Abstract
Sensitivity and specificity of serological assays are key parameters for the accurate estimation of SARS-CoV-2 sero-prevalence. The aim of this study was to compare 8 readily available IgG antibody tests using a panel of well-defined serum samples of prepandemic and pandemic origin. A cross-reaction panel included samples of patients with recent infection with either of the endemic Coronaviruses 229E, NL63, HKU1, or OC43. Additionally, samples with high antibody levels against influenza virus, adenovirus, and during acute EBV infection were included. Previous infection with endemic coronaviruses caused a significant amount of cross-reactivity in two of the assays. In contrast, the confidence intervals for the assays of Abbott, DiaSorin, Euroimmun and Roche encompassed the value of 98% for samples with a previous endemic HCoV infection. For all assays, sensitivities were between 91.3% and 98.8%. Assay performance was independent of the usage of either nucleocapsid or spike proteins.
Keywords: Covid-19, serology, Sensitivity, Cross-reactivity, SARS-CoV-2
1. Introduction
Since its emergence in December 2019 in the Chinese province of Hubei (Zhu et al., 2020), the novel coronavirus SARS-CoV-2 caused significant mortality and morbidity leading to more than 100 million proven infections and 2.300.000 deaths in more than 200 countries world-wide (Di et al., 2020; World Health Organization). Coronaviruses are enveloped and have a large positive-sense, single-stranded RNA-genome (Cui et al., 2019). The disease resulting from SARS-CoV-2 infection, COVID-19, is mild in the majority of cases. However, infection may also result in severe disease courses with acute respiratory distress, multi-organ failure, and death (Di et al., 2020). In acute infections, diagnosis is confirmed by nucleic acid amplification testing (NAAT) of nasal or oropharyngeal swabs as well as deep respiratory specimen. Specific antibodies appear in sufficient amounts not earlier than 10 to 14 days after infection (Kellam and Barclay, 2020; Okba et al., 2020).
Asymptomatic infections as well as patients having rather mild symptoms may result in underestimating the total disease burden. Although ongoing testing for acute infections will provide a point prevalence of acute infections, false negative PCR results, i.e. due to inadequate sampling or low viral load, should be taken into account. The detection of specific IgG directed against SARS-CoV-2 allows the identification of previously infected persons and patients and thus helps to establish more precisely the true prevalence of SARS-CoV-2 in a population.
High specificity and sensitivity of serological tests are of paramount importance. Different antigenic sites on the structural proteins show a varying degree of homology to other members of the Orthocoronavirinae, which could cause false positive test results. Back in 2002/2003, cross-reactivity was noted in the SARS epidemic (Che et al., 2005). Notably, 4 other human Coronaviruses (HCoV), namely the alpha-coronaviruses HCoV-229E and HCoV-NL63 and the beta-coronaviruses HCoV-OC43 and HCoV-HKU1, circulate in the human population (Su et al., 2016). The endemic human coronaviruses are widespread causing predominantly mild infections of the upper respiratory tract (Gaunt et al., 2010; Walsh et al., 2013). Infections could give rise to cross-reactive antibodies depending on their affinity to the specific antigen and thus may influence the performance of SARS-CoV-2 serological assays.
The aim of the study was to compare different commercial assays on a panel of selected serum samples with a focus on an in-depth analysis of specificity and cross-reactivity.
2. Methods
2.1. Specimen
A panel of anonymized leftover serum samples (samples; n = 257) were tested for SARS-CoV-2 IgG antibodies. The panel included 140 pre-pandemic samples from patients with a high probability of cross-reactivity due to a prior infection with endemic coronaviruses (HCoV 229E (n = 17), HCoV HKU1 (n = 14), HCoV OC43 (n = 16), HCoV NL63 (n = 27)), an acute Epstein-Barr virus infection (EBV; n = 25) or high antibody levels against either influenza viruses A and B (n = 21) or adenovirus (n = 20). Furthermore, the panel included 117 samples of patients that were tested for an infection with SARS-CoV-2 by NAAT in 2020. Of these, 37 were randomly chosen samples of in-patients with no history of contact to SARS-CoV-2 infected individuals and a negative SARS-CoV-2 NAAT. Additionally, 80 samples of patients with a laboratory confirmed SARS-CoV-2 infection were included.
2.2. SARS-CoV-2 IgG testing
Samples were tested for SARS-CoV-2 specific-IgG with the following assays according to the manufacturer's recommendations: (1) SARS-CoV-2 IgG (Abbott; Chicago, IL), (2) LIAISON SARS-CoV-2 IgG (DiaSorin, Saluggia, Italy), (3) EDI Novel Coronavirus COVID-19 IgG ELISA (Epitope Diagnostics, Inc., San Diego, California, USA), (4) Anti-SARS-CoV-2-ELISA (IgG) and (5) Anti-SARS-CoV-2-NCP-ELISA (IgG) (Euroimmun, Lübeck, Germany), (6) recomWell SARS-CoV-2 IgG (Mikrogen GmbH, Neuried, Germany), (7) COVID-19 ELISA IgG (Vircell, Granada, Spain), (8) Elecsys Anti-SARS-CoV-2 assay (Roche, Basel, Switzerland). Assay specifications are provided in the manufacturer's manuals (Table 1 ).
Table 1.
