Abstract
In this paper, we present FlyBy CNN, a novel deep learning based approach for 3D shape segmentation. FlyByCNN consists of sampling the surface of the 3D object from different view points and extracting surface features such as the normal vectors. The generated 2D images are then analyzed via 2D convolutional neural networks such as RUNETs. We test our framework in a dental application for segmentation of intra-oral surfaces. The RUNET is trained for the segmentation task using image pairs of surface features and image labels as ground truth. The resulting labels from each segmented image are put back into the surface thanks to our sampling approach that generates 1-1 correspondence of image pixels and triangles in the surface model. The segmentation task achieved an accuracy of 0.9.
Keywords: intra oral surface, mesh, segmentation, shape analysis
1. INTRODUCTION
Shape analysis has become of increasing interest in dentistry due to advances in 3D acquisition devices such as intra-oral surface (IOS) scanners which are being used to fabricate ceramic dental crowns,1 or to classify shape morphology. Analyzing and understanding 3D shapes for segmentation, classification, and retrieval, has become more and more important for many graphics and vision applications. This manuscript presents FlyBy CNN, a novel approach for the segmentation of 3D shapes. We test our framework in IOS segmentation. IOS help reveal hidden and overlooked defects in teeth and other parts of the cavity and it allows clinicians to render care effectively by adding information to plan treatments or to communicate with patients and experts regarding diagnosis, decisions, and protoco1s. Tooth segmentation on IOS is an essential step for treatment planning.
Fast and accurate segmentation of the IOS remains a challenge due to various geometrical shapes of teeth, complex tooth arrangements, different dental model qualities, and varying degrees of crowding problems.2
In this paper, we present FlyByCNN an approach for 3D shape analysis and segmentation.
In summary, our approach consists of generating 2D images of the 3D surface from different view points. The generated images are used to train a RUNETs.3,4 The resulting labels are put back into the IOS thanks to our sampling approach which uses octrees to locate triangles in the surface model and generate a relationship of pixels in the 2D image to triangles in the 3D surface. The following section describes the algorithms implementation and accuracy tests.
2. RELATED WORK
Several approaches have been proposed for shape analysis and they may be divided into two broad categories, hand-crafted based methods and learning based methods. Hand-crafted methods may be subdivided into local and global methods. Some examples of local methods include fundamental solutions to wave equations;5 heat equations;6,7 or modes of vibration of a sphere;8 global methods aim to describe objects as a whole and include approaches such as geometric moments.9
Alternatively, learning-based methods use vast amounts of data to learn descriptors directly from the data using state of the art deep learning algorithms, however, adapting those algorithms to work on 3D shapes is a challenging task. The main impediment is the arbitrary structures of 3D models which are usually represented by point clouds or triangular meshes, whereas deep learning algorithms use the regular grid-like structures found in 2D/3D images. For this particular reason, learning-based methods on 3D shapes could be subdivided into multi-view, volumetric, and parametric. Multi-view approaches render the 3D object from different view-points and capture snapshots that can be used to extract 2D image features, finally, the features are aggregated using pooling,10,11 or long-short term memory networks (LSTM);12 volumetric approaches work directly on the input point-cloud,13 or use 3D voxel grids to represent the shape and apply 3D convolutions to learn shape features,14 speed ups using octrees are proposed to reduce the number of operations performed during training;15,16 finally, parametric approaches transform the 3D object to a known manifold and implement 2D convolutions on the parametric space.17
Our approach FlyByCNN falls in the multi-view category. The main difference from the approaches described above is that the 1-1 relation that exists from the generated images and the triangles in the 3D object is lost using the rendering techniques. We solve this issue by generating images using octrees, i.e., we intersect line segments from a plane as shown in Figure 1. This approach allows us to create a relationship of the image pixel and a triangle in the 3D surface, which we use to extract 3D surface features such as normals and a depth map or distance from the view point to the 3D surface. Preserving relationship of each pixel in the generated image and the corresponding intersected cell in the 3D object is crucial for our segmentation task. The segmentation task aims to make predictions on the generated images and put this information back in the 3D object.
Figure 1.
