Abstract
Robertsonian translocations (RTs) result from fusion of 2 acrocentric chromosomes (e.g., 13, 14, 15, 21, 22) and consequential losses of segments of the p arms containing 47S rDNA clusters and transcription factor binding sites. Depending on the position of the breakpoints, the size of these losses vary considerably between types of RTs. The prevalence of RTs in the general population is estimated to be around 1 per 800 individuals, making RTs the most common chromosomal rearrangement in healthy individuals. Based on their prevalence, RTs are classified as “common,” rob(13;14) and rob(14;21), or “rare” (the 8 remaining nonhomologous combinations). Carriers of RTs are at an increased risk for offspring with chromosomal imbalances or with uniparental disomy. RTs are generally regarded as phenotypically neutral, although, due to RTs formation, 2 of the 10 ribosomal rDNA gene clusters, several long noncoding RNAs, and in the case of RTs involving chromosome 21, several mRNA encoding genes are lost. Nevertheless, recent evidence indicates that RTs may have a significant phenotypic impact. In particular, rob(13;14) carriers have a significantly elevated risk for breast cancer. While RTs are easily spotted by routine karyotyping, they may go unnoticed if only array-CGH and NextGen sequencing methods are applied. This review first discusses possible molecular mechanisms underlying the particularly high rates of RT formation and their incidence in the general population, and second, likely causes for the elevated cancer risk of some RTs will be examined.
Keywords: Breast cancer, Interchromosomal effect, Nucleolus organizer region, Robertsonian translocation, Single molecule optical mapping
Introduction
Robertsonian translocations (RTs) involve fusion of 2 acrocentric chromosomes (i.e., 13, 14, 15, 21, 22) during which parts of their short arms will be lost in subsequent cell divisions. They represent the most common chromosome rearrangement and occur at a rate of approximately 1 in 800 in the general population [Hamerton et al., 1975; Nielson and Wohlert, 1991; Hochstenbach et al., 2009]. RTs occur in 2 classes and arise predominantly during female meiosis [Page et al., 1996; Page and Shaffer, 1997; Bandyopadhyay et al., 2002]. The common (Class I) RTs, rob(13;14) and rob(14;21), result from recombination between microsatellite arrays, while the rare, Class II, RTs (all others) show heterogeneous breakpoints and consequently variable losses [Page et al., 1996; Bandyopadhyay et al., 2002]. Textbook knowledge assumes that the loss of the short arms of 2 of the acrocentric chromosomes will be phenotypically neutral, notwithstanding that each short arm carries variable numbers of 47S rDNA gene clusters, associated transcription factor binding sites, mRNA-encoding and long noncoding RNA genes [Lyle et al., 2007; Gardner and Amor, 2018; van Sluis et al., 2019]. Thus, carriers of RTs are presumed to show no phenotypic abnormalities, but to be at increased risk for miscarriages, infertility, uniparental disomy, and aneuploid offspring because of production of unbalanced gametes [Yamazawa et al., 2010; Bertini et al., 2017; Gardner and Amor, 2018]. Hence, other mechanisms by which structural genome variation may exert a phenotypic impact are thus neglected [Poot and Haaf, 2015; Lupiáñez et al., 2016]. A recent cohort study shows that certain types of RTs predispose to elevated risks for breast cancer or hematologic malignancies in their carriers [Schoemaker et al., 2019]. Here, we review the evidence for these phenotypic effects and discuss the potential pathogenic mechanisms of RTs.
Properties of Robertsonian Translocations
RTs are defined as the product of a fusion of the long arms (the q arms) of a pair of 2 of the same or of 2 different acrocentric chromosomes; for instance, an RT of 2 chromosomes 13 results in a rob(13;13), while a fusion of the q arm of a chromosome 13 with one from chromosome 14 produces a rob(13;14). Since in all but one case studied, all breaks leading to the formation of an RT occurred in the p arms of the participant chromosomes, proximal to 47S rRNA repeat arrays, they are dicentric chromosomes [Page et al., 1996; Jarmuz-Szymczak et al., 2014]. In 8 out of the 10 types of RTs studied, both centromeres displayed CENP-C immunofluorescence, which suggests that both centromeres were active and that the RT was functionally dicentric [Page and Shaffer, 1998]. Conceivably, the close proximity of 2 functional centromeres on RTs allows them to remain stable [Page and Shaffer, 1998]. The short arms of the affected acrocentric chromosomes are acentric and will consequently be lost during meiosis and mitosis.
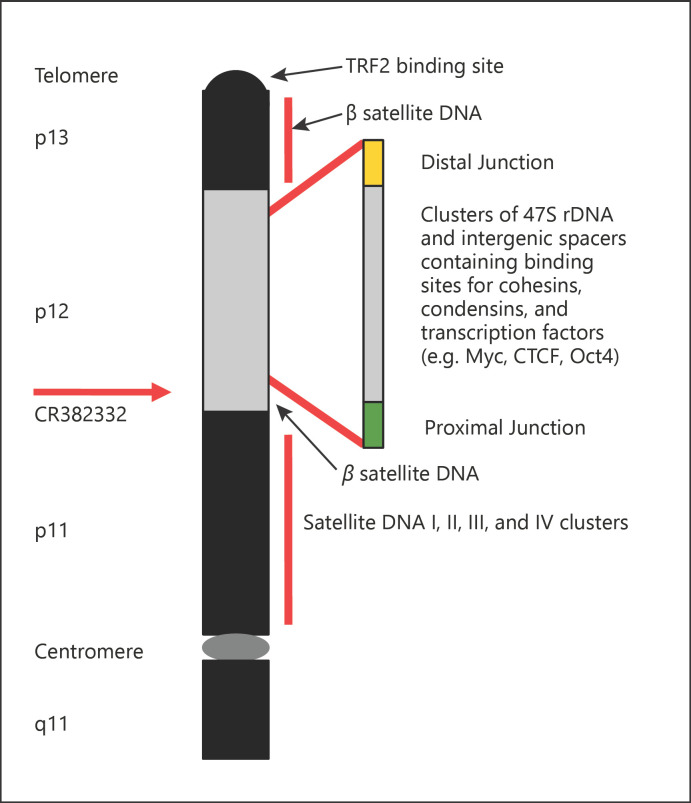
The short arms of the acrocentrtic chromosomes contain satellite DNA I, II, III, and IV clusters (in chromosome band p11), hundreds of transcription factor binding sites (e.g., for CTCF, Oct4, Myc), sites for DNA-binding proteins such as cohesins, condensins, and arrays of an rDNA operon encoding a 47S pre-rRNA (in band p12) as well as β satellite clusters (in bands p12 and p13) (Fig. 1) [McStay and Grummt, 2008; Floutsakou et al., 2013; Jarmuz-Szymczak et al., 2014; McStay, 2016; Mangan et al., 2017; Ramos et al., 2019; van Sluis et al., 2019]. The short arms of the acrocentric chromosomes are all clustered in the nucleolus, in which rDNA transcription and processing of 47S pre-RNA takes place [McStay and Grummt, 2008; McStay, 2016; Picart-Picolo et al., 2019; Wiland et al., 2019]. While the 47S rDNA arrays located on the short arms clustering in the nucleolus are transcriptionally active, this clustering is independent of the number of rDNA blocks [van Sluis et al., 2020]. The rDNA operons are transcribed by RNA polymerase I [Yuan et al., 2007]. The 47S pre-rRNA is posttranscriptionally processed into the 18S rRNA of the small ribosomal subunit and the 5.8 and 28S rRNAs of the large ribosomal subunit [Grozdanov et al., 2003; Stults et al., 2008]. The 47S rDNA clusters range in length from 50 kb to >6 Mb, are almost completely heterozygous for each individual, and manifest as unique rDNA electrophoretic fingerprints in pulsed-field gel electrophoresis after analyses with rare-cutting restriction enzymes [Stults et al., 2008]. The organization of 47S rDNA clusters and the distal junction sequence blocks is rather similar on all acrocentric chromosomes, but the proximal HERV-K and CER satellite blocks diverge considerably [van Sluis et al., 2019]. An array of tens to hundreds of copies of 5S rRNA, the third rRNA component of the large subunit of the ribosome, is located on chromosome 1 [Gibbons et al., 2015]. Loss of clusters of 47S pre-rRNA-encoding genes is balanced by a concerted copy number variation of the 5S RNA-encoding genes [Gibbons et al., 2015].