Specifications of the analyzed anti-SARS-CoV-2 IgG assays according to the manufacturers.
| Manufacturer | Assay | Used Platforma | Antigen | Test Principle | Measurement | Interpretation | Specificity (according to manufacturer) | Sensitivity (according to manufacturerb) | |
|---|---|---|---|---|---|---|---|---|---|
| Abbott | SARS-CoV-2 IgG | Abbott ARCHITECT i2000SR Sytsem | rN | CMIA | index (S/Co) | negative | < 1.4 | 99.63% | 100% (after 14 days) |
| positive | ≥ 1.4 | ||||||||
| DiaSorin | Liaison SARS-CoV-2 S1/S2 IgG | DiaSoin LIAISON | rS1/rS2 | CLIA | AU/ml | negative | < 12 | 98.5% | 97.4% after 15 days |
| indeterminate | 12 - < 15 | ||||||||
| positive | ≥ 15 | ||||||||
| Epitope Diagnostics | EDI Novel Coronavirus COVID-19 IgG ELISA | manual | rN | ELISA | OD | negative | ≤ negative cutoff | 100% | 100% |
| indeterminate | > negative cutoff < positive cutoff | ||||||||
| positive | ≥ positive cutoff | ||||||||
| Euroimmun | Anti-SARS-CoV-2 ELISA (IgG) | EUROIMMUN Analyzer I | rS1 | ELISA | ratio | negative | < 0.8 | 99% | 93.8% (after 20 days) |
| indeterminate | 0.8 - < 1.1 | ||||||||
| positive | ≥ 1.1 | ||||||||
| Euroimmun | Anti-SARS-Cov-2 NCP ELISA (IgG) | EUROIMMUN Analyzer I | rN | ELISA | ratio | negative | < 0.8 | 99.8% | 94.6% (after 10 days) |
| indeterminate | 0.8 - < 1.1 | ||||||||
| positive | ≥ 1.1 | ||||||||
| Mikrogen | recomWell SARS-CoV-2 IgG | manual | rN | ELISA | U/ml | negative | < 20 | 98.7% | 98% |
| indeterminate | 20 - 24 | ||||||||
| positive | > 24 | ||||||||
| Vircell | COVID-19 ELISA IgG | Vircell VIRCLIA | rS/rN | ELISA | antibody index | negative | < 4 | 98% | 70% (after 7 days) |
| indeterminate | 4 - 6 | ||||||||
| positive | > 6 | ||||||||
| Roche | Elecsys Anti-SARS-CoV-2c | Roche 8000/e602 | rN | ECLIA | COI | negative | < 1.0 | 99.81% | 100% (after 14 days) |
| positive | ≥ 1.0 | ||||||||
r = recombinant; N = nucleocapsid protein; S = spike protein; S1 = subunit 1 of S; S2, Subunit 2 of S; CMIA = chemiluminescent microparticle immunoassay; ECLIA = electrochemiluminescence immunoassay, CLIA = chemiluminescence immunoassay; S/Co, Signal/Cutoff; AU = arbitrary units; OD = optical density; COI, cutoff index.
Platform that was used for the conduction of this study.
Classification of the timeframe after infection varied between the manufacturers. Total or last timeframe is given as indicated.
IgA, IgM, and IgG are detected.
2.3. SARS-CoV-2 detection
A published qRT-PCR targeting the viral E gene was used (Corman et al., 2020).
2.4. Endemic Coronavirus detection
Testing was done with a multiplex NAAT for respiratory viruses (NxTAG RPP, Luminex Corporation; Austin, TX) according to the manufacturer's instructions.
2.5. Endemic Coronavirus IgG antibody testing
IgG antibodies to endemic coronaviruses were detected using the recomLine SASR-CoV-2 IgG blot (Mikrogen GmbH, Neuried, Germany), according to the manufacturer's recommendations.
2.5. EBV testing
Acute EBV infection was diagnosed using EBNA-1 IgG, VCA IgG, and IgM CMIA (Architect System; Abbott, Chicago, IL) with subsequent confirmation by recomLine EBV IgG, IgA, and IgM blots (Mikrogen GmbH, Neuried, Germany), according to the manufacturer's recommendations.
2.6. Influenza virus A and B IgG antibody testing
IgG antibodies to influenza viruses were detected using the Anti-Influenza A Virus ELISA and the Anti-Influenza B Virus ELISA (Euroimmun, Lübeck, Germany) according to the manufacturer's recommendations. Serum samples with reactivity above 100 relative units/ml (RU/mL) for both viruses were selected (assay cutoff: negative < 16 RU/mL; positive ≥ 22 RU/mL).
2.7. Adenovirus IgG antibody testing
IgG antibodies to adenovirus were detected using the Anti-Adenovirus ELISA (Euroimmun, Lübeck, Germany) according to the manufacturer's recommendations. Serum samples with reactivity above 150 RU/mL were selected (assay cutoff: negative < 16 RU/mL; positive ≥ 22RU/mL).
2.8. Statistical analysis
Statistical analysis was performed using IBM SPSS Statistics for Windows, Version 25.0 (Armonk, NY: IBM Corp.). For the analysis of sensitivity and specificity, samples with equivocal results were considered positive. The 95% confidence interval (CI) was calculated using the Wilson score method. Positive (PPV) and negative (NPV) predictive values were calculated using Bayes’ theorem with an assumed prevalence of 5%. The correlation analysis was done using the Spearman-Rho test. The scatterplot matrix was built with R (R Core Team 2018) using the GGally package (Schloerke et al., 2018).
3. Results
3.1. Study population
In total, 257 samples of 222 individual patients were included into the analysis. The median age of the patients was 50.8 years with a range from 12 to 91. Of all individuals, 38.3% were female. The median time of detection of endemic coronaviruses before sample collection was 34 days (interquartile range 6.25–96.25 days, range 0–478). An upper respiratory tract infection was reported in 51% (38/74) and a lower respiratory tract infection in 38% (28/74) of the cases. Eleven percent (8/74) did not have a respiratory tract infection. Of the SARS-CoV-2 infected individuals, 15 samples were collected within the first 15 days after symptom onset, the remaining samples thereafter. An upper respiratory tract infection was reported in 26.3% (21/80) and a lower respiratory tract infection in 72.5% (58/80) of the cases. No respiratory symptoms were reported for one case.
