Abstract
Detailed phylogenies of tumor populations can recount the history and chronology of critical events during cancer progression, such as metastatic dissemination. We applied a Cas9-based, single-cell lineage tracer to study the rates, routes, and drivers of metastasis in a lung cancer xenograft mouse model. We report deeply resolved phylogenies for tens of thousands of cancer cells traced over months of growth and dissemination. This revealed stark heterogeneity in metastatic capacity, arising from pre-existing and heritable differences in gene expression. We demonstrate that these identified genes can drive invasiveness, and uncovered an unanticipated suppressive role for KRT17. We also show that metastases disseminated via multidirectional tissue routes and complex seeding topologies. Overall, we demonstrate the power of tracing cancer progression at subclonal resolution and vast scale.
One Sentence Summary:
Single-cell lineage tracing and RNA-seq capture diverse metastatic behaviors and drivers in lung cancer xenografts in mice.
Cancer progression is governed by evolutionary principles (reviewed in (1)), which leave clear phylogenetic signatures upon every step of this process (2, 3), from early acquisition of oncogenic mutations (i.e., the relationships between normal and malignantly transformed cells (4)), to metastatic colonization of distant tissues (i.e., the relationship between a primary tumor and metastases (5)), and finally adaptation to therapeutic challenges (i.e., the relationship between drug-sensitive or -resistant populations (6)). Metastasis is a particularly critical step in cancer progression to study because it is chiefly responsible for cancer-related mortality (7). Yet because metastatic events are intrinsically rare, transient, and stochastic (8, 9), they have proved challenging to monitor in real time. Analogous to the cell fate maps that have deepened our understanding of organismal development and cell type differentiation (10, 11), accurately reconstructed phylogenetic trees of tumors and metastases can reveal key features of this process, such as the clonality, timing, frequency, origins, and destinations of metastatic seeding (12).
Lineage tracing techniques allow one to map the genealogy of related cells, providing a crucial tool for exploring the phylogenetic principles of biological processes like cancer progression and metastasis. Classical lineage tracing strategies can infer tumor ancestry from the pattern of shared sequence variations across tumor subpopulations (e.g., naturally occurring mutations, like single-nucleotide polymorphisms or copy-number variations) (13, 14). These “retrospective” tracing approaches are particularly valuable for studying the subclonal dynamics of cancer in patient-derived samples, such as elucidating which mutations contribute to metastasis and when they occur (15-18). However, the resolution of these approaches is limited by the number of distinguishing natural mutations, and the conclusions can be confounded by incomplete or impure bulk tumor sampling (19), sequencing artifacts (20), varying levels of intratumor heterogeneity, and non-neutral mutations (1, 5). Alternatively, so-called “prospective” lineage tracing approaches – wherein cells are marked with a static label, like a genetic barcode or fluorescent tag – can measure gross population dynamics at clonal resolution (21), but cannot resolve important and fine subclonal features of cancer biology, like evolution and the rate, order, and directionality of metastatic events.
The recent development of Cas9-enabled lineage tracing techniques with single-cell RNA-sequencing readouts (22-26) provides the potential to explore cancer progression at vastly larger scales and finer resolution than has been previously possible with classical prospective or retrospective tracing approaches. These methods rely on similar technical principles (reviewed in (27, 28)). Briefly, Cas9 cuts a defined genomic locus (hereafter “Target Site”), resulting in a stable insertion/deletion (indel) “allele” that is inherited over subsequent generations; as the cells divide, they accrue more Cas9-induced indels at additional sites that further distinguish successive clades of cells (Figs. 1A and S1). At the end of the lineage tracing experiment, the indel alleles are collected from individual cells by sequencing and paired with single-cell expression profiles of the cell state (22, 23). Then, as in retrospective tracing approaches, computational approaches (29-34) can reconstruct a phylogenetic tree that best models subclonal cellular relationships (e.g., by maximum-parsimony) from the observed shared or distinguishing alleles. Thus far, Cas9-enabled tracing has been successfully applied to study the cellular progenitor landscape in early mammalian embryogenesis (23, 35), hematopoiesis (36), and neural development in zebrafish (22). Additionally, resources now exist for studying other phylogenetic processes in murine models (23, 35), and analytical tools are available for computationally reconstructing and benchmarking trees from large lineage tracing datasets (33, 37).
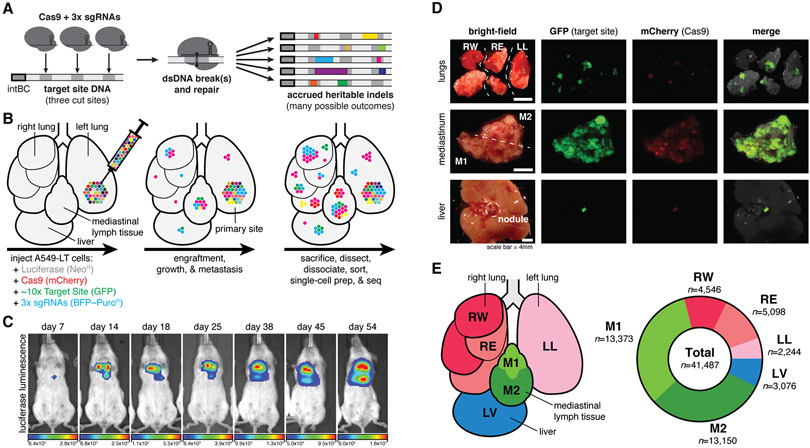
Fig. 1. Lineage tracing in a lung cancer xenograft model in mice.
(A) Our Cas9-enabled lineage tracing technology. Cas9 and three sgRNAs bind and cut cognate sequences on genomically integrated Target Sites, resulting in diverse indel outcomes (multicolored rectangles), which act as heritable markers of lineage. (B) Xenograft model of lung cancer metastasis. Approximately 5,000 A549-LT cells were surgically implanted into the left lung of immunodeficient mice. The cells engrafted at the primary site, proliferated, and metastasized within the five lung lobes, mediastinal lymph, and liver. (C) In vivo bioluminescence imaging of tumor progression over 54 days of lineage recording, from early engraftment to widespread growth and metastasis. (D) Fluorescent imaging of collected tumorous tissues. (E) Anatomical representation of the six tumorous tissue samples (left), and the number of cells collected with paired single-cell transcriptional and lineage datasets (right).
Tracing metastasis in a mouse xenograft model
Here we apply lineage tracing to explore the subclonal dynamics of metastatic dissemination in mouse cancer model (38). We used a human KRAS-mutant lung adenocarcinoma line (A549 cells) in an orthotopic xenograft model in mice because this system is characterized by aggressive metastases (38) and orthotopic xenografting experiments are useful for modeling cancer progression in vivo (39). We engineered A549 cells with a refined version of our molecular recorder technology (23) (Fig. S2, (40)). Specifically, the engineered cells contained: (i) luciferase for live imaging; (ii) Cas9 for generating heritable indels; (iii) ~10 uniquely barcoded copies of the Target Site for recording lineage information, which can be captured as expressed transcripts by single-cell RNA-sequencing; and finally (iv) triple-sgRNAs to direct Cas9 to the Target Sites, thereby initiating lineage recording (Figs. 1A and S2A-C). To enable tracing over long timescales, we designed the sgRNAs with nucleotide mismatches to the Target Sites, thereby decreasing their affinity (41, 42) and tuning the lineage recording rate (23, 43). Approximately 5,000 engineered cells (“A549-LT”) were then embedded in matrigel and surgically implanted into the left lung of an immunodeficient (C.B-17 SCID) mouse (Fig. 1B). We followed bulk tumor progression by live luciferase-based imaging (Fig. 1C): early bioluminescent signal was modest and restricted to the primary site (left lung), consistent with engraftment; with time, the signal progressively increased and spread throughout the thoracic cavity, indicating tumor growth and metastasis. After 54 days, the mouse was sacrificed and tumors were identified in the five lung lobes, throughout the mediastinal lymph tissue, and on the liver (Fig. 1D), in a pattern consistent with this model (38). From these tumorous tissues, we collected six samples, including one from the left lung (i.e., including the primary site; Fig. 1E, left). The tumor samples were dissociated, fluorescence-sorted to exclude normal mouse cells, and finally processed for single-cell RNA-sequencing. To simultaneously measure the transcriptional states and phylogenetic relationships of the cells, we prepared separate RNA expression and Target Site amplicon libraries, respectively, resulting in 41,487 paired single-cell profiles from six tissue samples (Figs. 1E, right and S3 (40)).
In addition to the mouse described above (hereafter “M5k”), we also performed lineage tracing in three other mice (called “M10k”, “M100k”, and “M30k”), using A549-LT cells engineered with slightly different versions of the lineage tracing technology (Fig. S4 (40)). Unless otherwise noted, we focus our primary discussion of the results on mouse M5k because it yielded the richest lineage tracing dataset with the most cells and distinct lineages.
Distinguishing clonal cancer populations
Our lineage recorder “Target Site” (23) carries two orthogonal units of lineage information: (i) a static 14bp-randomer barcode (“intBC”) that is unique and distinguishes between multiple integrated Target Site copies within each cell; and (ii) three independently evolving Cas9 cut-sites per Target Site that record heritable indel alleles and are used for subclonal tree reconstruction (Fig. 1A). Each Target Site is expressed from a constitutive promoter allowing it to be captured by single-cell RNA-sequencing. After amplifying and sequencing the Target Site mRNAs, the reads were analyzed using the Cassiopeia processing pipeline (33, 44). Briefly, this pipeline leverages unique molecular identifier (UMI) information and redundancy in sequencing reads to confidently call intBCs and indel alleles from the lineage data, which inform subsequent phylogenetic reconstruction (Fig. S1 (40)).
