 15 |
| 9 | Alpha-enolase | P17182 | 47.1 | 6.3 | 50 | 6.4 | 83 | 44 | 18 |
| 10 | Alpha-enolase | P17182 | 47.1 | 6.3 | 50 | 6.5 | 92 | 50 | 18 |
| 11 | Crk-like protein | P47941 | 33.8 | 6.2 | 37 | 6.2 | 59 | 29 | 9 |
| 12 | PDZ domain-containing pr. GIPC1o | Q9Z0G0 | 36.1 | 5.6 | 35 | 5.8 | 76 | 52 | 18 |
| 13 | High mobility group 1 protein B1 | P63158 | 24.9 | 5.7 | 27 | 6.4 | 68 | 51 | 15 |
| 14p | ATP synthase subunit beta mithoc.q,o | P56480 | 51.7 | 4.9 | 55 | 4.6 | 160 | 44 | 21 |
| 15p | Protein disulfide-isomerase (PDI) | P09103 | 55.1 | 4.7 | 50 | 4.5 | 99 | 39 | 17 |
| 16p | UV excission rep. pr. RAD 23 h Br | P54728 | 43.5 | 4.8 | 50 | 4.5 | 60 | 30 | 14 |
| 17p | Lymphocyte-specific protein 1 | P19973 | 36.7 | 4.8 | 40 | 4.6 | 86 | 30 | 8 |
| 19p | Nuclease sensitive element bind.pr.1s | P62960 | 35.7 | 9.9 | 30 | 6.0 | 114 | 49 | 16 |
| 20p | Hepatoma-derived growth factor | P51859 | 26.2 | 4.8 | 25 | 5.0 | 66 | 34 | 10 |
Proteins 1–13 correspond to the numbered spots shown in Fig. 1.
Accession number (UniProKB/Swiss-Prot).
Calculated Mr.
Calculated pI.
Observed Mr.
Observed pI.
Coverage.
Peptides.
Transitional endoplasmic reticulum ATPase.
Mixture.
5-aminoimidazole-4carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase.
Heterogeneous Nuclear Ribonuclease H.
PDZ domain-containing protein GAIP interacting protein C1.
Gel not shown, first dimension electrophoresis performed on pH 3–10 IPG strips.
ATP synthase subunit beta mithocondrial.
UV excision repair protein RAD 23 homologue B.
Nuclease sensitive element binding protein 1.
In a second set of experiments proteins from IL-7 stimulated cells were separated by 2D gel electrophoresis in a wider range of pH (pI 3–10), subjected to immunoblotting and probed with 4G10. Spots 14–20 (Table 1), displaying a marked 4G10 immunoreactivity (data not shown) were excised from 2D Coomassie stained gel and analysed by mass spectrometry. We identified 6 proteins (Table 1). Their level of phosphorylation in unstimulated cells was not analysed, but, interestingly, only one of these proteins (ATP synthase subunit beta mitochondrial) was previously known to be tyrosine phosphorylated.
3.2. Tyrosine phosphorylation of CrkL in murine and human T cells
CrkL is involved in multiple signal transduction pathways induced by hematopoietic cytokines, thus we studied the effect of IL-7 on CrkL phosphorylation in D1 cells and in primary cells using an antibody recognizing CrkL phosphorylation at the 207 tyrosine residue. The soluble protein fractions from unstimulated and IL-7 stimulated D1 cells were separated by 2D gel electrophoresis and subjected to immunoblotting. D1 cells showed a basal level of phosphorylation that was increased by IL-7 stimulation (Fig. 2). We assessed whether this occurred in primary T cells. The basal level of phospho-CrkL in freshly isolated murine or human T cells was somewhat variable, possibly reflecting variable exposure to IL-7 or other stimuli in vivo before isolation. In addition, different basal levels of phospho CrkL expression were observed in T cells cultured with different lots of bovine serum (data not shown). It is known that murine and human serum contain IL-7 [5,42]. We therefore adopted a previously published method used to assess the induction of CrkL phosphorylation in human T cells [43]: cells cultured in medium containing IL-7 for 5 days were deprived of IL-7 for 48 h. Cells (> 90% viable) were then stimulated with IL-7 (50 ng/ml) for 3, 15 min and other indicated time points (Fig. 3 A, B). Cell lysates from unstimulated or IL-7 stimulated cells were subjected to Western blot analysis and probed with anti-phospho-CrkL antibody. In unstimulated murine T cells CrkL showed a basal level of phosphorylation at all time points. IL-7 increased phosphorylation, this was still evident after 45 min (Fig. 3 A), persisted for 6 h (data not shown), and was lost by 40 h (Fig. 3 A). In human T cells CrkL phosphorylation increased within 3 min, persisted at high level up to 7 h and declined at 48 h. In the absence of IL-7 a basal level of CrkL phosphorylation was always evident (Fig. 3 B).
Fig. 2. CrkL tyrosine phosphorylation in IL-7-stimulated D1 cells.
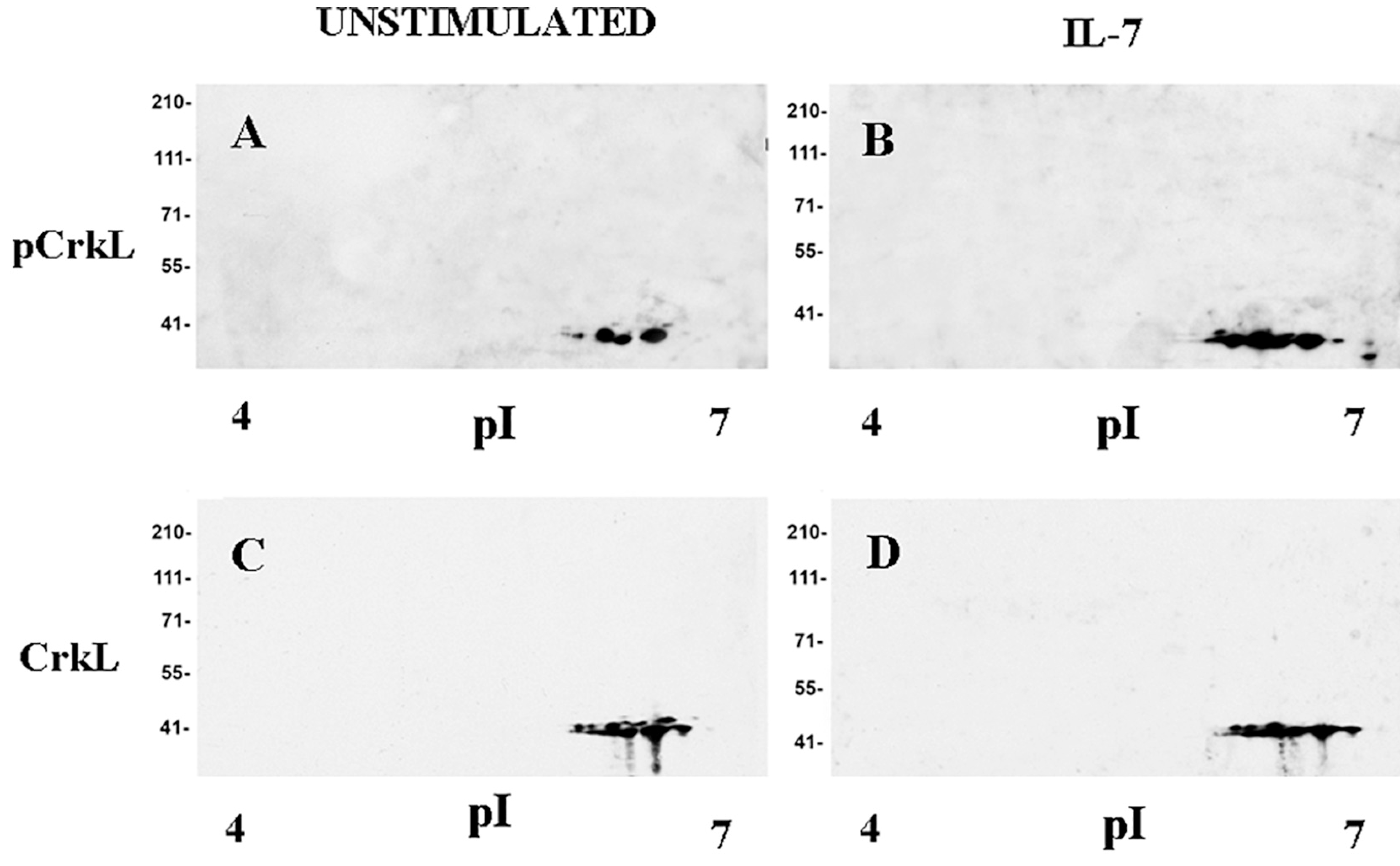
Soluble protein fractions from unstimulated or IL-7-stimulated D1 cells were separated and 2D Western blot analysis was performed as described in Materials and Methods. A, Unstimulated cells, anti-phospho-CrkL antibody; B, IL-7-stimulated cells anti-phospho-CrkL antibody; C, Unstimulated cells, anti-CrkL antibody; D, IL-7-stimulated cells, anti-CrkL antibody. Representative of 3 experiments.
Fig. 3. Tyrosine phosphorylation of CrkL in IL-7-stimulated murine and human T cells and in murine pro-B cells, and association of CrkL with Stat-5 in IL-7 stimulated human T cells.
