Abstract
Our understanding of NK biology is increased dramatically, a product of improved flow-cytometric techniques for analyzing these cells. NK cells undergo significant changes in repertoire during differentiation. A repeating stimulus, such as a cytomegalovirus infection, may result in accumulation of certain types of highly differentiated NK cells designated as memory-like, or adaptive NK cells. Adaptive NK cells are capable of rapid expansion and effective response to the recall stimulus. These cells differ significantly from conventional NK cells both functionally and phenotypically. Here we describe an approach for identification and analysis of adaptive NK cells in human peripheral blood. CD57-positive cells with high expression of activating-receptor NKG2C, increased expression of KIR receptors, lack of co-expression with inhibitory receptor NKG2A, and decreased expression of activating receptor NCR3 (NKp30) all characterize this cell type. The flow-cytometric method described below can identify this NK cell subset on a relatively simple flow cytometer.
Keywords: memory-like NK cells, adaptive NK cells, NK cell repertoire, differentiation, CD57, NKG2A, NKG2C
INTRODUCTION
NK cells are a large group of innate lymphoid cells producing IFNγ. They participate in antitumor response and defense against infections and coordinate interactions of innate and adaptive immune cells to modulate immune responses. The main mechanism NK cells use for eliminating their targets is contact cytolysis. Cytolytic activity of NK cells is controlled by an arsenal of activating and inhibitory receptors interacting with their relevant ligands. Stimulation of NK cells by contact interactions or/and soluble mediators (including IL-2, IL-15, IL-12, IL-18, and IL-21) leads to their proliferation, cytokine production, and an increase in cytotoxic activity.
Several types of NK cells are distinguished by the origin and site of their development, phenotypic characteristics and functional purpose. Conventional human NK cells considered in this unit comprise 5% to 20% of peripheral blood lymphocytes in normal individuals and can be distinguished phenotypically as CD3−CD56+/CD16+cells. The majority of circulating NK cells is CD56dim (90% to 95%). They express CD16, the low-affinity Fc receptor FcγRIII that determines their ability for antibody-dependent cell-mediated cytotoxicity (ADCC). Signal transduction via this receptor requires association withCD3ζ orFcɛRIγ (also known as FcRγ) adaptor units. CD56brightcells, good producers of cytokines, are considered to be earlier stage-differentiated NK cells.
The differentiation mechanism resulting in the generation heterogeneous mature mixture of various NK cell subsets expressing surface markers and signaling proteins typical for both myeloid and lymphoid cells are still not clear. During the differentiation process, NK cells lose expression of NKG2Adimerized CD94, and acquire expression of the cell maturity (or senescence) marker CD57 and killer cell immunoglobulin-like receptors (KIRs). The KIR family contains a number of inhibitory receptors that recognize various alleles of MHC class I molecules (MHC-I) expressed on the surface of normal cells. Strikingly, most of inhibitory KIRs have an activating counterpart. Activating KIRs have been shown to emerge on the NK cell surface on later differentiation stages (Beziat et al., 2014). Nevertheless, expression of NKG2A, CD57, and different KIRs can also be found on CD56dim NK cells in vivo (Bjorkstrom et al., 2010).
A new concept of NK cell memory now considers adaptive features for these innate immune cells. In a number of models, recurrent stimulation of NK cells leads to a more rapid and effective response (O’Sullivan et al., 2015). This response was associated with generation and maintenance of adaptive NK cell subsets, resulting in reconfiguration of the NK cell repertoire. Memory-like NK cells were induced by a combination of cytokines IL-12, IL-15, and IL-18 (Berrien-Elliott et al., 2015). These cytokine-stimulated NK cells displayed increased sensitivity to IL-2, elevated IFNγ production and cytotoxic activity. They actively proliferated while preserving their functional properties. Another type of memory-like NK cells was described for human patients prior to infection with human cytomegalovirus (HCMV). This subset typically expressed high level of activating receptor NKG2C, a counterpart of inhibitory receptor NKG2A, which decreased in these cells (Guma et al., 2004). HLA-E molecules are the ligands for both these receptors.
NK cell differentiation during memory generation is associated with epigenetic changes that result in stable marker expression and signaling pathways. It may lead, in turn, to changes in final “executive” steps of NK cell activation, including proliferation, cytotoxic activity and cytokine production. Long-lived memory-like NK cells are often deficient for some signaling units expressed typically in myeloid cells, including transmembrane adaptor protein FcRγ, tyrosine kinase SYK, and intracellular signaling adaptor EAT-2. At the same time, the expression of signaling proteins CD3ζ, ZAP-70, and SAP distinctive for T lymphocytes remains unchanged (Schlums et al., 2015). Epigenetic remodeling of regulatory regions for IFNG has been shown to mediate increased IFNγ production in cytokine-induced and HCMV-associated memory-like NK cells (Luetke-Emersloh et al., 2014). All these modifications lead to changes in the hierarchy of receptors controlling NK cell functions. Adaptive NK cells are therefore specialized for a highly effective immune response to certain stimuli, while their reaction to other stimuli decreases. The main functional characteristics of memory-like NK cells include decreased natural cytotoxicity and responsivity to cytokines produced by innate immune cells (IL-12, IL-18), increased ADCC and IFNγ production in response to appropriate stimuli, and long-time persistence in the organism (from 4 months to 1 year, according to various data).
