Abstract
Mitotic chromosome movements are orchestrated by interactions between spindle microtubules and chromosomes. It is well known that kinetochore is the major site where microtubule-chromosome attachment occurs. However, the functions of other domains of chromosome such as chromosome periphery have remained elusive. Our previous studies show that PinX1 distributes to chromosome periphery and kinetochore during mitosis, and harbors the microtubule binding activity. Here we report that PinX1 interacts with Nucleolin, a chromosome periphery protein, through its C-termini. Deconvolution microscopic analyses show PinX1 mainly co-localizes with Nucleolin at chromosome periphery in prometaphase. Moreover, depletion of Nucleolin abolishes chromosome periphery localizations of PinX1, suggesting a functional interrelationship between PinX1 and Nucleolin. Importantly, repression of PinX1 and Nucleolin abrogates chromosome segregation in real-time mitosis, validating the functional importance of PinX1–Nucleolin interaction. We propose PinX1 is recruited to chromosome periphery by Nucleolin and a complex of PinX1 and Nucleolin is essential for faithful chromosome congression.
Keywords: Mitotic chromosome periphery, Chromosome congression, Nucleolin, PinX1
Introduction
A complex layer existing around chromosomes during mitosis, hereafter referred to as the chromosome periphery, has been observed early in 1929 [1]. Cryo-electron microscopy studies showed that it is composed of closely packed fibrils associated with dense granules surrounding mitotic chromosomes [2]. This meshwork of fibrils and granules extends a short distance from the chromosomes, but is distinct from the surrounding cytoplasm [3,4]. After decades of researches, numerous proteins have been identified as constituents of chromosome periphery. Some of these proteins, such as topoisomerase II, Ki-67, Ku and H1.X, etc., are involved in maintaining chromosome structure [5–8] while others (e.g. RCC1, Kid and Kif4, etc.) participate in spindle formation and chromosome movement [9–11].
Nucleolin, which belongs to a RNA binding protein family, is a 707 amino acid protein. It is present in abundance at the dense fibrillar and granular regions of nucleolus, and appears to participate in many fundamental aspects of transcriptional regulation, cell proliferation and growth [12]. Recently, Nucleolin has been reported to localize to chromosome periphery during mitosis. Depletion of Nucleolin leads to a prolonged cell cycle with misaligned chromosomes. In addition, cold treatment experiments indicated that kinetochore microtubules become unstable in the absence of Nucleolin. Furthermore, it is likely that Nucleolin recruit several other nucleolar proteins such as fibrillarin and B23 to chromosome periphery, as depletion of Nucleolin abolishes the signals of these proteins from chromosome periphery [13].
PinX1 was first identified as a Pin2/TRF1 interacting protein in a yeast two-hybrid screen [14]. It serves as a natural telomerase inhibitor by interacting with the hTERT and hTR [15], and sequestering them in nucleoli [16,17]. Although the ultrastructure of chromosome periphery has been characterized and lots of components have been identified, our understandings about the architecture of these components and the functions of this specialized chromosome domain in mitosis are far to be complete. Here we report that the new identified chromosome peripheral protein PinX1 interacts with Nucleolin in vivo. Reconstitution of the PinX1–Nucleolin interaction using recombinant fusion proteins reveals that the interaction is direct and mediated by the C-termini of PinX1. PinX1 colocalizes with Nucleolin at chromosome periphery. More interestingly, depletion of Nucleolin abolishes the chromosome periphery localization of PinX1. Live cell imaging studies indicate that PinX1–Nucleolin participates in chromosome congression during mitosis. At last, we proposed a model of these chromosome periphery proteins in facilitating chromosome congression.
Materials and methods
Cell culture and synchronization.
HeLa cells (American Type Culture Collection, Rockville, MD) were maintained as sub-confluent monolayers in Dulbecco’s modified Eagle’s medium (Invitrogen) with 10% fetal bovine serum (Hyclone, Logan, UT) and 100 units/ ml penicillin plus 100 μg/ml streptomycin (Invitrogen) at 37 °C with 8% CO2. Cells were synchronized at G1/S with 5 mM thymidine for 12~16 h and then washed with phosphate-buffered saline (PBS) 5 times and cultured in thymidine-free medium for 10 h. To collect mitotic cells for in vitro studies, cells were synchronized with 0.1 μg/ml nocodazole for 16 h.
Plasmids construction and protein expression.