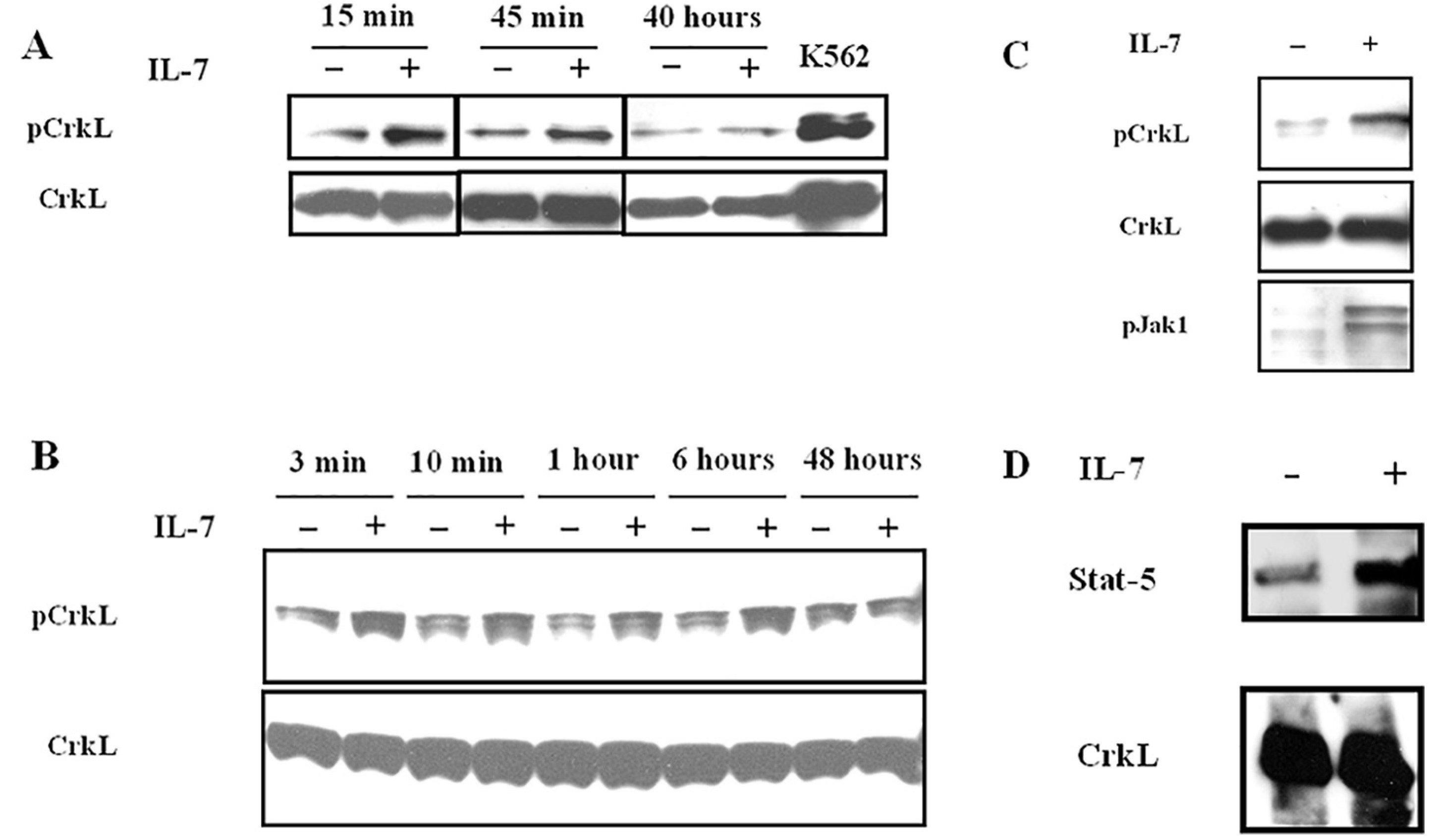
Western blot analysis with anti-phospho-CrkL, anti-phospho-Jak1, and anti-CrkL polyclonal antibodies. A, Murine T cells purified from lymph nodes as described in Materials and Methods were incubated with or without IL-7 for the indicated times. Representative of 3 experiments; B, Human T lymphocytes purified as described in Materials and Methods were incubated with or without IL-7 for the indicated times. Representative of 3 experiments; C, Pro-B cells purified from spleens of Rag1-deficient mice as described in Materials and Methods were incubated with or without IL-7 for 15 min. Representative of 3 experiments; D, Coimmunprecipitation on cell lysates from unstimulated and IL-7 stimulated human T cells was performed with anti-CrkL antibody as described in Materials and Methods. Blots were probed with anti-Stat5 and anti-CrkL antibodies. Representative of 3 experiments.
3.3. Tyrosine phosphorylation of CrkL in murine pro-B cells
In mice IL-7 in combination with Flt3 ligand is indispensable for B cell development [48], which is impaired in IL-7- and IL-7R-deficient mice [49,50]. In the absence of IL-7 B cell development is arrested at the pre-pro B stage [51]. Moreover, the development of committed lymphoid B cells within the common lymphoid progenitor (CLP) compartment is impaired [52]. IL-7 induces proliferation of pro-B cells and protects them from apoptosis [53,54]. In Rag1−/− mice, mature lymphocytes of either B (B220 + IgM+) or T (CD3+) lineage are not detectable [55], providing a convenient source to purify pro-B cells. Pro-B cells, negatively selected from Rag1−/− mice spleens [40], were deprived of IL-7 for 48 h in vitro, then cultured in medium or in medium plus IL-7 for 15 min. Cell lysates were analysed by Western blot analysis. Unstimulated cells showed a basal level of phosphorylation. IL-7 stimulation clearly increased CrkL phosphorylation, and, as expected, induced Jak1 phosphorylation (Fig. 3 C). The experiment was also performed utilizing splenic pro-B cells from C57BL/6 mice with similar results (data not shown).
3.4. IL-7 increases the association of CrkL with Stat-5 in human T cells
IFN-α, IL-2 and IL-5 induce or increase the formation of CrkL-Stat-5 heterodimers [25,27,28]. The IL-7 effect on the formation of Stat-5-CrkL heterodimers was investigated. Human T cells cultured in medium containing IL-7 for 5 days and deprived of IL-7 for 48 h were stimulated with IL-7 for 15 min as described in Section 3.2 [43]. Immunoprecipitation with anti-CrkL antibody was followed by Western blot analysis with anti-Stat5 antibody. Stat-5 was found to be associated with CrkL in unstimulated cells and IL-7 stimulation increased its amount (Fig. 3D).
3.5. Induction of apoptosis by Imatinib in D1 cells
In CML cells the reciprocal chromosomal translocation t (9; 22) (q34;11) generates the Bcr-Abl fusion protein, a constitutively active tyrosine kinase that confers enhanced proliferative activity and resistance to apoptosis. Bcr-Abl constitutively phosphorylates CrkL and its activity correlates with the level of phosho-CrkL [33]. Imatinib is a potent and selective inhibitor of tyrosine kinases including Abl, PDGF α and β receptors, SCF receptor and Src by competing with ATP, required for the kinase activity [56]. Imatinib decreases CrkL phosphorylation by Abl kinase, induces apoptosis of CML cells, and is used to treat CML, Philadelphia chromosome positive ALL, myeloproliferative disorders due to PDGF receptor chromosomal rearrangement and gastrointestinal stromal tumors with SCF receptor mutations [56]. Moreover, it induced apoptosis of memory T cells in vitro and decreased the percentage of memory cells expressing IL-7R α chain in vivo [57,58].
To investigate whether Imatinib could counteract the anti-apoptotic effect of IL-7 we used the IL-7 dependent D1 cells [10]. Cells were deprived of IL-7 for 6 h in the presence of Imatinib (0.5–10 μM) or of DMSO, then IL-7 (50 ng/ml) or medium alone were added to the cell cultures. Twenty four hours later the percentage of apoptotic cells was determined. In the absence of IL-7 > 70% of D1 cells underwent apoptosis (Fig. 4A). Imatinib counteracted the anti-apoptotic effect of IL-7 in a dose dependent manner, with a significant effect observed at 1 μM(Fig. 4A). Cells from parallel cultures were lysed after 15 min of incubation. CrkL phosphorylation, evaluated by Western blot analysis, was markedly inhibited by Imatinib, and a decrease of the basal level of phosphorylation was also observed (Fig. 4 B). Thus CrkL phosphorylation and cell survival were both inhibited by Imatinib, although this does not prove that the former causes the latter. This led us to a more specific method for inhibiting Crks using genetic deletion.
Fig. 4. Effect of Imatinib on cell survival and CrkL phosphorylation in IL-7-dependent cells.
