Abstract
Advances in computerized image analysis and the use of artificial intelligence–based approaches for image-based analysis and construction of prediction algorithms represent a new era for noninvasive biomarker discovery. In recent literature, it has become apparent that radiologic images can serve as mineable databases that contain large amounts of quantitative features with potential clinical significance. Extraction and analysis of these quantitative features is commonly referred to as texture or radiomic analysis. Numerous studies have demonstrated applications for texture and radiomic characterization methods for assessing brain tumors to improve noninvasive predictions of tumor histologic characteristics, molecular profile, distinction of treatment-related changes, and prediction of patient survival. In this review, the current use and future potential of texture or radiomic-based approaches with machine learning for brain tumor image analysis and prediction algorithm construction will be discussed. This technology has the potential to advance the value of diagnostic imaging by extracting currently unused information on medical scans that enables more precise, personalized therapy; however, significant barriers must be overcome if this technology is to be successfully implemented on a wide scale for routine use in the clinical setting.
Keywords: Adults and Pediatrics, Brain/Brain Stem, CNS, Computer Aided Diagnosis (CAD), Computer Applications-General (Informatics), Image Postprocessing, Informatics, Neural Networks, Neuro-Oncology, Oncology, Treatment Effects, Tumor Response
Supplemental material is available for this article.
© RSNA, 2020
Summary
Radiomic-based approaches and artificial intelligence can be used to analyze medical images and construct prediction algorithms with the potential to advance medical imaging into a new era of noninvasive biomarkers and predictive analytics toward more personalized, precision medical diagnostics for the evaluation of brain tumors.
Key Points
■ Radiologic images can serve as mineable databases containing quantitative features with potential clinical significance.
■ Texture or radiomic analysis combined with machine learning can be used to extract image quantitative features and combine them with other clinical information to construct “intelligent” prediction algorithms that improve with algorithm use and “experience.”
■ Early studies suggest the potential for better characterization of brain tumors including prediction of tumor histologic characteristics, molecular characteristics, and patient survival.
■ Wide-scale application of radiomic-based approaches in the clinical setting will require overcoming scientific, workflow automation, and regulatory approval barriers.
Introduction
Cross-sectional imaging techniques have a central role in the initial diagnosis and surveillance of brain tumors. CT and MRI are used for tumor identification, mapping, and determination of tumor extent and posttreatment surveillance as a routine part of standard of care. However, beyond the traditional, largely anatomic and qualitative- oriented role of imaging for the evaluation of brain tumors, there is increasing interest in more specific, noninvasive biomarkers that can be used for enhanced characterization of the tumor phenotype, prediction of response to therapy, prediction of survival, and distinction of treatment-induced changes or complications from tumor recurrence. Interest in discovering and developing better tumor biomarkers is partly fueled by the recognition of the increased complexity of these tumors, as evident in the incorporation of molecular and histologic features for defining many tumor entities in the 2016 World Health Organization Classification of Tumors of the Central Nervous System (1). There is also increasing complexity of different therapeutic regimens, including various systemic therapies, that are either in current use or under investigation, with interest in therapies targeting specific molecular pathways (2). These therapeutic advances hold the promise of more effective, personalized therapy for brain tumors. At the same time, they highlight the need for more precise biomarkers that enable earlier and optimal patient stratification and treatment selection. This represents both an opportunity and a challenge for medical imaging as it takes an even more central role in the decision-making process and optimal treatment selection in the future.
To meet these challenges, new MRI sequences that improve diagnostic performance have been developed. As a result, there has been significant progress in advanced imaging of brain tumors using a combination of standard sequences and more advanced MRI techniques, such as diffusion-weighted and tensor imaging, susceptibility-weighted imaging, perfusion and permeability imaging, and MR spectroscopy (3). There has also been progress in developing more standardized and reliable approaches for assessment of response to treatment, such as the use of the Response Assessment in Neuro-Oncology criteria (4,5). However, despite these advances, the imaging criteria currently used in routine clinical practice rely on qualitative assessment and relatively basic quantitative evaluation of the sequences performed, perhaps with the exception of some advanced techniques such as perfusion and permeability imaging, potentially ignoring or underusing large amounts of available quantitative data. To this end, there has been increasing interest in the use of advanced computerized image analysis approaches that enable objective, high-level quantitative evaluation of image features and pixel-level relationships for tumor characterization, commonly referred to as texture or radiomic analysis (Table 1). The interest in these types of analyses has been further fueled by recent advances in artificial intelligence (AI), especially machine learning (ML), for image analysis and prediction algorithm development.