The cDNA of PinX1 (NM_017884) was kindly provided by Dr. Kunping Lu (Harvard). To generate green fluorescent protein (GFP)-tagged and bacterial expression constructs of PinX1 and deletions, PCR-amplified cDNAs were cloned into pEGFP-C1 (Clontech) and pGEX-5X-3 (Amersham Bioscience) vectors by EcoRI and SalI. The bacterial expression construct of Nucleolin, MBP-1234-RGG, was a generous gift from Dr. Nancy Maizels (Yale).
The GST-tagged proteins and the MBP-1234-RGG were expressed and purified as previously reported [18,19].
Antibodies and RNA interference.
The following antibodies were used: anti-PinX1 (Abnova), anti-Nucleolin (Abcam), anti-cyclin B (BD). siRNA targeting PinX1 (SI00684971) and Nucleolin (SI02654925) were purchased from Qiagen. All the siRNAs and plasmids were transfected into HeLa cells using Lipofectamine 2000 (Invitrogen).
In vitro pulldown and immunoprecipitation.
GST-PinX1-bound Sepharose beads were used as an affinity matrix to isolate interacting proteins from mitotic HeLa cell lysate. Briefly, 5 × 107 HeLa cells were synchronized by nocodazole and collected by mitotic shake off. The mitotic cells were washed once with PBS and lysed in lysis buffer (50 mM Hepes, pH 6.9, 150 mM NaCl, 2 mM EGTA, 0.1% Triton X-100, 1 mM phenylmethylsulfonyl fluoride, 10 g/ml leupeptin and 10 g/ml pepstatin A). After centrifugation, the supernatant was first pre-cleared by incubating with GST-bound Sepharose beads for 2 h, and then incubated with GST-PinX1-bound Sepharose beads for 4 h. All the incubations were carried out at 4 °C to avoid protein degradation. Beads were washed 4 times with lysis buffer and boiled in SDS sample buffer, followed by fractionation of bound proteins on 8% SDS–PAGE gel. The protein band of interest was removed for mass spectrometric analyses as described previously [20].
For immunoprecipitation, control IgG and Nucleolin mAb were first conjugated to protein A/G (Pierce) according to the manufacturer’s protocol, and then incubated with mitotic HeLa cell lysate for 4 h. After extensive washes, beads were boiled in SDS sample buffer. The bound proteins were fractionated on 10% SDS–PAGE gel and transferred to nitrocellulose membrane. The membrane was divided into three strips and probed with antibodies against Nucleolin, PinX1 and tubulin, respectively.
Immunofluorescence microscopy and time-lapse microscopy.
Cells were seeded onto sterile, acid-treated 12-mm coverslips in 24 well plates (Corning Glass Works, Corning, NY). The next day, the cells were transfected with 1 μl of Lipofectamine 2000 pre-mixed with plasmids or siRNAs described above. If not specified, 48 h after transfection, cells were rinsed with PHEM buffer (100 mM PIPES, 20 mM Hepes, pH 6.9, 5 mM EGTA, 2 mM MgCl2 and 4 M glycerol) and fixation in freshly prepared 3.7% formaldehyde for 5 min. After permeabilization and rinsed three times in PBS, cells were blocked with PBST (0.05% Tween 20 in PBS) with 1% bovine serum albumin (Sigma), followed by incubation with various primary antibodies in a humidified chamber for 1 h. After three washes in PBST, primary antibodies were visualized by FITC (fluorescein isothiocyanate) or rhodamine conjugated goat anti-mouse or rabbit IgG. DNA was stained with DAPI (4′,6-diamidino-2-phenylindole). Slides were examined with a DeltaVision Deconvolution microscopy (Applied Precision Inc.). Images were processed using DeltaVision SoftWorx software. Images for display were generated by projecting the sum of the optical sections using the maximum-intensity method.
For time-lapse microscopy, cells were cultured in a glass-bottom culture dish (MatTek, MA) with Leibovitz’s L-15 medium (Invitrogen) at 37 °C and examined with a DeltaVision microscopy system. Images were acquired at ~2 min intervals and presented in Photoshop.