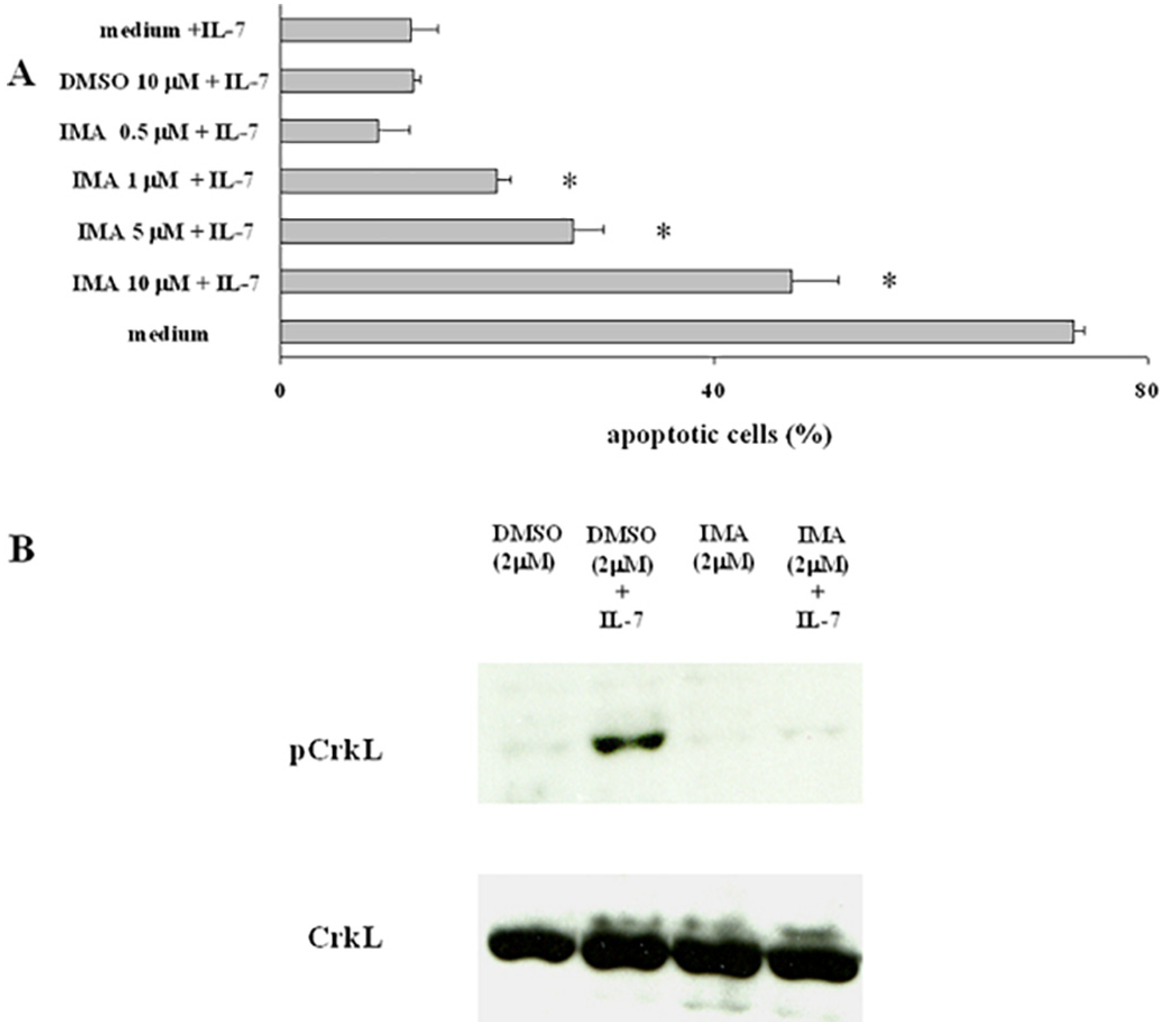
D1 cells were deprived of IL-7 in the presence of Imatinib or DMSO at the indicated concentrations for 6 h. A, Nuclear hypodiploidy and condensation were evaluated by propidium iodide staining following culture with IL-7 for 24 h. Representative of 5 experiments performed in triplicate, data are expressed as the mean ± SD; B, In parallel cultures following stimulation with IL-7 for 15 min, CrkL phosphorylation was evaluated by Western blot analysis. Representative of 3 experiments.
3.6. Apoptosis of Crk/CrkL conditionally-deficient pro-B cells cultured with IL-7
Redundant functions of Crk family proteins have been revealed in studies employing mice with floxed alleles. Thus deletion of single family members did not show a phenotype, whereas deletion of both CrkL and CrkII was necessary to detect functional defects in T cells, fibroblasts, muscle cells and neurons [17]. Whether pro-B cells express CrkII has not been previously reported. Antibodies that cross-react with CrkL and CrkII are available but antibodies specifically recognizing phospho CrkII or specifically immunoprecipitating CrkII are not available. A MoAb specifically recognizing CrkL, however, was available. We therefore investigated by Western blot analysis whether pro-B cells following the conditional deletion of CrkL expressed CrkII using the anti-CrkL specific MoAb and an anti-Crk moAb. The latter recognizes aminoacid sequences present in CrkL, CrkII, and CrkI, as reported by the antibody manufacturer and by previous studies [16]. As will be shown CrkL protein has a very long half life, thus we expected a sufficient CrkL depletion in conditionally-deficient cells would require extended culture, which we performed for 67 h. Pro-B cell-enriched bone marrow cells bearing loxP-flanked CrkL alleles cultured with IL-7 were transduced with pMSCV-Cre recombinase-IRES-GFP vector, sorted in GFP+ and GFP− cells and analysed by Western blot analysis. CrkL protein expression was markedly inhibited in Cre-recombinase transduced GFP+ cells compared to non transduced GFP− cells (Fig. 5A). In contrast, the anti-Crk MoAb showed positive immunoreactivity in both cell samples: two bands of 40 and 28 kDa respectively were observed, suggesting the presence of CrkII and CrkI in CrkL depleted cells (Fig. 5 A). These results pointed to the presence of CrkII and CrkI in pro-B cells that could functionally compensate for the absence of CrkL. We therefore deleted the whole family to investigate an effect on the pro-survival activity of IL-7 and found that survival was markedly impaired. Pro-B cells bearing loxP-flanked Crk/CrkL alleles, cultured in IL-7, were transduced with Cre-recombinase or with the empty vector and their survival was assessed after 5 days (Fig. 5 B). In two experiments the percentage of apoptotic cells was 17.54 and 38.9 in cells transduced with the empty vector, and 43.19 and 63.8% in cells transduced with Cre recombinase. These results are consistent with Crk/CrkL proteins performing a pro-survival role in IL-7-stimulated pro-B cells.
Fig. 6. IL-7-regulated transcripts in bone marrow pro-B cells.
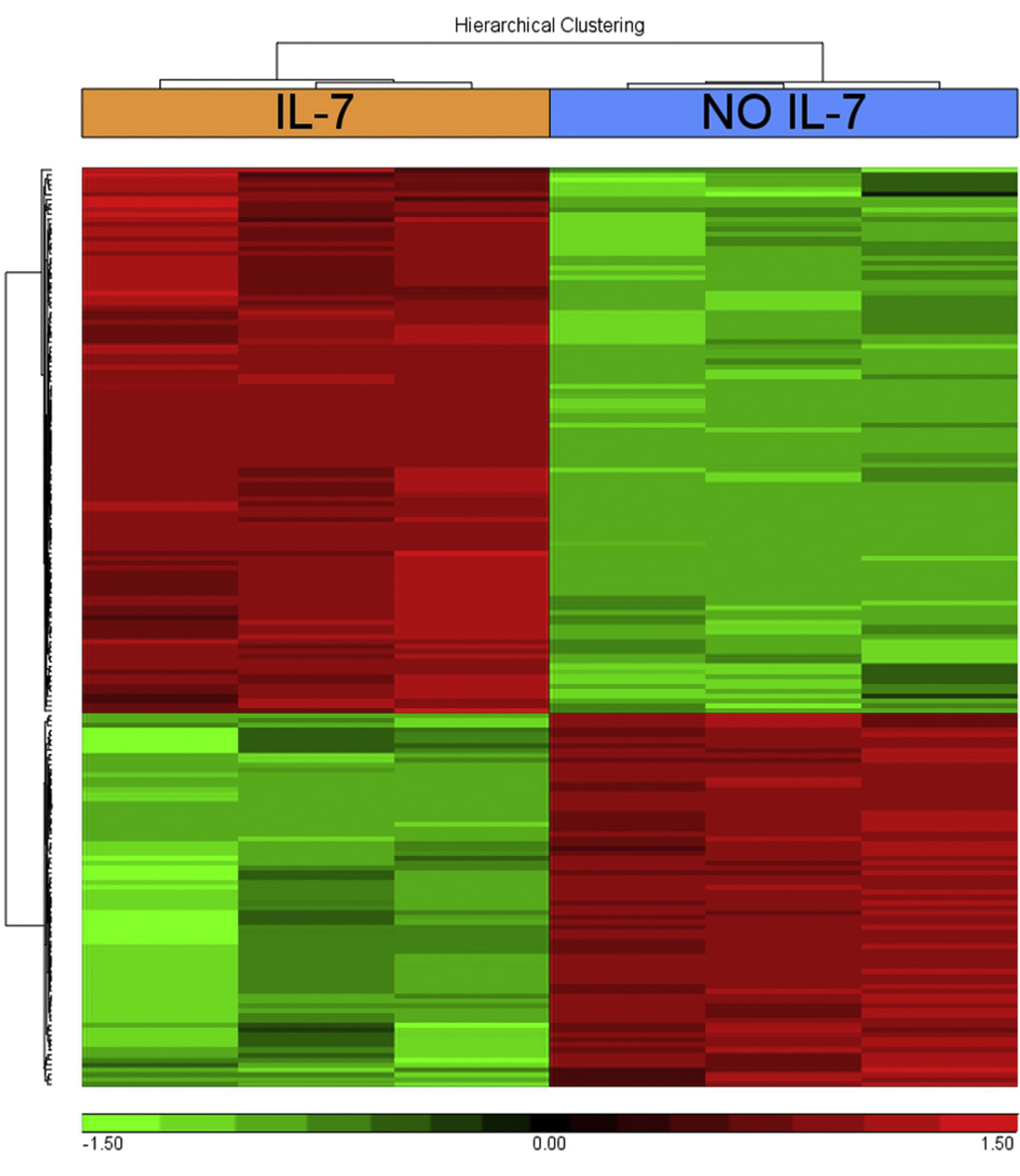
Hyerarchical clustering of genes that differentially showed a > 4 fold increase (red) or a < −4 fold decrease (green), gene titles are reported in Table S1. Bone marrow cells enriched in pro-B cells from C57BL/6 mice were cultured with or without IL-7 for 67 h. RNA from purified pro-B cells was analysed as described in Materials and Methods. Representative of 3 experiments.