Table 1:
Summary of Handcrafted Quantitative Features
An increasingly large body of evidence suggests that radiomics and AI applications can leverage existing information on scans that are routinely obtained as part of a patient’s workup, including information available on standard sequences and more advanced techniques, to enhance diagnostic evaluation of tumors and develop clinically useful noninvasive biomarkers for tumor characterization. These noninvasive applications will be discussed throughout this review with a focus on brain tumor imaging. An overview of texture and radiomic approaches for tumor analysis and the central role of ML in constructing prediction algorithms or classifiers will be discussed. The radiomic workflow and the use of “handcrafted” versus “deep” extracted features will be reviewed, followed by a general discussion of current applications for brain tumor analysis to demonstrate the potential of these approaches for tumor-specific biomarker development. The review will conclude with a discussion of the barriers and challenges for the adoption of this technology in the clinical setting and a brief discussion of potential next steps and early applications for deployment in the clinical setting.
Overview of Texture Analysis and Radiomics
Texture Analysis
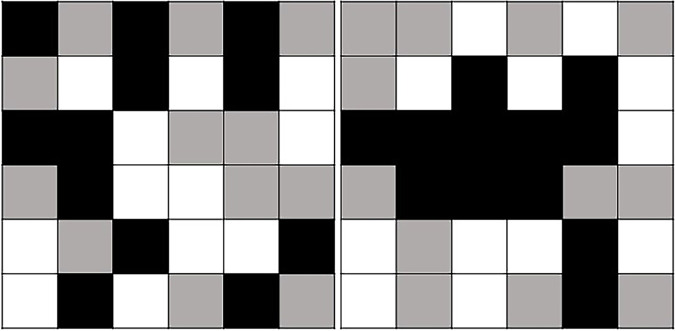
There are variations in the use and definition of medical image texture analysis in the medical and computer science literature. Broadly defined, texture analysis refers to computerized analysis of pixel position and fine pixel density or intensity variations on an image, with extraction of mathematically derived quantitative parameters that reflect those variations (6–12) (Fig 1). Texture analysis provides a quantitative map of different pixel variations and their relationships on the image. The goal of texture analysis is to extract and analyze fine variations that are not observed or consistently incorporated into the diagnostic decision making on evaluation by the naked eye during routine qualitative image interpretation performed in clinical practice. Interest in texture analysis has emerged partly from early studies of human perception that demonstrated that despite its impressive performance, the human visual system may have difficulty in effortlessly discriminating certain textural characteristics, for example those related to higher-order statistical features of an object or image (8,13). Interest in the use of computer-assisted analytic methods for image analysis is also driven partly by the increasing amount and complexity of the available information on a patient’s scan and electronic medical record.
Figure 1:
Texture or radiomic analysis for extraction of quantitative features reflecting higher-order pixel positions and relationships. Simplified diagram demonstrates two squares containing an identical number of white, gray, and black pixels. Using basic first-order statistical quantitative parameters, such as average or standard deviation, frequently used in traditional region of interest analysis, the boxes would have identical values, even though on visual inspection the patterns are clearly different. The objective of computerized image analysis approaches like texture or radiomic analysis is to extract quantitative parameters or features that capture the more complex, higher-order characteristics, such as those reflecting pixel positions and relationships.
The main objective and rationale behind texture analysis of tumors is to provide a noninvasive quantitative map of tumor heterogeneity, which in turn is used to predict a molecular or clinical end point of interest (9–12). Interest in texture analysis is not new, and reports of potential applications date back to the early days of computerized image analysis (7,8,14). However, there has been a revival of interest in texture analysis applications in the past decade, fueled by the impressive advances in computational power, increasing interest in quantitative biomarkers, and a general trend toward digital health applications and more precise, personalized medicine.
There is not a uniform definition for texture analysis in the literature. In the medical and radiology literature, texture analysis has been used to refer to a range of quantitative features that include primary (or first-order) statistical features, secondary statistical features, and higher-order statistical features or more complex relationships that are derived from model-based and transform-based methods (9–12) (Table 1). However, texture has also been defined more narrowly as the second-order determinants of spatial interrelationships of pixel (or voxel) gray-level values or texture matrix–based features (15–17). For simplicity and clarity, the broader range of quantitative features that can be extracted—including the previously mentioned—will all be included under the umbrella of handcrafted radiomic features in this review. For the rest of this review, the term radiomics will be used to describe this process or articles performing this type of analysis. Table 1 provides a broad overview of handcrafted texture or radiomic features. A more detailed discussion of these features is beyond the scope of this article but can be found in a number of reviews or reference manuals on this topic (6–10,12,15,16).
Radiomics
The term radiomics was first introduced in the medical literature in 2012, defined as “high throughput extraction of quantitative imaging features with the intent of creating mineable databases from radiological images” (18,19). In a more recent review, the definition of radiomics was expanded to “high-throughput extraction of quantitative features that result in the conversion of images into mineable data and the subsequent analysis of these data for decision support” (15). So far, the majority of published radiomic studies have been based on analysis of CT, MRI, or PET scans, but there is no reason to restrict radiomic analysis to these or any specific imaging modality. The exact definition and application of the term radiomics is likely to further expand with newly developed and increasingly sophisticated image analytic methods, an example of which will be discussed in the following sections when discussing the use of features extracted using deep learning (DL).
