Abstract
We describe a novel method for robust identification of common brain networks and their corresponding temporal dynamics across subjects from asynchronous functional MRI (fMRI) using tensor decomposition. We first temporally align asynchronous fMRI data using the orthogonal BrainSync transform, allowing us to study common brain networks across sessions and subjects. We then map the synchronized fMRI data into a 3D tensor (vertices × time × subject/session). Finally, we apply Nesterov-accelerated adaptive moment estimation (Nadam) within a scalable and robust sequential Canonical Polyadic (CP) decomposition framework to identify a low rank tensor approximation to the data. As a result of CP tensor decomposition, we successfully identified twelve known brain networks with their corresponding temporal dynamics from 40 subjects using the Human Connectome Project’s language task fMRI data without any prior information regarding the specific task designs. Seven of these networks show distinct subjects’ responses to the language task with differing temporal dynamics; two show sub-components of the default mode network that exhibit deactivation during the tasks; the remaining three components reflect non-task-related activities. We compare results to those found using group independent component analysis (ICA) and canonical ICA. Bootstrap analysis demonstrates increased robustness of networks found using the CP tensor approach relative to ICA-based methods.
Keywords: Brain network identification, Functional MRI, Tensor decomposition, Optimization
1. Introduction
Characterization and identification of brain networks from functional MRI provides important insights into brain organization and the influence of development, aging and disease on large-scale communication in the brain. The most widely used tools for identification of these networks across subjects are based on independent component analysis (ICA). One approach to group analysis is to perform ICA individually on each subject and then combine components across groups (Calhoun et al., 2001a; Esposito et al., 2005). Alternatively, group ICA can be performed directly across subjects through either temporal or spatial concatenation (Calhoun et al., 2001b; Guo and Pagnoni, 2008; Schmithorst and Holland, 2004; Svensén et al., 2002). Temporal concatenation produces components with unique time series for each subject but shared spatial maps. Spatial concatenation produces common time-series but unique spatial maps for each subject.
Although meaningful components can be found using these ICA-based approaches (Calhoun et al., 2009), by concatenating multi-subject data into a 2-dimensional representation, we lose the higher-dimensional (space × time × subject) low-rank structure that may be inherent in the data. This low-rank structure can be captured and efficiently represented in a tensor format as we illustrate below in Fig. 1. Another limitation of group ICA is that it requires an additional assumption of either spatial or temporal independence, which may not be realistic as brain networks can overlap and be correlated in both space and time (Karahanoğlu and Van De Ville, 2015). Further, it has been shown that stability or robustness of the solutions is a well-known issue associated with ICA (Himberg and Hyvärinen, 2003), although several variants have been developed to improve its stability (Du and Fan, 2013; Varoquaux et al., 2010). Significantly different independent components may be obtained from bootstrap resamples of the data, or even as a result of different initializations with the same data.
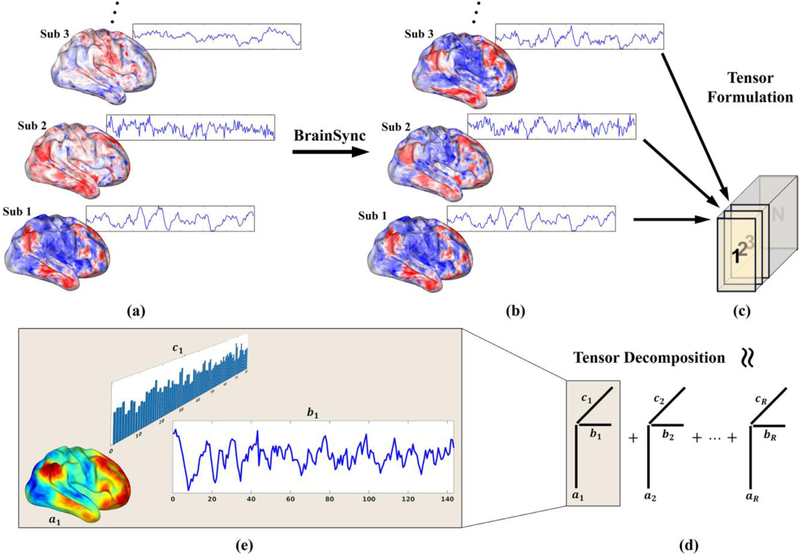
Fig. 1.
Brain network identification pipeline: (a) multi-subject fMRI data: the spatial maps show single-time brain activity at all vertices on a surface representation and the time courses show single-vertex brain activity through the entire fMRI recording; (b) The counterpart to (a) but after synchronization using BrainSync; (c) Tensor formation by arranging the subjects along the third dimension; (d) Tensor decomposition into R rank-1 components. (e) Each rank-1 component represents a brain network (or response to tasks) with a spatial map (ai), a temporal dynamic (bi), and a subject participation level (ci).
In order to address these issues, we use a tensor decomposition of the multi-subject fMRI data. Higher-order tensor decomposition is a generalization of matrix factorization. Application of tensor decomposition to fMRI data for brain network identification has previously been explored. The Tucker and canonical polyadic (CP) models (Cichocki et al., 2015; Kolda and Bader, 2009) are the two most commonly used tensor representations. Leonardi and Van De Ville applied a Tucker model to sliding-window-based dynamic connectivity tensors derived from task fMRI data and predicted the task design paradigm for unseen subjects (Leonardi and Van De Ville, 2013). Al-sharoa et al. performed a 4D Tucker decomposition by adding one extra dimension over the Leonardi model via random sampling over time and subjects, allowing them to identify five different brain states using k-means clustering (Al-sharoa et al., 2019). In comparison with the CP model, Tucker allows interactions between different modes through its core tensor which can have an impact on interpretability. Furthermore, the orthogonality constraints imposed on the Tucker solutions may not be realistic for brain networks. For these reasons, here we focus on the CP decomposition.
CP decomposition is most frequently performed using an alternating least square algorithm for a group-level fMRI study to find common networks among subjects. For example, Andersen and Rayens applied a third-order CP decomposition to finger-tapping task fMRI (Andersen and Rayens, 2004). Beckmann and Smith extended ICA to higher-order tensors by imposing an independence constraint in the spatial dimension (Beckmann and Smith, 2005). Instead of adding an independence constraint, Sen and Parhi imposed an orthogonality constraint in the spatial dimension as with PCA (Sen and Parhi, 2017). However, CP decomposition on fMRI data is not as popular as other methods because of several issues limiting its applicability to fMRI studies as discussed below.
Multi-subject group analysis on asynchronous fMRI data:
Almost all fMRI studies using CP decomposition were performed on task fMRI data, with the assumption that the temporal dynamics were synchronized across subjects based on alignment of the response stimulus timing (Andersen and Rayens, 2004). This temporal synchrony across multiple subjects is a strict requirement for CP decomposition to work well with low-rank models. However, the assumption may not be satisfied even when an identical task design is used across all subjects because individual responses to tasks may differ in latency, sometimes significantly for higher-level cognitive tasks (Friston et al., 1998). Low-rank CP decomposition cannot be performed when using different task designs across scans or when no task is present as in the case of resting-state fMRI. Moreover, any brain processes independent of the task cannot be identified using a CP decomposition because this task-independent activity is not synchronized even when the data are aligned to stimulus presentation.
Here we combine the tensor decomposition with a temporal alignment step based on an orthogonal transform, BrainSync (Joshi et al., 2018; Akrami et al., 2019). This transform exploits similarity in temporal correlation structure across subjects to produce alignment at homologous locations across subjects in the sense that the resulting time series are highly correlated. By applying this transformation prior to computing the tensor decomposition we are able to produce approximate synchronization of the time series for each component or network across subjects, as we show below.
Robustness against local minima and scalability to large dataset:
The most common method for CP decomposition uses alternating least squares (ALS) for optimization. It has been shown that ALS is not guaranteed to converge to a global minimum or a stationary point for a CP model, even with multi-start (Cichocki et al., 2015; Kolda and Bader, 2009). Although adding additional constraints, such as independence (Beckmann and Smith, 2005) or orthogonality (Sen and Parhi, 2017), may help avoid local minima, those constraints may not be physiologically reasonable for brain network identification. Indeed, specific concerns (Helwig and Hong, 2013; Stegeman, 2007) have been raised against imposing those constraints when applying the CP model to fMRI data. Moreover, the naïve ALS algorithm does not scale well to large datasets. As we have shown in (Li et al., 2018, 2017), computational complexity is approximately quadratically proportional to the largest dimension of the tensor. In fact, most of the studies cited above heavily downsampled the data in the spatial domain in order to have a tractable CP decomposition. For robustness, a multi-start strategy needs to be employed, resulting in an even higher complexity.