We determined the number of clonal populations (that is, groups of related cells that descended from a single clonogen at the beginning of the xenograft experiment), which are each associated with a set of intBCs. Importantly, the A549-LT cells were prepared such that clones carry distinct intBC sets. By sampling the A549-LT cells before implantation, we estimate that the implanted pool of 5,000 cells initially contained 2,150 distinguishable clones (Fig. S2D). From these intBC sets, we assigned most of the cancer cells collected from the mouse (97.7%) to 100 clonal populations (Figs. S5A-B), ranging in size from >11,000 (Clone #1, “CP001”) to ~30 cells (CP100) (Fig. S5C). Though there were some smaller clonal populations, we focused on these largest 100 because lineage tracing in small populations is less informative. Furthermore, despite initially implanting ~2,150 distinct clones, only ~100 clones successfully engrafted and proliferated, suggesting that only a small minority of cells were competent for engraftment and survival in vivo (Fig. S2D). Moreover, we find minimal correlation between initial (pre-implantation) and final (post-sacrifice) clonal population size (Spearman’s ρ=−0.026; Fig. S2E), suggesting that clone-intrinsic characteristics that confer greater fitness in vitro do not necessarily confer greater fitness in the in vivo environment (45, 46).
Features that influence the lineage recording capacity and tree reconstructability differed between clonal populations, such as the copy-number of Target Sites, the percentage of recording sites bearing indel alleles, and allele diversity (Figs. S6A-C and S7). While most clonal populations exceeded parametric standards for confident phylogenetic reconstruction, some had slow recording kinetics or low allele diversity and failed to pass quality-control filters (17 clones, 7.3% of total cells in mouse M5k, Figs. S6D and S7B); these clones were excluded from tree reconstruction and downstream analyses (40, 44).
We observed that the clonal populations exhibited distinct distributions across the six tissues (Fig. S8A-C), ranging from exclusively residing in the primary site (e.g., CP029, CP046), to overrepresented in a tissue (e.g., CP003, CP020), or distributed broadly over all sampled tissues (e.g., CP002, CP013). The level of tissue dispersal is logically a consequence of metastatic dissemination and thus can inform on the frequency of past metastatic events. To quantify the relationship between tissue distribution and metastatic dissemination, we defined a statistical measure of the observed-versus-expected tissue distributions of cells (termed “Tissue Dispersion Score” (40)) to operate as a coarse, tissue-resolved approximation of the dissemination frequency. Across the 100 clonal populations in this mouse, we observed a wide range of Tissue Dispersion Scores (Fig. S8D), suggesting broad metastatic heterogeneity across the tumor populations. We next explored this heterogeneity more directly and at far greater resolution using the evolving lineage information.
Single-cell-resolved cancer phylogenies
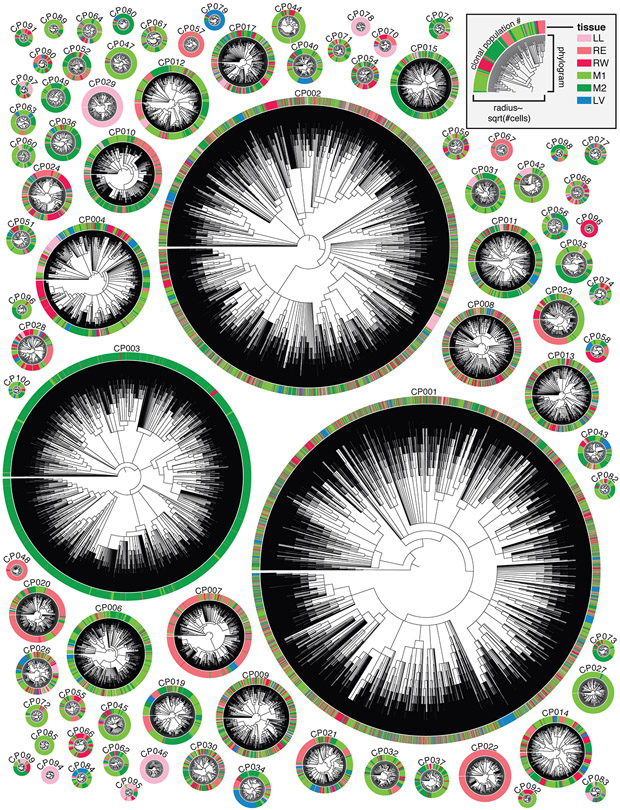
The key advantage of our lineage tracer is not in following clonal lineage dynamics (i.e. from cells’ static intBCs, as described above) but rather in reconstructing subclonal lineage dynamics (i.e. from cells’ continuously evolving indel alleles). As such, we reconstructed high-resolution phylogenetic trees using the Cassiopeia suite of phylogenetic inference algorithms (33) with parameters tailored to this dataset’s complexity and scale (40, 44). Each of the resulting trees comprehensively describes the phylogenetic relationships between all cells within the clonal population and summarizes their history of metastatic dissemination between tissues (Fig. 2). The trees are intricately complex (mean tree depth of 7.25; Fig. S6E) and highly resolved (consisting of 37,888 cells with 33,266 (87.8%) unique lineage states; Fig. S6C).
Fig. 2. High-resolution phylogenetic trees capture the histories of clonal cancer populations.
Highly detailed phylogenetic reconstructions for each clonal population, represented as radial phylograms. Each cell is represented along the circumference and colored by tissue, as in Fig. 1E and legend. Trees differ in size, tissue distribution, and frequency of tissue transitions. Each tree is scaled by the square-root of the number of cells.
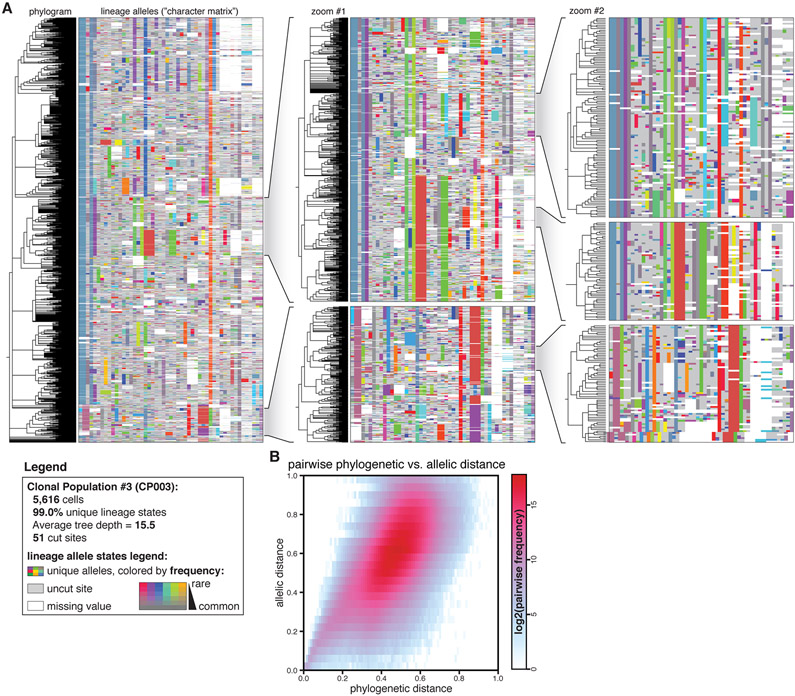
To illustrate the intricate complexity of the trees in this dataset, we present the reconstructed phylogram and lineage alleles for a representative clonal population of 5,616 cells (CP003; Fig. 3A) with 99.0% (5,560) unique cell lineage states, mean tree depth of 10.0, and maximum tree depth of 20. Intuitively, cells that are more closely related to one another ought to share more lineage alleles, which is evident from the patterns of shared alleles within clades and distinguishing alleles between clades (Fig. 3A, zoomed inlays). Indeed, we find systematic agreement between phylogenetic distance (i.e., the distance between two cells in the tree) and allelic distance (the difference between two cells’ lineage alleles) for this example (Fig. 3B) and across all other trees (Fig. S10). The high diversity of distinguishable Cas9-induced indels (9,936 unique alleles across all M5k cells; evident in the array of unique allele colors in Fig. 3A) also reduces the probability of homoplasy, an issue which complicates tree reconstruction and impairs tree accuracy (33, 47). Altogether, these features indicate that the reconstructed trees accurately model the true phylogenetic relationships between cells.
Fig. 3. Phylogenetic reconstructions are detailed and accurate.
(A) Phylogenetic tree and lineage alleles of one clonal population (CP003; N=5,616 cells). The phylogram (left) represents cell–cell relationships and the matrix (right) represents the lineage alleles for each cell. Alleles are uniquely colored, where saturation indicates allele rarity (legend). (A, inlays) Nested zooms of individual clades show the patterns of shared and distinguishing indel alleles, and highlight indel diversity, tree depth, and tree complexity. (B) Correspondence between phylogenetic distance (the normalized pairwise tree distance between two cells) and allelic distance (the normalized pairwise difference in alleles between two cells) for CP003, indicating that the tree accurately models phylogenetic relationships.
