Abstract
Background:
LBX2 antisense RNA 1 (LBX2-AS1), a long noncoding RNA, has been identified to be closely associated with the progression of various cancers. However, the role of LBX2-AS1 in colorectal cancer (CRC) is still poorly understood. In this study, we aimed to investigate the expression and function of LBX2-AS1 in CRC.
Material and Methods:
Expression data from the Gene Expression Omnibus (GEO) and Gene Expression Profiling Interactive Analysis (GEPIA) databases and results obtained from clinical samples/patients were used to determine the correlation between LBX2-AS1 expression and pathological stages, overall survival (OS). Furthermore, knockdown of LBX2-AS1 in CRC cells using the short interfering RNA (siRNA) technique, and observed its biological functions using western blotting, quantitative reverse transcription-polymerase chain reaction (qRT-PCR), cell counting kit-8 (CCK-8) and flow cytometry assay in the CRC cell line.
Results:
Our study demonstrated that the expression levels of LBX2-AS1 were higher in CRC cell lines than in normal colon mucosal cell lines. Bioinformatics analysis revealed that CRC patients with high LBX2-AS1 expression levels had poor OS. Furthermore, knockdown of LBX2-AS1 in CRC cells could attenuate the proliferative ability of CRC cells in vitro, which is associated with decreased expression of cyclin-dependent kinase (CDK) 3, CDK6, and CCND1 and enhanced expression of cyclin-dependent kinase inhibitor 1A.
Conclusions:
LBX2-AS1 plays a crucial role in the tumorigenesis of CRC, providing a potential therapeutic target for CRC patients.
Keywords: long noncoding RNA, LBX2 antisense RNA 1, colorectal cancer, prognosis, therapy
Introduction
Colorectal cancer (CRC), one of the most common and aggressive human malignancies, is the third leading cause of cancer-induced death worldwide.1,2 In 2015, approximately 0.8 million new cases and 0.4 million deaths from CRC in developed countries were reported.3 With the improvement of systemic therapy for CRC, in the past decades, substantial progress has been achieved in the treatment of CRC. However, the mortality rate of CRC caused by recurrence has not evidently changed.4 The progression and development of CRC may involve a multistep process that is closely associated with inherited factors and environmental factors, which may result in a series of changes in important molecules, thereby inducing cancer cell apoptosis, proliferation, and differentiation.5,6 With the popularity of molecular targeting therapy, some studies have focused on the molecular pathogenesis of CRC by analyzing molecular abnormalities in the tumorigenesis of CRC.7,8
Long noncoding RNAs (lncRNAs), a class of noncoding RNAs, have been recently identified.9,10 LncRNAs have a transcribing length of greater than 200 nt and have an insufficient complete functional open reading frame.11-13 In recent years, lncRNAs have been gradually gaining significant attention owing to their effects on cell biological behavior, specifically in tumor cells. Accumulating studies have revealed that lncRNAs are aberrantly expressed in several cancers, such as nasopharyngeal carcinoma,14 breast cancer,15 non-small cell lung cancer (NSCLC),16 and CRC.17 These abnormally expressed lncRNAs act as biomarkers for the diagnosis and therapy of cancers.
LBX2 antisense RNA 1 (LBX2-AS1) is a member of the lncRNA. It is located in the chr2p13.1 region and is 1786 bp in length. Emerging evidence indicates that LBX2-AS1 is associated with tumorigenesis in various cancers, including hepatocellular carcinoma (HCC),18 esophageal squamous cell carcinoma (ESCC),19 gastric cancer (GC),20 and NSCLC.21 However, their specific effects on the tumorigenesis of CRC are still poorly understood. In this study, we found that LBX2-AS1 was overexpressed in CRC cell lines and CRC samples, and CRC patients with high expression levels showed poor overall survival (OS). Furthermore, knockdown of the expression of LBX2-AS1 suppressed the proliferative ability of CRC cells in vitro. These results indicated that lncRNA LBX2-AS1 may be a novel target for cervical cancer therapy.
Materials and Methods
Bioinformatics Analysis
Microarray expression date was deposited in the Gene Expression Omnibus (GEO) database: GSE41328 (Affymetrix Human Genome U133 Plus 2.0 platform). The GSE41328 dataset contains 10 pairs of CRC and adjacent non tumor tissues.