Discrimination and isolation of memory-like NK cells will help to better characterize these cells and uncover mechanisms of expansion and elimination of the adaptive cell subsets. Various cytometric approaches for identification of NK cells with adaptive features have been described based on immunolabeling cell surface and intracellular molecules. These include inhibitory and activating receptors like ADCC receptor CD16, adhesion molecules, cytokine receptors, activation, and differentiation markers, granzyme B, intracellular cytokines (IFNγ and TNFα), phosphorylated signaling subunits and adaptor molecules. The phenotypic “signature” of memory-like NK cells is defined by their late differentiation stage, induction of activating counterpart receptors, such as NKG2C and activating KIRs, increase in expression of inhibitory KIRs specific for self MHC-I in “licensed” NK cells, and deficiency for several transcription factors and signaling units leading to phenotype alterations. In numerous studies, changes in expression levels of NKG2C, CD57, LILRB1, various KIRs, NKG2A, CD161, IL-18Rα, FcRγ, NKp30, NKp44, CD2,CD7, FAS, and Siglec-7 were assessed for identification and characterization of adaptive NK cells (Guma et al., 2004; Hwang et al., 2012; Beziat et al., 2013; Wu et al., 2013; Lee et al., 2015; Schlums et al., 2015; Muccio et al., 2016; Muntasell et al., 2016). In this unit we describe a simple method for identification HCMV-associated subset of adaptive NK cells expressing NKG2C.
SIGNIFICANCE STATEMENT
Stimulation of NK cells under specific conditions can lead to their differentiation into memory-like cells, which remain in the organism for a long time. There is now considerable data characterizing NK cells with adaptive features as a functionally different cell type with a distinct phenotype. Cytometric approaches for analyzing this adaptive reconfiguration of circulating human NK cell repertoire are now available. The method for identification of memory-like NK cells based on cell surface immunolabeling described here allows both identification and quantitation of these cells in both normal and pathological conditions, and allows isolation these cells by cell sorting for their further investigation.
BASIC PROTOCOL
ANALYSIS OF MEMORY-LIKE NK CELLS ASSOCIATED WITH HCMV INFECTION CIRCULATING IN HUMAN BLOOD
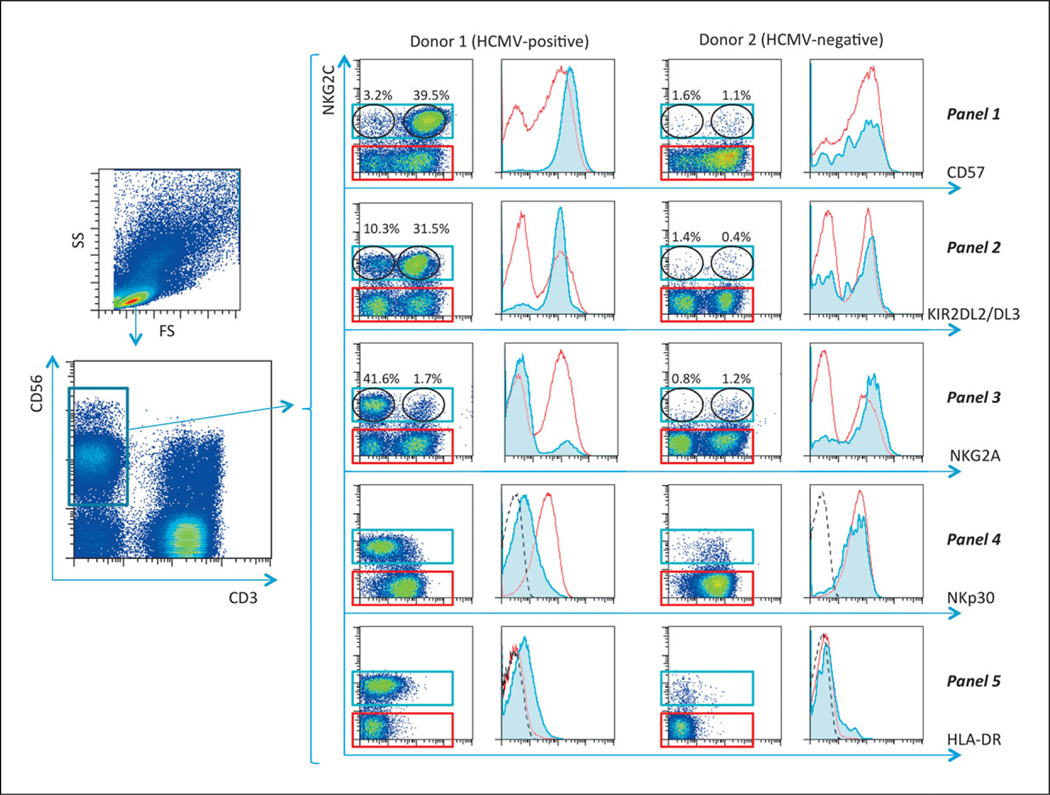
The most extensively studied type of NK cell with adaptive features is observed in individuals infected with HCMV (life-long latent infection in the majority of human populations). These memory-like NK cells include several subtypes of adaptive cells and are believed to be protective against the virus. HCMV reactivation drives expansion of this adaptive NK cell pool in irradiated recipients of hematopoietic cell-based transplantation (Foley et al., 2012; Muccio et al., 2016). The purpose of this method is to identify these memory-like NK cells circulating in human blood by immunolabeling in several panels for differentiation markers and receptors expressed on NK cell surface followed by flow cytometry analysis. These panels initially identify the entire NK cell population as CD3−CD56+. Detection of adaptive NK cells is then based mainly on NKG2C expression measurement. Degree