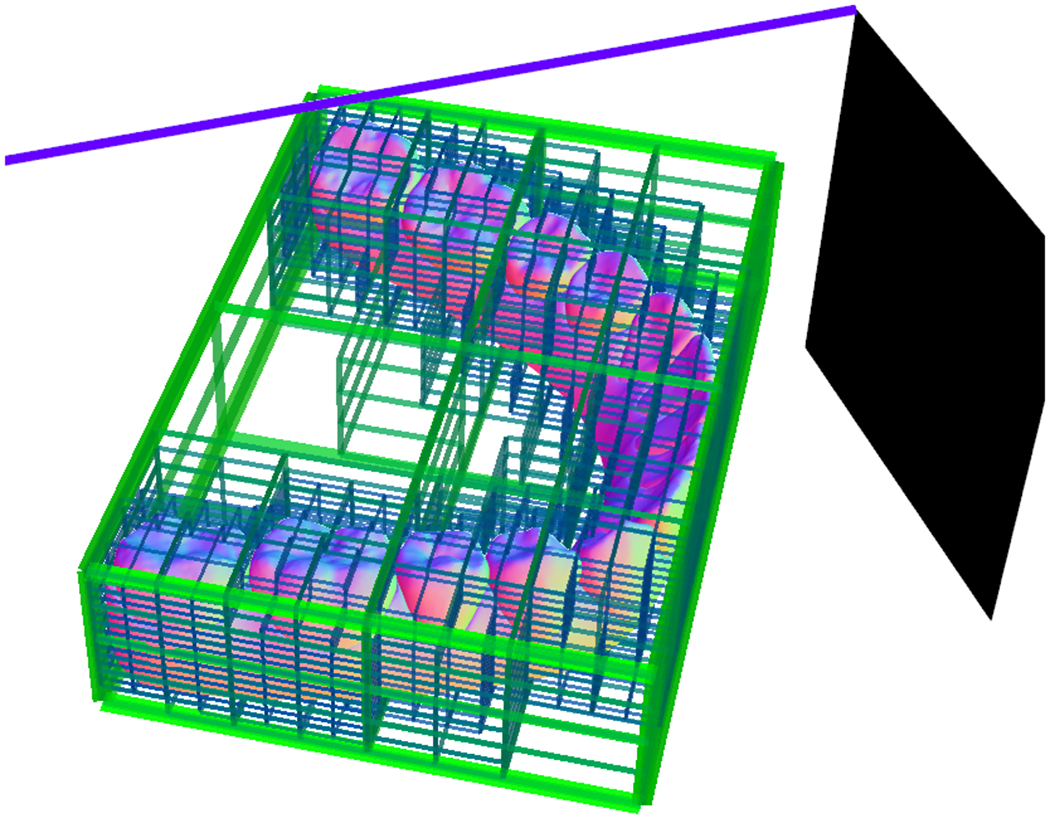
Octree rendering. A line segment is intersected with the octree to quickly locate the corresponding triangles of the 3D surface.
3. MATERIALS
40 IOS were acquired with the TRIOS 3D scanner (software version: TRIOS 1.3.4.5) using the ultrafast optical sectioning and confocal microscopy to generate 3D images from multiple 2-dimensional images with accuracy of 6.9 ± 0.9 μm. All scans were obtained according to the manufacturer’s instructions, by 1 trained operator.
4. METHODS
4.1. Surface segmentation via UNET
The ground truth labeling of the dental surface is done with a region growing algorithm that uses the minimum curvature as stopping criteria plus manually correcting the miss-classified regions directly on the 3D surface. Figure 2 shows the projection plane in blue that is used to probe the IOS surface and extract the associated shape features plus the corresponding label map (background, gum, boundary between gum and teeth).
Figure 2.