Cell Culture and Transfection
CRC cell lines (HT29, LoVo, SW620 and HCT116) and the human normal colon mucosal cell line NCM460 were used in this study were obtained from the American Type Culture Collection (ATCC, Manassas, USA). These cell lines(NCM460, LoVo, SW620, HT29) were cultured in RPMI-1640 medium (Invitrogen, USA) with 10% fetal bovine serum (FBS, Invitrogen, USA), and HCT116 cells were maintained in DMEM media (Invitrogen, USA) with 10% FBS. When the cells covered 60% of the bottom of the bottle, 50nM siRNA oligos were transfected by Lipofectamine 3000 (Invitrogen, USA), according to the manufacturer’s instructions. The sequences of the LBX2-AS1 targeting siRNAs were: LBX2-AS1-si-1: 5’-AGGAATGTTTGCTGAATTAATGG -3’; LBX2-AS1-si-2: 5’-CCCAAGTTATAAAACTATAATGC-3’; Sequences of non-target scramble controls were provided by RiboBio(Guangzhou, China).
Subcellular Fractionation Analysis
The cytoplasmic and nuclear RNAs of CRC cell lines were separated using the PARIS Kit (Invitrogen, USA), and the computing method was according to described previously.22
Quantitative Real-Time Polymerase Chain Reaction
RNA isolation and amplification were performed as described previously.23 SYBR_Premix ExTaq II kit (Takara, China) was used to execute the quantitative real-time polymerase chain reaction (qRT-PCR) assay in the CFX96 Real-Time PCR Detection System (Bio-Rad, USA), thereby analyzing the relative expression levels of target genes. The sequences of qRT-PCR primers: LBX2-AS1 forward: 5’-AGTTTGTCCCAGGTTTGGCA-3’, reverse: 5’-CATGCCAGG GTCCTTGTTCT-3’; Human CCND1 forward: 5’-TCGTTGCCCTCTGTGCCACA-3’, reverse: 5’-GCAGTCCGGGTCACACTTGA-3’; Human CDK3 forward: 5’-CCAGCTCTTTCGTATCTTTCGT-3’, reverse: 5’-CCAGCTCTTTCGTATCTTTCGT-3’; Human CDK6 forward: 5’-CCAGATGGCTCTAACCTCAGT-3’, reverse: 5’-AACTTCCACGAAAAAGAGGCTT -3’; Human CDKN1A forward: 5’-CGATGGAACTTCGACTTTGTCA-3’, reverse: 5’-GCACAAGGGTACAAGACAGTG -3’; Human GAPDH forward 5’-CCACATCGCTCAGACACCAT-3’, reverse 5’-TGACAAGCTTCCCGTTCTCA-3’; Human β-actin forward 5’-TCACCAACTGGGACGACATG-3’, reverse 5’-GTCACCGGAGTCCATCACGAT-3’; Human U6 (nuclear) forward 5’-CTCGCTTCGGCAGCACA-3’, reverse 5’-AACGCTTCACGAATTTGCGT-3’.
Western Blot Assays
Western blot assays were performed as previously described.23 Primary anti-CCND1, CDK3, CDK6, CDKN1A and GAPDH antibody (Cell Signaling Technology, Danvers, MA, USA) and horseradish peroxidase-conjugated secondary antibodies (Santa Cruz Biotechnology) were used.
CCK8 Proliferation Assay
CCK8 proliferation assay was used to detect the cell proliferative ability, the operating steps carried out as described previously.24 Each experiment was repeated for 3 times independently.
Cell Cycle Detected by Flow Cytometry
After transfection with si-NC or si-LBX2-AS1 36 h, approximately 1 × 106 HCT116 cells were collected and fixed with 70% precooled ethanol for flow cytometry assay. DNA content was detected using propidium iodide (PI) (Sigma, San Antonio, USA) staining, and cell cycle distribution were detected by flow cytometry(Beckman Coulter, South Kraemer, USA), according to the methods described in a previous study.25 Each experiment was repeated for 3 times independently.
Statistical Analysis
SPSS 20.0 software was utilized for analysis. The measurement data were expressed by mean± standard deviation. The differences among groups were analyzed by the t test was adopted for comparison between groups. p < 0.05 suggested that the difference was statistically significant.
