Abstract
Breast cancer angiogenesis is elicited and regulated by a number of factors including the Notch signaling. Notch receptors and ligands are expressed in breast cancer cells as well as in the stromal compartment and have been implicated in carcinogenesis. Signals exchanged between neighboring cells through the Notch pathway can amplify and consolidate molecular differences, which eventually dictate cell fates. Notch signaling and its crosstalk with many signaling pathways play an important role in breast cancer cell growth, migration, invasion, metastasis and angiogenesis, as well as cancer stem cell (CSC) self-renewal. Therefore, significant attention has been paid in recent years toward the development of clinically useful antagonists of Notch signaling. Better understanding of the structure, function and regulation of Notch intracellular signaling pathways, as well as its complex crosstalk with other oncogenic signals in breast cancer cells will be essential to ensure rational design and application of new combinatory therapeutic strategies. Novel opportunities have emerged from the discovery of Notch crosstalk with inflammatory and angiogenic cytokines and their links to CSCs. Combinatory treatments with drugs designed to prevent Notch oncogenic signal crosstalk may be advantageous over λ secretase inhibitors (GSIs) alone. In this review, we focus on the more recent advancements in our knowledge of aberrant Notch signaling contributing to breast cancer angiogenesis, as well as its crosstalk with other factors contributing to angiogenesis and CSCs.
Keywords: Notch, Breast cancer, Tumor angiogenesis, Oncogenesis, Breast cancer stem cells
1. Introduction
The formation of new blood vessels from existing ones (angiogenesis) is a crucial requirement for the growth, progression and metastatic spread of a tumor [1]. Low oxygen microenvironment triggers angiogenesis in normal and pathological conditions, i.e., tumor growth [2]. The malignant cells undergo an angiogenic switch leading to secretion of angiogenic factors and proteolytic enzymes in response to hypoxia culminating in the activation of endothelial cell (EC) proliferation, migration and establishment of a robust capillary network. This irregular and ill-organized network is capable of providing the growing tumor mass with all the required metabolites. In addition, the tumor angiogenesis network also provides tumor cells with the opportunity to enter the circulation and the opportunity to form distant metastases [3]. Tumor angiogenesis is elicited and regulated by several factors. Among these factors, Notch signaling plays an important role. Notch is essential for a variety of cell fate decisions and can regulate diverse cellular biological processes especially during embryogenesis. To signal, membrane-bound Notch receptors and ligands need to be co-expressed in adjacent cells. Notch receptors and ligands are expressed in tumor cells as well as in the stromal compartment and have been implicated in tumorigenesis [4,5]. Notch genes encode transmembrane receptors that are highly conserved from invertebrates to mammals. Notch-mediated signals regulate cell-fate decisions in a large number of developmental systems [6,7]. Such signals are mainly transmitted through direct contact between adjacent cells expressing Notch receptors and their ligands. Notch receptors activated in response to ligand expressed by adjacent cells have the potential to regulate cell fate specification, differentiation, proliferation, or survival [8]. Notch signaling pathway is frequently dysregulated in several human malignancies. Over expression of Notch receptors and their ligands has been found in cervical, colon, head and neck, lung, renal carcinoma, pancreatic cancer, acute myeloid, Hodgkin, Large-cell lymphomas, as well as breast cancer [9–11]. Overall, it is well-established that Notch signaling plays an important role in tumor progression [5,12]. Signals exchanged between neighboring cells through the Notch pathway can amplify and consolidate molecular differences, and influence how cells respond to intrinsic or extrinsic developmental cues that are necessary to unfold specific developmental programs [13]. Because the same signaling pathways within different contexts can trigger a variety of cellular activities, cancer progression activities induced by Notch and its crosstalk with other signaling pathways are also context dependent. In light of several valuable reviews published on the role of Notch signaling in several types of cancer [14–17], we wish to focus this review on the more recent advancements in understanding how aberrant Notch signaling and its crosstalk with other factors contribute to breast cancer angiogenesis and CSC.
2. Structure, activation and function of Notch receptors and ligands
The Notch system in vertebrates comprises four receptors (Notch1–Notch4) and at least five ligands from the families Delta and JAG/Serrate (DSL):JAG1,JAG2, Delta-like (Dll)-1, Dll-3, and Dll-4 [10,11,13]. Ligands of Notch receptors can be divided into several groups based on their domain composition. Canonical DSL ligands (JAG1,JAG2 and Dll-1) are type I cell surface proteins, consisting of the Delta/Serrate/LAG-2 (DSL), Delta and OSM-11-like proteins [DOS, which is specialized tandem EGF repeats] and EGF motifs. The other subtypes of DSL canonical ligands include Dll-3 and Dll-4 that lack the DOS motif [18–20]. Both the DSL and DOS domains are crucial for physical binding with Notch receptor [9]. However some membrane-tethered and secreted noncanonical ligands lacking DSL and DOS domains have also been documented to activate Notch signaling both in vitro and in vivo [19,21–26], which may explain the diverse and frequent effects of Notch signaling with the small number of canonical DSL ligands and receptors in vertebrate genomes [19].
Notch receptors belong to a large single-pass type 1 transmembrane protein family; the extracellular domain consists of 29–36 tandem arrays of EGF (epidermal growth factor)-like repeats, followed by a conserved negative regulatory region (NRR or LNR) consisting of three cysteine-rich Notch Lin12 repeats (N/Lin 12) and a heterodimerization (HD) domain [27]. Notch family members differ in the number of EGF-like repeats, however they share many similarities in structure [9,28]. EGF-like repeats mediate ligand binding, whereas NRR functions to prevent both ligand-dependent and -independent signaling [28]. The cytoplasmic portion of Notch is composed of a DNA binding protein (RBP-Jk associated molecule or RAM) domain and six ankyrin (ANK) repeats, which are flanked by two nuclear localization signals (NLS), followed by a transactivation domain (TAD) and a domain rich in proline, glutamine, serine and threonine residues (PESTs) that controls the receptor half life [9,29,30] (Fig. 1).
Fig. 1.
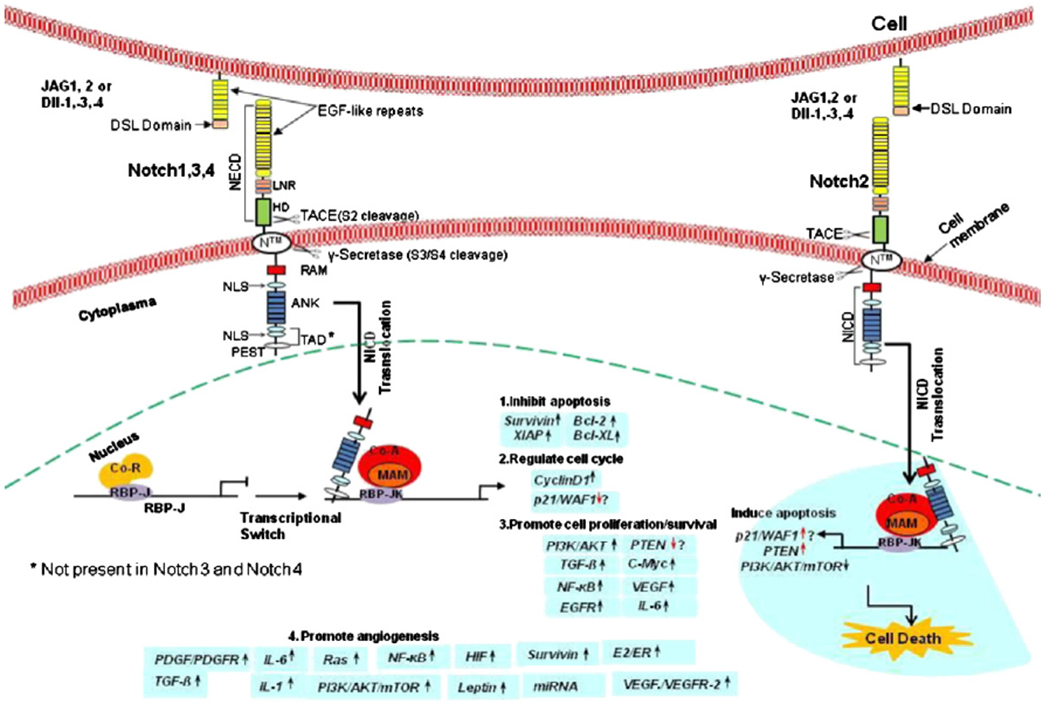
Notch signaling and its possible downstream targets in human cancers and angiogenesis. Mammalian ligands of Notch are membrane-bound proteins containing an extracellular NH2-terminal Delta/Serrate/LAG2 (DSL) motif followed by epidermal growth factor (EGF)-like repeats. Notch receptors are broadly expressed on the cell surface as heterodimers containing a Notch extracellular domain (NECD) composed by multiple extracellular EGF-like repeats and three Lin12/Notch repeats (LNR). Notch receptor cytoplasmatic region or Notch intra-cellular domain (NICD) contains one nuclear localization signals (NLS) linking RAM domain to six ankyrin (ANK) repeats (ANK domain) followed by an additional bipartite NLS, a loosely defined transactivation domain (TAD), and a conserved proline/glutamic acid/ser/threo-rich domain (PEST domain). In the absence of activated Notch signaling, the DNA binding protein RBP-Jk (CSL/CBF1/Su (H)/Lag1, a transcription factor) forms a complex with corepressor molecules that represses transcription of target genes. Ligand binding to NECD triggers successive proteolytic cleavages of Notch cytoplasmatic region by ADAM and γ-secretase, proteases resulting in the release of NICD, which translocates into nucleus and removes corepressors from RBP-Jk. This allows RBP-Jk to recruit a coactivator complex composed of Mastermind (MAM) and several transcription factors to transcriptionally activate Notch target genes. Activation of Notch could impact on the following processes in human cancer: 1) inhibition of apoptosis through upregulation of Survivin [86,87] and Bcl-2 protein family [342]; 2) activation of the cell cycle through upregulation of Cyclin D1 [95]; 3) promotion of cell proliferation/survival through upregulation of PI-3K/Akt [265], TGF-ß [184], c-myc [80], NF-κB [109], EGFR[165] and IL-6 [343] pathways; 4) stimulation of angiogenesis and VEGF/VEGFR-2 autocrine/paracrine loop by upregulation of IL-1 system and VEGF/VEGFR-2 [190,220]; 5) suppression of cancer growth in some cellular situations. For example, Notch2 signaling may function as a tumor suppressor through upregulation of PTEN or down-regulation of PI-3K/Akt/mTOR [344]. Coordinated actions of Notch affect cancer cell growth, migration, invasion, metastasis and angiogenesis, as well as CSC self-renewal.
Membrane localization of Notch requires S1 cleavage of precursor of the Notch receptor. This event occurs in the Golgi network by the action of a furin-like convertase. Then, the two fragments are re-assembled as a non-covalently linked heterodimeric receptor at the cell surface [6]. Mature Notch receptors are heterodimers made up of an extracellular subunit, a transmembrane subunit (N™) and a cytoplasmic subunit. Activation of Notch consists of two consecutive cleavages of the transmembrane receptor upon the binding of a Notch ligand, which triggers S2 cleavage. This process takes places at the cell surface. N™ subunit is cleaved by ADAM/Tumor necrosis factor-α-converting enzyme (TACE) metalloprotease family at Site 2 (located ~12 amino acids before the transmembrane domain). S2 cleavage releases the Notch extracellular domain (NECD) from the heterodimer and creates a membrane-tethered Notch extracellular truncation (NEXT), which becomes a substrate for γ-secretase. S3 is cleaved by γ-secretase at Sites 3 and 4 [31]. This last cleavage occurs on the plasma membrane and/or in endosome. The new mobile cytoplasmic subunit [Notch intracellular domain (NICD or NIC)] is translocated to the nucleus, where it interacts with members of the DNA-binding protein, recombination signal binding protein for immunoglobulin kappa J (RBP-Jk) or CBF1/Su(H)/Lag-1 (CSL) family of transcription factors [8]. Activated NICD–RBP-Jk complex displaces co-repressors and recruits coactivator (co-A) mediating the transcription of target genes such as Hes-1 (hairy enhancer of split), cyclin D, Hey-1 (hairy/enhancer-of-split related with YRPW motif) and others [10,11]. In the absence of NICD, CSL may interplay with the ubiquitous corepressor (Co-R) proteins and histone deacetylases (HDACs) to repress transcription of some target genes [32,33].
3. Notch signaling in vascular development
Gain- and loss-of-function mutations in humans, mice, and zebrafish have demonstrated the involvement of Notch signaling in multiple aspects of vascular development [34–37]. The vascular system comprises arteries, veins, and lymphatics which separated subdivisions into large vessels, small vessels and capillaries. These vessels primarily consist of endothelial cells, supporting cells (smooth muscle cells, SMCs and pericytes), and surrounding matrix. Mounting evidence suggests that Notch ligands (Dll-4, Jagged-1, and Jagged-2), receptors (Notch1, 2, 3, and 4) and effectors (HERP1, 2, and 3) are involved in the development of the vascular system [37–39]. In this process Notch pathway is to identify distinct cell subpopulations from bipotential precursor cells, a process known as lateral specification or lateral inhibition [13,40,41]. Mice homozygous for null mutations of Notch (including Notch1, plus Notch4, and Jagged1) show embryonic lethality together with vascular remodeling defects [35,42]. Both Dll-1-deficient and homozygous Notch2 mutant mouse embryos showed hemorrhage, possibly resulting from poor development of vascular structures. However, Dll-1 and Notch2 were not detected in large vessels of mutant embryos [43,44]. EC-specific expression of Notch4 activated form in transgenic mice led to embryonic lethality with abnormal vessel structure and patterning. This phenotype was similar to that seen in Notch1- and Notch1/Notch4-deficient mice [45]. The similarities between the vascular phenotypes observed in knockout mice (loss-of-function) and transgenic mice (gain-of-function) suggest that a window of appropriate Notch expression levels might be needed for proper development of the embryonic vasculature.
4. Activation of Notch signaling and its target genes in breast cancer angiogenesis
Roles of Notch1 and Notch4 in angiogenesis have been established using mutant mice. Notch1 and Notch1/Notch4 double mutant embryos displayed severe defects in angiogenic vascular remodeling [35]. Among Notch receptors with potential roles in carcinogenesis and angiogenesis, Notch1 (MW: 272 kDa and NICD MW: 110–120 kDa) is relatively the best studied [46–48]. It was found earlier that Notch1, a putative collaborator of c-myc, was mutated in high proportion (52%) in CD4+CD8+ T-cell tumors [49]. These mutations led to high expression of truncated Notch1 proteins. The Notch1 gene was identified as a novel target for mouse mammary tumor virus (MMTV) provirus insertional activation. MMTV insertion in the Notch1 gene induced the overexpression of 5′ truncated ~7 kb RNA (280 kDa mutant protein: Notch1 ectodomain) and truncated 3′ Notch1 transcripts (3.5–4.5 kb) and proteins (86–110 kDa) that can transform HC11 mouse mammary epithelial cells in vitro [46]. Notch1 was further found to be a mediator of oncogenic Ras (retrovirus-associated DNA sequence kinase, small cytoplasmic GTP-binding proteins) [50]. Moreover, Notch1 and JAG1 were co-upregulated upon estrogen treatment not only in MCF-7 breast cancer cells, but also in ECs, suggesting a role of Notch1-JAG1 in angiogenesis [51].
The exact roles of Notch2 (MW: 205 kDa) and NICD (MW:110 kDa) in angiogenesis have not been determined. Notch2−/− mice develop normally until E9.5, and then around E11.5 massive cell death occurs [52]. These results suggest that Notch2 plays an essential role in post implantation development in mice. The role of Notch 2 is dependent on its ankyrin repeats and is probably linked to some aspects of cell specification and/or differentiation [52]. However, Notch2 (unlike Notch1) is not essential for generating hematopoietic stem cells from ECs [53]. The development of hematopoietic cells is closely related to angiogenesis, indicating the existence of hemangioblasts and hemogenic ECs. A recent report shows Notch2 has a role in EC and vascular dysfunction. Inflammatory cytokines can elicit a switch toward Notch2 expression over Notch4, leading to reduced Notch activity and increased apoptosis [54].
Mutations in human Notch3 (MW:244 and NICD MW:86 kDa) cause CADASIL (cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy), a late-onset disorder causing stroke and dementia, which arises from slowly developing systemic vascular lesions ultimately resulting in the degeneration of vascular smooth muscle cell [55,56]. Notch3 expression maintains a differentiated phenotype of mural cells (smooth muscle cells, pericytes, or fibroblasts) through an autoregulatory loop that requires endothelial-expressed Jagged1 [57]. Thus, Notch3 is considered to be critical for proper angiogenesis and mural cell investment [58]. ECs and mural cell interactions support fully functional blood vessels and regulate vessel assembly and differentiation or maturation. Alterations in mural cell density and attachment to the endothelium are associated with several human diseases such as diabetic retinopathy, venous malformation, and hereditary stroke. In addition, mural cells are implicated in regulating tumor growth and have thus been suggested as potential targets in tumor antiangiogenic therapy [59]. Notch4 actions in breast cancer are also cell-context dependent. Early studies indicated that Notch4 is an EC specific homologue of Notch and it may play a crucial role in vasculogenesis and angiogenesis [60]. However, in contrast to Notch1, constitutive Notch4 activation in ECs inhibits angiogenesis in part by promoting β1-integrin-mediated adhesion to the underlying matrix [61]. Such inhibition of angiogenesis requires the ankyrin repeats and appears to involve RBP-Jκ-dependent and independent signaling [62]. On the other hand, mammary carcinogenesis is related to gain-of-function mutations of Notch4 leading to dysregulated levels of the Notch4 NICD [63,64]. Transgenic expression of the 1.8 Kb Notch4 RNA species in non-malignant human mammary epithelial cell line MCF-10A enabled these cells to grow in soft agar, suggesting Notch4 can transform MCF-10A cells [65]. Notch4 was also found to subvert normal epithelial morphogenesis and to promote invasion of the extracellular matrix. Moreover, Notch4 significantly increased the tumorigenic potential in vitro of mammary epithelial cells by changing the morphogenetic properties [66,67].
The best-characterized Notch targets are transcriptional repressors of the Hes (Hes1-7) and Hey subfamilies (Hey1, Hey2, HeyL, HesL/HelT, Dec1/BHLHB2, Dec2/BHLHB3) [68–70]. Both Hes and Hey proteins contain a basic domain which determines DNA binding specificity, and a helix-loop-helix domain which allows the proteins to form homo- or heterodimers. In contrast, Hes6 is a novel estrogen-regulated gene and a potential oncogene overexpressed in breast cancer, with tumor-promoting and proliferative functions [71]. Other Notch target genes include proteins and factors involved in the control of the cell cycle and survival processes such as p21WAF1/Cip1, a cyclin-dependent kinase inhibitor that acts as both a sensor and an effector of multiple antiproliferative signals. Notch activation contributes to contact inhibition of ECs, in part through repression of p21Cip1 expression [72]. Deltex, including Deltex1, Deltex2, Deltex4 [73], acts as a positive regulator of Notch signaling through interactions with the Notch ankyrin repeats [74]. Deltex family also functions as a transcriptional regulator downstream of the Notch receptor, but has no direct role in angiogenesis [75]. Nuclear factor-kappa B (NF-κB) (a transcriptional factor) was identified as a Notch target gene early on [76], and was later found to collaborate with Notch signaling in angiogenesis [77,78]. Dysregulation of cyclin D1 (a mitogenic sensor and allosteric activator of cyclin dependent kinase CDK4/6) [79] and c-myc (an oncogene and cell cycle regulator) is considered one of the hallmarks of many cancers [10–13,80]. Both factors are involved in angiogenesis and have been identified as Notch target genes [79,81].
Notch signaling can modulate apoptosis. NICD interacts with, and can inactivate p53 through phosphorylation [82]. Recently, Survivin, a member of the inhibitor of apoptosis family of proteins (IAP) that induces cell proliferation, was identified as a novel Notch target gene [83,84]. Notch stimulation resulted in direct activation of Survivin gene transcription through at least one RPB-Jκ site in the Survivin promoter [85]. Activation of Notch directly induced the transcriptional up-regulation of Survivin in ER-breast cancer cells [86,87]. Accumulated evidence suggests that lack of Survivin in EC causes embryonic defects in angiogenesis [88]. Moreover, the knockdown of Survivin in ECs could inhibit angiogenesis [89,90].