3.7. IL-7-regulated gene expression of pro-B cells from wild type and Crk/CrkL conditionally-deficient mice
To further evaluate the role of Crk family proteins, gene expression studies were performed in wild type and in Crk/CrkL conditionally-deficient bone marrow pro-B cells. Pro-B cells from wild type C57BL/6 mice were cultured with or without IL-7 for 67 h. Differentially expressed genes with an increase or a decrease greater than four fold are listed in Table S1, and their hierarchical clustering is represented in Fig. 6. This analysis showed that IL-7 has numerous effects on pro-B cells. A significant differential expression of 187 genes was observed, 111 were upregulated and 76 downregulated. Some genes showing pronounced upregulation were known to be induced by IL-7 and to be related to B cell proliferation and differentiation: lymphotoxin α (24-fold) [59,60], regulatory myosin light chain 10 (13.50 fold) [61], protein kinase C η (8.40-fold) [62], and myc (6.83-fold) [63]. Other genes were expressed by B220+ bipotent common lymphoid progenitors that have the capacity to become thymic immigrants: Thy1 (18-fold), T cell receptor (TCR) constant 1 γ chain, (306-fold) and TCR variable 4 γ chain 4 (48.10-fold) [64–66]. Others were not previously known to be expressed by pro-B cells: suppressor of cytokine signaling 2 (SOCS-2) (12.89-fold), or to be induced by IL-7: Purkinje cell protein 4 (Pcp4) (185 fold), brain expressed gene 1 (BEX1) (8.14-fold), and neurofilament heavy peptide (9.5-fold).
Fig. 5. Expression of Crk I and II proteins by CrkL conditionally-deficient pro-B cells and apoptosis of conditionally-deficient Crk/CrkL pro-B cells.
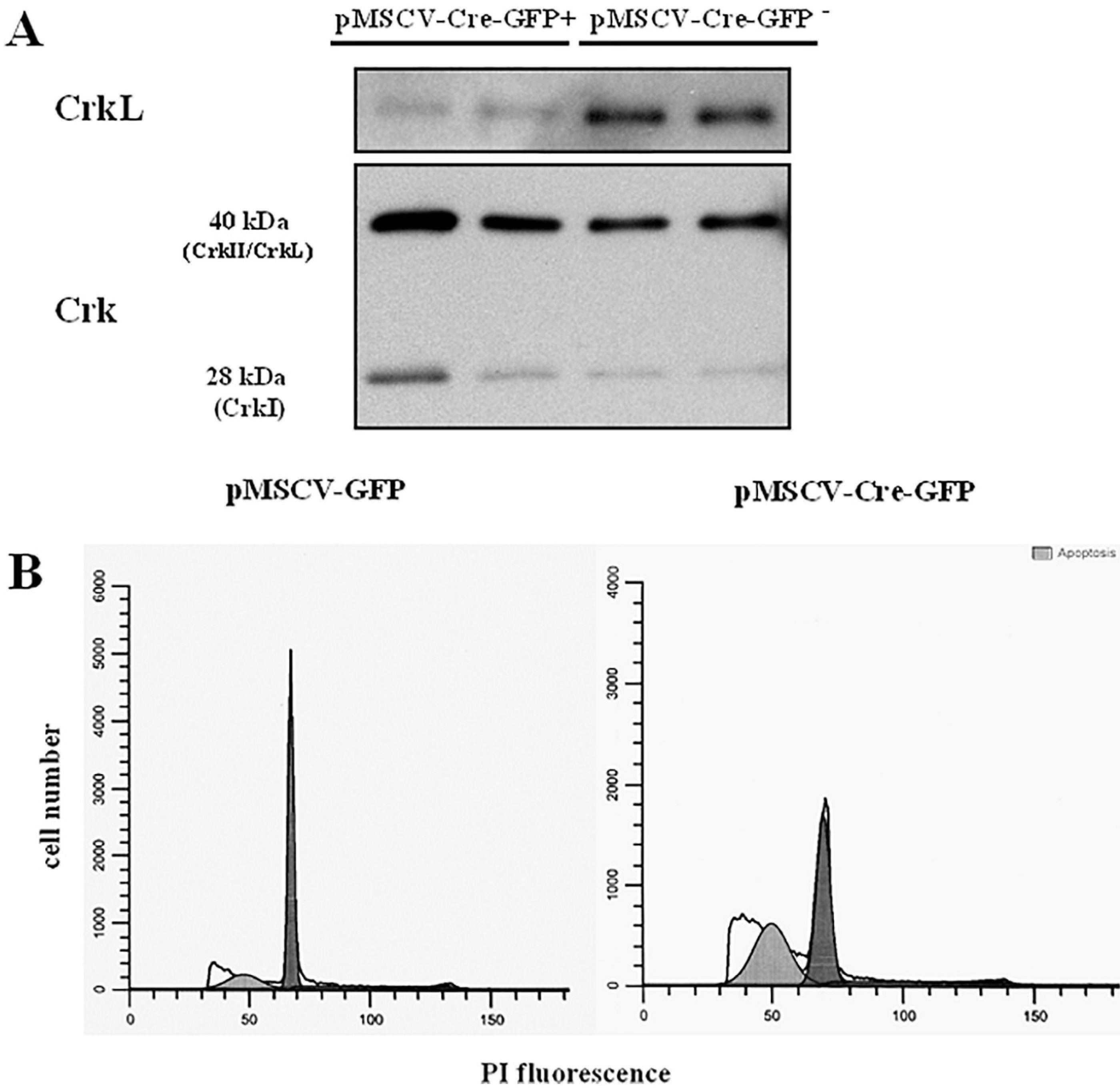
A: Western blot analysis with anti-CrkL and anti-Crk MoAb performed in duplicate. Purified pro-B cell bearing loxP-flanked CrkL alleles transduced with pMSCV-Cre recombinase-IRES-GFP in the presence of IL-7 for 67 h were sorted in GFP+ and GFP− cells. Representative of 3 experiments;B: Pro-B cells bearing loxP-flanked Crk/CrkL alleles were transduced with pMSCV-Cre recombinase-IRES-GFP or with the pMSCV-IRES-GFP empty vector in the presence of IL-7 and analysed after 5 days. The percentage of apoptotic cells was 43.19 and 17.54%, respectively. Representative of two experiments.
Down regulated genes included several receptors: CD209 a (−15.07-fold) and d (−7.06-fold), CD8 α (−8.03-fold) and β (−8.23-fold), CD7 (−6.15-fold) and CD28 (−5.03-fold), the FMS-like tyrosine kinase 3 receptor (Flt3) (−4.59-fold), expressed by dendritic cells, myeloid dendritic cells, myeloid cells, monocytes, and T cells [67–70] and an inhibitory receptor; the leukocyte-associated Ig-like receptor1 (LAIR1) (−4.41 fold).
To investigate the role of the Crk family of proteins in IL-7-regulated transcription we studied the gene expression profile of compound homozygous Crk/CrkL conditionally-deficient pro-B cells cultured in IL-7 for 67 h (≥90%viable). Confirming this conditional deletion at the protein level, in pro-B cell enriched bone marrow cells bearing loxP-flanked Crk/CrkL alleles transduced with Cre recombinase the expression of CrkL and Crk isoforms was markedly inhibited whereas no inhibition was observed in cells transduced with the control empty vector (Fig. 7A). The microarray analysis showed that CrkL and Crk RNA expression was significantly lower in pro-B cells transduced with Cre recombinase than in cells transduced with the control vector (Fig. 7 B and C). No significant difference was observed between the gene expression profile of pro-B cells transduced with the control vector and that of wild type pro-B cells (Methods, Section 2.8), indicating that the control vector did not modify pro-B cell gene expression.
Fig. 7. IL-7-regulated transcripts in Crk/CrkL conditionally-deficient pro-B cells.