Results and discussion
Identification of Nucleolin as a novel PinX1-binding partner
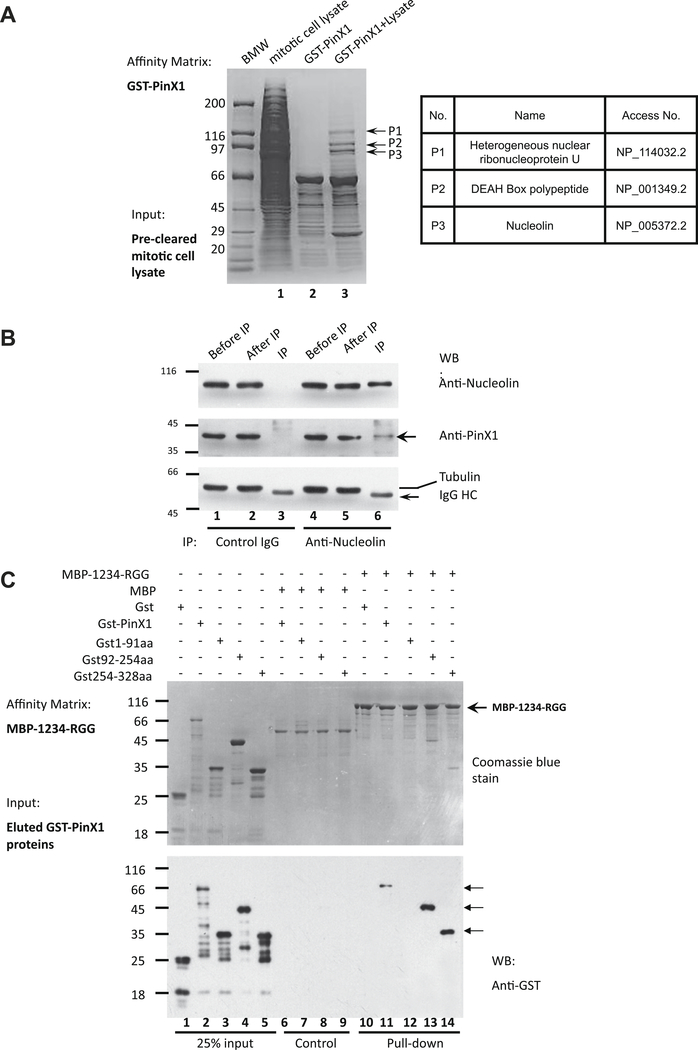
Our previous studies showed that PinX1 localizes to chromosome periphery in mitosis (Kai Yuan et al. submitted for publication). To explore the molecular mechanism, we carried out a pull-down assay using GST-PinX1 bound glutathione beads as an affinity matrix to incubate pre-cleared mitotic HeLa cell lysate. After extensive washing, the beads were boiled in SDS–PAGE sample buffer and the components separated on SDS–PAGE. As shown on the coomassie-stained gel (Fig. 1A, left panel, lane 3), GST-PinX1 absorbed several proteins from the lysate. The indicated bands were removed and digested with trypsin, and the resulting tryptic peptides were submitted for mass spectrographic analyses and database search. The three proteins identified were heterogeneous nuclear ribonucleoprotein U (P1), the DEAH box polypeptide (P2) and Nucleolin (P3). The nucleolar protein, Nucleolin was recently reported to distribute to chromosome periphery and have an important role in chromosome congression [13].
Fig. 1.
PinX1 interacts with chromosome periphery protein Nucleolin in vitro and in vivo. (A) Identification of PinX1–Nucleolin interaction. Mitotic HeLa cell Triton X-100 extracts were applied to GST-PinX1 affinity column. After binding, columns were extensively washed, and bound proteins were eluted with SDS sample buffer and separated by SDS–PAGE. The indicated protein (P1–P3) was extracted from the gel and digested with trypsin, and the amino acid sequence of peptides was determined by MALDI-TOF mass spectrometry. Table shown on the right illustrated the candidate proteins identified by mass spectrometric analyses. (B) PinX1 forms a cognate complex with Nucleolin in vivo. Extracts from nocodazole-synchronized mitotic HeLa cells were incubated with antibodies against Nucleolin (lanes 4–6) and control IgG (lanes 1–3), and immunoprecipitants (IP) were resolved by SDS–PAGE. Western blotting verified co-immunoprecipitation of PinX1 (middle panel) and Nucleolin (upper panel). No tubulin was detected in Nucleolin immunoprecipitants (lower panel). Lanes 2 and 5 are non-binding materials. (C) Reconstitution of the PinX1–Nucleolin interaction using recombinant fusion proteins. Aliquots of MBP-tagged recombinant Nucleolin fragment (1234-RGG) purified on maltose-agarose beads were used as affinity matrixes to absorb purified GST-tagged Pinx1 protein and its deletion mutant proteins purified from bacteria (lane 2–5). Aliquots of MBP protein were used as controls (lanes 6–9). Western blotting analyses using GST antibody show that the C-terminal PinX1 (aa92–328) is responsible for this direct binding.