Fig. 1.
Schematic representation of the p arm of a generic acrocentric chromosome with tentative localization of the satellite DNA clusters (p11), the 47S rDNA clusters, binding sites for cohesins, condensins, transcription factors (e.g., CTCF, Oct4, Myc) (p12), the β satellite clusters (p12 and p13), and of the TRF2 protein binding site (telomere). The chromosomal regions are depicted on the left hand side; the red arrow indicates the position of the breakpoint and the BAC clone that is either lost or disrupted in the Class I RTs. This figure is based on McStay [2016], Floutsakou et al. [2013], Jarmuz-Szymczak et al. [2014], van Sluis et al. [2019], and Ramos et al. [2019]. For further details, see text.
Breakpoint analyses by FISH revealed 2 classes of RTs [Page et al., 1996]. By FISH the common (Class I) RTs, rob(13;14) and rob(14; 21), consistently show breakpoints within the satellite I and satellite III arrays in the p11 regions of chromosomes 13, 14, and 21 [Page et al., 1996]. Thus, in all Class I RTs, the rDNA clusters and the nucleolus organizer regions (NORs) are lost [Page et al., 1996]. With FISH using 8 BAC clones, the location of the breakpoints of RTs on chromosome 21 was further refined to locate in or close to BAC clone CR382332 [Jarmuz-Szymczak et al., 2014]. It should be noted that these BAC clones also encompassed mRNA-encoding genes, some of which may be lost after RT-formation [Lyle et al., 2007]. By reverse-transcriptase-PCR loci identical to the 21pGM15 gene were found to be expressed from human chromosomes 13, 15, 21, and 22 in monochromosomal somatic cell hybrids [Lyle et al., 2007].
The 7 out of the 8 rarer (Class II) RTs being investigated showed highly variable breakpoints, which were not even consistent within a particular type of RT [Page et al., 1996]. For instance, 5 cases with rob(14;15) showed 4 different breakpoints on chromosome 15. Two out of the 3 rob(14;22) studied were dicentric, and 1 was monocentric.
Loss of clusters of 47S pre-rRNA-encoding genes causes hypersensitivity to DNA double-strand breaks induced by bleomycin, ionizing radiation, methyl methanesulfonate, or hydroxyurea [Xu et al., 2017]. Conceivably, carriers of the RTs, in which the 47S rDNA clusters were lost, may be more sensitive to DNA double-strand breaks than noncarriers or those with RTs in which the 47S rDNA clusters were retained. The rDNA genes are in general arranged as transcribed regions interrupted by intervening sequences [Caburet et al., 2005; Kim et al., 2018]. Up to one third of the 47S rDNA gene clusters are rearranged as, presumably nonfunctional, palindromic structures. The rate of 47S rDNA gene cluster rearrangement varies highly among individuals, but is independent of the age of the individuals, and clearly elevated in primary fibroblast cultures, SV40-transformed fibroblasts and lymphoblastoid cell lines from patients with Werner syndrome [Caburet et al., 2005]. This chromosomal instability disorder is characterized by a variegated translocation mosaicism, spontaneous deletion formation, and hypersensitivity to DNA double-strand break inducing drugs [Hoehn et al., 1975; Salk et al., 1981; Poot et al., 2001, 2004; Yang et al., 2002; Dhillon et al., 2007]. These cellular phenotypes reflect defective DNA repair by chromosomal recombination and lack of resolution of Holliday junctions due to loss of the 3′→5′ helicase and 3′→5′ exonuclease activities encoded by the WRN gene [Shen and Loeb, 2000; Yang et al., 2002; Dhillon et al., 2007]. In contrast to the strongly elevated rates of palindrome formation of 47S rDNA gene clusters and the hypersensitivity of Werner syndrome cells to bleomycin and hydroxyurea, frequencies of RTs were not increased in cells of Werner syndrome patients [Salk et al., 1981; Poot et al., 2001, 2004; Melcher et al., 2009; Shimamoto et al., 2014].
Yet, the pseudodiploid male fibrosarcoma cell line HT1080, the highly aneuploid female cervical carcinoma cell line HeLa, and the female bone osteosarcoma epithelial cell line U2OS all carry RTs [van Sluis et al., 2020]. HeLa cells are defective for a TP53-encoded 3′→5′ exonuclease and U2OS cells are defective for the alpha thalassemia/mental retardation X-linked (ATRX) gene. Although these observations point toward a role for DNA damage, the precise nature of the cell cycle/DNA damage response pathway(s) protecting against RT-formation is hitherto unknown.
In multigenerational human families, the rDNA clusters are transmitted according to mendelian rules, yet they prove to be subject to meiotic rearrangement at a frequency >10% per cluster, per meiosis [Stults et al., 2008]. The rDNA clusters are flanked by proximal and telomeric sequence blocks that are roughly 95 and 99% identical between all 5 acrocentric chromosomes for the proximal and the telomeric sequences, respectively [Floutsakou et al., 2013]. The proximal sequences are almost entirely segmentally duplicated, similar to the regions bordering the centromeres. In contrast, the distal sequence is predominantly unique to the acrocentric short arms and contains a very large inverted repeat [Floutsakou et al., 2013]. This high intrinsic recombinational instability makes rDNA clusters the most plastic segments of the human genome.
In the genomes of healthy individuals, the total rDNA copy numbers range between 120 and 670 as measured in lymphocytes from peripheral blood [Parks et al., 2018; Porokhovnik and Lyapunova, 2019]. In addition, the number of rDNA clusters and distal junction blocks vary considerably between acrocentric chromosomes and between individuals [van Sluis et al., 2019, 2020]. This wide range of rDNA copy numbers found in both mouse and healthy humans represent differences of approximately 66 million nucleotides of rDNA between individuals at the extremes. In agreement with the rDNA residing only on autosomes, rDNA copy numbers do not stratify by sex. Together with the observed population stratification, this is consistent with a high rate of meiotic rDNA recombination. In a comparison of blood samples of healthy individuals of advanced age (>72 years) with younger subjects, no difference in copy number of rDNA clusters was observed (272–541 in older vs. 200–711 copies in younger subjects) [Malinovskaya et al., 2018]. Surprisingly, in primary fibroblasts the number of hypermethylated transcribed rDNA copies declined during replicative senescence in culture. This is in stark contrast with the increase during aging in global genomic DNA methylation in peripheral blood lymphocytes of healthy individuals and patients with Werner syndrome [Maierhofer et al., 2017; Horvath and Raj, 2018; Wang and Lemos, 2019]. In the latter syndrome, genes linked to transcription factor activity and sequence-specific DNA binding to promoters transcribed by RNA polymerase II were affected by differential methylation, which suggests that dysregulation of mRNA transcription may contribute to this syndrome [Maierhofer et al., 2019]. In contrast, RNA polymerase I rDNA transcription rates are not affected by differential methylation or loss of rDNA copies during aging [Malinovskaya et al., 2018].
How Do Robertsonian Translocations Arise?
From the above, it is evident that the p arms of the acrocentic chromosomes exhibit a peculiar high degree of genomic instability. A probable mechanism for this instability and consequent RT formation may be a block in premeiotic replication due to DNA double-strand breaks or stalled replication forks, which then interfere with meiotic recombination [Borde et al., 2000; Murakami and Nurse, 2001]. Repeat-rich regions, such as the satellite III regions of the short arms of the acrocentric chromosomes, may subsequently be repaired such that chromosomal rearrangements, including RTs, result [Richardson et al., 1998; Warmerdam and Wolthuis, 2019]. Particularly high rates of palindrome formation within the 47S rDNA clusters were found in cells defective for canonical nonhomologous end joining (NHEJ) of DNA double-strand breaks (Werner syndrome), which suggests that this pathway likely protects against RT formation [Bandyopadhyay et al., 2002; Caburet et al., 2005; Hustedt and Durocher, 2016]. If this is true, RTs would arise as a specific manifestation of the variegated translocation mosacism of this quintessential DNA double-strand break repair disorder. In cells from Werner syndrome patients, the most extensively karyotyped chromosomal instability disorder, frequencies of RTs were not increased [Salk et al., 1981; Melcher et al., 2009; Shimamoto et al., 2014]. Although this is not conclusive proof that loss of canonical NHEJ of DNA double-strand breaks is not involved in the formation of RTs, it does not support this potential mechanism either.