It is important to note that traditional radiomic feature extraction neither requires nor is based on AI, or more specifically ML. However, ML approaches such as convolutional neural networks (CNNs; a type of DL) can be used for direct image analysis and feature extraction (Table 1). Regardless of the method used for extracting radiomic features, ML is a powerful approach for developing algorithms based on extracted quantitative features to construct clinically useful prediction models or classifiers. It is worthwhile to distinguish the radiomics process from the traditional computer-aided diagnosis and detection systems. As discussed by Gillies et al (15), computer-aided diagnosis and detection systems are usually stand-alone systems designated by the Food and Drug Administration for use in either the detection or diagnosis of disease and are typically designed to deliver a single answer (eg, presence or absence of an abnormality). On the other hand, radiomics is a process designed to extract vast amounts of quantitative features from digital images that can be mined for hypothesis generation, testing, or both. These data can then be combined with other patient characteristics and clinically available information to develop decision support tools, and therefore some radiomics applications could be implemented as an extension of computer-aided diagnosis and detection.
When discussing radiomic features, one may make a distinction between semantic and agnostic features as the two broad categories of features that can be extracted using radiomics, as discussed by Gillies et al (15). Semantic features refer to characteristics or features commonly used in the radiology lexicon to describe a lesion of interest (ie, size, shape, and the presence of necrosis), which may be quantified for incorporation into radiomic models. The other broad category of features, agnostic features, can be characterized as mathematically extracted quantitative descriptors that are generally not part of radiologists’ lexicons (15). Examples of agnostic features include the first-, second-, and higher-order statistical features that were discussed earlier. Many of the published texture and radiomics studies are based primarily on agnostic features, but there is no reason to impose any such limitation as the ultimate goal is to take advantage of all available clinically useful information. Having a component that includes or reflects semantic features also has a potential advantage in the sense that making any algorithm more “explainable” (20,21) may improve user acceptance as well as regulatory approval.
Traditional Handcrafted versus Deep Radiomics Features
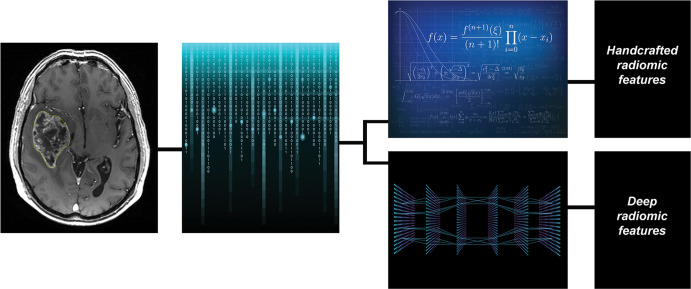
There is an increasing use of CNNs for image analysis and tumor characterization. In the preceding sections, texture or radiomic features were described as including first-, second-, and higher-order statistical features as well as other complex features, including those based on more complex relationships derived using model-based and transform-based methods (6–12,15,16). Regardless of the variety of features that can be extracted and their complexity, these approaches share the characteristic that they are derived using clearly defined or explicit mathematical formulas designed by experts, often independent of or prior to the experiment. These features can be collectively referred to as “handcrafted” or “hand-engineered” features (Fig 2). In contradistinction to handcrafted features extracted using traditional radiomics, features extracted based on image analysis with DL approaches such as CNNs are not explicitly defined by an expert. Instead, they are learned from data through a learning algorithm, such as backpropagation. These may be referred to as deep (extracted) features and the process as “deep radiomics” (Fig 2).
Figure 2:
Texture or radiomic feature extraction: handcrafted feature versus deep features. During the process of texture or radiomic analysis, quantitative imaging features are extracted with the potential to serve as quantitative biomarkers that can be used to predict a clinical or molecular end point of interest. Broadly, traditional radiomic features may be defined as those derived using clearly defined or explicit mathematical formulas designed by experts, often independently and prior to the experiment, which may in turn be referred to as handcrafted or hand-engineered features. In contradistinction, features extracted based on image analysis with deep learning approaches, such as convolutional neural networks, are not clearly definable or derived using expert-designed explicit mathematical formulas. Instead, they are learned from data through a learning algorithm. These may be referred to as deep (extracted) features and the process as “deep radiomics.”
Compared with DL, handcrafted features that include semantic features may be more limited in scope because they are based on a finite set of mathematically derived relations. Therefore, their predictive ability may be potentially inferior to DL in very large data sets. Handcrafted features, by definition, are explainable, which may be an advantage when compared with DL approaches. However, handcrafted features are prone to technical variations and noise, and they likely require image preprocessing and standardization to be generalizable and successfully applied on scans acquired using different techniques, scanners, or sites. Typically, handcrafted features work better on smaller data sets, which explains the preponderance of studies using this approach in the literature. DL is not constrained by a predetermined number or type of features and has the significant advantage of having the potential to “learn” any imaging feature(s) predictive of a given end point of interest. There is debate on whether the same extent of preprocessing would be needed for DL, and with sufficiently large and varied data sets, image standardization may not be required for DL. It may even be argued that not only is standardization unnecessary, but it would have a negative impact by unduly removing information. However, these questions are unsettled at this time. In general, DL is more likely to have a poor or misleading performance on smaller data sets. Although many of the current texture or radiomic studies use handcrafted features for prediction, it is likely that deep radiomics will play an increasing role for this purpose. Early studies may report an advantage for either approach; however, at this time, there is insufficient evidence to conclude that deep radiomics will completely replace traditional radiomics or whether a combination of the two will yield the most robust biomarkers (22).