3.2. Sero-reactivity
3.2.1. Specificity analysis
The majority of antibody assays delivered negative test results. However, sero-reactivity in the pre-pandemic samples differed considerably between the commercial assays (Table 2 ). Lowest specificities were seen for the HCoV NL63 subgroup (Epitope Diagnostics) and the HCoV HKU1 subgroup (Vircell). Reactive samples were mostly reactive in one of the assays, except for six samples that reacted in both the assays of Epitope Diagnostics and Mikrogen. The confidence interval for the assays of Abbott, DiaSorin, Euroimmun, and Roche encompassed a value of at least 98% for the samples with a known previous endemic HCoV infection. While high levels of antibodies against influenza viruses A and B and adenovirus were only associated with an increased cross-reactivity in one assay (Epitope Diagnostics), EBV infection was associated with specificities below 70% for two assays (Epitope diagnostics and Vircell). Again, reactive samples were mostly reactive in one of the assays with exception of one influenza sample that reacted in four of the assays (Epitope diagnostics, Abbott, Mikrogen, and Euroimmun (NCP)). Two other samples were reactive in the assays of Epitope diagnostics, Mikrogen, and Euroimmun (NCP). For pandemic samples (Table 3 , lower panel), the confidence interval included values of at least 98% for all but two assays (Epitope Diagnostics and Mikrogen). Different samples showed a positive reactivity in different assays except for two samples that were tested positive in the assays of Epitope diagnostics and Mikrogen. The specificities of all negative samples were 99.4% (CI 69.9%–99.9%, PPV 89.4) for Abbott, 97.7% (CI 94.3%–99.1%, PPV 67.6) for DiaSorin, 74% (CI 67.1%–79.9%, PPV 16.5) for Epitope Diagnostics, 97.7% (CI 94.3%–99.1%, PPV 68.5) for Euroimmun (S1), 97.7% (CI 94.3%–99.1%, PPV 69.3) for Euroimmun (NCP), 92.7% (CI 87.8%–95.7%, PPV 41) for Mikrogen, 84.2% (CI 78.1%–88.8%, PPV 24.8) for Vircell and 100% (CI 97.9%–100%, PPV 100) for Roche.
Table 2.
Test performance using prepandemic sera.
| Manufacturer | Abbott | DiaSorin | EpitopeDiagnostics | Euroimmun (S1) | Euroimmun (NCP) | Mikrogen | Vircell | Roche |
|---|---|---|---|---|---|---|---|---|
| HCoV 229E infection [n = 17] | ||||||||
| positive | 0 | 1 | 4 | 0 | 0 | 3 | 1 | 0 |
| indeterminate | NA | 0 | 0 | 1 | 0 | 0 | 1 | NA |
| negative | 17 | 16 | 13 | 16 | 17 | 14 | 15 | 17 |
| specificity [%] | 100 | 94.1 | 76.5 | 94.1 | 100 | 82.4 | 88.2 | 100 |
| HCoV NL63 infection [n = 27] | ||||||||
| positive | 0 | 0 | 2 | 0 | 0 | 1 | 4 | 0 |
| indeterminate | NA | 0 | 6 | 1 | 1 | 0 | 1 | NA |
| negative | 27 | 27 | 19 | 26 | 26 | 26 | 22 | 27 |
| specificity [%] | 100 | 100 | 70.4 | 96.3 | 96.3 | 96.3 | 81.5 | 100 |
| HCoV HKU1 infection [n = 14] | ||||||||
| positive | 0 | 0 | 1 | 1 | 0 | 1 | 4 | 0 |
| indeterminate | NA | 0 | 0 | 0 | 0 | 0 | 3 | NA |
| negative | 14 | 14 | 13 | 13 | 14 | 13 | 7 | 14 |
| specificity [%] | 100 | 100 | 92.9 | 92.9 | 100 | 92.9 | 50 | 100 |
| HCoV OC43 infection [n = 16] | ||||||||
| positive | 0 | 0 | 3 | 0 | 0 | 2 | 2 | 0 |
| indeterminate | NA | 0 | 0 | 0 | 0 | 0 | 2 | NA |
| negative | 16 | 16 | 13 | 16 | 16 | 14 | 12 | 16 |
| specificity [%] | 100 | 100 | 81.3 | 100 | 100 | 87.5 | 75.0 | 100 |
| total HCoV samples [n = 74] | ||||||||
| positive | 0 | 1 | 10 | 1 | 0 | 7 | 11 | 0 |
| indeterminate | NA | 0 | 6 | 2 | 1 | 0 | 7 | NA |
| negative | 74 | 73 | 58 | 71 | 73 | 67 | 56 | 74 |
| specificity [%] | 100 | 98.7 | 78.4 | 96 | 98.7 | 90.5 | 75.7 | 100 |
| CI [%] | 99.1-100 | 92.7-99.8 | 67.7-86.2 | 88.8-98.7 | 92.7-99.8 | 81.2-95.3 | 64.8-84 | 99.1-100 |
| PPV [%] | 100 | 78.7 | 19.2 | 55.6 | 80 | 34.8 | 17.6 | 100 |
| acute EBV infection [n = 25] | ||||||||
| positive | 0 | 0 | 12 | 0 | 0 | 0 | 5 | 0 |
| indeterminate | NA | 0 | 2 | 0 | 0 | 0 | 3 | NA |
| negative | 25 | 25 | 11 | 25 | 25 | 25 | 17 | 25 |
| specificity [%] | 100 | 100 | 44.0 | 100 | 100 | 100 | 68.0 | 100 |
| Influenza virus A/B IgG positive [n = 21] | ||||||||
| positive | 0 | 0 | 2 | 0 | 2 | 2 | 0 | 0 |
| indeterminate | NA | 2 | 2 | 0 | 0 | 0 | 1 | NA |
| negative | 21 | 19 | 17 | 21 | 19 | 19 | 20 | 21 |
| specificity [%] | 100 | 90.5 | 80.1 | 100 | 90.5 | 90.5 | 95.2 | 100 |
| Adenovirus IgG positive [n = 20] | ||||||||
| positive | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 0 |
| indeterminate | NA | 0 | 1 | 0 | 0 | 0 | 0 | NA |
| negative | 19 | 20 | 18 | 19 | 19 | 19 | 20 | 20 |
| specificity [%] | 95 | 100 | 90 | 95 | 95 | 95 | 100 | 100 |
| total pre-pandemic samples [n = 140] | ||||||||
| positive | 1 | 1 | 25 | 2 | 3 | 10 | 16 | 0 |
| indeterminate | NA | 2 | 11 | 2 | 1 | 0 | 11 | NA |
| negative | 139 | 137 | 104 | 136 | 136 | 130 | 113 | 140 |
| specificity [%] | 99.3 | 97.9 | 74.3 | 97.1 | 97.1 | 92.9 | 80.1 | 100 |
| CI [%] | 96.1-99.9 | 93.9-99.3 | 66.5-80.1 | 92.9-98.9 | 92.9-98.9 | 87.4-96.1 | 87.4-96.1 | 97.1-100 |
| PPV [%] | 87.9 | 69.6 | 16.6 | 63.3 | 64.2 | 41.7 | 20.7 | 100 |
CI = 95% confidence interval; PPV = positive predictive value.