To test if PinX1 forms a cognate complex with Nucleolin, we carried out an immunoprecipitation in which Nucleolin antibody and control IgG were pre-conjugated to the protein A/G Sepharose beads and incubated with mitotic HeLa cells lysate. After extensive washes, the immunoprecipitants in the Protein A/G beads were solubilized in SDS–PAGE sample buffer and fractionated in SDS–PAGE. As shown in Fig. 1B, PinX1 was present in the anti-Nucleolin immunoprecipitant (lane 6, middle panel), but not the control IgG immunoprecipitant (lane 3, middle panel). Neither Nucleolin nor PinX1 were precipitated by control IgG (lane 3; middle and upper panels). In addition, no tubulin was precipitated by Nucleolin antibody, indicating the immunoprecipitation was specific (lanes 3 and 6). Thus, we conclude that PinX1 forms a cognate complex with Nucleolin in mitosis.
To validate if PinX1 physically interacts with Nucleolin, we performed an in vitro pull-down assay in which recombinant Nucleolin and PinX1 proteins were utilized. To this end, MBP tagged Nucleolin lacking the N-terminal histone 1 like domain (named MBP-1234-RGG), was purified as described [18,21] and used as an affinity matrix to incubate with the GST-PinX1 and its deletion mutants. After extensive washes, the absorbed proteins on the MBP-1234-RGG affinity matrix were fractionated in SDS–PAGE followed by Western blotting analysis using GST antibody. As shown in Fig. 1C, full-length, 92–254aa and 254–328aa, but not 1–91aa of PinX1 can be pulled down by MBP-1234-RGG (lanes 11–14). As control, GST was not absorbed by the Nucleolin affinity matrix, indicating the specificity of PinX1–Nucleolin interaction. Thus, we conclude that PinX1 physically binds to Nucleolin.
PinX1 co-localizes with Nucleolin to the mitotic chromosome periphery
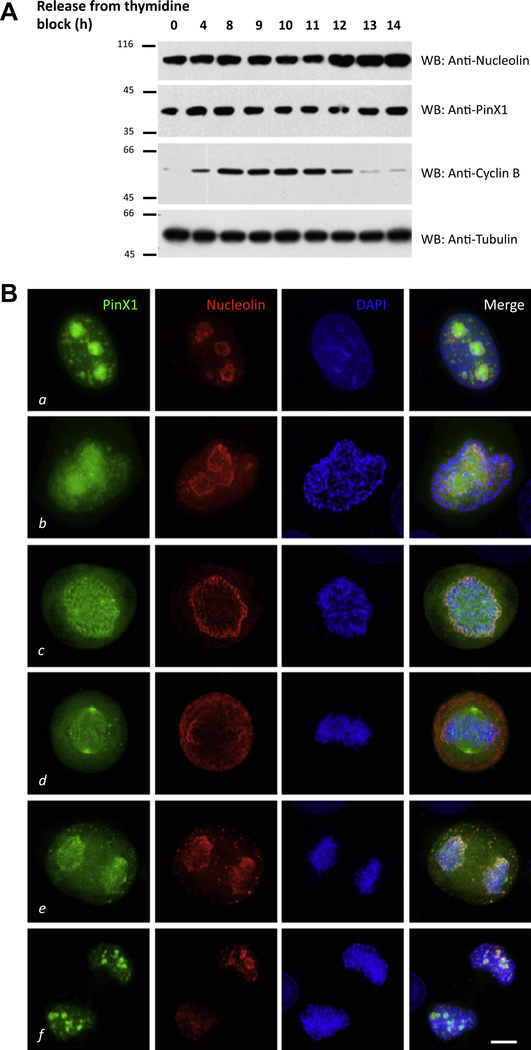
Having demonstrated the bona fide interaction between PinX1 and Nucleolin, we wanted to investigate their cell cycle expressions and spatiotemporal distribution profiles. To study the cell cycle expressions of these two proteins, HeLa cells were released from thymidine block to the indicated time points, and harvested for Western blot. The expression levels of PinX1 and Nucleolin remain almost constant during the cell cycle (Fig. 2A).
Fig. 2.
PinX1 co-localizes with Nucleolin in mitosis. (A) Cell cycle expression of PinX1 and Nucleolin. HeLa cells were harvested at the indicated time points after releasing from thymidine block and boiled in SDS sample buffer. The samples were analyzed by Western blot with anti-PinX1, anti-Nucleolin, anti-cyclin B and antitubulin. (B) This montage represents optical images collected from PinX1 transfected HeLa cells triply stained for GFP-PinX1 (green), Nucleolin (red) and DAPI (blue). In prometaphase, PinX1 co-distributes with Nucleolin mainly on the perichromosomal regions with light deposition on kinetochore (c). Bar: 10 μm.