Actively transcribed genes, such as rDNA clusters, pose a risk for genomic stability [Ide et al., 2010]. Since rDNA clusters are very actively transcribed during oogenesis, but not during spermatogenesis, it is more likely that RTs arise in the maternal rather than in the paternal germline [Page et al., 1996]. The genomes of yeasts and the p arms of the human acrocentric chromosomes contain many untranscribed rDNA clusters, which when they are lost, render the cells more sensitive to DNA damage induced by mutagens [Ide et al., 2010]. This DNA damage sensitivity is dependent upon rDNA transcriptional activity, which interferes with cohesion between rDNA loci of sister chromatids. The extra untranscribed rDNA copies promote condensin association and sister-chromatid cohesion, thereby facilitating recombinational repair. Actively transcribed rDNA clusters may be subject to recombination due to a collision between the rDNA transcription machinery and the DNA replication fork [Takeuchi et al., 2003]. In yeast, the availability of rDNA clusters to DNA recombination upon DNA double-strand breaks after replication fork stalling is curtailed by the NAD+-dependent histone deacetylase Sir2 (silent information regulator 2; sirtuin 7 in humans) [Kobayashi et al., 2004]. Since sirtuin 7 activity is regulated by NAD+ levels, it links rDNA stability, nucleolar activity and rDNA transcription with the energy status of the cell. It is presently not known whether sirtuin 7 is involved in the prevention of RT formation.
As was demonstrated by differential psoralen-UV crosslinking accessibility assays, only a small fraction of the rRNA genes in a human cell are transcriptionally active [Conconi et al., 2002]. In mammals, methylation of CpG dinucleotides (meCpG) within the rRNA genes proved not to be the sole mechanism of rDNA silencing [Grummt and Pikaard, 2003]. In mammalian cells, rRNA genes occur in 1 of at least 4 different populations: first, silenced via CpG methylation and probably constitutively heterochromatic; second, inactive and nucleosomal but not silenced via CpG methylation; third, transcriptionally inactive but non-nucleosomal and poised for activity, and fourth, transcriptionally active [Santoro et al., 2002; Moss et al., 2019]. In Arabidopsis thaliana, active rRNA genes are present within sorted nucleoli, whereas silenced rRNA genes exist in the nucleoplasm outside of the nucleoli, as shown by fluorescence-activated sorting [Pontvianne et al., 2013]. In cells with mutated DNA methyltransferase (met1), histone deacetylase (hdac6), or chromatin assembly (caf1), in which rDNA transcription was silenced, this nucleoplasmic-partitioning was abrogated. Bisulfite sequencing indicated that active nucleolar rRNA genes are nearly completely demethylated at their promoter CpGs, whereas silenced genes are nearly fully methylated [Pontvianne et al., 2013]. It is currently not known whether formation of RTs is preceded by demethylation of the promoter regions of rDNA clusters or altered transcription levels and potentially recombination between the p arms of acrocentric chromosomes. Similar experiments have not yet been performed with human cells.
To explain the consistent breakpoints within the satellite I and satellite III arrays in the p11 regions of chromosomes 13, 14, and 21 of the Class I RTs, an alternative hypothesis has been proposed [Shaffer et al., 1996]. During the dictyotene arrest of oogenesis the satellite arrays come in close contact to each other for a prolonged period of time. The size of these arrays is sufficient to allow for recombination between these homologous sequences on the nonhomologous chromosomes by nonallelic homologous recombination (NAHR) [Shaffer and Lupski, 2000; Bandyopadhyay et al., 2002]. Since NAHR involves blocks of repetitive sequences, this process will generate chromosome exchanges with consistent localization of breakpoints in agreement with the breakpoint patterns of Class I RTs [Page et al., 1996; Bandyopadhyay et al., 2002].
In contrast, the 8 rarer (Class II) RTs showed a high variability in breakpoint locations, parental origin, and timing of formation [Page et al., 1996; Bandyopadhyay et al., 2002]. Among the 5 cases of rob(14;15), 4 different breakpoints on chromosome 15p were found, which makes this RT the most variable with regard to breakpoint location [Page et al., 1996]. Of the rob(14;15), 1 was maternal, 1 paternal, and 1 maternal and paternal in origin [Bandyopadhyay et al., 2002]. Therefore, the latter case must have arisen postzygotically, while all others are consistent with RT formation during meoisis. The high variability among the rare RTs suggests that they arose by a more “random” process than their Class I counterparts [Page et al., 1996; Bandyopadhyay et al., 2002]. A mechanism of formation of RTs with “random” breakpoints may involve telomere stress [Stimpson et al., 2014]. The p arms of acrocentric chromosomes are generally located in the nucleolus and their telomeres coated by the telomere-binding protein TRF2. RNAi knockdown of TRF2 or introduction of an inducible dominant-negative form of TRF2 causes disruption of nucleoli, chromatin decondensation, and RNA polymerase I-mediated rDNA transcription [Stimpson et al., 2014]. Such telomere stress has also been documented as a mechanism underlying a specific form of chromothripsis [Maciejowski et al., 2015; Maciejowski and de Lange, 2017]. Upon damage to telomeric repeat sequences, 2 chromosomes may undergo end-to-end fusion [Maciejowski et al., 2015]. The resulting double chromosome will lag during anaphase [Maciejowski et al., 2015]. Subsequently, the TREX1-encoded 3′→5′exonuclease will resect the anaphase bridge, and the chromosomes may undergo several breakage-fusion-bridge cycles [Maciejowski et al., 2015]. After each round of breakage, the chromosome ends will be repaired by either canonical or alternative NHEJ [Maciejowski and de Lange, 2017]. The latter pathway will introduce extensive deletions at the breakpoints [Poot, 2018]. This form of chromothripsis, which affects a single arm of each of the 2 involved chromosomes, is consistent with the highly variable breakpoints found in Class II RTs. In summary, Class I RTs likely arise through NAHR between blocks of repetitive sequences between the short arms of 2 acrocentric chromosomes, which leads to loss of the distal chromosome segments. In contrast, Class II RTs may result from a form of chromothripsis that likely involves a telomere crisis. Thus, the breakpoints of the remaining RTs are probably unique, and no generalizations regarding lost chromosomal segments and possibly other rearrangements can be made.
Detection and Incidence of Both Class I and Class II RTs
Even before chromosome banding techniques became available, RTs were detected reliably. In 1975, the first study of the prevalence of chromosome abnormalities in newborns was published [Hamerton et al., 1975]. In 13 out of 14,069 (0.092%) consecutive newborns, an RT was discovered. Combining these results with 5 other comparable studies, comprising a total of 46,150 newborn infants, indicates a frequency of RTs close to 0.1% in live-born babies. Subsequent studies have largely corroborated the work of Hamerton et al. [1975] by yielding estimates of 1 in 812 in a Danish cohort of 34,910 newborns and 1 in 826 in a Dutch cohort of 36,325 cases referred for developmental delay and/or malformations (Table 1) [Nielsen and Wohlert, 1991; Hochstenbach et al., 2009].
Table 1.