Inferring and quantifying past metastatic events from phylogenies
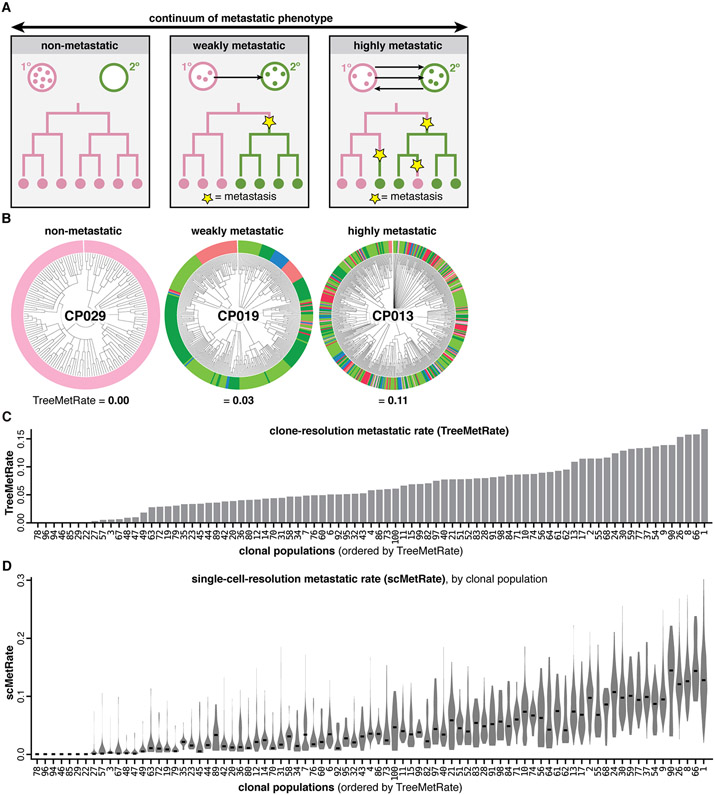
A striking feature revealed by the reconstructed phylogenies is the varying extent to which closely related cells reside in different tissues (Fig. 2), patterns which directly result from ancestor cells having physically transited from one tissue to another in the past (i.e., metastatic seeding). Varying rates of metastasis produce different patterns of concordance between phylogeny and tissue (Fig. 4A). For example, non-metastatic populations result in all clades remaining within a single tissue (Fig. 4A-B, left); conversely, highly metastatic populations result in closely related cells residing in different tissues (Fig. 4A-B, right). Finally, intermediate levels of metastasis can similarly lead to a dispersed tissue distribution as in the highly metastatic regime, though with fewer metastatic transitions, thus supporting the need to reconstruct trees in order to distinguish such cases (Fig. 4A-B, middle).
Fig. 4. Quantifying the diverse metastatic phenotypes of clonal populations directly from cell lineages.
(A) Theoretical continuum of metastatic phenotypes, spanning non-metastatic (never exiting the primary site) to highly metastatic (frequently transitioning between tumors; arrows). Ancestral metastatic events between tissues leave clear phylogenetic signatures (yellow stars). (B) Example clonal populations that illustrate the wide range of metastatic phenotypes observed: a non-metastatic population that never exits the primary site (CP029); a moderately metastatic population that infrequently transitions between different tissues (CP019); and a frequently metastasizing population with closely related cells residing in different tissues (CP013). Cells colored by tissue as in Fig. 1E; metastatic phenotypes scored by the TreeMetRate. (C) The distribution of TreeMetRates for each clonal population. (D) The distributions of single-cell-resolution metastatic phenotypes (scMetRates) for each clonal population, rank-ordered by TreeMetRate; median scMetRate indicated in black.
To quantitatively study the relationship between metastatic phenotype and phylogenetic topology, we used the Fitch-Hartigan maximum parsimony algorithm (48, 49). Our implementation of this algorithm provides the minimal number of ancestral (i.e., not directly observed) metastatic transitions that are needed to explain the final (i.e., observed) tissue location of each cell in a given tree. We defined a score of the metastatic potential (termed “TreeMetRate”) by dividing the inferred minimal number of metastatic transitions by the total number of possible transitions (i.e., edges in the tree). Empirically, we observe a distribution of clonal populations that spans the full spectrum of metastatic phenotypes between low (non-metastatic) and high (very metastatic) TreeMetRates (Fig. 4B,C). The TreeMetRate is stable across bootstrapping experiments in simulated trees (Fig. S9E-F) and when using an alternative phylogenetic reconstruction method (Neighbor-Joining (29)) on empirical data (Fig. S11A; Pearson’s ρ=0.94), indicating that the TreeMetRate is a robust measurement of metastatic behavior – though, notably, Cassiopeia trees are more parsimonious than those reconstructed by Neighbor-Joining (Fig. S11B). Empirically, the Tissue Dispersal Score agrees with the TreeMetRate at low metastatic rates (Fig. S12A,C), however, the TreeMetRate more accurately captures the underlying metastatic rate over a broad range of simulated metastatic rates because it can distinguish between moderate and high metastatic rates (Fig. S9D), which both result in broad dispersion across tissues (Fig. 4A), whereas the Tissue Dispersal Score saturates at intermediate metastatic rates (Fig. S9B). Furthermore, the TreeMetRate agrees with an alternate measure that does not depend on tree reconstruction (termed “AlleleMetRate” (40); Fig. S12B, D), though again simulations indicate that the TreeMetRate best reflects the underlying metastatic rate (Fig. S9A-D).
We further extended our parsimony-based approach to quantify the metastatic phenotype at the resolution of individual cells (termed the “scMetRate”) by averaging the TreeMetRate for all subclades containing a given cell (40, 44). This measurement is sensitive to subclonal differences in metastatic behavior (Fig. 4C), and highlighted intriguing bimodal metastatic behavior for clone CP007 (discussed below). Additionally, we find that the scMetRate is uncorrelated to clonal population size, proliferation signatures (50, 51), or cell cycle stage (52) (Fig. S13), indicating that it can measure metastatic potential uncoupled from proliferative capacity. Overall, these results indicate that cancer cells in this dataset exhibit diverse metastatic phenotypes both between and within clonal populations, which can be meaningfully distinguished and quantified by virtue of the lineage tracer, but would have otherwise been hidden from classical barcoding approaches.
Transcriptional drivers of differences in metastatic phenotype
We next explored the extent to which single-cell transcriptional states underlied metastatic capacity (53). By comparing the paired transcriptional and lineage datasets, we found that different metastatic behaviors corresponded to differential expression of genes, many with known roles in metastasis. First, after filtering and normalizing the scRNA-sequencing data, we applied Vision (54), a tool for assessing the extent to which the variation in cell-level quantitative phenotypes can be explained by transcriptome-wide variation in gene expression. While we found little transcriptional effect attributable to clonal population assignment, we observed a modest association between a cell’s transcriptional profile and its tissue sample or metastatic rate (Fig. S14). We next performed pairwise differential expression analyses comparing cells from completely non-metastatic clonal populations (i.e., four clones that never metastasized from the primary tissue in the left lung, like CP029) to metastatic clones in the same tissue (Fig. S15). This clone-resolution analysis identified several genes with significant expression changes which were also consistent across each non-metastatic clone (log2 fold-change > 1.5, FDR < 0.01), such as IFI6. These initial results suggested that differences in metastatic phenotype may manifest in characteristic differences in gene expression, and motivated deeper analysis.
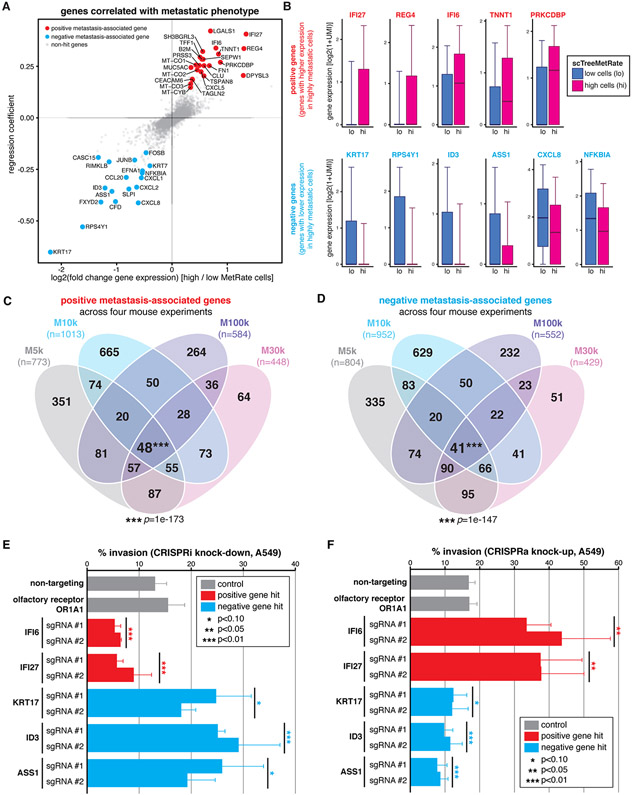
Next, we sought to comprehensively identify genes that are associated with metastatic behavior by regressing single-cell gene expression against the scMetRates (over all observed cells, clonal populations, and tissues; Fig. 5A (40)), thereby leveraging both the scRNA-seq dataset and the single-cell phylogenies. Many of the identified positive metastasis-associated candidates (i.e., genes with significantly higher expression in highly metastatic cells) have known roles in potentiating tumorigenicity (Fig. 5B, top), like IFI27 (55, 56), REG4 (57) (58), and TNNT1 (59). Reciprocally, many negative metastasis-associated candidates have known roles in attenuating metastatic potential (Fig. 5B, bottom), like NFKBIA (60), ID3 (61), and ASS1 (62). The gene whose expression we identified as most strongly and significantly anticorrelated with metastatic capacity, KRT17, has paradoxically been implicated in promoting invasiveness in lung adenocarcinoma (63) and its overexpression has been associated with poor prognosis in some cancers (64); we follow-up on this unexpected finding below. Additionally, many of the identified genes were significantly reproduced across every mouse in this study (Figs. 5C-D and S17). And more generally, the gene-level expression trends are broadly supported by significant correlation between the TreeMetRate and several gene expression signatures (65), like cancer invasiveness (66) and epithelial-mesenchymal transition (67) (Fig. S16).
Fig. 5. Divergent metastatic phenotypes are driven by differences in gene expression.