To investigate their spatiotemporal distribution profiles, HeLa cells transfected with GFP-PinX1 were fixed and counter-stained with anti-Nucleolin antibody and DAPI. As shown in Fig. 2B, PinX1 resided mainly within nucleoli while Nucleolin was prominently localized to the outer layer of the nucleoli of in interphase cells (a); In prophase, parts of PinX1 were relocated to the kinetochores while Nucleolin still enriched in nucleoli structures (b); In prometaphase, PinX1 and Nucleolin strongly co-localized at the chromosome periphery (c); In metaphase, chromosome periphery localizations of PinX1 were reduced, and spindle localization of PinX1 became obvious (d); In anaphase, some PinX1 and Nucleolin remained associated with chromosome periphery, while some of them were packaged into nucleolus-derived foci (DNFs) [22] in cytoplasm (e); In telophase, PinX1 and Nucleolin were recycled back to the forming Nucleoli (f).
Nucleolin recruits PinX1 to the chromosome periphery
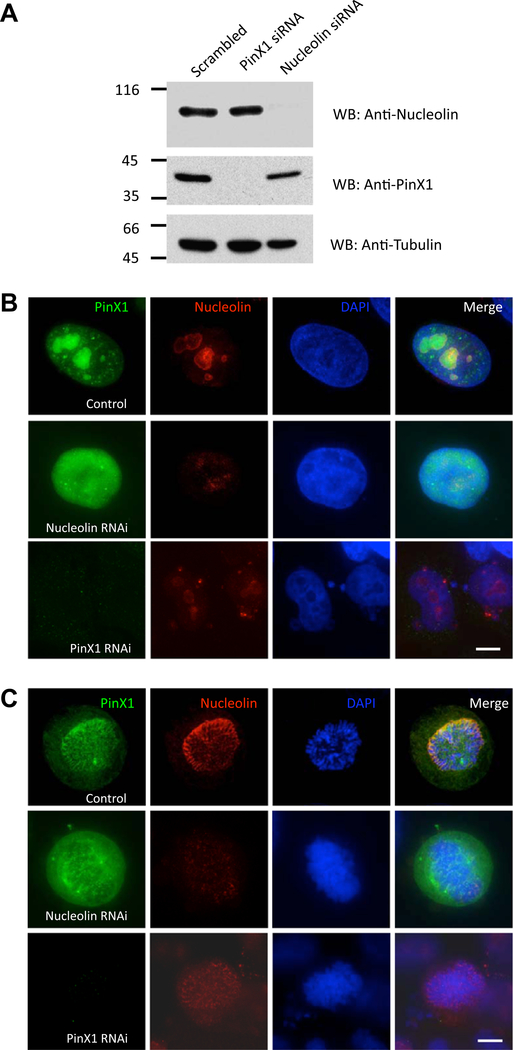
Next, we wanted to examine the localization inter-dependency of PinX1–Nucleolin. To this end, control, Nucleolin-depleted and PinX1-depleted cells were stained with anti-Nucleolin, anti-PinX1 and DAPI respectively. The efficiency and specificity of PinX1 and Nucleolin depletion judged by the Western blotting analyses is shown in Fig. 3A. In interphase cells (Fig. 3B), nucleolus distribution of PinX1 was disrupted in the absence of Nucleolin (Nucleolin RNAi panel); while consistent with our recent report (Kai Yuan et al. submitted for publication), depletion of PinX1 generated several micronuclei in interphase, and these micronuclei were Nucleolin positive (PinX1 RNAi panel). In prometaphase cells (Fig. 3C), depletion of Nucleolin abolished chromosome periphery localization of PinX1; however, the kinetochore and spindle localization of PinX1 were not affected (Nucleolin RNAi panel). While chromosome periphery distribution of Nucleolin was also slightly reduced in PinX1 eliminated cells (PinX1 RNAi panel).
Fig. 3.
