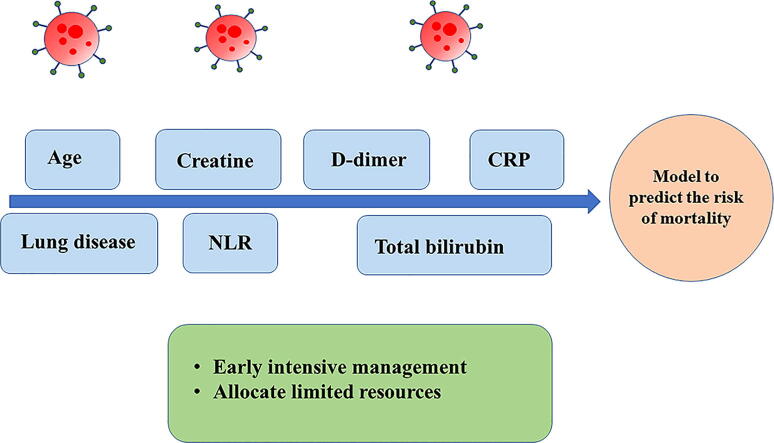
Graphical abstract
Keywords: COVID-19, Severe, Critical, Risk, Mortality, Predictive model
Abstract
Background
To investigate and select the useful prognostic parameters to develop and validate a model to predict the mortality risk for severely and critically ill patients with the coronavirus disease 2019 (COVID-19).
Methods
We established a retrospective cohort of patients with laboratory-confirmed COVID-19 (≥18 years old) from two tertiary hospitals: the People’s Hospital of Wuhan University and Leishenshan Hospital between February 16, 2020, and April 14, 2020. The diagnosis of the cases was confirmed according to the WHO interim guidance. The data of consecutive severely and critically ill patients with COVID-19 admitted to these hospitals were analyzed. A total of 566 patients from the People’s Hospital of Wuhan University were included in the training cohort and 436 patients from Leishenshan Hospital were included in the validation cohort. The least absolute shrinkage and selection operator (LASSO) and multivariable logistic regression were used to select the variables and build the mortality risk prediction model.
Results
The prediction model was presented as a nomograph and developed based on identified predictors, including age, chronic lung disease, C-reactive protein (CRP), D-dimer levels, neutrophil-to-lymphocyte ratio (NLR), creatinine, and total bilirubin. In the training cohort, the model displayed good discrimination with an AUC of 0.912 [95% confidence interval (CI): 0.884–0.940] and good calibration (intercept = 0; slope = 1). In the validation cohort, the model had an AUC of 0.922 [95% confidence interval (CI): 0.891–0.953] and a good calibration (intercept = 0.056; slope = 1.161). The decision curve analysis (DCA) demonstrated that the nomogram was clinically useful.
Conclusion
A risk score for severely and critically ill COVID-19 patients' mortality was developed and externally validated. This model can help clinicians to identify individual patients at a high mortality risk.
1. Background
The city of Wuhan witnessed a large outbreak of an epidemic respiratory disease of unknown cause since December 2019. This worldwide epidemic disease represents the third coronavirus outbreak within 20 years, following the severe acute respiratory syndrome (SARS) and the Middle East respiratory syndrome (MERS). A novel coronavirus was isolated from the infected patients and identified to be the causative agent; it was named the severe acute respiratory syndrome coronavirus 2 (SARS-CoV2; previously known as 2019-nCoV) and the disease was called the COVID-19 disease.
Most patients with COVID-19 had mild symptoms, while severe and critical cases were reported in 19% of the infected patients [1]. A severe COVID-19 infection may lead to fatal consequences, such as respiratory failure and even death in the absence of timely diagnosis and treatment. Predicting the risk of death for severe COVID-19 patients and ensuring that they receive timely and appropriate management represent difficult challenges for health care providers. Identifying the risk factors for severe patients is of great importance, as it may reduce the mortality rate and facilitate efficient utilization of the medical resources that might be insufficient. Although some risk factors have been reported to be associated with the mortality of COVID-19 patients, the earlier studies may be biased since they included mildly ill patients because, almost no death incidents were reported among non-severe patients. Based on existing studies and our initial analysis of the data and clinical experience, we developed and validated a nomogram to predict the mortality of patients with severe and critical COVID-19. The proposed predictive model will help to identify COVID-19 patients at high risk of death for early intensive management and assist in allocating limited medical resources.