Chromosomal spectrum of RTs found in diagnostic screening
| Warburton [1991] |
Zhao et al. [2015] |
Schoemaker et al. [2019] |
||||
|---|---|---|---|---|---|---|
| Number of cases | % | Number of cases | % | Number of cases | % | |
| 13;13 | 0 | 0 | 5 | 0.9 | 2 | 0.1 |
| 13;14 | 51 | 69.9 | 344 | 59.0 | 1,248 | 62.8 |
| 13;15 | 2 | 2.7 | 24 | 4.1 | 55 | 2.8 |
| 13;21 | 1 | 1.4 | 16 | 2.7 | 20 | 1.0 |
| 13;22 | 0 | 0 | 14 | 2.4 | 20 | 1.0 |
| 14;14 | 0 | 0 | 4 | 0.7 | 5 | 0.3 |
| 14;15 | 3 | 4.1 | 23 | 4.0 | 52 | 2.6 |
| 14;21 | 12 | 16.4 | 86 | 14.8 | 390 | 19.6 |
| 14;22 | 1 | 1.4 | 14 | 2.4 | 57 | 2.9 |
| 15;15 | 1 | 1.4 | 5 | 0.9 | 2 | 0.1 |
| 15;21 | 0 | 0 | 16 | 2.7 | 35 | 1.8 |
| 15;22 | 0 | 0 | 9 | 1.5 | 35 | 1.8 |
| 21;21 | 1 | 1.4 | 6 | 1.0 | 6 | 0.3 |
| 21;22 | 0 | 0 | 14 | 2.4 | 38 | 1.9 |
| 22;22 | 1 | 1.4 | 3 | 0.5 | 10 | 0.5 |
| Other | 0 | 0 | 0 | 0 | 12 | 0.6 |
| Total | 73 | 100.0 | 583 | 100.0 | 1,987 | 100.0 |
In a recent analysis of diagnostic cytogentic screening in the UK, a total of 1,987 individuals with RTs were reported [Schoemaker et al., 2019]. The Class I RTs, rob(13;14) and rob(14;21), made up 62.8 and 19.6% of cases, respectively; the other RTs occurred at less than 2% each (Table 1). With the frequencies of rob(13;14) ranging between 59 and 69% and of rob(14;21) ranging between 14.8 and 19.6% in 2 other studies, the data by Schoemaker et al. [2019] largely corroborated previous studies [Warburton, 1991; Zhao et al., 2015].
While RTs can be unequivocally distinguished by classical karyotyping, genome-wide copy number screening and whole-genome sequencing techniques do not reliably detect this most common type of chromosome rearrangement [Hochstenbach et al., 2009, 2019]. Because of the highly repetitive nature of the sequence blocks on the short arms of the acrocentric chromosomes and the very similar sequence organisation of the p arms of all acrocentric chromosomes, those are refractory to current sequencing methods [McStay, 2016]. Consequently, no markers are available for oligonucleotide or SNP arrays, which are commonly used for genome-wide copy number screening [Miller et al., 2010]. For a NextGen-based diagnostic method, sequence reads need to be mapped onto a reference genome. Since the currently available human reference genome sequence does not cover the short arms of the acrocentric chromosomes, these are uncharted territory for NextGen-based approaches [Hochstenbach et al., 2019].
Nanochannel-based single molecule optical mapping allows to resolve structural genome variation on a megabase-scale [Lam et al., 2012]. In contrast to paired-end and mate-pair sequencing, this approach allows to bridge repeat-rich regions such as segmental duplications, the Duchenne muscular dystrophy gene, and D4Z4 arrays in patients with facioscapulohumeral muscular dystrophy 1 [Kloosterman et al., 2011; Barseghyan et al., 2017; Zhang et al., 2019; Zheng et al., 2020]. Structural variations detected by single molecule optical mapping are in good accordance with those detected by other state-of-the-art methods [Chaisson et al., 2019; Neveling et al., 2020]. Yet, this method can only map structural variations within genomic regions covered by the human reference genome sequence. Thus, the centromeres, some pericentromeric regions, and the short arms of the acrocentic chromosomes are beyond reach for single molecule optical mapping [Basreghyan et al., 2017].
Evidence for Phenotypic Effects
Since RTs are the product of a chromosomal rearrangement and may themselves provoke further chromosomal instability (see below), the life expectancy and the cancer incidence of RT carriers has been investigated. To do so, a cohort of 1,987 RT carriers diagnosed in Great Britain were followed over an average of 24.1 years [Schoemaker et al., 2019]. Overall mortality was higher for carriers diagnosed before age 15 years (standardized mortality ratio (SMR) = 2.00, 95% confidence interval (CI): 1.09, 3.35), similar for those diagnosed aged 15–44 years (SMR = 1.06, 95% CI: 0.86, 1.28), and lower for those diagnosed aged 45–84 years (SMR = 0.81, 95% CI: 0.68, 0.95). The increased mortality rates for RTs carriers diagnosed during childhood possibly reflects age-specific reasons for referral to medical genetic evaluation. The lower SMR for RT carriers above 45 years of age contradicts a role for RTs in ageing-related mortality. This is in stark contrast to the DNA double-strand repair and chromosomal instability disorder Werner syndrome, which characteristically shows premature, segmental aging.
In this cohort, cancer incidence was higher for non-Hodgkin lymphoma (standardized incidence ratio (SIR) = 1.90, 95% CI: 1.01, 3.24) and childhood leukemia (SIR = 14.5, 95% CI: 1.75, 52.2) (Table 2) [Schoemaker et al., 2019]. rob(13;14) carriers showed a higher breast cancer risk (SIR = 1.58, 95% CI: 1.12, 2.15). In case-control studies, the number of cases limits the power to detect statistically significant associations. Therefore, it is not surprising that the rob(13;14) (1,248 cases) showed an elevated rate of individuals with breast cancer, while the rob(14;21) (390 cases) did not. Taken together, these results indicate that RTs confer a greater risk of childhood leukemia and non-Hodgkin lymphoma, and of breast cancer in individuals with a rob(13;14). This does not exclude an elevated risk for any of the rarer Class II RTs. Since these occur at much lower frequencies in the general population, and assuming increases in disease risk due to these rarer RTs to be similar to those found with the Class I RTs, much larger cohorts would be needed to estimate their potential pathogenic impact. In addition, the breakpoints within some types of Class II RTs are heterogeneous, which precludes generalizations regarding their putative phenotypic effects.
Table 2.
Significantly increased risk for malignant disease in carriers of a robertsonian translocation
| Disease type | RT | SIR | 95% CI |
|---|---|---|---|
| Leukemia diagnosed in childhood (age 0–14 years) | rob(13;14) rob(15;21) | 14.5 | 1.75, 52.2 |
| Non-Hodgkin lymphoma | 9 out of 13 cases had rob(13;14) | 1.90 | 1.01, 3.24 |
| Breast cancer | rob(13;14) | 1.58 | 1.12, 2.15 |
Potential Pathogenetic Mechanisms
Searching for potential pathogenic mechanisms of the elevated cancer risk of RTs carriers, we will first consider the possible impact of lost segments of the p arms, and second, the effects of potential intranuclear mislocalization of the q arms of the chromosomes affected by RTs.
During formation of all Class I and some Class II RTs, some rDNA clusters will be lost, which may conceivably affect 47S RNA production and consequently ribosome biogenesis. During RT formation, also clusters of transcription factors such as the CCCTC-binding factor CTCF will be lost. These losses will be discussed with regard to the impact of CTCF on breast cancer [Oh et al., 2017; Kentepozidou et al., 2020].
Furthermore, losses of rDNA clusters may alter chromosomal contacts in interphase nuclei and in particular in nucleoli [McStay, 2016; Picart-Picolo et al., 2019; Wiland et al., 2019]. We will consider whether such altered chromosomal contacts in nuclei with an RT may affect mitotic stability of the nuclei and the recombinational repair of genes related to breast cancer, in particular in carriers of a rob(13;14). Finally, we will discuss a possible interchromosomalsomal effect of RTs [Alfarawati et al., 2012].
Insufficient rRNA Synthesis and Decreased Ribosome Biogenesis
Conceivably, loss of 47S rDNA clusters after formation of RTs may lead to insufficient rRNA production and ribosome formation. In such cases, a ribosomopathy will arise. Ribosomopathies resulting from impaired ribosome biogenesis due to mutations in the ribosomal protein-encoding genes have been described extensively [Narla and Ebert, 2010; Warmerdam and Wolthuis, 2019]. These disorders involve impaired hematopoiesis, for example, macrocytic anemia, neutropenia, hypoplastic anemia, with concomitant skeletal and skin abnormalities. For instance, the autosomal recessive Schwachman-Diamond syndrome, which is due to mutations in the SBDS gene, presents with exocrine pancreatic insufficiency in addition to infections as a result of neutropenia. All ribosomopathies share an elevated risk for malignant disease, such as osteosarcoma and myelodysplastic syndrome (MDS) in Diamond-Blackfan anemia, and MDS and acute myeloid leukemia (AML) in Schwachman-Diamond syndrome.
Contrary to the ribosomopathies, no specific rDNA transcription-related disorders have as yet been described. In contrast to patients with ribosomopathies, carriers with RTs do not show osteosarcoma, MDS or AML. Therefore, an insufficient number of active ribosomes due to loss of rDNA clusters in RT carriers seems highly unlikely.
