Abstract
Anti-epithelial growth factor receptor or anti-vascular endothelial growth factor agents combined with chemotherapy were the standard of treatment for metastatic colorectal cancer (CRC). However, increasing evidence of molecularly stratified treatment makes the complexity of treatment. Anaplastic lymphoma kinase (ALK) gene alternation is one of potential target for biomarker-guided therapy for CRC. We present a case of a 56-year-old man who suffered from advanced ascending colon cancer, harboring echinoderm microtubule associated protein-like 4 (EML4)-ALK fusion gene E21; A20 variant, a rare variant in EML4-ALK fusion gene in lung cancer. We also detected this fusion gene from different tissue types including circulating tumor DNA (ctDNA) and ascites fluid. The patient was offered alectinib, an ALK inhibitor, with partial response in lung, liver, and peritoneal metastasis for 8 months. Tumor heterogeneity, especially in gastrointestinal tract cancer, raise our interest in comprehensive genetic profiling in clinical practice. Convenience and reliability of next-generation sequencing, including using ctDNA, help physicians deal with clinical dilemma. ALK-positive CRC is rare. However, advanced CRC with ALK gene alteration responds to ALK inhibitor. It is reasonable to check ALK gene alteration in clinical practice for CRC.
Keywords: ALK gene alternation, Colorectal cancer, Next-generation sequencing
Introduction
The treatment response of patients with advanced colorectal cancer (CRC) has been improved by combining chemotherapy with anti-vascular endothelial growth factor (VEGF) or anti-epidermal growth factor receptor (EGFR) agents. Worldwide practice guidelines recommend the use of RAS mutation testing to select target therapeutic agents. The use of other genetic mutation profiles such as BRAF or HER2 was also suggested, and the test results influenced the choice of treatment and patient outcomes [1]. However, some patients still experience a poor response to these standard treatments in clinical practice. Anaplastic lymphoma kinase (ALK) fusion genes could be detected in various tumor types, including lymphoma, lung cancer, inflammatory myofibroblastic tumor, and CRC [2, 3]. Reports have demonstrated that few patients with colorectal adenocarcinoma harbor the ALK fusion gene. Because of a very low incidence of 0.1–2% in different study populations, ALK-based testing is not routinely practiced in advanced CRC [4, 5, 6]. However, owing to improvements in treating patients with ALK-positive lung cancer, ALK inhibitors could be an effective treatment for CRC patients with ALK gene alternation [5, 7, 8, 9]. We present a case report where a patient with advanced colon cancer with ALK gene alternation obtained clinical response to ALK inhibitor and methods how to detect ALK gene alternation in CRC.
Case Report
A 56-year-old man visited our clinics in December 2017 because of progressive abdominal fullness and tarry stool passages over the previous month. An evaluation through colonoscopy and computed tomography (CT) scanning revealed an ascending colon tumor with multiple small pulmonary nodules, liver metastasis, and peritoneal carcinomatosis (Fig. 1). A pathology review of colon tumor specimens indicated disorganized and crowded tubule-glandular structures. Immunohistochemical staining revealed positive results for CK20 and negative results for CK7, which was consistent with a moderately differentiated adenocarcinoma of the colon. Molecular analysis for KRAS, NRAS, and BRAF genes demonstrated no additional genetic mutations. Immunohistochemical testing for mismatch repair proteins demonstrated no loss of nuclear expression of such proteins, including MLH1, MSH2, MSH6, and PMS2. HER2 expression, assessed by immunohistochemical staining, was also negative. The patient was treated with panitumumab combined with a FOLFOX regimen after the diagnosis of stage IV colon cancer. However, partial response was only maintained for 3 months before the disease progressed with increases in liver and lung metastasis. Subsequently, treatment was modified to FOLFOXIRI combined with bevacizumab, which still led to disease progression with elevated expression of tumor marker carcinoembryonic antigen (CEA) (Fig. 2).
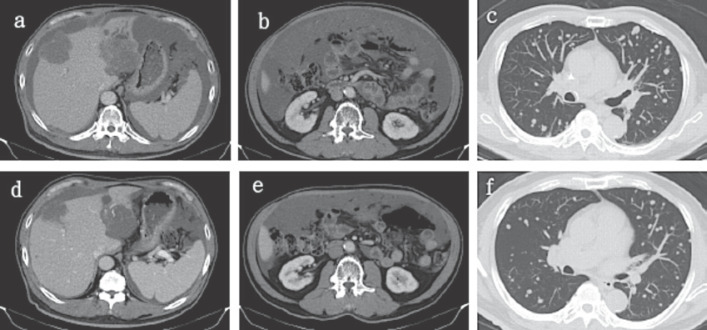
Fig. 1.