2. Methods
2.1. Study cohort and population
This is a retrospective single-center cohort study of adult inpatients (≥18 years old) with laboratory-confirmed COVID-19 from the People’s Hospital of Wuhan University and Leishenshan Hospital between February 16, 2020, and April 14, 2020. Patients who were treated at the People’s Hospital of Wuhan University were included in the derivation cohort, whereas patients from Leishenshan Hospital were included in the validation cohort.
The diagnosis of the cases was confirmed according to the WHO interim guidance. The types of the novel coronavirus pneumonia were classified as mild, common, severe and critically severe, such that the classification was defined according to the guideline of the National Health Committee of the People's Republic of China (version 7.0, http://www.nhc.gov.cn/). Severe cases should meet any of the following criteria: 1. Respiratory distress, a respiratory rate of 30 beats/min; 2. Oxygen saturation ≤93% in the resting state; 3. Arterial blood oxygen partial pressure (PaO2) / oxygen concentration (FiO2) ≤ 300 mmHg (1 mmHg = 0.133 kPa); 4. Lung involvement on imaging >50% within 24 to 48 h. Critically severe cases were defined as at least one of the followings: the patients who met any of the following criteria: 1. Respiratory failure requiring mechanical ventilation; 2. Shock; 3. Other forms of organ failure requiring admission to the intensive care unit (ICU).
In this study, only severe and critical patients older than 18 years old with laboratory-confirmed COVID-19 were included, while the patients with missing data on relevant predictors or follow-up data were excluded. Based on the inclusion and exclusion criteria, 1002 patients with COVID-19 were included in the study. All the enrolled patients had a definite outcome.
2.2. Predictors and outcomes
All COVID-19 cases were confirmed with a positive result of real-time reverse-transcriptase polymerase-chain-reaction (RT-PCR) assay for nasal and pharyngeal swab specimens. The data of the patients were retrospectively collected from the electronic medical records, including the following data: demographic information (age, sex, date of hospital admission, date of development of severe and critical illness, hospital discharge date or date of death), previous comorbidities (diabetes mellitus, hypertension, cerebrovascular disease, malignancy, chronic lung disease, cardiovascular disease, chronic kidney disease), laboratory test results and chest computed tomography (CT) findings within 24 h of admission, vital signs at the time of admission and treatment details. For the patients who had two relapses during the study period, only the first data were included. The endpoint was all-cause inhospital mortality.
2.3. Model development and validation
All the clinical data were already anonymized before being cleaned and analyzed. We used the reported studies and systematic reviews to identify the predictors of death in severe and critical COVID-19 cases, which could be easily ascertained by clinicians with clinical experience and part of patients' routine tests [2], [3], [4], [5], [6]. In order to identify any potential novel risk factors that were not previously reported, we also used the least absolute shrinkage and selection operator (LASSO). For small datasets with a low events per variables (EPV) ratio, LASSO is more appropriate than the traditional stepwise regression [7], and it is suitable for regression models with high-dimensional predictors [8]. The final prediction model was constructed by a logistic regression model with predictors identified from LASSO regression and presented as a nomograph.
The discrimination of the model was evaluated using the area under the receiver-operator characteristic curve (AUC), which evaluates the ability of the model to predict future outcomes. The closer the AUC is to 1, the better the model performance, and a value of 0.5 indicates that the ability of the model to discriminate is due to chance.
The calibration of the model was measured using calibration curve, and expressed as the calibration intercept and slope. A calibration intercept of 0 and a calibration slope of 1 indicates perfect calibration.
Furthermore, decision curve analysis (DCA) was detected to evaluate potential clinical effects of the nomogram and the scope of application.
Statistical AnalysisContinuous variables were reported as means (±SDs) or medians (with interquartile ranges [IQRs]) based on the data distribution, while categorical variables were expressed as frequencies (with percentages). The baseline characteristics between groups were compared using the χ2 test for categorical variables and the analysis of variance or Kruskal–Wallis tests for continuous variables where appropriate.
All the tests were two-sided, and a P value of < 0.05 was considered to be statistically significant. All Statistical analysis was performed using the R statistical software (R version 3.6.2).