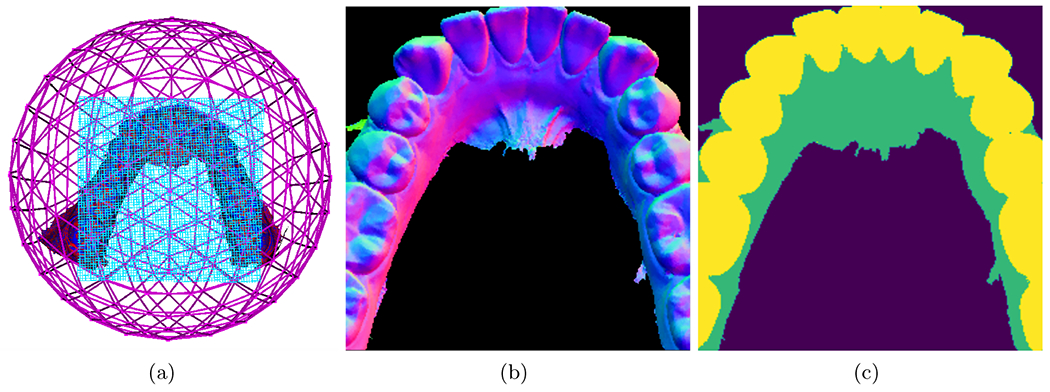
a) Icosahedron subdivision and the tangent plane. b) Mesh features. c) Labeled regions.
We use 32 IOS to generate our training samples and leave 10 for testing our segmentation approach. Only lower arches are used to create our training set images. We leave out some upper arches to test our segmentation framework. Each IOS is randomly rotated 50 times and samples are generated using an icosahedron linearly subdivided at level 5 (252 points on the sphere), i.e., in total, we have 378000 (252 x 50 x 30) image samples to train our RUNET.3,4 Since the IOS are randomly rotated, this allows our method to work independently of the original orientation of the IOS which may vary across patients and/or scanners. We use the Intersection over Union (IoU) metric as loss function. The training minimizes 1 – IoU, the learning rate is set to 1e−4 using the Adam optimizer.
Figure 3 shows the steps to label the IOS after the segmentation by the UNET. The labels are put back into the surface by using the intersected triangles found during the sampling process and the output label maps of the RUNET as there is a 1-1 relationship from the images and the sampled triangles. A post-processing step is needed as there will be triangles that are not captured during the sampling process. The following section describes the post-processing approach.
Figure 3.
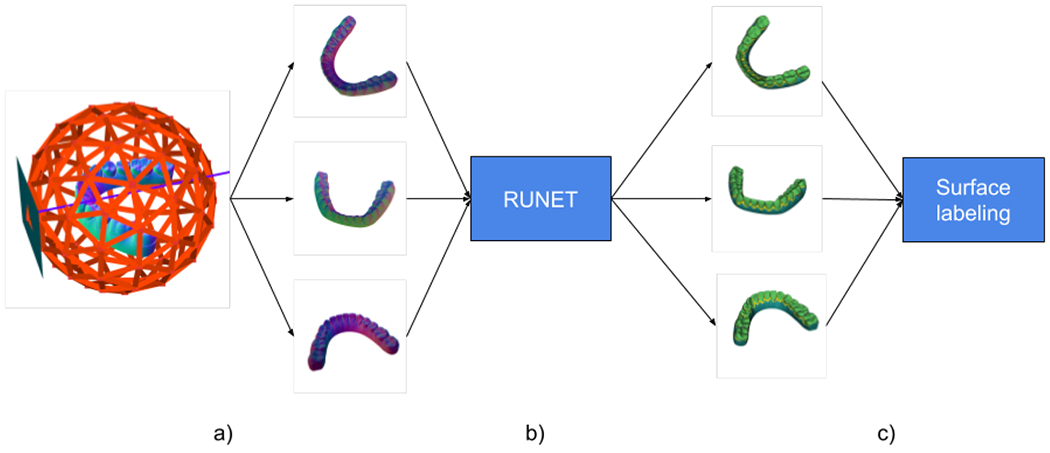
a) Feature image generation via octrees and surface sampling. b) Image segmentation via RUNET. c) The resulting labels from the segmentation are put back in the 3D surface by using the intersections found during the sampling.
4.2. Post process
The resulting labeled surfaces as shown in Figure 4 may contain triangles that are not labeled, i.e., the sampling process does not capture all triangles in the IOS. The unlabeled regions or islands are removed by detecting the closest connected region that has been labeled. Once the islands are removed, a region growing algorithm is performed to label the connected teeth regions with a unique id. The boundary region between each tooth allows labeling with a unique id each connected region. Finally, the boundary region is eroded. At the end of this process, the IOS has a unique label for each tooth and gum.