Results
LBX2-AS1 Overexpression in Colorectal Cancer (CRC)
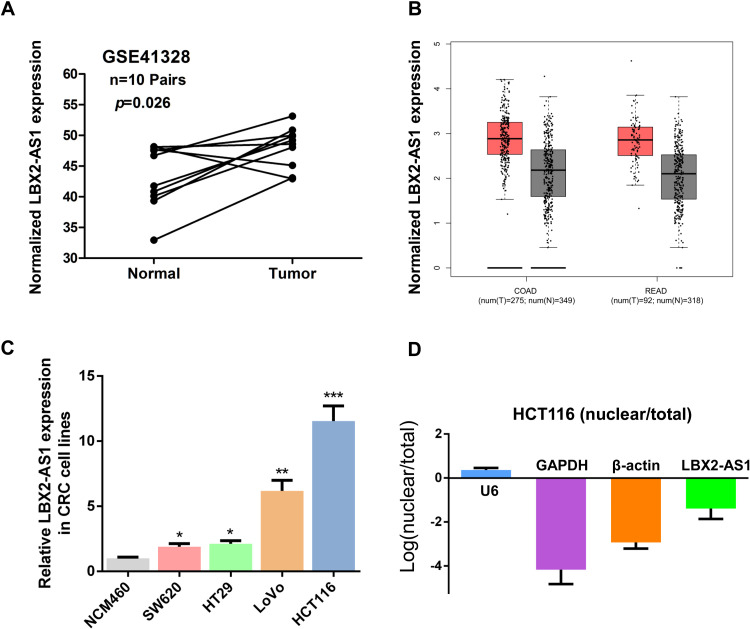
To analyze the expression of lncRNA LBX2-AS1 in CRC, we first analyzed GEO datasets GSE41328 using Affymetrix HG_U133 Plus 2 arrays to identify dysregulated lncRNAs in CRC. We found that LBX2-AS1 was significantly upregulated in GSE41328 (p < 0.001, Figure 1A). Meanwhile, LBX2-AS1 was significantly upregulated in Colon adenoma (COAD) and Rectum adenoma (READ) tissues of The Cancer Genome Atlas (TCGA) database by analyzing the Gene Expression Profiling Interactive Analysis (GEPIA) public platform26 (Figure 1B). Subsequently, we detected the expression of LBX2-AS1 in CRC cell lines by qRT-PCR and found that LBX2-AS1 expression was higher in the CRC cell lines (HT29, LoVo, SW620, and HCT116) than in the normal colon mucosal cell line NCM460 (all p < 0.05, Figure 1C). Additionally, LBX2-AS1 expression was the highest in HCT116 cells. Moreover, we analyzed the cellular localization of LBX2-AS1 in HCT116 cells and found that the cytoplasm proportion of LBX2-AS1 was higher than the nuclear proportion of LBX2-AS1, indicating that the subcellular localization of LBX2-AS1 in the CRC cells was mainly located at the cytoplasm (Figure 1D).
Figure 1.
LBX2 antisense RNA 1 (LBX2-AS1) is overexpressed in colorectal cancer (CRC). (A) LBX2-AS1 expression, as measured by Affymetrix microarray, was more upregulated in CRC tissues compared with normal colon mucosal tissues in #GSE41328 (containing 10 pairs of CRC tissues and corresponding normal colorectal tissues) from the Gene Expression Omnibus database. (B) The Gene Expression Profiling Interactive Analysis database was used to analyze the expression of LBX2-AS1 in CRC tissues using the display form of box plots. (C) LBX2-AS1 expression was significantly higher in CRC cell lines (HT29, LoVo, SW620, and HCT116) than in the normal colon mucosal cell line NCM460. (D) Nucleic and cytoplasmic RNA were analyzed using quantitative real-time polymerase chain reaction to detect the expression of LBX2-AS1 in HCT116 cells. U6 was used as a nucleic RNA control; glyceraldehyde 3-phosphate dehydrogenase and β-actin were used as cytoplasmic RNA controls. Data are shown as mean ± standard error of the mean.
* p < 0.05, ** p < 0.01, *** p < 0.001 compared with control
Upregulated LBX2 Antisense RNA 1 (LBX2-AS1) Is Associated With the Clinicopathological Features of CRC
Poor prognosis of CRC patients
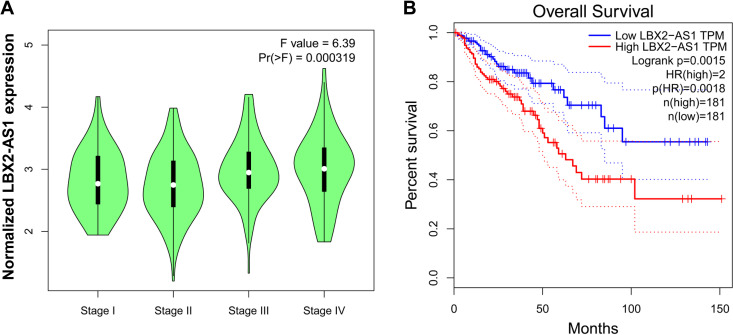
Next, we assessed the association between LBX2-AS1 expression and the pathological stages of CRC by analyzing the GEPIA public platform. We found that the higher expression of LBX2-AS1 was significantly associated with the pathological stages of CRC (Figure 2A). To assess the prognostic value of LBX2-AS1 expression in CRC patients, we examined the association between LBX2-AS1 expression levels and OS using Kaplan-Meier analysis with the log-rank test by analyzing the GEPIA public platform. The results revealed that patients with high LBX2-AS1 expression levels had lower OS (p = 0.0018, Figure 2B). Taken together, these results indicate that high LBX2-AS1 expression is an independent risk factor for CRC patients.