(A) Poisson regression analysis of single-cell gene expression and scMetRate for all cells and all tissues; fold-change and coefficient of regression shown. The strongest and most significant positive and negative genes are annotated (red and blue, respectively; Methods). (B) Expression level of several positive and negative metastasis-associated gene candidates (top and bottom rows, respectively) in cells with low or high scMetRate (blue and magenta box-plots, respectively). Boxes: first, second, and third quartiles; whiskers: 9th and 91st percentiles of expression distribution. (C and D) Overlap of identified positive and negative metastasis-associated genes, respectively, from the four mouse experiments; number of genes indicated. Four-way intersections between gene sets are significant by SuperExactTest (85) multi-set intersection test. (E and F) In vitro transwell invasion assays following CRISPRi or CRISPRa gene perturbation, respectively, in A549 cells. Perturbations were performed using two independent sgRNAs per gene. Differences in invasion phenotype relative to two negative control guides (non-targeting and olfactory receptor) were significant by two-tailed t-test; error bars show standard deviation across triplicates.
While we identified many interesting and reproducible gene candidates in our regression analysis, it was unclear whether they were directly driving the metastatic phenotype or were merely associated with it. To address this point, we next explored the functional impact on metastatic behavior of modulating the expression of five high-scoring gene candidates (IFI6, IFI27, KRT17, ID3, and ASS1). First, we engineered A549 cells to enable CRISPR-inhibition or -activation perturbations (CRISPRi/a; activity validated in Fig. S18C,D), then increased or decreased expression, respectively, of the five gene targets using two independent sgRNAs per gene. Finally, we measured the perturbed cells’ invasion phenotype in vitro using a transwell invasion assay (Fig. 5E,F (40)). As hypothesized, CRISPRi knock-down resulted in decreased invasiveness for positive metastasis-associated genes (IFI6 and IFI27; p=0.001, 0.005, respectively, by two-tailed t-test) and increased invasiveness for negative metastasis-associated genes (KRT17, ID3, and ASS1; p=0.054, 0.003, and 0.062, respectively; Fig. 5E). Conversely, we found that elevating candidate gene expression by CRISPRa produced the exact opposite results (Fig. 5F), indicating that the invasion phenotype can be quantitatively altered by both increased or decreased expression for each of the five candidate genes tested, including notably KRT17, in agreement with the results of the lineage tracing experiments. We confirmed that the modulation of expression of each of these genes strongly and significantly modulated invasiveness (p<0.01, by two-tailed t-test) in a separate human lung cancer cell line (H1299 cells, which are KRAS wild-type, TP53-mutant, and harbor endogenous NRASQ61K; Fig. S18A,B); though, for two of the genes (IFI27, IFI6), CRISPRa had a significant effect (p<0.01) while CRISPRi did not. Taken together, these results indicate that (i) the lineage tracer can meaningfully identify metastasis-associated genes in vivo, (ii) some of these gene candidates are sufficient to drive differences in metastatic phenotype, and (iii) these genes’ roles in mediating invasiveness extend beyond the one A549 cancer model and across different oncogenic backgrounds.
Heterogeneity and heritability of metastatic behavior in pre-implantation cells
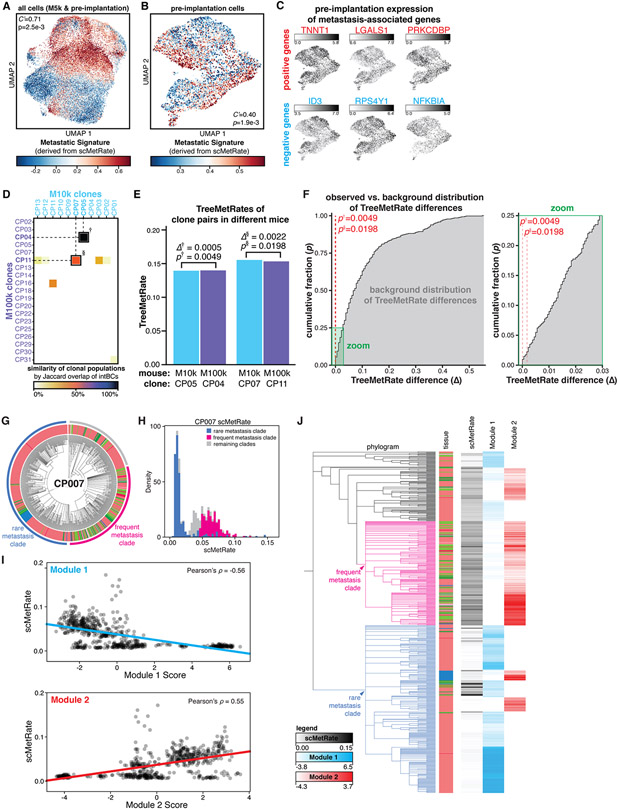
We next used the positive and negative metastasis-associated genes identified above (Fig. 5A) to define a de novo transcriptional signature (hereafter, “Metastasis Signature”; Figs. 6A and S19A). Even prior to implantation into the mice, the cells already exhibited meaningful heterogeneity in the Metastatic Signature (Fig. 6B), and metastasis-associated genes like ID3 and TNNT1 were similarly heterogeneously expressed pre-implantation (Fig. 6C). Next, we used the lineage barcodes to map cells from the in vitro pre-implantation pool to the clonal populations that engrafted in vivo (Fig. S19B). We then segregated these mapped cells into the top and bottom halves by their corresponding TreeMetRate, and queried their pre-implantation Metastatic Signatures. We found that cells from more metastatic clones in the mouse had modestly, yet significantly, higher metastatic signatures prior to implantation, and vice versa (Fig. S19C). This indicates that the pre-implantation transcriptional signature is mildly predictive of in vivo metastatic phenotype (Fig. S19D), though the distinction becomes more amplified in vivo (Fig. S19C, D). This result suggests that even before cells were xenografted into the mouse, they were primed for greater or lesser metastatic capacity in vitro.
Fig. 6. Metastatic phenotype is predetermined, heritable, and reproducible.
(A and B) Projections of transcriptional states of M5k cancer cells and pre-implantation cells (A) or pre-implantation cells alone (B), colored by Metastatic Signature. Association between transcriptional state and Metastatic Signature is measured by inverted Geary’s C’ and significance by false discovery rate (p). (C) Pre-implantation cells exhibit heterogeneity in expression of metastasis-associated genes. (D) Jaccard overlap of intBC sets between clonal populations in M10k and M100k mice. Two pairs of clonal populations (indicated by † and §) were related between the two mouse experiments (Jaccard overlap>50%). (E) Comparison of TreeMetRates from related clones implanted in M10k and M100k, showing minimal difference in metastatic rate (∆) between clone pairs. (F) Cumulative distribution plot of the background distribution of all possible pairwise TreeMetRate differences between M10k and M100k clones (gray), with zoom to show low-∆ regime. Both of the observed differences are statistically smaller than expected (p†=0.0049 and p§=0.0198; red dashes). (G) Divergent subclonal metastatic behavior exhibited in the phylogenetic tree of clonal population #7, with annotated subclades; cells colored by tissue as in Fig. 1E. (H) The bimodal distribution of scMetRates for cells in CP007, with cells from the divergent subclades indicated. (I) Comparison of single-cell metastatic phenotype and Hotspot transcriptional module scores. (J) Overlay illustrating concordance between CP007 phylogeny, scMetRates, and Hotspot Module scores.
While the pre-existing transcriptional heterogeneity in the pre-implantation cells was noteworthy, it remained unclear whether these differences were stochastic or intrinsic properties of the cells that could be robustly propagated in vitro and in vivo. One way to address this question is by implanting two cells from the same clone into two distinct mice and querying how well their metastatic phenotype is reproduced. Using the cells’ intBCs, which stably mark clones, we identified two such instances where cells from the same clonal population seeded tumors in two different mice (Figs. 6D and S20). Strikingly, for each of the two pairs of clonal populations, the TreeMetRates were nearly identical (Fig. 5E). In fact, one of these pairs had the most similar TreeMetRates across all pairs of clones in the two mouse experiments (∆(TreeMetRate)=0.0005, p=0.0049; Fig. 6F). Taken together, these results indicate that (i) the diverse metastatic phenotypes in vivo are determined before implantation (also Fig. 6B, C), (ii) the metastatic phenotype is reproducible over generations and is thus heritable (Figs. 5E,F and S19C,D), and (iii) our analytical approaches for quantifying the metastatic rate, including reconstruction of the phylogenies, are experimentally robust (Fig. 6E).
Evolution of metastatic phenotype
Though we have thus far discussed how metastatic phenotype is clone-intrinsic and stably inherited, we identified a clear example within the dataset that was the exception to this general rule. Specifically, Clone #7 (CP007) exhibited distinct subclonal metastatic behaviors, wherein one clade metastasized frequently to other tissues while another clade remained predominantly in the right lung (Fig. 6G). This distinction is reflected in a bimodal distribution of scMetRates (Figs. 4C and 6H). We used the Hotspot (68) algorithm to explore the relationship between subclonal structure and gene expression, and identified two modules of correlated genes that exhibit heritable expression programs (Fig. S21A). Strikingly, the cumulative expression of genes in Module 1 is correlated with lower metastatic rates, while the opposite holds for Module 2 (Figs. 6I and S21B,C). Consistently, the two modules broadly correspond to the two clades with diverging metastatic phenotypes (Fig. 6J). This result is reproduced even in a control analysis of CP007 cells from the right lung only (Fig. S21D-G), indicating that these differences in gene expression indeed reflect differences in metastatic phenotype rather than tissue-specific effects. This example illustrates that although the metastatic rate is stably inherited, it can also evolve – albeit rarely – within a clonal population, alongside concordant changes in transcriptional signature. Importantly, this finding could only be appreciated by virtue of the subclonal resolution of the lineage tracer.