Nucleolin recruits PinX1 to the chromosome periphery. (A) Specificity and efficiency of siRNA treatments in HeLa cells. Aliquots of HeLa cells were transfected with 50 nM siRNA oligonucleotide duplexes for PinX1, Nucleolin and a control scrambled oligonucleotide for 96 h and subjected to SDS–PAGE and immunoblotting. Upper panel: Nucleolin; middle panel: PinX1; lower panel: tubulin. (B) In interphase, depletion of Nucleolin destroyed the nucleolus structure and abolished nucleolus localization of PinX1 (Nucleolin RNAi panel). Depletion of PinX1 yielded several Nucleolin-positive micronuclei cells and slightly diminished the nucleolus localization of Nucleolin (PinX1 RNAi panel). Bar: 10 μm. (C) In mitosis, chromosome peripheral localization of PinX1 almost disappeared in the absence of Nucleolin (Nucleolin RNAi panel); chromosome peripheral localization of Nucleolin was also weakened in the absence of PinX1 (PinX1 RNAi panel). Bar: 10 μm.
PinX1 cooperates with Nucleolin for chromosome congression
Given the interaction between PinX1 and Nucleolin and their specific distribution on chromosome periphery, we sought to probe the functional relevance of PinX1 and Nucleolin in mitotic progression.
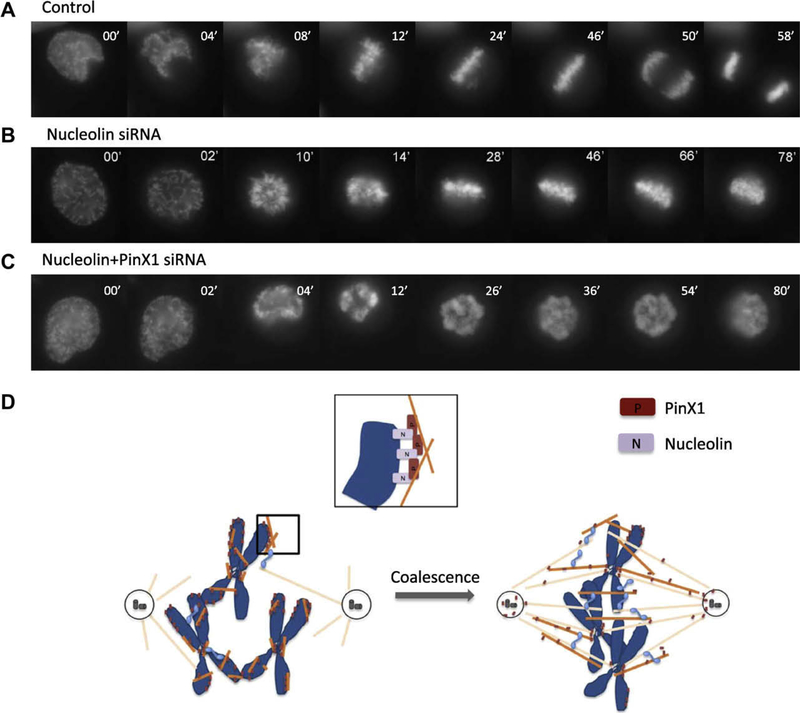
To this end, HeLa cells were transfected with mock, Nucleolin or Nucleolin plus PinX1 siRNA, along with GFP-H2B to visualize the chromosomes [19]. Chromosome movements of these cells were monitored by live cell imaging. As shown in Fig. 4A, in control-transfected cells, chromosome congression was finished ~26 min after NEB. The anaphase onset was observed ~47 min after NEB, and the complete segregation of sister chromatids was achieved ~58 min after NEB. However, in Nucleolin siRNA transfected cells (Fig. 4B), chromosome congression was not well-achieved and sister chromatids failed to separate even at ~78 min after NEB, which is consistent with a previous study [13]. Moreover, in the Nucleolin and PinX1 double depleted cells (Fig. 4C), defects in chromosome congression were more severe. Combined with the aforementioned results, we propose that Nucleolin recruits PinX1 to chromosome periphery and participate in chromosome congression.
Fig. 4.
Manipulation of Nucleolin–PinX1 expression levels with siRNA affects mitotic chromosome congression. (A–C) Live cell imaging of chromosome movements in control, Nucleolin or Nucleolin/PinX1 depleted cells. Chromosomes were marked by GFP-H2B. Elimination of Nucleolin or Nucleolin/PinX1 leads to chromosome congression defects. (D) A model demonstrating the function of Nucleolin–PinX1 in mitosis.
A model demonstrating the function of PinX1-Nucleolin in prometaphase
Decades of researches have focused on the intrinsic properties of mitotic chromosomes. In 1975, McGill and Brinkley have reported that human chromosomes and centrioles function as the microtubule nucleating sites [23]. It is noteworthy that although kinetochores maybe the prominent sites attached to these microtubules [24], lots of short microtubules were also found at the chromosome periphery region [23]. Another elegant experiments, done by Heald and colleagues in 1996, have demonstrated that DNA-coated beads can induce the formation of bipolar spindle in Xenopus egg extracts, in the absence of centrosomes and kinetochores [25]. This observation emphasizes the role of chromosome in bipolar spindle formation, and it is also yield a possibility that the chromosome peripheral proteins may involve in this process.