Specificity is provided as percent for the negative samples in total and stratified by subgroups.
Table 3.
Test performance using pandemic sera.
| Manufacturer | Abbott | DiaSorin | EpitopeDiagnostics | Euroimmun (S1) | Euroimmun (NCP) | Mikrogen | Vircell | Roche |
|---|---|---|---|---|---|---|---|---|
| pandemic samples of SARS-CoV-2 NAAT positive patients [n = 80] | ||||||||
| positive | 77 | 72 | 78 | 71 | 78 | 75 | 76 | 73 |
| indeterminate | NA | 1 | 0 | 5 | 1 | 2 | 3 | NA |
| negativea | 3(2) | 7(4) | 2(1) | 4(3) | 1(1) | 3(1) | 1(0) | 7(6) |
| sensitivity [%] | 96.3 | 91.3 | 97.5 | 95 | 98.8 | 96.3 | 98.8 | 91.3 |
| CI [%] | 89.6-98.7 | 83-95.7 | 91.3-99.3 | 87.8-98 | 93.3-99.8 | 89.6-98.7 | 93.3-99.8 | 83-95.7 |
| pandemic samples from SARS-CoV-2 NAAT negative patients of 2020 [n = 37] | ||||||||
| positive | 0 | 1 | 8 | 0 | 0 | 2 | 0 | 0 |
| indeterminate | NA | 0 | 2 | 0 | 0 | 1 | 1 | NA |
| negative | 37 | 36 | 27 | 37 | 37 | 34 | 36 | 37 |
| specificity [%] | 100 | 97.3 | 73 | 100 | 100 | 91.9 | 97.3 | 100 |
| CI [%] | 90.6-100 | 86.2-99.5 | 57-84.6 | 90.6-100 | 90.6-100 | 78.7-97.2 | 86.2-99.5 | 90.6-100 |
| PPV [%] | 100 | 64 | 16 | 100 | 100 | 38.5 | 65.8 | 100 |
CI = 95% confidence interval; PPV = positive predictive value.
The number of samples that were tested negative in the respective assay are given. Numbers in brackets indicate serum samples collected < 15 days after symptom onset.
To further characterize the samples that showed cross-reactivity in any of the assays, all reactive samples were tested for IgG antibodies to endemic coronaviruses, adenovirus, influenza virus A and B, and for the EBV status. Comparison was done to the preselected samples for each chosen pathogen and to the pandemic samples of SARS-CoV-2 NAAT negative patients of 2020 (Table 4 ). The highest seroprevalence was detected for the preselected samples for all endemic Coronaviruses except for HCoV-HKU1 when compared to all cross-reactive samples. Cross-reactive samples with a prior infection with HCoV 229E showed the highest seroprevalence for any endemic coronavirus. The seroprevalence for any endemic coronavirus of the samples with a prior infection with endemic coronaviruses was 78.4 % (63.5 % for multiple HCoVs). A high seroprevalence was seen for adenovirus, influenza virus A and B and EBV. However, the mean sero-reactivity was highest in the preselected samples. No additional acute EBV infection could be detected.
Table 4.
Serological characterization of cross-reactive samples.