Tissue routes and topologies of metastasis
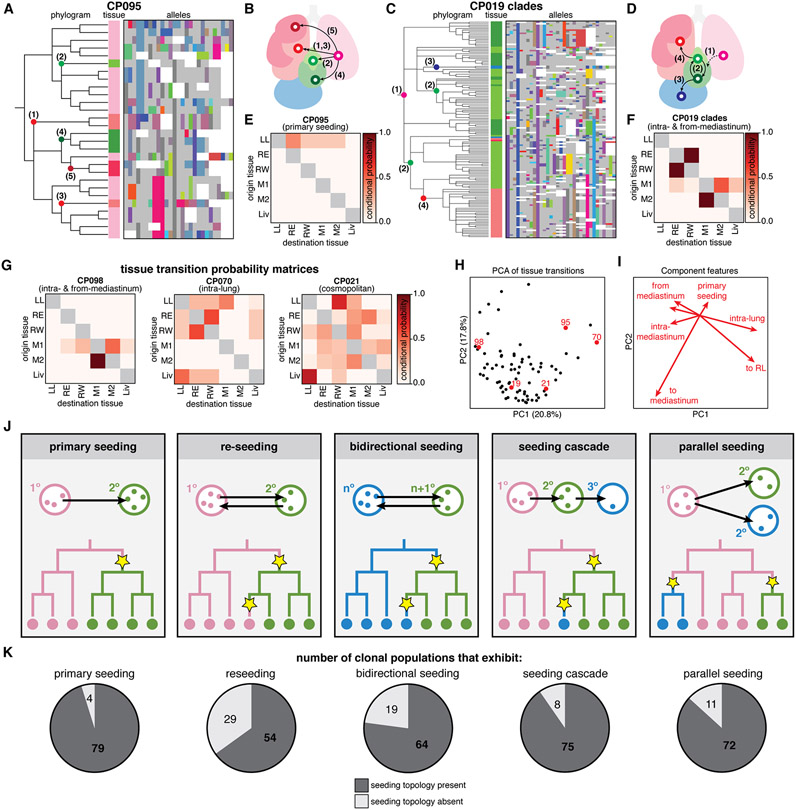
The phylogenetic reconstructions also made it possible to describe detailed histories about the tissue routes and the directionality of metastatic seeding. For example, the phylogenetic tree for CP095 reveals five distinct metastatic events from the left lung to different tissues, in a paradigmatic example of simple primary seeding (Fig. 7A-B). Other phylogenies revealed more complicated trajectories, such as CP019, wherein early primary seeding to the mediastinum was likely followed by intra-mediastinal transitions and later seeding from the mediastinum to the liver and right lung (Fig. 7C-D). To more systematically characterize the tissue transition routes revealed by the phylogenetic trees, we extended the Fitch-Hartigan algorithm (48, 49) to infer the directionality of each tissue transition (i.e., the origin and destination of each metastatic event) along a clonal population’s ancestry. Our algorithm, called FitchCount, builds on other ancestral inference algorithms like MACHINA (69) by scaling to large inputs and providing tissue transitions frequencies that are aggregated across all ancestries that satisfy the maximum parsimony criterion (40, 44). Through simulation we show that FitchCount can accurately recover underlying transition probabilities better than a naive application of the Fitch-Hartigan algorithm (Fig. S9G-H (40)), likely because the naive approach summarizes only a single optimal assignment solution, whereas FitchCount summarizes all optimal solutions. The resulting conditional probabilities of metastasis to and from each tissue are summarized in a tissue transition probability matrix (Fig. 7E-F). Notably, we found that these transition matrices are varied and distinct to each clone (Figs. 7G and S22).
Fig. 7. Metastases were seeded via complex tissue routes and multidirectional topologies.
(A and D) Phylogenetic trees and lineage alleles for clonal population #95 and #19 clades, respectively. Notable metastatic events are annotated in the phylogram and represented graphically as arrows (B and E); cells colored by tissue as in Fig. 1E; lineage alleles colored as in Fig. 3A; dashed arrow indicates an assumed transition. (C and F) Tissue transition matrices representing the conditional probability of metastasizing from and to tissues, defining the tissue routes of metastasis for each clonal population. CP095 solely exhibits primary seeding from the left lung, whereas CP019 shows more complex seeding routes. (G) Tissue transition matrices illustrating the diversity of tissue routes, including metastasis from and within the mediastinum (left), between the lung lobes (middle), or amply to and from all tissues (right). (H) Principal component analysis (PCA) of tissue transition probabilities for each clonal population. Displayed clones are annotated in red; percentage of variance explained by components indicated on axes. (I) Component vectors of PCA with descriptive features. (J) Possible phylogenetic topologies of metastatic seeding, represented as in Fig. 4A. (K) Number of clonal populations that exhibit each metastatic seeding topology.
We next used principal component analysis (PCA) to stratify clones by their transition matrices (Fig. 7H) and identified descriptive features that capture differences in the metastatic tissue routes traversed by each clone (Figs. 7I and S23). These descriptive features include primary seeding from the left lung (as in CP095, Fig. 7A-B), metastasis from and within the mediastinum (CP098, Fig. 7G, left), or metastasis between lung lobes (CP070, Fig. 7G, middle), and may reflect intrinsic differences in tissue tropism. From this feature analysis we also note that many clones primarily metastasized via the mediastinal lymph tissue (Fig. 7H-I), suggesting that the mediastinum may act as a nexus for seeding in this mouse model, perhaps because the mediastinal lymph is a favorable niche with extensive tissue connections (70). This observation is consistent with previous experiments in this model (38), bulk live imaging during tumor progression in this experiment wherein tumors appear to quickly colonize the mediastinum (Fig. 1C), and the terminal disease state wherein the mediastinum harbors the majority of the tumor burden (Fig. S8). This illustrates how the lineage tracer can capture subtle differences in tissue tropism for different tumor populations.
Many models of metastatic seeding topology (i.e, the sequence and directionality of metastatic transitions) have been described in cancer (1), including reseeding, seeding cascades, parallel seeding, and others; and each is characterized by a distinct phylogenetic signature (Fig. 7J). These different metastatic topologies can critically influence the progression, relapse, and treatment of cancers (9, 71-73); for example, reseeding of metastatic cells returning to the primary tumor site can contribute genetic diversity, resistance to treatment, and metastatic potential to tumors (74, 75). Within this single dataset, we find numerous examples of all of these topologies (Fig. 7K); in fact, we most often observe examples of all topologies within every clone (Fig. S24), as well as more complex topologies that defy simple classifications (e.g., Fig. 7D,G right), further underscoring the aggressive metastatic nature of A549 cells in this xenograft model. Extending beyond this model, these findings suggest that metastatic seeding patterns can be highly complex or patient-specific.
Discussion
By applying our next-generation, Cas9-based lineage tracer to a mouse model of metastasis, we observed meaningful features of metastatic biology that were only apparent by virtue of subclonal lineage information. Among these key insights were the broad range of metastatic rates for different tumor populations, the pre-existence and stable heritability of these heterogeneous metastatic phenotypes, and the complex, multidirectional tissue routes by which cancer cells disseminate in this model.
The heterogeneity we observed may have intriguing implications for understanding the biology of cancer metastasis. First, rather than being a simple binary process, there appear to exist multiple distinct cell states that have characteristic and graded differences in metastatic potential, and these differences are orthogonal to proliferative potential. Second, there are characteristic transcriptional differences underlying the different metastatic states, and multiple genes involved in these differences are individually sufficient to modulate the degree of cell invasion; this suggests that coherent transcriptional programs drive these different metastatic states. Third, although these transcriptional differences can be detected in vitro, they are muted in that context and are amplified in vivo, suggesting an interplay between tissue environment and cell phenotype. Finally, these phenotypes are stably inherited over cell generations but are capable of evolution, as we document in one clear example. Understanding the genetic and/or epigenetic bases for these phenotypic differences – how they arise, how they change, how they affect cell biology – could broadly inform our understanding of how cancer disseminates and progresses.
As a first report, this work by necessity focuses on a single model of metastasis. Nonetheless, multiple distinct steps underlie the metastatic process – including extravasation, transit between tissues, intravasation, and colonization – and the approaches described here can be broadly applied to study each of these steps and indeed other aspects of cancer progression in future work. The lineage tracing approach could be applied to models of inducible tumor initiation (76) or patient-derived xenografts (77, 78), which we anticipate may provide a window into earlier stages of cancer progression, such as slower or less complex metastatic dynamics than the aggressively metastatic behavior observed here by A549 cells. Lineage tracing in syngeneic cancer lines or autochthonous models of cancer could chart how an intact immune system may influence cancer progression (79-81). It will also be of interest to investigate the roles that other gene candidates identified here play in metastasis, as well as to elucidate the molecular mechanism by which KRT17 suppresses metastatic phenotype in vitro and in vivo – an unexpected role that this work uncovered. Merging lineage tracing with recent high-resolution spatial sequencing approaches (24, 82-84) would enable the exploration of cancer biology at higher spatial resolution (e.g., resolving individual tumors, rather than resolving tumorous tissues as here) to distinguish the clonality of micrometastases, monophyletic versus polyphyletic dissemination (12), intercellular interactions between cancer cells and the microenvironment, and the spatial constraints of tumor growth and metastasis.
Our work establishes that it is now possible to uniquely distinguish tens of thousands of cells over several months of growth in vivo, reconstruct deeply resolved and accurate cell phylogenies, and then interpret them to identify rare, transient events in the cells’ ancestry (here, metastasis) revealing otherwise unapparent distinctions in cellular phenotypes. Extending beyond metastasis, this approach can inform many other facets of cancer biology, like the timing or order of genetic mutations during malignant transformation, adaptation to different tumor microenvironments, or the origin and mechanism by which tumor cells acquire resistance to therapeutic agents. And beyond cancer, our approach has the potential to empower the study of the phylogenetic foundations of biological processes that transpire over many cell generations at unprecedented resolution and scale.