5. Notch signaling, breast cancer stem cell and angiogenesis
A new theory about the initiation and progression of cancer is emerging from the idea that tumors, like normal adult tissues, contain stem cells (called cancer stem cells, or CSCs) and more importantly, could arise from them [91]. Genetic mutations in genes encoding proteins involved in critical signaling pathways for stem cells such as BMP (bone morphogenetic protein), Notch, Hedgehog and Wnt would allow cells to undergo uncontrolled proliferation and form tumors. Notch receptors and/or ligands were demonstrated to correlate with CSCs in several cancer types (Table 1). Notch activity had increasingly been investigated in breast CSC (BCSC) subpopulation [92–94]. Upregulated Notch expression was found in BCSC and initiating cell populations characterized by phenotypic markers CD44+/CD24− [14], and was linked to tumor-initiating properties and CSC-like invasive features [14]. Notch1 NICD impairs mammary stem cell (CD24+CD29high) self-renewal and facilitates their transformation through a cyclin D1-dependent pathway [95]. Moreover, Notch1 is related to BCSC self-renewal. The ErbB2 (HER2) promoter contains Notch–RBP-Jκ binding sequences [96] that can be activated by Notch1 signaling and increase HER2 transcription in both mammary stem/progenitor cells [15,93] and BCSC. These Notch1 effects could impact the self-renewal properties of BCSC [97]. Expression of erythropoietin receptor (EpoR) on the surface of BCSC has been reported [98]. It has been shown that Notch1 interacts with erythropoietin (Epo) to maintain the self-renewing capacity of BCSC. In addition, recombinant human Epo (rhEpo) increased the numbers of BCSC and self-renewing activity in a Notch-dependent manner through induction of JAG1 [98].
Table 1.
Function of Notch signaling in cancer stem cells (CSCs).
| Cancer types | CSC marker | Notch/ligand | Function or changes | References |
|---|---|---|---|---|
| Bladder cancer | K5, p63, BMI-1, OCT-4 | Notch1 | Activation | [110] |
| Brain medulloblastoma | CD133, CD15 | DLL1 | Cell proliferation | [111] |
| Brain glioblastoma | CD133 | Jagged1, DLL1, DLL4 | Cancer stem-like cell self-renewal | [112] |
| Brain glioblastoma | CD133 | Notch1 | Cell proliferation | [113] |
| Brain glioblastoma | CD133, NESTIN, BMI1 | Notch2 | Tumor growth | [114] |
| Brain medulloblastoma | CD133 | HES1 | Cell proliferation and anchorage | [115] |
| Brain medulloblastoma | CD133 | Notch pathway | Stem-like cells self-renewal | [116] |
| Breast cancer | CD44Hi/CD24Low | Notch1 | CSC self-renewal | [117] |
| Breast cancer | CD44Hi/CD24Low | Notch1 | Brain metastases | [118] |
| Breast cancer | CD24(−) CD44(+) | Notch1 | Cell proliferation | [119] |
| Breast cancer | ESA(+)/CD44(+)/CD24(low) | Notch4 | Stem cell activity & tumor formation | [92] |
| Colon cancer | CD133, CD44, ESA, ALDH1 | Notch | Prevention of apoptosis | [120] |
| Lung cancer | ALDH(+) | Notch3 | CSC maintenance | [121] |
| Liver cancer | Oct3/4, OV6, CD133, EpCAM | Notch | Self-renewal, extensive proliferation | [122] |
| Liver cancer | EpCAM | Jagged1 | Tumorigenesis | [123] |
| Ovary cancer | CD44 High/CD24 Low | Notch1 | Proliferation/division and survival | [124] |
| Pancreatic cancer | CD44 and EpCAM | Notch1 | Acquisition of EMT phenotype | [125] |
Notch1 mRNA is primarily expressed in luminal cells of normal breast epithelium [99]. In contrast, Notch4 is mainly present in the basal cell population and in the BCSC-enriched population [92]. These data suggest that Notch1 and Notch4 may impact different subpopulation cells via distinct roles in BCSC. Secretase inhibitors, DAPT and DBZ, which preferentially affect Notch1 activity, only partially abrogated mammosphere-forming units (MFUs) and tumor formation, whereas Notch4 knockdown caused a significantly greater inhibition in MFUs than Notch1 [92]. Therefore, it was suggested that Notch4 signaling regulates the route from BCSC into progenitor populations. In contrast, Notch1 activity regulates the progenitor proliferation and luminal differentiation. Then, the single activation of Notch1 receptor gene might not be sufficient to generate mammary carcinogenesis in mice. Conversely, activation of the Notch4 receptor inhibited mammary epithelial cell differentiation and could be sufficient for mammary carcinogenesis in mice [100].
Notch2 and Notch3 have been also linked to BCSC. Recently, a single nucleotide polymorphism (SNP) rs11249433 in the 1p11.2 region has been identified as a novel risk factor for breast cancer that was strongly associated with ER+ but not ER− cancer [101]. Notch2 expression was particularly enhanced in carriers of the risk genotypes (AG/GG) of rs11249433, that may favor development of ER+ luminal tumors and affect tumor-initiating cells [101]. Notch3 is a poor activator of Hairy/Enhancer of split 1 and 5 (Hes-1 and Hes-5), in contrast to that of Notch1 [102]. The Notch 3 intracellular domain represses Notch1-mediated activation through Hes promoters. Notch3 is critical for the differentiation of human progenitor cells to luminal lineage in vitro [99]. Notch activation leads to the formation of dimers of Hes and/or Hey proteins that repress the transcription of a variety of genes by interacting with co-repressors or sequestering transcriptional activators. Moreover, activation of canonical Notch signaling induces the maintenance of stem or progenitor cells through the inhibition of normal cell differentiation [13,103,104]. Several oncogenes, such as HER2 [105,106], Akt [107] as well as transcriptional factors, such as STAT3 [108], NF-κB [109] were recently found to be associated with BCSC. Since Notch signaling crosstalks with these oncogenic pathways (discussed below), it could impact BCSC and breast cancer development through such crosstalks.
CSCs also have critical roles in promoting tumor angiogenesis. Strong evidence comes from studies of correlation of CSCs and VEGF/VEGFR. The VEGF expression in CD133+ glioma CSCs was up-regulated by 10–20 folds, combined with a dramatically increased vascular density identified by CD31 staining [126]. In addition, VEGF neutralizing antibody (bevacizumab) can deplete glioma CSCs-induced vascular EC migration and tube formation [126]. Recent studies further established that CXCL12 and its receptor CXCR4 may promote glioma CSCs growth and angiogenesis by stimulating VEGF production [127]. Malignant melanoma-initiating cells (MMICs) were identified by an ATP-binding cassette (ABC) member ABCB5 [128,129]. ABCB5 (+) melanoma cells have been shown to overexpress the vasculogenic differentiation markers CD144 (VE-cadherin) and TIE1 and are associated with CD31 (−) vasculogenic mimicry (VM), an established biomarker associated with tumor angiogenesis and increased patient mortality [130,131]. Induced VEGF was found in ABCB5 (+) cells that constitutively expressed VEGFR-1 but not in ABCB5 (−) bulk populations that were predominantly VEGFR-1 (−). In vivo, melanoma-specific shRNA-mediated knockdown of VEGFR-1 inhibited the development of ABCB5 (+) VM morphology and ABCB5 (+) VM-associated production of the secreted melanoma mitogen laminin [131]. Their results support the notion that not only VEGF, but also VEGFR-1 in MMIC regulates VM and is associated with laminin production and tumor angiogenesis. Additional evidence was provided in a report in which tumor with larger CSC population was found to recruit a substantially higher number of endothelial progenitor cells (EPCs), indicating that CSCs promote local angiogenesis and EPC mobilization via stimulating proangiogenic factors such as VEGF and SDF-1 [132].
To date, our understanding of the interplay between CSCs and angiogenesis is limited and continues to evolve with intense investigations. Although we believe Notch signaling is critical for both CSCs and angiogenesis, other signaling pathways which may have direct or indirect interactions with Notch are also important as discussed below.
6. Crosstalk between Notch signaling and other oncogenic pathways in breast cancer angiogenesis
In breast cancer, activation of Notch signaling can upregulate several factors that in turn transmit bidirectional signals among cancer cells expressing both ligands and receptors. Notch could also transmit signals among cancer, stroma and endothelium cells [10,133]. Therefore, it is not surprising that extensive crosstalks exist between Notch signaling and many others such as Wnt and Hedgehog signaling, growth factors, cytokines, oncogenic kinases as well as transcriptional factors.
6.1. Developmental signaling
6.1.1. Hedgehog signaling
Hedgehog is a developmental signaling pathway that plays key roles in embryogenesis, maintenance of adult tissue homeostasis, tissue repair during chronic persistent inflammation, and carcinogenesis [134–136]. More recently, hedgehog signaling has also been implicated in angiogenesis. While hedgehog signaling in adult angiogenesis may constitute a simple recapitulation of that in embryonic development, it should be appreciated that Hedgehog signaling occurs in embryonic angiogenesis in different developmental contexts [137]. Hedgehog family ligands, Sonic hedgehog (Shh), Indian hedgehog (Ihh) and Desert hedgehog (Dhh), undergo autoprocessing and lipid modification to generate mature peptides [138–140]. Genetic evidence in mice as well as molecular biological studies in human cells clearly indicate that dysregulated Hedgehog signaling can lead to mammary hyperplasia and tumor formation [141]. Notch ligand, JAG2 is induced by Hedgehog signaling during carcinogenesis [142].
Notch–Hedgehog crosstalk induces the expression of Hes3 and Shh through rapid activation of cytoplasmic signals, including Akt, STAT3 and the mammalian target of rapamycin (mTOR), promoting the survival of neural stem cells [143]. Hedgehog signals could induce Hes1 in both C3H/10T1/2 mesodermal and MNS70 neural cells [144]. In human breast cancer, dysregulated Hedgehog, together with Notch and Wnt signals, could also regulate the self-renewal and differentiation ability of BCSC [145].
6.1.2. Wnt signaling
Dysregulation of the Wnt pathway has been extensively studied in multiple diseases, including some angiogenic disorders. Wnt signaling activation is a major stimulator in pathological angiogenesis and thus, Wnt antagonists are considered to have a therapeutic role for neovascular disorders [146,147]. Some Wnt antagonists have been identified directly from the anti-angiogenic factor family [148–150]. Wnt1 expression led to subsequent activation of Notch signaling in human mammary epithelial cells (HMECs) [151]. Concomitant upregulation of the Wnt target genes Lef1 and Axin2 along with Notch ligand Dll-3 and Dll-4 was found in breast carcinomas, suggesting that the same process takes place in tumors [151]. The blockade of expression of Notch ligands abrogated HMEC transformation by Wnt1, demonstrating the requirement for Notch–Wnt crosstalk during mammary tumorigenesis [151]. Notch-regulated ankyrin repeat protein (Nrarp) acts as a molecular link between Notch- and Wnt signaling in ECs to control stability of new vessel connections in mouse and zebrafish [152]. Dll4/Notch-induced expression of Nrarp limits Notch signaling and promotes Wnt signaling in endothelial stalk cells through interactions with Lymphoid enhancer-binding factor-1 (Lef1) [152]. These results suggest that the balance between Notch and Wnt signaling determines whether to make or break new vessel connections.
6.2. Growth factors
6.2.1. HER/ErbB
HER/ErbB genes (HER1/EGFR, HER3 and HER4) encode for receptor tyrosine kinase (RTK)-transmembrane proteins that, upon binding of several ligands (epidermal growth factor, EGF; amphiregulin; heregulin or neu-mouse and transforming growth factor alpha, TGF-α) regulate cell proliferation, differentiation and survival [153,154]. Dysregulation of EGF receptor (EGFR) by over-expression or constitutive activation can promote tumor angiogenesis and metastasis. Moreover, EGFR overexpression is associated with poor prognosis in many human malignancies including breast cancer [155,156].
Growth factors and their receptors play an essential role in regulating the proliferation of epithelial cells [157,158]. Over-expression of HER2 in human tumor cells is closely associated with increased angiogenesis and expression of VEGF-A. HER2 signaling may increase the rate of hypoxia-inducible factor 1 α (HIF-1α) synthesis which in turn mediates VEGF-A expression [159]. On the other hand, inhibition of the VEGF pathway leads to suppression of tumor growth. The anti-HER2 antibody trastuzumab has been shown to inhibit tumor cell growth and VEGF expression [160,161].
Notch signaling could regulate HER2 activity since the HER2 promoter contains Notch-binding sequences [96]. Yamaguchi et al. [162] observed that down-regulation of Notch3 significantly suppressed proliferation and promoted apoptosis of the ErbB2-negative tumor cell lines. Magnifico et al. demonstrated that HER2-overexpressing cells displayed activated Notch1signaling [97], and that inhibition of Notch1 signaling by small interfering RNA or γ-secretase inhibitor down-regulated HER2 expression and reduced sphere formation [97].
In contrast to HER2, the long-standing relationships between the EGFR and Notch signaling pathways, and the opposing effects exerted by these signal transduction cascades, have been well documented in various developmental settings and organisms [163,164]. Dai et al. found that forced overexpression of Notch1 by transfection increased EGFR expression in human breast cancer cells [165]. Moreover, overexpression of Notch1 reversed EGFR inhibitor-induced cell toxicity, suggesting that Notch and EGFR signaling may be positively cross-linked in human breast cancer. Dong et al. [166] further observed that inhibition of either EGFR or Notch signaling alone was insufficient to suppress basal-like breast tumor cell survival and proliferation, whereas simultaneous inhibition of EGFR and Notch signaling uncovered a lethal relationship between these two oncogenic pathways [166].
6.2.2. PDGF/PDGFR signaling
Platelet-derived growth factor (PDGF) is a potent angiogenic family of molecules comprised of four polypeptide chains encoded by different genes. PDGF-A and PDGF-B were identified earlier whereas PDGF-C and PDGF-D were discovered more recently [167–169]. The PDGF isoforms exert their cellular effects by specific binding to two structurally related tyrosine kinase receptors (α and β PDGFR). PDGF is a potent mitogen and chemoattractant for mesenchymal cells, neutrophils and monocytes [170]. Therefore, the expression of PDGF correlates with advanced tumor stages and unfavorable prognosis in human breast carcinomas [171]. PDGF produced in carcinomas is generally thought to act on the non-epithelial tumor stroma promoting angiogenesis [172].
The growing body of literature strongly suggests that a crosstalk between PDGF-D and Notch signaling occurs in cancer [173]. Dr. Sarkar’s group demonstrated that down-regulation of PDGF-D leads to the inactivation of Notch1 and NF-κB DNA-binding activity, as well as down-regulation of its target genes, such as VEGF and MMP-9 in pancreatic cancer cells [78]. Therefore, the inactivation of PDGF-D-mediated cell invasion and angiogenesis could in part be attributable to inactivation of Notch1 [78]. Additionally, down-regulation of PDGF-D also inhibited the Notch1 expression in breast cancer cells [174]. Interestingly, mRNA and protein expressions of Notch1–4, Dll-1, Dll-3, Dll-4, JAG2 as well as Notch downstream targets, such as Hes and Hey were significantly higher in PC3 prostate cancer cells expressing PDGF-D, indicating PDGF-D was correlated to Notch signaling [173].
6.2.3. TGF-β signaling
Genes encoding components of TGF-β signaling pathway, including ligands TGF-β1 and TGF-β2 and receptor TGFBRI are functionally polymorphic in humans [175–177]. TGF-β can regulate such diverse processes as cell proliferation, differentiation, motility, adhesion, organization, and apoptosis. Both in vitro and in vivo experiments suggest that TGF-β can utilize these diverse programs to promote cancer metastasis through its effects on the tumor microenvironment, enhanced invasive properties, and inhibition of immune cell function [178,179]. Recent knockout studies of factors in TGF-β signaling have shown that this pathway is also indispensable for angiogenesis [180–182].
TGF-β signaling is linked to Notch in many processes. First, TGF-β can upregulate Notch ligands. JAG1 has been shown to be a TGF-β target gene in multiple types of mammalian cells. JAG1 and Hey1 are critical for TGF-β-induced epithelial–mesenchymal transformation (EMT) in cells derived from several organs [183]. In addition, JAG1 upregulation also contributes to TGF-β effects on cell cycle by stimulating p21 expression and cytostasis in epithelial cells [184]. Second, TGF-β and Notch can synergistically regulate common target genes in many cell types, for example, Smad3, a downstream transcription factor of TGF-β and Notch1 NICD can directly interact and form a complex with CSL that binds to specific DNA sequences as those found in the promoter of Hes-1 [185]. Notch1 NICD not only interacts with activated Smad3 and facilitates its nuclear translocation [186], but also remains bound with pSmad3 in the nucleus where they jointly upregulate the transcription factor Forkhead box P3 (Foxp3) that is involved in immune processes [187].
6.2.4. VEGF/VEGFR-2 signaling
Vascular endothelial growth factor (VEGF) is the major angiogenic factor in physiological and pathological angiogenesis [188,189]. The expression of the VEGF gene is enhanced in a variety of angiogenic tumors [188,189]. VEGFR-2, receptor type 2 (KDR or flk-1) is generally recognized to have a principal role in mediating VEGF-induced responses and is considered as the earliest marker for EC development [190]. Moreover, VEGFR-2 directly regulates tumor angiogenesis [189–191]. In addition to its angiogenic actions in ECs, the VEGF/VEGFR-2 signaling paracrine–autocrine loop functions as an important survival process in breast cancer cells [190,192].
VEGF was first shown to act upstream of Notch in determining arterial cell fate in vascular development [193]. VEGF was demonstrated to increase Dll-4 and Notch expression, in turn leading to the activation of Notch signaling and arterial specification (expression of a set of arterial genes). Further studies in several systems established that VEGF regulates the expression of Notch signaling components [194–196]. Blocking VEGF, by intravitreal injection of soluble VEGF receptors, results in decreased sprouting and reduced expression of Dll-4 in retinal vessels [197]. Similar interactions among VEGF signaling, growing vessels and Notch components expression were found in tumor vessels [195,198–200].
Providing a feedback mechanism, Notch signaling in turn can alter expression levels of all three VEGF receptors. For example, VEGFR-2 was down-regulated by either Notch1,4 or Hey1 in ECs [201]. Reciprocally, VEGFR-2 expression increased in vessels of Dll-4 heterozygous mice or as a result of Dll-4 blockade [197]. Thus, Notch signaling can provide negative feedback to reduce the activity of the VEGF/VEGFR-2 axis in ECs. Taken together, VEGF pathway acts as a potent upstream activating stimulus for angiogenesis, whereas Notch pathway helps to shape that action appropriately [202,203]. Thus, an important feature of angiogenesis is the manifold ways in which the VEGF and Notch pathways interact [203].
6.3. Inflammatory cytokines
6.3.1. IL-6
IL-6, a multifunctional cytokine, produced by various types of cells, including macrophages and cancer cells, is an important factor for immune responses, cell survival, apoptosis, proliferation and angiogenesis [204–206]. IL-6 signals via a heterodimeric IL-6R/gp130 receptor complex, whose engagement triggers the activation of Janus (JAK) kinases, and the downstream effectors STAT proteins [204]. A number of studies implicated IL-6 and STAT3 as pro-tumorigenic and pro-angiogenic agents in many cancers including breast cancer [207–211].
Sansone et al. first determined that Notch pathway was a critical downstream target of IL-6 [212]. IL-6 treatment triggered Notch3-dependent upregulation of the Notch ligand JAG1 and promotion of primary human mammospheres and MCF-7-derived spheroid growth. Moreover, autocrine IL-6 signaling relied upon Notch3 activity to sustain the aggressive features of MCF-7-derived hypoxia-selected cells. These data support the hypothesis that IL-6 induces malignant features in Notch3-expressing stem/progenitor cells from human ductal breast carcinoma and normal mammary gland. It was also shown that the hypoxia resistance gene carbonic anhydrase (CA-IX) was activated in breast cancer cells by IL-6/Notch/JAG action and provided survival advantages under hypoxic conditions. Very recently, within the IL-6 gene promoter region, the signature binding motif of CSL was identified and found to overlap with a consensus of NF-κB-binding site [213]. These authors demonstrated that Notch1 positively regulates IL-6 expression via NF-κB in activated macrophages [213].
Lee et al. established a HeLa/rtTAA/TRE-N1-IC cell line capable of doxycycline-induced expression of human Notch1 NICD [214]. They found that the induction of Notch signaling activated HIF-1α and its target gene expression in the above cells. Interestingly, HIF-1α expression was required for Notch signaling enhanced STAT3 phosphorylation required under hypoxia conditions. Furthermore, Src (a proto-oncogenic tyrosine kinase) was also required for the enhanced STAT3 phosphorylation in response to Notch signaling. Notch signaling activated Src/STAT3 pathway was dependent on the Notch effector Hes1 transcription factor. However, the treatment of Trichostatin A (TSA) that interferes with Hes1 transcriptional regulation did not affect STAT3 phosphorylation, and dominant negative Hes1 failed to interfere with Hes1-dependent Src/STAT3 pathway and induction of HIF-1α. These observations indicate that Hes1-dependent activation of Src/STAT3 pathway is independent of Hes1 transcription regulation. Therefore, Hes1-dependent Src/STAT3 pathway provides a functional link between Notch signaling and hypoxia pathway.