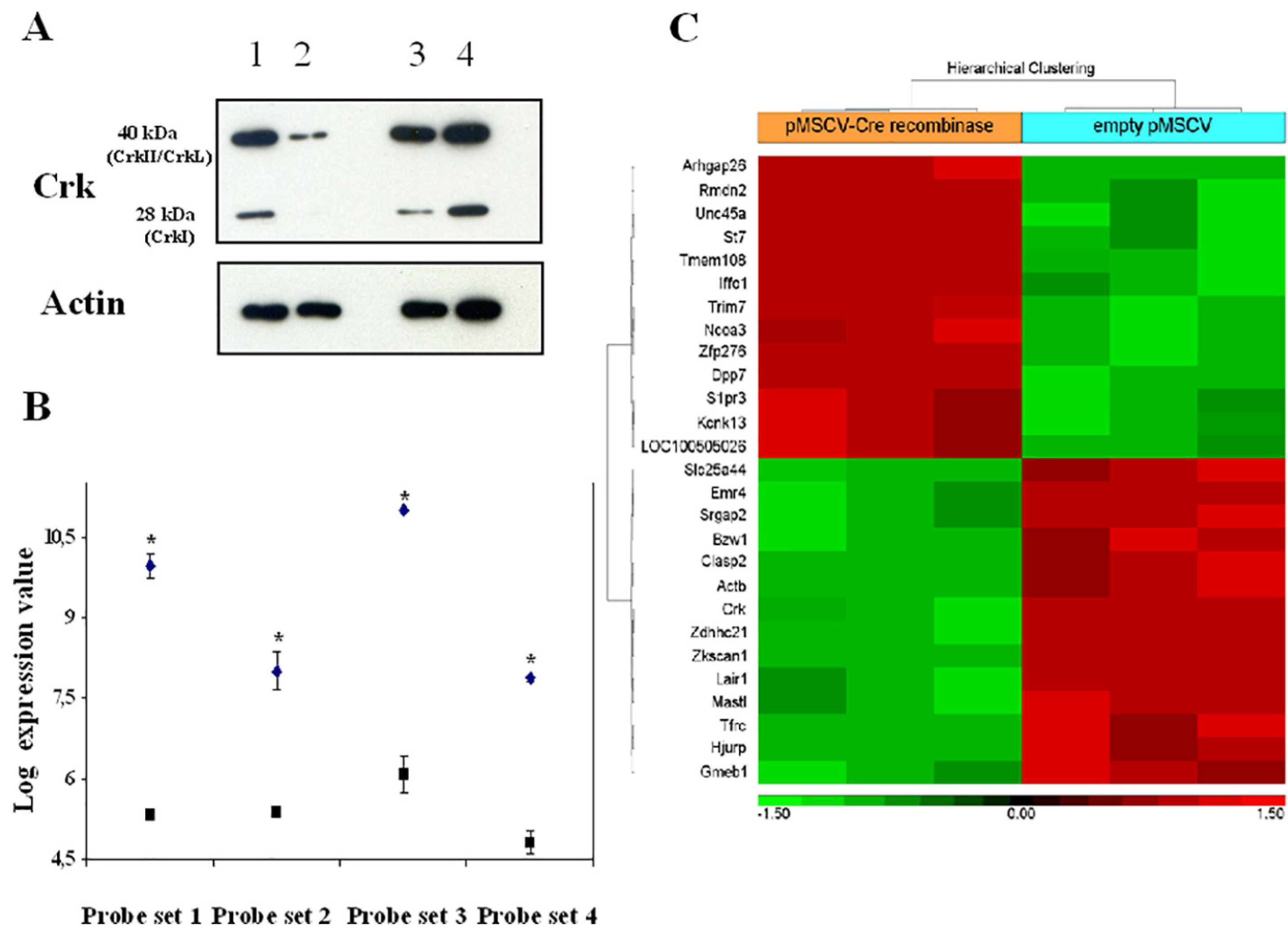
Cells from mice bearing loxP-flanked Crk/CrkL alleles cultured with IL-7 for 67 h were transduced with the retroviral vector pMSCV-Cre recombinase-IRES-GFP or with the control empty vector (pMSCV-IRES-GFP). A, Bone marrow cells enriched in pro-B cells: Western blot analysis with anti-Crk moAb. Lane 1: no vector; lane 2: pMSCV-Cre recombinase-IRES-GFP; lane 3: no vector; lane 4: empty vector pMSCV-IRES-GFP; B, RNA purified from pro-B cells was analysed by microarray as described in Materials and Methods. The 4 probe sets (1,416,201, 1,460,176 and 1,448,248) at FDR P-val < 0.05 (*), and 1,425,855 at nominal P-val < 0.05 (*) recognized homologous regions of Crk and CrkL RNA. Closed squares: cells transduced with pMSCV-Cre recombinase-IRES-GFP; closed diamonds: cells transduced cells with the control empty vector pMSCV-IRES-GFP. Representative of 3 experiments; C, Hyerarchical clustering of genes that differentially showed upregulation (> 2 fold-increase, red) or downregulation (<−2 fold-decrease, green). Bone marrow cells enriched in pro-B cells from C57BL/6 mice bearing loxP-flanked Crk/CrkL alleles cultured with IL-7 for 67 h were transduced with the retroviral vector pMSCV-Cre recombinase-IRES-GFP or with the control vector pMSCV-IRES-GFP. RNA from purified pro-B cells was analysed as described in Materials and Methods. Representative of 3 experiments.
In Crk/CrkL-deficient cells cultured with IL-7 for 67 h there was a significant differential expression of 26 genes, 13 upregulated and 13 downregulated. The hierarchical clustering of the Crk/CrkL regulated genes that significantly showed an increase or a decrease greater than two-fold is shown in Fig. 7 C. IL-7 inhibited LAIR1 expression (−4.41 fold, P < 0.0007) as also confirmed by qPCR (Table S1 and Fig. 8). Interestingly, Crk/CrkL deficiency inhibited the effect of IL-7 on LAIR1 (−1.55 fold P < 6.81 E-5, Fig. 7 C). Thus, of the IL-7-regulated genes (Table S1), LAIR1 was the most dependent on Crk/CrkL.
Fig. 8. Decrease of LAIR1 RNA expression induced by IL-7 evaluated by quantitative real time PCR.
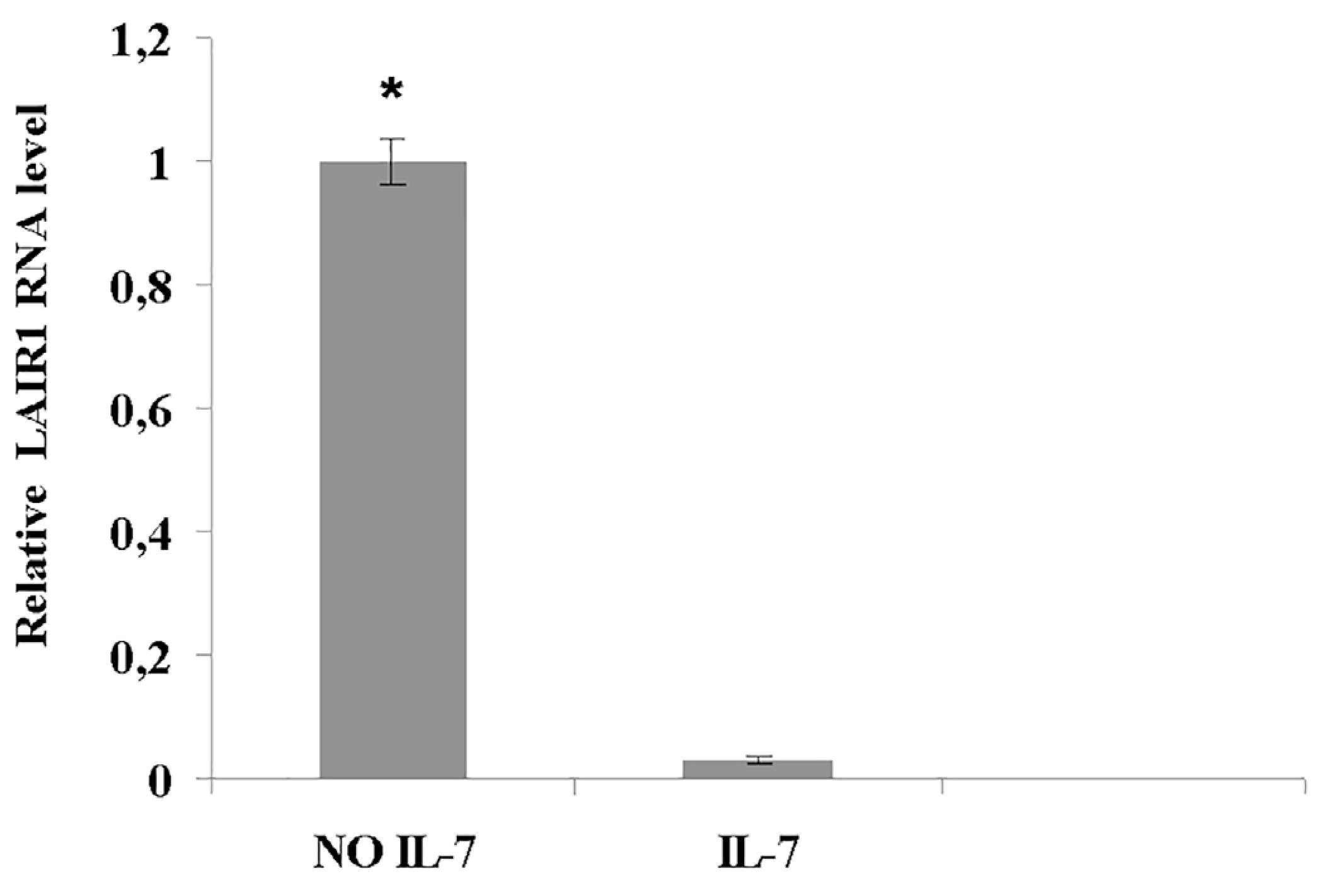
Pro-B cells were cultured with or without IL-7 for 67 h. Expression of LAIR1 was analysed by quantitative real time PCR as described in Materials and Methods. Samples were run in triplicate. P < 0.05 (*). Representative of two experiments.