3. Results
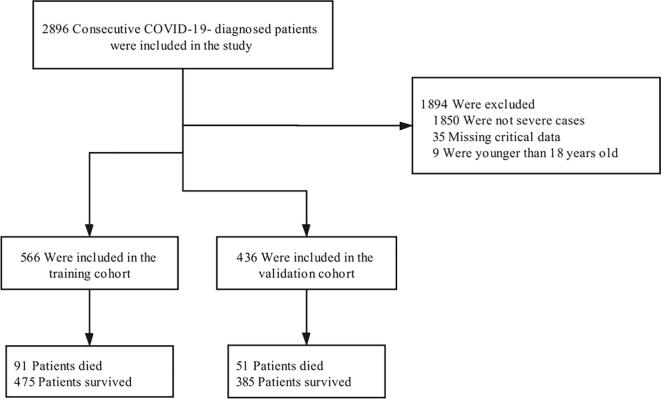
A total of 2896 patients with COVID-19 between February 16, 2020 and April 14, 2020 were included in this retrospective cohort study (Supplementary Fig. 1). The analysis included 1002 severe and critical cases from two hospitals to be evaluated. The factors of gender, comorbidities except for cardiovascular disease, platelet count, white blood cell count, neutrophil count, procalcitonin level, ALT (alanine aminotransferase) level and D-dimer level did not significantly differ between the training cohort and validation cohort. The characteristics of the patients with severe COVID-19 in the training and validation cohorts are shown in Table 1 and Supplementary Table 1.
Table 1.
Demographic and clinical features of study population by mortality.
| Training cohort (n = 566) |
Validation cohort (n = 436) |
|||||
|---|---|---|---|---|---|---|
| Characteristic | Survivors (n = 475) | Non-survivors (n = 91) | p value | Survivors (n = 385) | Non-survivors (n = 51) | p value |
| Age, y | 58.6 ± 14.0 | 71.6 ± 13.8 | <0.001 | 64.4 ± 13.1 | 72.7 ± 11.4 | <0.001 |
| Gender | ||||||
| Female | 241 (50.7%) | 32 (35.2%) | 0.006 | 171 (44.42%) | 22 (43.14%) | 0.863 |
| Male | 234 (49.3%) | 59 (64.8%) | 214 (55.58%) | 29 (56.86%) | ||
| SBP, mmHg | 130.01 ± 18.30 | 133.24 ± 23.12 | 0.141 | 133.72 ± 18.43 | 133.88 ± 18.36 | 0.953 |
| DBP, mmHg | 77.76 ± 11.70 | 79.10 ± 14.38 | 0.338 | 81.06 ± 11.74 | 77.53 ± 12.31 | 0.061 |
| Symptoms | ||||||
| Fever | 388 (81.68%) | 72 (79.12%) | 0.566 | 242 (62.86%) | 30 (58.82%) | 0.576 |
| Cough | 305 (64.21%) | 53 (58.24%) | 0.279 | 228 (59.22%) | 23 (45.10%) | 0.055 |
| Fatigue | 152 (32.00%) | 35 (38.46%) | 0.230 | 167 (43.38%) | 14 (27.45%) | 0.030 |
| Headache | 24 (5.05%) | 1 (1.10%) | 0.093 | 19 (4.94%) | 2 (3.92%) | 0.751 |
| Diarrhea | 78 (16.42%) | 11 (12.09%) | 0.298 | 37 (9.61%) | 2 (3.92%) | 0.181 |
| Dyspnea | 168 (35.37%) | 42 (46.15%) | 0.051 | 89 (23.12%) | 21 (41.18%) | 0.005 |
| Comorbidities | ||||||
| Hypertension | 153 (32.2%) | 47 (51.7%) | <0.001 | 144 (37.40%) | 24 (47.06%) | 0.183 |
| Diabetes | 66 (13.9%) | 15 (16.5%) | 0.518 | 62 (16.10%) | 10 (19.61%) | 0.527 |
| Chronic lung disease | 15 (3.2%) | 13 (14.3%) | <0.001 | 17 (4.42%) | 6 (11.76%) | 0.027 |
| Cardiovascular disease | 36 (7.6%) | 17 (18.7%) | <0.001 | 71 (18.44%) | 15 (29.41%) | 0.064 |
| Malignancy | 12 (2.53%) | 5 (5.49%) | 0.129 | 6 (1.56%) | 5 (9.80%) | <0.001 |
| Laboratory findings on admission | ||||||
| White blood cell, × 109/L | 6.26 ± 3.63 | 9.94 ± 6.98 | <0.001 | 6.39 ± 3.03 | 10.16 ± 5.17 | <0.001 |
| Lymphocyte, × 109/L | 1.28 ± 0.67 | 0.66 ± 0.34 | <0.001 | 1.33 ± 0.57 | 0.71 ± 0.38 | <0.001 |
| Neutrophil, × 109/L | 4.39 ± 3.06 | 8.85 ± 6.81 | <0.001 | 4.37 ± 2.86 | 8.74 ± 5.01 | <0.001 |
| NLR | 4.87 ± 5.49 | 16.19 ± 11.71 | <0.001 | 4.33 ± 4.41 | 18.22 ± 19.30 | <0.001 |