6.3.2. IL-1 signaling
IL-1 family belongs to pro-inflammatory/-angiogenesis cytokines that is represented by two ligands: IL-1α, IL-1β, an antagonist: interleukin-1 receptor antagonist (IL-1Ra) and two receptors: IL-1R tI (type I receptor) and IL-1R tII (type II receptor) [215]. IL-1 plays a key role in the onset and development of the host reaction to invasion, being an important factor in the initiation of the inflammatory response and immune functions. IL-1 is also abundant at tumor sites, where it may affect the process of carcinogenesis, tumor growth and invasiveness, the patterns of tumor–host interactions and tumor angiogenesis [216]. There is also convincing evidence that IL-1 family and leptin (the major adipocytokine) crosstalk represents a major link among obesity, inflammation, angiogenesis and cancer progression [217–220].
IL-1 activates Notch signaling pathway probably through the NF-κB pathway [221–223], which is present as a latent, inactive, light polypeptide gene enhancer (I-κB, inhibitor of NF-κB)-bound complex in the cytoplasm in majority of cells. IL-1 activates NF-κB via IL-1 receptor-associated kinase (IRAK) and mitogen-activated protein kinase (MAPK) dependent inhibition of I-κB [224,225]. c-Rel (an NF-κB subunit) can trigger Notch1 signaling pathway by inducing expression of JAG1 [226,227]. Results from our laboratory suggest that leptin is an important inducer of IL-1 system in breast cancer cells [218]. Moreover, IL-1, Notch and leptin-induced upregulation of their gene components and NF-κB, HIF-1α and VEGF/VEGFR-2 are interconnected [190,228].
6.3.3. Leptin signaling
Leptin, a pluripotent cytokine secreted primarily not only by adipocytes but also by breast cancer cells, plays key roles in regulating energy intake and energy expenditure, including appetite and metabolism [229]. In the past decade, accumulating evidence indicates that leptin actions are related not only to energy metabolism, but also to reproduction, proliferation, inflammation and angiogenesis [230,231]. More recently, leptin signaling was also demonstrated to associate with BCSCs [232]. Breast cancer cells express higher levels of leptin and leptin receptor, OB-R, than normal mammary cells. Importantly, higher levels of leptin/OB-R levels correlated with metastasis and lower survival of breast cancer patients [233–235]. In vitro, leptin was demonstrated to stimulate the proliferation of breast cancer cell lines [233,236,237]. In vivo studies clearly demonstrated a role for leptin in mammary tumor initiation and development as evidenced by the fact that mutant mice deficient in leptin (LepobLepob), or with non-functioning leptin receptors (LeprdbLeprdb) do not develop transgene-induced mammary tumors [238,239]. The disruption of leptin signaling using pegylated leptin peptide receptor antagonist (PEG-LPrA2) markedly reduced the growth of tumors in mouse models of syngeneic and human breast cancer xenografts [240,241]. These effects were accompanied by a significant decrease in VEGF/VEGFR-2, IL-1 R tI, cyclin D1 and PCNA levels. Moreover, tumor angiogenesis was also impaired [240,241].
Leptin and IL-1 are associated in several pathological situations [161,242], suggesting an interplay between them. Indeed, leptin regulates IL-1 family members in a diabetic context [243] and in endometrial cancer cells [244]. Leptin was found to increase protein and mRNA levels of all components of the IL-1 system in a mouse mammary cancer cell line. Leptin-induced canonical signaling pathways (JAK2/STAT3, MAPK/ERK 1/2 and PI-3K/Akt1) were mainly involved in IL-1 upregulation. In addition, leptin upregulation of IL-1α promoter involved the activation of SP1 and NF-κB transcription factors [218].
Little information on leptin–Notch interactions is available. An earlier report shows that leptin regulates the expression of JAG1 and Notch4 in human cord blood CD34+ cells and early differentiated ECs (HUVEC) where leptin promotes cell differentiation [245]. We and others recently observed that leptin was able to activate the Notch signaling pathway in breast cancer cells [220,246]. Moreover, leptin increased the expression of both Notch receptors and ligands [220]. In these cells leptin also up-regulated Notch-target genes Hey2 and Survivin [190,220]. Leptin-induced non-canonical signaling pathways (PKC, p38 and JNK) differentially impacted on CSL promoter activity and on the expression of IL-1 system in mouse 4T1 mammary cancer cell line [218]. Interestingly, effects of leptin upregulation on pro-angiogenic factors IL-1, VEGF/VEGFR-2 and Notch were significantly abrogated by a γ-secretase inhibitor, DAPT as well as siRNA against CSL in 4T1 cells [220].
6.4. Oncogenic kinases and transcription factors
6.4.1. Ras signaling pathway
Ras signaling plays an important role in transmitting signaling from RTKs to ser/threo kinases. Among the effector molecules connected with the group of cell surface receptors, Ras transduces extracellular signals to diverse intracellular events by controlling the activities of multiple signaling pathways [247]. The multifunctional signal transducer Ras is a proto-oncogene that is frequently mutated in human cancers, including angiosarcomas [248]. Because Ras signaling impacts many cellular functions, including cell cycle regulation, apoptosis, cell survival, EC function and angiogenesis, it is a major target for the development of novel cancer treatments [248,249]. The signaling networks regulated by Ras are very complex due to their multi-faceted functions and crosstalks [250].
Crosstalk between Ras and Notch pathways has been described in pancreatic ductal adenocarcinoma [251], colorectal tumors [252], astrocytic gliomas [253], leukemia [254,255], as well as breast cancer [256,257]. In an early report [50], Weijzen et al. demonstrated that oncogenic Ras activates Notch signaling. Notch1 was necessary to maintain the neoplastic phenotype in Ras-transformed human cells in vitro and in vivo [50]. Ras increased the expression and activity of Notch1 NICD and upregulated Notch ligand Dll-1 and presenilin-1, a protein involved in Notch processing, through a p38-mediated pathway [50]. These observations established that Notch signals were among the key downstream effectors of oncogenic Ras. Gustafson et al. [256] observed that transformation of MCF-10A cells by Harvey-Ras (Ha-Ras) induced CCAAT/enhance binding protein beta (C/EBPβ), a transcriptional factor, and activated the Notch signaling pathway to block SIM2s (a transcriptional factor) gene expression. High expression level of Notch receptors, ligands and their cooperation with the Ras/MAPK pathway in several breast cancers and early precursors place Notch signaling as a key player in breast cancer pathogenesis. This offers combined inhibition of the two pathways as a new modality for breast cancer treatment [257]. Given the regulation of Ras is important for EC function, angiogenesis and activated Ras signaling is critical for vascular malformations and angiosarcoma, crosstalk between Ras and Notch pathways might occur in ECs. However, the exact role and mechanism of these two pathways in ECs need to be determined.
6.4.2. PI-3K/Akt signaling pathway
The phosphatidylinositol 3-kinase (PI-3K/Akt) pathway is a central player in a variety of cellular processes including cell growth, proliferation, motility, survival, angiogenesis, as well as EMT in tumor cells [258–261]. PI-3K/Akt pathway acts upon tumor cells in both autocrine and paracrine manners [262–264]. Notch has been shown to regulate the Akt (ser/threo) or Protein kinase B (PKB) pathway. Liu et al. [265] reported that Notch1 activation enhanced melanoma cell survival via activation of the Akt pathway. Palomero et al. [266] found that Notch1 induced up-regulation of the PI-3K/Akt pathway via Hes1, which negatively controlled the expression of phosphatase and tensin homolog on chromosome 10 (PTEN) in T-cell acute lymphoblastic leukemia (T-ALL). Additional reports also demonstrated that Notch1 crosstalks with Akt pathway in T-ALL, melanoma as well as breast epithelial cells [264,267,268]. On one hand, activation of Akt was necessary for Notch-induced protection against apoptosis in MCF-10A. On the other hand, inhibiting Notch signaling in breast cancer cells induced a decrease in Akt activity and an increase in apoptotic sensitivity [264]. Down-regulation of Notch1 or JAG1 mediated the inhibition of cell growth, migration and invasion, and the induction of apoptosis in prostate cancer. These effects were in part due to inactivation of Akt, mTOR, and NF-κB signaling pathways [269]. In an early report [270], activated Notch1 synergizes with papillomavirus oncogenes in the transformation of immortalized epithelial cells, leading to the generation of resistance to anoikis, an apoptotic response induced by matrix withdrawal. This resistance to anoikis by activated Notch1 is mediated through the activation of PKB/Akt. The cellular responsiveness to Notch signaling dependent PI-3K/Akt pathway has also been observed in other types of cells, such as Chinese hamster ovary (CHO) cells, primary T-cells and hippocampal neurons [271].
PI3-K/Akt pathway also regulates Noch1 and DLL4 in ECs. VEGF can induce gene expression of Notch1 and DLL4 in human arterial ECs. Furthermore, the VEGF-induced specific signaling is mediated through VEGFR-1 & 2 and is transmitted via the PI3-K/Akt pathway [272]. Other reports confirmed that PI3-K/Akt pathway can regulate Notch signaling in ECs [273,274].
6.4.3. mTOR signaling
mTOR, a key protein kinase, controls signal transduction from various growth factors and upstream proteins to the level of mRNA translation and ribosome biogenesis. mTOR is a ser/threo kinase that is often a downstream effector of PI-3K/Akt signaling pathway in breast and many other types of cancer cells. However, MAPK pathway was identified as the preferential upstream regulator of mTOR in the induction of inflammatory/pro-angiogenic molecules in endometrial cancer cells [244]. mTOR can also phosphorylate Akt [275]. mTOR has been intensely studied for over a decade as a central regulator of cell growth, proliferation, differentiation, autophagy, angiogenesis and survival [276–278]. mTOR functions as two distinct multiprotein complexes, mTORC1 and mTORC2 [275,279]. mTORC1 phosphorylates p70 S6 kinase (S6K1), eukaryotic initiation factor 4E (eIF4E) binding protein 1 (4E-BP1) and integrates hormones, growth factors, nutrients, stressors and energy signals. In contrast, mTORC2 is insensitive to nutrients or energy conditions. However, in response to hormones or growth factors, mTORC2 phosphorylates Akt, and regulates actin cytoskeleton and cell survival [275]. Aberrant activation of mTOR pathway is frequently found in cancer and its role in breast cancer cell proliferation and anti-cancer drug resistance has been implicated [280–282]. mTOR signaling has been reported to crosstalk with the Notch signaling pathway in several malignant cell lines [143,283–285].
Inhibition of p53 by Notch1 NICD mainly occurs through mTOR linked to PI-3K/Akt pathway. Moreover, rapamycin treatment abrogated NICD inhibition of p53 and reversed the chemoresistance [285]. Chemoresistant MCF-7 and MOLT4 (T-cell acute lymphoblastic leukemia) cells have aberrant Notch1 that can be reversed by using both PI-3K and mTOR inhibitors [285]. Efferson et al. [284] used an ERbB2-transgenic mouse model of breast cancer (neuT) to show that Notch signaling plays a critical role in tumor maintenance. Inhibition of the Notch pathway with a γ-secretase inhibitor (GSI) decreased both the Notch and mTOR/Akt pathways. Antitumor activity resulting from GSI treatment was associated with decreased cell proliferation [285]. Since mTOR is closely linked to the PI-3K/Akt pathway in ECs [286–288], it is reasonable to speculate that mTOR should also be a regulator of Notch pathway in ECs.
6.4.4. NF-κB signaling pathway
The family of NF-κB transcription factors is involved in the expression of key genes for innate and adaptive immunity, cell proliferation and survival, and lymphoid organ development. NF-κB is activated in a variety of cancers [289,290] linked to tumor angiogenesis [228]. NF-κB family, RelA (p65), RelB, c-Rel, p105/p50 and p100/p52 are evolutionarily conserved molecules that form hetero- or homodimers. The p65/p50 heterodimer, the most abundant form of NF-κB is regulated by the so-called canonical pathway [289,291].
Numerous reports have described the bidirectional regulation of Notch and NF-κB through different context-dependent mechanisms. First, Oswald et al. [76] clearly demonstrated that Notch was able to transcriptionally regulate NF-κB members. RBP-Jk is a strong transcriptional repressor of p100/p52 whose effects can be overcome by activated Notch1, suggesting that p100/p52 is a Notch target gene. Cheng et al. [292] further observed that Notch1 upregulated the expression of p65, p50, RelB, and c-Rel subunits in hemopoietic progenitor cells using Notch1 antisense transgenic (Notch-AS-Tg) mice. Second, NF-κB subunits are also able to transcriptionally regulate Notch family members. This is supported by the findings of Bash et al. [226] that demonstrated c-Rel can activate Notch signaling pathway by up-regulating JAG1 gene expression in lymphocytes. A role for JAG1 in B-cell activation, differentiation or function was also suggested [226]. Lastly, members from Notch and NF-κB family could physically interact with each other. Wang et al. [293] demonstrated that the N-terminal portion of Notch1 NICD interacted specifically with p50 subunit and inhibited p50 DNA binding in human NTera-2 embryonal carcinoma cells. In contrast, in T-cells Notch1 NICD was found to activate NF-κB by directly interacting with NF-κB and competing with IκBα. These processes lead to the retention of NF-κB in the nucleus. It seems that in T-cells there are two ‘waves’ of NF-κB activation: an initial, Notch-independent phase, and a later, sustained activation of NF-κB, which is Notch dependent [294]. Two recent reports also confirmed that Notch activation was required for NF-κB activation in ECs [54,77].
6.4.5. HIF signaling pathway
A critical aspect of tumor biology is the sensation of oxygen in the microenvironment. In response to hypoxia, a hallmark of most solid tumors, cells adapt by regulating metabolism, erythropoiesis, and angiogenesis and by modulating pathways that result in survival or cell death. HIF is a key molecule upregulated in response to oxygen deficiency, as it acts as a master regulator of genes involved in tissue reoxygenation [295]. Additionally, HIF has been known to facilitate cancer progression by promoting tumor neoangiogenesis, cell motility, and invasion [296]. HIF is a heterodimer consisting of a constitutively expressed HIF-1β subunit and an oxygen-regulated, unstable HIF-1α subunit. HIF interactions with DNA are mediated through hypoxia-responsive elements (HRE) [295]. Several studies have demonstrated that HIF-1 plays important roles in the development and progression of cancer through activation of various genes involved in crucial aspects of cancer biology, including energy metabolism, vasomotor function, erythropoiesis, cell survival and angiogenesis [297].
Gustafsson et al. [298] showed evidence that hypoxia promotes the undifferentiated cell state in various stem and precursor cell populations. In this process, hypoxia blocks neuronal and myogenic differentiation in a Notch-dependent manner. Upon Notch activation under hypoxic conditions, Notch1 NICD can interact with HIF-1α, and the complex is recruited to Notch1-responsive promoters. Sahlgren et al. [299] further demonstrated that a hypoxia/Notch/EMT axis exists in tumor cells, where Notch serves as a critical intermediate in conveying the hypoxic response into EMT. Hypoxia-induced increased motility and invasiveness of the tumor cells require Notch signaling, and activated Notch mimicked hypoxia in the induction of EMT. In this process, Notch signaling acts in synergy to control the expression of Snail-1, a zinc-finger transcriptional factor repressor of E-cadherin and a critical regulator of EMT. First, NICD could interact with the Snail-1 promoter, and second, Notch potentiated HIF-1α recruitment to the lysyl oxidase (LOX; a copper-dependent amine oxidase) promoter and elevated the hypoxia-induced up-regulation of LOX, which stabilizes the Snail-1 protein [299]. Hypoxia increased Notch1 mRNA and protein level as well as Notch activity, measured as Hes1 and Hey1 expression and Hes1 promoter activity. This effect was dependent on HIF-1α [267]. These results suggest that Notch1 is under the control of oncogenes and the tissue microenvironment. Therefore, HIF-1α and Notch signaling pathways play a critical role in the regulation of EMT and open up perspectives for pharmacological intervention within hypoxia-induced EMT, cell invasiveness and angiogenesis in tumors.
6.5. Other crosstalk signaling
6.5.1. ER signaling
Estrogens, in particular 17beta-estradiol (E2), play a pivotal role in sexual development and reproduction and are also implicated in a large number of physiological processes, including the cardiovascular system. The recognized risk factors for breast cancer are ages at: menarche, first pregnancy, and menopause. This suggests that endogenous ovarian steroids may profoundly affect initiation, promotion, and progression of carcinogenesis through a cascade of reactions initiated by activation of the ER [300,301]. ERs are known to regulate a huge number of genes affecting cancer proliferation and vascular function [302,303].
Soares et al. [51] first demonstrated that a crosstalk between estrogen and Notch signaling occurs in breast cancer and EC. The authors observed that E2 promoted 8-fold and 6-fold increases in Notch1 and JAG1 expression, respectively, in MCF-7 breast cancer cells. A similar up-regulation of both Notch1 receptor and JAG1 ligand was also found in EC. Notch gene expression was required for tubule-like structure formation in EC. Moreover, Notch gene expression, together with HIF-1α, was upregulated by E2. In another report, E2 and parathion (an organophosphate compound and potent insecticide) alone and in combination also led to the activation of Notch signaling in MCF-I0F, in the process of malignant transformation as indicated by anchorage independency and in vitro invasive capabilities [304]. Notch and ERα crosstalk in breast cancer suggests that combinations of antiestrogens and Notch inhibitors maybe more effective in treating ERα (+) breast cancers [305]. Overall, the crosstalk between Notch and estrogen signaling pathway has a significant role in human breast carcinogenesis and angiogenesis.
6.5.2. miRNA actions
There are several reports on the crosstalk between miRNA and Notch signaling pathways [306–308]. Yoo and Greenwald first reported that Notch activation leads to miR-61 mediated down-regulation of Vav, a proto-oncogene in Caenorhabditis elegans [308]. Interestingly, miR-61 could control the expression of oncogene orthologues Ras and Vav, indicating miRNA capacity to act as tumor suppressors [309]. Therapeutic potential of let-7 in cancer (initially identified as a timing developmental regulator in C. elegans) was recently reviewed [310]. In various human cell lines, Notch activation up-regulates miRNA let-7 [307]. Let-7 regulates self renewal and tumorigenicity of breast cancer cells [311], as well as ERα signaling in ER positive breast cancer [312].
On the other hand, miRNAs can regulate Notch pathways. miR-34a down-regulated the expression of Notch1 and Notch2 proteins in glioma cells [306]. miR-34 down-regulated JAG1 and Notch1 in cervical carcinoma and choriocarcinoma cells [313]. miR-34 was required for a normal cellular response to DNA damage in vivo. Therefore, a potential therapeutic use for anti-miR-34 as a radiosensitizing agent in p53-mutant breast cancer is predicted [314]. In addition, altered miRNA signatures including miR-34 may be associated with breast carcinogenesis and metastasis [315]. Loss of miR-8/200 has been commonly observed in advanced tumors [316] and correlates with their invasion [317–319] and acquisition of stem-like properties [320,321]. Recently, miR-8/200 was identified to have the ability to inhibit Jagged1, thus attenuating Notch signaling and impeding proliferation of human metastatic prostate cancer cells [322].
7. Notch signaling as a therapeutic drug target in breast cancer
The prevailing new strategy for rationally targeted cancer treatment is aimed at the development of target-selective “smart” drugs on the basis of characterized mechanisms of action. The connection between Notch signaling, carcinogenesis and angiogenesis, as well as its crosstalk with many oncogenic signaling pathways suggest that Notch signaling may be such a candidate for multi-target drugs. The major therapeutic targets in the Notch pathway are the Notch receptors, in which GSIs prevent the generation of the oncogenic NICD and suppress the Notch activity [323,324].