Liver (a), peritoneum (b), and lung (c) metastasis before alectinib and post alectinib (d–f).
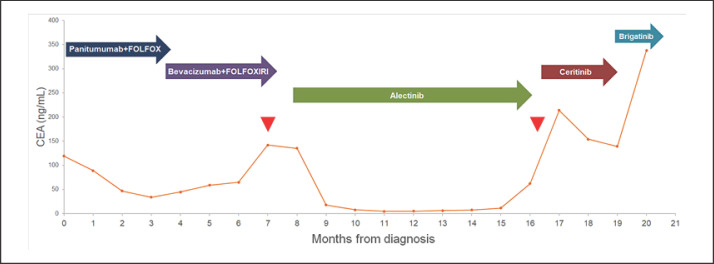
Fig. 2.
CEA change during different lines of treatment. Colored arrows indicate different lines of treatment. Red arrowheads point out two times of comprehensive genetic profiling.
Because of the patient's poor response to the standard treatment for advanced CRC, we used biopsy specimens from the colonoscopy to obtain a comprehensive genetic profiling using the FoundationOne® CDx assay. The sequencing did not detect disease-relevant gene mutations (KRAS, NRAS, and BRAF mutations) and demonstrated microsatellite stable and low tumor mutational burden (5 Muts/Mb). However, we detected fusion rearrangement between echinoderm microtubule associated protein-like 4 (EML4) and ALK, with breakpoints at EML4 intron 21 and ALK intron 19. Other non-targetable mutations were also detected, including GATA6 amplification, KDM6A splice site (443+2T>C), NOTCH2 truncation intron 22, and TP53 splice site (783-2A>G) (Table 1).
Table 1.
Next-generation sequencing of different tissue types in this patient
| Genetic finding | Colon tumor biopsy | Ascites fluid cell block | Liquid biopsy | |
|---|---|---|---|---|
| ALK | EML4-ALK fusion (E21; A20) | Present | Present | Present L1319V (VUS*) |
| KRAS | Q61H | Not detected | Not detected | 0.32% |
| Amplification | Not detected | Present | Not detected | |
| TP53 | Splice site 783–2A>G | Present (32.8%) | Present (3.75%) | Present (39.1%) |
| NF1 | Q20 | Not detected | Not detected | 0.71% |
| GATA6 | Amplification | Present | Not detected | Not tested |
| KDM6A | Splice site 443+2T>C | Present | Not detected | Not tested |
| NOTCH2 | Truncation intron 22 | Present | Not detected | Not tested |
| Tumor mutational burden | Low (5 Muts/Mb) | Can not be determined | Not tested | |
| Microsatellite status | MS-stable | MSI-can not be determined | Not tested | |
VUS, variants of unknown significance.
According to previously published data and literature reviews, alectinib, which can block the activity of ALK and is used for non-small cell lung cancer with ALK positivity, was prescribed after careful discussion with the patient regarding the indications of ALK inhibitors not approved by the Federal Drug Administration (FDA) in CRC. After a month of alectinib treatment, CEA decreased considerably (Fig. 2) and clinical symptoms were alleviated with decreases in ascites. Repeat CT scanning revealed a partial response per Response Evaluation Criteria in Solid Tumors (RECIST) 1.1 criteria with shrinkage of pulmonary metastasis nodules and liver metastatic tumors (Fig. 1). Clinical and biochemical improvements were maintained for 8 months followed by further disease progression.
We subsequently obtained another genetic profiling from ascites fluid using FoundationOne® CDx assay. In parallel, peripheral blood sampling was submitted for FoundationOne® Liquid assay. The sequencing results from the ascites cell block revealed EML4-ALK fusion genes, KRAS amplification, and a TP53 splice site (738-2A>G) (Table 1). Microsatellite status and tumor mutational burden could not be determined because of the low tumor purity. The sequencing results from peripheral blood sampling also revealed the presence of an EML4-ALK fusion gene. KRAS Q61H mutation, NF1 Q20 and TP53 splice site (783-2A>G) mutations were detected. Within the variants of unknown significance, an ALK L1319V mutation was detected from this blood sampling (Table 1). No other ALK mutations were reported in these two sequencing results. We performed immunohistochemical staining using anti-ALK antibodies for the ascites cell block; tumor cells displayed cytoplasmic granular staining for ALK (Fig. 3).