| HCOV |
EBV |
Influenza virus A |
Influenza virus B |
Adenovirus |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n | 229E P [%] |
NL63 P [%] |
HKU1 P [%] |
OC43 P [%] |
any HCoVa P [%] | multiple HCoVb P [%] |
P [%] | acute infection [%] | P [%] | Mean [RU/mL] | P [%] | Mean [RU/mL] | P [%] | Mean [RU/mL] | ||
| A | HCoV 229E infection | 7 | 85.7 | 100 | 85.7 | 57.1 | 100 | 100 | 100 | 0 | 85.7 | 40.9 | 85.7 | 80.6 | 100 | 68.1 |
| HCoV NL63 infection | 11 | 54.5 | 63.3 | 63.6 | 72.7 | 90.9 | 72.7 | 81.8 | 0 | 81.8 | 49.4 | 81.8 | 89.5 | 90.9 | 67 | |
| HCoV HKU1 infection | 8 | 25 | 37.5 | 25 | 12.5 | 50 | 25 | 87.5 | 0 | 37.5 | 34.1 | 75 | 107.3 | 62.5 | 91.5 | |
| HCoV OC43 infection | 7 | 71.4 | 85.7 | 85.7 | 71.4 | 85.7 | 85.7 | 100 | 0 | 100 | 65.9 | 85.7 | 106.3 | 85.7 | 67 | |
| total HCoV samples | 33 | 57.6 | 69.7 | 63.3 | 54.5 | 81.8 | 69.7 | 90.9 | 0 | 75.8 | 47.4 | 81.8 | 95.5 | 84.8 | 73.1 | |
| acute EBV infection | 17 | 29.4 | 29.4 | 35.3 | 23.5 | 52.9 | 41.2 | 100 | 100 | 76.5 | 52.8 | 76.5 | 77.6 | 82.4 | 114.8 | |
| Influenza virus A/B IgG positive | 6 | 33.3 | 66.7 | 50 | 33.3 | 83.3 | 50 | 100 | 0 | 100 | 144.2 | 100 | 174.5 | 66.7 | 62.7 | |
| Adenovirus IgG positive | 3 | 66.7 | 66.7 | 33.3 | 33.3 | 66.7 | 66.7 | 100 | 0 | 100 | 80.7 | 100 | 82.3 | 100 | 182 | |
| pandemic samples from SARS-CoV-2 NAAT negative patients of 2020 | 13 | 53.8 | 46.2 | 61.5 | 46.2 | 76.9 | 69.2 | 84.6 | 0 | 84.6 | 79.2 | 100 | 115.2 | 84.6 | 79.2 | |
| all cross-reactive samples | 72 | 48.6 | 55.6 | 54.2 | 43.1 | 73.6 | 61.1 | 93.1 | 25 | 81.9 | 61.3 | 86.1 | 100.8 | 83.3 | 87.7 | |
| B | HCoV 229E infection | 17 | 64.7 | 82.4 | 64.7 | 47.1 | 94.1 | 82.4 | 100 | 0 | 53 | 28.5 | 82.4 | 79 | 88.2 | 46.2 |
| HCoV NL63 infection | 27 | 40.7 | 63 | 44.4 | 44.4 | 77.8 | 55.6 | 81.4 | 0 | 74.1 | 68.8 | 81.5 | 74.4 | 74.1 | 68.8 | |
| HCoV HKU1 infection | 14 | 35.7 | 42.9 | 42.9 | 28.6 | 57.1 | 35.7 | 85.7 | 0 | 50 | 83.3 | 71.4 | 86.5 | 64.3 | 83.3 | |
| HCoV OC43 infection | 16 | 56.3 | 62.5 | 81.3 | 75 | 81.3 | 81.3 | 93.8 | 0 | 87.5 | 70.6 | 81.3 | 103.2 | 87.5 | 70.6 | |
| total HCoV samples | 74 | 48.6 | 63.5 | 41.9 | 48.6 | 78.4 | 63.5 | 89.2 | 0 | 78.4 | 66.8 | 79.7 | 84 | 78.4 | 66.8 | |
| acute EBV infection | 25 | 28 | 24 | 28 | 45.3 | 56 | 40 | 100 | 100 | 76 | 48.4 | 80 | 92.6 | 76 | 109.4 | |
| Influenza virus A/B IgG positive | 21 | 19 | 42.9 | 42.9 | 33.3 | 61.9 | 38.1 | 100 | 0 | 100 | 150.9 | 100 | 169 | 66.7 | 75.4 | |
| Adenovirus IgG positive | 20 | 30 | 45 | 25 | 20 | 46 | 35 | 90 | 0 | 60 | 56.3 | 85 | 97 | 100 | 189.9 | |
| C | pandemic samples from SARS-CoV-2 NAAT negative patients of 2020 | 37 | 22.2 | 30.6 | 38.9 | 25 | 52.8 | 33.3 | 94,6 | 0 | 97.2 | 78.9 | 100 | 118.8 | 88,9 | 86.4 |
The seroprevalence of cross-reactive samples (A) was characterized for the indicated pathogens. For EBV the amount of acute infections is given. For Influenza virus A and B and adenovirus the mean reactivity is given. All parameters are also shown for the samples which were specifically selected for the respective pathogen (B) and for the samples of the pandemic samples of SARS-CoV-2 NAAT negative patients of 2020 (C). P, seroprevalence.
Reactive for at least one endemic coronavirus.
Reactive for at least two endemic coronaviruses.
3.2.2. Sensitivity analysis
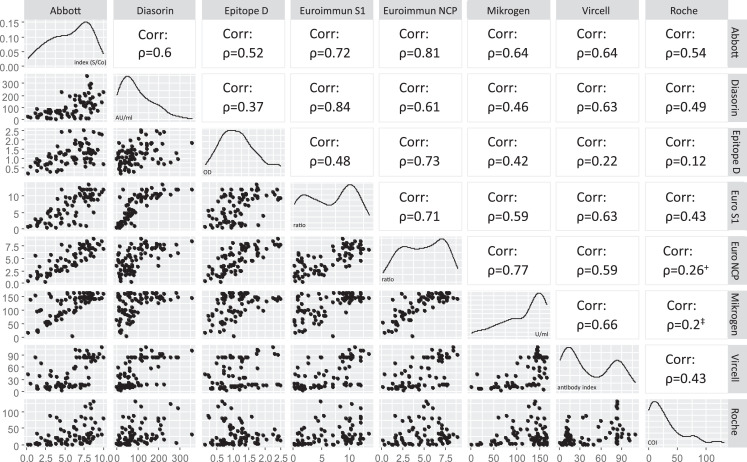
The sensitivity of all tested assays was above 90% with highest sensitivities seen for the assays of Euroimmun (NCP) and Vircell (Table 3, upper panel). Fifteen samples, collected before day 15 after symptom onset included the majority of false negative test results. However, each of the tested samples was reactive in at least 2 of the assays. If only the samples taken ≥ 15 days after symptom onset were considered, the sensitivities were 100% [CI 94.4%–100%] for Euroimmun (NCP), 96.9% [89.5%–99.2%] for Mikrogen, and 98.5% [91.8%–99.7%] for the remaining assays. The negative predictive value was above 99% for each of the tested assays. Quantitative results were compared pairwise for each of the used assays (Fig. 1 ). Spearman-Rho correlation coefficients were highest for the assays of Euroimmun (S1) and Diasorin (ρ = 0.84), and Euroimmun (NCP) and Abbott (ρ = 0.81).