Gamma-secretase is a large membrane-integral multisubunit protease complex, which is essential for Notch receptor activation [325]. Rasul et al. [326] tested the effects of three different GSIs in breast cancer cells. One inhibitor (GSI1) was lethal to breast cancer cell lines including MCF-7, MDA-MB-231, ZR-75–1, T47D and CAL-51 cells (range of IC50 values: 0.6–0.9 μM). No effect on the non-tumorigenic 226-L-U19 and 226-L-TS4 cell lines was seen in the range 0.5–40 μM, which showed IC50 values around 50 μM. GSI1 treatment resulted in a marked decrease in γ-secretase activity and down-regulation of the Notch signaling pathway with no effects on expression of the γ-secretase components or ligands. Differential responses between tumourigenic and non-tumourigenic cell lines may be explained by the differential expression of Numb, a negative regulator of the Notch pathway, and NICD [326,327]. Non-tumourigenic cells express Numb but not NICD and the Notch pathway is not activated, in contrast, cancer cells have Numb downregulated, NICD upregulated and the Notch pathway activated, thus are sensitive to the cytotoxic effect of GSI1 by its effect on the Notch pathway [48]. In a recent report [284], the authors observed that inhibition of the Notch pathway with a GSI decreased both the Notch and mTOR/Akt pathways. Antitumor activity resulting from GSI treatment was associated with decreased cell proliferation as measured by Ki67 and decreased expression of glucose transporter Glut1 [284]. GSI effects are much higher in HER2/neu-positive cell lines where HER2 is amplified and/or over-expressed (ZR-75-1 and MDA-MB-453) compared with HER2-negative cells (MCF-7 and MDA-MB-231) that lack ERbB2 amplification and show low HER2 expression [86,305,326]. Since HER2 can influence the activity of Notch [96] and inhibition of HER2 via trastuzumab can activate Notch signaling [328], it will be important to consider GSI as a monotherapy or in combination with trastuzumab or lapatinib in HER2 breast cancer patients.
Triple-negative breast cancer (TNBC) is characterized by the lack of expression of ER, PgR, and HER-2. This difficult-to-treat form of breast cancer shows an undesirable tendency to overcome drug effectiveness [329,330]. In TNBC there is higher intratumoural expression of VEGF than non-TNBC [331]. BCSCs are thought to be responsible for the development of drug-resistance and relapse of TNBC [332]. Basal-like breast cancer (BLBC) frequently expresses a CD44+/CD24− phenotype, which has been associated with a ‘stem-cell’ phenotype. These cells show resistance to conventional treatment and allow repopulation of the cancer. BCSC shows some specific molecular alterations including activation of the Notch pathway [333,334]. Therefore, there is strong evidence to suggest that the Notch pathway is a key event in TNBC etiology, and that targeting the Notch pathway may improve patient outcomes by targeting angiogenesis and the hormone-insensitive chemoresistant BCSCs. As expected, GSIs can reduce the growth and dissemination of MDA-MB-231TNBC xenografts [335]. However, a recent report shows that GSIs only reduced sphere formation and xenograft growth from TNBC CD44+/CD24low+ cells, but CD44+/CD24neg were resistant to GSI treatment [336]. Thus, while GSIs hold promise for targeting BCSCs, stem cell heterogeneity could limit GSI efficacy.
Although several GSIs have been developed into clinical trials [324], GSIs fail to distinguish individual Notch receptors. In addition, GSIs inhibit other signaling pathways [337] and cause intestinal toxicity [338], probably attributable to dual inhibition of Notch1 and Notch2 [339]. Very recently, Wu et al. [340] utilized phage display technology to generate highly specialized antibodies that specifically antagonize each receptor paralogue, enabling the discrimination of Notch1 versus Notch2 function in rodent models as well as in humans. Their results showed that inhibition of either receptor alone reduces or avoids toxicity, demonstrating a clear advantage over pan-Notch inhibitors. The gastrointestinal toxicity and abnormalities in the thymus and spleen are major side-effects with GSI use [341], likely resulting from inhibition of Notch cleavage in regulating cell-fate decisions. Therefore, close attention needs be paid to the therapeutic window so that the minimally active dose needed to inhibit Notch is employed, thereby reducing adverse side effects. In addition, development of a practical combination therapy [133] should minimize problematic side-effects.
8. Conclusion and overall perspectives
Notch signaling and its crosstalk with many signaling pathways play an important role in breast cancer cell growth, migration, invasion, metastasis and angiogenesis, as well as CSC self-renewal (see Fig. 1). Therefore, increasing attention has been paid in recent years to the development of clinically useful antagonists of Notch signaling. Better understanding of the structure, function and regulation of Notch intracellular signaling pathways, as well as its complex crosstalk with other oncogenic signals in cancer cells will be essential to ensure rational use of treatment and development of new combinatory therapeutic possibilities. Emerging novel opportunities arise from the discovery of Notch crosstalk with inflammatory and angiogenic cytokines and their links to obesity-related cancers. Combination therapy with drugs designed to prevent Notch oncogenic signal crosstalk may be advantageous over GSIs alone.
Acknowledgements
The authors’ work cited in this review was funded by grants from National Natural Science Foundation of China (81172509) (ZW); NIH 3G12MD007595 (GW); Women’s Cancer Society grant 66253L (GS); and Facilities and support services at Morehouse School of Medicine (NIH RR03034 and 1C06 RR18386).
Glossary
- 4T1 cells
mouse mammary cancer cell line
- ADAM
a disintegrin and metalloprotease
- Akt
protein kinase B
- ALDH
aldehyde dehydrogenase
- ANK
ankyrin
- Axin2
the Axin-related protein
- Bcl-2
B-cell lymphoma 2
- BCSCs
breast cancer stem cells
- BMP
bone morphogenetic protein
- CBF1
centromere-binding factor 1
- CD4
cluster of differentiation 4
- CD8
cluster of differentiation 8
- c-myc
Myc proto-oncogene protein
- Co-A
recruits coactivator
- Co-R
co-repressor
- CSC
cancer stem cell
- CSL
CBF1/Su(H)/Lag-1
- Cyclin D1
kinase and regulator of cell cycle D1
- DAPT
N-[N-(3,5-Difluorophenacetyl)-l-alanyl]-S-phenylglycine t-butyl ester
- DBZ
dibenzazepine
- DLL-1
Delta-like 1
- DOS
Delta and OSM-11-like proteins
- DSL
Delta/Serrate/LAG-2
- E2
17β-estradiol
- EC
endothelial cell
- EGF
epidermal growth factor
- EGFR
epidermal growth factor receptor
- EMT
epithelial–mesenchymal transformation
- ER
estrogen receptor
- ERK 1/2
extracellular regulated kinase 1 and 2
- GSI
a γ-secretase inhibitor
- HD
heterodimerization
- HDAC
histone deacetylases
- HIF-1α
hypoxia regulated factor-1 α
- HUVECs
human umbilical vein ECs
- ICN
intracellular region of Notch
- IL-1
interleukin-1
- IL-1R tI
interleukin-1 type I receptor
- IL-6
interleukin-6
- IL-6R
interleukin-6 receptor
- JAK2
Janus kinase 2
- MAPK
mitogen activated protein kinase
- MCF-7
ER positive human breast cancer cell line
- MDA-MB-231
ER negative human breast cancer cell line
- MFE
Mammosphere-forming efficiency
- miRNA
MicroRNA
- mTOR
mammalian target of rapamycin
- NECD
Notch extracellular domain
- NEXT
Notch extracelluar truncation
- NF-κB
eukaryotic nuclear transcription factor kappa B
- NICD
Notch intracellular domain
- NRR
negative regulatory region
- OB-R
leptin receptor
- PDGF
platelet-derived growth factor
- PEST
proline, glutamine, serine and threonine residue
- PI-3K
phosphoinositide 3-kinase
- RhoC
Ras homolog gene family, member C
- Src
a proto-oncogenic tyrosine kinase
- STAT3
signal transducer and activator of transcription 3
- TACE
tumor necrosis factor-α-converting enzyme
- TAD
transactivation domain
- TAM
tamoxifen
- T-ALL
T-cell acute lymphoblastic leukemia
- TGF-β
transforming growth factor beta
- TNBC
Triple-negative breast cancer
- TNF-α
tumor necrosis factor alpha
- TSA
Trichostatin A
- VEGF
vascular endothelial growth factor
- VEGFR-2
vascular endothelial growth factor receptor 2 or KDR or Flk-1
References
- [1].Hanahan D, Weinberg RA, Hallmarks of cancer: the next generation, Cell 144 (2011) 646–674. [DOI] [PubMed] [Google Scholar]
- [2].Adams RH, Alitalo K, Molecular regulation of angiogenesis and lymphangiogenesis, Nat. Rev. Mol. Cell Biol 8 (2007) 464–478. [DOI] [PubMed] [Google Scholar]
- [3].Prager GW, Poettler M, Angiogenesis in cancer. Basic mechanisms and therapeutic advances, Hamostaseologie 32 (2011). [DOI] [PubMed] [Google Scholar]
- [4].Bridges E, Oon CE, Harris A, Notch regulation of tumor angiogenesis, Future Oncol. 7 (2011) 569–588. [DOI] [PubMed] [Google Scholar]
- [5].Guo S, Liu M, Gonzalez-Perez RR, Role of Notch and its oncogenic signaling crosstalk in breast cancer, Biochim. Biophys. Acta 1815 (2011) 197–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Lewis J, Notch signalling and the control of cell fate choices in vertebrates, Semin. Cell Dev. Biol 9 (1998) 583–589. [DOI] [PubMed] [Google Scholar]
- [7].Simpson P, Developmental genetics. The Notch connection, Nature 375 (1995) 736–737. [DOI] [PubMed] [Google Scholar]
- [8].Borggrefe T, Oswald F, The Notch signaling pathway: transcriptional regulation at Notch target genes, Cell. Mol. Life Sci 66 (2009) 1631–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Kopan R, Ilagan MX, The canonical Notch signaling pathway: unfolding the activation mechanism, Cell 137 (2009) 216–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Miele L, Notch signaling, Clin. Cancer Res 12 (2006) 1074–1079. [DOI] [PubMed] [Google Scholar]
- [11].Miele L, Miao H, Nickoloff BJ, Notch signaling as a novel cancer therapeutic target, Curr. Cancer Drug Targets 6 (2006) 313–323. [DOI] [PubMed] [Google Scholar]
- [12].Miele L, Osborne B, Arbiter of differentiation and death: Notch signaling meets apoptosis, J. Cell. Physiol 181 (1999) 393–409. [DOI] [PubMed] [Google Scholar]
- [13].Artavanis-Tsakonas S, Rand MD, Lake RJ, Notch signaling: cell fate control and signal integration in development, Science 284 (1999) 770–776. [DOI] [PubMed] [Google Scholar]
- [14].Farnie G, Clarke RB, Mammary stem cells and breast cancer—role of Notch signalling, Stem Cell Rev. 3 (2007) 169–175. [DOI] [PubMed] [Google Scholar]
- [15].Politi K, Feirt N, Kitajewski J, Notch in mammary gland development and breast cancer, Semin. Cancer Biol 14 (2004) 341–347. [DOI] [PubMed] [Google Scholar]
- [16].Wang Z, Li Y, Sarkar FH, Notch signaling proteins: legitimate targets for cancer therapy, Curr. Protein Pept. Sci 11 (6) (2010) 398–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Wu F, Stutzman A, Mo YY, Notch signaling and its role in breast cancer, Front. Biosci 12 (2007) 4370–4383. [DOI] [PubMed] [Google Scholar]
- [18].Cordle J, Johnson S,Tay JZ, Roversi P, Wilkin MB, de Madrid BH, Shimizu H, Jensen S, Whiteman P, Jin B, Redfield C, Baron M, Lea SM, Handford PA, A conserved face of the Jagged/Serrate DSL domain is involved in Notch trans-activation and cis-inhibition, Nat. Struct. Mol. Biol 15 (2008) 849–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].D’Souza B, Miyamoto A, Weinmaster G, The many facets of Notch ligands, Oncogene 27 (2008) 5148–5167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Komatsu H, Chao MY, Larkins-Ford J, Corkins ME, Somers GA, Tucey T, Dionne HM, White JQ, Wani K, Boxem M, Hart AC, OSM-11 facilitates LIN-12 Notch signaling during Caenorhabditis elegans vulval development, PLoS Biol. 6 (2008) e196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Albig AR, Becenti DJ, Roy TG, Schiemann WP, Microfibril-associate glycoprotein-2 (MAGP-2) promotes angiogenic cell sprouting by blocking notch signaling in endothelial cells, Microvasc. Res 76 (2008) 7–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Cui XY, Hu QD, Tekaya M, Shimoda Y, Ang BT, Nie DY, Sun L, Hu WP, Karsak M, Duka T, Takeda Y, Ou LY, Dawe GS, Yu FG, Ahmed S, Jin LH, Schachner M, Watanabe K, Arsenijevic Y, Xiao ZC, NB-3/Notch1 pathway via Deltex1 promotes neural progenitor cell differentiation into oligodendrocytes, J. Biol. Chem 279 (2004) 25858–25865. [DOI] [PubMed] [Google Scholar]
- [23].Gupta R, Hong D, Iborra F, Sarno S, Enver T, NOV (CCN3) functions as a regulator of human hematopoietic stem or progenitor cells, Science 316 (2007) 590–593. [DOI] [PubMed] [Google Scholar]
- [24].Heath E, Tahri D, Andermarcher E, Schofield P, Fleming S, Boulter CA, Abnormal skeletal and cardiac development, cardiomyopathy, muscle atrophy and cataracts in mice with a targeted disruption of the Nov (Ccn3) gene, BMC Dev. Biol 8 (2008) 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Leask A, Abraham DJ, All in the CCN family: essential matricellular signaling modulators emerge from the bunker, J. Cell Sci 119 (2006) 4803–4810. [DOI] [PubMed] [Google Scholar]
- [26].Lu L, Chen X, Zhang CW, Yang WL, Wu YJ, Sun L, Bai LM, Gu XS, Ahmed S, Dawe GS, Xiao ZC, Morphological and functional characterization of predifferentiation of myelinating glia-like cells from human bone marrow stromal cells through activation of F3/Notch signaling in mouse retina, Stem Cells 26 (2008) 580–590. [DOI] [PubMed] [Google Scholar]
- [27].Milner LA, Bigas A, Notch as a mediator of cell fate determination in hematopoiesis: evidence and speculation, Blood 93 (1999) 2431–2448. [PubMed] [Google Scholar]
- [28].Weng AP, Ferrando AA, Lee W, Morris J.P.t., Silverman LB, Sanchez-Irizarry C, Blacklow SC, Look AT, Aster JC, Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia, Science 306 (2004) 269–271. [DOI] [PubMed] [Google Scholar]
- [29].Okuyama R, Tagami H, Aiba S, Notch signaling: its role in epidermal homeostasis and in the pathogenesis of skin diseases, J. Dermatol. Sci 49 (2008) 187–194. [DOI] [PubMed] [Google Scholar]
- [30].Tien AC, Rajan A, Bellen HJ, A Notch updated, J. Cell Biol 184 (2009) 621–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Mumm JS, Schroeter EH, Saxena MT, Griesemer A, Tian X, Pan DJ, Ray WJ, Kopan R, A ligand-induced extracellular cleavage regulates gamma-secretase-like proteolytic activation of Notch1, Mol. Cell 5 (2000) 197–206. [DOI] [PubMed] [Google Scholar]
- [32].Fiuza UM, Arias AM, Cell and molecular biology of Notch, J. Endocrinol 194 (2007) 459–474. [DOI] [PubMed] [Google Scholar]
- [33].Fortini ME, Artavanis-Tsakonas S, The suppressor of hairless protein participates in notch receptor signaling, Cell 79 (1994) 273–282. [DOI] [PubMed] [Google Scholar]
- [34].Duarte A, Hirashima M, Benedito R, Trindade A, Diniz P, Bekman E, Costa L, Henrique D, Rossant J, Dosage-sensitive requirement for mouse Dll4 in artery development, Genes Dev. 18 (2004) 2474–2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Krebs LT, Xue Y, Norton CR, Shutter JR, Maguire M, Sundberg JP, Gallahan D, Closson V, Kitajewski J, Callahan R, Smith GH, Stark KL, Gridley T, Notch signaling is essential for vascular morphogenesis in mice, Genes Dev. 14 (2000) 1343–1352. [PMC free article] [PubMed] [Google Scholar]
- [36].Phng LK, Gerhardt H, Angiogenesis: a team effort coordinated by notch, Dev. Cell 16 (2009) 196–208. [DOI] [PubMed] [Google Scholar]
- [37].Roca C, Adams RH, Regulation of vascular morphogenesis by Notch signaling, Genes Dev. 21 (2007) 2511–2524. [DOI] [PubMed] [Google Scholar]
- [38].Gridley T, Notch signaling in vascular development and physiology, Development 134 (2007) 2709–2718. [DOI] [PubMed] [Google Scholar]
- [39].Gridley T, Notch signaling in the vasculature, Curr. Top. Dev. Biol 92 (2010) 277–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Egan SE, St-Pierre B, Leow CC, Notch receptors, partners and regulators: from conserved domains to powerful functions, Curr. Top. Microbiol. Immunol 228 (1998) 273–324. [DOI] [PubMed] [Google Scholar]
- [41].Greenwald I, LIN-12/Notch signaling: lessons from worms and flies, Genes Dev. 12 (1998) 1751–1762. [DOI] [PubMed] [Google Scholar]
- [42].Xue Y, Gao X, Lindsell CE, Norton CR, Chang B, Hicks C, Gendron-Maguire M, Rand EB, Weinmaster G, Gridley T, Embryonic lethality and vascular defects in mice lacking the Notch ligand Jagged1, Hum. Mol. Genet 8 (1999) 723–730. [DOI] [PubMed] [Google Scholar]
- [43].Hrabe de Angelis M, McIntyre II J, Gossler A, Maintenance of somite borders in mice requires the Delta homologue DII1, Nature 386 (1997) 717–721. [DOI] [PubMed] [Google Scholar]
- [44].McCright B, Gao X, Shen L, Lozier J, Lan Y, Maguire M, Herzlinger D, Weinmaster G, Jiang R, Gridley T, Defects in development of the kidney, heart and eye vasculature in mice homozygous for a hypomorphic Notch2 mutation, Development 128 (2001)491–502. [DOI] [PubMed] [Google Scholar]
- [45].Uyttendaele H, Ho J, Rossant J, Kitajewski J, Vascular patterning defects associated with expression of activated Notch4 in embryonic endothelium, Proc. Natl. Acad. Sci. U. S. A 98 (2001) 5643–5648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Dievart A, Beaulieu N, Jolicoeur P, Involvement of Notch1 in the development of mouse mammary tumors, Oncogene 18 (1999) 5973–5981. [DOI] [PubMed] [Google Scholar]
- [47].Kiaris H, Politi K, Grimm LM, Szabolcs M, Fisher P, Efstratiadis A, Artavanis-Tsakonas S, Modulation of notch signaling elicits signature tumors and inhibits hras1-induced oncogenesis in the mouse mammary epithelium, Am. J. Pathol 165 (2004) 695–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Stylianou S, Clarke RB, Brennan K, Aberrant activation of notch signaling in human breast cancer, Cancer Res. 66 (2006) 1517–1525. [DOI] [PubMed] [Google Scholar]
- [49].Girard L, Hanna Z, Beaulieu N, Hoemann CD, Simard C, Kozak CA, Jolicoeur P, Frequent provirus insertional mutagenesis of Notch1 in thymomas of MMTVD/myc transgenic mice suggests a collaboration of c-myc and Notch1 for oncogenesis, Genes Dev. 10 (1996) 1930–1944. [DOI] [PubMed] [Google Scholar]