Figure 2.
Upregulated LBX2 antisense RNA 1 (LBX2-AS1) is associated with the clinicopathological features of colorectal cancer (CRC). (A) The Gene Expression Profiling Interactive Analysis (GEPIA) database was used to analyze the association between LBX2-AS1 expression and the pathological stages of CRC. (B) The GEPIA database was used to analyze the clinical impact of LBX2-AS1 expression patterns on CRC patients’ survival in a CRC specimen expression profile dataset, group 1 = low expression of LBX2-AS1, n = 181; group 2 = high expression of LBX2-AS1, n = 181.
LBX2-AS1 knockdown suppresses the proliferative ability of CRC cells
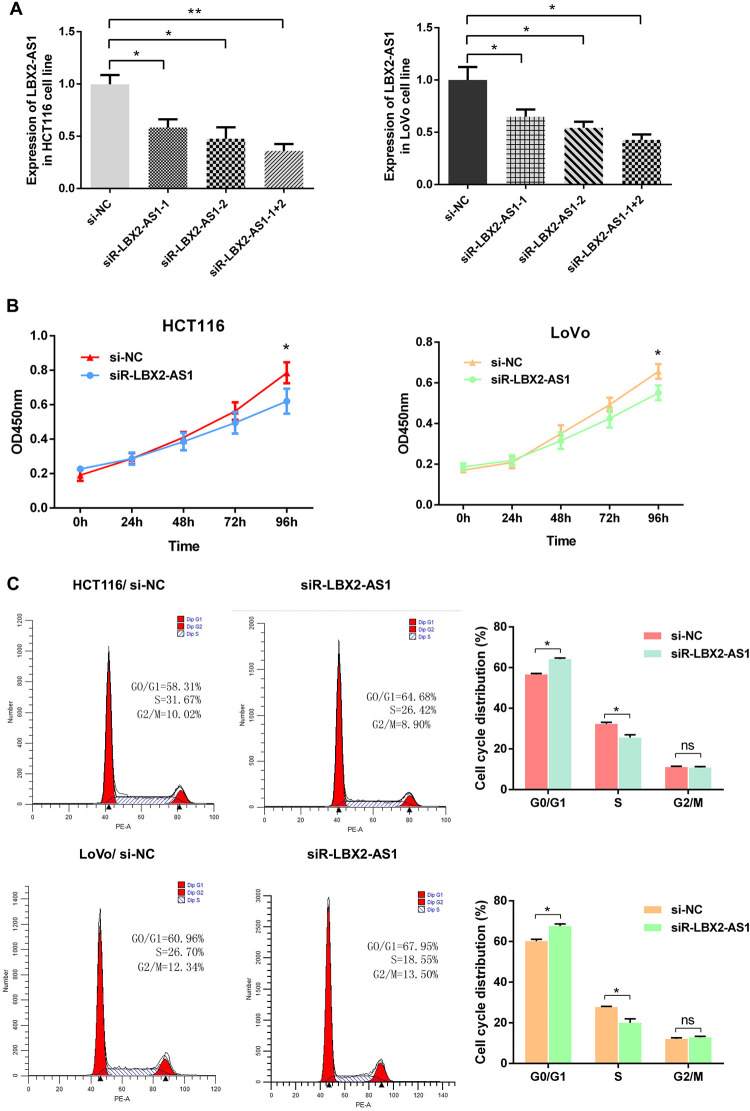
To verify LBX2-AS1 function in colon cancer cells, we first measured the efficiency of the short interfering RNA (siRNA) siRNA-LBX2-AS1 (si-LBX2-AS1). The si-LBX2-AS1-1+2 group showed the higher interfering efficiency in HCT116 and LoVo cells than the si-LBX2-AS1-1 and si-LBX2-AS1-2 groups (Figure 3A). Therefore, we selected si-LBX2-AS1-1+2 to knockdown LBX2-AS1 expression in CRC cells and subsequently assessed the function of LBX2-AS1 in CRC cells. We investigated the effect of LBX2-AS1 knockdown on the cell proliferative ability of CRC cells. The cell growth rates were determined by performing the CCK-8 proliferation assay. Knockdown of LBX2-AS1 expression significantly inhibited HCT116 and LoVo cell proliferation, relative to control cells (p < 0.001, Figure 3B). Meanwhile, flow cytometry analysis demonstrated that knockdown of LBX2-AS1 expression had lower percentage of cells in the S phase and higher percentage of cells in the G1 phase in CRC cells than control cells (all p < 0.05, Figure 3C). These results indicated that LBX2-AS1 knockdown suppresses CRC cell proliferation.