Prediction Model Construction and ML Classifiers
Radiomic features need to be incorporated into a prediction (classification) algorithm, which is called a “classifier,” to be used for the determination of a molecular or clinical end point of interest. For example, analysis of a region of interest, such as a brain tumor, may yield hundreds or thousands of handcrafted radiomic features or deep radiomic features. There may also be nonradiomic features, such as clinical patient characteristics, biochemical results, or pathologic findings or molecular data obtained through biopsy that can be included as features in the classification model. Among the large number of features available, some will have variable associations (strong, weak, or insignificant) with the clinical end point of interest. Furthermore, among the different features, many may be redundant or highly correlated. Last, certain features may have a weak association with the outcome of interest when used in isolation but could increase in importance when used in combination with other features through interactions with those features. To predict a clinical end point of interest, an algorithm should be designed to analyze all the available features and optimize the set to the smallest number of characteristics that can accurately predict the outcome of interest.
Prediction algorithms or classifiers can be constructed using a purely classic statistical approach or ML. ML is particularly well suited for development of prediction algorithms with the potential for future translation into clinical decision support tools. One of the major advantages of ML over traditional software is that ML algorithms, as the name implies, can “learn” from experience and improve over time. There are many types of ML classification algorithms, and broadly, these can be divided into classic ML and DL (17,23–31). An example of DL that is widely used for medical image analysis is a type of artificial neural network called the CNN, as was discussed earlier.
When using handcrafted radiomic features, the feature extraction and prediction model construction steps are achieved in different steps (Fig 3). For example, features extracted from a lesion of interest are used as an input for a ML classifier for predicting an end point of interest. DL approaches, such as CNNs, can be used for direct image analysis and construction of a prediction model or classifier, combining the two steps and executing them using the same process. There is also a fundamental difference between classic ML and DL approaches. Generally, classic ML approaches are poorly suited to perform sophisticated image analysis, practically excluding them from the image analysis part of the radiomic process. However, the robust performance and complex architecture of DL makes DL-based approaches very attractive for image analysis and feature extraction applications. Although DL can be used both for image analysis and prediction algorithm reconstruction, it is not absolutely necessary, and one can combine DL and deep extracted features with other ML methods, including classic methods, for classification (32,33).
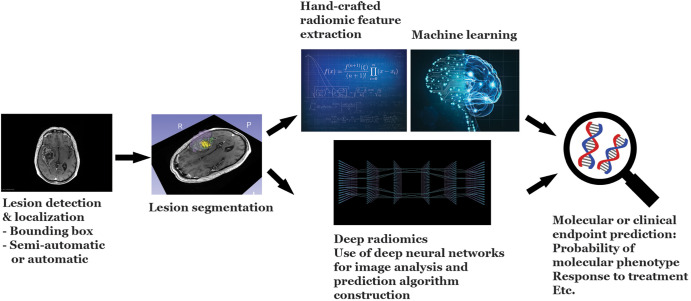
Figure 3:
Overview of the radiomic workflow. The major steps in the radiomic workflow consist of lesion identification and localization, segmentation, feature extraction, and prediction model construction. In the short term, lesion detection and localization may be facilitated either by directing the algorithm using a bounding box approach by the radiologist or, alternatively, by the radiologist pointing at the lesion of interest using a cursor. However, in the long term, lesion identification may also be performed automatically by the algorithm. It should be noted that regardless of the degree of automation, these steps would be performed under the supervision of the expert radiologist, with the ability to make adjustments or modifications as needed.
As a general rule, a traditional radiomic approach with classic ML may perform better with smaller data sets because DL typically requires larger data sets for algorithm training and development (17). Early studies suggest that there may be an advantage to combining the two approaches, but this requires further investigation. Whether assessments of very large data sets with DL approaches will completely replace the traditional radiomic approach, or if the two will be combined for an optimal biomarker and classifier development, is not a settled question and will have to be determined in future investigations. A detailed discussion of different ML methods is beyond the scope of this article but can be found in various publications and review articles on the topic (17,23–31).
Summary of the Radiomic Workflow
The major steps in the radiomic workflow are illustrated in Figure 3 and include lesion identification and localization, segmentation, feature extraction, and prediction model construction. Additional complexities and consideration related to various steps in the workflow are discussed in Table 2. Familiarity with the radiomic flow is important both for understanding the process as well as the multiple levels of complexity involved and challenges that must be overcome for eventual translation into a clinical decision support tool. As should be evident based on the earlier discussion, DL can in theory be used to perform every major step in the radiomic workflow, including localization and segmentation of a tumor (34,35), extraction of deep features, and construction of a prediction algorithm or classifier. It is therefore likely that DL will play an important role in radiomic-based clinical decision support tools of the future. However, one should not write off other approaches such as those based on traditional computer vision or combination of classic ML with DL for certain processes. On the basis of early studies, there may be a need or at least an advantage of combining different approaches (22). Whether with sufficiently large data sets the DL can completely replace the other components will have to be determined in future investigations.