of maturity of the cells is determined by surface CD57 expression. Analysis of KIR receptor expression (KIR2DL2/DL3 in this protocol), which typically increases in adaptive NK cells is included in this panel set. Loss of NKG2A and decreased surface expression of natural cytotoxicity receptor NKp30 (CD337) in NKG2C-positive cells are also important in identifying adaptive NK cells. Measurement of HLA-DR that is often observed on proliferating NK cells (Evans et al., 2011) is also included.
This method is based on analysis of cell surface molecules only and is useful for cytometers equipped with a minimum of two lasers. An advantage of this method is that it can be carried out on a relatively simple four-color analyzer.
Materials
Lymphocyte separation medium 1.077 (Lonza, cat. no. 17-829E)
Human blood collected in heparin- or EDTA-containing tubes
Phosphate-buffered saline, pH 7.2 (PBS; Fisher Scientific, cat. no. 20-012-027)
Staining solution (see recipe)
Ice
Monoclonal antibodies:
Anti-human CD3-PE-Cy7 (Beckman Coulter, clone UCHT1, cat. no. 6607100)
Anti-human CD56-APC [Beckman Coulter, clone N901 (HLDA6), cat. no. IM2474]
Anti-human NKG2C-AlexaFluor (AF) 488 (R&D Systems, clone 134591, cat. no. FAB138G-100)
Anti-human NKG2A-PE (R&D Systems, clone no. 131411, cat. no. FAB1059P-100)
Anti-human KIR2DL2/DL3 (CD158b)-PE (Miltenyi Biotech, clone DX27, cat. no. 130-092-618)
Anti-human CD57-PE (eBioscience, clone TB01, cat. no. 12-0577-42)
Anti-human NKp30-PE (eBioscience, clone AF29-4D12, cat. no. 12-3379-42)
Anti-human HLA-DR-FITC (Beckman Coulter, clone B8.12.2, cat. no. IM0463U)
50-ml conical centrifuge tubes (Fisher Scientific, cat. no. 14-432-22)
Centrifuge
Automatic pipets
Cell counter or hemacytometer
5-ml polystyrene round-bottom tubes (Fisher Scientific, cat. no. 352008)
Multi-laser flow cytometer with appropriate filters for collection of fluorescence emission
NOTE: For this protocol two 9-ml tubes with blood should be enough. For the Support Protocol (including NK cells separation) minimum 36 ml (four 9-ml tubes) of blood should be taken.
PBMC isolation
Warm the lymphocyte separation medium to room temperature before use.
Dilute 15 to 20 ml of the whole blood sample with equal volume of PBS in a 50-mlFalcon conical centrifuge tube. Use blood samples collected on the same day.
Carefully dispense 10 ml of lymphocyte separation medium under the diluted blood. Maintain the phase separation between blood and separation medium. Do not mix the solutions.
When using an automatic pipet accidental mixing of the solutions may occur. It is preferable to use a medical syringe with 100 to 150-mm needle or other manual dispensing device to slowly dispense the separation medium under the blood.
4. Centrifuge for 20 min at 500 × g, 25°C, with the centrifuge brake off.
5. Draw off the upper layer containing plasma and platelets using an automatic pipet, leaving the underlying layer of mononuclear cells undisturbed.
6. Remove the mononuclear cell layer using a pipet.
7. Transfer the layer of mononuclear cells into a new 50-ml conical centrifuge tube.
8. Add PBS solution to reach 50-ml and centrifuge for 15 min at 300 × g, 25°C, with the brake on.
This wash step will remove any remaining lymphocyte separation medium.