In this paper we describe a method that uses tensor decomposition to robustly identify common brain networks, (that is, it computes both spatial maps and temporal dynamics simultaneously) across multiple subjects from potentially asynchronous fMRI data without imposing unrealistic constraints on the networks. We approach this problem using the Nesterov-accelerated adaptive moment estimation (Nadam) method (Dozat, 2016) applied to a full-gradient search to simultaneously estimate all components of the tensor. As with our earlier work, we use a sequential canonical polyadic decomposition framework (Li et al., 2018, 2017), in which we compute a sequence of solutions, increasing the rank by one at each step and using the lower rank solution as a warm-start to initialize the search. As noted above, we first apply the BrainSync algorithm (Joshi et al., 2018), which uses a temporally orthogonal transform to align time-series across subjects.
We refer to our tensor decomposition algorithm as Nadam-Accelerated SCAlable and Robust (NASCAR) CP decomposition. Using this pipeline, as illustrated in Fig. 1, we show that spatially overlapped and temporally correlated brain networks can be successfully identified from multi-subject task fMRI data using NASCAR. We also explore robustness of the resulting networks relative to group ICA (Calhoun et al., 2001b) and canonical ICA (CanICA) (Varoquaux et al., 2010) using multi-start and bootstrapping.
An outline and some preliminary work of the idea described here have been previously presented in (Li et al., 2019). The current paper provides a more detailed description of the method and novel experimental results using language task fMRI data.
2. Methods
2.1. Time-series synchronization
Functional MRI time series from two different subjects are often not directly comparable. This is clearly the case for resting-state fMRI studies where spontaneous activity varies over subjects and time. Even in stimulus-locked event-related studies, brain activity can vary due to differing latencies in response (Friston et al., 1998). However, in order to usefully represent multi-subject fMRI data as a third-order tensor, they must be temporally aligned. We address this problem before computing the tensor decomposition using the recently developed BrainSync method for temporal synchronization (Joshi et al., 2018). This method exploits similarity in correlation structure across subjects to perform an orthogonal rotation of time series data between two or more subjects to induce a high correlation between time series at homologous locations.
Let Xi and Xj be matrices representing fMRI data for any two subjects, each of size V × T, where V is the number of vertices and T is the number of time points with V ≫ T. We assume in the following that these data have been mapped onto a tessellated representation of the mid-cortical surface, non-rigidly aligned and resampled onto a common mesh (Glasser et al., 2013). BrainSync finds an orthogonal transform OS that minimizes the total squared error:
where O(T) represents the group of T × T orthogonal matrices. The problem is well-posed when V ≫ T and has the closed form solution (Joshi et al., 2018):
where is the singular value decomposition of the cross-correlation matrix between Xi and Xj and “T” is the transpose operator. After applying this transform, the time series at homologous locations in the two subjects will be aligned in the sense that they will be highly correlated as illustrated in Fig. 1 (a) and (b). An extension of BrainSync from pair-wise to group alignment is described in (Akrami et al., 2019).
2.2. Tensor representation and decomposition of fMRI data
The Canonical Polyadic (CP) decomposition approximates a third-order tensor as a sum of rank-1 tensors:
| (1) |
where , , have unit norm; λr represents the scale factor for each component; “∘” is the vector outer product and R is the rank or number of components. For fMRI data, the tensor is formed by arranging spatiotemporal data for a group of subjects as illustrated in Fig. 1 (c). Each component obtained from the decomposition, λr ar °br °cr, can be viewed as representing a distinct brain network, where ar, br, cr are the spatial map, temporal dynamics, and relative amplitude of each subject’s involvement in that network, respectively, as illustrated in Fig. 1 (d) and (e).
If we combine the ar to form a matrix and similarly for and , then we can express a least-squares fit of the tensor model to the data in terms of the following three equivalent cost functions (Kolda and Bader, 2009):
| (2) |
where X (i) is the matricized or unfolded version of tensor along the ith dimension (Kolda and Bader, 2009); “⊙” is the Khatri-Rao product between two matrices.
A common approach to fitting the tensor is to use Alternating Least Squares (ALS): first solving for A with B and C fixed, then solving for B with A and C fixed, and so on until convergence. We previously used an ALS approach for tensor decompositions of EEG data in combination with a warm start procedure. The warm start approach, Scalable and Robust Sequential CP Decomposition (SRSCPD), builds a model of successively higher order, r, with each search initialized using the results from the previous order, r − 1 (Li et al., 2018, 2017). Here we use a full gradient-based method, again in combination with a warm start, which we found to converge substantially faster than ALS.
2.3. Gradient of the CP Model, Adam and Nadam
If we treat the variables in a CP model as a high-dimensional vector lying in the space of , where N = I × R + J × R + K × R, then objective function f(A, B, C) in Eq. (2) is a scalar-valued cost function . Solutions can be obtained using gradient-based optimization. The partial gradient of the objective function f with respect to the loading matrix A is:
and like-wise for B and C, where “*” is the Hadamard product between two matrices (Acar et al., 2011). A gradient-based search on the unregularized cost function will not produce a unique solution because all solutions in the form of {η1A, η2B, η3C } with η1η2η3 = 1 are equivalent. To resolve this ambiguity, we use the Tikhonov regularizer:
| (3) |
where μA is the regularization parameter and similarly for B and C. The role of the regularization term is to prevent individual terms in the outer product becoming arbitrarily large. For this reason, the minimizer of Eq. (3) is relatively insensitive to the choice of regularization parameter μA provided the value is not too large (see Section 3.2). In this regularized case, the gradient becomes (Acar et al., 2011):
| (4) |
and similarly for B and C. Moreover, in real application to in-vivo fMRI data below, we imposed an additional non-negativity constraint on the subject mode (C ≽ 0) since we assume each subject could either participate or not in a network but could not negatively participate.
Adaptive moment estimation (Adam) (Kingma and Ba, 2014) is a popular first-order solver used in the deep learning community (Ruder, 2016). Its superior performance is achieved using momentum-based acceleration together with an adaptive learning rate, which allows small step sizes for parameters with large accumulative gradients and large step sizes for parameters with small accumulative gradients. Recently, Dozat described a modified algorithm, Nadam, in which Nesterov acceleration is incorporated into Adam (Dozat, 2016). In the following, we use Nadam to update all modes simultaneously using the gradients above. The default values for the parameters are chosen to be α = 0.001, β1 = 0.9, β2 = 0.999 and ϵ = 10−8 per (Kingma and Ba, 2014).
We use a warm initialization from lower rank solutions to improve convergence rates. We demonstrate below that this full gradient-based approach is also more robust than ALS. The Nadam-Accelerated SCAlable and Robust (NASCAR) CP decomposition algorithm is outlined in Algorithm 1. Starting with a rank-1 solution using ALS, we then use Nadam to solve the main decomposition problem at each rank from 2 to R with warm initializations {A*, B*, C*}, where f in line 7 is the Tikhonov regularized objective function shown in Eq. (3) with gradient shown in Eq. (4), and similar forms for B and C, except that the non-negativity constraint on C is tackled by a projected version of Nadam to the non-negative orthant. Note that the model in Eq. (1) constrains the components of {A*, B*, C*} to have unit norm. Rather than integrate this constraint into the Nadam search, we simply normalize and then re-scale the components after and before the Nadam procedure as shown in lines 8 and 6 respectively.