The CTCF Transcription Factor and Breast Cancer
On the short arms of the acrocentric chromosomes, multiple clusters of transcription factor binding sites reside. The CCCTC-binding transcription factor CTCF is a conserved 11-zinc finger DNA-binding protein that regulates chromosomal architecture [van de Nobelen et al., 2010]. CTCF binding sites constitute approximately 7.5% of the human genome. Between 29 and 37% of all contacts between 47S rDNA clusters and the remainder of the genome overlapped with a CTCF binding site, as was demonstrated by Hi-C sequencing [Yu and Lemos, 2018]. Upstream of the rDNA promoter reside clusters of CTCF binding sites, which on the one hand regulate rDNA expression and on the other hand control 3D chromatin organization via cohesin binding [Pugacheva et al., 2020]. Conceivably, the latter affects the intranuclear localization of the RTs and may interfere with their mitotic pairing with the intact homologues. Thus, losses of CTCF and cohesin binding sites may lead to the nucleolar disruption as observed in cells with RTs [Pontvianne et al., 2013; Wiland et al., 2019].
CTCF has been related to gene expression in breast cancer [Kemp et al., 2014; Oh et al., 2017; Aitken et al., 2018]. Mouse tissues with a hemizygous loss of CTCF exhibit increased variability in genome-wide CpG methylation [Kemp et al., 2014]. This loss of epigenetic stability removes a major barrier to neoplastic progression [Kemp et al., 2014; Aitken et al., 2018]. In addition, loss of CTCF binding sites may interfere with higher order genome organization and epigenetic control, which in turn prevents epithelial to mesenchymal transition of cells in organs such as the breast [Fritz et al., 2019].
An Interchromosomal Effekt of RTs?
In studies of early embryonic development within the context of in vitro fertilization, Alfarawati et al. [2012] observed frequent losses of one or even several of the normal chromosomes in carriers of RTs. The oocytes of these probands show, apart from the RT chromosome, a normal karyotype. During the first mitotic divisions, up to the blastomere stage, the RT chromosome is retained, while a normal chromosome is lost at a significantly elevated rate as compared to early embryos from normal donors or with an inversion or a reciprocal translocation. Similar findings were obtained in a study of sperm nuclei of male carriers of RTs, reciprocal translocations, an inversion and a control cohort [Balasar and Acar, 2020]. In 4 out of 5 RT carriers and in one proband with an inversion, aneuploidy for one of the noninvolved chromosomes was demonstrated by FISH, while all other samples did not show any aneuploidies [Balasar and Acar, 2020].
To explain this finding, termed interchromosomal effect, the authors put forward 2 hypotheses [Alfarawati et al., 2012]. First, the RT chromosome may not be able to pair with their structurally normal homologues counterparts, thus disrupting the positioning and segregation of the other, normal chromosomes during mitosis. Second, in cleavage-stage embryos, a high frequency of double-strand breaks in chromosomes occurs, which are subsequently repaired. Conceivably, mislocalization of RT chromosomes may hamper the chromosome pairing needed for this type of DNA repair. This may in particular be detrimental for damaged chromosomes carrying genes related to cancer. For the case of breast cancer, these may be chromosome 1 (RAD54L on 1p34.1), 2 (CASP8 on 2q33.1 and BARD1 on 2q35), 3 (PIK3CA on 3q26.32), 5 (HMMR on 5q34), 6 (NQO2 on 6p25.2 and ESR1 on 6q25.1–25.2), 8 (RB1CC1 on 8q11.23), 11 (SLC22A18 on 11p15.4 and ATM on 11q22.3), 12 (KRAS on 12p12.1), 13 (BRCA2 on 13q13.1), 14 (AKT1 on 14q32.33 and XRCC3 on 14q32.33), 15 (RAD51 on 15q15.1), 16 (PALB2 on 16p12.2 and CDH1 on 16q22.1), 17 (TP53 on 17p13.1, BRCA1 on 17q21.31, PPM1D on 17q23.2, PHB on 17q21.33, and BRIP1 on 17q23.2), and finally, chromosome 22 (CHEK2 on 22q12.1). Chromosome 17, with BRCA1, PPM1D, PHB, and BRIP1 may be particularly vulnerable to this mechanism and breast cancer a probable outcome for carriers of RTs. Although no studies of the relative number of germline chromosome aberrations, let alone germline RT carriers, in cohorts of breast cancer patients have been published, sorted nuclei of breast tumors from young high-risk patients showed a strikingly high number of chromosome breaks, with chromosome 17 (containing BRCA1, PPM1D, PHB, and BRIP1) being the most vulnerable [Przybytkowski et al., 2014].
Ramifications, Open Questions, and Future Avenues of Research
Several ramifications of our current knowledge regarding losses of variable parts of short arms of the acrocentric chromosomes due to RT formation need to be considered. It is now clear that some RTs may exert a significant risk for somatic disease in their carriers. In particular, the rob(13;14) confers a significantly elevated risk for breast cancer (SIR of 1.58) [Schoemaker et al., 2019]. Since the lifetime risk for breast cancer is roughly 12% for women and the rob(13;14) occurs approximately once per 1,250 screenings of female newborns for this RT, it appears warranted [Ascha et al., 2019; Schoemaker et al., 2019]. In a mouse model for leukemia, the malignant cells have paradoxically lower rDNA copy numbers than normal tissue, despite their higher proliferation rate, and consequentially increased rRNA and protein synthesis level [Xu et al., 2017]. If in breast cancer patients with an RT the cancer cells also have a lower number of rDNA clusters, these may be less vulnerable to bleomycin, methyl methanosulfonate, ionizing radiation, and hydroxyurea than the normal cells in the surrounding tissue. Thus, testing breast cancer patients for the presence of an RT appears indicated before any of these treatments are being considered. Nevertheless, current molecular genome analytical methods in the diagnostic laboratory fail to detect them. Therefore, priority should be given to the development of novel high-throughput methods to detect RTs.
Because of the very similar sequence organisation of the p arms of all acrocentric chromosomes and the highly repetitive nature of the sequence blocks on the short arms of the acrocentric chromosomes, these are refractory to current sequencing methods [McStay, 2016]. Consequently, no markers are available for oligonucleotide or SNP arrays. Yet, a few bacterial artificial chromosome (BAC) probes have been localized to unique positions on the short arm of chromosome 21 [Lyle et al., 2007; Jarmuz-Szymczak et al., 2014]. In addition, BAC clones mapping proximally and telomerically to 47S rDNA clusters have been identified [Floutsakou et al., 2013]. With such BAC probes, either interphase FISH or arrays to perform array-CGH may become an optional molecular cytogenetic test for RTs in medical genetic institutions.
The recently discovered BAC-based probes, which proved useful to analyze the organization of the short arms of human acrocentric chromosomes, may also be helpful to study altered nucleolar topology in cases with RTs [Lyle et al., 2007; Floutsakou et al., 2013; Jarmuz-Szymczak et al., 2014; McStay, 2016; Picart-Picolo et al., 2019; Wiland et al., 2019]. In particular, the effects of losses of CTCF and cohesin binding sites due to RT formation are now amenable to detailed study [Pontvianne et al., 2013; Wiland et al., 2019; Khoury et al., 2020; Pugacheva et al., 2020]. In addition, a plethora of monoclonal antibodies for chromatin modifications allow study of altered epigenetics in carriers of RTs [Bartova et al., 2010]. Recent studies of model organisms have generated both valuable insights into and tools to examine the nucleoli and the NORs of cells from carriers of RTs [McStay, 2016]. Although RT carriers do not experience accelerated aging, it may still be worthwhile to study methylation of rDNA clusters, which have been identified as an “epigenetic aging clock” [Maierhofer et al., 2017; Horvath and Raj, 2018; Schoemaker et al., 2019]. With single cell array-CGH of cells from RT carriers, putative mitotic copy number instability of cells carrying RTs can be studied. Finally, an interchromosomal effect as a possible cause of chromosomal instability in RT carriers and subsequent carcinogenesis merits further study [Alfarawati et al., 2012; Balasar and Acar, 2020]. Given the clinical importance and its prevalence in the general population, RTs are clearly an underappreciated genomic phenomenon in need of “illumination” [McStay, 2016].
Conflict of Interest Statement
The authors have no conflict of interests to declare.
Funding Sources
No funding was obtained for this study.