Figure 4.
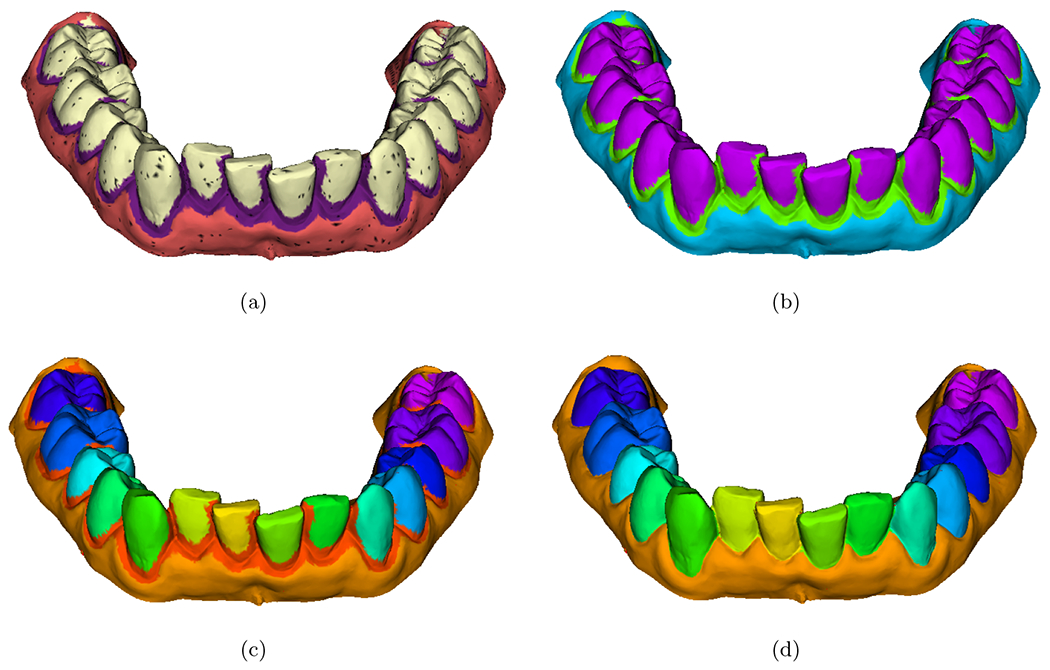
Post processing of the mesh to isolate dental crowns. The sampling via FlyBy approach may not sample all triangles of the mesh. a) Majority voting output for each individual snapshot. b) Removal of unlabeled triangles. c) Region growing to label each tooth with a unique label. d) Erosion of the boundary between teeth and gum.
5. RESULTS
Figure 5 shows the result of the segmentation task. The segmentation is accurate for the gum and teeth regions, while the boundary has lower accuracy, it is satisfactory because we are able to assign a unique label to each tooth as shown in Figure 6 Figure 5.a shows the number of individual pixels classified. Figure 5.b shows a normalized confusion matrix. Even though, the boundary region has low accuracy when compared to the teeth and gum regions, Figure 6 shows that is enough to separate the teeth and provide unique labels for each one of them. Figure 6 shows results for 6 of our test scans. Each tooth has been given a unique label. The RUNET segmentation task was trained using lower arches only, however, the segmentation of the upper arches is satisfactory.
Figure 5.