Algorithm 1:
NASCAR.
| s | Algorithm NASCAR (, R) |
| 1 | a1, b1, c1, λ1 ← CP-ALS (, 1) |
| 2 | ← – Tensor_Recon (a1, b1, c1, λ1) |
| 3 | a′, b′, c′, λ′ ← CP-ALS (, 1) |
| 4 | A* ← [a1 a′]; B* ← [b1 b′]; C* ← [c1 c′]; |
| 5 | For r = 2, 3, …, R |
| 6 | Scale the ith components of A*, B*, C* by |
| 7 | Ar, Br, Cr ← Nadam (f, {A*, B*, C*}) |
| 8 | Normalize the ith components of Ar, Br, Cr and store the norm product into λr |
| 9 | ← – Tensor_Recon (Ar, Br, Cr, λr) |
| 10 | a′, b′, c′, λ′ ← CP-ALS (, 1) |
| 11 | A* ← [Ar a′]; B* ← [Br b′]; C* ← [Cr c′]; |
| 12 | End For |
| 13 | Return a set of solutions {a1, b1, c1, λ1}, {A2, B2, C2, λ2}, …, {AR, BR, CR, λR] |
| e | End Algorithm |
3. Materials and experiments
3.1. Simulation
Third-order tensors , with rank R varying from 1 to 10, were simulated from the outer product of factors randomly sampled from a standard normal distribution. We added Gaussian white noise to the simulated tensor with a SNR of 2. We then performed tensor decomposition with rank R on using NASCAR (Nadam inside) as well as SRSPCD (ALS inside) (Li et al., 2018, 2017). For a fair comparison, we used the same random initializations for both methods. We evaluated the performance using the averaged congruence product (ACP) (Tomasi and Bro, 2005). ACP is a measure of correlation between components defined as
where A, B, C are the ground truth loading matrices and , , their estimated counterparts, P is a permutation matrix accounting for the ambiguity of the ordering of the solutions (Harshman, 1970) and tr (·) is the trace of a matrix. We evaluated ACP of the solutions obtained from both NASCAR and SRSCPD as a function of R. For each R, we ran 100 Monte Carlo trials and generated box plots of the ACP for visualization. For each simulated trial above, we also recorded the run time for each of the methods.
3.2. In-vivo language task fMRI data
The minimally preprocessed language task fMRI data from 40 randomly selected subjects (16 male and 24 female, age 26–30, all right-handed) in the publicly available Human Connectome Project (HCP) database (Glasser et al., 2013; Van Essen et al., 2013) were used. The list of subject IDs is shown in the supplementary materials. These data were acquired for two independent sessions with opposite phase encoding direction (LR, RL) using a gradient-echo EPI sequence (2 mm × 2 mm × 2 mm voxels, TR = 720 ms, TE = 33.1 ms), where each session ran 3 mins and 57 secs with 316 frames in total.
Task fMRI was used here, instead of resting-state fMRI, because the task designs and the results from the generalized linear regression model (GLM) (Barch et al., 2013) can be used for validation purpose. However, we note that, through the use of BrainSync alignment, NASCAR can be applied to a range of multi-subject fMRI recording paradigms including resting-state fMRI and self-paced event-related fMRI studies.
The language processing task was selected because it consists of several spatially overlapped networks that span a substantial fraction of the cortical surface. The design of the language processing task, developed by (Binder et al., 2011), consists of four interleaved blocks of a story task and a math task. In each story block, the subjects were presented with a brief auditory story (Present Story) followed by a forced-choice question about the topic of the story (Question Story). Then, the subjects chose one answer from two alternatives by pressing a button (Respond Story). In each math block, the subjects were asked to perform some addition or subtraction calculation (Question Math) after listening to a series of arithmetic operations (Present Math). Finally, similar to the story blocks, the subjects selected one answer from two alternative choices (Respond Math). The order of task blocks is identical within each session but different between the two sessions.
The results below used language task fMRI data resampled onto the cortical surface extracted from each subject’s T1-weighted MRI and co-registered to a common surface atlas as described in Glasser et al., 2013). Each session was represented as a V × T matrix, where V ≈ 22 K is the number of vertices across the two hemispheres and T = 316 is the number of time points. The time series at each vertex was normalized to have zero mean and unit norm. We applied the BrainSync algorithm to temporally align all sessions of task fMRI datasets to the first session of the first subject (this reference was HCP subject 100307). Although we did not find significant difference in the choice of reference subject in this study, we note that a pair-wise group alignment could be used to avoid potential bias towards one specific subject (Akrami et al., 2019). The temporally aligned task fMRI data were then combined along the third dimension, forming a third-order data tensor , where S = 80 is the number of subjects (40) by sessions (2). Analogous to rank-reduction preprocessing methods used in ICA, we performed a greedy CP decomposition (Acar et al., 2011) to the tensor to reduce its rank to 20. Specifically, we recursively fit a rank-1 component to the data tensor and then subtracted this from the residual data tensor until we had found 20 components in total. Next, we applied the NASCAR algorithm to the rank-reduced tensor to extract brain networks with a desired rank of 20. The rank 20 here is chosen to match the rank used in the group ICA method (Calhoun et al., 2001b) against which we compare below. The regularization parameter μ in Eq. (3) and ((4) was chosen to be 0.001. We found in practice that this value works reasonably well across different fMRI datasets, possibly because we normalize each time-series to zero mean and unit magnitude before applying the decomposition. Our experiments have also shown that results are robust to a wide choice of parameter values as illustrated in Fig. S1 in the supplementary materials. Smaller values, or even no regularization, produced very similar results. Note that despite the negligible difference in results without regularization, using a non-zero regularization parameter is still prudent as theoretically the magnitude ambiguity issue (Section 2.3) can still occur. Large values, however, should be avoided to prevent the regularizer reducing the fit of the tensor to the data in Eq. (3).
We also ran group ICA on the same language task fMRI dataset as a comparison. Following the procedure described in (Calhoun et al., 2001b), the (temporally) PCA-denoised (to rank 40) individual language task fMRI data was temporally concatenated, then the temporal dimensionality was further reduced to 20 using PCA again and a spatial group ICA performed to extract independent components. We also compared with the Canonical ICA (CanICA) method (Varoquaux et al., 2010), where a canonical correlation decomposition was applied in the second stage instead of the PCA as in (Calhoun et al., 2001b). Parameters were identical to those used in (Calhoun et al., 2001b) as a fair comparison.
3.3. Reproducibility analysis
We investigated the reproducibility of NASCAR across two sessions of the same set of 40 subjects’ language task fMRI data. NASCAR was applied to each session independently and an averaged cross-correlation between matched pair-wise components in two sessions was computed as a measure of reproducibility or consistency (Varoquaux et al., 2010). Specifically, a cross-correlation matrix was first computed:
where A1 and A2 are the spatial modes of the solutions obtained from the two sessions, respectively. Then the rows and columns of Q was reordered into such that the spatial maps in A1 and A2 are optimally matched to each other using the stable matching algorithm (Gale and Shapley, 1962). Next, the absolute values of the diagonal elements of were sorted in a descending order, denoted as . The “t” reproducibility measure used in (Varoquaux et al., 2010) was then equivalent to the average of the elements of q:
where R = 20 is the total number of components in our experiment. Since we do not expect the subject-specific components shown below to be consistent across sessions, we generalized the “t” measure to a “tr” measure:
| (5) |
indicating the change of consistency as a function of rank r. Hence, the “t” measure is the special case of the “tr” measure when r = R.
We repeated the analysis above and computed reproducibility using both group ICA and CanICA as a comparison. We also explored reproducibility of the subject mode across the two sessions in NASCAR as the subject mode is a unique property available in NASCAR but absent from the ICA methods.
3.4. Stability analysis
We investigated the stability of NASCAR, group ICA and CanICA by running them on the language task fMRI data 100 times, each time with different random initializations. Thus, 2000 (20 networks/run × 100 runs) brain networks were obtained in total. We then projected the spatial map of these 2000 networks non-linearly onto a 2D plane using the curvilinear component analysis (CCA) algorithm (Demartines and Hérault, 1997) provided by the ICASSO software (Himberg and Hyvärinen, 2003) for easy visualization. For the tensor decomposition we color-coded the projected spatial maps with the number of participating subjects for each network. A subject is defined to participate in a particular network if the values of the normalized session mode (the third dimension of the tensor) exceeded 0.05 in any of the two sessions for that subject. The threshold was chosen heuristically based on the overall histogram of the session modes from all decomposition results.
Quantitatively, for each method, we randomly selected a pair of solutions from the 100 runs with replacement and computed the “tr” in Eq. (5) as a measure of consistency across runs. We repeated the random selection 1000 times and visualized the results using box plots.
We also investigated stability of group ICA, CanICA, and NASCAR using bootstrap analysis. We repeated the decompositions for each of 100 bootstrap resamples and used the CCA embedding and color coding as described above to visualize the results.