| Hemoglobin, g/L | 124.43 ± 16.13 | 120.74 ± 22.40 | 0.360 | 115.18 ± 21.14 | 114.29 ± 26.89 | 0.786 |
| Platelet, × 109/L | 233.07 ± 88.19 | 179.40 ± 105.07 | <0.001 | 238.20 ± 96.46 | 187.02 ± 111.46 | <0.001 |
| PCT, ng/mL | 1.24 ± 17.29 | 2.50 ± 9.66 | <0.001 | 0.37 ± 2.59 | 1.71 ± 6.27 | 0.006 |
| AST, U/L | 35.58 ± 76.31 | 49.59 ± 38.58 | <0.001 | 28.58 ± 35.31 | 149.38 ± 597.64 | <0.001 |
| Total bilirubin, μmol/L | 12.29 ± 7.00 | 18.12 ± 11.82 | <0.001 | 10.09 ± 5.84 | 14.71 ± 10.74 | <0.001 |
| Albumin, g/L | 37.62 ± 4.74 | 34.46 ± 5.67 | <0.001 | 34.83 ± 4.34 | 29.92 ± 4.07 | <0.001 |
| Creatinine, μmol/L | 67.08 ± 59.47 | 122.32 ± 190.02 | <0.001 | 111.32 ± 177.38 | 133.48 ± 178.21 | 0.022 |
| CRP, mg/L | 35.35 ± 47.24 | 99.88 ± 66.75 | <0.001 | 15.21 ± 27.87 | 65.80 ± 51.70 | <0.001 |
| D-dimer, μg/mL | ||||||
| ≤1 | 292 (61.5%) | 17 (18.7%) | <0.001 | 227 (58.96%) | 9 (17.65%) | <0.001 |
| >1 | 183 (38.5%) | 74 (81.3%) | 158 (41.04%) | 42 (82.35%) | ||
Abbreviation: SBP, systolic blood pressure; DBP, diastolic blood pressure; RR, respiratory rate; HR, heart rate; NLR, neutrophil-to-lymphocyte ratio; PCT, procalcitonin; ALT, alanine aminotransferase; AST, aspartate aminotransferase; CRP, C-reactive protein.
3.1. Characteristics of the training and validation cohorts
We screened 566 severe COVID-19 cases out of 1096 cases from the People’s Hospital of Wuhan University to be included in the training cohort. All the patients’ discharge status had been recorded, and the overall mortality rate of the training cohort was 8.3%, reaching 16.1% in severely ill cases. The median age was 60.7 (±14.7) years, the age of non-survivors was higher than that of the survivors (71.6 vs. 58.6 years, P < 0.001). Among the 566 severely ill cases, 273 (48.2%) were females and 293 (51.8%) were males. No significant difference was observed in the symptoms between the survivors and non-survivors, but comorbidities, including hypertension, chronic lung disease and cardiovascular disease, were more common in the non-survivors compared with the survivors. As for the laboratory findings, non-survivors had higher white blood cell count and NLR and lower lymphocyte and platelet count compared with the survivors. Regarding the inflammation markers, such as PCT (procalcitonin) and CRP (C-reactive protein), they were higher in non-survivors. As for the liver function tests, non-survivors had significantly higher levels of AST (aspartate aminotransferase), total bilirubin and creatinineand a remarkably lower level of albumin. The elevated D-dimer level was higher in non-survivors.
The validation cohort included 436 patients with a mean age of 65.3 (±13.2) years, among which 193 (44.27%) were females and 243 (55.73%) were males. A total of 51 patients eventually died, the overall mortality rate of the validation cohort was 5.5%, and the mortality rate of the severely ill cases was 11.7%.