4. Discussion
IL-7 induces the proliferation of double negative thymocytes and pro-B cells, and promotes their survival [3–5]. Mature T cells require IL-7 for survival and for homeostatic proliferation, whereas mature B cells do not express IL-7 receptor [3–6]. To identify tyrosine phosphorylated molecules involved in IL-7 signaling we utilized the IL-7 dependent cell line D1, which proliferates in response to IL-7, and undergoes apoptosis in its absence [10]. Sixteen proteins were identified as possible novel substrates of tyrosine phosphorylation in response to IL-7 stimulation. Most of these showed increased phosphorylation over a constitutive background, whereas in three proteins there was no prior detectable phosphorylation: STI1, ATIC, and hnRNP.
IL-7 function in the brain is largely unexplored. IL-7R is expressed by neurons, astrocytes, and oligodendrocytes [71,72]. It is controversial whether IL-7 is neurotrophic [71,73]. Secreted STI1 promotes the survival of neurons and astrocytes [74,75]. Intracellular STI-1 has been shown to be tyrosine phosphorylated in fibroblasts [76].The induction of STI1 phosphorylation could suggest an IL-7-mediated neurotrophic function.
ATIC catalyzes the last two steps of the de novo synthesis of purines, necessary for building nucleic acids. Inhibitors of purine biosynthesis (i.e. methotrexate) are used in cancer chemotherapy, and a specific ATIC inhibitor showed anti-proliferative activity [77]. Thus, ATIC phosphorylation may contribute to IL-7 signaling leading to proliferation.
The family of hnRNPs is a group of RNA-binding proteins that regulate alternative splicing, RNA capping, polyadenylation, and translation [78]. hnRNP H has been identified among other tyrosine phosphorylated substrates in a human promyelocytic leukemia cell line [79]. Its deregulated splicing activity promotes cancer cell survival and resistance to chemotherapy [80,81].
Thus, phosphorylation of STI1, ATIC, and hnRNP H may contribute to the proliferative and pro-survival effects of IL-7 in lymphoid and nonlymphoid cells.
The adaptor Crk-L, highly expressed in hematopoietic cells, integrates multiple signaling pathways and plays a relevant role in carcinogenesis [32,33,35,36], but its function in response to cytokine stimulation remains unclear. We demonstrated an increased CrkL phosphorylation at tyrosine 207 in the D1 cell line, and in mature murine and human T lymphocytes and this persisted for several hours. IL-7 stimulation also increased CrkL phosphorylation in murine pro-B cells, as in T cells, suggesting the activation of a common pathway. A basal level of phosphorylation was observed in unstimulated cells, and it was slightly variable between experiments. This was possibly due to the presence of IL-7 in murine and human serum, necessary for regulating survival and homeostatic proliferation (5, 42).
In T cells, T cell receptor triggering induced CrkL phosphorylation via Abl kinase [82]. The identity of the kinase activated by cytokine receptor triggering is unknown. Our experiments implicate the involvement of one of the Imatinib-inhibited kinases, activated downstream Jak1, which is not inhibited by Imatinib [83]. In pro-B cells, the kinase involved in CrkL phosphorylation might be the src family kinase Lyn, which is activated by IL-7 [84], and phosphorylates CrkL in hematopoietic cells [85]. The observation that Imatinib induced apoptosis in IL-7 stimulated D1 cells suggested that this drug could counteract IL-7-dependent pathways leading to survival, perhaps involving CrkL activity. In line with this hypothesis, Crk/CrkL conditionally-deleted pro-B underwent apoptosis, counteracting the survival effect of IL-7.
IL-7 stimulation, like other cytokine pathways, increased the association of CrkL with Stat-5. Structural studies have shown that phosphorylated tyrosine 207 binds to its own SH2 within CrkL itself, so that the SH2 would not be available for other molecules [18,86]. However, in contrast to CrkII, in CrkL the binding of the phosphorylated Y207 to its own SH2 does not reduce the affinity of the SH3 domain for another ligand. Thus, CrkL tyrosine phosphorylation is unlikely to inhibit CrkL function [18].
We investigated whether Crk family proteins could play a role in IL-7-regulated gene expression. Our data confirmed that IL-7 upregulated the transcription of cMyc, involved in proliferation, survival and lymphomagenesis [63], and of genes related to pro-B cell differentiation, such as the cytoskeletal regulatory myosin light chain [61], lymphotoxin α, involved in the formation secondary lymphoid organs [59,60], and protein kinase C η which induces Ig k rearrangement [62]. Other genes were not known to be expressed by pro-B cells: suppressor of cytokine signaling 2 (SOCS-2) and genes expressed in neurons: Pcp4, a calcium signaling regulator [87], the cytoskeletal neurofilament heavy peptide and BEX1 involved in axonal regeneration [88].
IL-7 downregulated the expression of LAIR1, a transmembrane inhibitory molecule expressed by committed hematopoietic progenitors, pro-B cells, thymocytes and all mature leukocytes [89]. LAIR1 induces phosphorylation of tyrosine-based inhibitory motifs and the recruitment of SH2 domain containing phosphatases, negatively regulating cellular signaling [89]. It inhibits BCR-mediated proliferation and cytokine production [90,91]. Downregulation of LAIR1 expression by IL-7 may promote pro-B cell proliferation and/or influence differentiation, since some mature B cell subsets do not express LAIR1 [90]. Interestingly, LAIR1 binds all types of collagens, including the N-terminal collagen-like sequence of C1q, whose deficiency is associated with the development of systemic lupus erythematosus which results from overactivation of autoimmune B cells [92]. In Crk/CrkL-deficient cells the IL-7-dependent inhibition of LAIR1 expression was reduced, suggesting that these adaptors partly mediated this inhibitory effect. The reduction of LAIR1 inhibition in our study may be underestimated, since the deletion of Crk/CrkL proteins was not complete, possibly due to their long half-life.
5. Conclusions
This study identified new tyrosine phosphorylated substrates following activation of the IL-7 receptor that may contribute to the proliferative and pro-survival effects of IL-7 and suggests a pro-survival role of Crk family proteins in IL-7 signaling. In pro-B cells the RNA expression of LAIR1, a receptor which inhibits B cell proliferation, was downregulated by IL-7, and this was at least partly mediated by Crk/CrkL in the signaling pathway. Our results suggest pathways in which tyrosine kinase inhibitors could be used to counteract pro-leukemogenic and autoimmune effects of IL-7.
Supplementary Material
Acknowledgments
We thank A. Imamoto for the conditionally Crk/CrkL-deficient mice, K. Noer and R. Matthai for flow cytometry, D. Mc Vicar, M. Gadina and J. Willette Brown for protocols and helpful discussion, J. J. Oppenheim, T. Musso and R. Mariani Costantini for critical review of the manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.cellsig.2018.03.008.