Table 2:
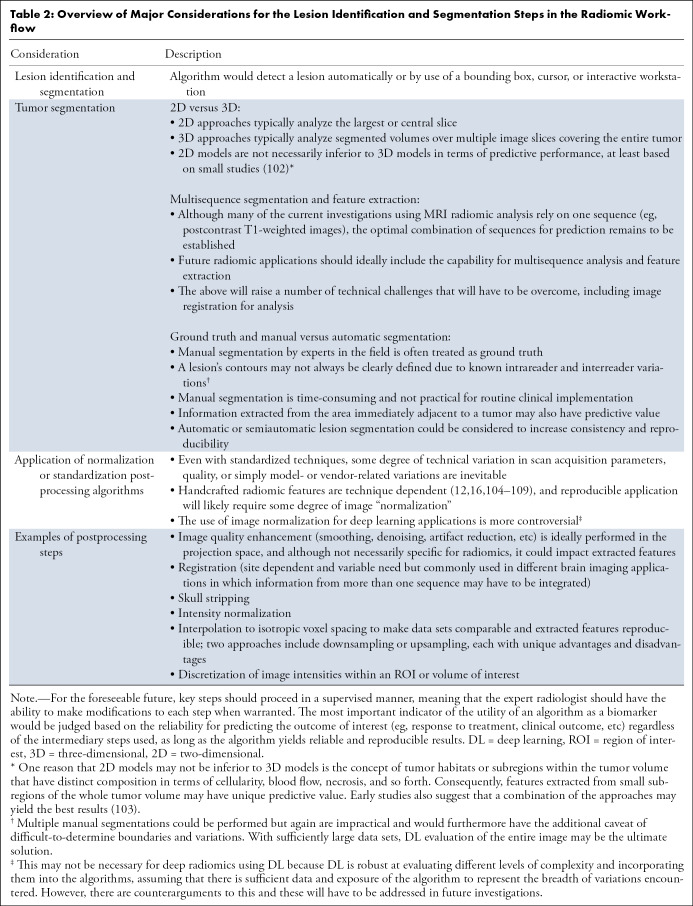
Overview of Major Considerations for the Lesion Identification and Segmentation Steps in the Radiomic Workflow
Radiomic Models for Tumor Evaluation
Brain tumor evaluation applications consist of a mixture of traditional texture or radiomic studies using handcrafted features as well as the use of DL for tumor evaluation. Because MRI is the advanced imaging modality most commonly used for tumor evaluation, the majority of the radiomic studies performed for brain tumor evaluation use MRI, so this section will focus on radiomics applications based on MRI scans. However, some studies also show potential for application of radiomics to brain CT scans (36) or other modalities (37) for brain tumor evaluation. Although many of the studies are based solely on radiomic features, it is important to note that when planning investigation and development of clinical decision support tools, analysis should not be limited to radiomic features alone. On the contrary, all available information should be used, including clinical, biopsy, and molecular data, when available (34,38) for prediction model construction as part of a patient care pathway. There is also an important secondary consideration of ensuring that radiomic features have additional value and are not simply surrogates or redundant features to more basic parameters such as tumor size or other routinely obtained information. Studies are beginning to incorporate and demonstrate the additive value of radiomics (38) to more routinely obtained image-based or clinical parameters.