3.5. Are BrainSync and Nadam essential for successful brain network identification?
For task fMRI, data are typically aligned with respect to the stimulus timing. Additional synchronization across subjects or sessions is usually not performed. However, as discussed earlier, temporal synchronization may not be strictly satisfied across subjects even when an identical task design is used. Responses to tasks from individual subjects can differ in their latencies, especially for higher-level cognitive tasks. Further, any brain networks that are independent from the task designs cannot be found without temporal synchronization. To explore the additional benefit that might be gained from synchronizing time series across subjects using BrainSync, we repeated our experiments as described above on one session of the 40 subjects’ language task fMRI data, where identical task design was used to all subjects, with and without BrainSync.
Furthermore, to investigate the impact of using Nadam relative to ALS, we repeated the tensor-decomposition composition using our earlier SRSCPD framework (Li et al., 2018, 2017) on the same language task fMRI data as describe above.
4. Results and discussion
4.1. Simulation
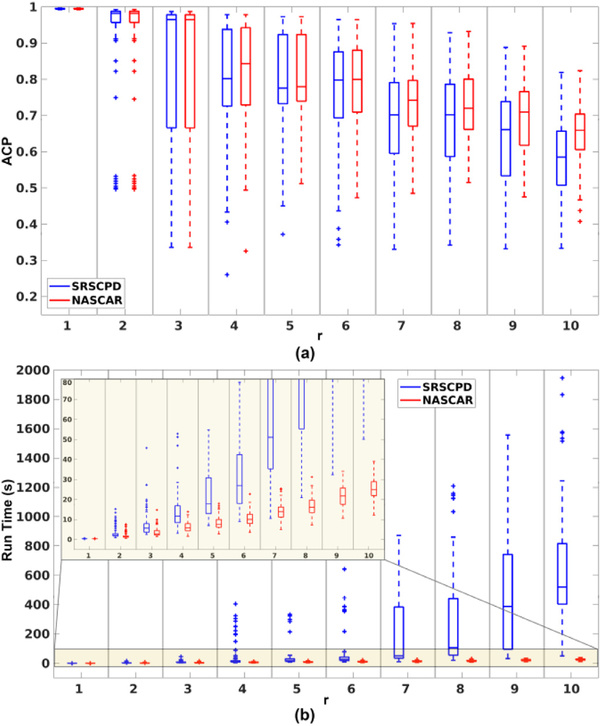
Fig. 2 (a) shows box plots of the ACP over 100 Monte Carlo trials as a function of the rank r using NASCAR (red) and SRSCPD (blue). When r is small, they perform approximately equally well. However, NASCAR outperforms SRSCPD by a margin that increases with r, indicating an improved robustness of NASCAR over SRSCPD.
Fig. 2.
Simulation results. (a) Box plots of ACP as a function of r for NASCAR in red and SRSCPD in blue; (b) box plots of the run time corresponding to the experiments in (a). The bottom part is magnified and shown on the top left corner for easy visualization.
Fig. 2 (b) shows box plots of the run time corresponding to the simulated trials in (a). The run time is substantially lower using NASCAR than that using SRSCPD and the difference becomes larger and larger as r increases, indicating a significant improvement of the scalability to large dataset and higher rank decomposition. The difference in run time is largely due to the substantially fewer total iterations through the data required with NASCAR relative to SRSCPD.
4.2. In-vivo language task fMRI data
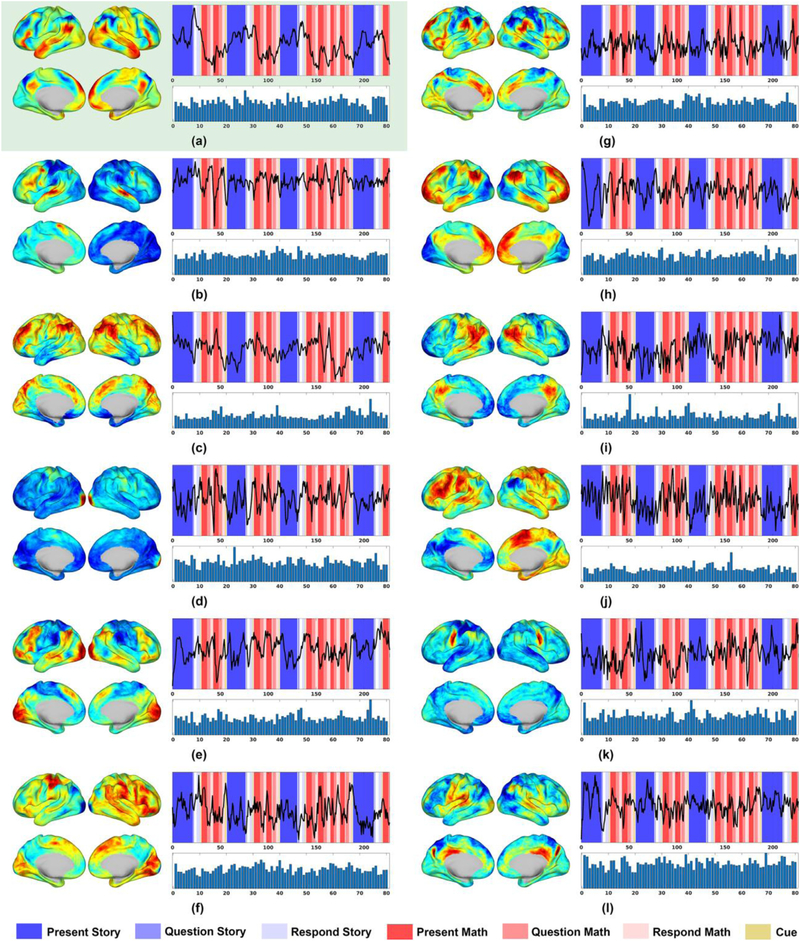
Fifteen components were identified by the NASCAR method that could plausibly represent networks or other physiological components in the sense that the spatial maps are smooth and are found in most subjects. Of these, twelve appear consistent with known networks as shown in Fig. 3 (the remaining three are shown in Fig. S2 in supplementary materials). For each component, the left sub-figure shows the spatial map, the top right sub-figure shows the temporal dynamics overlaid with color-coded task design (math tasks shown in red and story tasks shown in blue) and the bottom right sub-figure shows the subject and session participation mode.
Fig. 3.
Identified and recognized components using NASCAR on language task fMRI data. (a) Classic language network in response to Story-Math contrast (St-Ma); (b) Auditory network (AN); (c) Frontoparietal attentional control network (FPACN); (d) Visual network (VN); (e) Extended language network (LN); (f) Right-hand-visual co-activation (RH-V); (g) Cingulo-opericular network (CON); (h) First sub-component of the default mode network (DMN); (i) Second sub-component of the DMN; (j) Respiratory effect (Resp); (k) Sensorimotor network (SMN) near the tongue area; (l) Memory-retrieval-related network (Mem). In each component, the left sub-figure shows the spatial map, the top right sub-figure shows the temporal dynamics overlaid with color-coded task designs, and the bottom right sub-figure shows the subject/session participation mode.
Recall that task-timing is different between the two sessions but the BrainSync transform brings them into alignment as we have shown previously (Joshi et al., 2018). Similarly, BrainSync should also align not only responses to the stimuli across subjects but also underlying brain activity independent of the stimulus. The results here are aligned to the timing for the first session and the first (reference) subject. Applying the appropriate inverse BrainSync transform we can obtain the corresponding network dynamics for each subject.
Fig. 3 (a) shows a classic language network where activity in Broca’s area, Wernicke’s area, and the anterior temporal lobe are strongly inter-correlated. The spatial map of this network is consistent with the result shown in Fig. 8 in (Barch et al., 2013), which is the spatial response to the Story-Math task contrast (St-Ma) obtained using a GLM from 77 subjects. Unlike in that case, here the component is obtained directly from the data without knowledge of the task. The associated temporal mode shows a strong correlation to the Story-Math contrast with a maximum lagged correlation of r = 0.77 (uncorrected p-value < 10−60) at a lag of d = 3.6 s from the design to the response, consistent with the latency associated with the hemodynamic response function.
Fig. 8.
Stability analysis results using bootstrapped data with different random initializations for each run; (a) and (d): Group ICA; (b) and (e) CanICA; (c) and (f) NASCAR. See caption for Fig. 6.
Fig. 3 (b) shows a clear auditory network in response to the auditory task stimuli (note again that both story and math descriptions in the first phase of the tasks were given as auditory presentations), where spatially bilateral auditory cortex was activated and temporally it is significantly correlated with the combined Presentation blocks from both story and math tasks, i.e. Present Story + Present Math (r = 0.43, p = 9 × 10−16, d = 5.8 s).