Recent studies have identified Nucleolin as a chromosome periphery protein and participate in chromosome congression [13]. Our latest results suggested PinX1 localizes to chromosome periphery and harbors the microtubule binding activity (Kai Yuan et al. submitted for publication). Here we characterized the interaction between PinX1 and Nucleolin, revealed the molecular mechanism underlying the chromosome periphery localization of PinX1, and explored the roles of these chromosome periphery components in chromosome congression.
Based on the observations mentioned above, we proposed a model here to illustrate how chromosome peripheral protein PinX1 cooperates with Nucleolin in chromosome capture and congression (Fig. 4D). In prometaphase, Nucleolin recruits PinX1 to the chromosome periphery, at where PinX1 grasps several short microtubules, which may be generated by kinetochore or chromosome mediated microtubule nucleation. Meanwhile, the centrosome-nucleated microtubules start to elongate. With the help of motor proteins and microtubule-associated proteins, the microtubules enriched around chromosomes coalesce with the centrosomal microtubules, and greatly facilitate the “search and capture” process [26].
Taken together, in this study we identified Nucleolin as a binding partner of PinX1 and specifies PinX1’s chromosome peripheral localization. This PinX1–Nucleolin complex participates in chromosome congression. We believe these new findings will help us to understand the architecture of chromosome periphery, and also shed new light on the functions of chromosome periphery during mitosis.
Acknowledgments
We thank Dr. Kunping Lu and Dr. Nancy Maizel for reagents. This work was supported by grants from the Chinese Academy of Science (KSCX1-YW-R65, KSCX2-YW-H10), Chinese 973 project (2002CB713700, 2007CB914503, and 2006CB943600), Chinese 863 project (2006AA02A247), Chinese Natural Science Foundation (30270654, 30070349, 90508002, and 30121001), a Georgia Cancer Coalition Breast Cancer Research grant and an Atlanta Clinical and Translational Science Award Chemical Biology grant (to X.Y.). The facilities used were partially supported by NIH/NCRR/RCM1 Grant G-12-RR03034.
References
- [1].Sharp LW, Structure of large somatic chromosomes, Bot. Gaz. 88 (1929) 349– 382. [Google Scholar]
- [2].Gautier T, Masson C, Quintana C, Arnoult J, Hernandez-Verdun D, The ultrastructure of the chromosome periphery in human cell lines. An in situ study using cryomethods in electron microscopy, Chromosoma 101 (1992) 502–510. [DOI] [PubMed] [Google Scholar]
- [3].Hernandez-Verdun D, Gautier T, The chromosome periphery during mitosis, Bioessays 16 (1994) 179–185. [DOI] [PubMed] [Google Scholar]
- [4].Van Hooser AA, Yuh P, Heald R, The perichromosomal layer, Chromosoma 114 (2005) 377–388. [DOI] [PubMed] [Google Scholar]
- [5].Earnshaw WC, Halligan B, Cooke CA, Heck MM, Liu LF, Topoisomerase II is a structural component of mitotic chromosome scaffolds, J. Cell. Biol. 100 (1985) 1706–1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Takagi M, Matsuoka Y, Kurihara T, Yoneda Y, Chmadrin: a novel Ki-67 antigen-related perichromosomal protein possibly implicated in higher order chromatin structure, J. Cell. Sci. 112 (Pt 15) (1999) 2463–2472. [DOI] [PubMed] [Google Scholar]
- [7].Bertuch AA, Lundblad V, The Ku heterodimer performs separable activities at double-strand breaks and chromosome termini, Mol. Cell. Biol. 23 (2003) 8202–8215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Takata H, Matsunaga S, Morimoto A, Ono-Maniwa R, Uchiyama S, Fukui K, H1.X with different properties from other linker histones is required for mitotic progression, FEBS Lett. 581 (2007) 3783–3788. [DOI] [PubMed] [Google Scholar]