References
- [1].Puel A, Ziegler SF, Buckley RH, Defective IL7R expression in T(−)B(+)NK(+) severe combined immunodeficiency, Nat. Genet 20 (1998) 394–397. [DOI] [PubMed] [Google Scholar]
- [2].Noguchi M, Yi H, Rosenblatt HM, Interleukin-2 receptor gamma chain mutation results in X-linked severe combined immunodeficiency in humans, Cell 73 (1993) 147–157. [DOI] [PubMed] [Google Scholar]
- [3].R Ceredig R, Rolink AG, The key role of IL-7 in lymphopoiesis, Semin. Immunol (3) (2012) 159–164. [DOI] [PubMed] [Google Scholar]
- [4].Jiang Q, Li WQ, Aiello FB, Cell biology of IL-7, a key lymphotrophin, Cytokine Growth Factor Rev 16 (2005) 513–533. [DOI] [PubMed] [Google Scholar]
- [5].Mazzucchelli R, Durum SK, Interleukin-7 receptor expression: intelligent design, Nat. Rev. Immunol 7 (2007) 144–154. [DOI] [PubMed] [Google Scholar]
- [6].Mackall CL, Fry TJ, Gress RE, Harnessing the biology of IL-7 for therapeutic application, Nat. Rev. Immunol 11 (2011) 330–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Yao Z, Cui Y, Watford WT, Stat5a/b are essential for normal lymphoid development and differentiation, Proc. Natl. Acad. Sci. U. S. A 103 (2006) 1000–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Goetz CA, Harmon IR, O’Neil JJ, STAT5 activation underlies IL7 receptor-dependent B cell development, J. Immunol 172 (2004) 4770–4778. [DOI] [PubMed] [Google Scholar]
- [9].Dai X, Chen Y, Di L, Stat5 is essential for early B cell development but not for B cell maturation and function, J. Immunol 179 (2007) 1068–1079. [DOI] [PubMed] [Google Scholar]
- [10].Kim K, Khaled AR, Reynolds D, Characterization of an interleukin-7-dependent thymic cell line derived from a p53(−/−) mouse, J. Immunol. Methods 274 (2003) 177–184. [DOI] [PubMed] [Google Scholar]
- [11].Feller SM, Crk family adaptors-signalling complex formation and biological roles, Oncogene 20 (2001) 6348–6371. [DOI] [PubMed] [Google Scholar]
- [12].Gelkop S, Babichev Y, Kalifa R, Involvement of crk adapter proteins in regulation of lymphoid cell functions, Immunol. Res 28 (2003) 79–91. [DOI] [PubMed] [Google Scholar]
- [13].Shigeno-Nakazawa Y, Kasai T, Ki S, A pre-metazoan origin of the CRK gene family and co-opted signaling network, Sci. Rep 6 (2016) 34349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Guris DL, Fantes J, Tara D, Mice lacking the homologue of the human 22q11.2 gene CRKL phenocopy neurocristopathies of DiGeorge syndrome, Nat. Genet 27 (2001) 293–298. [DOI] [PubMed] [Google Scholar]
- [15].Park TJ, Boyd K, Curran T, Cardiovascular and craniofacial defects in Crk-null mice, Mol. Cell. Biol 26 (2006) 6272–6282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Ingham RJ, Krebs DL, Barbazuk SM, B cell antigen receptor signaling induces the formation of complexes containing the Crk adapter proteins, J. Biol. Chem 271 (1996) 32306–32314. [DOI] [PubMed] [Google Scholar]
- [17].Park TJ, Curran T, Essential roles of Crk and CrkL in fibroblast structure and motility, Oncogene 33 (2014) 5121–5132. [DOI] [PubMed] [Google Scholar]
- [18].Jankowski W, Saleh T, Pai MT, Domain organization differences explain Bcr-Abl’s preference for CrkL over CrkII, Nat. Chem. Biol 8 (2012) 590–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Giacomelli M, Kumar R, Soresina A, Reduction of CRKL expression in patients with partial DiGeorge syndrome is associated withimpairment of T-cell functions, J. Allergy Clin. Immunol 138 (2016) 229–240. [DOI] [PubMed] [Google Scholar]
- [20].Ahmad S, Alsayed YM, Druker BJ, The type I interferon receptor mediates tyrosine phosphorylation of the CrkL adaptor protein, J. Biol. Chem 272 (1997) 29991–29994. [DOI] [PubMed] [Google Scholar]
- [21].Alsayed Y, Uddin S, Ahmad S, IFN-gamma activates the C3G/Rap1 signaling pathway, J. Immunol 164 (2000) 1800–1806. [DOI] [PubMed] [Google Scholar]
- [22].Gesbert F, Garbay C, Bertoglio J, Interleukin-2 stimulation induces tyrosine phosphorylation of p120-Cbl and CrkL and formation of multimolecular signalling complexes in T lymphocytes and natural killer cells, J. Biol. Chem 273 (1998) 3986–3993. [DOI] [PubMed] [Google Scholar]
- [23].Sattler M, Salgia R, Shrikhande G, Steel factor induces tyrosine phosphorylation of CRKL and binding of CRKL to a complex containing c-kit, phosphatidylinositol 3-kinase, and p120(CBL), J. Biol. Chem 273 (1997) 10248–10253. [DOI] [PubMed] [Google Scholar]
- [24].Du J, Alsayed YM, Xin F, Engagement of the CrkL adapter in interleukin-5 signaling in eosinophils, J. Biol. Chem 275 (2000) 33167–33175. [DOI] [PubMed] [Google Scholar]
- [25].Vallejo-Illarramendi A, Zang K, Reichardt LF, Focal adhesion kinase is required for neural crest cell morphogenesis during mouse cardiovascular development, J. Clin. Invest 119 (2009) 2218–2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Yokote K, Hellman U, Ekman S, Identification of Tyr-762 in the platelet-derived growth factor alpha-receptor as the binding site for Crk proteins, Oncogene 16 (1998) 1229–1239. [DOI] [PubMed] [Google Scholar]
- [27].Oda A, Wakao H, Fujihara M, Thrombopoietin and interleukin-2 induce association of CRK with STAT5, Biochem. Biophys. Res. Commun 278 (2000) 299–305. [DOI] [PubMed] [Google Scholar]
- [28].Fish EN, Uddin S, Korkmaz M, Activation of a CrkL-stat5 signaling complex by type I interferons, J. Biol. Chem 274 (1999) 571–573. [DOI] [PubMed] [Google Scholar]
- [29].Nautiyal J, Kumar PG, Laloraya M, 17Beta-estradiol induces nuclear translocation of CrkL at the window of embryo implantation, Biochem. Biophys. Res. Commun 318 (2004) 103–112. [DOI] [PubMed] [Google Scholar]
- [30].Lekmine F, Sassano A, Uddin S, The CrkL adapter protein is required for type I interferon-dependent gene transcription and activation of the small G-protein Rap1, Biochem. Biophys. Res. Commun 291 (2002) 744–750. [DOI] [PubMed] [Google Scholar]
- [31].Laloraya M, Davoodi-Semiromi A, Kumar GP, Impaired Crkl expression contributes to the defective DNA binding of Stat5b in nonobese diabetic mice, Diabetes 55 (2006) 734–741. [DOI] [PubMed] [Google Scholar]
- [32].Guo C, Liu S, Sun MZ, The role of CT10 regulation of kinase-like in cancer, Future Oncol 10 (2014) 2687–2697. [DOI] [PubMed] [Google Scholar]
- [33].Nichols GL, Raines MA, Vera JC, Identification of CRKL as the constitutively phosphorylated 39-kD tyrosine phosphoprotein in chronic myelogenous leukemia cells, Blood 84 (1994) 2912–2918. [PubMed] [Google Scholar]
- [34].K Shuai K, Halpern J, ten Hoeve J, Constitutive activation of STAT5 by the BCR-ABL oncogene in chronic myelogenous leukemia, Oncogene 13 (1996) 247–254. [PubMed] [Google Scholar]
- [35].Johnson KJ, Griswold IJ, O’Hare T, A BCR-ABL mutant lacking direct binding sites for the GRB2, CBL and CRKL adapter proteins fails to induce leukemia in mice, PLoS One 4 (2009) e7439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Seo JH, Wood LJ, Agarwal A, A specific need for CRKL in p210BCR-ABL-induced transformation of mouse hematopoietic progenitors, Cancer Res 70 (2010) 7325–7335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Hoelbl A, Schuster C, Kovacic B, Stat5 is indispensable for the maintenance of bcr/abl-positive leukaemia, EMBO Mol. Med 2 (2010) 98–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Lopez-Rivera E, Liu YP, Verbitsky M, Genetic drivers of kidney defects in the DiGeorge syndrome, N. Engl. J. Med 376 (2017) 742–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Haller M, Mo Q, Imamoto A, Murine model indicates 22q11.2 signaling adaptor CRKL is a dosage-sensitive regulator of genitourinary development, Proc. Natl. Acad. Sci. U. S. A 114 (2017) 4981–4986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Aiello FB, Keller JR, Klarmann KD, IL-7 induces myelopoiesis and erythropoiesis, J. Immunol 178 (2007) 1553–1563. [DOI] [PubMed] [Google Scholar]
- [41].Xu LL, McVicar DW, Ben-Baruch A, Monocyte chemotactic protein-3 (MCP3) interacts with multiple leukocyte receptors: binding and signaling of MCP3 through shared as well as unique receptors on monocytes and neutrophils, Eur. J. Immunol 25 (1995) 2612–2617. [DOI] [PubMed] [Google Scholar]
- [42].Lundtoft C, Afum-Adjei Awuah A, Aberrant plasma IL-7 and soluble IL-7 receptor levels indicate impaired T-cell response to IL-7 in human tuberculosis, PLoS Pathog 13 (2017) e1006425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Yip-Schneider MT, Horie M, Broxmeyer HE, Characterization of interleukin-7-induced changes in tyrosine phosphorylation and c-myc gene expression in normal human T cells, Exp. Hematol 21 (1993) (1648–1456. [PubMed] [Google Scholar]
- [44].Massignan T, Biasini E, Lauranzano E, Mutant prion protein expression is associated with an alteration of the Rab GDP dissociation inhibitor alpha (GDI)/Rab11 pathway, Mol. Cell. Proteomics 9 (2010) 611–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Pappin DJ, Hojrup P, Bleasby AJ, Rapid identification of proteins by peptidemass fingerprinting, Curr. Biol 3 (1993) 327–332. [DOI] [PubMed] [Google Scholar]
- [46].Li WQ, Jiang Q, Khaled AR, Interleukin-7 inactivates the pro-apoptotic protein bad promoting T cell survival, J. Biol. Chem 279 (2004) 29160–29166. [DOI] [PubMed] [Google Scholar]