9. Remove the supernatant by decanting or with an automatic pipet. Resuspend the pellet in 3 to 5 ml PBS by gently mixing using an automatic pipet and then adding PBS solution to 50 ml.
10. Centrifuge for 10 min at 200 × g, 25°C, with the brake on. Remove the supernatant by decanting or using an automatic pipet.
This wash step will remove the platelets.
Antibody labeling
11. Resuspend the pellet in 3 to 5 ml of staining solution using automatic pipet and then add staining solution to 50 ml. Count the cells using a hemacytometer or cell counter.
12. Centrifuge for 4 min at 300 × g, 4°C.
13. Resuspend the pellet in staining solution to the concentration 1.5 × 107 PBMC per ml (or 2.5 × 106 NK cells per millilter; for NK cell isolation, see Support Protocol). Prepare the cell samples by transferring 100 μl of cell suspension into 5-ml Falcon polystyrene 12 × 75–mm round-bottom tubes [including experimental samples, nonlabeled, single-stained, and Fluorescence Minus One (FMO) controls]. Pellet the cells by centrifuging for 4 min at 300 × g, 4°C, and discard the supernatant completely. Tap the tubes gently to disperse the pellets, and place the tubes on ice. Further manipulations should be performed on ice.
-
14. Add the antibodies into the tubes with experimental, single-stained, and FMO control samples. Final staining volume should be 100 μl. The following experimental samples should be prepared:
Panel 1: CD3-PE-Cy7, CD56-APC, anti-NKG2C-AF488, CD57-PE
Panel 2: CD3-PE-Cy7, CD56-APC, anti-NKG2C-AF488, anti-KIR2DL2/DL3-PE
Panel 3: CD3-PE-Cy7, CD56-APC, anti-NKG2C-AF488, anti-NKG2A-PE
Panel 4: CD3-PE-Cy7, CD56-APC, anti-NKG2C-AF488, anti-NKp30-PE
Panel 5: CD3-PE-Cy7, CD56-APC, anti-NKG2C-PE, anti-HLA-DR-FITC (optional).
Fluorescence Minus One (FMO) controls should include:
FMO control 1: CD3-PE-Cy7, CD56-APC, CD57-PE
FMO control 2: CD3-PE-Cy7, CD56-APC, anti-NKG2C-AF488
FMO control 3: CD3-PE-Cy7, CD56-APC, anti-NKG2C-PE
Mix gently by vortexing and incubate for 30 min on ice.
Antibodies may be replaced by other fluorochrome conjugates depending on the capability f the flow cytometer. For example, if cytometer is equipped with a laser of 405 nm and the appropriate detector, anti-CD56Brilliant Violet 421 (BioLegend, clone HCD56, cat. no. 318327) can be substituted.
Antibodies should be titrated to determine the appropriate concentration prior to use in experiments.
15. Add 1 ml staining solution, centrifuge for 4 min at 300 × g, 4°C, and decant the supernatant. Add 1 ml staining solution, centrifuge for 4 min at 300 × g, 4°C, and decant the supernatant.
16. Resuspend the pellet in 300 to 500 μl PBS. Analyze promptly using a multi-laser flow cytometer with appropriate filters for collection of fluorescence emission.
A flow cytometer with a minimum of two lasers (blue-green 488 nm and red 640 nm) is required for this analysis. Cytometers with higher multicolor analysis capability can also be used, and fluorochrome assignments changed as needed. For example, instrument with violet lasers can use Pacific Blue or Brilliant Violet conjugates.
SUPPORT PROTOCOL
IDENTIFICATION OF HCMV INFECTION-ASSOCIATED ADAPTIVE NK CELL SUBSET IN NK CELLS EX VIVO-PURIFIED BY MAGNETIC SEPARATION
This protocol supplements the previous one by isolating purified NK cells from PBMC by magnetic separation prior to staining with antibodies. The use of this protocol allows to easily identify the subpopulations with low level of expression of certain surface markers, such as HLA-DR, and to shorten the sorting procedure duration (if used). However, the precise proportion value of memory-like NK cells in human PBMCs cannot be determined.
Additional Materials (also see Basic Protocol)
Separation buffer (see recipe)
NK Cell Isolation Kit, human (MiltenyiBiotec, cat. no. 130-092-657) containing: NK Cell Biotin-Antibody Cocktail
NK Cell MicroBead Cocktail
15-ml conical centrifuge tubes (Fisher Scientific, cat. no. 14-959-53A)
4°C incubator
MACS LD Columns (MiltenyiBiotec, cat. no. 130-042-901)
MidiMACS Separator (MiltenyiBiotec, cat. no. 130-042-302)
PBMC isolation
Warm lymphocyte separation medium to room temperature before use.
Dilute 36 to 40 ml of the whole blood sample with equal volume of PBS in two50-ml Falcon conical centrifuge tubes. Use blood samples collected on the same day.
Carefully dispense 10 ml of the lymphocyte separation medium under the diluted blood in each tube. Maintain the phase separation between blood and separation medium. Do not mix the solutions.