- [50].Weijzen S, Rizzo P, Braid M, Vaishnav R, Jonkheer SM, Zlobin A, Osborne BA, Gottipati S, Aster JC, Hahn WC, Rudolf M, Siziopikou K, Kast WM, Miele L, Activation of Notch-1 signaling maintains the neoplastic phenotype in human Ras-transformed cells, Nat. Med 8 (2002) 979–986. [DOI] [PubMed] [Google Scholar]
- [51].Soares R, Balogh G, Guo S, Gartner F, Russo J, Schmitt F, Evidence for the notch signaling pathway on the role of estrogen in angiogenesis, Mol. Endocrinol 18 (2004) 2333–2343. [DOI] [PubMed] [Google Scholar]
- [52].Hamada Y, Kadokawa Y, Okabe M, Ikawa M, Coleman JR, Tsujimoto Y, Mutation in ankyrin repeats of the mouse Notch2 gene induces early embryonic lethality, Development 126 (1999) 3415–3424. [DOI] [PubMed] [Google Scholar]
- [53].Kumano K, Chiba S, Kunisato A, Sata M, Saito T, Nakagami-Yamaguchi E, Yamaguchi T, Masuda S, Shimizu K, Takahashi T, Ogawa S, Hamada Y, Hirai H, Notch1 but not Notch2 is essential for generating hematopoietic stem cells from endothelial cells, Immunity 18 (2003) 699–711. [DOI] [PubMed] [Google Scholar]
- [54].Quillard T, Devalliere J, Coupel S, Charreau B, Inflammation dysregulates Notch signaling in endothelial cells: implication of Notch2 and Notch4 to endothelial dysfunction, Biochem. Pharmacol 80 (2010) 2032–2041. [DOI] [PubMed] [Google Scholar]
- [55].Joutel A, Corpechot C, Ducros A, Vahedi K, Chabriat H, Mouton P, Alamowitch S, Domenga V, Cecillion M, Marechal E, Maciazek J, Vayssiere C, Cruaud C, Cabanis EA, Ruchoux MM, Weissenbach J, Bach JF, Bousser MG, Tournier-Lasserve E, Notch3 mutations in CADASIL, a hereditary adult-onset condition causing stroke and dementia, Nature 383 (1996) 707–710. [DOI] [PubMed] [Google Scholar]
- [56].Ruchoux MM, Guerouaou D, Vandenhaute B, Pruvo JP, Vermersch P, Leys D, Systemic vascular smooth muscle cell impairment in cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy, Acta Neuropathol. 89 (1995) 500–512. [DOI] [PubMed] [Google Scholar]
- [57].Liu H, Kennard S, Lilly B, NOTCH3 expression is induced in mural cells through an autoregulatory loop that requires endothelial-expressed JAGGED1, Circ. Res 104 (2009) 466–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Liu H, Zhang W, Kennard S, Caldwell RB, Lilly B, Notch3 is critical for proper angiogenesis and mural cell investment, Circ. Res 107 (2010) 860–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Gaengel K, Genove G, Armulik A, Betsholtz C, Endothelial–mural cell signaling in vascular development and angiogenesis, Arterioscler. Thromb. Vasc. Biol 29 (2009) 630–638. [DOI] [PubMed] [Google Scholar]
- [60].Shirayoshi Y, Yuasa Y, Suzuki T, Sugaya K, Kawase E, Ikemura T, Nakatsuji N, Proto-oncogene of int-3, a mouse Notch homologue, is expressed in endothelial cells during early embryogenesis, Genes Cells 2 (1997) 213–224. [DOI] [PubMed] [Google Scholar]
- [61].Leong KG, Hu X, Li L, Noseda M, Larrivee B, Hull C, Hood L, Wong F, Karsan A, Activated Notch4 inhibits angiogenesis: role of beta 1-integrin activation, Mol. Cell. Biol 22 (2002) 2830–2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].MacKenzie F, Duriez P, Larrivee B, Chang L, Pollet I, Wong F, Yip C, Karsan A, Notch4-induced inhibition of endothelial sprouting requires the ankyrin repeats and involves signaling through RBP-Jkappa, Blood 104 (2004) 1760–1768. [DOI] [PubMed] [Google Scholar]
- [63].Gallahan D, Jhappan C, Robinson G, Hennighausen L, Sharp R, Kordon E, Callahan R, Merlino G, Smith GH, Expression of a truncated Int3 gene in developing secretory mammary epithelium specifically retards lobular differentiation resulting in tumorigenesis, Cancer Res. 56 (1996) 1775–1785. [PubMed] [Google Scholar]
- [64].Jhappan C, Gallahan D, Stahle C, Chu E, Smith GH, Merlino G, Callahan R, Expression of an activated Notch-related int-3 transgene interferes with cell differentiation and induces neoplastic transformation in mammary and salivary glands, Genes Dev. 6 (1992) 345–355. [DOI] [PubMed] [Google Scholar]
- [65].Imatani A, Callahan R, Identification of a novel NOTCH-4/INT-3 RNA species encoding an activated gene product in certain human tumor cell lines, Oncogene 19 (2000) 223–231. [DOI] [PubMed] [Google Scholar]
- [66].Soriano JV, Uyttendaele H, Kitajewski J, Montesano R, Expression of an activated Notch4(int-3) oncoprotein disrupts morphogenesis and induces an invasive phenotype in mammary epithelial cells in vitro, Int. J. Cancer 86 (2000) 652–659. [DOI] [PubMed] [Google Scholar]
- [67].Callahan R, Raafat A, Notch signaling in mammary gland tumorigenesis, J. Mammary Gland Biol. Neoplasia 6 (2001) 23–36. [DOI] [PubMed] [Google Scholar]
- [68].Iso T, Kedes L, Hamamori Y, HES and HERP families: multiple effectors of the Notch signaling pathway, J. Cell. Physiol 194 (2003) 237–255. [DOI] [PubMed] [Google Scholar]
- [69].Schwanbeck R, Schroeder T, Henning K, Kohlhof H, Rieber N, Erfurth ML, Just U, Notch signaling in embryonic and adult myelopoiesis, Cells Tissues Organs 188 (2008) 91–102. [DOI] [PubMed] [Google Scholar]
- [70].Zanotti S, Canalis E, Notch and the skeleton, Mol. Cell. Biol 30 (2010) 886–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Hartman J, Lam EW, A Gustafsson J, Strom A, Hes-6, an inhibitor of Hes-1, is regulated by 17 beta-estradiol and promotes breast cancer cell proliferation, Breast Cancer Res. 11 (2009) R79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Noseda M, Chang L, McLean G,Grim JE, Clurman BE, Smith LL, Karsan A, Notch activation induces endothelial cell cycle arrest and participates in contact inhibition: role of p21Cip1 repression, Mol. Cell. Biol 24 (2004) 8813–8822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Lehar SM, Bevan MJ, T cells develop normally in the absence of both Deltex1 and Deltex2, Mol. Cell. Biol 26 (2006) 7358–7371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Matsuno K, Diederich RJ, Go MJ, Blaumueller CM, Artavanis-Tsakonas S, Deltex acts as a positive regulator of Notch signaling through interactions with the Notch ankyrin repeats, Development 121 (1995) 2633–2644. [DOI] [PubMed] [Google Scholar]
- [75].Yamamoto N, Yamamoto S, Inagaki F, Kawaichi M, Fukamizu A, Kishi N, Matsuno K, Nakamura K, Weinmaster G, Okano H, Nakafuku M, Role of Deltex-1 as a transcriptional regulator downstream of the Notch receptor, J. Biol. Chem 276 (2001) 45031–45040. [DOI] [PubMed] [Google Scholar]
- [76].Oswald F, Liptay S, Adler G, Schmid RM, NF-kappaB2 is a putative target gene of activated Notch-1 via RBP-Jkappa, Mol. Cell. Biol 18 (1998) 2077–2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].A Johnston D, Dong B, Hughes CC, TNF induction of jagged-1 in endothelial cells is NFkappaB-dependent, Gene 435 (2009) 36–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Wang Z, Kong D, Banerjee S, Li Y, Adsay NV, Abbruzzese J, Sarkar FH, Down-regulation of platelet-derived growth factor-D inhibits cell growth and angiogenesis through inactivation of Notch-1 and nuclear factor-kappaB signaling, Cancer Res. 67 (2007) 11377–11385. [DOI] [PubMed] [Google Scholar]
- [79].Ronchini C, Capobianco AJ, Induction of cyclin D1 transcription and CDK2 activity by Notch(ic): implication for cell cycle disruption in transformation by Notch(ic), Mol. Cell. Biol 21 (2001) 5925–5934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Efstratiadis A, Szabolcs M, Klinakis A, Notch, Myc and breast cancer, Cell Cycle 6 (2007)418–429. [DOI] [PubMed] [Google Scholar]
- [81].Weng AP,Millholland JM, Yashiro-Ohtani Y, Arcangeli ML, Lau A, Wai C, Del Bianco C, Rodriguez CG, Sai H, Tobias J, Li Y, Wolfe MS, Shachaf C, Felsher D, Blacklow SC, Pear WS, Aster JC, c-Myc is an important direct target of Notch1 in T-cell acute lymphoblastic leukemia/lymphoma, Genes Dev. 20 (2006) 2096–2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Kim SB, Chae GW, Lee J, Park J, Tak H, Chung JH, Park TG, Ahn JK, Joe CO, Activated Notch1 interacts with p53 to inhibit its phosphorylation and transactivation, Cell Death Differ. 14 (2007) 982–991. [DOI] [PubMed] [Google Scholar]
- [83].Altieri DC, New wirings in the survivin networks, Oncogene 27 (2008) 6276–6284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Ryan BM, O’Donovan N, Duffy MJ, Survivin: a new target for anti-cancer therapy, Cancer Treat. Rev 35 (2009) 553–562. [DOI] [PubMed] [Google Scholar]
- [85].Bray SJ, Notch signalling: a simple pathway becomes complex, Nat. Rev. Mol. Cell Biol 7 (2006) 678–689. [DOI] [PubMed] [Google Scholar]
- [86].Lee CW, Raskett CM, Prudovsky I, Altieri DC, Molecular dependence of estrogen receptor-negative breast cancer on a notch–survivin signaling axis, Cancer Res. 68 (2008) 5273–5281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Lee CW, Simin K, Liu Q, Plescia J, Guha M, Khan A, Hsieh CC, Altieri DC, A functional Notch–survivin gene signature in basal breast cancer, Breast Cancer Res. 10 (2008) R97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Zwerts F, Lupu F, De Vriese A, Pollefeyt S, Moons L, Altura RA, Jiang Y, Maxwell PH, Hill P, Oh H, Rieker C, Collen D, Conway SJ, Conway EM, Lack of endothelial cell survivin causes embryonic defects in angiogenesis, cardiogenesis, and neural tube closure, Blood 109 (2007) 4742–4752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Coma S, Noe V, Lavarino C,Adan J, Rivas M, Lopez-Matas M, Pagan R, Mitjans F, Vilaro S, Piulats J, Ciudad CJ, Use of siRNAs and antisense oligonucleotides against survivin RNA to inhibit steps leading to tumor angiogenesis, Oligonucleotides 14 (2004) 100–113. [DOI] [PubMed] [Google Scholar]
- [90].Mesri M, Morales-Ruiz M, Ackermann EJ, Bennett CF, Pober JS, Sessa WC, Altieri DC, Suppression of vascular endothelial growth factor-mediated endothelial cell protection by survivin targeting, Am. J. Pathol 158 (2001) 1757–1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Ischenko I, Seeliger H, Schaffer M, Jauch KW, Bruns CJ, Cancer stem cells: how can we target them? Curr. Med. Chem 15 (2008) 3171–3184. [DOI] [PubMed] [Google Scholar]
- [92].Harrison H, Farnie G, Howell SJ, Rock RE, Stylianou S, Brennan KR, Bundred NJ, Clarke RB, Regulation of breast cancer stem cell activity by signaling through the Notch4 receptor, Cancer Res. 70 (2010) 709–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Dontu G, Jackson KW, McNicholas E, Kawamura MJ, Abdallah WM, Wicha MS, Role of Notch signaling in cell-fate determination of human mammary stem/progenitor cells, Breast Cancer Res. 6 (2004) R605–R615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Sansone P, Storci G, Giovannini C, Pandolfi S, Pianetti S, Taffurelli M, Santini D, Ceccarelli C, Chieco P, Bonafe M, p66Shc/Notch-3 interplay controls self-renewal and hypoxia survival in human stem/progenitor cells of the mammary gland expanded in vitro as mammospheres, Stem Cells 25 (2007) 807–815. [DOI] [PubMed] [Google Scholar]
- [95].Ling H, Sylvestre JR, Jolicoeur P, Notch1-induced mammary tumor development is cyclin D1-dependent and correlates with expansion of pre-malignant multipotent duct-limited progenitors, Oncogene 29 (32) (2010) 4543–4554. [DOI] [PubMed] [Google Scholar]
- [96].Chen Y, Fischer WH, Gill GN, Regulation of the ErBB-2 promoter by RBPJkappa and NOTCH, J. Biol. Chem 272 (1997) 14110–14114. [DOI] [PubMed] [Google Scholar]
- [97].Magnifico A, Albano L, Campaner S, Delia D, Castiglioni F, Gasparini P, Sozzi G, Fontanella E, Menard S, Tagliabue E, Tumor-initiating cells of HER2-positive carcinoma cell lines express the highest oncoprotein levels and are sensitive to trastuzumab, Clin. Cancer Res 15 (2009) 2010–2021. [DOI] [PubMed] [Google Scholar]
- [98].Phillips TM, Kim K, Vlashi E, McBride WH, Pajonk F, Effects of recombinant erythropoietin on breast cancer-initiating cells, Neoplasia 9 (2007) 1122–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Raouf A, Zhao Y, To K, Stingl J, Delaney A, Barbara M, Iscove N, Jones S, McKinney S, Emerman J, Aparicio S, Marra M, Eaves C, Transcriptome analysis of the normal human mammary cell commitment and differentiation process, Cell Stem Cell 3 (2008) 109–118. [DOI] [PubMed] [Google Scholar]
- [100].Gallahan D, Callahan R, Mammary tumorigenesis in feral mice: identification of a new int locus in mouse mammary tumor virus (Czech II)-induced mammary tumors, J. Virol 61 (1987) 66–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Fu YP, Edvardsen H, Kaushiva A, Arhancet JP, Howe TM, Kohaar I, Porter-Gill P, Shah A, Landmark-Hoyvik H, Fossa SD, Ambs S, Naume B, Borresen-Dale AL, Kristensen VN, Prokunina-Olsson L, NOTCH2 in breast cancer: association of SNP rs11249433 with gene expression in ER-positive breast tumors without TP53 mutations, Mol. Cancer 9 (2010) 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Beatus P, Lundkvist J, Oberg C, Lendahl U, The notch 3 intracellular domain represses notch 1-mediated activation through Hairy/Enhancer of split (HES) promoters, Development 126 (1999) 3925–3935. [DOI] [PubMed] [Google Scholar]
- [103].Leong KG, Karsan A, Recent insights into the role of Notch signaling in tumorigenesis, Blood 107 (2006) 2223–2233. [DOI] [PubMed] [Google Scholar]
- [104].Radtke F, Raj K, The role of Notch in tumorigenesis: oncogene or tumour suppressor? Nat. Rev. Cancer 3 (2003) 756–767. [DOI] [PubMed] [Google Scholar]
- [105].Korkaya H, Paulson A, Iovino F, Wicha MS, HER2 regulates the mammary stem/progenitor cell population driving tumorigenesis and invasion, Oncogene 27 (2008) 6120–6130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Korkaya H, Wicha MS, HER-2, notch, and breast cancer stem cells: targeting an axis of evil, Clin. Cancer Res 15 (2009) 1845–1847. [DOI] [PubMed] [Google Scholar]
- [107].Korkaya H, Paulson A, Charafe-Jauffret E, Ginestier C, Brown M, Dutcher J, Clouthier SG, Wicha MS, Regulation of mammary stem/progenitor cells by PTEN/Akt/ beta-catenin signaling, PLoS Biol. 7 (2009) e1000121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Zhou J, Wulfkuhle J, Zhang H, Gu P, Yang Y, Deng J, Margolick JB, Liotta LA, Petricoin E III, Zhang Y, Activation of the PTEN/mTOR/STAT3 pathway in breast cancer stem-like cells is required for viability and maintenance, Proc. Natl. Acad. Sci. U. S. A 104 (2007) 16158–16163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Pratt MA, Tibbo E, Robertson SJ, Jansson D, Hurst K, Perez-Iratxeta C, Lau R, Niu MY, The canonical NF-kappaB pathway is required for formation of luminal mammary neoplasias and is activated in the mammary progenitor population, Oncogene 28 (2009) 2710–2722. [DOI] [PubMed] [Google Scholar]
- [110].Tokar EJ, BA Diwan MP Waalkes, Arsenic exposure transforms human epithelial stem/progenitor cells into a cancer stem-like phenotype, Environ. Health Perspect 118 (2010) 108–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].de Antonellis P, Medaglia C, Cusanelli E, Andolfo I, Liguori L, De Vita G, Carotenuto M, Bello A, Formiggini F, Galeone A, De Rosa G, Virgilio A, Scognamiglio I, Sciro M, Basso G, Schulte JH, Cinalli G, Iolascon A, Zollo M, MiR-34a targeting of Notch ligand Delta-like 1 impairs CD15+/CD133+ tumor-propagating cells and supports neural differentiation in medulloblastoma, PLoS ONE 6 (2011) e24584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Zhu TS, Costello MA, Talsma CE, Flack CG, Crowley JG, Hamm LL, He X, Hervey-Jumper SL, Heth JA, Muraszko KM, DiMeco F, Vescovi AL, Fan X, Endothelial cells create a stem cell niche in glioblastoma by providing NOTCH ligands that nurture self-renewal of cancer stem-like cells, Cancer Res. 71 (2011) 6061–6072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Wang J, Wang C, Meng Q, Li S, Sun X, Bo Y, Yao W, siRNA targeting Notch-1 de-creases glioma stem cell proliferation and tumor growth, Mol. Biol. Rep 39 (3) (2011) 2497–2503. [DOI] [PubMed] [Google Scholar]
- [114].Fan X, Khaki L, Zhu TS, Soules ME, Talsma CE, Gul N, Koh C, Zhang J, Li YM, Maciaczyk J, Nikkhah G, Dimeco F, Piccirillo S, Vescovi AL, Eberhart CG, NOTCH pathway blockade depletes CD133-positive glioblastoma cells and inhibits growth of tumor neurospheres and xenografts, Stem Cells 28 (2010) 5–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Garzia L, Andolfo I, Cusanelli E, Marino N, Petrosino G, De Martino D, Esposito V, Galeone A, Navas L, Esposito S, Gargiulo S, Fattet S, Donofrio V, Cinalli G, Brunetti A, Vecchio LD, Northcott PA, Delattre O, Taylor MD, Iolascon A, Zollo M, MicroRNA-199b-5p impairs cancer stem cells through negative regulation of HES1 in medulloblastoma, PLoS ONE 4 (2009) e4998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Fan X, Matsui W, Khaki L, Stearns D, Chun J, Li YM, Eberhart CG, Notch pathway inhibition depletes stem-like cells and blocks engraftment in embryonal brain tumors, Cancer Res. 66 (2006) 7445–7452. [DOI] [PubMed] [Google Scholar]
- [117].Sharma A, Paranjape AN, Rangarajan A, Dighe RR, A monoclonal antibody against human notch1 ligand binding domain depletes subpopulation of breast cancer stem-like cells, Mol. Cancer Ther 11 (1) (2011) 77–86. [DOI] [PubMed] [Google Scholar]
- [118].McGowan PM, Simedrea C, Ribot EJ, Foster PJ, Palmieri D, Steeg PS, Allan AL, Chambers AF, Notch1 inhibition alters the CD44hi/CD24lo population and reduces the formation of brain metastases from breast cancer, Mol. Cancer Res 9 (2011) 834–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Du Z, Li J, Wang L, Bian C, Wang Q, Liao L, Dou X, Bian X, Zhao RC, Overexpression of DeltaNp63alpha induces a stem cell phenotype in MCF7 breast carcinoma cell line through the Notch pathway, Cancer Sci. 101 (2010) 2417–2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Sikandar SS, Pate KT, Anderson S, Dizon D, Edwards RA, Waterman ML, Lipkin SM, NOTCH signaling is required for formation and self-renewal of tumor-initiating cells and for repression of secretory cell differentiation in colon cancer, Cancer Res. 70 (2010) 1469–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Sullivan JP, Spinola M, Dodge M, Raso MG, Behrens C, Gao B, Schuster K, Shao C, Larsen JE, Sullivan LA, Honorio S, Xie Y, Scaglioni PP, DiMaio JM, Gazdar AF, Shay JW, Wistuba II JD Minna, Aldehyde dehydrogenase activity selects for lung adenocarcinoma stem cells dependent on notch signaling, Cancer Res. 70 (2010) 9937–9948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Cao L, Zhou Y, Zhai B, Liao J, Xu W, Zhang R, Li J, Zhang Y, Chen L, Qian H, Wu M, Yin Z, Sphere-forming cell subpopulations with cancer stem cell properties in human hepatoma cell lines, BMC Gastroenterol. 11 (2011) 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Nishina S, Shiraha H, Nakanishi Y, Tanaka S, Matsubara M, Takaoka N, Uemura M, Horiguchi S, Kataoka J, Iwamuro M, Yagi T, Yamamoto K, Restored expression of the tumor suppressor gene RUNX3 reduces cancer stem cells in hepatocellular carcinoma by suppressing Jagged1-Notch signaling, Oncol. Rep 26 (2011) 523–531. [DOI] [PubMed] [Google Scholar]