Figure 3.
LBX2 antisense RNA 1 (LBX2-AS1) knockdown suppressed the proliferative ability of colorectal cancer cells. (A) The interference efficiency of si-LBX2-AS1 was verified in HCT116 and LoVo cells. HCT116 and LoVo cells were transfected with either si-NC or si-LBX2-AS1 (1#, 2#, 1+2#) for 48 h, and subsequently, LBX2-AS1 expression was analyzed by quantitative real-time polymerase chain reaction. (B-C) Cell Counting Kit-8 proliferation assay and flow cytometry assay were used to detect the cell proliferative ability after transfection with si-NC or si-LBX2-AS1 for 48 h in HCT116 and LoVo cells. Data are shown as mean ± standard error of the mean. * p < 0.05, ** p < 0.01, *** p < 0.001 compared with control.
LBX2-AS1 knockdown inhibits the expression of proliferation-associated markers in CRC cells
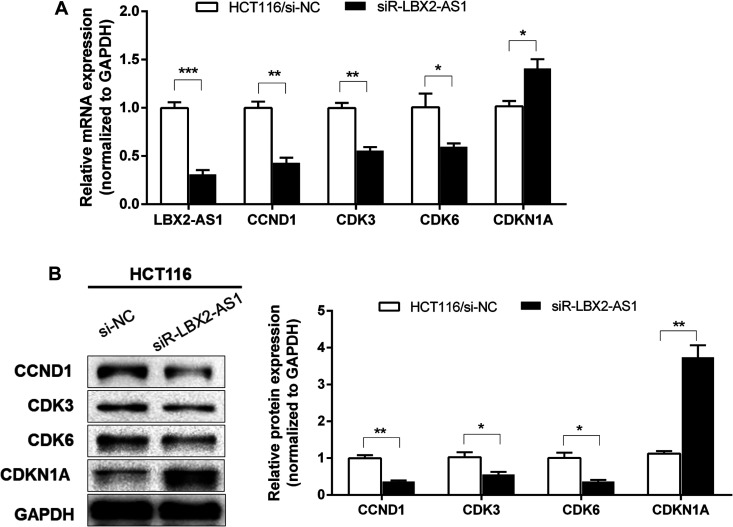
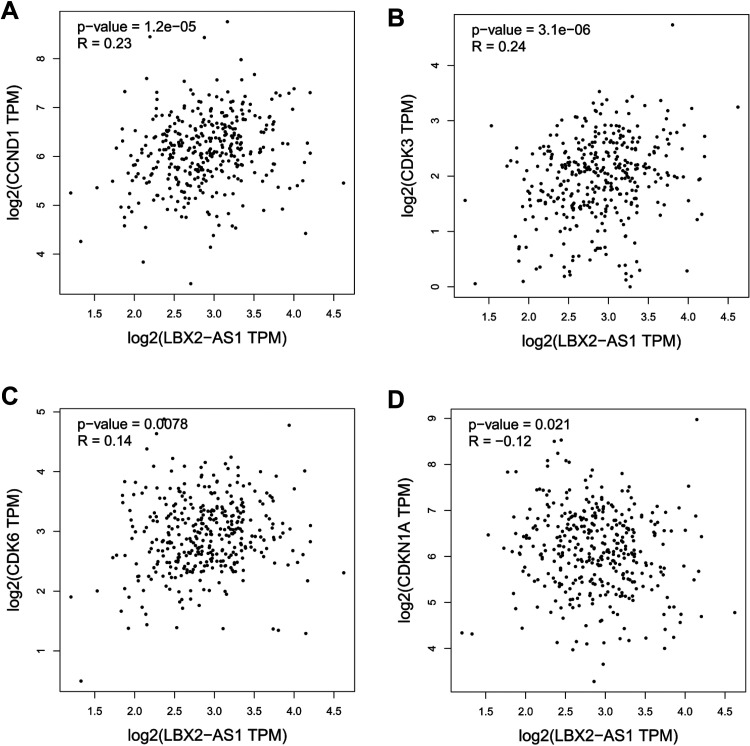
To further study the molecular mechanism of LBX2-AS1 in CRC cells, qRT-PCR and western blotting were used to assess the mRNA and protein levels of the proliferation-associated markers CCND1, cyclin-dependent kinase (CDK) 3, CDK6, and cyclin-dependent kinase inhibitor 1A (CDKN1A) in HCT116 cells. Knockdown of LBX2-AS1 expression significantly decreased the mRNA and protein expression levels of CCND1, CDK3, and CDK6 and increased the mRNA and protein expression levels of cell cycle cyclin-dependent kinase inhibitor CDKN1A (all p < 0.05, Figure 4A-B). Furthermore, GEPIA database analysis showed that LBX2-AS1 expression was positively associated with the expression of CCND1, CDK3, and CDK6 and negatively associated with the expression of CDKN1A in CRC (all p < 0.05, Figure 5). This indicates that LBX2-AS1 contributes to the regulation of proliferated marker expression in CRC cells.