When using an automatic pipet accidental mixing of the solutions may occur. It is preferable to use a medical syringe with 100- to 150-mm needle or other manual dispensing device to slowly dispense the separation medium under the blood.
4. Centrifuge for 20 min at 500 × g, 25°C, with the centrifuge brake off.
5. Remove the upper layer containing plasma and platelets using an automatic pipet, leaving the underlying layer of mononuclear cells undisturbed.
6. Remove the mononuclear cell layer using a pipet.
7. Transfer the layer of mononuclear cells into a new 50-ml conical centrifuge tube.
8. Add PBS solution to 50 ml and centrifuge for 15 min at 300 × g, 25°C, with the brake on.
This wash step will remove any remaining lymphocyte separation medium.
9. Remove the supernatant by decanting or with an automatic pipet. Resuspend the pellet in 5 ml of PBS. Then add PBS solution to 50 ml.
10. Centrifuge at 200 × g for 10 min at 25°C, with the brake on. Remove the supernatant by decanting or using automatic pipette.
This wash step will remove the platelets.
NK cells magnetic separation
11. Resuspend the pellet and add separation buffer to make the final volume 50 ml. Determine the cell number using a cell counter or hemacytometer.
12. Centrifuge for 10 min at 200 × g, 25°C. Remove the supernatant by decanting or with an automatic pipet.
13. Resuspend the pellet in 40 μl separation buffer per 107 total cells and transfer the cell suspension into a new 15-ml polypropylene conical centrifuge tube.
14. Add 10 μl NK Cell Biotin-Antibody Cocktail per 107 total cells and mix gently by vortexing.
15. Incubate for 5 min at 4°C.
16. Add 30 μl separation buffer and 20 μl of NK Cell MicroBead Cocktail per 107 total cells.
17. Mix gently by vortexing and incubate for 10 min at 4°C.
18. Add separation buffer to a final volume of 15 ml and centrifuge for 5 min at 300 × g, 25°C.
19. Remove the supernatant and resuspend the pellet in 500 μl separation buffer per 108 total cells.
20. Place MACS LD column in the magnetic field and prepare it for the separation procedure by rinsing with 3 ml separation buffer.
21. Place a new 15-ml tube under the column and apply cell suspension onto the column.
22. Wash the column three times, each time with 3 ml separation buffer. It is important to add a new portion of the buffer when the previous one is passed through the column.
23. Take the tube out of the column, mix the cell suspension well by inverting repeatedly, and determine cell number using a cell counter or hemacytometer. Then label separated NK cells as described in the Basic Protocol, steps 11 to 16.
REAGENTS AND SOLUTIONS
Use Milli-Q purified water in all recipes.
Separation buffer
Prepare 0.5% bovine serum albumin (BSA; Serva, cat. no. 11934) and 2 mM EDTA (Serva, cat. no. 11278) in phosphate-buffered saline (PBS). Adjust pH to 7.2 using NaOH solution. Use the same day or sterilize by filtering through 0.22-μm Millipore filter and store up to 2 months at 4°C.
For staining of cells in sterile conditions (e.g., on purpose of sterile sorting), use sterile separation buffer instead of staining solution.
Staining solution
Prepare 0.1% bovine serum albumin (BSA; Serva, cat. no. 11934) and 0.1% sodium azide (Amresco, cat. no. 0639-250G) in phosphate-buffered saline (PBS). Adjust pH to 7.2–7.4 using NaOH solution. Store up to 2 months at 4°C.
COMMENTARY
Background Information
Memory-like NK cells possess specific functionality and unique transcriptional and phenotypic features. One of first described phenotypic characteristics of human adaptive NK cells was high expression of NKG2C in a CD57+NK cell subset found in peripheral blood of seropositive for HCMV individuals (Guma et al., 2004). These NK cells were preferentially negative for NKG2A. This is the subset induced to rapid expansion by HCMV reactivation in recipients after hematopoietic cell transplantation. However, no specific functional interaction of the activating receptor CD94/NKG2C with HCMV-derived proteins was confirmed until recently. It is now suggested that signaling via NKG2C is causally linked to NK cell differentiation (Muntasell et al., 2016). Presence in this subset of the carbohydrate antigen CD57 is another piece of the adaptive NK cell “signature” indicating the late stage of differentiation of these cells (Bjorkstrom et al., 2010). NKG2C+ CD57+ NKG2A− NK cells also express particular self-specific KIRs, including activating KIRs (KIR2DS4, KIR2DS2 or KIR3DS1), imprinting clonal-like expansion of the adaptive cells (Foley et al., 2012; Beziat et al., 2013). One of specific feature of HCMV-induced memory-like NK cells is the deficiency for FcRγ, important for the signal transduction pathways of a number of NK cell receptors including NKp30, NKp46 and CD16. A consequence of this FcRγ deficiency is decreased expression of NKp30 and NKp46.This may partially explain an observed decrease in the natural cytotoxicity of adaptive NK cells although CD16 expression is mostly unchanged (Guma et al., 2004; Hwang et al., 2012; Beziat et al., 2013).Association of NKG2C and KIR expression with the loss of FcRγ expression in highly differentiated cells is considered a common pattern of adaptive NK cell response to HCMV. These signature elements are acquired independently during HCMV-induced NK cell differentiation (Lee et al., 2015; Muntasell et al., 2016). Evaluation of different expression of the cell markers in NKG2C+ subset can be used for identification of memory-like NK cells.