The temporal dynamics of Fig. 3 (c), (d), (e), (f), and (g) show significant correlations with the combined Response blocks from both story and math tasks, i.e. Respond Story + Respond Math, with a short but variable delay, suggesting that they correspond to brain activity during the Response period of the tasks. Indeed, the spatial pattern of these components indicates a frontoparietal attentional control network (FPACN) (Hopfinger et al., 2000; Marek and Dosenbach, 2018) (r = 0.26, p = 1.5 × 10−6, d = 7.2 s) for (c), a visual network (VN) (r = 0.29, p = 1.3 × 10−7, d = 2.9 s) for (d), an extended language network or reading network (LN) (Dehaene et al., 2010; Fedorenko and Thompson-Schill, 2014) (r = 0.28, p = 3.3 × 10−7, d = 1.4 s) for (e), a right-hand-visual co-activation (RH-V) (r = 0.36, p = 1.7 × 10−11, d = 2.2 s) for (f), and a cingulo-opercular network (CON) (Sylvester et al., 2012) (r = 0.28, p = 2.4 × 10−7, d = 5 s) for (g), all reflecting the subjects’ response or brain networks activated in order to answer the task questions.
The spatial maps for Fig. 3 (h) and (i) show two sub-components of a typical default mode network (DMN) with classic areas, such as medial prefrontal cortex, precuneus and posterior cingulate cortex, temporalparietal junction, highly activated (Raichle, 2015; Simony et al., 2016). The DMN was first known as a task-negative network (Fox et al., 2005; Raichle et al., 2001) and in fact a strong negative correlation between the temporal mode of (h) and (i) and the task blocks can be clearly observed both visually (dips during tasks and peaks between tasks) and quantitatively (r = −0.3, p = 3.5 × 10−8, d = 4.3 s for (h) and r = −0.22, p = 4.2 × 10−5, d = 7.9 s for (i)).
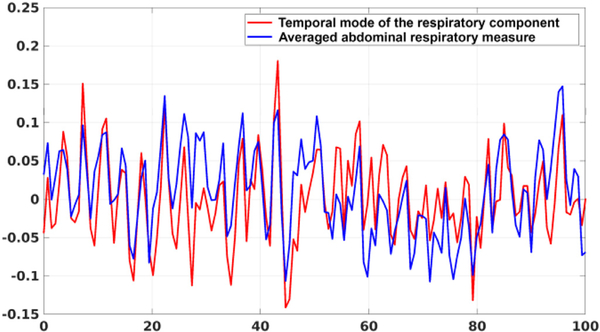
Fig. 3 (j) shows a spatially global, oscillatory (~0.3 Hz) and non-task-related activity, suggesting that it may represent a residual respiration effect (Resp) common across subjects. Further investigation shows that this component, as shown in Fig. 4, is strongly correlated (r = 0.67) with the actual abdominal respiratory measure (also provided as part of the physiological data in the HCP dataset). To obtain the average abdominal respiratory signal, we applied the BrainSync transformation matrices obtained from the fMRI data from each subject to that subject’s respiratory data and then averaged across subjects. The resulting time series, overlaid with the temporal mode from Fig. 3 (j), is shown in Fig. 4.
Fig. 4.
Plot of the respiratory component (red) obtained from the tensor decomposition (temporal mode of Fig. 3 (j)) and the averaged abdominal respiratory measure across subjects (blue). Only the first 100 seconds are plotted and both signals were normalized to have zero mean and unit norm for easy visualization and comparison.
Fig. 3 (k) shows a sensorimotor network (SMN) around the tongue area. Fig. 3 (l) shows a brain network potentially related to memory retrieval (Mem), although it is not well reported in the literature (Power et al., 2011). Both of these two networks are non-task-related as they do not correlate with any of the tasks or sub-tasks. Note that the synchronization of non-task-related activity across subjects using BrainSync allows us to identify components (j), (k), and (l) from the third-order tensor in addition to task-related networks.
All twelve identified networks show strong subject modes in almost all 40 subjects, indicating that these networks are indeed common across subjects. However, considerable differences in the subject mode among these subjects are also observed. For example, in the extended language network shown in Fig. 3 (e), the values of the subject mode span the range from 0.04 to 0.174. Similarly, the counterpart for the DMN shown in Fig. 3 (h) has a range from 0.065 to 0.166. These differences in the subject mode are presumably indicative of the degree of activity in that particular network for each subject, so that they could be used as features to study inter-subject variability of participation of networks in specific tasks or to study how the participation level of networks are altered during development and aging, or by neurological disease (Li et al., 2020).
The remaining three plausible but un-recognized components also exhibit smooth spatial maps as shown in Fig. S2 (a)–(c) in the supplementary materials. For example, Fig. S2 (a) shows bilateral activations in the sensorimotor foot area. As with the components in Fig. 3, the subject mode shows participation across all subjects, which is consistent with these networks being common to all 40 subjects. Inspection of the remaining five components of the rank-20 decomposition reveals that their subject modes have a large value for a single subject, indicating that these are likely artifacts originating from that subject. Fig. S2 (d) shows one such component out of the five as an example. Further exploration of components with higher rank (R > 20) on this dataset only revealed additional subject-specific or noisy components.
When applying group ICA to the same language fMRI dataset, we were able to identify seven components as shown in Fig. S3 in the supplementary materials. Of the seven components, (a) VN and (b) AN can be clearly recognized. Another five networks exhibit spatial maps similar to those identified by the NASCAR method in Fig. 3, suggesting a FPACN for (c), a DMN for (d), a RH-V for (e), a SMN for (f), and a Mem for (g). The other components found using the NASCAR method, such as St-Ma, LN, and Resp, were not obviously identifiable in the ICA results, as shown in Fig. S4 for the remaining 13 components. When using CanICA, we also identified seven components, which were almost identical to those using group ICA as described above.
4.3. Reproducibility analysis
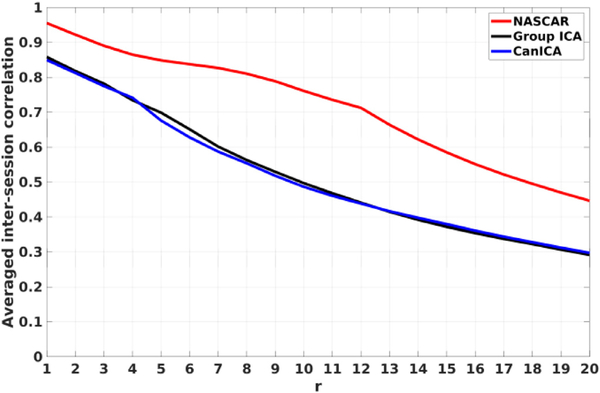
Fig. 5 shows the averaged reproducibility metric tr between two sessions as a function of r. Overall, for all three methods, the top few components are fairly consistent across sessions with high reproducibility values (> 0.75). As r increases, tr exhibits a decreasing trend indicating that more inconsistent subject-specific or noisy components were discovered during the decomposition.
Fig. 5.
Averaged inter-session correlation tr as a function of r.
The results for group ICA and CanICA are comparable and a slightly higher correlation was observed in CanICA than group ICA in higher ranks. In contrast, NASCAR outperforms both group ICA and CanICA by a large margin through the entire range of r.
We also observed a similar trend in reproducibility of the subject mode from the NASCAR results, as shown in Fig. S5.