Figure 4.
LBX2 antisense RNA 1 (LBX2-AS1) knockdown inhibited the expression of the proliferation-associated markers in colorectal cancer (CRC) cells. After transfection with si-NC or si-LBX2-AS1 for 48 h in HCT116 cells, mRNA expression of CCND1, cyclin-dependent kinase 3 (CDK3), CDK6, and cyclin-dependent kinase inhibitor 1A (CDKN1A) in CRC cells were analyzed by quantitative real-time polymerase chain reaction (A); meanwhile, the protein expressions of CCND1, CDK3, CDK6, and CDKN1A in CRC cells were analyzed by western blotting and densitometry (B). Data are shown as mean ± standard error of the mean. * p < 0.05, ** p < 0.01, *** p < 0.001 compared with control.
Figure 5.
Scatter plots showing the statistical association between LBX2 antisense RNA 1 (LBX2-AS1) expression and proliferation marker expression. The Gene Expression Profiling Interactive Analysis database was used to analyze the association between LBX2-AS1 expression and the expression of CCND1, cyclin-dependent kinase 3 (CDK3), CDK6, and cyclin-dependent kinase inhibitor 1A in colorectal cancer.
Discussion
CRC is one of the most common gastrointestinal tumors worldwide,27 specifically in developed countries. CRC is the fifth major cause of cancer-associated deaths in China and has a substantial effect on human health.28 Despite significant advancements in the diagnosis and treatment of CRC, the mortality and morbidity rates of CRC remain high, possibly due to postsurgical recurrence of primary tumors.29 Therefore, determining novel therapeutic targets to better monitor recurrence, metastasis, and other diseases is significantly required.
Increasing studies have indicated that lncRNAs play a significant role in various biological processes by regulating the mechanisms of cellular processes, including cell apoptosis, proliferation, and differentiation.30,31 Recently, several studies have demonstrated that disruption or disability expression of lncRNAs is closely associated with the progression of malignant tumors and is involved in cancer cell proliferation, inflammation, epithelial-mesenchymal transition (EMT), and drug resistance.11,32 LncRNAs have the following characteristics: they can be easily extracted, are detected with higher sensitivity and specificity, and exist constantly in the blood and tissue.33 LncRNAs are considered potential novel biomarkers for cancer diagnosis, recurrence, and prognosis. In CRC, some lncRNAs have been shown to be differentially expressed, indicating poor prognosis, including LINC00467,34 LINC01638,35 CCAT1,36 LINC00152,37 and GAS5.38
With the development of high-throughput sequencing, a series of public databases (e.g. TCGA, GEO) have been widely used to predict and identify valuable lncRNAs.17 In this study, we analyzed the dysregulated lncRNAs in CRC using the GEO dataset GSE41328 using the Affymetrix Human Genome U133 Plus 2.0 platform. We found that lncRNA LBX2-AS1 was significantly overexpressed in CRC datasets. In previous studies, LBX2-AS1 was overexpressed in various human cancers, and patients with higher expression of LBX2-AS1 had unfavorable OS, including patients diagnosed with HCC,18 GC,20 and NSCLC.21 However, the expression and function of LBX2-AS1 in CRC have not been reported. In our study, we demonstrated that LBX2-AS1 expression in CRC tissues was significantly higher than that in matched adjacent normal tissues. Meanwhile, the overexpression of LBX2-AS1 was significantly associated with shorter OS of CRC patients, which may be potentially developed as a novel biomarker for CRC diagnosis and prognosis.
Regarding the effect of LBX2-AS1 in cancers, convincing evidence has shown that it plays critical roles in regulating the development and progression of cancer. For sexample, Yang et al. demonstrated that LBX2-AS1 was overexpressed in ESCC, and it could interact with the RNA-binding protein heterogeneous nuclear ribonucleoprotein C to regulate the expression of ZEB1 and ZEB2, thereby promoting cell migration and EMT of ESCC cells.19 Moreover, Wang et al. showed that LBX2-AS1 could sponge with miR-384 to promote the expression of insulin receptor substrate 1, thereby accelerating cell proliferation, migration, and invasion of HCC cells and inducing apoptosis in vitro.18 However, the effect of LBX2-AS1 on CRC proliferation has been rarely understood. Our results demonstrated that knockdown of LBX2-AS1 expression suppressed cell proliferation in HCT116 cell lines, which was associated with decreased CCND1, CDK3, and CDK6 expression, but enhanced CDKN1A expression. This indicates that LBX2-AS1 regulates the expression of proliferation markers in CRC cell lines.