- [124].Mine T, Matsueda S, Gao H, Li Y, Wong KK, Peoples GE, Ferrone S, Ioannides CG, Created Gli-1 duplex short-RNA (i-Gli-RNA) eliminates CD44 Hi progenitors of taxol-resistant ovarian cancer cells, Oncol. Rep 23 (2010) 1537–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Bao B, Wang Z, Ali S, Kong D, Li Y, Ahmad A, Banerjee S, Azmi AS, Miele L, Sarkar FH, Notch-1 induces epithelial–mesenchymal transition consistent with cancer stem cell phenotype in pancreatic cancer cells, Cancer Lett. 307 (2011) 26–36. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [126].Bao S, Wu Q, Sathornsumetee S, Hao Y, Li Z, Hjelmeland AB, Shi Q, McLendon RE, Bigner DD, Rich JN, Stem cell-like glioma cells promote tumor angiogenesis through vascular endothelial growth factor, Cancer Res. 66 (2006) 7843–7848. [DOI] [PubMed] [Google Scholar]
- [127].Ping YF, Yao XH, Jiang JY, Zhao LT, Yu SC, Jiang T, Lin MC, Chen JH, Wang B, Zhang R, Cui YH, Qian C, Wang J, Bian XW, The chemokine CXCL12 and its receptor CXCR4 promote glioma stem cell-mediated VEGF production and tumour angiogenesis via PI3K/AKT signalling, J. Pathol 224 (2011) 344–354. [DOI] [PubMed] [Google Scholar]
- [128].Zabierowski SE, Herlyn M, Learning the ABCs of melanoma-initiating cells, Cancer Cell 13 (2008) 185–187. [DOI] [PubMed] [Google Scholar]
- [129].Gazzaniga P, Cigna E, Panasiti V, Devirgiliis V, Bottoni U, Vincenzi B, Nicolazzo C, Petracca A, Gradilone A, CD133 and ABCB5 as stem cell markers on sentinel lymph node from melanoma patients, Eur. J. Surg. Oncol 36 (2010) 1211–1214. [DOI] [PubMed] [Google Scholar]
- [130].Maniotis AJ, Folberg R, Hess A, Seftor EA, Gardner LM, Pe’er J, Trent JM, Meltzer PS, Hendrix MJ, Vascular channel formation by human melanoma cells in vivo and in vitro: vasculogenic mimicry, Am. J. Pathol 155 (1999) 739–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Frank NY, Schatton T, Kim S, Zhan Q, Wilson BJ, Ma J, Saab KR, Osherov V, Widlund HR, Gasser M, Waaga-Gasser AM, Kupper TS, Murphy GF, Frank MH, VEGFR-1 expressed by malignant melanoma-initiating cells is required for tumor growth, Cancer Res. 71 (2011) 1474–1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Folkins C, Shaked Y, Man S, Tang T, Lee CR, Zhu Z, Hoffman RM, Kerbel RS, Glioma tumor stem-like cells promote tumor angiogenesis and vasculogenesis via vascular endothelial growth factor and stromal-derived factor 1, Cancer Res. 69 (2009) 7243–7251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Rizzo P, Osipo C, Foreman K, Golde T, Osborne B, Miele L, Rational targeting of Notch signaling in cancer, Oncogene 27 (2008) 5124–5131. [DOI] [PubMed] [Google Scholar]
- [134].Hooper JE, Scott MP, Communicating with Hedgehogs, Nat. Rev. Mol. Cell Biol 6 (2005)306–317. [DOI] [PubMed] [Google Scholar]
- [135].Lum L, PA Beachy, The Hedgehog response network: sensors, switches, and routers, Science 304 (2004) 1755–1759. [DOI] [PubMed] [Google Scholar]
- [136].Pasca di Magliano M, Hebrok M, Hedgehog signalling in cancer formation and maintenance, Nat. Rev. Cancer 3 (2003) 903–911. [DOI] [PubMed] [Google Scholar]
- [137].Nagase T, Nagase M, Machida M, Fujita T, Hedgehog signalling in vascular development, Angiogenesis 11 (2008) 71–77. [DOI] [PubMed] [Google Scholar]
- [138].Katoh Y, Katoh M, Identification and characterization of rat Desert hedgehog and Indian hedgehog genes in silico, Int. J. Oncol 26 (2005) 545–549. [PubMed] [Google Scholar]
- [139].Katoh Y, Katoh M, Comparative genomics on Sonic hedgehog orthologs, Oncol. Rep 14 (2005) 1087–1090. [PubMed] [Google Scholar]
- [140].Marigo V, Roberts DJ, Lee SM, Tsukurov O, Levi T, Gastier JM, Epstein DJ, Gilbert DJ, Copeland NG, Seidman CE, et al. , Cloning, expression, and chromosomal location of SHH and IHH: two human homologues of the Drosophila segment polarity gene hedgehog, Genomics 28 (1995) 44–51. [DOI] [PubMed] [Google Scholar]
- [141].Visbal AP, Lewis MT, Hedgehog signaling in the normal and neoplastic mammary gland, Curr. Drug Targets 11 (9) (2010) 1103–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Katoh M, Networking of WNT, FGF, Notch, BMP, and Hedgehog signaling pathways during carcinogenesis, Stem Cell Rev. 3 (2007) 30–38. [DOI] [PubMed] [Google Scholar]
- [143].Androutsellis-Theotokis A, Leker RR, Soldner F, Hoeppner DJ, Ravin R, Poser SW, Rueger MA, Bae SK, Kittappa R, McKay RD, Notch signalling regulates stem cell numbers in vitro and in vivo, Nature 442 (2006) 823–826. [DOI] [PubMed] [Google Scholar]
- [144].Ingram WJ, McCue KI, Tran TH, Hallahan AR, Wainwright BJ, Sonic Hedgehog regulates Hes1 through a novel mechanism that is independent of canonical Notch pathway signalling, Oncogene 27 (2008) 1489–1500. [DOI] [PubMed] [Google Scholar]
- [145].Zhao X, Malhotra GK, Lele SM, Lele MS, West WW,Eudy JD, Band H, Band V, Telomerase-immortalized human mammary stem/progenitor cells with ability to self-renew and differentiate, Proc. Natl. Acad. Sci. U. S. A 107 (2010) 14146–14151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Dejana E, The role of Wnt signaling in physiological and pathological angiogenesis, Circ. Res 107 (2010) 943–952. [DOI] [PubMed] [Google Scholar]
- [147].Zhang B, Ma JX, Wnt pathway antagonists and angiogenesis, Protein Cell 1 (2010) 898–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Barcelos LS, Duplaa C, Krankel N, Graiani G, Invernici G, Katare R, Siragusa M, Meloni M, Campesi I, Monica M, Simm A, Campagnolo P, Mangialardi G, Stevanato L, Alessandri G, Emanueli C, Madeddu P, Human CD133+ progenitor cells promote the healing of diabetic ischemic ulcers by paracrine stimulation of angiogenesis and activation of Wnt signaling, Circ. Res 104 (2009) 1095–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Hu J, Dong A, Fernandez-Ruiz V, Shan J, Kawa M, Martinez-Anso E, Prieto J, Qian C, Blockade of Wnt signaling inhibits angiogenesis and tumor growth in hepatocellular carcinoma, Cancer Res. 69 (2009) 6951–6959. [DOI] [PubMed] [Google Scholar]
- [150].Zhang B, Abreu JG, Zhou K, Chen Y, Hu Y, Zhou T, He X, Ma JX, Blocking the Wnt pathway, a unifying mechanism for an angiogenic inhibitor in the serine proteinase inhibitor family, Proc. Natl. Acad. Sci. U. S. A 107 (2010) 6900–6905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151].Ayyanan A, Civenni G, Ciarloni L, Morel C, Mueller N, Lefort K, Mandinova A, Raffoul W, Fiche M, Dotto GP, Brisken C, Increased Wnt signaling triggers oncogenic conversion of human breast epithelial cells by a Notch-dependent mechanism, Proc. Natl. Acad. Sci. U. S. A 103 (2006) 3799–3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Phng LK, Potente M, Leslie JD, Babbage J, Nyqvist D, Lobov I, Ondr JK, Rao S, Lang RA, Thurston G, Gerhardt H, Nrarp coordinates endothelial Notch and Wnt signaling to control vessel density in angiogenesis, Dev. Cell 16 (2009) 70–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Koutras AK, Fountzilas G, Kalogeras KT, Starakis I, Iconomou G, Kalofonos HP, The upgraded role of HER3 and HER4 receptors in breast cancer, Crit. Rev. Oncol. Hematol 74 (2010) 73–78. [DOI] [PubMed] [Google Scholar]
- [154].Tai W, Mahato R, Cheng K, The role of HER2 in cancer therapy and targeted drug delivery, J. Control. Release 146 (3) (2010) 264–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Flynn JF, Wong C, Wu JM, Anti-EGFR therapy: mechanism and advances in clinical efficacy in breast cancer, J. Oncol 2009 (2009) 526963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156].Jimeno A, Hidalgo M, Pharmacogenomics of epidermal growth factor receptor (EGFR) tyrosine kinase inhibitors, Biochim. Biophys. Acta 1766 (2006) 217–229. [DOI] [PubMed] [Google Scholar]
- [157].Kumar R, Yarmand-Bagheri R, The role of HER2 in angiogenesis, Semin. Oncol 28 (2001)27–32. [DOI] [PubMed] [Google Scholar]
- [158].Press MF, Lenz HJ, EGFR, HER2 and VEGF pathways: validated targets for cancer treatment, Drugs 67 (2007) 2045–2075. [DOI] [PubMed] [Google Scholar]
- [159].Laughner E, Taghavi P, Chiles K, Mahon PC, Semenza GL, HER2 (neu) signaling increases the rate of hypoxia-inducible factor1alpha (HIF-1alpha) synthesis: novel mechanism for HIF-1-mediated vascular endothelial growth factor expression, Mol. Cell. Biol 21 (2001) 3995–4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [160].Koukourakis MI, Simopoulos C, Polychronidis A, Perente S, Botaitis S, Giatromanolaki A, Sivridis E, The effect of trastuzumab/docatexel combination on breast cancer angiogenesis: dichotomus effect predictable by the HIFI alpha/VEGF pre-treatment status? Anticancer Res. 23 (2003) 1673–1680. [PubMed] [Google Scholar]
- [161].Kumar S, Kishimoto H, Chua HL, Badve S, Miller KD, Bigsby RM, Nakshatri H, Interleukin-1 alpha promotes tumor growth and cachexia in MCF-7 xenograft model of breast cancer, Am. J. Pathol 163 (2003) 2531–2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [162].Yamaguchi N, Oyama T, Ito E, Satoh H, Azuma S, Hayashi M, Shimizu K, Honma R, Yanagisawa Y, Nishikawa A, Kawamura M, Imai J, Ohwada S, Tatsuta K, Inoue J, Semba K, Watanabe S, NOTCH3 signaling pathway plays crucial roles in the proliferation of ErbB2-negative human breast cancer cells, Cancer Res. 68 (2008) 1881–1888. [DOI] [PubMed] [Google Scholar]
- [163].Hasson P, Paroush Z, Crosstalk between the EGFR and other signalling pathways at the level of the global transcriptional corepressor Groucho/TLE, Br. J. Cancer 94 (2006) 771–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [164].Sundaram MV, The love-hate relationship between Ras and Notch, Genes Dev. 19 (2005) 1825–1839. [DOI] [PubMed] [Google Scholar]
- [165].Dai J, Ma D, Zang S, Guo D, Qu X, Ye J, Ji C, Cross-talk between Notch and EGFR signaling in human breast cancer cells, Cancer Invest. 27 (2009) 533–540. [DOI] [PubMed] [Google Scholar]
- [166].Dong Y, Li A, Wang J, Weber JD, Michel LS, Synthetic lethality through combined Notch-epidermal growth factor receptor pathway inhibition in basal-like breast cancer, Cancer Res. 70 (2010) 5465–5474. [DOI] [PubMed] [Google Scholar]
- [167].Bergsten E, Uutela M, Li X, Pietras K, Ostman A, Heldin CH, Alitalo K, Eriksson U, PDGF-D is a specific, protease-activated ligand for the PDGF beta-receptor, Nat. Cell Biol 3 (2001) 512–516. [DOI] [PubMed] [Google Scholar]
- [168].LaRochelle WJ, Jeffers M, McDonald WF, Chillakuru RA, Giese NA, Lokker NA, Sullivan C, Boldog FL, Yang M, Vernet C, Burgess CE, Fernandes E, Deegler LL, Rittman B, Shimkets J, Shimkets RA, Rothberg JM, Lichenstein HS, PDGF-D, a new protease-activated growth factor, Nat. Cell Biol 3 (2001) 517–521. [DOI] [PubMed] [Google Scholar]
- [169].Li X, Eriksson U, Novel PDGF family members: PDGF-C and PDGF-D, Cytokine Growth Factor Rev. 14 (2003) 91–98. [DOI] [PubMed] [Google Scholar]
- [170].Li M, Jendrossek V, Belka C, The role of PDGF in radiation oncology, Radiat. Oncol 2 (2007) 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [171].Seymour L, Bezwoda WR, Positive immunostaining for platelet derived growth factor (PDGF) is an adverse prognostic factor in patients with advanced breast cancer, Breast Cancer Res. Treat 32 (1994) 229–233. [DOI] [PubMed] [Google Scholar]
- [172].Pietras K, Sjoblom T, Rubin K, Heldin CH, Ostman A, PDGF receptors as cancer drug targets, Cancer Cell 3 (2003) 439–443. [DOI] [PubMed] [Google Scholar]
- [173].Wang Z, Ahmad A, Li Y, Kong D, Azmi AS, Banerjee S, Sarkar FH, Emerging roles of PDGF-D signaling pathway in tumor development and progression, Biochim. Biophys. Acta 1806 (2010) 122–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [174].Ahmad A, Wang Z, Kong D, Ali R, Ali S, Banerjee S, Sarkar FH, Platelet-derived growth factor-D contributes to aggressiveness of breast cancer cells by up-regulating Notch and NF-kappaB signaling pathways, Breast Cancer Res. Treat 126 (1) (2010) 15–25. [DOI] [PubMed] [Google Scholar]
- [175].Beisner J, Buck MB, Fritz P, Dippon J, Schwab M, Brauch H, Zugmaier G, Pfizenmaier K, Knabbe C, A novel functional polymorphism in the transforming growth factor-beta2 gene promoter and tumor progression in breast cancer, Cancer Res. 66 (2006) 7554–7561. [DOI] [PubMed] [Google Scholar]
- [176].Cambien F, Ricard S, Troesch A, Mallet C, Generenaz L, Evans A, Arveiler D, Luc G, Ruidavets JB, Poirier O, Polymorphisms of the transforming growth factor-beta 1 gene in relation to myocardial infarction and blood pressure. The Etude Cas-Temoin de l’Infarctus du Myocarde (ECTIM) study, Hypertension 28 (1996) 881–887. [DOI] [PubMed] [Google Scholar]
- [177].Pasche B, Kolachana P, Nafa K, Satagopan J, Chen YG, Lo RS, Brener D, Yang D, Kirstein L, Oddoux C, Ostrer H, Vineis P, Varesco L, Jhanwar S, Luzzatto L, Massague J, Offit K, TbetaR-I(6A) is a candidate tumor susceptibility allele, Cancer Res. 59 (1999) 5678–5682. [PubMed] [Google Scholar]
- [178].Barcellos-Hoff MH, Akhurst RJ, Transforming growth factor-beta in breast cancer: too much, too late, Breast Cancer Res. 11 (2009) 202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [179].Padua D, Massague J, Roles of TGFbeta in metastasis, Cell Res. 19 (2009) 89–102. [DOI] [PubMed] [Google Scholar]
- [180].Pardali E, Goumans MJ, ten Dijke P, Signaling by members of the TGF-beta family in vascular morphogenesis and disease, Trends Cell Biol. 20 (2010) 556–567. [DOI] [PubMed] [Google Scholar]
- [181].Pardali E, ten Dijke P, Transforming growth factor-beta signaling and tumor angiogenesis, Front. Biosci 14 (2009) 4848–4861. [DOI] [PubMed] [Google Scholar]
- [182].van Meeteren LA, Goumans MJ, Ten Dijke P, TGF-beta receptor signaling pathways in angiogenesis; emerging targets for anti-angiogenesis therapy, Curr. Pharm. Biotechnol 12 (12) (2011) 2108–2120. [DOI] [PubMed] [Google Scholar]
- [183].Zavadil J, Cermak L, Soto-Nieves N, Bottinger EP, Integration of TGF-beta/Smad and Jagged1/Notch signalling in epithelial-to-mesenchymal transition, EMBO J. 23 (2004) 1155–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [184].Niimi H, Pardali K, Vanlandewijck M, Heldin CH, Moustakas A, Notch signaling is necessary for epithelial growth arrest by TGF-beta, J. Cell Biol 176 (2007) 695–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [185].Blokzijl A, Dahlqvist C, Reissmann E, Falk A, Moliner A, Lendahl U, Ibanez CF, Cross-talk between the Notch and TGF-beta signaling pathways mediated by interaction of the Notch intracellular domain with Smad3, J. Cell Biol 163 (2003) 723–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [186].Asano N, Watanabe T, Kitani A, Fuss IJ, Strober W, Notch1 signaling and regulatory T cell function, J. Immunol 180 (2008) 2796–2804. [DOI] [PubMed] [Google Scholar]
- [187].Samon JB, Champhekar A, Minter LM, Telfer JC, Miele L, Fauq A, Das P, Golde TE, Osborne BA, Notch1 and TGFbeta1 cooperatively regulate Foxp3 expression and the maintenance of peripheral regulatory T cells, Blood 112 (2008) 1813–1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [188].Ferrara N, Vascular endothelial growth factor: molecular and biological aspects, Curr. Top. Microbiol. Immunol 237 (1999) 1–30. [DOI] [PubMed] [Google Scholar]
- [189].Ferrara N, Gerber HP, LeCouter J, The biology of VEGF and its receptors, Nat. Med 9 (2003) 669–676. [DOI] [PubMed] [Google Scholar]
- [190].Guo S, Colbert LS, Fuller M, Zhang Y, Gonzalez-Perez RR, Vascular endothelial growth factor receptor-2 in breast cancer, Biochim. Biophys. Acta 1806 (2010) 108–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [191].Shibuya M, Claesson-Welsh L, Signal transduction by VEGF receptors in regulation of angiogenesis and lymphangiogenesis, Exp. Cell Res 312 (2006) 549–560. [DOI] [PubMed] [Google Scholar]
- [192].Guo S, Colbert L, McGlothen T, Gonzalez-Perez R, Regulation of angiogenesis in human cancer via vascular endothelial growth factor receptor-2 (VEGFR-2), in: Sophia Ran (Ed.), Tumor Angiogenesis, 2012, (www.intechweb.org). [Google Scholar]
- [193].Lawson ND, Vogel AM, Weinstein BM, sonic hedgehog and vascular endothelial growth factor act upstream of the Notch pathway during arterial endothelial differentiation, Dev. Cell 3 (2002) 127–136. [DOI] [PubMed] [Google Scholar]
- [194].Hainaud P, Contreres JO, Villemain A, Liu LX, Plouet J, Tobelem G, Dupuy E, The role of the vascular endothelial growth factor-Delta-like 4 ligand/Notch4-ephrin B2 cascade in tumor vessel remodeling and endothelial cell functions, Cancer Res. 66 (2006) 8501–8510. [DOI] [PubMed] [Google Scholar]
- [195].Patel NS, Li JL, Generali D, Poulsom R, Cranston DW, Harris AL, Up-regulation of Delta-like 4 ligand in human tumor vasculature and the role of basal expression in endothelial cell function, Cancer Res. 65 (2005) 8690–8697. [DOI] [PubMed] [Google Scholar]
- [196].Ridgway J, Zhang G, Wu Y, Stawicki S, Liang WC, Chanthery Y, Kowalski J, Watts RJ, Callahan C, Kasman I, Singh M, Chien M, Tan C, Hongo JA, de Sauvage F, Plowman G, Yan M, Inhibition of Dll4 signalling inhibits tumour growth by deregulating angiogenesis, Nature 444 (2006) 1083–1087. [DOI] [PubMed] [Google Scholar]
- [197].Suchting S, Freitas C, le Noble F, Benedito R, Breant C, Duarte A, Eichmann A, The Notch ligand Delta-like 4 negatively regulates endothelial tip cell formation and vessel branching, Proc. Natl. Acad. Sci. U. S. A 104 (2007) 3225–3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [198].Mailhos C, Modlich U, Lewis J, Harris A, Bicknell R, Ish-Horowicz D, Delta4, an endothelial specific notch ligand expressed at sites of physiological and tumor angiogenesis, Differentiation 69 (2001) 135–144. [DOI] [PubMed] [Google Scholar]