4.4. Stability analysis
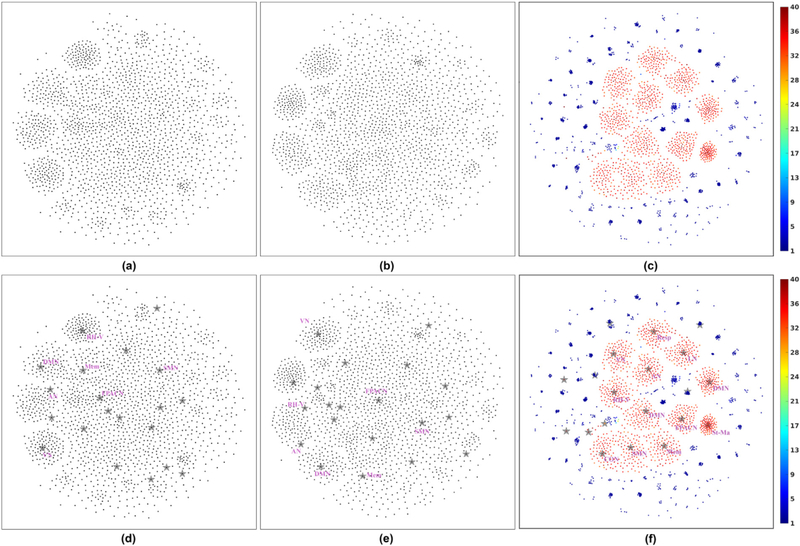
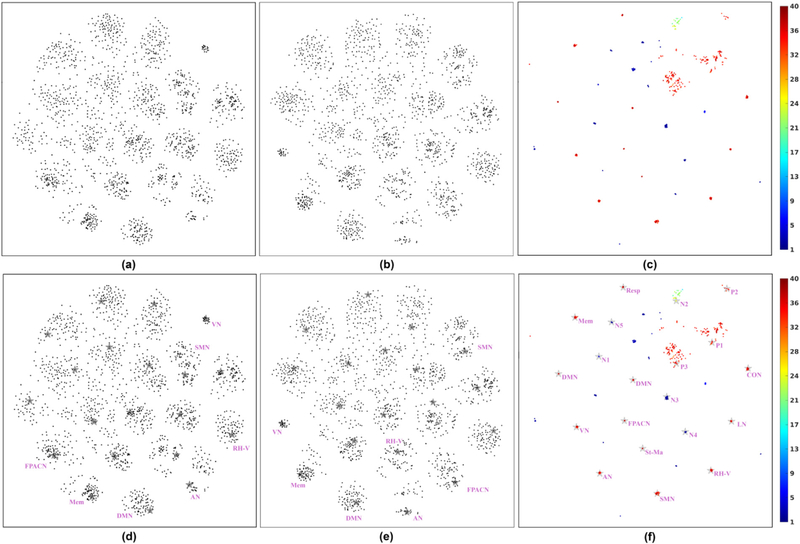
Fig. 6 (a)–(c) show scatter plots of the projected spatial maps obtained using group ICA, CanICA and NASCAR, respectively, with different random initializations. Each dot represents a single component. The NASCAR results are color-coded to indicate the number of subjects that exhibit that component (session mode value >.05). Fig. 6 (d)–(f) show copies of (a)-(c), respectively, with the addition as gray stars of the components from the original decomposition (results in Fig. 3 and Figs. S1–S3), which we used to identify the brain networks described above. We also annotate each of these components (7 for ICA and 12 for NASCAR).
Fig. 6.
Stability analysis results with different random initializations. (a) Scatter plot of the projected spatial maps obtained using group ICA; (b) The counterpart to (a) when using CanICA; (c) The counterpart to (a) when using NASCAR; (d) Same as (a) but with single run results (Fig. 3 and Figs. S1–S3) plotted as stars and annotated; (e) The counterpart to (d) when using CanICA with single run results; (f) The counterpart to (d) when using NASCAR with single run results. For (c) and (f), the color of each dot (component) represents the number of subjects participating in that component. Acronyms and abbreviations of the identified components are given in the caption of Fig. 3. P1-P3: three plausible but un-recognized components (Fig. S2 (a)–(c)); N1-N5: five subject-specific components (Fig. S2 (d)). The color bar indicates the number of subjects that participate in each of the components found and is used to color code the points (components) in (c) and (f).
In these results, tight clusters indicate a strong similarity in components for different initializations, and hence less dependence of decomposition method on initialization. While all three methods clearly exhibit clustering behavior (Fig. 6 (a)–(c)), the NASCAR clusters are consistently tighter than those for ICA and CanICA. By overlaying the results from Fig. 3 and Figs. S1–S2, we see how these clusters map to identified networks (Fig. 6 (d)–(f)). As shown in Fig. 6 (f), the color of the components in the clusters that contain the 12 recognized brain networks found using NASCAR indicate a strong participation in each network across subjects. Direct examination of the subject modes for each run and network revealed that all 12 networks were found for all subjects in each of the 100 runs using NASCAR. Of the three plausible but un-recognized components, P2 is found consistently across subjects and runs with little variability. P1 and P3 are also found consistently across subjects but are more sensitive to initialization leading to more spread in the clusters. Finally, the subject-specific components, N1, N3, N4 and N5, are also found consistently across runs, with the blue coloring indicating that they are unique to a single subject. The ICA and CanICA results also show clear clustering behavior for each of the identified networks, Fig. 6 (d) and (e), but with an increased sensitivity to initialization compared to NASCAR.
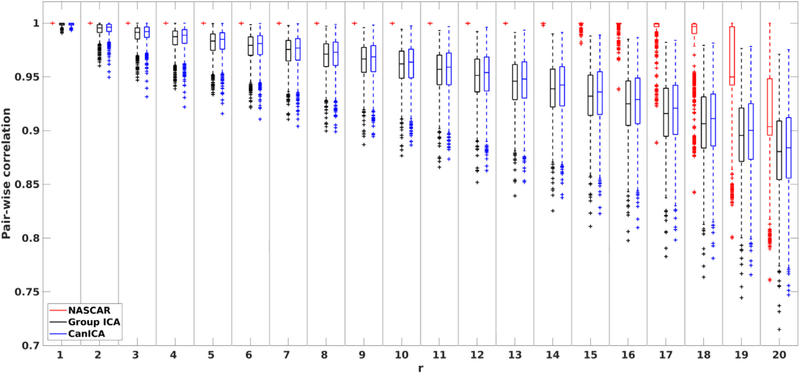
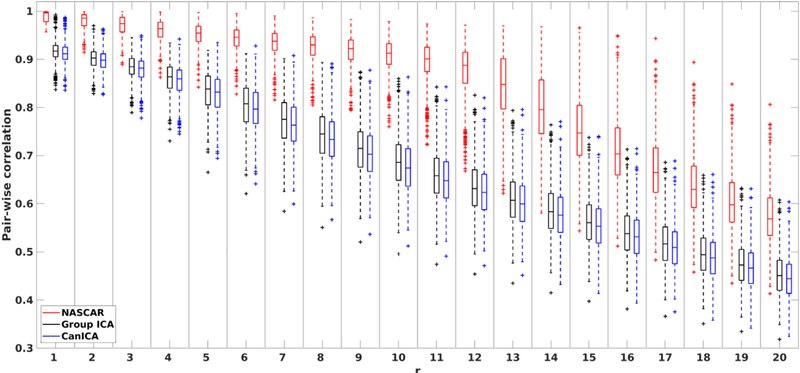
Quantitatively, Fig. 7 shows the pair-wise consistency measures tr as a function of r. Similar to Fig. 5, the correlation between pairs of runs decreases as r increases for all three methods. The results using NASCAR confirmed our observation in Fig. 6 that the identified 12 common networks (r = 1, …, 12) are highly consistent (tr ≈ 1) across runs and that tr starts decreasing only as more subject-specific components are identified. The correlation values are substantially higher in NASCAR than that in either CanICA or group ICA through the entire range of r, although CanICA exhibits an improved robustness over group ICA.
Fig. 7.
Pair-wise correlations over 1000 randomly selected pairs of solutions in Fig. 6 as a function of r.
Results obtained by decomposing 100 different bootstrap resamples of the data together with randomized initialization are shown in Fig. 8. As might be expected, ICA, CanICA and NASCAR all show increased variability relative to the case where only the initialization is changed (Fig. 6). Visual inspection of Fig. 8 (a) reveals three or four clear clusters in the group ICA case (similarly in the CanICA case) whereas multiple clusters remain visible for NASCAR in Fig. 8 (c). There are exactly 12 clusters with dense centroids that correspond to the 12 recognized components in Fig. 3, indicating that they are robustly identified in each of the bootstrap runs as shown in Fig. 8 (f). Furthermore, the red or orange color of most points in these 12 clusters indicates that almost all subjects participate in these networks in each of the bootstrap runs. Conversely the blue clusters in the peripheral area correspond to networks that are specific to a single subject and reproduced for that subject in multiple bootstrap runs. The corresponding quantitative measures tr are shown in Fig. 9 where a similar trend is observed as Fig. 7 but with overall lower correlation values (note the difference in the scale of the y-axis).
Fig. 9.
Pair-wise correlations over 1000 randomly selected pairs of solutions in Fig. 8 as a function of r.