Fig. 1.
Correlation of quantitative assay results of serum samples of patients with a known SARS-CoV-2 infection. Each scatter plot represents the test results of two assays to show interassay correlation (left part). Each dot represents one sample (pandemic samples of SARS-CoV-2 NAAT positive patients). Spearman-Rho correlation coefficient (ρ) is shown on the right part. All correlations are significant (P≤ 0.01), except for +(significant; 0.01> P≤ 0.05) and ‡(not significant). Variable distribution for each assay and test specific units are displayed on the diagonal. (Epitope D, Epitope Diagnostics; Euro, Euroimmun.)
4. Discussion
Antibody tests are generally suitable (1) to determine seroprevalence in a cohort of patients to be used for defining a denominator of the overall magnitude of regional past infections, (2) to identify blood donors which may serve for the preparation of convalescent plasma, and (3) to investigate rare complications of a SARS-CoV-2 infection, e.g., in children (Viner and Whittaker, 2020).
A pivotal quality of a serological test is high specificity in order to exclude false positive results. However, large discrepancies to the manufacturers’ specifications and to previous studies were determined for some of the assays when challenged with potential cross-reactive samples. In line with previous studies, the highest specificities were seen for the assays of Roche and Abbott, which were reported to be above 99% (Bryan et al., 2020; Espejo et al., 2020; Pflüger et al., 2020). Both were the most reliable assays in the cross-reaction panel. Of the two Euroimmun assays, the one using the spike protein (S1) was previously reported with specificities between 96% and 100% (Espejo et al., 2020; Haselmann et al.2020; Kohmer et al., 2020). However, one study used a cross-reaction panel of 37 samples reporting a specificity of 91.9% (Jääskeläinen et al., 2020). Evaluation of the Euroimmun (NCP) assay is scarce, however one study reported a specificity of 98.2% (Herroelen et al., 2020). Studies on the assay of Epitope Diagnostics mostly report low specificities between 78 and 86% (Haselmann et al., 2020; Krüttgen et al., 2020; Wechselberger et al., 2020; Whitman et al., 2020). However, one study used a modified threshold to obtain a specificity of 98.4% (Bundschuh et al., 2020). For the assays of Mikrogen, Vircell, and Diasorin specificities were reported to range between 97% and 100% (Krüttgen et al., 2020; Wechselberger et al., 2020), 94 and 98% (Alharbi et al., 2020; Wechselberger et al.2020), and 97% and 99.1% (Bonelli et al., 2020; Espejo et al., 2020; Kohmer et al., 2020), respectively. The choice of the antigen, either as complete protein or as a recombinant subunit, could profoundly influence the results of the respective assay. The investigated test systems used either the SARS-CoV-2 spike protein and/or the nucleocapsid protein. The spike protein is expected to be more specific while the nucleocapsid protein is expected to be more sensitive, because of the large abundance during an infection (Motley et al., 2020; Ou et al., 2020; Pflüger et al., 2020). Both proteins show a variable degree of homology to other members of the Orthocoronavirinae (Cui et al., 2019; Lu et al., 2020; Okba et al., 2020; Zuwała et al., 2015). Thus, a recent infection with a coronavirus other than SARS-CoV-2 may lead to false positive test results. Accordingly, high rates of reactive samples were detected for three of the eight tested assays (Epitope Diagnostics, Mikrogen, and Vircell). Interestingly, the alpha-coronaviruses HCoV 229E and HCoV NL63 appeared to have a similar impact on cross-reactivity, in spite of the fact that SARS-CoV-2 is a member of beta-coronaviruses. However, this could be a random effect of the small sample size for the individual coronavirus subgroups. Depending on the rate of coronavirus infections in a respiratory season and occurrence in a given geographical area, the positive predictive values of a given serological SARS-CoV-2 assay may therefore be influenced to varying extents. Performance of the assays was test specific and not dependent on the used antigen as assays using either the nucleocapsid or spike-protein (i.e., both Euroimmun assays) were able to demonstrate an equally high specificity. An acute infection with EBV did not lead to cross-reactivation except in two assays (Epitope Diagnostics and Vircell). Additionally, the presence of high levels of antibodies against influenza viruses A and B and adenovirus were only associated with a marked cross-reactivation in one of the assays (Epitope Diagnostics). The specificity of the assays was closest to the ones stated by the manufacturers when randomly selected samples from the diagnostic routine were used, equivalent to test validations using random blood donor samples. However, we show that a cross-reaction panel is necessary to thoroughly characterize the performance of a serological assay.