- [199].Noguera-Troise I, Daly C, Papadopoulos NJ, Coetzee S, Boland P, Gale NW, Lin HC, Yancopoulos GD, Thurston G, Blockade of Dll4 inhibits tumour growth by promoting non-productive angiogenesis, Nature 444 (2006) 1032–1037. [DOI] [PubMed] [Google Scholar]
- [200].Sainson RC, Johnston DA, Chu HC, Holderfield MT, Nakatsu MN, Crampton SP, Davis J, Conn E, Hughes CC, TNF primes endothelial cells for angiogenic sprouting by inducing a tip cell phenotype, Blood 111 (2008) 4997–5007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [201].Taylor KL, Henderson AM, Hughes CC, Notch activation during endothelial cell network formation in vitro targets the basic HLH transcription factor HESR-1 and downregulates VEGFR-2/KDR expression, Microvasc. Res 64 (2002) 372–383. [DOI] [PubMed] [Google Scholar]
- [202].Siekmann AF, Covassin L, Lawson ND, Modulation of VEGF signalling output by the Notch pathway, Bioessays 30 (2008) 303–313. [DOI] [PubMed] [Google Scholar]
- [203].Thurston G,Kitajewski J, VEGF and Delta–Notch: interacting signalling pathways in tumour angiogenesis, Br. J. Cancer 99 (2008) 1204–1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [204].Kishimoto T, Interleukin-6: from basic science to medicine—40 years in immunology, Annu. Rev. Immunol 23 (2005) 1–21. [DOI] [PubMed] [Google Scholar]
- [205].Neurath MF, Finotto S, IL-6 signaling in autoimmunity, chronic inflammation and inflammation-associated cancer, Cytokine Growth Factor Rev. 22 (2011) 83–89. [DOI] [PubMed] [Google Scholar]
- [206].Tawara K,Oxford JT, L Jorcyk C, Clinical significance of interleukin (IL)-6 in cancer metastasis to bone: potential of anti-IL-6 therapies, Cancer Manag. Res. 3 (2011) 177–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [207].Anglesio MS, George J, Kulbe H, Friedlander M, Rischin D, Lemech C, Power J, Coward J, Cowin PA, House CM, Chakravarty P, Gorringe KL, Campbell IG, Okamoto A, Birrer MJ, Huntsman DG, de Fazio A, Kalloger SE, Balkwill F, Gilks CB, Bowtell DD, IL6-STAT3-HIF signaling and therapeutic response to the angiogenesis inhibitor sunitinib in ovarian clear cell cancer, Clin. Cancer Res 17 (2011) 2538–2548. [DOI] [PubMed] [Google Scholar]
- [208].Liu Q, Li G, Li R, Shen J, He Q, Deng L, Zhang C, Zhang J, IL-6 promotion of glioblastoma cell invasion and angiogenesis in U251 and T98G cell lines, J. Neurooncol 100 (2010) 165–176. [DOI] [PubMed] [Google Scholar]
- [209].Nilsson MB, Langley RR, Fidler IJ, Interleukin-6, secreted by human ovarian carcinoma cells, is a potent proangiogenic cytokine, Cancer Res. 65 (2005) 10794–10800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [210].Shinriki S, Jono H, Ota K, Ueda M, Kudo M, Ota T, Oike Y, Endo M, Ibusuki M, Hiraki A, Nakayama H, Yoshitake Y, Shinohara M, Ando Y, Humanized anti-interleukin-6 receptor antibody suppresses tumor angiogenesis and in vivo growth of human oral squamous cell carcinoma, Clin. Cancer Res 15 (2009) 5426–5434. [DOI] [PubMed] [Google Scholar]
- [211].Wani AA, Jafarnejad SM, Zhou J, Li G, Integrin-linked kinase regulates melanoma angiogenesis by activating NF-kappaB/interleukin-6 signaling pathway, Oncogene 30 (2011) 2778–2788. [DOI] [PubMed] [Google Scholar]
- [212].Sansone P, Storci G, Tavolari S, Guarnieri T, Giovannini C, Taffurelli M, Ceccarelli C, Santini D, Paterini P, Marcu KB, Chieco P, Bonafe M, IL-6 triggers malignant features in mammospheres from human ductal breast carcinoma and normal mammary gland, J. Clin. Invest 117 (2007) 3988–4002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [213].Wongchana W, Palaga T, Direct regulation of interleukin-6 expression by Notch signaling in macrophages, Cell. Mol. Immunol 9 (2) (2011) 155–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [214].Lee JH, Suk J, Park J, Kim SB, Kwak SS, Kim JW, Lee CH, Byun B, Ahn JK, Joe CO, Notch signal activates hypoxia pathway through HES1-dependent SRC/signal transducers and activators of transcription 3 pathway, Mol. Cancer Res 7 (2009) 1663–1671. [DOI] [PubMed] [Google Scholar]
- [215].Dinarello CA, Interleukin-1, interleukin-1 receptors and interleukin-1 receptor antagonist, Int. Rev. Immunol 16 (1998) 457–499. [DOI] [PubMed] [Google Scholar]
- [216].Apte RN, Dotan S, Elkabets M, White MR, Reich E, Carmi Y, Song X, Dvozkin T, Krelin Y, Voronov E, The involvement of IL-1 in tumorigenesis, tumor invasiveness, metastasis and tumor–host interactions, Cancer Metastasis Rev. 25 (2006) 387–408. [DOI] [PubMed] [Google Scholar]
- [217].Perrier S, Caldefie-Chezet F, Vasson MP, IL-1 family in breast cancer: potential interplay with leptin and other adipocytokines, FEBS Lett. 583 (2009) 259–265. [DOI] [PubMed] [Google Scholar]
- [218].Zhou W, Guo S, Gonzalez-Perez RR, Leptin pro-angiogenic signature in breast cancer is linked to IL-1 signalling, Br. J. Cancer 104 (1) (2010) 128–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [219].Zhou W, Guo S, Gonzalez-Perez RR, Leptin pro-angiogenic signature in breast cancer is linked to IL-1 signalling, Br. J. Cancer 104 (2011) 128–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [220].Guo S, Gonzalez-Perez RR, Notch, IL-1 and leptin crosstalk outcome (NILCO) is critical for leptin-induced proliferation, migration and VEGF/VEGFR-2 expression in breast cancer, PLoS One 6 (6) (2011) e21467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [221].Cao Z, Tanaka M, Regnier C, Rothe M, Yamit-hezi A,Woronicz JD, Fuentes ME, Durnin MH, Dalrymple SA, Goeddel DV, NF-kappa B activation by tumor necrosis factor and interleukin-1, Cold Spring Harb. Symp. Quant. Biol 64 (1999) 473–483. [DOI] [PubMed] [Google Scholar]
- [222].Vallabhapurapu S, Karin M, Regulation and function of NF-kappaB transcription factors in the immune system, Annu. Rev. Immunol 27 (2009) 693–733. [DOI] [PubMed] [Google Scholar]
- [223].Verstrepen L, Bekaert T, Chau TL, Tavernier J, Chariot A, Beyaert R, TLR-4, IL-1R and TNF-R signaling to NF-kappaB: variations on a common theme, Cell. Mol. Life Sci 65 (2008) 2964–2978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [224].Renard P, Raes M, The proinflammatory transcription factor NFkappaB: a potential target for novel therapeutical strategies, Cell Biol. Toxicol 15 (1999) 341–344. [DOI] [PubMed] [Google Scholar]
- [225].Yao J, Kim TW, Qin J, Jiang Z, Qian Y, Xiao H, Lu Y, Qian W, Gulen MF, Sizemore N, DiDonato J, Sato S, Akira S, Su B, Li X, Interleukin-1 (IL-1)-induced TAK1-dependent versus MEKK3-dependent NFkappaB activation pathways bifurcate at IL-1 receptor-associated kinase modification, J. Biol. Chem 282 (2007) 6075–6089. [DOI] [PubMed] [Google Scholar]
- [226].Bash J, Zong WX, Banga S, Rivera A, Ballard DW, Ron Y, Gelinas C, Rel/NF-kappaB can trigger the Notch signaling pathway by inducing the expression of Jagged1, a ligand for Notch receptors, EMBO J. 18 (1999) 2803–2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [227].Osipo C, Golde TE, Osborne BA, Miele LA, Off the beaten pathway: the complex cross talk between Notch and NF-kappaB, Lab. Invest. 88 (2008) 11–17. [DOI] [PubMed] [Google Scholar]
- [228].Gonzalez-Perez RR, Xu Y, Guo S, Watters A, Zhou W, Leibovich SJ, Leptin upregulates VEGF in breast cancer via canonic and non-canonical signalling path-ways and NFkappaB/HIF-1alpha activation, Cell. Signal. 22 (2010) 1350–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [229].Brennan AM, Mantzoros CS, Drug insight: the role of leptin in human physiology and pathophysiology—emerging clinical applications, Nat. Clin. Pract. Endocrinol. Metab 2 (2006) 318–327. [DOI] [PubMed] [Google Scholar]
- [230].Dardeno TA, Chou SH, Moon HS, Chamberland JP, Fiorenza CG, Mantzoros CS, Leptin in human physiology and therapeutics, Front. Neuroendocrinol 31 (2010) 377–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [231].Fernandez-Riejos P, Najib S, Santos-Alvarez J, Martin-Romero C, Perez-Perez A, Gonzalez-Yanes C, Sanchez-Margalet V, Role of leptin in the activation of immune cells, Mediat. Inflamm 2010 (2010) 568343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [232].Guo S, Liu M, Wang G, Torroella-Kouri M, Gonzalez-Perez RR, Oncogenic role and therapeutic target of leptin signaling in breast cancer and cancer stem cells, Biochim. Biophys. Acta 1825 (2012) 207–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [233].Hu X, Juneja SC, Maihle NJ, Cleary MP, Leptin—a growth factor in normal and malignant breast cells and for normal mammary gland development, J. Natl. Cancer Inst 94 (2002) 1704–1711. [DOI] [PubMed] [Google Scholar]
- [234].Laud K, Gourdou I, Pessemesse L, Peyrat JP, Djiane J, Identification of leptin receptors in human breast cancer: functional activity in the T47-D breast cancer cell line, Mol. Cell. Endocrinol 188 (2002) 219–226. [DOI] [PubMed] [Google Scholar]
- [235].Tessitore L, Vizio B, Jenkins O, De Stefano I, Ritossa C, Argiles JM, Benedetto C, Mussa A, Leptin expression in colorectal and breast cancer patients, Int. J. Mol. Med 5 (2000) 421–426. [DOI] [PubMed] [Google Scholar]
- [236].Chen C, Chang YC, Liu CL, Chang KJ, Guo IC, Leptin-induced growth of human ZR-75-1 breast cancer cells is associated with up-regulation of cyclin D1 and c-Myc and down-regulation of tumor suppressor p53 and p21WAF1/CIP1, Breast Cancer Res. Treat 98 (2006) 121–132. [DOI] [PubMed] [Google Scholar]
- [237].Dieudonne MN, Machinal-Quelin F, Serazin-Leroy V, Leneveu MC, Pecquery R, Giudicelli Y, Leptin mediates a proliferative response in human MCF7 breast cancer cells, Biochem. Biophys. Res. Commun 293 (2002) 622–628. [DOI] [PubMed] [Google Scholar]
- [238].Cleary MP, Juneja SC, Phillips FC, Hu X, Grande JP, Maihle NJ, Leptin receptor-deficient MMTV-TGF-alpha/Lepr(db)Lepr(db) female mice do not develop oncogene-induced mammary tumors, Exp. Biol. Med. (Maywood) 229 (2004) 182–193. [DOI] [PubMed] [Google Scholar]
- [239].Cleary MP, Phillips FC, Getzin SC, Jacobson TL, Jacobson MK, Christensen TA, Juneja SC, Grande JP, Maihle NJ, Genetically obese MMTV-TGF-alpha/Lep(ob) Lep(ob) female mice do not develop mammary tumors, Breast Cancer Res. Treat 77 (2003) 205–215. [DOI] [PubMed] [Google Scholar]
- [240].Gonzalez RR, Cherfils S, Escobar M, Yoo JH, Carino C, Styer AK, Sullivan BT, Sakamoto H, Olawaiye A, Serikawa T, Lynch MP, Rueda BR, Leptin signaling promotes the growth of mammary tumors and increases the expression of vascular endothelial growth factor (VEGF) and its receptor type two (VEGF-R2), J. Biol. Chem 281 (2006) 26320–26328. [DOI] [PubMed] [Google Scholar]
- [241].Rene Gonzalez R, Watters A, Xu Y, Singh UP, Mann DR, Rueda BR, Penichet ML, Leptin-signaling inhibition results in efficient anti-tumor activity in estrogen receptor positive or negative breast cancer, Breast Cancer Res. 11 (2009) R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [242].Johnston A, Arnadottir S, Gudjonsson JE, Aphale A, Sigmarsdottir AA, Gunnarsson SI, Steinsson JT, Elder JT, Valdimarsson H, Obesity in psoriasis: leptin and resistin as mediators of cutaneous inflammation, Br. J. Dermatol 159 (2008) 342–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [243].Maedler K, Sergeev P, Ehses JA, Mathe Z, Bosco D, Berney T, Dayer JM, Reinecke M, Halban PA, Donath MY, Leptin modulates beta cell expression of IL-1 receptor antagonist and release of IL-1beta in human islets, Proc. Natl. Acad. Sci. U. S. A 101 (2004) 8138–8143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [244].Carino C, Olawaiye AB, Cherfils S, Serikawa T, Lynch MP, Rueda BR, Gonzalez RR, Leptin regulation of proangiogenic molecules in benign and cancerous endometrial cells, Int. J. Cancer 123 (2008) 2782–2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [245].Polus GJA, Piatkowska E, Dembinska-Kiec A, Differences in leptin, VEGF, and bFGF-induced angiogenic differentiation of HUVEC and human umbilical blood CD34+ progenitor cells, Eur. J. Biochem (2003) 1–67(Abstract number: P4). [Google Scholar]
- [246].Knight BB, Oprea-Ilies GM, Nagalingam A, Yang L, Cohen C, Saxena NK, Sharma D, Survivin upregulation, dependent on leptin-EGFR-Notch1 axis, is essential for leptin induced migration of breast carcinoma cells, Endocr. Relat. Cancer 18 (4) (2011)413–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [247].DeNicola GM, Tuveson DA, RAS in cellular transformation and senescence, Eur. J. Cancer 45 (Suppl.1) (2009) 211–216. [DOI] [PubMed] [Google Scholar]
- [248].Bajaj A, Zheng Q, Adam A, Vincent P, Pumiglia K, Activation of endothelial ras signaling bypasses senescence and causes abnormal vascular morphogenesis, Cancer Res. 70 (2010) 3803–3812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [249].Young A, Lyons J, Miller AL, Phan VT, Alarcon IR, McCormick F, Ras signaling and therapies, Adv. Cancer Res 102 (2009) 1–17. [DOI] [PubMed] [Google Scholar]
- [250].Zebisch A, Czemilofsky AP, Keri G, Smigelskaite J, Sill H, Troppmair J, Signaling through RAS-RAF-MEK-ERK: from basics to bedside, Curr. Med. Chem 14 (2007) 601–623. [DOI] [PubMed] [Google Scholar]
- [251].Hanlon L, Avila JL, Demarest RM, Troutman S, Allen M, Ratti F, Rustgi AK, Stanger BZ, Radtke F, Adsay V, Long F, Capobianco AJ, Kissil JL, Notch1 functions as a tumor suppressor in a model of K-ras-induced pancreatic ductal adenocarcinoma, Cancer Res. 70 (2010) 4280–4286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [252].Veenendaal LM, Kranenburg O, Smakman N, Klomp A, Borel Rinkes IH, van Diest PJ, Differential Notch and TGFbeta signaling in primary colorectal tumors and their corresponding metastases, Cell. Oncol 30 (2008) 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [253].Xu P, Qiu M, Zhang Z, Kang C, Jiang R, Jia Z, Wang G, Jiang H, Pu P, The oncogenic roles of Notch1 in astrocytic gliomas in vitro and in vivo, J. Neurooncol 97 (2010) 41–51. [DOI] [PubMed] [Google Scholar]
- [254].Chiang MY, Xu L, Shestova O, Histen G, L’Heureux S, Romany C, Childs ME, Gimotty PA, Aster JC, Pear WS, Leukemia-associated NOTCH1 alleles are weak tumor initiators but accelerate K-ras-initiated leukemia, J. Clin. Invest 118 (2008)3181–3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [255].Kindler T, Cornejo MG, Scholl C, Liu J, Leeman DS, Haydu JE, Frohling S, Lee BH, Gilliland DG, K-RasG12D-induced T-cell lymphoblastic lymphoma/leukemias harbor Notch1 mutations and are sensitive to gamma-secretase inhibitors, Blood 112 (2008) 3373–3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [256].Gustafson TL, Wellberg E, Laffin B, Schilling L, Metz RP, Zahnow CA, Porter WW Ha-Ras transformation of MCF10A cells leads to repression of singleminded-2s through NOTCH and C/EBPbeta, Oncogene 28 (2009) 1561–1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [257].Mittal S, Subramanyam D, Dey D, Kumar RV, Rangarajan A, Cooperation of Notch and Ras/MAPK signaling pathways in human breast carcinogenesis, Mol. Cancer 8 (2009) 128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [258].Sabbah M, Emami S, Redeuilh G, Julien S, Prevost G, Zimber A, Ouelaa R, Bracke M, De Wever O, Gespach C, Molecular signature and therapeutic perspective of the epithelial-to-mesenchymal transitions in epithelial cancers, Drug Resist. Updat 11 (2008) 123–151. [DOI] [PubMed] [Google Scholar]
- [259].Steelman LS, Stadelman KM, Chappell WH, Horn S, Basecke J, Cervello M, Nicoletti F, Libra M, Stivala F, Martelli AM, McCubrey JA, Akt as a therapeutic target in cancer, Expert Opin. Ther. Targets 12 (2008) 1139–1165. [DOI] [PubMed] [Google Scholar]
- [260].Wickenden JA, Watson CJ, Key signalling nodes in mammary gland development and cancer. Signalling downstream of PI3 kinase in mammary epithelium: a play in 3 Akts, Breast Cancer Res. 12 (2010) 202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [261].Dillon RL, Muller WJ, Distinct biological roles for the akt family in mammary tumor progression, Cancer Res. 70 (2010) 4260–4264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [262].Ju X, Katiyar S, Wang C, Liu M, Jiao X, Li S, Zhou J, Turner J, Lisanti MP, Russell RG, Mueller SC, Ojeifo J, Chen WS, Hay N, Pestell RG, Akt1 governs breast cancer progression in vivo, Proc. Natl. Acad. Sci. U. S. A 104 (2007) 7438–7443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [263].Nteliopoulos G, Marley SB, Gordon MY, Influence of PI-3K/Akt pathway on Wnt signalling in regulating myeloid progenitor cell proliferation. Evidence for a role of autocrine/paracrine Wnt regulation, Br. J. Haematol 146 (2009) 637–651. [DOI] [PubMed] [Google Scholar]
- [264].Meurette O, Stylianou S, Rock R, Collu GM, Gilmore AP, Brennan K, Notch activation induces Akt signaling via an autocrine loop to prevent apoptosis in breast epithelial cells, Cancer Res. 69 (2009) 5015–5022. [DOI] [PubMed] [Google Scholar]
- [265].Liu ZJ, Xiao M, Balint K, Smalley KS, Brafford P, Qiu R, Pinnix CC, Li X, Herlyn M, Notch1 signaling promotes primary melanoma progression by activating mitogen-activated protein kinase/phosphatidylinositol 3-kinase-Akt pathways and up-regulating N-cadherin expression, Cancer Res. 66 (2006) 4182–4190. [DOI] [PubMed] [Google Scholar]
- [266].Palomero T, Dominguez M, Ferrando AA, The role of the PTEN/AKT Pathway in NOTCH1-induced leukemia, Cell Cycle 7 (2008) 965–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [267].Bedogni B, Warneke JA, Nickoloff BJ, Giaccia AJ, Powell MB, Notch1 is an effector of Akt and hypoxia in melanoma development, J. Clin. Invest 118 (2008) 3660–3670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [268].Gutierrez A, Look AT, NOTCH and PI3K–AKT pathways intertwined, Cancer Cell 12 (2007) 411–413. [DOI] [PubMed] [Google Scholar]
- [269].Wang Z, Li Y, Banerjee S, Kong D, Ahmad A, Nogueira V, Hay N, Sarkar FH, Down-regulation of Notch-1 and Jagged-1 inhibits prostate cancer cell growth, migration and invasion, and induces apoptosis via inactivation of Akt, mTOR, and NF-kappaB signaling pathways, J. Cell. Biochem 109 (2010) 726–736. [DOI] [PubMed] [Google Scholar]