Author Contributions
Both authors equally took part in design, research and interpretation of the literature, and writing of the paper.
References
- Aitken SJ, Ibarra-Soria X, Kentepozidou E, Flicek P, Feig C, Marioni JC, et al. CTCF maintains regulatory homeostasis of cancer pathways. Genome Biol. 2018;19((1)):106. doi: 10.1186/s13059-018-1484-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfarawati S, Fragouli E, Colls P, Wells D. Embryos of robertsonian translocation carriers exhibit a mitotic interchromosomal effect that enhances genetic instability during early development. PLoS Genet. 2012;8((10)):e1003025. doi: 10.1371/journal.pgen.1003025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ascha MS, Ostrom QT, Wright J, Kumthekar P, Bordeaux JS, Sloan AE, et al. Lifetime Occurrence of Brain Metastases Arising from Lung, Breast, and Skin Cancers in the Elderly: A SEER-Medicare Study. Cancer Epidemiol Biomarkers Prev. 2019;28((5)):917–25. doi: 10.1158/1055-9965.EPI-18-1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasar Ö, Acar H. Investigation of the interchromosomal effects in male carriers with structural chromosomal abnormalities using FISH. Turk J Urol. 2020;46(3):178–85. doi: 10.5152/tud.2020.19255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandyopadhyay R, Heller A, Knox-DuBois C, McCaskill C, Berend SA, Page SL, et al. Parental origin and timing of de novo Robertsonian translocation formation. Am J Hum Genet. 2002;71((6)):1456–62. doi: 10.1086/344662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barseghyan H, Tang W, Wang RT, Almalvez M, Segura E, Bramble MS, et al. Next-generation mapping: a novel approach for detection of pathogenic structural variants with a potential utility in clinical diagnosis. Genome Med. 2017;9((1)):90. doi: 10.1186/s13073-017-0479-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bártová E, Horáková AH, Uhlírová R, Raska I, Galiová G, Orlova D, et al. Structure and epigenetics of nucleoli in comparison with non-nucleolar compartments. J Histochem Cytochem. 2010;58((5)):391–403. doi: 10.1369/jhc.2009.955435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertini V, Fogli A, Bruno R, Azzarà A, Michelucci A, Mattina T, et al. Maternal Uniparental Disomy 14 (Temple Syndrome) as a Result of a Robertsonian Translocation. Mol Syndromol. 2017;8((3)):131–8. doi: 10.1159/000456062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borde V, Goldman AS, Lichten M. Direct coupling between meiotic DNA replication and recombination initiation. Science. 2000;290((5492)):806–9. doi: 10.1126/science.290.5492.806. [DOI] [PubMed] [Google Scholar]
- Caburet S, Conti C, Schurra C, Lebofsky R, Edelstein SJ, Bensimon A. Human ribosomal RNA gene arrays display a broad range of palindromic structures. Genome Res. 2005;15((8)):1079–85. doi: 10.1101/gr.3970105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaisson MJP, Sanders AD, Zhao X, Malhotra A, Porubsky D, Rausch T, et al. Multi-platform discovery of haplotype-resolved structural variation in human genomes. Nature Commun. 2019;10:1784. doi: 10.1038/s41467-018-08148-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conconi A, Bespalov VA, Smerdon MJ. Transcription-coupled repair in RNA polymerase I-transcribed genes of yeast. Proc Natl Acad Sci USA. 2002;99((2)):649–54. doi: 10.1073/pnas.022373099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhillon KK, Sidorova J, Saintigny Y, Poot M, Gollahon K, Rabinovitch PS, et al. Functional role of the Werner syndrome RecQ helicase in human fibroblasts. Aging Cell. 2007;6((1)):53–61. doi: 10.1111/j.1474-9726.2006.00260.x. [DOI] [PubMed] [Google Scholar]
- Floutsakou I, Agrawal S, Nguyen TT, Seoighe C, Ganley AR, McStay B. The shared genomic architecture of human nucleolar organizer regions. Genome Res. 2013;23((12)):2003–12. doi: 10.1101/gr.157941.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritz AJ, Gillis NE, Gerrard DL, Rodriguez PD, Hong D, Rose JT, et al. Higher order genomic organization and epigenetic control maintain cellular identity and prevent breast cancer. Genes Chromosomes Cancer. 2019;58((7)):484–99. doi: 10.1002/gcc.22731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner RJ, Amor DJ. 5 ed. Oxford University Press; 2018. Gardner and Sutherland's Chromosome Abnormalities and Genetic Counseling. [Google Scholar]
- Gibbons JG, Branco AT, Godinho SA, Yu S, Lemos B. Concerted copy number variation balances ribosomal DNA dosage in human and mouse genomes. Proc Natl Acad Sci USA. 2015;112((8)):2485–90. doi: 10.1073/pnas.1416878112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grozdanov P, Georgiev O, Karagyozov L. Complete sequence of the 45-kb mouse ribosomal DNA repeat: analysis of the intergenic spacer. Genomics. 2003;82((6)):637–43. doi: 10.1016/s0888-7543(03)00199-x. [DOI] [PubMed] [Google Scholar]
- Grummt I, Pikaard CS. Epigenetic silencing of RNA polymerase I transcription. Nat Rev Mol Cell Biol. 2003;4((8)):641–9. doi: 10.1038/nrm1171. [DOI] [PubMed] [Google Scholar]
- Hamerton JL, Canning N, Ray M, Smith S. A cytogenetic survey of 14,069 newborn infants. I. Incidence of chromosome abnormalities. Clin Genet. 1975;8((4)):223–43. doi: 10.1111/j.1399-0004.1975.tb01498.x. [DOI] [PubMed] [Google Scholar]
- Hochstenbach R, van Binsbergen E, Engelen J, Nieuwint A, Polstra A, Poddighe P, et al. Array analysis and karyotyping: workflow consequences based on a retrospective study of 36,325 patients with idiopathic developmental delay in the Netherlands. Eur J Med Genet. 2009;52((4)):161–9. doi: 10.1016/j.ejmg.2009.03.015. [DOI] [PubMed] [Google Scholar]
- Hochstenbach R, van Binsbergen E, Schuring-Blom H, Buijs A, Ploos van Amstel HK. A survey of undetected, clinically relevant chromosome abnormalities when replacing postnatal karyotyping by Whole Genome Sequencing. Eur J Med Genet. 2019;62((9)):103543. doi: 10.1016/j.ejmg.2018.09.010. [DOI] [PubMed] [Google Scholar]
- Hoehn H, Bryant EM, Au K, Norwood TH, Boman H, Martin GM. Variegated translocation mosaicism in human skin fibroblast cultures. Cytogenet Cell Genet. 1975;15((5)):282–98. doi: 10.1159/000130526. [DOI] [PubMed] [Google Scholar]
- Horvath S, Raj K. DNA methylation-based biomarkers and the epigenetic clock theory of ageing. Nat Rev Genet. 2018;19((6)):371–84. doi: 10.1038/s41576-018-0004-3. [DOI] [PubMed] [Google Scholar]
- Hustedt N, Durocher D. The control of DNA repair by the cell cycle. Nat Cell Biol. 2016;19((1)):1–9. doi: 10.1038/ncb3452. [DOI] [PubMed] [Google Scholar]
- Ide S, Miyazaki T, Maki H, Kobayashi T. Abundance of ribosomal RNA gene copies maintains genome integrity. Science. 2010;327((5966)):693–6. doi: 10.1126/science.1179044. [DOI] [PubMed] [Google Scholar]