3.2. Development of the prediction model
A total of 35 variables measured at admission to the hospital were collected for analysis, among which potential predictor variables were included in the LASSO regression after excluding irrelevant variables. After conducting the LASSO regression selection (Supplementary Fig. 2A and Supplementary Fig. 2B), the following 7 variables with nonzero coefficients were chosen as risk predictors: age, chronic lung disease, NLR, creatinine, CRP, D-dimer and total bilirubin. Combined with the reported studies and clinical experience, these factors were finally included to build the prediction model using logistic regression, which showed that 5 variables were statistically significant predictors of the risk of mortality [age (OR, 1.065; 95%CI, 1.037–1.094; P < 0.001), NLR (OR, 1.110; 95%CI, 1.069–1.153; P < 0.001), creatinine (OR, 1.003; 95%CI, 1.000–1.005; P = 0.032), CRP (OR, 1.011; 95%CI, 1.006–1.016; P < 0.001), total bilirubin (OR, 1.035; 95%CI, 1.000–1.071; P = 0.047)] (Supplementary Table 2).
3.3. Model performance in the derivation cohort
A nomogram was created to predict the individual probability of mortality in severely ill COVID-19 patients (Fig. 1). Higher total points, calculated by the sum of the assigned number of points for each variable in the nomogram, were associated with a higher mortality. The discrimination of the model was measured with the area under the ROC curve (AUC), such that the closer the AUC to 1, the better the discrimination. The AUC for the prediction nomogram was 0.912 (95% CI: 0.884–0.940) for the training cohort (Fig. 2A), and 0.903 as calculated through bootstrapping validation (1000 bootstrap resamples). In order to assess the calibration of the model, the calibration curve was plotted (intercept = 0; slope = 1), which measures the relationship between the risk of mortality predicted by the model and the observed risk of mortality in the training cohort (Fig. 3A). The 45° line indicates the best calibration, while a predicted line above or below the 45° line indicates an underprediction or overprediction of the patients’ actual risk. Fig. 4 shows the decision curve analysis (DCA) for the nomogram, which demonstrated that the nomogram was clinically useful at a threshold probability of >1%.
Fig. 1.
The nomogram to predict the risk of mortality in severely ill COVID-19 patients was created based on seven independent prognostic factors.
Fig. 2.
ROC for predicting the mortality among severely ill COVID-19 patients in the training cohort (A) and validation cohort (B). ROC, receiver operator characteristic; AUC, the area under the receiver operator characteristic curve.
Fig. 3.
The calibration curve for the prediction of the mortality risk in severely ill COVID-19 patients in the training cohort (A) and validation cohort (B).
Fig. 4.

Decision curve analysis for the non-adherence nomogram.
3.4. Model performance in the validation cohort
The external validation was performed using data from the Leishenshan Hospital representing the external validation set. In this set, the final logistic regression model for mortality showed a strong external validity, with a discrimination AUC of 0.922 (95% CI, 0.891–0.953) (Fig. 2B). Fig. 3B shows the calibration curve, which visually assesses the performance of the model, and it shows a good calibration (intercept = 0.056; slope = 1.161). The overall accuracy was measured using the Brier Score, such that the lower the Brier score, the more accurate the overall performance of the model. The Brier Score of the model was 0.07, showing a good overall performance.