- [270].Rangarajan A, Syal R, Selvarajah S, Chakrabarti O, Sarin A, Krishna S, Activated Notch1 signaling cooperates with papillomavirus oncogenes in transformation and generates resistance to apoptosis on matrix withdrawal through PKB/Akt, Virology 286 (2001) 23–30. [DOI] [PubMed] [Google Scholar]
- [271].McKenzie G, Ward G, Stallwood Y, Briend E, Papadia S, Lennard A, Turner M, Champion B, Hardingham GE, Cellular Notch responsiveness is defined by phosphoinositide 3-kinase-dependent signals, BMC Cell Biol. 7 (2006) 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [272].Liu ZJ, Shirakawa T, Li Y, Soma A, Oka M, Dotto GP, Fairman RM, Velazquez OC, Herlyn M, Regulation of Notch1 and Dll4 by vascular endothelial growth factor in arterial endothelial cells: implications for modulating arteriogenesis and angiogenesis, Mol. Cell. Biol 23 (2003) 14–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [273].Kikuchi R, Takeshita K, Uchida Y, Kondo M, Cheng XW, Nakayama T, Yamamoto K, Matsushita T, Liao JK, Murohara T, Pitavastatin-induced angiogenesis and arteriogenesis is mediated by Notch1 in a murine hindlimb ischemia model without induction of VEGF, Lab. Invest 91 (2011) 691–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [274].Zhang J, Fukuhara S, Sako K, Takenouchi T, Kitani H, Kume T, Koh GY, Mochizuki N, Angiopoietin-1/Tie2 signal augments basal Notch signal controlling vascular quiescence by inducing Delta-like 4 expression through AKT-mediated activation of beta-catenin, J. Biol. Chem 286 (2011) 8055–8066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [275].Sparks CA, Guertin DA, Targeting mTOR: prospects for mTOR complex 2 inhibitors in cancer therapy, Oncogene 29 (2010) 3733–3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [276].Ciuffreda L, Di Sanza C, Incani UC, Milella M, The mTOR pathway: a new target in cancer therapy, Curr. Cancer Drug Targets 10 (2010) 484–495. [DOI] [PubMed] [Google Scholar]
- [277].Jung CH, Ro SH, Cao J, Otto NM, Kim DH, mTOR regulation of autophagy, FEBS Lett. 584 (2010) 1287–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [278].Menon S, Manning BD, Common corruption of the mTOR signaling network in human tumors, Oncogene 27 (Suppl. 2) (2008) S43–S51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [279].Zhou H, Huang S, The complexes of mammalian target of rapamycin, Curr. Protein Pept. Sci 11 (6) (2010) 409–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [280].Ghayad SE, A Cohen P, Inhibitors of the PI3K/Akt/mTOR pathway: new hope for breast cancer patients, Recent Pat. Anticancer Drug Discov 5 (2010) 29–57. [DOI] [PubMed] [Google Scholar]
- [281].Hadad SM, Fleming S, Thompson AM, Targeting AMPK: a new therapeutic opportunity in breast cancer, Crit. Rev. Oncol. Hematol 67 (2008) 1–7. [DOI] [PubMed] [Google Scholar]
- [282].Noh WC, Kim YH, Kim MS, Koh JS, Kim HA, Moon NM, Paik NS, Activation of the mTOR signaling pathway in breast cancer and its correlation with the clinicopathologic variables, Breast Cancer Res. Treat 110 (2008) 477–483. [DOI] [PubMed] [Google Scholar]
- [283].Chan SM, Weng AP, Tibshirani R, Aster JC, Utz PJ, Notch signals positively regulate activity of the mTOR pathway in T-cell acute lymphoblastic leukemia, Blood 110 (2007) 278–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [284].Efferson CL, Winkelmann CT, Ware C, Sullivan T, Giampaoli S, Tammam J, Patel S, Mesiti G, Reilly JF, Gibson RE, Buser C, Yeatman T, Coppola D, Winter C, Clark EA, Draetta GF, Strack PR, Majumder PK, Downregulation of Notch pathway by a gamma-secretase inhibitor attenuates AKT/mammalian target of rapamycin signaling and glucose uptake in an ERBB2 transgenic breast cancer model, Cancer Res. 70 (2010) 2476–2484. [DOI] [PubMed] [Google Scholar]
- [285].Mungamuri SK, Yang X, Thor AD, Somasundaram K, Survival signaling by Notch1: mammalian target of rapamycin (mTOR)-dependent inhibition of p53, Cancer Res. 66 (2006) 4715–4724. [DOI] [PubMed] [Google Scholar]
- [286].Minhajuddin M, Bijli KM, Fazal F, Sassano A, Nakayama KI, Hay N, Platanias LC, Rahman A, Protein kinase C-Delta and phosphatidylinositol 3-kinase/Akt activate mammalian target ofrapamycin to modulate NF-kappaB activation and intercellu-lar adhesion molecule-1 (ICAM-1) expression in endothelial cells, J. Biol. Chem 284 (2009) 4052–4061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [287].Rafiee P, Binion DG, Wellner M, Behmaram B, Floer M, Mitton E, Nie L, Zhang Z, Otterson MF, Modulatory effect of curcumin on survival of irradiated human intestinal microvascular endothelial cells: role of Akt/mTOR and NF-{kappa}B, Am. J. Physiol. Gastrointest. Liver Physiol 298 (2010) G865–G877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [288].Zhang H, Han Y, Tao J, Liu S, Yan C, Li S, Cellular repressor of E1A-stimulated genes regulates vascular endothelial cell migration by The ILK/AKT/mTOR/VEGF(165) signaling pathway, Exp. Cell Res 317 (2011) 2904–2913. [DOI] [PubMed] [Google Scholar]
- [289].Hayden MS, Ghosh S, Shared principles in NF-kappaB signaling, Cell 132 (2008) 344–362. [DOI] [PubMed] [Google Scholar]
- [290].Prasad S, Ravindran J, Aggarwal BB, NF-kappaB and cancer: how intimate is this relationship, Mol. Cell. Biochem 336 (2010) 25–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [291].Hayden MS, Ghosh S, Signaling to NF-kappaB, Genes Dev. 18 (2004) 2195–2224. [DOI] [PubMed] [Google Scholar]
- [292].Cheng P, Zlobin A, Volgina V, Gottipati S, Osborne B, Simel EJ, Miele L, Gabrilovich DI, Notch-1 regulates NF-kappaB activity in hemopoietic progenitor cells, J. Immunol 167 (2001) 4458–4467. [DOI] [PubMed] [Google Scholar]
- [293].Wang J, Shelly L, Miele L, Boykins R, Norcross MA, Guan E, Human Notch-1 inhibits NF-kappa B activity in the nucleus through a direct interaction involving a novel domain, J. Immunol 167 (2001) 289–295. [DOI] [PubMed] [Google Scholar]
- [294].Shin HM, Minter LM, Cho OH, Gottipati S, Fauq AH, Golde TE, Sonenshein GE, Osborne BA, Notch1 augments NF-kappaB activity by facilitating its nuclear retention, EMBO J. 25 (2006) 129–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [295].Ke Q, Costa M, Hypoxia-inducible factor-1 (HIF-1), Mol. Pharmacol 70 (2006) 1469–1480. [DOI] [PubMed] [Google Scholar]
- [296].Hirota K, Semenza GL, Regulation of angiogenesis by hypoxia-inducible factor 1, Crit. Rev. Oncol. Hematol 59 (2006) 15–26. [DOI] [PubMed] [Google Scholar]
- [297].Li Y, Ye D, Cancer therapy by targeting hypoxia-inducible factor-1, Curr. Cancer Drug Targets 10 (7) (2010) 782–796. [DOI] [PubMed] [Google Scholar]
- [298].Gustafsson MV, Zheng X, Pereira T, Gradin K, Jin S, Lundkvist J, Ruas JL, Poellinger L, Lendahl U, Bondesson M, Hypoxia requires notch signaling to maintain the undifferentiated cell state, Dev. Cell 9 (2005) 617–628. [DOI] [PubMed] [Google Scholar]
- [299].Sahlgren C, Gustafsson MV, Jin S, Poellinger L, Lendahl U, Notch signaling mediates hypoxia-induced tumor cell migration and invasion, Proc. Natl. Acad. Sci. U. S. A 105 (2008) 6392–6397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [300].Bernstein L, Ross RK, Endogenous hormones and breast cancer risk, Epidemiol. Rev 15 (1993) 48–65. [DOI] [PubMed] [Google Scholar]
- [301].Hankinson SE, Endogenous hormones and risk of breast cancer in postmenopausal women, Breast Dis. 24 (2005) 3–15. [DOI] [PubMed] [Google Scholar]
- [302].Mendelsohn ME, Karas RH, The protective effects ofestrogen on the cardiovascular system, N. Engl. J. Med 340 (1999) 1801–1811. [DOI] [PubMed] [Google Scholar]
- [303].Welboren WJ, Stunnenberg HG, Sweep FC, Span PN, Identifying estrogen receptor target genes, Mol. Oncol 1 (2007) 138–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [304].Calaf GM, Roy D, Cell adhesion proteins altered by 17beta estradiol and parathion in breast epithelial cells, Oncol. Rep 19 (2008) 165–169. [PubMed] [Google Scholar]
- [305].Rizzo P, Miao H, D’Souza G, Osipo C, Song LL, Yun J, Zhao H, Mascarenhas J, Wyatt D, Antico G, Hao L, Yao K, Rajan P, Hicks C, Siziopikou K, Selvaggi S, Bashir A, Bhandari D, Marchese A, Lendahl U, Qin JZ, Tonetti DA, Albain K, Nickoloff BJ, Miele L, Cross-talk between notch and the estrogen receptor in breast cancer suggests novel therapeutic approaches, Cancer Res. 68 (2008) 5226–5235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [306].Li Y, Guessous F, Zhang Y, Dipierro C, Kefas B, Johnson E, Marcinkiewicz L, Jiang J, Yang Y, Schmittgen TD, Lopes B, Schiff D, Purow B, Abounader R, MicroRNA-34a inhibits glioblastoma growth by targeting multiple oncogenes, Cancer Res. 69 (2009) 7569–7576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [307].Solomon A, Mian Y, Ortega-Cava C, Liu VW, Gurumurthy CB, Naramura M, Band V, Band H, Upregulation of the let-7 microRNA with precocious development in lin-12/Notch hypermorphic Caenorhabditis elegans mutants, Dev. Biol 316 (2008) 191–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [308].Yoo AS, Greenwald I, LIN-12/Notch activation leads to microRNA-mediated down-regulation of Vav in C. elegans, Science 310 (2005) 1330–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [309].Jannot G, Simard MJ, Tumour-related microRNAs functions in Caenorhabditis elegans, Oncogene 25 (2006) 6197–6201. [DOI] [PubMed] [Google Scholar]
- [310].Barh D, Malhotra R, Ravi B, Sindhurani P, Microrna let-7: an emerging next-generation cancer therapeutic, Curr. Oncol 17 (2010) 70–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [311].Yu F, Yao H, Zhu P, Zhang X, Pan Q, Gong C, Huang Y, Hu X, Su F, Lieberman J, Song E, let-7 regulates self renewal and tumorigenicity of breast cancer cells, Cell 131 (2007) 1109–1123. [DOI] [PubMed] [Google Scholar]
- [312].Zhao Y, Deng C, Wang J, Xiao J, Gatalica Z, Recker RR, Xiao GG, Let-7 family miRNAs regulate estrogen receptor alpha signaling in estrogen receptor positive breast cancer, Breast Cancer Res. Treat 127 (1) (2010) 69–80. [DOI] [PubMed] [Google Scholar]
- [313].Pang RT, Leung CO, Ye TM, Liu W, Chiu PC, Lam KK, Lee KF, Yeung WS, MicroRNA-34a suppresses invasion through downregulation of Notch1 and Jagged1 in cervical carcinoma and choriocarcinoma cells, Carcinogenesis 31 (2010) 1037–1044. [DOI] [PubMed] [Google Scholar]
- [314].Kato M, Paranjape T, Muller RU, Nallur S, Gillespie E, Keane K, Esquela-Kerscher A, Weidhaas JB, Slack FJ, The mir-34 microRNA is required for the DNA damage response in vivo in C. elegans and in vitro in human breast cancer cells, Oncogene 28 (2009) 2419–2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [315].O’Day E, Lal A, MicroRNAs and their target gene networks in breast cancer, Breast Cancer Res. 12 (2010) 201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [316].Valastyan S, Weinberg RA, MicroRNAs: crucial multi-tasking components in the complex circuitry of tumor metastasis, Cell Cycle 8 (2009) 3506–3512. [DOI] [PubMed] [Google Scholar]
- [317].Hurteau GJ, Carlson JA, Spivack SD, Brock GJ, Overexpression of the microRNA hsa-miR-200c leads to reduced expression of transcription factor 8 and increased expression of E-cadherin, Cancer Res. 67 (2007) 7972–7976. [DOI] [PubMed] [Google Scholar]
- [318].Bracken CP, Gregory PA, Kolesnikoff N, Bert AG, Wang J, Shannon MF, Goodall GJ, A double-negative feedback loop between ZEB1-SIP1 and the microRNA-200 family regulates epithelial–mesenchymal transition, Cancer Res. 68 (2008) 7846–7854. [DOI] [PubMed] [Google Scholar]
- [319].Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y, Goodall GJ, The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1, Nat. Cell Biol 10 (2008) 593–601. [DOI] [PubMed] [Google Scholar]
- [320].Shimono Y, Zabala M, Cho RW, Lobo N, Dalerba P, Qian D, Diehn M, Liu H, Panula SP, Chiao E, Dirbas FM, Somlo G, Pera RA, Lao K, Clarke MF, Downregulation of miRNA-200c links breast cancer stem cells with normal stem cells, Cell 138 (2009) 592–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [321].Wellner U,Schubert J, Burk UC, Schmalhofer O, Zhu F, Sonntag A, Waldvogel B, Vannier C, Darling D, zur Hausen A, Brunton VG, Morton J, Sansom O, Schuler J, Stemmler MP, Herzberger C, Hopt U, Keck T, Brabletz S, Brabletz T, The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs, Nat. Cell Biol 11 (2009) 1487–1495. [DOI] [PubMed] [Google Scholar]
- [322].Vallejo DM, Caparros E, Dominguez M, Targeting Notch signalling by the conserved miR-8/200 microRNA family in development and cancer cells, EMBO J. 30 (2011) 756–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [323].Imbimbo BP, Therapeutic potential of gamma-secretase inhibitors and modulators, Curr. Top. Med. Chem 8 (2008) 54–61. [DOI] [PubMed] [Google Scholar]
- [324].Shih Ie M, Wang TL, Notch signaling, gamma-secretase inhibitors, and cancer therapy, Cancer Res. 67 (2007) 1879–1882. [DOI] [PubMed] [Google Scholar]
- [325].Bergmans BA, De Strooper B, gamma-secretases: from cell biology to therapeutic strategies, Lancet Neurol. 9 (2010) 215–226. [DOI] [PubMed] [Google Scholar]
- [326].Rasul S, Balasubramanian R, Filipovic A, Slade MJ, Yague E, Coombes RC, Inhibition of gamma-secretase induces G2/M arrest and triggers apoptosis in breast cancer cells, Br. J. Cancer 100 (2009) 1879–1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [327].Pece S, Confalonieri S, Romano PR, Di Fiore PP, NUMB-ing down cancer by more than just a NOTCH, Biochim. Biophys. Acta 1815 (2011) 26–43. [DOI] [PubMed] [Google Scholar]
- [328].Osipo C, Patel P, Rizzo P, Clementz AG, Hao L, Golde TE, Miele L, ErbB-2 inhibition activates Notch-1 and sensitizes breast cancer cells to a gamma-secretase inhibitor, Oncogene 27 (2008) 5019–5032. [DOI] [PubMed] [Google Scholar]
- [329].Jaspers JE, Rottenberg S, Jonkers J, Therapeutic options for triple-negative breast cancers with defective homologous recombination, Biochim. Biophys. Acta 1796 (2009) 266–280. [DOI] [PubMed] [Google Scholar]
- [330].Chen JQ, Russo J, ERalpha-negative and triple negative breast cancer: molecular features and potential therapeutic approaches, Biochim. Biophys. Acta 1796 (2009) 162–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [331].Linderholm BK, Hellborg H, Johansson U, Elmberger G, Skoog L, Lehtio J, Lewensohn R, Significantly higher levels of vascular endothelial growth factor (VEGF) and shorter survival times for patients with primary operable triple-negative breast cancer, Ann. Oncol 20 (2009) 1639–1646. [DOI] [PubMed] [Google Scholar]
- [332].Fillmore CM, Kuperwasser C, Human breast cancer cell lines contain stem-like cells that self-renew, give rise to phenotypically diverse progeny and survive chemotherapy, Breast Cancer Res. 10 (2008) R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [333].Berrada N, Delaloge S, Andre F, Treatment of triple-negative metastatic breast cancer: toward individualized targeted treatments or chemosensitization? Ann. Oncol 21 (Suppl. 7) (2010) vii30–vii35. [DOI] [PubMed] [Google Scholar]
- [334].Zhu H, Bhaijee F, Ishaq N, Pepper DJ, Backus K, Brown AS, Zhou X, Miele L, Correlation of Notch1, pAKT and nuclear NF-kappaB expression in triple negative breast cancer, Am. J. Cancer Res 3 (2013) 230–239. [PMC free article] [PubMed] [Google Scholar]
- [335].Zhang CC, Pavlicek A, Zhang Q, Lira ME, Painter CL, Yan Z, Zheng X, Lee NV, Ozeck M, Qiu M, Zong Q, Lappin PB, Wong A, Rejto PA, Smeal T, Christensen JG, Biomarker and pharmacologic evaluation of the gamma-secretase inhibitor PF-03084014 in breast cancer models, Clin. Cancer Res 18 (2012) 5008–5019. [DOI] [PubMed] [Google Scholar]
- [336].Azzam DJ, Zhao D, Sun J, Minn AJ, Ranganathan P, Drews-Elger K, Han X, Picon-Ruiz M, Gilbert CA, Wander SA, Capobianco AJ, El-Ashry D, Slingerland JM, Triple negative breast cancer initiating cell subsets differ in functional and molecular characteristics and in gamma-secretase inhibitor drug responses, EMBO Mol. Med 5 (2013) 1502–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [337].Beel AJ, Sanders CR, Substrate specificity of gamma-secretase and other intramembrane proteases, Cell. Mol. Life Sci 65 (2008) 1311–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [338].van Es JH, van Gijn ME, Riccio O, van den Born M, Vooijs M, Begthel H, Cozijnsen M, Robine S, Winton DJ, Radtke F, Clevers H, Notch/gamma-secretase inhibition turns proliferative cells in intestinal crypts and adenomas into goblet cells, Nature 435 (2005) 959–963. [DOI] [PubMed] [Google Scholar]
- [339].Riccio O, van Gijn ME, Bezdek AC, Pellegrinet L, van Es JH, Zimber-Strobl U, Strobl LJ, Honjo T, Clevers H, Radtke F, Loss of intestinal crypt progenitor cells owing to inactivation of both Notch1 and Notch2 is accompanied by derepression of CDK inhibitors p27Kip1 and p57Kip2, EMBO Rep. 9 (2008) 377–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [340].Wu Y, Cain-Hom C, Choy L, Hagenbeek TJ, de Leon GP, Chen Y, Finkle D, Venook R, Wu X, Ridgway J, Schahin-Reed D, Dow GJ, Shelton A, Stawicki S, Watts RJ, Zhang J, Choy R, Howard P, Kadyk L, Yan M, Zha J, Callahan CA, Hymowitz SG, Siebel CW, Therapeutic antibody targeting of individual Notch receptors, Nature 464 (2010) 1052–1057. [DOI] [PubMed] [Google Scholar]
- [341].Barten DM,Meredith JE Jr., Zaczek R, Houston JG, Albright CF, Gamma-secretase inhibitors for Alzheimer’s disease: balancing efficacy and toxicity, Drugs R D 7 (2006) 87–97. [DOI] [PubMed] [Google Scholar]
- [342].Sade H, Krishna S, Sarin A, The anti-apoptotic effect of Notch-1 requires p56lck-dependent, Akt/PKB-mediated signaling in T cells, J. Biol. Chem 279 (2004) 2937–2944. [DOI] [PubMed] [Google Scholar]
- [343].Knupfer H, Preiss R, Significance of interleukin-6 (IL-6) in breast cancer (review), Breast Cancer Res. Treat 102 (2007) 129–135. [DOI] [PubMed] [Google Scholar]
- [344].Graziani I, Eliasz S, De Marco MA, Chen Y, Pass HI, De May RM, Strack PR, Miele L, Bocchetta M, Opposite effects of Notch-1 and Notch-2 on mesothelioma cell survival under hypoxia are exerted through the Akt pathway, Cancer Res. 68 (2008) 9678–9685. [DOI] [PubMed] [Google Scholar]