The sensitivity of all investigated assays was estimated to be above 90%. Especially, when only samples after 15 days of symptom onset were considered, the sensitivity was similar between the different assays. Additionally, based on an assumed 5% prevalence of SARS-CoV-2 infected individuals, the negative predictive value of all assays was above 99%. The highest sensitivities were calculated for the assays of Euroimmun (NCP) and Vircell. The comparison to other studies, however, is difficult due to the wide spectrum of collection time points after symptom onset. Means to increase sensitivity, may include the use of two assays as reported previously (Gudbjartsson et al., 2020). However, these results underline the recommendation by the World Health Organization not to apply serological assays for the determination of acute infections (WHO). Spearman-Rho correlation coefficients were moderate for most of the compared assay combinations. The highest coefficient was detected for the S-based assays of Diasorin and Euroimmun, which was also reported before (Pflüger et al., 2020), followed by the N-based assays of Abbott and Euroimmun. The best correlation of two assays using different proteins was calculated for both assays of Euroimmun. Interestingly, the assay of Roche only showed a single correlation that was above 0.5 with one of the other assays (Abbott). It is the only pan-Ig assay included in this study and thus, this may be due to an unknown proportion of IgA and IgM reactivity of the assay result. This may be an indication for different areas of application for different test systems: assays for the detection of (formerly infected) individuals and assays that allow the estimation of the level of the immune response after infection or upon vaccination (Kohmer et al., 2020; Weidner et al., 2020). For the later, S1-based assays may be most suitable since the neutralizing immunity is directed towards the spike protein. Interassay correlation may be a requirement for the establishment of international standards. However, correlation of a positive result to immunity is still not well understood. In order to infer humoral immunity, a virus neutralization assay needs to be applied, but a variable degree of correlation was reported (Bonelli et al., 2020; Jiang et al., 2020; Peterhoff et al., 2020). Furthermore, even neutralization tests may not allow a reliable prediction of the immunity against SARS-CoV-2 and its duration.
There are several limitations of this study. The seroprevalence for each of the endemic coronaviruses did not reach 100% in the preselected samples. Unfortunately, there is a lack of thoroughly validated test systems for the detection of the serological response towards endemic coronaviruses, which is also acknowledged by the manufacturer of the used assay. Thus, the results need to be interpreted with caution as neither false negative results nor cross-reactivity can be ruled out. However, the time frame of samples after the infection with an endemic coronavirus used in this study is similar to ones described before (Okba et al., 2020). Additionally, no seroconversion panel was included in this study to adequately describe the sensitivity in different periods after symptom onset. Nevertheless, the analysis of eighty samples allowed a valuable comparison of the investigated assays.
In conclusion, high specificity is of paramount importance and should be the main criteria for the choice of a respective test system. The investigated assays by Abbott, DiaSorin, Euroimmun, and Roche showed the highest estimated specificity, especially with regard to other human Coronaviruses. Other assays may be applied with a subsequent validation by the aforementioned tests.
Author contribution
Study design: UGL; Data collection and testing: MH, CL, MN, MM, CP, TB, ND, TG, VN; Data analysis: MH, UGL; Data interpretation: MH, MM, CL, AD, UGL; Writing: MH, MM, CP, TB, TG, AD, UGL.
Funding
Parts of this study were supported by a grant from the State Parliament of the Free State of Saxony to A.D.
Conflict of interest
All authors have no conflict of interest to declare.
References
- Alharbi SA, Almutairi AZ, Jan AA, Alkhalify AM. Enzyme-linked immunosorbent assay for the detection of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) IgM/IgA and IgG antibodies among healthcare workers. Cureus. 2020;12:e10285. doi: 10.7759/cureus.10285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonelli F, Sarasini A, Zierold C, Calleri M, Bonetti A, Vismara C, et al. Clinical and analytical performance of an automated serological test that identifies S1/S2-Neutralizing IgG in COVID-19 patients semiquantitatively. J Clin Microbiol. 2020;58 doi: 10.1128/JCM.01224-20. e01224-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryan A, Pepper G, Wener MH, Fink SL, Morishima C, Chaudhary A, et al. Performance characteristics of the abbott architect SARS-CoV-2 IgG assay and seroprevalence in boise, Idaho. J Clin Microbiol. 2020;58 doi: 10.1128/JCM.00941-20. e00941-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bundschuh C, Egger M, Wiesinger K, Gabriel C, Clodi M, Mueller T, et al. Evaluation of the EDI enzyme linked immunosorbent assays for the detection of SARS-CoV-2 IgM and IgG antibodies in human plasma. Clin Chim Acta. 2020;509:79–82. doi: 10.1016/j.cca.2020.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Che X-Y, Qiu L-W, Liao Z-Y, Y-d Wang, Wen K, Pan Y-X, et al. Antigenic cross-reactivity between severe acute respiratory syndrome-associated coronavirus and human coronaviruses 229E and OC43. J Infect Dis. 2005;191:2033–2037. doi: 10.1086/430355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman VM, Landt O, Kaiser M, Molenkamp R, Meijer A, Chu DK, et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25 doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J, Li F, Shi Z-L. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol. 2019;17:181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Di, T Wu, Liu Q, Yang Z. The SARS-CoV-2 outbreak: what we know. Int J Infect Dis. 2020;94:44–48. doi: 10.1016/j.ijid.2020.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espejo AP, Akgun Y, Al Mana AF, Tjendra Y, Millan NC, Gomez-Fernandez C, et al. Review of current advances in serologic testing for COVID-19. Am J Clin Pathol. 