4. Discussion
The global outbreak of the novel coronavirus disease COVID-19 caused a global threat, which led to an urgent increased demand for medical resources, especially hospital beds and medical equipment. In China, in order to tackle the COVID-19 outbreak crisis, the patients with mild to moderate COVID-19 infection were isolated from their families and communities and sent to Fangcang shelter hospitals, which are large temporary hospitals converted from existing public venues and rapidly built to provide numerous resources for the patients with non-severe disease. These large-scale hospitals were considered to be a major reason for the successful COVID-19 control in China as suggested by the Chinese experience [9]. However, only basic medical care for mild and moderate patients can be offered in these hospitals, while severely ill patients should be referred to higher-level hospitals. Prognostic models can help to reduce mortality and support medical decision when allocating healthcare resources. Most patients with COVID-19 did not have a severe infection, as a mild infection was reported in 81% of the cases, while 14% were severely ill and 5% were critically ill, and the overall mortality rate was 2.3% [1]. In contrast to the previously published mortality-prediction models [3], [10], [11], our study focused on the severely ill patients, since almost no deaths were reported among non-severe cases. In addition, previous prognostic models are considered to be at high risk of bias because they included non-representative control patients, excluded the patients who did not experience the endpoint for the clinical event by the end of the study, suffered from overfitting and no external validation was performed in most of them due to the small number of cases [12]. Considering these issues, we developed and externally validated a novel practical model to identify severely ill COVID-19 patients who had a high risk of mortality. The development and validation of the model followed the established recommendations of the transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD) statement [13]. A list of candidate predictors consisting of the clinical and demographic features of the patients were carefully selected, such that we excluded the parameters with large missing values or that are not routinely assessed in clinical practice. We did not select predictors only using LASSO, but also based on previous literature and experts’ opinion. The model consists of seven variables at admission that are easily obtained: the patient age, history of chronic lung disease, NLR, CRP, creatinine, total bilirubin and D-dimer level. All of the seven selected predictors were driven from our data and were also reported by previous studies. The model exhibited good discriminatory performance and calibration, it was then successfully externally validated and showed satisfying discrimination and calibration as well. Future studies might refine the predictors by selecting other features driven form the CT scans, lymphocyte subtypes, organ damage markers and cytokines.
The other particular strength of our study is that all of our patients had experienced the event of interest by the end of the present study and there were no COVID-19 patients at the two hospitals at which the patients were recruited any more, which decreased the risk of bias. The good performance of the model in external validation and the inclusion of a large sample size supports the clinical use of this nomogram for the prediction of mortality risk in severely ill COVID-19 patients.
All the predictors were routinely available, quickly measured and relatively inexpensive. We also took into account several variables that were previously reported to be associated with the outcome of the COVID-19 disease for their inclusion in the formal nomograms. A lot of risk factors were reported by recent studies to be associated with the prognosis of the disease severity, but it’s difficult to evaluate the factors that determine the outcome of disease. Some studies reported that older age, high SOFA score, D-dimer >1 µg/mL, lymphopenia, LDH and C-reactive protein were associated with a poor prognosis [2], [14], [15], [16]. A meta-analysis reported that comorbidities in COVID-19 patients, including hypertension, chronic lung disease and cardiovascular disease, may represent a risk factor for a severe infection [17]. We also found that the factors of age, comorbidities, white blood cell count, neutrophil count, lymphocyte count, NLR, creatinine, total bilirubin and D-dimer were strongly linked to death. Different from previous studies, our findings showed that hypertension or cardiovascular disease did not represent risk factors for death in severe and critical COVID-19 patients.
In this study, the lymphocyte count was significantly reduced in non-survivors, which suggested that the immune response might contribute to the disease progression. Accumulating evidence suggested that the cytokine storm syndrome played an important role in the disease progression to a severe COVID-19 infection, mortality may be caused by a virally induced hyperinflammation [15], [18]. However, in this study, we did not analyse cytokine because of the missing data. Age was known to be a risk factor for death in COVID-19 patients, which is maybe because older patients had more comorbidities due to age-related immune dysfunctions. The therapeutic options of immunosuppression, such as corticosteroid, may be effective to prevent the viral induced hyperinflammation and decrease mortality in selective severe patients [19].
The data of this study were derived from severely ill patients, and the model is thus appropriate to be used in severely ill adult patients. Patients with low and medium risk of death are suitable for treatment in a general ward, while patients with high risk of death may require intensive care treatment, including corticosteroid and mechanical ventilation. Our model is useful to identify patients at high risk of death; early identification and consequently timely intervention will lead to an appropriate treatment strategy and may improve the outcome of these patients. Accurate prognostic assessment is also critical to support decisions regarding hospital admission and triaging patients, while allocating limited medical resources and alleviating stress on the health care system.
There were several limitations to our study. Firstly, this is a retrospective cohort study based on Chinese datasets, which may not be representative of COVID-19 patients in other countries due to the racial/ethnic differences; thus, international datasets are needed to externally validate the proposed nomogram. Secondly, this is a retrospective study, and the data driven from electronic medical records were incomplete. However, we tried to overcome this by imputing the missing data, which were already limited, with the random forest method. Thirdly, we didn’t collect data on drugs used for treatment, such as antiviral, antibiotics and traditional Chinese herbs. The effect of different treatments was not considered in the nomogram development. Fourthly, some factors, such as LDH, SOFA score, cytokines and CT findings, were also reported as prognostic markers but were not assessed in our nomogram due to the missing data. Finally, the lab data may change as the disease progresses, inclusion of onset to analysis in the model is not possible due to the nature of retrospective study.