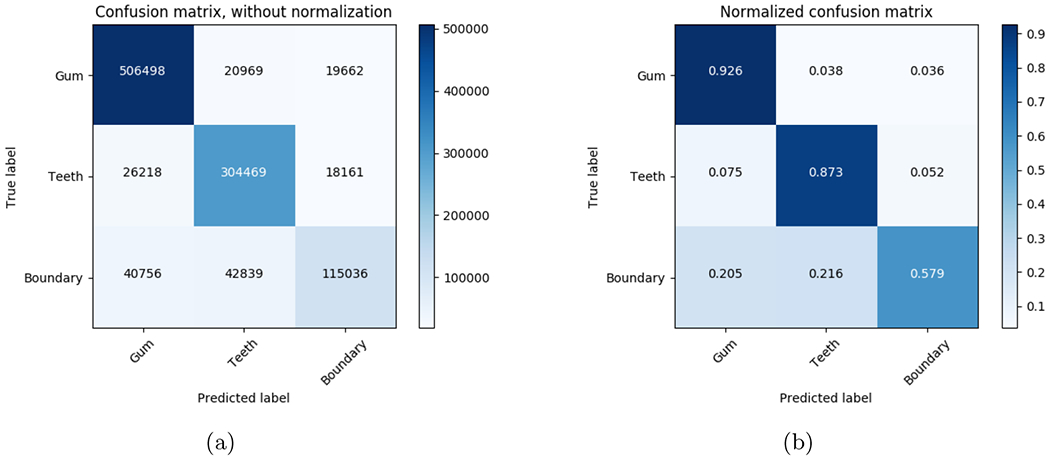
Prediction on 10 evaluation scans for the gum, teeth, and boundary between gum and teeth. The predicted boundary is used to split each tooth and produce a single labeled output for each crown.
Figure 6.
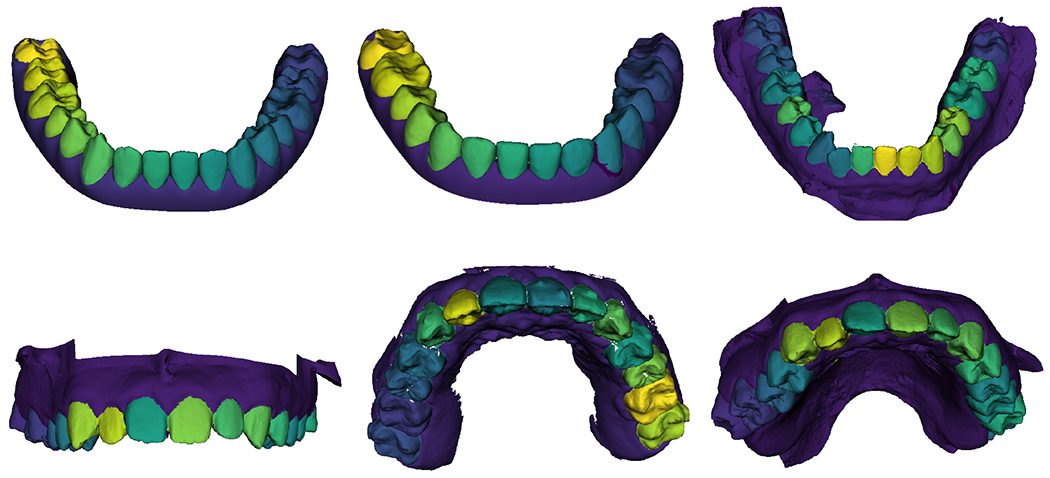
Resulting labeled IOS from our test set. Top row, lower arches. Bottom row, upper arches.
6. CONCLUSIONS
We presented a novel framework to analyze 3D surfaces via sampling of the surface of the object. The flexibility of the framework is demonstrated in a segmentation task of IOS. Our framework differentiates from existing approaches in the features that are used to generate the 2D images which are purely geometric. Additionally, our generated image samples preserve a 1-1 relationship of pixels in the image and triangles in the 3D object. This relationship allows us to put information back in the 3D object after the 2D image samples are analyzed by state of the art convolutional neural networks. Our framework may be used for task such as classification of 3D objects or segmentation tasks as demonstrated in this paper. Future work aims to speed up the 2D image generation process and improve the IOS segmentation by assigning unique labels to each tooth following a universal id scheme across patients.