Supplementary Material
Acknowledgements:
We thank R. Weinberg, D. Yang, A. Khodaverdian, and R. Zhang for discussions; M. Jost and J.K. Nuñez for plasmids; and UCSF Center for Advanced Technology (E. Chow and S. Elmes) and UCSF Preclinical Therapeutics Core (B. Hann).
Funding: This work was supported by NIH-NIGMS F32GM125247 (J.J.Q.); UCSF Discovery Fellowship (M.G.J.); NIH K08CA222625 (R.A.O.); NIH R01DA036858 and 1RM1HG009490 (J.S.W.); NIH R01CA231300, U54CA224081, R01CA204302, R01CA211052, and R01CA169338, and the Pew and Stewart Foundations (T.G.B.); NIH-NIAID U19AI090023 (N.Y.), and Chan-Zuckerberg Initiative 2018-184034. J.S.W. is a Howard Hughes Medical Institute Investigator. M.M.C. is a Gordon and Betty Moore fellow of the Life Sciences Research Foundation.
Footnotes
Materials and methods
A detailed version of the materials and methods is provided in the supplementary materials.
Competing interests: J.S.W. is on the editorial board of Cell and Molecular Cell, and is an advisor and/or has equity in KSQ Therapeutics, Maze Therapeutics, Amgen, Tenaya, and 5 AM Ventures. T.G.B. is an advisor to Novartis, Astrazeneca, Revolution Medicines, Array/Pfizer, Springworks, Strategia, Relay, Jazz, Rain, and EcoR1, and receives research funding from Novartis, Revolution Medicines, and Strategia. N.Y. is an advisor and/or has equity in Cellarity, Celsius Therapeutics, and Rheos Medicines. All other authors declare no competing interests.
Data and materials availability: The vectors for generating the A549-LT cell line will be made available via Addgene; parental A549 line is available via ATCC. Raw sequencing reads and processed lineage tracing data files are available via GEO (accession #GSE161363). Lineage tracer processing pipeline, phylogenetic reconstruction algorithm, analysis scripts, notebooks, and FitchCount are publicly available via Zenodo (accession #4243162)(44).
References and Notes:
- 1.Turajlic S, Swanton C, Metastasis as an evolutionary process. Science. 352, 169–175 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Navin NE, Hicks J, Tracing the tumor lineage. Mol. Oncol 4, 267–283 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nowell PC, The clonal evolution of tumor cell populations. Science. 194, 23–28 (1976). [DOI] [PubMed] [Google Scholar]
- 4.Stratton MR, Campbell PJ, Futreal PA, The cancer genome. Nature. 458, 719–724 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brannon AR, Vakiani E, Sylvester BE, Scott SN, McDermott G, Shah RH, Kania K, Viale A, Oschwald DM, Vacic V, Emde A-K, Cercek A, Yaeger R, Kemeny NE, Saltz LB, Shia J, D’Angelica MI, Weiser MR, Solit DB, Berger MF, Comparative sequencing analysis reveals high genomic concordance between matched primary and metastatic colorectal cancer lesions. Genome Biol. 15, 454 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bhang H-EC, Ruddy DA, Krishnamurthy Radhakrishna V, Caushi JX, Zhao R, Hims MM, Singh AP, Kao I, Rakiec D, Shaw P, Balak M, Raza A, Ackley E, Keen N, Schlabach MR, Palmer M, Leary RJ, Chiang DY, Sellers WR, Michor F, Cooke VG, Korn JM, Stegmeier F, Studying clonal dynamics in response to cancer therapy using high-complexity barcoding. Nat. Med 21, 440–448 (2015). [DOI] [PubMed] [Google Scholar]
- 7.Chaffer CL, Weinberg RA, A perspective on cancer cell metastasis. Science. 331, 1559–1564 (2011). [DOI] [PubMed] [Google Scholar]
- 8.Lambert AW, Pattabiraman DR, Weinberg RA, Emerging Biological Principles of Metastasis. Cell. 168, 670–691 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Massagué J, Obenauf AC, Metastatic colonization by circulating tumour cells. Nature. 529, 298–306 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sulston JE, Schierenberg E, White JG, Thomson JN, The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev. Biol 100, 64–119 (1983). [DOI] [PubMed] [Google Scholar]
- 11.Han X, Wang R, Zhou Y, Fei L, Sun H, Lai S, Saadatpour A, Zhou Z, Chen H, Ye F, Huang D, Xu Y, Huang W, Jiang M, Jiang X, Mao J, Chen Y, Lu C, Xie J, Fang Q, Wang Y, Yue R, Li T, Huang H, Orkin SH, Yuan G-C, Chen M, Guo G, Mapping the Mouse Cell Atlas by Microwell-Seq. Cell. 173, 1307 (2018). [DOI] [PubMed] [Google Scholar]
- 12.Birkbak NJ, McGranahan N, Cancer Genome Evolutionary Trajectories in Metastasis. Cancer Cell. 37, 8–19 (2020). [DOI] [PubMed] [Google Scholar]
- 13.Wu S-HS, Lee J-H, Koo B-K, Lineage Tracing: Computational Reconstruction Goes Beyond the Limit of Imaging. Mol. Cells 42, 104–112 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schwartz R, Schäffer AA, The evolution of tumour phylogenetics: principles and practice. Nat. Rev. Genet 18, 213–229 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jamal-Hanjani M, Hackshaw A, Ngai Y, Shaw J, Dive C, Quezada S, Middleton G, de Bruin E, Le Quesne J, Shafi S, Falzon M, Horswell S, Blackhall F, Khan I, Janes S, Nicolson M, Lawrence D, Forster M, Fennell D, Lee S-M, Lester J, Kerr K, Muller S, Iles N, Smith S, Murugaesu N, Mitter R, Salm M, Stuart A, Matthews N, Adams H, Ahmad T, Attanoos R, Bennett J, Birkbak NJ, Booton R, Brady G, Buchan K, Capitano A, Chetty M, Cobbold M, Crosbie P, Davies H, Denison A, Djearman M, Goldman J, Haswell T, Joseph L, Kornaszewska M, Krebs M, Langman G, MacKenzie M, Millar J, Morgan B, Naidu B, Nonaka D, Peggs K, Pritchard C, Remmen H, Rowan A, Shah R, Smith E, Summers Y, Taylor M, Veeriah S, Waller D, Wilcox B, Wilcox M, Woolhouse I, McGranahan N, Swanton C, Tracking genomic cancer evolution for precision medicine: the lung TRACERx study. PLoS Biol. 12, e1001906 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu Z, Ding J, Ma Z, Sun R, Seoane JA, Scott Shaffer J, Suarez CJ, Berghoff AS, Cremolini C, Falcone A, Loupakis F, Birner P, Preusser M, Lenz H-J, Curtis C, Quantitative evidence for early metastatic seeding in colorectal cancer. Nat. Genet 51, 1113–1122 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gerlinger M, Rowan AJ, Horswell S, Math M, Larkin J, Endesfelder D, Gronroos E, Martinez P, Matthews N, Stewart A, Tarpey P, Varela I, Phillimore B, Begum S, McDonald NQ, Butler A, Jones D, Raine K, Latimer C, Santos CR, Nohadani M, Eklund AC, Spencer-Dene B, Clark G, Pickering L, Stamp G, Gore M, Szallasi Z, Downward J, Futreal PA, Swanton C, Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med 366, 883–892 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shih DJH, Nayyar N, Bihun I, Dagogo-Jack I, Gill CM, Aquilanti E, Bertalan M, Kaplan A, D’Andrea MR, Chukwueke U, Ippen FM, Alvarez-Breckenridge C, Camarda ND, Lastrapes M, McCabe D, Kuter B, Kaufman B, Strickland MR, Martinez-Gutierrez JC, Nagabhushan D, De Sauvage M, White MD, Castro BA, Hoang K, Kaneb A, Batchelor ED, Paek SH, Park SH, Martinez-Lage M, Berghoff AS, Merrill P, Gerstner ER, Batchelor TT, Frosch MP, Frazier RP, Borger DR, Iafrate AJ, Johnson BE, Santagata S, Preusser M, Cahill DP, Carter SL, Brastianos PK, Genomic characterization of human brain metastases identifies drivers of metastatic lung adenocarcinoma. Nat. Genet (2020), doi: 10.1038/s41588-020-0592-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hong WS, Shpak M, Townsend JP, Inferring the Origin of Metastases from Cancer Phylogenies. Cancer Res. 75, 4021–4025 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reiter JG, Makohon-Moore AP, Gerold JM, Bozic I, Chatterjee K, Iacobuzio-Donahue CA, Vogelstein B, Nowak MA, Reconstructing metastatic seeding patterns of human cancers. Nat. Commun 8, 14114 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Woodworth MB, Girskis KM, Walsh CA, Building a lineage from single cells: genetic techniques for cell lineage tracking. Nat. Rev. Genet 18, 230–244 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Raj B, Wagner DE, McKenna A, Pandey S, Klein AM, Shendure J, Gagnon JA, Schier AF, Simultaneous single-cell profiling of lineages and cell types in the vertebrate brain. Nat. Biotechnol 36, 442–450 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chan MM, Smith ZD, Grosswendt S, Kretzmer H, Norman TM, Adamson B, Jost M, Quinn JJ, Yang D, Jones MG, Khodaverdian A, Yosef N, Meissner A, Weissman JS, Molecular recording of mammalian embryogenesis. Nature. 570, 77–82 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frieda KL, Linton JM, Hormoz S, Choi J, Chow K-HK, Singer ZS, Budde MW, Elowitz MB, Cai L, Synthetic recording and in situ readout of lineage information in single cells. Nature. 541, 107–111 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alemany A, Florescu M, Baron CS, Peterson-Maduro J, van Oudenaarden A, Whole-organism clone tracing using single-cell sequencing. Nature. 556, 108–112 (2018). [DOI] [PubMed] [Google Scholar]
- 26.Spanjaard B, Hu B, Mitic N, Olivares-Chauvet P, Janjuha S, Ninov N, Junker JP, Simultaneous lineage tracing and cell-type identification using CRISPR-Cas9-induced genetic scars. Nat. Biotechnol 36, 469–473 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baron CS, van Oudenaarden A, Unravelling cellular relationships during development and regeneration using genetic lineage tracing. Nat. Rev. Mol. Cell Biol 20, 753–765 (2019). [DOI] [PubMed] [Google Scholar]