- Jarmuz-Szymczak M, Janiszewska J, Szyfter K, Shaffer LG. Narrowing the localization of the region breakpoint in most frequent Robertsonian translocations. Chromosome Res. 2014;22((4)):517–32. doi: 10.1007/s10577-014-9439-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp CJ, Moore JM, Moser R, Bernard B, Teater M, Smith LE, et al. CTCF haploinsufficiency destabilizes DNA methylation and predisposes to cancer. Cell Rep. 2014;7((4)):1020–9. doi: 10.1016/j.celrep.2014.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kentepozidou E, Aitken SJ, Feig C, Stefflova K, Ibarra-Soria X, Odom DT, et al. Clustered CTCF binding is an evolutionary mechanism to maintain topologically associating domains. Genome Biol. 2020;21((1)):5. doi: 10.1186/s13059-019-1894-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoury A, Achinger-Kawecka J, Bert SA, Smith GC, French HJ, Luu PL, et al. Constitutively bound CTCF sites maintain 3D chromatin architecture and long-range epigenetically regulated domains. Nat Commun. 2020;11((1)):54. doi: 10.1038/s41467-019-13753-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Dilthey AT, Nagaraja R, Lee HS, Koren S, Dudekula D, et al. Variation in human chromosome 21 ribosomal RNA genes characterized by TAR cloning and long-read sequencing. Nucleic Acids Res. 2018;46((13)):6712–25. doi: 10.1093/nar/gky442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloosterman WP, Guryev V, van Roosmalen M, Duran KJ, de Bruijn E, Bakker SC, et al. Chromothripsis as a mechanism driving complex de novo structural rearrangements in the germline. Hum Mol Genet. 2011;20((10)):1916–24. doi: 10.1093/hmg/ddr073. [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Horiuchi T, Tongaonkar P, Vu L, Nomura M. SIR2 regulates recombination between different rDNA repeats, but not recombination within individual rRNA genes in yeast. Cell. 2004;117((4)):441–53. doi: 10.1016/s0092-8674(04)00414-3. [DOI] [PubMed] [Google Scholar]
- Lam ET, Hastie A, Lin C, Ehrlich D, Das SK, Austin MD, et al. Genome mapping on nanochannel arrays for structural variation analysis and sequence assembly. Nat Biotechnol. 2012;30((8)):771–6. doi: 10.1038/nbt.2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupiáñez DG, Spielmann M, Mundlos S. Breaking TADs: How Alterations of Chromatin Domains Result in Disease. Trends Genet. 2016;32((4)):225–37. doi: 10.1016/j.tig.2016.01.003. [DOI] [PubMed] [Google Scholar]
- Lyle R, Prandini P, Osoegawa K, ten Hallers B, Humphray S, Zhu B, et al. Islands of euchromatin-like sequence and expressed polymorphic sequences within the short arm of human chromosome 21. Genome Res. 2007;17((11)):1690–6. doi: 10.1101/gr.6675307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maciejowski J, de Lange T. Telomeres in cancer: tumour suppression and genome instability. Nat Rev Mol Cell Biol. 2017;18((3)):175–86. doi: 10.1038/nrm.2016.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maciejowski J, Li Y, Bosco N, Campbell PJ, de Lange T. Chromothripsis and kataegis induced by telomere crisis. Cell. 2015;163((7)):1641–54. doi: 10.1016/j.cell.2015.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maierhofer A, Flunkert J, Oshima J, Martin GM, Haaf T, Horvath S. Accelerated epigenetic aging in Werner syndrome. Aging (Albany NY) 2017;9((4)):1143–52. doi: 10.18632/aging.101217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maierhofer A, Flunkert J, Oshima J, Martin GM, Poot M, Nanda I, et al. Epigenetic signatures of Werner syndrome occur early in life and are distinct from normal epigenetic aging processes. Aging Cell. 2019;18((5)):e12995. doi: 10.1111/acel.12995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinovskaya EM, Ershova ES, Golimbet VE, Porokhovnik LN, Lyapunova NA, Kutsev SI, et al. Copy Number of Human Ribosomal Genes With Aging: Unchanged Mean, but Narrowed Range and Decreased Variance in Elderly Group. Front Genet. 2018;9:306. doi: 10.3389/fgene.2018.00306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangan H, Gailin MO, McStay B. Integrating the genomic architecture of human nucleolar organizer regions with biophysical properties of nucleoli. FEBS J. 2017;284:3977–985. doi: 10.1111/febs.14108. [DOI] [PubMed] [Google Scholar]
- McStay B. Nucleolar organizer regions: genomic ‘dark matter’ requiring illumination. Genes Dev. 2016;30:1598–610. doi: 10.1101/gad.283838.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McStay B, Grummt I. The epigenetics of rRNA genes: from molecular to chromosome biology. Annu Rev Cell Dev Biol. 2008;24:131–57. doi: 10.1146/annurev.cellbio.24.110707.175259. [DOI] [PubMed] [Google Scholar]
- Melcher R, von Golitschek R, Steinlein C, Schindler D, Neitzel H, Kainer K, et al. Spectral karyotyping of Werner syndrome fibroblast cultures. Cytogenet Cell Genet. 2009;91((1-4)):180–5. doi: 10.1159/000056841. [DOI] [PubMed] [Google Scholar]
- Miller DT, Adam MP, Aradhya S, Biesecker LG, Brothman AR, Carter NP, et al. Consensus statement: chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. Am J Hum Genet. 2010;86((5)):749–64. doi: 10.1016/j.ajhg.2010.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss T, Mars JC, Tremblay MG, Sabourin-Felix M. The chromatin landscape of the ribosomal RNA genes in mouse and human. Chromosome Res. 2019;27((1-2)):31–40. doi: 10.1007/s10577-018-09603-9. [DOI] [PubMed] [Google Scholar]
- Murakami H, Nurse P. Regulation of premeiotic S phase and recombination-related double-strand DNA breaks during meiosis in fission yeast. Nat Genet. 2001;28((3)):290–3. doi: 10.1038/90142. [DOI] [PubMed] [Google Scholar]
- Narla A, Ebert BL. Ribosomopathies: human disorders of ribosome dysfunction. Blood. 2010;115((16)):3196–205. doi: 10.1182/blood-2009-10-178129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neveling K, Mantere T, Vermeulen S, Oorsprong M, van Beek R, et al. Next generation cytogenetics: comprehensive assessment of 48 leukemia genomes by genome imaging. bioRxiv. Preprint. 2020. [DOI] [PMC free article] [PubMed]
- Nielsen J, Wohlert M. Chromosome abnormalities found among 34,910 newborn children: results from a 13-year incidence study in Arhus, Denmark. Hum Genet. 1991;87((1)):81–3. doi: 10.1007/BF01213097. [DOI] [PubMed] [Google Scholar]
- Oh S, Oh C, Yoo KH. Functional roles of CTCF in breast cancer. BMB Rep. 2017;50((9)):445–53. doi: 10.5483/BMBRep.2017.50.9.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page SL, Shaffer LG. Nonhomologous Robertsonian translocations form predominantly during female meiosis. Nat Genet. 1997;15((3)):231–2. doi: 10.1038/ng0397-231. [DOI] [PubMed] [Google Scholar]
- Page SL, Shaffer LG. Chromosome stability is maintained by short intercentromeric distance in functionally dicentric human Robertsonian translocations. Chromosome Res. 1998;6((2)):115–22. doi: 10.1023/a:1009286929145. [DOI] [PubMed] [Google Scholar]
- Page SL, Shin JC, Han JY, Choo KH, Shaffer LG. Breakpoint diversity illustrates distinct mechanisms for Robertsonian translocation formation. Hum Mol Genet. 1996;5((9)):1279–88. doi: 10.1093/hmg/5.9.1279. [DOI] [PubMed] [Google Scholar]
- Parks MM, Kurylo CM, Dass RA, Bojmar L, Lyden D, Vincent CT, et al. Variant ribosomal RNA alleles are conserved and exhibit tissue-specific expression. Sci Adv. 2018;4((2)):eaao0665. doi: 10.1126/sciadv.aao0665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picart-Picolo A, Picault N, Pontvianne F. Ribosomal RNA genes shape chromatin domains associating with the nucleolus. Nucleus. 2019;10((1)):67–72. doi: 10.1080/19491034.2019.1591106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontvianne F, Blevins T, Chandrasekhara C, Mozgová I, Hassel C, Pontes OM, et al. Subnuclear partitioning of rRNA genes between the nucleolus and nucleoplasm reflects alternative epiallelic states. Genes Dev. 2013;27((14)):1545–50. doi: 10.1101/gad.221648.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poot M, Haaf T. Mechanisms of Origin, Phenotypic Effects and Diagnostic Implications of Complex Chromosome Rearrangements. Mol Syndromol. 2015;6((3)):110–34. doi: 10.1159/000438812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poot M, Yom JS, Whang SH, Kato JT, Gollahon KA, Rabinovitch PS. Werner syndrome cells are sensitive to DNA cross-linking drugs. FASEB J. 2001;15((7)):1224–6. doi: 10.1096/fj.00-0611fje. [DOI] [PubMed] [Google Scholar]