- [47].Lebbink RJ, de Ruiter T, Verbrugge A, The mouse homologue of the leukocyte-associated Ig-like receptor-1 is an inhibitory receptor that recruits Src homology region 2-containing protein tyrosine phosphatase (SHP)-2, but not SHP-1, J. Immunol 172 (2004) 5535–5543. [DOI] [PubMed] [Google Scholar]
- [48].Sitnicka E, Brakebusch C, Martensson IL, Complementary signaling through flt3 and interleukin-7 receptor alpha is indispensable for fetal and adult B cell genesis, J. Exp. Med 198 (2003) 1495–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Peschon JJ, Morrissey PJ, Grabstein KH, Early lymphocyte expansion is severely impaired in interleukin 7 receptor-deficient mice, J. Exp. Med 180 (1994) 1955–1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].von Freeden-Jeffry U, Vieira P, Lucian LA, Lymphopenia in interleukin (IL)-7 gene-deleted mice identifies IL-7 as a nonredundant cytokine, J. Exp. Med 181 (1995) 1519–1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Kikuchi K, Lai AY, Hsu CL, IL-7 receptor signaling is necessary for stage transition in adult B cell development through up-regulation of EBF, J. Exp. Med 201 (2005) 1197–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Tsapogas P, Zandi S, Åhsberg J, IL-7 mediates Ebf-1-dependent lineage restriction in early lymphoid progenitors, Blood 118 (2011) 1283–1290. [DOI] [PubMed] [Google Scholar]
- [53].Namen AE, Lupton S, Hjerrild K, Stimulation of B-cell progenitors by cloned murine interleukin-7, Nature 333 (1988) 571–573. [DOI] [PubMed] [Google Scholar]
- [54].Lu L, Chaudhury P, Osmond DG, Regulation of cell survival during B lymphopoiesis: apoptosis and Bcl-2/Bax content of precursor B cells in bone marrow of mice with altered expression of IL-7 and recombinase-activating gene-2, J. Immunol 162 (1999) 1931–1940. [PubMed] [Google Scholar]
- [55].Chen J, Shinkai Y, Young F, Probing immune functions in RAG-deficient mice, Curr. Opin. Immunol 6 (2) (1994. April) 313–319. [DOI] [PubMed] [Google Scholar]
- [56].Waller CF, Imatinib mesylate, Cancer Res 184 (2010) 3–20. [DOI] [PubMed] [Google Scholar]
- [57].Seggewiss R, Loré K, Greiner E, Imatinib inhibits T-cell receptor-mediated T-cell proliferation and activation in a dose-dependent manner, Blood 105 (2005) 2473–2479. [DOI] [PubMed] [Google Scholar]
- [58].Sinai P, Berg RE, Haynie JM, Imatinib mesylate inhibits antigen-specific memory CD8 T cell responses in vivo, J. Immunol 178 (2007) 2028–2037. [DOI] [PubMed] [Google Scholar]
- [59].Rennert PD, Browning JL, Mebius R, Surface lymphotoxin alpha/beta complex is required for the development of peripheral lymphoid organs, J. Exp. Med 184 (1996) 1999–2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Tumanov AV, Kuprash DV, Mach JA, Lymphotoxin and TNF produced by B cells are dispensable for maintenance of the follicle-associated epithelium but are required for development of lymphoid follicles in the Peyer’s patches, J. Immunol 173 (2004) 86–91. [DOI] [PubMed] [Google Scholar]
- [61].Oltz EM, Yancopoulos GD, Morrow MA, A novel regulatory myosin light chain gene distinguishes pre-B cell subsets and is IL-7 inducible, EMBO J 7 (1992) 2759–2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Oda A, Ono T, Yamamoto M, Goitsuka R, Kitamura D, PKC eta directs induction of IRF-4 expression and Ig kappa gene rearrangement in pre-BCR signaling pathway, Int. Immunol 20 (2008) 1417–1426. [DOI] [PubMed] [Google Scholar]
- [63].Morrow MA, Lee G, Gillis S, Interleukin-7 induces N-myc and c-myc expression in normal precursor B lymphocytes, Genes Dev 6 (1992) 61–70. [DOI] [PubMed] [Google Scholar]
- [64].McCubrey JA, Steelman LS, Risser RG, Structure and expression of the T cell receptor gamma locus in pre-B and early hemopoietic cells, Eur. J. Immunol (12) (1989) 2303–2308. [DOI] [PubMed] [Google Scholar]
- [65].Durum SK, Candèias S, Nakajima H, Interleukin 7 receptor control of T cell receptor gamma gene rearrangement: role of receptor-associated chains and locus accessibility, J. Exp. Med 188 (1998) 2233–2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Martin CH, Aifantis I, Scimone ML, Efficient thymic immigration of B220+ lymphoid-restricted bone marrow cells with T precursor potential, Nat. Immunol 4 (2003) 866–873. [DOI] [PubMed] [Google Scholar]
- [67].Zhang F, Ren S, Zuo Y, DC-SIGN, DC-SIGNR and LSECtin: C-type lectins for infection, Int. Rev. Immunol 33 (2014) 54–66. [DOI] [PubMed] [Google Scholar]
- [68].Geginat J, Nizzoli G, Paroni M, Immunity to pathogens taught by specialized human dendritic cell subsets, Front. Immunol 6 (2015) 527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Parcells BW, Ikeda AK, Simms-Waldrip T, FMS-like tyrosine kinase 3 in normal hematopoiesis and acute myeloid leukemia, Stem Cells 24 (2006) 1174–1184. [DOI] [PubMed] [Google Scholar]
- [70].Stillwell R, Bierer BE, T cell signal transduction and the role of CD7 in costimulation, Immunol. Res 24 (2001) 31–52. [DOI] [PubMed] [Google Scholar]
- [71].Michaelson MD, Mehler MF, Xu H, Interleukin-7 is trophic for embryonic neurons and is expressed in developing brain, Dev. Biol 179 (1996) 251–263. [DOI] [PubMed] [Google Scholar]
- [72].Ashbaugh JJ, Brambilla R, Karmally SA, IL7Rα contributes to experimental autoimmune encephalomyelitis through altered T cell re sponses and non-hematopoietic cell lineages, J. Immunol 190 (2013) 4525–4534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Nunnari G, Xu Y, Acheampong EA, Exogenous IL-7 induces Fas-mediated human neuronal apoptosis potential effects during human immunodeficiency virus type 1 infection, J. Neuro-Oncol 11 (2005) 319–328. [DOI] [PubMed] [Google Scholar]
- [74].Lopes MH, Hajj GN, Muras AG, Interaction of cellular prion and stress-inducible protein 1 promotes neuritogenesis and neuroprotection by distinct signaling pathways, J. Neurosci 25 (2005) 11330–11339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Arantes C, Nomizo R, Lopes MH, Prion protein and its ligand stress inducible protein 1 regulate astrocyte development, Glia 57 (2009) 1439–1449. [DOI] [PubMed] [Google Scholar]
- [76].Longshaw VM, Chapple JP, Balda MS, Nuclear translocation of the Hsp70/Hsp90 organizing protein mSTI1 is regulated by cell cycle kinases, J. Cell Sci 117 (2004) 701–710. [DOI] [PubMed] [Google Scholar]
- [77].Spurr IB, Birts CN, Cuda F, Targeting tumour proliferation with a small-molecule inhibitor of AICAR transformylase homodimerization, Chembiochem 13 (2012) 1628–1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Krecic AM, Swanson MS, hnRNP complexes: composition, structure, and function, Curr. Opin. Cell Biol 11 (1999) 363–371. [DOI] [PubMed] [Google Scholar]
- [79].Navakauskiene R, Treigyte G, Gineitis A, Identification of apoptotic tyrosine-phosphorylated proteins after etoposide or retinoic acid treatment, Proteomics 4 (2004) 1029–1041. [DOI] [PubMed] [Google Scholar]
- [80].Rauch J, O’Neill E, Mack B, Heterogeneous nuclear ribonucleoprotein H blocks MST2-mediated apoptosis in cancer cells by regulating A-Raf transcription, Cancer Res 70 (2010) 1679–1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Stark M, Bram EE, Akerman M, Heterogeneous nuclear ribonucleoprotein H1/H2-dependent unsplicing of thymidine phosphorylase results in anticancer drug resistance, J. Biol. Chem 286 (2011) 3741–3754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Zipfel PA, Zhang W, Quiroz M, Requirement for Abl kinases in T cell receptor signaling, Curr. Biol 14 (2004) 1222–1231. [DOI] [PubMed] [Google Scholar]
- [83].Fabian MA, Biggs WH 3rd, Treiber DK, A small molecule-kinase interaction map for clinical kinase inhibitors, Nat. Biotechnol 23 (2005) 329–336. [DOI] [PubMed] [Google Scholar]
- [84].Seckinger P, Fougereau M, Activation of src family kinases in human pre-B cells by IL-7, J. Immunol 153 (1994) 97–109. [PubMed] [Google Scholar]
- [85].Arai A, Kanda E, Nosaka Y, CrkL is recruited through its SH2 domain to the erythropoietin receptor and plays a role in Lyn-mediated receptor signalling, J. Biol. Chem 276 (2001) 33282–33290. [DOI] [PubMed] [Google Scholar]
- [86].Kobashigawa Y, Sakai M, Naito M, Structural basis for the transforming activity of human cancer-related signaling adaptor protein CRK, Nat. Struct. Mol. Biol 14 (2007) 503–510. [DOI] [PubMed] [Google Scholar]
- [87].Jacobson AC, Weis JJ, Weis JH, CD21 signaling via C3 regulates Purkinje cell protein 4 expression, Mol. Immunol 46 (2009) 1488–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Khazaei MR, Halfter H, Karimzadeh F, Bex1 is involved in the regeneration of axons after injury, J. Neurochem 115 (2010) 910–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Meyaard L, The inhibitory collagen receptor LAIR-1 (CD305), J. Leukoc. Biol 83 (2008) 799–803. [DOI] [PubMed] [Google Scholar]
- [90].Tang X, Tian L, Esteso G, Leukocyte-associated Ig-like receptor-1-deficient mice have an altered immune cell phenotype, J. Immunol 188 (2012) 548–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Merlo A, Tenca C, Fais F, Inhibitory receptors CD85j, LAIR-1, and CD152 downregulate immunoglobulin and cytokine production by human B lymphocytes, Clin. Diagn. Lab. Immunol (6) (2005) 705–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Son M, Santiago-Schwarz F, Al-Abed, C1q limits dendritic cell differentiation and activation by engaging LAIR-1, Proc. Natl. Acad. Sci. U. S. A 109 (2012) E3160–3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