- 28.Wagner DE, Klein AM, Lineage tracing meets single-cell omics: opportunities and challenges. Nat. Rev. Genet (2020), doi: 10.1038/s41576-020-0223-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saitou N, Nei M, The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol 4, 406–425 (1987). [DOI] [PubMed] [Google Scholar]
- 30.Camin JH, Sokal RR, A Method for Deducing Branching Sequences in Phylogeny. Evolution. 19, 311 (1965). [Google Scholar]
- 31.Sugino K, Lee T, Robust Reconstruction of CRISPR and Tumor Lineage Using Depth Metrics. bioRxiv (2019). [Google Scholar]
- 32.Feng J, DeWitt WS III, McKenna A, Simon N, Willis A, Matsen FA IV, Estimation of cell lineage trees by maximum-likelihood phylogenetics. bioRxiv (2019), p. 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jones MG, Khodaverdian A, Quinn JJ, Chan MM, Hussmann JA, Wang R, Xu C, Weissman JS, Yosef N, Inference of single-cell phylogenies from lineage tracing data using Cassiopeia. Genome Biol. 21, 64 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zafar H, Lin C, Bar-Joseph Z, Single-cell lineage tracing by integrating CRISPR-Cas9 mutations with transcriptomic data. Nat. Commun 11, 3055 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kalhor R, Kalhor K, Mejia L, Leeper K, Graveline A, Mali P, Church GM, Developmental barcoding of whole mouse via homing CRISPR. Science. 361 (2018), doi: 10.1126/science.aat9804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bowling S, Sritharan D, Osorio FG, Nguyen M, Cheung P, Rodriguez-Fraticelli A, Patel S, Yuan W-C, Fujiwara Y, Li BE, Orkin SH, Hormoz S, Camargo FD, An Engineered CRISPR-Cas9 Mouse Line for Simultaneous Readout of Lineage Histories and Gene Expression Profiles in Single Cells. Cell. 181, 1693–1694 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McKenna A, Findlay GM, Gagnon JA, Horwitz MS, Schier AF, Shendure J, Whole-organism lineage tracing by combinatorial and cumulative genome editing. Science. 353, aaf7907 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Okimoto RA, Breitenbuecher F, Olivas VR, Wu W, Gini B, Hofree M, Asthana S, Hrustanovic G, Flanagan J, Tulpule A, Blakely CM, Haringsma HJ, Simmons AD, Gowen K, Suh J, Miller VA, Ali S, Schuler M, Bivona TG, Inactivation of Capicua drives cancer metastasis. Nat. Genet 49, 87–96 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Francia G, Cruz-Munoz W, Man S, Xu P, Kerbel RS, Mouse models of advanced spontaneous metastasis for experimental therapeutics. Nat. Rev. Cancer 11, 135–141 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Materials, methods, and supplemental text are available as supplementary material on Science Online.
- 41.Boyle EA, Andreasson JOL, Chircus LM, Sternberg SH, Wu MJ, Guegler CK, Doudna JA, Greenleaf WJ, High-throughput biochemical profiling reveals sequence determinants of dCas9 off-target binding and unbinding. Proc. Natl. Acad. Sci. U. S. A 114, 5461–5466 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jones SK Jr, Hawkins JA, Johnson NV, Jung C, Hu K, Rybarski JR, Chen JS, Doudna JA, Press WH, Finkelstein IJ, Massively parallel kinetic profiling of natural and engineered CRISPR nucleases. Nat. Biotechnol (2020), doi: 10.1038/s41587-020-0646-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jost M, Santos DA, Saunders RA, Horlbeck MA, Hawkins JS, Scaria SM, Norman TM, Hussmann JA, Liem CR, Gross CA, Weissman JS, Titrating gene expression using libraries of systematically attenuated CRISPR guide RNAs. Nat. Biotechnol (2020), doi: 10.1038/s41587-019-0387-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Quinn JJ, Jones MG, Okimoto RA, Nanjo S, Chan MM, Yosef N, Bivona TG, Weissman JS, Single-cell lineages reveal the rates, routes, and drivers of metastasis in cancer xenografts. Zenodo (2020), doi: 10.5281/zenodo.4243162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McGranahan N, Swanton C, Clonal Heterogeneity and Tumor Evolution: Past, Present, and the Future. Cell. 168, 613–628 (2017). [DOI] [PubMed] [Google Scholar]
- 46.Hata AN, Niederst MJ, Archibald HL, Gomez-Caraballo M, Siddiqui FM, Mulvey HE, Maruvka YE, Ji F, Bhang H-EC, Krishnamurthy Radhakrishna V, Siravegna G, Hu H, Raoof S, Lockerman E, Kalsy A, Lee D, Keating CL, Ruddy DA, Damon LJ, Crystal AS, Costa C, Piotrowska Z, Bardelli A, Iafrate AJ, Sadreyev RI, Stegmeier F, Getz G, Sequist LV, Faber AC, Engelman JA, Tumor cells can follow distinct evolutionary paths to become resistant to epidermal growth factor receptor inhibition. Nat. Med 22, 262–269 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Salvador-Martínez I, Grillo M, Averof M, Telford MJ, Is it possible to reconstruct an accurate cell lineage using CRISPR recorders? Elife. 8 (2019), doi: 10.7554/eLife.40292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fitch WM, Toward Defining the Course of Evolution: Minimum Change for a Specific Tree Topology. Syst. Zool 20, 406 (1971). [Google Scholar]
- 49.Hartigan JA, Minimum Mutation Fits to a Given Tree. Biometrics. 29 (1973), p. 53. [Google Scholar]
- 50.Ben-Porath I, Thomson MW, Carey VJ, Ge R, Bell GW, Regev A, Weinberg RA, An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat. Genet 40, 499–507 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rosty C, Sheffer M, Tsafrir D, Stransky N, Tsafrir I, Peter M, de Crémoux P, de La Rochefordière A, Salmon R, Dorval T, Thiery JP, Couturier J, Radvanyi F, Domany E, Sastre-Garau X, Identification of a proliferation gene cluster associated with HPV E6/E7 expression level and viral DNA load in invasive cervical carcinoma. Oncogene. 24, 7094–7104 (2005). [DOI] [PubMed] [Google Scholar]
- 52.Whitfield ML, Sherlock G, Saldanha AJ, Murray JI, Ball CA, Alexander KE, Matese JC, Perou CM, Hurt MM, Brown PO, Botstein D, Identification of genes periodically expressed in the human cell cycle and their expression in tumors. Mol. Biol. Cell 13, 1977–2000 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Celià-Terrassa T, Kang Y, Distinctive properties of metastasis-initiating cells. Genes Dev. 30, 892–908 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.DeTomaso D, Jones MG, Subramaniam M, Ashuach T, Ye CJ, Yosef N, Functional interpretation of single cell similarity maps. Nat. Commun 10, 4376 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang H, Qiu X, Lin S, Chen X, Wang T, Liao T, Knockdown of IFI27 inhibits cell proliferation and invasion in oral squamous cell carcinoma. World J. Surg. Oncol 16, 64 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li S, Xie Y, Zhang W, Gao J, Wang M, Zheng G, Yin X, Xia H, Tao X, Interferon alpha-inducible protein 27 promotes epithelial-mesenchymal transition and induces ovarian tumorigenicity and stemness. J. Surg. Res 193, 255–264 (2015). [DOI] [PubMed] [Google Scholar]
- 57.Guo Y, Xu J, Li N, Gao F, Huang P, RegIV potentiates colorectal carcinoma cell migration and invasion via its CRD domain. Cancer Genet. Cytogenet 199, 38–44 (2010). [DOI] [PubMed] [Google Scholar]
- 58.Sun S, Hu Z, Huang S, Ye X, Wang J, Chang J, Wu X, Wang Q, Zhang L, Hu X, Yu H, REG4 is an indicator for KRAS mutant lung adenocarcinoma with TTF-1 low expression. J. Cancer Res. Clin. Oncol 145, 2273–2283 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hao Y-H, Yu S-Y, Tu R-S, Cai Y-Q, TNNT1, a prognostic indicator in colon adenocarcinoma, regulates cell behaviors and mediates EMT process. Biosci. Biotechnol. Biochem 84, 111–117 (2020). [DOI] [PubMed] [Google Scholar]
- 60.Bredel M, Scholtens DM, Yadav AK, Alvarez AA, Renfrow JJ, Chandler JP, Yu ILY, Carro MS, Dai F, Tagge MJ, Ferrarese R, Bredel C, Phillips HS, Lukac PJ, Robe PA, Weyerbrock A, Vogel H, Dubner S, Mobley B, He X, Scheck AC, Sikic BI, Aldape KD, Chakravarti A, Harsh GR 4th, NFKBIA deletion in glioblastomas. N. Engl. J. Med 364, 627–637 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen F-F, Liu Y, Wang F, Pang X-J, Zhu C-D, Xu M, Yu W, Li X-J, Effects of upregulation of Id3 in human lung adenocarcinoma cells on proliferation, apoptosis, mobility and tumorigenicity. Cancer Gene Ther. 22, 431–437 (2015). [DOI] [PubMed] [Google Scholar]