4.5. The necessity of BrainSync and Nadam in brain network identification from asynchronous multi-subject fMRI data
Table 1 (3rd column, yellow) summarizes tensor decomposition results on the full BrainSync synchronized dataset using SRSCPD, our previously described method based on alternating least squares (ALS) in combination with a warm start (Li et al., 2018, 2017). This method performed similarly to NASCAR, which assumes an identical model but replaces the ALS approach with a full gradient-based method. Most of the networks found were visually equivalent to those found using NASCAR. However, as listed in Table 1 (2nd column, red), the FPACN, CON, DMN, and Mem components were not found using SRSCPD. Also, St-Ma and VN were each split into two components during the decomposition. These results are indicative that NASCAR provides a more robust decomposition than SRSCPD.
Table 1.
Summary of brain networks identified in language task fMRI data.
| Decomposition method | NASCAR (Nadam inside) | SRSCPD (ALS inside) | NASCAR (Nadam inside) | NASCAR (Nadam inside) | ||||
|---|---|---|---|---|---|---|---|---|
| BrainSync | Yes | No | ||||||
| Number of sessions | 80 (two sessions, 40 subjects, two different task designs) | 40 (one session, 40 subjects, one task design) | ||||||
| Recognizable | 12 | St-Ma | 9* | St-Ma* | 12 | St-Ma | 2 | St-Ma† |
| AN | AN | AN | AN† | |||||
| FPACN | FPACN | |||||||
| VN | VN* | VN | ||||||
| LN | LN | LN | ||||||
| RH-V | RH-V | RH-V | ||||||
| CON | CON | |||||||
| DMN* | DMN* | |||||||
| SMN | SMN | SMN | ||||||
| Resp | Resp | Resp | ||||||
| Mem | Mem | |||||||
| Plausible but un-recognized | 3 | 2 | 0 | 0 | ||||
| Subject-specific | 5 | 9 | 8 | 18 | ||||
| Total | 20 | |||||||
The component is split into two sub-components during the decomposition.
Only a subset of subjects shows participation in this network.
We also examined what happens without synchronization. In this case we could only use one of the two sessions since timing is different for the tasks in the two sessions. As shown in Table 1 (4th column, blue), when using NASCAR on synchronized single-session dataset, we are still able to identify the 12 common brain networks, although those plausible but un-recognized components (Fig. S2 (a)–(c)) are missing from the decomposition. However, when applying NASCAR to the unsynchronized dataset (Table 1, last column, green) only the St-Ma and AN were found using NASCAR, but even then, only in a subset of subjects. We believe the difference is due to the differing latencies of the subjects’ responses to the task. With the tensor representation, the time series are assumed to be approximately equal across subjects. Then temporal synchronization using BrainSync is a key factor in successful identification of brain networks. Note that this is not the case in group ICA or CanICA where time series are concatenated across subjects so that we do not assume temporal synchrony in the ICA decomposition. For this reason, ICA-based methods are able to identify multiple networks without synchronization, while NASCAR requires synchronization to work. However, as shown in the earlier figures, with the inclusion of BrainSync synchronization, it appears that NASCAR can more reliably identify task-related network components that group ICA and CanICA. Moreover, components that are independent of the task, such as the respiratory and sensory motor components in Fig. 3, can also be identified as a result of synchronization.
5. Conclusion
Using NASCAR with BrainSync, we identified and recognized twelve spatially overlapped and temporally correlated common networks across multiple subjects: seven task-related networks, two sub-components of the default mode network, respiratory effect, a sensorimotor activity, and a memory-retrieval-related network in the language task fMRI data. Although we did not use any prior information regarding the task designs, our results not only replicated the task timing, but also showed expected differences in the temporal dynamics of those networks. These networks were not all found using the group ICA and CanICA method or when BrainSync synchronization or Nadam was not used. The bootstrapping results show that NASCAR is potentially robust in identifying the spatial, temporal and subject-dependent behavior of brain networks that compares favorably with ICA-based approach. Furthermore, synchronization of time-series across subjects prior to decomposition can allow identification of components independent of task.
Supplementary Material
Acknowledgment
This work was supported by National Institutes of Health [grant number R01-EB009048, R01-EB026299, R01-NS089212, and K23-HD099309].
Footnotes
Disclosure of competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Data and code availability
The data used in this study are publicly available from the Human Connectome Project (HCP), Young Adult Study at https://www.humanconnectome.org/study/hcp-young-adult.
Open-access code is released for research purposes at https://neuroimageusc.github.io/NASCAR
Supplementary materials
Supplementary material associated with this article can be found, in the online version, at doi: 10.1016/j.neuroimage.2020.117615.
References
- Acar E, Dunlavy DM, Kolda TG, 2011. A scalable optimization approach for fitting canonical tensor decompositions. J. Chemom 25, 67–86. 10.1002/cem.1335. [DOI] [Google Scholar]
- Akrami H, Joshi AA, Li J, Leahy RM, 2019. Group-wise alignment of resting fMRI in space and time,. In: Proc. SPIE Medical Imaging 2019: Image Processing, SPIE, San Diego, CA, pp. 737–744. [Google Scholar]
- Al-sharoa E, Al-khassaweneh M, Aviyente S, 2019. Tensor based temporal and multilayer community detection for studying brain dynamics during resting state fMRI. IEEE Trans. Biomed. Eng 66, 695–709. 10.1109/TBME.2018.2854676. [DOI] [PubMed] [Google Scholar]
- Andersen AH, Rayens WS, 2004. Structure-seeking multilinear methods for the analysis of fMRI data. Neuroimage 22, 728–739. 10.1016/j.neuroimage.2004.02.026. [DOI] [PubMed] [Google Scholar]
- Barch DM, Burgess GC, Harms MP, Petersen SE, Schlaggar BL, Corbetta M, Glasser MF, Curtiss S, Dixit S, Feldt C, Nolan D, Bryant E, Hartley T, Footer O, Bjork JM, Poldrack R, Smith S, Johansen-Berg H, Snyder AZ, Van Essen DC, 2013. Function in the human connectome: Task-fMRI and individual differences in behavior. Neuroimage 80, 169–189. 10.1016/j.neuroimage.2013.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckmann CF, Smith SM, 2005. Tensorial extensions of independent component analysis for multisubject fMRI analysis. Neuroimage 25, 294–311. 10.1016/j.neuroimage.2004.10.043. [DOI] [PubMed] [Google Scholar]
- Binder JR, Gross WL, Allendorfer JB, Bonilha L, Chapin J, Edwards JC, Grabowski TJ, Langfitt JT, Loring DW, Lowe MJ, Koenig K, Morgan PS, Ojemann JG, Rorden C, Szaflarski JP, Tivarus ME, Weaver KE, 2011. Mapping anterior temporal lobe language areas with fMRI: a multicenter normative study. Neuroimage 54, 1465–1475. 10.1016/j.neuroimage.2010.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calhoun VD, Adali T, McGinty VB, Pekar JJ, Watson TD, Pearlson GD, 2001a. fMRI activation in a visual-perception task: network of areas detected using the general linear model and independent components analysis. Neuroimage 14, 1080–1088. 10.1006/nimg.2001.0921. [DOI] [PubMed] [Google Scholar]
- Calhoun VD, Adali T, Pearlson GD, Pekar JJ, 2001b. A method for making group inferences from functional MRI data using independent component analysis. Hum. Brain Mapp 14, 140–151. 10.1002/hbm.1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calhoun VD, Liu J, Adali T, 2009. A review of group ICA for fMRI data and ICA for joint inference of imaging, genetic, and ERP data. Neuroimage 45, S163–S172. 10.1016/j.neuroimage.2008.10.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cichocki A, Mandic D, De Lathauwer L, Zhou G, Zhao Q, Caiafa C, Phan HA, 2015. Tensor decompositions for signal processing applications: from two-way to multiway component analysis. IEEE Signal Process. Mag 32, 145–163. 10.1109/MSP.2013.2297439. [DOI] [Google Scholar]