Several methods are currently proposed for tracing the adaptation of NK cells to viral infection. One of the methods suggests simultaneous analysis of KIR repertoire in combination with other markers of memory-like NK cells (Beziat et al., 2014). However, determination of full repertoire of KIRs is complex, partly because of current unavailability of specific antibodies to certain KIRs. This analysis requires 13 to 15 color flow cytometry. The current protocol measurement of KIR2DL2/DL3 as one of phenotypic markers of adaptive NK cells is recommended as a more practical alternative because: (1) inhibitory receptors KIR2DL2 and KIR2DL3 (allelic forms of one gene) have rather broad HLA-I specificity (Locatelli et al., 2013); (2) the frequency of occurrence of this gene in human population is rather high (close to 100%) (Leung et al., 2005); (3) the KIR-specific antibody employed in this protocol (clone DX27) is cross-reactive for activating KIR2DS2, which is another signature element of memory-like NK cells. This marker is therefore likely to be expressed on adaptive NK cells in a significant numbers of individuals.
Another cytometric approach for identification of memory-like NK cells is based on the determination of FcRγ together with NKG2C and CD57.This method requires intracellular staining (Muntasell et al., 2016). The protocol described here includes only surface staining, simplifying and decreasing the time of the experiment. Instead of FcRγ, the described method measures the FcRγ-dependent surface level of NKp30 (NCR3, CD337). NKp30 recognizes BAG6, B7-H6, and galectin-3. Surface expression of NKp30 requires the association with FcRγ adaptor protein (Hwang et al., 2012).
Memory-like NK cell subsets have been shown to contain largely resting cells with no CD69 expression (Zhang et al., 2013). However, NKG2C+ adaptive NK cells are partially positive for HLA-DR, usually a marker of late NK cell activation. High proportions of HLADR-positive NK cells have been observed in some inflammatory conditions. HLA-DR expression in NK cells is associated with enhanced degranulation and proliferation (Evans et al., 2011). Hence, HLA-DR labeling may be included as both a phenotypic and functional characteristic of memory-like NK cells (Panel 5 in step 14 of the Basic Protocol).
Analysis of several panels described here is the integral convenient method for detection memory-like NK cells. This allows a comprehensive view of the dynamic expansion of adaptive NK cells. An example of this analysis for HCMV-seropositive and seronegative individuals is presented in Figure 9.50.1 for all five labeling panels.
Figure 9.50.1.
Gating strategy for identifying HCMV-induced memory-like NK cells.
Critical Parameters
High-quality fluorescent-labeled antibodies should be used for these panels. The described panels use fluorochrome conjugates with good spectral compatibility. If changes are to be made to the fluorochrome selection, they should be critically assessed for spectral compatibility and minimal fluorescence spillover (Mahnke and Roederer, 2007). An optimal concentration of each antibody should be determined by its titration prior multicolor labeling. For all multicolor panels, a set of single color compensation or spillover controls is essential. These samples can consist of cells or antibody capture beads.
Good cell viability is critical for immunophenotypic analysis, and should be assessed prior to labeling.
A minimum of 106 lymphocytes per sample should be labeled for each panel. This is necessary for measurement of rare cell subsets in the whole PBMC population. For NK cells purified by magnetic separation a minimum of 0.2 × 106 morphologically viable cells should be labeled per sample.
Alternative kits for NK cells magnetic separation can also be used. The EasySep Human NK Cell Enrichment Kit (StemCell Technologies, Dynabeads Untouched Human NK Cells Kit (ThermoFisher Scientific) and others can be substituted. However, it is important to note the compositions of antibody cocktails for NK cell separation. The Stem Cell Technologies and ThermoFisher kits include antibodies against HLA class II, leading to the removal of HLA-DR+ NK cells. The MiltenyiBiotec kit does not include this antibody.
Use sterile solutions and instruments for preparing samples for cell sorting to perform long-time (more than 8 hr) experiments.
Troubleshooting
As a rule, the entire NK population canbeidentifiedasCD3−CD56+,withsubsequent identification of NKG2C-positive cells as a subset of this group. For more precise NK identification, it is possible to use antibodies to lineage antigens CD19 and CD14 labeled by the same fluorochrome as CD3 antibody (a “dump” channel). In this case, NK cells will be determined as CD3−/CD14−/CD19−CD56+.