5. Conclusion
This study shows that the prediction of mortality among severely ill COVID-19 patients is possible using simple clinical parameters. Although further confirmation by future studies outside China is needed, it shows that a simple nomogram combining the factors of the patient age, history of chronic lung disease, NLR, CRP, creatinine, as well as the total bilirubin and D-dimer levels is able to calculate the mortality risk of severely ill COVID-19 patients.
CRediT authorship contribution statement
Bo Chen: Conceptualization, Software, Validation, Writing - original draft. Hongqiu Gu: Software, Visualization, Supervision. Yi Liu: Data curation. Guqin Zhang: Data curation. Hang Yang: Data curation. Huifang Hu: Data curation. Chenyang Lu: Data curation. Yang Li: Data curation, Investigation. Liyi Wang: Data curation. Yi Liu: Conceptualization, Methodology, Supervision. Yi Zhao: Conceptualization, Methodology. Huaqin Pan: Conceptualization, Methodology.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgements
The authors would like to express their gratitude to all the medical staff involved in the study for taking care of the patients and make medical records.
Authors’ contributions
Yi Zhao, Yi Liu and Huaqin Pan contributed in study concept and design. Bo Chen and Yang Li contributed in drafting of the article. Hongqiu Gu and Bo Chen contributed in statistical analysis. Yi Liu, Guqin Zhang, Hang Yang, Huifang Hu, Chenyang Lu, Liyi Wang and Yang Li contributed in data collecting, acquisition, and critical revision of the article for important intellectual content.
Funding
Yi Liu received funding from 1.3.5 project for disciplines of excellence, West China Hospital, Sichuan University (Project no. ZYGD18015). Huaqin Pan received funding from National Natural Science Foundation of China (Grant no. 81700493). The remaining authors have disclosed that they do not have any conflicts of interest.
Availability of data and materials
The datasets generated during the current study are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
This study was approved by the Research Ethics Committee of West China Hospital, who waived the requirement for informed consent due to the urgent need to collect the data and the retrospective nature of the study.
Consent for publication
Not applicable.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2021.03.012.
Contributor Information
Yi Liu (刘毅), Email: yi2006liu@163.com.
Yi Zhao, Email: zhao.y1977@163.com.
Huaqin Pan, Email: phq2012@whu.edu.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Supplementary figure 1.
Flowchart summarizing the selection of the training and validation cohorts.
Supplementary figure 2.

LASSO coefficient profiles of the selected features. A coefficient profile plot was produced against the log(λ) sequence, the optimal lambda resulted in seven features with nonzero coefficients.
Supplementary figure 3.
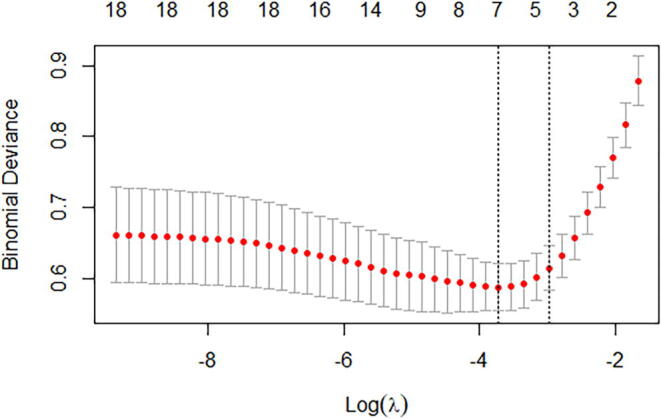
Selection of the optimal variable (λ) in the LASSO regression model used cross-validation via the minimum criteria. The partial likelihood deviance curve was plotted versus log(λ).