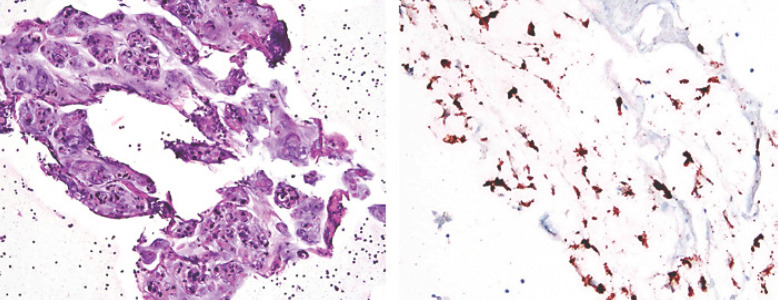
Fig. 3.
Histology and IHC stain with anti-ALK antibody of cell block from patient's ascites. Left: hematoxylin and eosin stain of ascites cell block. Right: IHC stain with anti-ALK antibody of ascites cell block. The tumor cells show cytoplasmic granular staining for ALK (brown color).
Owing to the repeated detection of ALK fusion genes in different tissue types and at different time-points of the disease, we attempted to shift to treatment using ceritinib for 3 months. Despite temporarily decreased CEA level (Fig. 2), the disease progressed nonetheless with enlarged metastatic liver tumors and peritoneal carcinomatosis. After ceritnib, brigatinib was administered as a monotherapy; however, the patient died of liver failure 22 months after the colon cancer diagnosis.
Discussion and Conclusion
Patients with RAS wild-type CRC are known to obtain response to anti-EGFR monoclonal antibodies. However, several patients still have poor clinical outcome after anti-EGFR monoclonal antibodies-based therapy. ALK receptor activation stimulates downstream signaling including MAPK/ERK, PI3K/AKT pathway, which is the potential mechanism of resistance to anti-EGFR monoclonal antibodies in RAS wild-type CRC [10]. About 5% of non-small cell lung cancer harbor ALK gene alternation. Patients with lung cancer are routinely tested for ALK genetic alternation in clinical practice. Because of development of several effective compounds for inhibiting ALK kinase activity, therapeutic effects have dramatic improvement in ALK-positive non-small cell lung cancer. Low incidence of ALK gene alternation was mentioned in CRC. However, obtaining tumor response to ALK inhibitors in CRC patients was demonstrated by several case reports (Table 2). Therefore, detection of ALK gene alternation gives rise to novel treatment options in these CRC patients.
Table 2.
Published case reports of treatment with ALK inhibitor in CRC patients harboring ALK gene alternation
| First author (year) | Age/gender | Clinical presentation | Fusion partner | First method of detecting ALK | Treatment and response | Follow-up** |
|---|---|---|---|---|---|---|
| Amatu (2015) [9] | 53/F | Right side CRC, stage IV with brain, thoracic lymph node and liver metastasis | CAD-ALK | IHC | Entrectinib: PR for 4.5 months | |
| Yakirevich (2016) [5] | 87/F | A-colon (cecum), lung metastasis, PD with peritoneal and umbilical metastasis | STRN-ALK | NGS | FOLFOX6: PR; FOLFIRI: SD at least; ceritinib SD for 9 months | KRAS G13D (post ceritinib) |
| Gambacorti-Passerini (2018) [8] | N/A | Mucinous type CRC | N/A | FaISH | Crizotinib: response duration 25 months | |
| Hsiao (2021)* | 56/M | A-colon, stage IV with lung, liver, and peritoneal metastasis | EML4-ALK | NGS | Anti-EGFR with FOLFOX: PR for 3 months; anti-VEGF with FOLFOXIRI: PD; alectinib: PR for 8 months; ceritinib and brigatinib: PD | KRAS Q61H (post alectinib) |
PR, partial response; SD, stable disease; PD, progressive disease; A-colon, ascending colon; IHC, immunohistochemistry; NGS, next-generation sequencing; FISH, fluorescence in situ hybridization; N/A, not available.
This report.
Suspicious emerging resistant mutation after ALK inhibitor.