Conclusions
In summary, our study indicated that the expression of LBX2-AS1 is upregulated in CRC tissues and CRC cell lines, and these patients with high LBX2-AS1 expression showed poor OS. Additionally, knockdown of LBX2-AS1 expression significantly suppressed the proliferative ability of CRC cells. Therefore, LBX2-AS1 may play a crucial role in regulating the tumorigenesis of CRC and provide a prospect for the development of novel CRC therapies after further investigation.
Footnotes
Authors’ Contributions: QL, HX, SW and ZZ performed all the experiments, analyzed all the data and were a major contributor in writing the manuscript. QL, HX, SW and ZZ also contributed to performing the experimental tasks and aided in the drafting of the manuscript. All authors read and approved the final manuscript and agree to be accountable for all aspects of the research in ensuring that the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Authors’ Note: Qing Li and Hui Xie are also affiliated to the Key Laboratory of Medical Imaging and Artifical Intelligence of Hunan Province.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This study was supported by The Science and Technology Funding Project of Hunan Province, China (No.2017SK4010), and Key laboratory of tumor precision medicine, Hunan colleges and university project (2019-379).”
ORCID iD: Zijian Zhang  https://orcid.org/0000-0002-2702-8025
https://orcid.org/0000-0002-2702-8025
References
- 1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70(1):7–30. [DOI] [PubMed] [Google Scholar]
- 2. Obuch JC, Ahnen DJ. Colorectal cancer: genetics is changing everything. Gastroenterol Clin North Am. 2016;45(3):459–476. [DOI] [PubMed] [Google Scholar]
- 3. Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127(12):2893–2917. [DOI] [PubMed] [Google Scholar]
- 4. Lee YC, Lee YL, Chuang JP, Lee JC. Differences in survival between colon and rectal cancer from SEER data. Plos One. 2013;8(11):e78709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nie H, Wang Y, Liao Z, Zhou J, Ou C. The function and mechanism of circular RNAs in gastrointestinal tumours. Cell Prolif. 2020;53(7):e12815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sun Z, Ou C, Liu J, et al. YAP1-induced MALAT1 promotes epithelial-mesenchymal transition and angiogenesis by sponging miR-126-5p in colorectal cancer. Oncogene. 2019;38(14):2627–2644. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 7. Yiu AJ, Yiu CY. Biomarkers in colorectal cancer. Anticancer Res. 2016;36(3):1093–1102. [PubMed] [Google Scholar]
- 8. Vaiopoulos AG, Athanasoula K, Papavassiliou AG. Epigenetic modifications in colorectal cancer: molecular insights and therapeutic challenges. Biochim Biophys Acta. 2014;1842(7):971–980. [DOI] [PubMed] [Google Scholar]
- 9. Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet. 2016;17(1):47–62. [DOI] [PubMed] [Google Scholar]
- 10. Orom UA, Derrien T, Beringer M, et al. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143(1):46–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Schmitt AM, Chang HY. Long Noncoding RNAs in Cancer Pathways. Cancer Cell. 2016;29(4):452–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. He X, Ou C, Xiao Y, Han Q, Li H, Zhou S. LncRNAs: key players and novel insights into diabetes mellitus. Oncotarget. 2017;8(41):71325–71341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bazzini AA, Johnstone TG, Christiano R, et al. Identification of small ORFs in vertebrates using ribosome footprinting and evolutionary conservation. Embo J. 2014;33(9):981–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhao CH, Bai XF, Hu XH. Knockdown of lncRNA XIST inhibits hypoxia-induced glycolysis, migration and invasion through regulating miR-381-3p/NEK5 axis in nasopharyngeal carcinoma. Eur Rev Med Pharmacol Sci. 2020;24(5):2505–2517. [DOI] [PubMed] [Google Scholar]
- 15. Zhang M, Wang F, Xiang Z, Huang T, Zhou WB. LncRNA XIST promotes chemoresistance of breast cancer cells to doxorubicin by sponging miR-200c-3p to upregulate ANLN. Clin Exp Pharmacol Physiol. 2020;47(8):1464–1472. [DOI] [PubMed] [Google Scholar]
- 16. Dong Z, Liu H, Zhao G. Long noncoding RNA SNHG6 promotes proliferation and inhibits apoptosis in non-small cell lung cancer cells by regulating miR-490-3p/RSF1 axis. Cancer Biother Radiopharm. 2020;35(5):351–361. [DOI] [PubMed] [Google Scholar]