Some samples have hypofunctional NK cells that are CD56-negative but express CD16. In blood samples from healthy donors these cells usually comprise no more than 1% to 3% of the entire NK population, but their level can increase in some pathologic conditions. Discrimination of these cells requires two additional antibodies—CD16 (conjugated with the same fluorochrome as the CD56 antibody) and CD7 with an additional fluorochrome. In this case, NK cells will be discriminated as CD3−CD56+/CD16+CD7+.
For accurate analysis, we recommend samples with a minimal level of dead cells. To check cell viability use quality control sample containing cells stained with propidium iodide (PI) (2 μl/ml) for 5 to 15 min. Analysis of experimental samples should be performed if percentage of PI-stained cells in the quality control sample does not exceed 3%.If a multi-laser flow cytometer is available, a fluorescent viability reagent can be included in the panel to gate out nonviable cells. Covalent viability probes including Live/Dead Violet or Aqua (Thermo Fisher Scientific) and Zombie Aqua or Yellow (BioLegend) can be used with the above panels on cytometers equipped with a violet laser.
Anticipated Results
The typical proportion of NKG2C-positive cells in the whole NK population of CD3−CD56+ cells in HCMV-seropositive donors without revealed pathologies varies widely from 1% to 2% up to 55%. In seronegative persons, this proportion constitutes no more than 5%. The proportion of NKG2C+NKG2A+ cells is less variable, usually consisting of 1% to 2% of whole NK cells.
The presence of adaptive NK cell subsets in an individual represents expansion of cells possessing several features of memory-like NK cells (NKG2C, CD57 or KIR2DL2/DL3 positivity, low NKG2A, low NKp30). This subset is typically more than 8% of CD3−CD56+cells. This threshold was calculated as mean + 2SD of NKG2C+NKG2A− NK cells in HCMV-seronegative donors (Muntasell et al., 2016). It should be noted that some individuals are homozygous for NKG2C gene deletions (~4% NKG2Cdel/del). This method is not applicable for these donors.
Time Considerations
The Basic Protocol requires about 4 hr broken down as follows:
Isolation of peripheral blood mononuclear cells requires about 1.5 to 2 hr. If you do not immediately use the cells, they can be transferred to RPMI-1640 medium supplemented with 10% fetal bovine serum (HyClone) and 2 mM L-glutamine, and incubated under standard conditions (37°C, 5% CO2) for 1 to 3 hr. NK cells and activity are lost rapidly after blood collection, and will be reduced in samples stored for longer periods.
Preparation of samples and labeling of cells requires about 1.5 hr.
Cytometric analysis requires 0.5 to1 hr.
Magnetic separation requires an additional 2 hr, increasing the total protocol length to 6 hr.
Acknowledgements
This work was supported by the Russian Science Foundation, grant no. 16-15-00309.
Footnotes
Conflict of Interest Authors declare no conflict of interests.
Literature Cited
- Berrien-Elliott MM, Wagner JA, and Fehniger TA 2015. Human cytokine-induced memory-like natural killer cells. J. Innate Immun. 7:563571. doi: 10.1159/000382019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beziat V, Traherne J, Malmberg JA, Ivarsson MA, Bjorkstrom NK, Retiere C, Ljunggren HG, Michaelsson J, Trowsdale J, and Malmberg KJ 2014. Tracing dynamic expansion of human NK-cell subsets by high-resolution analysis of KIR repertoires and cellular differentiation. Eur. J. Immunol. 44:2192–2196. doi: 10.1002/eji.201444464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beziat V, Liu LL, Malmberg JA, Ivarsson MA, Sohlberg E, Bjorklund AT, Retiere C, Sverremark-Ekstrom E, Traherne J, Ljungman P, Schaffer M, Price DA, Trowsdale J, Michaelsson J, Ljunggren HG, and Malmberg KJ 2013. NK cell responses to cytomegalovirus infection lead to stable imprints in the human KIR repertoire and involve activating KIRs. Blood 121:2678–2688. doi: 10.1182/blood-2012-10-459545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjorkstrom NK, Riese P, Heuts F, Andersson S, Fauriat C, Ivarsson MA, Bjorklund AT, Flodstrom-Tullberg M, Michaelsson J, Rottenberg ME, Guzman CA, Ljunggren HG, and Malmberg KJ 2010. Expression patterns of NKG2A, KIR, and CD57 define a process of CD56dim NK-cell differentiation uncoupled from NK-cell education. Blood 116:3853–3864. doi: 10.1182/blood-2010-04-281675. [DOI] [PubMed] [Google Scholar]