A higher incidence is reported if detection of ALK alternation is restricted to patients with right side colon cancer or mucinous type CRC [5]. We may perform ALK genetic testing restricting to patients with right side colon cancer with RAS wild type or obtaining resistant response to anti-EGFR agents. However, we should take into consideration that left side colon cancer also has the possibility of ALK gene alternation as mentioned by a previous report [4]. Several methods could be employed to detect ALK gene alternation, including fluorescence in situ hybridization (FISH) and next-generation sequencing (NGS). However, during routine clinical practice in lung cancer, immunohistochemical staining using anti-ALK antibodies is practical and affordable, which could also be used to detect ALK-positive CRC, such as staining of an ascites cell block performed in our patient and on patients detailed in other studies [4, 6]. Growing evidence suggests that in the gastrointestinal tract, cancer tumors are heterogeneous and that parallel molecularly stratified treatment strategies are effective [11, 12]. Not only RAS gene status but also other genes, such as BRAF, HER2, MET, and ALK, play an important role in treatment decision for CRC patients. NGS could help physicians capture whole genetic pictures of tumors and guide their clinical treatment decisions [13]. Using tumor tissue from a formalin-fixed paraffin-embedded (FFPE) sample for sequencing is the standard method. However, difficulty in tumor sampling sometimes leads to clinical dilemmas. Concurrent with a previous report, we determined that the use of circulating tumor DNA (ctDNA) was a convenient method for detecting ALK gene alternation in CRC [14, 15]. We genetically profiled different tissue types (including body fluid and ctDNA) other than FFPE samples. ALK alternation was detected in all three tissues. Genetic sequencing of different metastatic sites, combined with the use of ctDNA, may help physicians understand tumor heterogeneity and select the optimal treatment.
Because of the low incidence of ALK-positive CRC and no FDA approval for ALK inhibitors in CRC, reports on treatment responses to ALK inhibitor for colon cancer are rare. There is still no treatment guideline focusing on ALK-positive CRC. Some reports demonstrated a treatment response to standard chemotherapy with or without anti-VEGF agents [14]. Case reports of ALK-positive CRC patients treated with ALK inhibitors are summarized in Table 2. Several cases revealed advanced clinical stage and obtained a resistant response to standard treatment for CRC. As our case, treatment response to anti-EGFR agent combined with chemotherapy was only maintained for 3 months, also failure to a more toxic FOLFOXIRI regimen. We reported the first case of a patient with colon cancer with an EML4-ALK fusion gene who obtained a partial response by alectinib and was subsequently treated by different ALK inhibitor agents as the disease progressed. Reports have demonstrated that administering these ALK inhibitors, such as ceritinib or entrectinib, in patients with colon cancer with various ALK fusion genes can lead to clinical improvements [5, 9]. However, a secondary ALK inhibitor did not halt disease progression. One possible explanation was emerging resistant mutation and more potent ALK inhibitors, such as lorlatinib, were not introduced as subsequent line of treatment. Another possible explanation is that the detection of KRAS Q61H using ctDNA after disease progression indicated tumor heterogeneity of gastrointestinal cancer or obtaining secondary resistance mechanism. Standard treatment options such as regorafenib or lonsurf should be taken into consideration.
This paper is the first comprehensive case report on a patient with ascending colon adenocarcinoma harboring EML4-ALK fusion gene being treated with alectinib monotherapy for 8 months and then sequentially with ceritinib and brigatinib. As exemplified by this case, ALK fusion gene could be detected by the application of different tissue types, including FFPE samples, peripheral blood, and body fluid cell block. As shown by previous reports and our case experience, ALK inhibitor is instrumental for ALK-positive colon cancer. Therefore, it is important and necessary for detecting ALK gene alternation for patients with advanced CRC.
Statement of Ethics
All authors confirmed this report was conducted according to the Declaration of Helsinki. Informed consent for this report and permission to publish this article were obtained from the patient's next of kin.
Conflict of Interest Statement
The authors indicate no conflict of interest and financial relationships.
Funding Sources
No funding was received.
Author Contributions
Sheng-Yen Hsiao contributed to the conception, design, and manuscript writing. Sheng-Yen Hsiao, Hong-Lin He, Chao-Jung Tsao contributed to the study material and patient care. Cheng-Yao Lin, Wen-Tsung Huang collected and assembled the data. Chien-Ming Chao contributed to the data analysis. All authors have read and approved the final version of the manuscript.
Acknowledgments
We would like to thank Foundation Medicine for providing data of comprehensive genetic profiling.