- 17. Wang Y, Nie H, He X, et al. The emerging role of super enhancer-derived noncoding RNAs in human cancer. Theranostics. 2020;10(24):11049–11062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wang Y, Zhao Y, Zhang X, Zhang A, Ma J. Long noncoding RNA LBX2-AS1 drives the progression of hepatocellular carcinoma by sponging microRNA-384 and thereby positively regulating IRS1 expression. Pathol Res Pract. 2020;216(4): 152903. [DOI] [PubMed] [Google Scholar]
- 19. Zhang Y, Chen W, Pan T, Wang H, Zhang Y, Li C. LBX2-AS1 is activated by ZEB1 and promotes the development of esophageal squamous cell carcinoma by interacting with HNRNPC to enhance the stability of ZEB1 and ZEB2 mRNAs. Biochem Biophys Res Commun. 2019;511(3):566–572. [DOI] [PubMed] [Google Scholar]
- 20. Yang Z, Dong X, Pu M, et al. LBX2-AS1/miR-219a-2-3p/FUS/LBX2 positive feedback loop contributes to the proliferation of gastric cancer. Gastric Cancer. 2019;23(3):449–463. [DOI] [PubMed] [Google Scholar]
- 21. Tang LX, Su SF, Wan Q, He P, Xhang Y, Cheng XM. Novel long non-coding RNA LBX2-AS1 indicates poor prognosis and promotes cell proliferation and metastasis through Notch signaling in non-small cell lung cancer. Eur Rev Med Pharmacol Sci. 2019;23(17):7419–7429. [DOI] [PubMed] [Google Scholar]
- 22. Huarte M, Guttman M, Feldser D, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142(3):409–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ou C, Sun Z, Zhang H, et al. SPLUNC1 reduces the inflammatory response of nasopharyngeal carcinoma cells infected with the EB virus by inhibiting the TLR9/NF-kappaB pathway. Oncol Rep. 2015;33(6):2779–2788. [DOI] [PubMed] [Google Scholar]
- 24. Li Y, Zhao C, Yu Z, et al. Low expression of miR-381 is a favorite prognosis factor and enhances the chemosensitivity of osteosarcoma. Oncotarget. 2016;7(42):68585–68596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dang W, Qin Z, Fan S, et al. miR-1207-5p suppresses lung cancer growth and metastasis by targeting CSF1. Oncotarget. 2016;7(22):32421–32432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45(w1):W98–W102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bray F, Ren JS, Masuyer E, Ferlay J. Global estimates of cancer prevalence for 27 sites in the adult population in 2008. Int J Cancer. 2013;132(5):1133–1145. [DOI] [PubMed] [Google Scholar]
- 28. Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132. [DOI] [PubMed] [Google Scholar]
- 29. DeSantis CE, Miller KD, Dale W, et al. Cancer statistics for adults aged 85 years and older, 2019. CA Cancer J Clin. 2019;69(6):452–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Atianand MK, Caffrey DR, Fitzgerald KA. Immunobiology of long noncoding RNAs. Annu Rev Immunol. 2017;35:177–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Thomson DW, Dinger ME. Endogenous microRNA sponges: evidence and controversy. Nat Rev Genet. 2016;17(5):272–283. [DOI] [PubMed] [Google Scholar]
- 32. Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136(4):629–641. [DOI] [PubMed] [Google Scholar]
- 33. Ou C, Li G. Long non-coding RNA TUG1: a novel therapeutic target in small cell lung cancer. J Thorac Dis. 2017;9(7):E644–E645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. He X, Li S, Yu B, et al. Up-regulation of LINC00467 promotes the tumourigenesis in colorectal cancer. J Cancer. 2019;10(25):6405–6413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhuo W, Hu D, Chen X, Zhang T. LINC01638 silencing inhibits cancer cell proliferation in colorectal adenocarcinoma through interaction with RUNX2. Mol Med Rep. 2019;19(6):5275–5280. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 36. Wang N, Yu Y, Xu B, Zhang M, Li Q, Miao L. Pivotal prognostic and diagnostic role of the long noncoding RNA colon cancerassociated transcript 1 expression in human cancer (Review). Mol Med Rep. 2019;19(2):771–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ou C, Sun Z, He X, et al. Targeting YAP1/LINC00152/FSCN1 signaling axis prevents the progression of colorectal cancer. Adv Sci (Weinh). 2020;7(3):1901380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yin D, He X, Zhang E, Kong R, De W, Zhang Z. Long noncoding RNA GAS5 affects cell proliferation and predicts a poor prognosis in patients with colorectal cancer. Med Oncol. 2014;31(11):253. [DOI] [PubMed] [Google Scholar]