Histologic Classification and Grading of Tumors
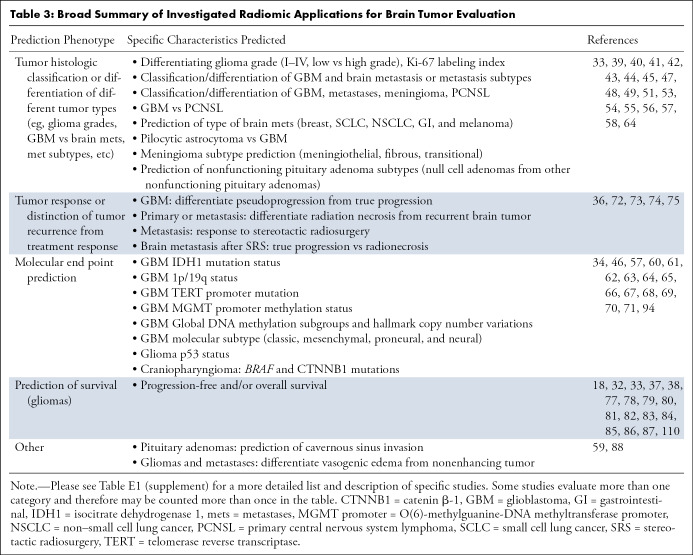
Despite the exquisite anatomic detail and functional information provided using advanced MRI techniques, distinction of tumor type or grades is not always possible using current approaches for image interpretation. Radiomic approaches have the potential to further enhance noninvasive tumor characterization by enabling histopathologic classification or grading (Table 3 and Table E1 [supplement]). Preliminary studies have shown that tumor radiomic features may be used to distinguish different tumor types, such as primary brain tumors from metastases (39–41), primary central nervous system lymphoma (41–44), or other tumor types (41). Radiomic analysis has also been reported to help the distinction of glioblastoma from pilocytic astrocytoma (45), distinction of different histologic types of craniopharyngiomas (46), discrimination of meningioma subtypes (47), or distinction of nonfunctioning pituitary adenomas subtypes (48). Radiomic analysis combined with basic clinical information can also help in distinguishing different types of brain metastasis, and for certain metastasis types, the performance has been reported to be superior to expert radiologist evaluation (49). Some studies also suggest that radiomic features can be used for predicting tumor grade, such as distinguishing different grade gliomas (50–58). Radiomic analysis may also be used to estimate tumor proliferation indexes such as Ki67 (57) or for the differentiation of infiltrating tumor from vasogenic edema (59).
Table 3:
Broad Summary of Investigated Radiomic Applications for Brain Tumor Evaluation
Classification of Molecular Characteristics of Tumors
Incorporation of molecular characteristics of tumors into diagnostic and treatment algorithms is key for optimal tumor therapy, explaining the incorporation of certain tumor molecular features into the most recent World Health Organization tumor classification. It is therefore no surprise that developing noninvasive biomarkers for predicting tumor molecular characteristics is an area of great interest and active investigation. Multiple investigations have demonstrated the potential of radiomic approaches for prediction of tumor molecular phenotype (Table 3 and Table E1 [supplement]). These include determination of isocitrate dehydrogenase 1 mutation status in gliomas (34,60–66), determination of nondeleted versus co-deleted 1p/19q status (62,64,67,68), prediction of O(6)-methylguanine-DNA methyltransferase promoter methylation status (69), isocitrate dehydrogenase 1/2-mutant with a telomerase reverse transcriptase promoter mutation (61), or prediction of p53 status or other molecular characteristics (70,71) in gliomas. One study also used radiomics for prediction of mutations in BRAF and catenin β-1 in craniopharyngiomas (46).
Posttreatment Change, Survival, and Other Studies of Interest
Distinguishing treatment-related changes in tumors continues to represent a challenge, even with the use of advanced MRI techniques. Preliminary studies suggest that radiomic analysis may be useful for distinction of progression or recurrence of primary or metastatic brain tumors from radiation necrosis and pseudoprogression (72–75). Multiple studies also suggest that radiomics can be used to predict patient survival (32,33,37,70,76–87). Radiomics has also shown potential for predicting the response of brain metastases to stereotactic radiosurgery (36). For pituitary adenomas, at least one study suggests that MRI-based radiomic features may be useful for predicting cavernous sinus invasion (88). As discussed earlier, the goal of these studies is to improve patient care by enabling more accurate diagnosis or optimizing treatment planning. Therefore, beyond prediction of probability for a specific molecular phenotype, the most promising and exciting application of these biomarkers is the establishment of a reliable and reproducible association with treatment response and outcomes. These associations may then enable better prediction of tumor response to different treatment, enabling earlier institution of the optimal therapy, with significant potential positive impact on patient care. Beyond prediction alone, quantitative and ML-based approaches also have the potential to optimize radiation therapy plans, potentially reducing toxicity to healthy tissues (89). Image-derived parameters may also be used to estimate tumor proliferation indexes such as Ki67, which potentially could be used to guide stereotactic biopsy (90).
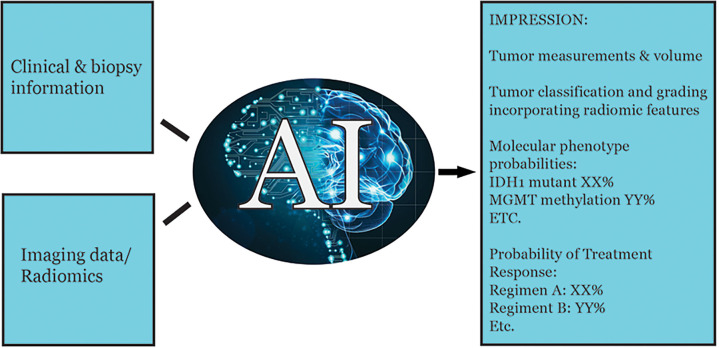
The preceding sections have provided examples of potential applications of radiomics and ML for brain tumor evaluation. In the future, radiomic features can potentially be incorporated into staging systems and used to provide predictions for key tumor characteristics of interest in the radiology report (Fig 4).