References
- 1.Wu Z.Y., Jennifer M.M. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: summary of a report of 72314 cases from the chinese center for disease control and prevention. JAMA. 2020;323(13):1239–1242. doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- 2.Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395(10229):1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cheng A., Hu L., Wang Y., Huang L., Zhao L., Zhang C. Diagnostic performance of initial blood urea nitrogen combined with D-dimer levels for predicting in-hospital mortality in COVID-19 patients. Int J Antimicrob Agents. 2020;56(3):106110. doi: 10.1016/j.ijantimicag.2020.106110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grasselli G., Greco M., Zanella A., Albano G., Antonelli M., Bellani G. Risk factors associated with mortality among patients with COVID-19 in intensive care units in Lombardy, Italy. JAMA Intern Med. 2020;180(10):1345. doi: 10.1001/jamainternmed.2020.3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gupta S., Hayek S.S., Wang W. Factors associated with death in critically ill patients with coronavirus disease 2019 in the US. JAMA Intern Med. 2020;180(11):1–12. doi: 10.1001/jamainternmed.2020.3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li X., Xu S., Yu M., Wang K.e., Tao Y.u., Zhou Y. Risk factors for severity and mortality in adult COVID-19 inpatients in Wuhan. J Allergy Clin Immunol. 2020;146(1):110–118. doi: 10.1016/j.jaci.2020.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Steyerberg E.W., Eijkemans M.J.C., Harrell F.E., Habbema J.D.F. Prognostic modelling with logistic regression analysis: a comparison of selection and estimation methods in small data sets. Stat Med. 2000;19(8):1059–1079. doi: 10.1002/(sici)1097-0258(20000430)19:8<1059::aid-sim412>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 8.Willi S., Patrick R., Harald B. Selection of important variables and determination of functional form for continuous predictors in multivariable model building. Stat Med. 2007;26(30):5512–5528. doi: 10.1002/sim.3148. [DOI] [PubMed] [Google Scholar]
- 9.Chen S., Zhang Z., Yang J., Wang J., Zhai X., Bärnighausen T. Fangcang shelter hospitals: a novel concept for responding to public health emergencies. Lancet. 2020;395(10232):1305–1314. doi: 10.1016/S0140-6736(20)30744-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dong YM, Sun J and Li YX, et al: Development and Validation of a Nomogram for Assessing Survival in Patients with COVID-19 Pneumonia. Clin Infect Dis. 2020 Jul 10: ciaa963. Epub ahead of print. [DOI] [PMC free article] [PubMed]
- 11.Zhang S., Guo M., Duan L., Wu F., Hu G., Wang Z. Development and validation of a risk factor-based system to predict short-term survival in adult hospitalized patients with COVID-19: a multicenter, retrospective, cohort study. Crit Care. 2020;24(1) doi: 10.1186/s13054-020-03123-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Laura W., Ben V.C., Gary S.C. Prediction models for diagnosis and prognosis of covid-19 infection: systematic review and critical appraisal. BMJ. 2020;369:1328. doi: 10.1136/bmj.m1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Collins G.S., Reitsma J.B., Altman D.G., Moons K.G.M. Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): the TRIPOD statement. Eur Urol. 2015;67(6):1142–1151. doi: 10.1016/j.eururo.2014.11.025. [DOI] [PubMed] [Google Scholar]
- 14.Wang D., Hu B.o., Hu C., Zhu F., Liu X., Zhang J. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA. 2020;323(11):1061. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ruan Q., Yang K., Wang W., Jiang L., Song J. Clinical predictors of mortality due to COVID-19 based on an analysis of data of 150 patients from Wuhan, China. Intens care med. 2020;46(5):846–848. doi: 10.1007/s00134-020-05991-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Giuseppe L., Mario P. Laboratory abnormalities in patients with COVID-2019 infection. Clinl Chem Lab Med. 2020;58(7):1131–1134. doi: 10.1515/cclm-2020-0198. [DOI] [PubMed] [Google Scholar]
- 17.Yang J., Zheng Y.a., Gou X.i., Pu K.e., Chen Z., Guo Q. Prevalence of comorbidities and its effects in patients infected with SARS-Cov-2: a systematic review and meta-analysis. Int J Infect Dis. 2020;94:91–95. doi: 10.1016/j.ijid.2020.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mehta P., McAuley D.F., Brown M., Sanchez E., Tattersall R.S., Manson J.J. COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet. 2020;395(10229):1033–1034. doi: 10.1016/S0140-6736(20)30628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.WHO Rapid Evidence Appraisal for COVID-19 Therapies (REACT) Working Group, Sterne JAC and Murthy S, et al: Association between administration of systemic corticosteroids and mortality among critically ill patients with COVID-19: a meta-ANALYSIS. JAMA 2020; 324(13):1330–1341. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during the current study are available from the corresponding author on reasonable request.