References
- 1.Dienstmann R, Vermeulen L, Guinney J, Kopetz S, Tejpar S, Tabernero J. Consensus molecular subtypes and the evolution of precision medicine in colorectal cancer. Nat Rev Cancer. 2017;17((2)):79–92. doi: 10.1038/nrc.2016.126. [DOI] [PubMed] [Google Scholar]
- 2.Mano H. ALKoma: a cancer subtype with a shared target. Cancer Discov. 2012;2((6)):495–502. doi: 10.1158/2159-8290.CD-12-0009. [DOI] [PubMed] [Google Scholar]
- 3.Marino-Enriquez A, Dal Cin P. ALK as a paradigm of oncogenic promiscuity: different mechanisms of activation and different fusion partners drive tumors of different lineages. Cancer Genet. 2013;206((11)):357–73. doi: 10.1016/j.cancergen.2013.07.001. [DOI] [PubMed] [Google Scholar]
- 4.Lee J, Kim HC, Hong JY, Wang K, Kim SY, Jang J, et al. Detection of novel and potentially actionable anaplastic lymphoma kinase (ALK) rearrangement in colorectal adenocarcinoma by immunohistochemistry screening. Oncotarget. 2015;6((27)):24320–32. doi: 10.18632/oncotarget.4462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yakirevich E, Resnick MB, Mangray S, Wheeler M, Jackson CL, Lombardo KA, et al. Oncogenic ALK Fusion in Rare and Aggressive Subtype of Colorectal Adenocarcinoma as a Potential Therapeutic Target. Clin Cancer Res. 2016;22((15)):3831–40. doi: 10.1158/1078-0432.CCR-15-3000. [DOI] [PubMed] [Google Scholar]
- 6.Houang M, Toon CW, Clarkson A, Sioson L, de Silva K, Watson N, et al. ALK and ROS1 overexpression is very rare in colorectal adenocarcinoma. Appl Immunohistochem Mol Morphol. 2015;23((2)):134–8. doi: 10.1097/PAI.0000000000000025. [DOI] [PubMed] [Google Scholar]
- 7.Medico E, Russo M, Picco G, Cancelliere C, Valtorta E, Corti G, et al. The molecular landscape of colorectal cancer cell lines unveils clinically actionable kinase targets. Nat Commun. 2015;6:7002. doi: 10.1038/ncomms8002. [DOI] [PubMed] [Google Scholar]
- 8.Gambacorti-Passerini C, Orlov S, Zhang L, Braiteh F, Huang H, Esaki T, et al. Long-term effects of crizotinib in ALK-positive tumors (excluding NSCLC): A phase 1b open-label study. Am J Hematol. 2018;93((5)):607–14. doi: 10.1002/ajh.25043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Amatu A, Somaschini A, Cerea G, Bosotti R, Valtorta E, Buonandi P, et al. Novel CAD-ALK gene rearrangement is drugable by entrectinib in colorectal cancer. Br J Cancer. 2015;113((12)):1730–4. doi: 10.1038/bjc.2015.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holla VR, Elamin YY, Bailey AM, Johnson AM, Litzenburger BC, Khotskaya YB, et al. ALK: a tyrosine kinase target for cancer therapy. Cold Spring Harb Mol Case Stud. 2017;3((1)):a001115. doi: 10.1101/mcs.a001115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu Y, Sethi NS, Hinoue T, Schneider BG, Cherniack AD, Sanchez-Vega F, et al. Comparative Molecular Analysis of Gastrointestinal Adenocarcinomas. Cancer Cell. 2018;33((4)):721–35.e8. doi: 10.1016/j.ccell.2018.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sveen A, Kopetz S, Lothe RA. Biomarker-guided therapy for colorectal cancer: strength in complexity. Nat Rev Clin Oncol. 2020;17((1)):11–32. doi: 10.1038/s41571-019-0241-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnson B, Cooke L, Mahadevan D. Next generation sequencing identifies ‘interactome’ signatures in relapsed and refractory metastatic colorectal cancer. J Gastrointest Oncol. 2017;8((1)):20–31. doi: 10.21037/jgo.2016.09.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lai AZ, Schrock AB, Erlich RL, Ross JS, Miller VA, Yakirevich E, et al. Detection of an ALK Fusion in Colorectal Carcinoma by Hybrid Capture-Based Assay of Circulating Tumor DNA. Oncologist. 2017;22((7)):774–9. doi: 10.1634/theoncologist.2016-0376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Siravegna G, Sartore-Bianchi A, Mussolin B, Cassingena A, Amatu A, Novara L, et al. Tracking a CAD-ALK gene rearrangement in urine and blood of a colorectal cancer patient treated with an ALK inhibitor. Ann Oncol. 2017;28((6)):1302–8. doi: 10.1093/annonc/mdx095. [DOI] [PubMed] [Google Scholar]