- [9].Lee YM, Lee S, Lee E, Shin H, Hahn H, Choi W, Kim W, Human kinesin superfamily member 4 is dominantly localized in the nuclear matrix and is associated with chromosomes during mitosis, Biochem. J. 360 (2001) 549–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Yajima J, Edamatsu M, Watai-Nishii J, Tokai-Nishizumi N, Yamamoto T, Toyoshima YY, The human chromokinesin Kid is a plus end-directed microtubule-based motor, EMBO J. 22 (2003) 1067–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Hood FE, Clarke PR, RCC1 isoforms differ in their affinity for chromatin, molecular interactions and regulation by phosphorylation, J. Cell. Sci. 120 (2007) 3436–3445. [DOI] [PubMed] [Google Scholar]
- [12].Srivastava M, Pollard HB, Molecular dissection of nucleolin’s role in growth and cell proliferation: new insights, FASEB J. 13 (1999) 1911–1922. [PubMed] [Google Scholar]
- [13].Ma N, Matsunaga S, Takata H, Ono-Maniwa R, Uchiyama S, Fukui K, Nucleolin functions in nucleolus formation and chromosome congression, J. Cell. Sci. 120 (2007) 2091–2105. [DOI] [PubMed] [Google Scholar]
- [14].Zhou XZ, Lu KP, The Pin2/TRF1-interacting protein PinX1 is a potent telomerase inhibitor, Cell 107 (2001) 347–359. [DOI] [PubMed] [Google Scholar]
- [15].Banik SS, Counter CM, Characterization of interactions between PinX1 and human telomerase subunits hTERT and hTR, J. Biol. Chem. 279 (2004) 51745– 51748. [DOI] [PubMed] [Google Scholar]
- [16].Lin J, Blackburn EH, Nucleolar protein PinX1p regulates telomerase by sequestering its protein catalytic subunit in an inactive complex lacking telomerase RNA, Genes Dev. 18 (2004) 387–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Lin J, Jin R, Zhang B, Yang PX, Chen H, Bai YX, Xie Y, Huang C, Huang J, Characterization of a novel effect of hPinX1 on hTERT nucleolar localization, Biochem. Biophys. Res. Commun. 353 (2007) 946–952. [DOI] [PubMed] [Google Scholar]
- [18].Hanakahi LA, Sun H, Maizels N, High affinity interactions of nucleolin with G-G-paired rDNA, J. Biol. Chem. 274 (1999) 15908–15912. [DOI] [PubMed] [Google Scholar]
- [19].Yuan K, Hu H, Guo Z, Fu G, Shaw AP, Hu R, Yao X, Phospho-regulation of HsCdc14A By Polo-like kinase 1 is essential for mitotic progression, J. Biol. Chem. 282 (2007) 27414–27423. [DOI] [PubMed] [Google Scholar]
- [20].Fang Z, Miao Y, Ding X, Deng H, Liu S, Wang F, Zhou R, Watson C, Fu C, Hu Q, Lillard JW Jr., Powell M, Chen Y, Forte JG, Yao X, Proteomic identification and functional characterization of a novel ARF6 GTPase-activating protein, ACAP4, Mol. Cell. Proteomics 5 (2006) 1437–1449. [DOI] [PubMed] [Google Scholar]
- [21].Cao X, Ding X, Guo Z, Zhou R, Wang F, Long F, Wu F, Bi F, Wang Q, Fan D, Forte JG, Teng M, Yao X, PALS1 specifies the localization of ezrin to the apical membrane of gastric parietal cells, J. Biol. Chem. 280 (2005) 13584–13592. [DOI] [PubMed] [Google Scholar]
- [22].Dundr M, Meier UT, Lewis N, Rekosh D, Hammarskjold ML, Olson MO, A class of nonribosomal nucleolar components is located in chromosome periphery and in nucleolus-derived foci during anaphase and telophase, Chromosoma 105 (1997) 407–417. [DOI] [PubMed] [Google Scholar]
- [23].McGill M, Brinkley BR, Human chromosomes and centrioles as nucleating sites for the in vitro assembly of microtubules from bovine brain tubulin, J. Cell. Biol. 67 (1975) 189–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Mitchison TJ, Kirschner MW, Properties of the kinetochore in vitro. I. Microtubule nucleation and tubulin binding, J. Cell. Biol. 101 (1985) 755–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Heald R, Tournebize R, Blank T, Sandaltzopoulos R, Becker P, Hyman A, Karsenti E, Self-organization of microtubules into bipolar spindles around artificial chromosomes in Xenopus egg extracts, Nature 382 (1996) 420–425. [DOI] [PubMed] [Google Scholar]
- [26].Wollman R, Cytrynbaum EN, Jones JT, Meyer T, Scholey JM, Mogilner A, Efficient chromosome capture requires a bias in the ‘search-and-capture’ process during mitotic-spindle assembly, Curr. Biol. 15 (2005) 828–832. [DOI] [PubMed] [Google Scholar]