- Dehaene S, Pegado F, Braga LW, Ventura P, Filho GN, Jobert A, Dehaene-Lambertz G, Kolinsky R, Morais J, Cohen L, 2010. How learning to read changes the cortical networks for vision and language. Science 330, 1359–1364. 10.1126/science.1194140. [DOI] [PubMed] [Google Scholar]
- Demartines P, Hérault J, 1997. Curvilinear component analysis: a self-organizing neural network for nonlinear mapping of data sets. IEEE Trans. Neural Netw 8, 148–154. 10.1109/72.554199. [DOI] [PubMed] [Google Scholar]
- Dozat T, 2016. Incorporating Nesterov momentum into Adam. ICLR Workshop. [Google Scholar]
- Du Y, Fan Y, 2013. Group information guided ICA for fMRI data analysis. Neuroimage 69, 157–197. 10.1016/j.neuroimage.2012.11.008. [DOI] [PubMed] [Google Scholar]
- Esposito F, Scarabino T, Hyvarinen A, Himberg J, Formisano E, Comani S, Tedeschi G, Goebel R, Seifritz E, Di Salle F, 2005. Independent component analysis of fMRI group studies by self-organizing clustering. Neuroimage 25, 193–205. 10.1016/j.neuroimage.2004.10.042. [DOI] [PubMed] [Google Scholar]
- Fedorenko E, Thompson-Schill SL, 2014. Reworking the language network. Trends Cogn. Sci 18, 120–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox MD, Snyder AZ, Vincent JL, Corbetta M, Van Essen DC, Raichle ME, 2005. The human brain is intrinsically organized into dynamic, anti-correlated functional networks. Proc. Natl. Acad. Sci 102, 9673–9678. 10.1073/pnas.0504136102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friston KJ, Fletcher P, Josephs O, Holmes A, Rugg MD, Turner R, 1998. Event-related fMRI: characterizing differential responses. Neuroimage 7, 30–40. 10.1006/nimg.1997.0306. [DOI] [PubMed] [Google Scholar]
- Gale D, Shapley LS, 1962. College admissions and the stability of marriage. Am. Math. Monthly 69, 9–15. 10.2307/2312726. [DOI] [Google Scholar]
- Glasser MF, Sotiropoulos SN, Wilson JA, Coalson TS, Fischl B, Andersson JL, Xu J, Jbabdi S, Webster M, Polimeni JR, Van Essen DC, Jenkinson M, 2013. The minimal preprocessing pipelines for the Human Connectome Project. Neuroimage 80, 105–124. 10.1016/j.neuroimage.2013.04.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Pagnoni G, 2008. A unified framework for group independent component analysis for multi-subject fMRI data. Neuroimage 42, 1078–1093. 10.1016/j.neuroimage.2008.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harshman RA, 1970. Foundations of the PARAFAC procedure: models and conditions for an “explanatory” multimodal factor analysis. UCLA Work. Papers Phonetics 16, 1–84. [Google Scholar]
- Helwig NE, Hong S, 2013. A critique of tensor probabilistic independent component analysis: implications and recommendations for multi-subject fMRI data analysis. J. Neurosci. Methods 213, 263–273. 10.1016/j.jneumeth.2012.12.009. [DOI] [PubMed] [Google Scholar]
- Himberg J, Hyvärinen A, 2003. ICASSO: software for investigating the reliability of ICA estimates by clustering and visualization. In: IEEE Workshop on Neural Networks for Signal Processing, pp. 259–268. [Google Scholar]
- Hopfinger JB, Buonocore MH, Mangun GR, 2000. The neural mechanisms of top-down attentional control. Nat. Neurosci 3, 284–291. 10.1038/72999. [DOI] [PubMed] [Google Scholar]
- Joshi AA, Chong M, Li J, Choi S, Leahy RM, 2018. Are you thinking what I’m thinking? Synchronization of resting fMRI time-series across subjects. Neuroimage 172, 740–752. 10.1016/j.neuroimage.2018.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karahanoğlu FI, Van De Ville D, 2015. Transient brain activity disentangles fMRI resting-state dynamics in terms of spatially and temporally overlapping networks. Nat. Commun 6, 7751. 10.1038/ncomms8751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingma DP, Ba J, 2014. Adam: A method for stochastic optimization. arXiv preprint arXiv: 1412.6980. [Google Scholar]
- Kolda TG, Bader BW, 2009. Tensor decompositions and applications. SIAM Rev 51, 455–500. 10.1137/07070111X. [DOI] [Google Scholar]
- Leonardi N, Van De Ville D, 2013. Identifying network correlates of brain states using tensor decompositions of whole-brain dynamic functional connectivity. In: 3rd International Workshop on Pattern Recognition in Neuroimaging, pp. 74–77. [Google Scholar]
- Li J, Haldar J, Mosher JC, Nair D, Gonzalez-Martinez J, Leahy RM, 2018. Scalable and robust tensor decomposition of spontaneous stereotactic EEG data. IEEE Trans. Biomed. Eng 10.1109/TBME.2018.2875467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Joshi AA, Leahy RM, 2020. A network-based approach to study of ADHD using tensor decomposition of resting fMRI data. IEEE 17th International Symposium on Biomedical Imaging. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Mosher JC, Nair DR, Gonzalez-Martinez J, Leahy RM, 2017. Robust tensor decomposition of resting brain networks in stereotactic EEG. In: IEEE 51th Asilomar Conference on Signals, Systems and Computers, Pacific Grove, pp. 1544–1548. [Google Scholar]
- Li J, Wisnowski JL, Joshi AA, Leahy RM, 2019. Brain network identification in asynchronous task fMRI data using robust and scalable tensor decomposition. In: Proc. SPIE Medical Imaging 2019: Image Processing, SPIE, San Diego, pp. 164–172. [Google Scholar]
- Marek S, Dosenbach NUF, 2018. The frontoparietal network: function, electrophysiology, and importance of individual precision mapping. Dialogues Clin. Neurosci 20, 133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power JD, Cohen AL, Nelson SM, Wig GS, Barnes KA, Church JA, Vogel AC, Laumann TO, Miezin FM, Schlaggar BL, 2011. Functional network organization of the human brain. Neuron 72, 665–678. 10.1016/j.neuron.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raichle ME, 2015. The brain’s default mode network. Annu. Rev. Neurosci 38, 433–447. 10.1146/annurev-neuro-071013-014030. [DOI] [PubMed] [Google Scholar]
- Raichle ME, MacLeod AM, Snyder AZ, Powers WJ, Gusnard DA, Shulman GL, 2001. A default mode of brain function. Proc. Natl. Acad. Sci 98, 676–682. 10.1073/pnas.98.2.676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruder S, 2016. An overview of gradient descent optimization algorithms. arXiv preprint arXiv: 1609.04747. [Google Scholar]
- Schmithorst VJ, Holland SK, 2004. Comparison of three methods for generating group statistical inferences from independent component analysis of functional magnetic resonance imaging data. J. Magn. Reson. Imaging 19, 365–368. 10.1002/jmri.20009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen B, Parhi KK, 2017. Extraction of common task signals and spatial maps from group fMRI using a PARAFAC-based tensor decomposition technique. In: IEEE International Conference on Acoustics, Speech and Signal Processing, pp. 1113–1117. [Google Scholar]
- Simony E, Honey CJ, Chen J, Lositsky O, Yeshurun Y, Wiesel A, Hasson U, 2016. Dynamic reconfiguration of the default mode network during narrative comprehension. Nat. Commun 7, 12141. 10.1038/ncomms12141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegeman A, 2007. Comparing independent component analysis and the PARAFAC model for artificial multi-subject fMRI data, Unpublished Technical report, University of Groningen. [Google Scholar]
- Svensén M, Kruggel F, Benali H, 2002. ICA of fMRI group study data. Neuroimage 16, 551–563. 10.1006/nimg.2002.1122. [DOI] [PubMed] [Google Scholar]
- Sylvester CM, Corbetta M, Raichle ME, Rodebaugh TL, Schlaggar BL, Sheline YI, Zorumski CF, Lenze EJ, 2012. Functional network dysfunction in anxiety and anxiety disorders. Trends Neurosci 35, 527–535. 10.1016/j.tins.2012.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasi G, Bro R, 2005. PARAFAC and missing values. Chemom. Intell. Lab. Syst 75, 163–180. 10.1016/j.chemolab.2004.07.003. [DOI] [Google Scholar]
- Van Essen DC, Smith SM, Barch DM, Behrens TEJ, Yacoub E, Ugurbil K, 2013. The WU-minn human connectome project: an overview. Neuroimage 80, 62–79. 10.1016/j.neuroimage.2013.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varoquaux G, Sadaghiani S, Pinel P, Kleinschmidt A, Poline J-B, Thirion B, 2010. A group model for stable multi-subject ICA on fMRI datasets. Neuroimage 51, 288–299. 10.1016/j.neuroimage.2010.02.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.