Figure 4:
Potential example of a future radiology report incorporating radiomic features and machine learning for predictive modeling. One could even envision a preliminary draft of the report being generated automatically based on automated image analysis combined with natural language processing, which can then be modified by the expert radiologist as required. IDH1 = isocitrate dehydrogenase 1, MGMT = O(6)-methylguanine-DNA methyltransferase.
Radiomics, Pathology, and Molecular Profiling
When discussing the potential for radiomic prediction of certain molecular features of tumors, it is worthwhile to step back and evaluate the broader potential implications and limitations of this approach. Even if proven to be reliable, in the absence of tumor-specific contrast agents, radiomic predictions will be based on associations of tumor macroscopic or microscopic image features with the molecular features and not direct confirmatory tests of the presence of a given mutation. Therefore, one should not take away from this review that radiomics will replace pathologic assessment and molecular profiling. However, radiomics has the potential to expand the frontiers of image-based noninvasive biomarkers further into the molecular realm. After all, even basic semantic image features have shown that imaging characteristics have an association with certain tumor phenotypes. It is also worth noting that just like imaging, AI will expand the frontiers of pathologic assessment, including in the semiautomated or automated analysis of pathologic slides and the use of ML approaches for incorporation of vast amounts of molecular data.
Notwithstanding the inherent indirect or statistical basis of radiomic approaches, there is true potential for radiomics and AI technology. This is because many treatment algorithms are far from perfect, some to the point of trial and error approach, especially for more advanced disease or disease that is unresponsive to standard, first-line therapy. As such, a good noninvasive predictive algorithm, even if not perfect, has significant potential for a clinical benefit in optimizing patient therapy. Furthermore, although radiomic markers cannot be expected to match molecular profiling in terms of accuracy or depth of detailed information provided, they do have the advantage of enabling whole-tumor analysis, in contradistinction to the sampling biases that are inherent in biopsy specimens due to tumor heterogeneity. By performing whole-tumor analysis, ML approaches can extract information from different tumor parts and surrounding regions, which may in turn be used to predict outcomes of interest or guide biopsy (50,90–92). Certain molecular analyses may also not be practical or cost-effective for routine clinical implementation, and radiomic-based approaches have the potential to fill that gap. Importantly, radiomics is based on studies already obtained as part of routine patient care, representing an additional added value without added inconvenience to the patient or new cost beyond that of the analytic platform. Although radiomics will not replace pathology or sophisticated molecular profiling, there is potential to reduce biopsies in select circumstances.
Radiomics and Advanced Imaging Techniques
Although many applications of radiomics so far are applied to conventional anatomic images, radiomics and ML can also be applied to advanced imaging techniques including, but not limited to, diffusion-weighted and tensor imaging, perfusion imaging, perfusion and permeability imaging, MR spectroscopy, and functional MRI (52,73,93–98) (Table 3, Table E1 [supplement]). The fusion of radiomics and advanced imaging techniques has the potential to enhance the use of information extracted from advanced imaging techniques in the same way that radiomics and ML can be used to extract additional information with predictive value from standard anatomic images. Indeed, overcoming the practical barriers in the radiomic workflow could by extension result in more workflow-friendly implementation and enhanced use of advanced imaging techniques.
Scientific and Practical Barriers to Radiomic Applications
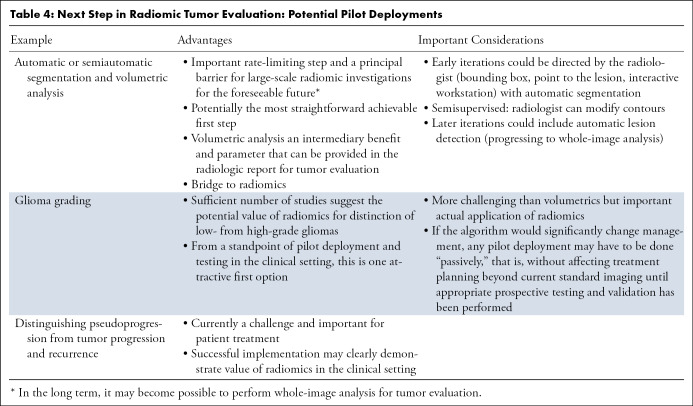
Thus far, a discussion of the many exciting potential applications of radiomics and AI has been provided. Notwithstanding the potential for this technology, it should be evident that significant barriers must be overcome before these applications can be implemented on a routine basis in clinical practice. Currently, image segmentation is often the rate-limiting step in the implementation of radiomic approaches and clinical deployment, at least in the foreseeable future until whole-image or scan analysis becomes possible on a routine basis using DL. Indeed, from a feasibility perspective, one may argue that segmentation would be the most achievable (Table 4). Deployment of a reliable and automated tumor-segmentation algorithm could also provide actual tumor volumes—an additional intermediary benefit—when assessing treatment response in both clinical and research contexts, which would provide supplemental information for assessing current disease response categories.