2020;154:293–304. doi: 10.1093/ajcp/aqaa112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaunt ER, Hardie A, Claas ECJ, Simmonds P, Templeton KE. Epidemiology and clinical presentations of the four human coronaviruses 229E, HKU1, NL63, and OC43 detected over 3 years using a novel multiplex real-time PCR method. J Clin Microbiol. 2010;48:2940–2947. doi: 10.1128/JCM.00636-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudbjartsson DF, Helgason A, Jonsson H, Magnusson OT, Melsted P, Norddahl GL, et al. Spread of SARS-CoV-2 in the Icelandic Population. N Engl J Med. 2020;382:2302–2315. doi: 10.1056/NEJMoa2006100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haselmann V, Kittel M, Gerhards C, Thiaucourt M, Eichner R, Costina V, et al. Comparison of test performance of commercial anti-SARS-CoV-2 immunoassays in serum and plasma samples. Clin Chim Acta. 2020;510:73–78. doi: 10.1016/j.cca.2020.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herroelen PH, Martens GA, Smet D de Swaerts K, Decavele A-S. Humoral immune response to SARS-CoV-2. Am J Clin Pathol. 2020;154:610–619. doi: 10.1093/ajcp/aqaa140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jääskeläinen AJ, Kekäläinen E, Kallio-Kokko H, Mannonen L, Kortela E, Vapalahti O, et al. Evaluation of commercial and automated SARS-CoV-2 IgG and IgA ELISAs using coronavirus disease (COVID-19) patient samples. Euro Surveill. 2020;25 doi: 10.2807/1560-7917.ES.2020.25.18.2000603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang C, Wang Y, Hu M, Wen L, Wen C, Wang Y, et al. Antibody seroconversion in asymptomatic and symptomatic patients infected with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Clin Transl Immunol. 2020;9:e1182. doi: 10.1002/cti2.1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellam P, Barclay W. The dynamics of humoral immune responses following SARS-CoV-2 infection and the potential for reinfection. J Gen Virol. 2020;101:791–797. doi: 10.1099/jgv.0.001439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohmer N, Westhaus S, Rühl C, Ciesek S, Rabenau HF. Brief clinical evaluation of six high-throughput SARS-CoV-2 IgG antibody assays. J Clin Virol. 2020;129 doi: 10.1016/j.jcv.2020.104480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krüttgen A, Cornelissen CG, Dreher M, Hornef M, Imöhl M, Kleines M. Comparison of four new commercial serologic assays for determination of SARS-CoV-2 IgG. J Clin Virol. 2020;128 doi: 10.1016/j.jcv.2020.104394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R, Zhao X, Li J, Niu P, Yang B, Wu H, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet North Am Ed. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motley MP, Bennett-Guerrero E, Fries BC, Spitzer ED. Review of viral testing (Polymerase Chain Reaction) and antibody/serology testing for severe acute respiratory syndrome-coronavirus-2 for the intensivist. Crit Care Explor. 2020;2:e0154. doi: 10.1097/CCE.0000000000000154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okba NMA, Müller MA, Li W, Wang C, GeurtsvanKessel CH, Corman VM, et al. Severe Acute Respiratory Syndrome Coronavirus 2-Specific antibody responses in Coronavirus Disease 2019 patients. Emerging Infect Dis. 2020;26 doi: 10.3201/eid2607.200841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou X, Liu Y, Lei X, Li P, Mi D, Ren L, et al. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat Commun. 2020;11:1620. doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterhoff D, Glück V, Vogel M, Schuster P, Schütz A, Neubert P, et al. A highly specific and sensitive serological assay detects SARS-CoV-2 antibody levels in COVID-19 patients that correlate with neutralization. Infection. 2020 doi: 10.1007/s15010-020-01503-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pflüger LS, Bannasch JH, Brehm TT, Pfefferle S, Hoffmann A, Nörz D, et al. Clinical evaluation of five different automated SARS-CoV-2 serology assays in a cohort of hospitalized COVID-19 patients. J Clin Virol. 2020;130 doi: 10.1016/j.jcv.2020.104549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team . R Foundation for Statistical Computing; Vienna, Austria: 2018. R: A language and environment for statistical computing.https://www.R-project.org/ Available from: [Google Scholar]
- B Schloerke, Di Cook, J Larmarange, F Briatte, M Marbach, E Thoen, et al. GGally: extension to 'ggplot2′. Available from: https://CRAN.R-project.org/package=GGally (2018).
- Su S, Wong G, Shi W, Liu J, Lai ACK, Zhou J, et al. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol. 2016;24:490–502. doi: 10.1016/j.tim.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viner RM, Whittaker E. Kawasaki-like disease: emerging complication during the COVID-19 pandemic. Lancet North Am Ed. 2020 doi: 10.1016/S0140-6736(20)31129-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh EE, Shin JH, Falsey AR. Clinical impact of human coronaviruses 229E and OC43 infection in diverse adult populations. J Infect Dis. 2013;208:1634–1642. doi: 10.1093/infdis/jit393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wechselberger C, Süßner S, Doppler S, Bernhard D. Performance evaluation of serological assays to determine the immunoglobulin status in SARS-CoV-2 infected patients. J Clin Virol. 2020;131 doi: 10.1016/j.jcv.2020.104589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidner L, Gänsdorfer S, Unterweger S, Weseslindtner L, Drexler C, Farcet M, et al. Quantification of SARS-CoV-2 antibodies with eight commercially available immunoassays. J Clin Virol. 2020;129 doi: 10.1016/j.jcv.2020.104540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitman JD, Hiatt J, Mowery CT, Shy BR, Yu R, Yamamoto TN, et al. Test performance evaluation of SARS-CoV-2 serological assays. medRxiv. 2020 [Google Scholar]
- WHO. Diagnostic testing for SARS-CoV-2 (Last accessd January 4 2021). Available from: https://www.who.int/publications/i/item/diagnostic-testing-for-sars-cov-2.
- World Health Organization. Coronaviurs Disease (COVID-19) Situation Report, Available https://www.who.int/publications/m/item/weekly-epidemiological-update—16-february-2021/situation-reports/20200810-covid-19-sitrep-203.pdf?sfvrsn=aa050308_2.
- Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, et al. A Novel Coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuwała K, Golda A, Kabala W, Burmistrz M, Zdzalik M, Nowak P, et al. The nucleocapsid protein of human coronavirus NL63. PLoS One. 2015;10 doi: 10.1371/journal.pone.0117833. [DOI] [PMC free article] [PubMed] [Google Scholar]