- Poot M, Jin X, Hill JP, Gollahon KA, Rabinovitch PS. Distinct functions for WRN and TP53 in a shared pathway of cellular response to 1-beta-D-arabinofuranosylcytosine and bleomycin. Exp Cell Res. 2004;296((2)):327–36. doi: 10.1016/j.yexcr.2004.02.011. [DOI] [PubMed] [Google Scholar]
- Porokhovnik LN, Lyapunova NA. Dosage effects of human ribosomal genes (rDNA) in health and disease. Chromosome Res. 2019;27((1-2)):5–17. doi: 10.1007/s10577-018-9587-y. [DOI] [PubMed] [Google Scholar]
- Przybytkowski E, Lenkiewicz E, Barrett MT, Klein K, Nabavi S, Greenwood CM, et al. Chromosome-breakage genomic instability and chromothripsis in breast cancer. BMC Genomics. 2014;15:579. doi: 10.1186/1471-2164-15-579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugacheva EM, Kubo N, Loukinov D, Tajmul M, Kang S, Kovalchuk AL, et al. CTCF mediates chromatin looping via N-terminal domain-dependent cohesin retention. Proc Natl Acad Sci USA. 2020;117((4)):2020–31. doi: 10.1073/pnas.1911708117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos S, Rodríguez R, Castro O, Grether P, Molina B, Frias S. Presence of 15p Marker D15Z1 on the Short Arm of Acrocentric Chromosomes is Associated with Aneuploid Offspring in Mexican Couples. Int J Mol Sci. 2019;20((21)):5251. doi: 10.3390/ijms20215251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson C, Moynahan ME, Jasin M. Double-strand break repair by interchromosomal recombination: suppression of chromosomal translocations. Genes Dev. 1998;12((24)):3831–42. doi: 10.1101/gad.12.24.3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salk D, Au K, Hoehn H, Martin GM. Cytogenetics of Werner's syndrome cultured skin fibroblasts: variegated translocation mosaicism. Cytogenet Cell Genet. 1981;30:92–107. doi: 10.1159/000131596. [DOI] [PubMed] [Google Scholar]
- Santoro R, Li J, Grummt I. The nucleolar remodeling complex NoRC mediates heterochromatin formation and silencing of ribosomal gene transcription. Nat Genet. 2002;32((3)):393–6. doi: 10.1038/ng1010. [DOI] [PubMed] [Google Scholar]
- Schoemaker MJ, Jones ME, Higgins CD, Wright AF. United Kingdom Clinical Cytogenetics Group; Swerdlow AJ. Mortality and Cancer Incidence in Carriers of Balanced Robertsonian Translocations: A National Cohort Study. Am J Epidemiol. 2019;188:500–8. doi: 10.1093/aje/kwy266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer LG, Lupski JR. Molecular mechanisms for constitutional chromosomal rearrangements in humans. Annu Rev Genet. 2000;34:297–329. doi: 10.1146/annurev.genet.34.1.297. [DOI] [PubMed] [Google Scholar]
- Shen JC, Loeb LA. Werner syndrome exonuclease catalyzes structure-dependent degradation of DNA. Nucleic Acids Res. 2000;28((17)):3260–8. doi: 10.1093/nar/28.17.3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimamoto A, Kagawa H, Zensho K, Sera Y, Kazuki Y, Osaki M, et al. Reprogramming suppresses premature senescence phenotypes of Werner syndrome cells and maintains chromosomal stability over long-term culture. PLoS One. 2014;9((11)):e112900. doi: 10.1371/journal.pone.0112900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stimpson KM, Sullivan LL, Kuo ME, Sullivan BA. Nucleolar Organization, Ribosomal DNA Array Stability, and Acrocentric Chromosome Integrity Are Linked to Telomere Function. PLoS One. 2014;9((3)):e92432. doi: 10.1371/journal.pone.0092432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stults DM, Killen MW, Pierce HH, Pierce AJ. Genomic architecture and inheritance of human ribosomal RNA gene clusters. Genome Res. 2008;18((1)):13–8. doi: 10.1101/gr.6858507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi Y, Horiuchi T, Kobayashi T. Transcription-dependent recombination and the role of fork collision in yeast rDNA. Genes Dev. 2003;17((12)):1497–506. doi: 10.1101/gad.1085403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Nobelen S, Rosa-Garrido M, Leers J, Heath H, Soochit W, Joosen L, et al. CTCF regulates the local epigenetic state of ribosomal DNA repeats. Epigenetics Chromatin. 2010;3((1)):19. doi: 10.1186/1756-8935-3-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Sluis M, Gailín MÓ, McCarter JGW, Mangan H, Grob A, McStay B. Human NORs, comprising rDNA arrays and functionally conserved distal elements, are located within dynamic chromosomal regions. Genes Dev. 2019;33((23-24)):1688–701. doi: 10.1101/gad.331892.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Sluis M, van Vuuren C, Mangan H, McStay B. NORs on human acrocentric chromosome p-arms are active by default and can associate with nucleoli independently of rDNA. Proc Natl Acad Sci USA. 2020;117((19)):10368–77. doi: 10.1073/pnas.2001812117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Lemos B. Ribosomal DNA harbors an evolutionarily conserved clock of biological aging. Genome Res. 2019;29((3)):325–33. doi: 10.1101/gr.241745.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburton D. De Novo Balanced Chromosome Rearrangements and Extra Marker Chromosomes Identified at Prenatal Diagnosis: Clinical Significance and Distribution of Breakpoints. Am J Hum Genet. 1991;49:995–1013. [PMC free article] [PubMed] [Google Scholar]
- Warmerdam DO, Wolthuis RMF. Keeping ribosomal DNA intact: a repeating challenge. Chromosome Res. 2019;27((1-2)):57–72. doi: 10.1007/s10577-018-9594-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiland E, Olszewska M, Huleyuk N, Chernykh VB, Kurpisz M. The effect of Robertsonian translocations on the intranuclear positioning of NORs (nucleolar organizing regions) in human sperm cells. Sci Rep. 2019;9((1)):2213. doi: 10.1038/s41598-019-38478-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B, Li H, Perry JM, Singh VP, Unruh J, Yu Z, et al. Ribosomal DNA copy number loss and sequence variation in cancer. PLoS Genet. 2017;13((6)):e1006771. doi: 10.1371/journal.pgen.1006771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazawa K, Ogata T, Ferguson-Smith AC. Uniparental disomy and human disease: an overview. Am J Med Genet C Semin Med Genet. 2010;154C((3)):329–34. doi: 10.1002/ajmg.c.30270. [DOI] [PubMed] [Google Scholar]
- Yang Q, Zhang R, Wang XW, Spillare EA, Linke SP, Subramanian D, et al. The processing of Holliday junctions by BLM and WRN helicases is regulated by p53. J Biol Chem. 2002;277((35)):31980–7. doi: 10.1074/jbc.M204111200. [DOI] [PubMed] [Google Scholar]
- Yu S, Lemos B. The long-range interaction map of ribosomal DNA arrays. PloS Genet. 2018;14((3)):e1007258. doi: 10.1371/journal.pgen.1007258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan X, Feng W, Imhof A, Grummt I, Zhou Y. Activation of RNA polymerase I transcription by cockayne syndrome group B protein and histone methyltransferase G9a. Mol Cell. 2007;27((4)):585–95. doi: 10.1016/j.molcel.2007.06.021. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Xu X, Ding L, Li H, Xu C, Gong Y, et al. Clinical application of single-molecule optical mapping to a multigeneration FSHD1 pedigree. Mol Genet Genomic Med. 2019;7((3)):e565. doi: 10.1002/mgg3.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W-W, Wu M, Chen F, Jiang S, Su H, Liang J, et al. Robertsonian Translocations: An Overview of 872 Robertsonian Translocations Identified in a Diagnostic Laboratory in China. PloS One. 2015;10((5)):e0122647. doi: 10.1371/journal.pone.0122647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Kong L, Xu H, Lu Y, Zhao X, Yang Y, et al. Rapid prenatal diagnosis of Facioscapulohumeral Muscular Dystrophy 1 by combined Bionano optical mapping and karyomapping. Prenat Diagn. 2020;40((3)):317–23. doi: 10.1002/pd.5607. [DOI] [PMC free article] [PubMed] [Google Scholar]