- 62.Rabinovich S, Adler L, Yizhak K, Sarver A, Silberman A, Agron S, Stettner N, Sun Q, Brandis A, Helbling D, Korman S, Itzkovitz S, Dimmock D, Ulitsky I, Nagamani SC, Ruppin E, Erez A, Diversion of aspartate in ASS1-deficient tumours fosters de novo pyrimidine synthesis. Nature. 527, 379–383 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liu J, Liu L, Cao L, Wen Q, Keratin 17 Promotes Lung Adenocarcinoma Progression by Enhancing Cell Proliferation and Invasion. Med. Sci. Monit 24, 4782–4790 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hobbs RP, Batazzi AS, Han MC, Coulombe PA, Loss of Keratin 17 induces tissue-specific cytokine polarization and cellular differentiation in HPV16-driven cervical tumorigenesis in vivo. Oncogene. 35, 5653–5662 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP, Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U. S. A 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Anastassiou D, Rumjantseva V, Cheng W, Huang J, Canoll PD, Yamashiro DJ, Kandel JJ, Human cancer cells express Slug-based epithelial-mesenchymal transition gene expression signature obtained in vivo. BMC Cancer. 11, 529 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jechlinger M, Grunert S, Tamir IH, Janda E, Lüdemann S, Waerner T, Seither P, Weith A, Beug H, Kraut N, Expression profiling of epithelial plasticity in tumor progression. Oncogene. 22, 7155–7169 (2003). [DOI] [PubMed] [Google Scholar]
- 68.DeTomaso D, Yosef N, Identifying Informative Gene Modules Across Modalities of Single Cell Genomics. bioRxiv (2020), p. 54. [DOI] [PubMed] [Google Scholar]
- 69.El-Kebir M, Satas G, Raphael BJ, Inferring parsimonious migration histories for metastatic cancers. Nat. Genet 50, 718–726 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pereira ER, Kedrin D, Seano G, Gautier O, Meijer EFJ, Jones D, Chin S-M, Kitahara S, Bouta EM, Chang J, Beech E, Jeong H-S, Carroll MC, Taghian AG, Padera TP, Lymph node metastases can invade local blood vessels, exit the node, and colonize distant organs in mice. Science. 359, 1403–1407 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Langley RR, Fidler IJ, The seed and soil hypothesis revisited--the role of tumor-stroma interactions in metastasis to different organs. Int. J. Cancer 128, 2527–2535 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fidler IJ, Kripke ML, The challenge of targeting metastasis. Cancer Metastasis Rev. 34, 635–641 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Oskarsson T, Batlle E, Massagué J, Metastatic stem cells: sources, niches, and vital pathways. Cell Stem Cell. 14, 306–321 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Heyde A, Reiter JG, Naxerova K, Nowak MA, Consecutive seeding and transfer of genetic diversity in metastasis. Proc. Natl. Acad. Sci. U. S. A 116, 14129–14137 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Comen E, Norton L, Self-seeding in cancer. Recent Results Cancer Res. 195, 13–23 (2012). [DOI] [PubMed] [Google Scholar]
- 76.DuPage M, Jacks T, Genetically engineered mouse models of cancer reveal new insights about the antitumor immune response. Curr. Opin. Immunol 25, 192–199 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhang X, Claerhout S, Prat A, Dobrolecki LE, Petrovic I, Lai Q, Landis MD, Wiechmann L, Schiff R, Giuliano M, Wong H, Fuqua SW, Contreras A, Gutierrez C, Huang J, Mao S, Pavlick AC, Froehlich AM, Wu M-F, Tsimelzon A, Hilsenbeck SG, Chen ES, Zuloaga P, Shaw CA, Rimawi MF, Perou CM, Mills GB, Chang JC, Lewis MT, A renewable tissue resource of phenotypically stable, biologically and ethnically diverse, patient-derived human breast cancer xenograft models. Cancer Res. 73, 4885–4897 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hidalgo M, Amant F, Biankin AV, Budinská E, Byrne AT, Caldas C, Clarke RB, de Jong S, Jonkers J, Mælandsmo GM, Roman-Roman S, Seoane J, Trusolino L, Villanueva A, Patient-derived xenograft models: an emerging platform for translational cancer research. Cancer Discov. 4, 998–1013 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gonzalez H, Hagerling C, Werb Z, Roles of the immune system in cancer: from tumor initiation to metastatic progression. Genes Dev. 32, 1267–1284 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Angelova M, Mlecnik B, Vasaturo A, Bindea G, Fredriksen T, Lafontaine L, Buttard B, Morgand E, Bruni D, Jouret-Mourin A, Hubert C, Kartheuser A, Humblet Y, Ceccarelli M, Syed N, Marincola FM, Bedognetti D, Van den Eynde M, Galon J, Evolution of Metastases in Space and Time under Immune Selection. Cell. 175, 751–765.e16 (2018). [DOI] [PubMed] [Google Scholar]
- 81.Binnewies M, Roberts EW, Kersten K, Chan V, Fearon DF, Merad M, Coussens LM, Gabrilovich DI, Ostrand-Rosenberg S, Hedrick CC, Vonderheide RH, Pittet MJ, Jain RK, Zou W, Howcroft TK, Woodhouse EC, Weinberg RA, Krummel MF, Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat. Med 24, 541–550 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rodriques SG, Stickels RR, Goeva A, Martin CA, Murray E, Vanderburg CR, Welch J, Chen LM, Chen F, Macosko EZ, Slide-seq: A scalable technology for measuring genome-wide expression at high spatial resolution. Science. 363, 1463–1467 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ståhl PL, Salmén F, Vickovic S, Lundmark A, Navarro JF, Magnusson J, Giacomello S, Asp M, Westholm JO, Huss M, Mollbrink A, Linnarsson S, Codeluppi S, Borg Å, Pontén F, Costea PI, Sahlén P, Mulder J, Bergmann O, Lundeberg J, Frisén J, Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science. 353, 78–82 (2016). [DOI] [PubMed] [Google Scholar]
- 84.Askary A, Sanchez-Guardado L, Linton JM, Chadly DM, Budde MW, Cai L, Lois C, Elowitz MB, In situ readout of DNA barcodes and single base edits facilitated by in vitro transcription. Nat. Biotechnol 38, 66–75 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wang M, Zhao Y, Zhang B, Efficient Test and Visualization of Multi-Set Intersections. Sci. Rep 5, 16923 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Jost M, Chen Y, Gilbert LA, Horlbeck MA, Krenning L, Menchon G, Rai A, Cho MY, Stern JJ, Prota AE, Kampmann M, Akhmanova A, Steinmetz MO, Tanenbaum ME, Weissman JS, Combined CRISPRi/a-Based Chemical Genetic Screens Reveal that Rigosertib Is a Microtubule-Destabilizing Agent. Mol. Cell 68, 210–223.e6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gilbert LA, Horlbeck MA, Adamson B, Villalta JE, Chen Y, Whitehead EH, Guimaraes C, Panning B, Ploegh HL, Bassik MC, Qi LS, Kampmann M, Weissman JS, Genome-Scale CRISPR-Mediated Control of Gene Repression and Activation. Cell. 159, 647–661 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Horlbeck MA, Gilbert LA, Villalta JE, Adamson B, Pak RA, Chen Y, Fields AP, Park CY, Corn JE, Kampmann M, Weissman JS, Compact and highly active next-generation libraries for CRISPR-mediated gene repression and activation. Elife. 5 (2016), doi: 10.7554/eLife.19760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Adamson B, Norman TM, Jost M, Cho MY, Nuñez JK, Chen Y, Villalta JE, Gilbert LA, Horlbeck MA, Hein MY, Pak RA, Gray AN, Gross CA, Dixit A, Parnas O, Regev A, Weissman JS, A Multiplexed Single-Cell CRISPR Screening Platform Enables Systematic Dissection of the Unfolded Protein Response. Cell. 167, 1867–1882.e21 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tak YE, Kleinstiver BP, Nuñez JK, Hsu JY, Horng JE, Gong J, Weissman JS, Joung JK, Inducible and multiplex gene regulation using CRISPR-Cpf1-based transcription factors. Nat. Methods 14, 1163–1166 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bergsma W, A bias-correction for Cramér’s and Tschuprow’s. Journal of the Korean Statistical Society. 42 (2013), pp. 323–328. [Google Scholar]
- 92.Lopez R, Regier J, Cole MB, Jordan MI, Yosef N, Deep generative modeling for single-cell transcriptomics. Nat. Methods 15, 1053–1058 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Liberzon A, Subramanian A, Pinchback R, Thorvaldsdóttir H, Tamayo P, Mesirov JP, Molecular signatures database (MSigDB) 3.0. Bioinformatics. 27, 1739–1740 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wolf FA, Angerer P, Theis FJ, SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 19, 15 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Joy JB, Liang RH, McCloskey RM, Nguyen T, Poon AFY, Ancestral Reconstruction. PLoS Comput. Biol 12, e1004763 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Slatkin M, Maddison WP, A cladistic measure of gene flow inferred from the phylogenies of alleles. Genetics. 123, 603–613 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.McPherson A, Roth A, Laks E, Masud T, Bashashati A, Zhang AW, Ha G, Biele J, Yap D, Wan A, Others, Divergent modes of clonal spread and intraperitoneal mixing in high-grade serous ovarian cancer. Nat. Genet 48, 758 (2016). [DOI] [PubMed] [Google Scholar]
- 98.Deshwar AG, Vembu S, Yung CK, Jang GH, Stein L, Morris Q, PhyloWGS: Reconstructing subclonal composition and evolution from whole-genome sequencing of tumors. Genome Biol. 16, 35 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.El-Kebir M, Oesper L, Acheson-Field H, Reconstruction of clonal trees and tumor composition from multi-sample sequencing data (2015) (available at https://academic.oup.com/bioinformatics/article-abstract/31/12/i62/216528). [DOI] [PMC free article] [PubMed]
- 100.Sankoff D, Minimal Mutation Trees of Sequences. SIAM J. Appl. Math 28, 35–42 (1975). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.