- Evans JH, Horowitz A, Mehrabi M, Wise EL, Pease JE, Riley EM, and Davis DM 2011. A distinct subset of human NK cells expressing HLA-DR expand in response to IL-2 and can aid immune responses to BCG. Eur. J. Immunol. 41:1924–1933. doi: 10.1002/eji.201041180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley B, Cooley S, Verneris MR, Pitt M, Curtsinger J, Luo X, Lopez-Verges S, Lanier LL, Weisdorf D, and Miller JS 2012. Cytomegalovirus reactivation after allogeneic transplantation promotes a lasting increase in educated NKG2C+ natural killer cells withpotentfunction. Blood 119:2665–2674.doi: 10.1182/blood-2011-10-386995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guma M, Angulo A, Vilches C, GomezLozano N, Malats N, and Lopez-Botet M. 2004. Imprint of human cytomegalovirus infection on the NK cell receptor repertoire. Blood 104:3664–3671. doi: 10.1182/blood-2004-05-2058. [DOI] [PubMed] [Google Scholar]
- Hwang I, Zhang T, Scott JM, Kim AR, Lee T, Kakarla T, Kim A, Sunwoo JB, and Kim S. 2012. Identification of human NK cells that are deficient for signaling adaptor FcRgamma and specialized for antibody-dependent immune functions. Int. Immunol. 24:793–802. doi: 10.1093/intimm/dxs080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Zhang T, Hwang I, Kim A, Nitschke L, Kim M, Scott JM, Kamimura Y, Lanier LL, and Kim S. 2015. Epigenetic modification and antibody-dependent expansion of memory-like NK cells in human cytomegalovirusinfected individuals. Immunity 42:431–442. doi: 10.1016/j.immuni.2015.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung W, Iyengar R, Triplett B, Turner V, Behm FG, Holladay MS, Houston J, and Handgretinger R. 2005. Comparison of killer Ig-like receptor genotyping and phenotyping for selection of allogeneic blood stem cell donors. J. Immunol. 174:6540–6545. doi: 10.4049/jimmunol.174.10.6540. [DOI] [PubMed] [Google Scholar]
- Locatelli F, Pende D, Mingari MC, Bertaina A, Falco M, Moretta A, and Moretta L. 2013.Cellularandmolecularbasisofhaploidentical hematopoietic stem cell transplantation in the successful treatment of high-risk leukemias: Role of alloreactive NK cells. Front. Immunol. 4:15. doi: 10.3389/fimmu.2013.00015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luetke-Eversloh M, Hammer Q, Durek P, Nordstrom K, Gasparoni G, Pink M, Hamann A, Walter J, Chang HD, Dong J, and Romagnani C. 2014. Human cytomegalovirus drives epigenetic imprinting of the IFNG locus in NKG2Chi natural killer cells. PLoS Pathog. 10:e1004441. doi: 10.1371/journal.ppat.1004441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahnke YD and Roederer M. 2007. Optimizing a multicolor immunophenotyping assay. Clin. Lab. Med. 27:469–485. doi: 10.1016/j.cll.2007.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muccio L, Bertaina A, Falco M, Pende D, Meazza R, Lopez-Botet M, Moretta L, Locatelli F, Moretta A, and Chiesa MD 2016. Analysis of memory-like natural killer cells in human cytomegalovirusinfected children undergoing alphabeta+T and B cell-depleted hematopoietic stem cell transplantation for hematological malignancies. Haematologica 101:371–381. doi: 10.3324/haematol.2015.134155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muntasell A, Pupuleku A, Cisneros E, Vera A, Moraru M, Vilches C, and Lopez-Botet M. 2016. Relationship of NKG2C copy number with the distribution of distinct cytomegalovirus-induced adaptive NK cell subsets. J. Immunol. pii:1502438. doi: 10.4049/jimmunol.1502438. [DOI] [PubMed] [Google Scholar]
- O’Sullivan TE, Sun JC, and Lanier LL 2015. Natural killer cell memory. Immunity 43:634–645. doi: 10.1016/j.immuni.2015.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlums H, Cichocki F, Tesi B, Theorell J, Beziat V, Holmes TD, Han H, Chiang SC, Foley B, Mattsson K, Larsson S, Schaffer M, Malmberg KJ, Ljunggren HG, Miller JS, and Bryceson YT 2015. Cytomegalovirus infection drives adaptive epigenetic diversification of NK cells with altered signaling and effector function. Immunity 42:443–456. doi: 10.1016/j.immuni.2015.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Sinzger C, Frascaroli G, Reichel J, Bayer C, Wang L, Schirmbeck R, and Mertens T. 2013. Human cytomegalovirus-induced NKG2C(hi) CD57(hi) natural killer cells are effectors dependent on humoral antiviral immunity. J. Virol. 87:7717–7725. doi: 10.1128/JVI.01096-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Scott JM, Hwang I, and Kim S. 2013. Cutting edge: Antibody-dependent memory-like NK cells distinguished by FcRgamma deficiency. J. Immunol. 190:1402–1406. doi: 10.4049/jimmunol.1203034. [DOI] [PMC free article] [PubMed] [Google Scholar]