REFERENCES
- [1].Berrendero S, Salido MP, Valverde A, Ferreiroa A, and Pradíes G. Influence of conventional and digital intraoral impressions on the fit of cad/cam-fabricated all-ceramic crowns. Clinical oral investigations, 20(9):2403–2410, 2016. [DOI] [PubMed] [Google Scholar]
- [2].Li Zhongyi and Wang Hao. Interactive tooth separation from dental model using segmentation field. PloS one, 11(8):e0161159, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Ronneberger Olaf, Fischer Philipp, and Brox Thomas. U-net: Convolutional networks for biomedical image segmentation. In International Conference on Medical image computing and computer-assisted intervention, pages 234–241. Springer, 2015. [Google Scholar]
- [4].He Kaiming, Zhang Xiangyu, Ren Shaoqing, and Sun Jian. Deep residual learning for image recognition. In Proceedings of the IEEE conference on computer vision and pattern recognition, pages 770–778, 2016. [Google Scholar]
- [5].Aubry Mathieu, Schlickewei Ulrich, and Cremers Daniel. The wave kernel signature: A quantum mechanical approach to shape analysis. In 2011 IEEE international conference on computer vision workshops (ICCV workshops), pages 1626–1633. IEEE, 2011. [Google Scholar]
- [6].Bronstein Michael M and Kokkinos Iasonas. Scale-invariant heat kernel signatures for non-rigid shape recognition. In 2010 IEEE Computer Society Conference on Computer Vision and Pattern Recognition, pages 1704–1711. IEEE, 2010. [Google Scholar]
- [7].Ribera Nina Tubau, De Dumast Priscille, Yatabe Marilia, Ruellas Antonio, Ioshida Marcos, Paniagua Beatriz, Styner Martin, Gonçalves João Roberto, Bianchi Jonas, Cevidanes Lucia, et al. Shape variation analyzer: a classifier for temporomandibular joint damaged by osteoarthritis. In Medical Imaging 2019: Computer-Aided Diagnosis, volume 10950, page 1095021. International Society for Optics and Photonics, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Styner Martin, Oguz Ipek, Xu Shun, Brechbühler Christian, Pantazis Dimitrios, Levitt James J, Shenton Martha E, and Gerig Guido. Framework for the statistical shape analysis of brain structures using spharmpdm. The insight journal, (1071):242, 2006. [PMC free article] [PubMed] [Google Scholar]
- [9].Hu Ming-Kuei. Visual pattern recognition by moment invariants. IRE Transactions on Information Theory, 8(2):179–187, 1962. [Google Scholar]
- [10].Su Hang, Maji Subhransu, Kalogerakis Evangelos, and Learned-Miller Erik. Multi-view convolutional neural networks for 3d shape recognition. In Proceedings of the IEEE international conference on computer vision, pages 945–953, 2015. [Google Scholar]
- [11].Kanezaki Asako, Matsushita Yasuyuki, and Nishida Yoshifumi. Rotationnet: Joint object categorization and pose estimation using multiviews from unsupervised viewpoints. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pages 5010–5019, 2018. [Google Scholar]
- [12].Ma Chao, Guo Yulan, Yang Jungang, and An Wei. Learning multi-view representation with lstm for 3-d shape recognition and retrieval. IEEE Transactions on Multimedia, 21(5):1169–1182, 2018. [Google Scholar]
- [13].Qi Charles R, Su Hao, Mo Kaichun, and Guibas Leonidas J. Pointnet: Deep learning on point sets for 3d classification and segmentation. In Proceedings of the IEEE conference on computer vision and pattern recognition, pages 652–660, 2017. [Google Scholar]
- [14].Wu Zhirong, Song Shuran, Khosla Aditya, Yu Fisher, Zhang Linguang, Tang Xiaoou, and Xiao Jianxiong. 3d shapenets: A deep representation for volumetric shapes. In Proceedings of the IEEE conference on computer vision and pattern recognition, pages 1912–1920, 2015. [Google Scholar]
- [15].Wang Peng-Shuai, Liu Yang, Guo Yu-Xiao, Sun Chun-Yu, and Tong Xin. O-cnn: Octree-based convolutional neural networks for 3d shape analysis. ACM Transactions on Graphics (TOG), 36(4):1–11, 2017. [Google Scholar]
- [16].Riegler Gernot, Ulusoy Ali Osman, and Geiger Andreas. Octnet: Learning deep 3d representations at high resolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pages 3577–3586, 2017. [Google Scholar]
- [17].Liu Min, Yao Fupin, Choi Chiho, Sinha Ayan, and Ramani Karthik. Deep learning 3d shapes using alt-az anisotropic 2-sphere convolution. In International Conference on Learning Representations, 2018. [Google Scholar]