Table 4:
Next Step in Radiomic Tumor Evaluation: Potential Pilot Deployments
Some of the other barriers have already been alluded to and pertain to reproducibility and standardization of the radiomic process with appropriate quality controls ranging from image processing prior to feature extraction to the mechanics and approaches of the actual feature extraction and prediction algorithm construction (Fig 3, Table 2). The technical variations in radiomic studies were highlighted in a recent systematic review (99) and must be overcome if these approaches are to be reliably deployed and used in the clinical setting. To this end, various important initiatives are being undertaken, such as the “image biomarker standardization initiative” (16). One approach for improving reproducibility of radiomic studies for which there is emerging evidence is the use of DL or CNNs as an image standardization or normalization approach that may then improve the reproducibility of handcrafted radiomic approaches (100,101).
Another fundamental requirement for developing reliable and generalizable DL algorithms is the use of large data sets that are varied and representative of the different techniques and variations that may be encountered at the time of independent testing or deployment of the algorithm. A survey of studies performed thus far for brain tumor characterization shows that the patient numbers used in these studies are invariably small (Table E1 [supplement]). A majority of the studies evaluated fewer than 200 patients, with only a few studies evaluating between 200 and 500 or 500 and 1000 patients. Furthermore, the majority of studies are based out of a single institution. There is a need for large-scale, multi-institutional studies to advance the field. In this sense, multi-institutional collaborations and data sharing will be key in the development of reliable and generalizable algorithms. Thus, there is a need for platforms that enable seamless, secure data sharing, annotation, and algorithm development. These platforms are currently being developed by different vendors. Such collaborative platforms will accelerate algorithm development and translation from a protype to a potentially deployable clinical tool. Ideally, such platforms will be auditable, which could facilitate future regulatory approval.
Although scientific considerations are obviously paramount, one should not ignore the more practical barriers related to workflow as well as regulatory requirements. Without addressing these barriers, radiomics and AI are unlikely to ever realize their true potential. There is an ever-increasing demand on imaging services, and these demands are a challenge for the radiologists who have to interpret the studies as well as the health care system in terms of cost and sustainability. Making the radiomic process workflow friendly and seamless is essential if this technology is to be used routinely in clinical practice. In the bigger picture, some of the challenges of AI implementation may be much more basic and related to the robustness of basic information technology infrastructure within an organization or health system and the accessibility and connectivity of different components that unfortunately may frequently be in silos. These barriers must be broken down to enable seamless and optimal use of the information in a patient’s medical chart and images for high-quality personalized care. For clinical implementation, the radiomic process needs to be automated and seamless. For example, image segmentation could be initiated by clicking on a region of interest, and then an algorithm would work in the “background” with as little radiologist intervention as possible. There is sufficient evidence for pilot deployments of some of these approaches in the clinical setting, establishing their feasibility and, more directly, evaluating their potential value to patient care (Table 4).
It is also important to demonstrate that the radiomic applications have added value, both in terms of quality and timeliness of patient care as well as in cost savings. For example, potential benefits of using radiomic applications could include enabling earlier institution of the optimal treatment regimen, avoiding harmful and potentially toxic costly therapies that have a low likelihood of success, and potentially reducing certain noninvasive procedures and biopsies. These benefits will increase the likelihood that payors and decision makers will facilitate the implementation and adoption of this technology. Last, implementation of radiomics and AI into clinical practice presents unique challenges, especially for the AI component. The current approach for regulatory approval of computer-aided diagnosis and detection may not be suitable for AI algorithms that are designed to “learn” and change for optimal utility. There is a need for development of regulatory processes that are tailored for AI application, which may be achieved by implementation of validated regulatory pathways and precertification of companies proven to be able to develop and monitor algorithm performance reliably, with periodic updates for optimizing performance. All of these represent important challenges that need to be overcome in the coming years for this technology to realize its full potential.
Conclusion
In this article, radiomic analysis and the use of ML for prediction algorithm construction was reviewed. An increasingly large body of literature suggests that radiomic features are useful for characterization of brain tumors, including prediction of tumor histologic characteristics, certain molecular characteristics, and patient survival. There remain significant challenges and barriers to routine implementation of radiomic analysis in clinical practice. However, these challenges are not insurmountable, and radiomics, powered by AI, represents a new horizon in medical imaging and noninvasive diagnostic evaluation of brain tumors. Implementation of these technologies represents an opportunity to further advance the essential role of medical imaging in the care of patients with brain tumors by enabling more precise, personalized tumor characterization, which in turn will direct optimal and personalized therapy.
SUPPLEMENTAL TABLES
R.F. is a clinical research scholar (chercheur-boursier clinicien) supported by the Fonds de Recherche en Santé du Québec (FRQS).
Disclosures of Conflicts of Interest: R.F. Activities related to the present article: disclosed no relevant relationships. Activities not related to the present article: author is consultant to GE Healthcare (FDA 510K review); institution receives research support and has a research agreement that includes beta testing with GE Healthcare; author a paid speaker for one event for GE Healthcare; author receives royalties from Thieme Medical on book edited on skull base imaging; author is founding partner of and has stock in 4intelligent. Other relationships: disclosed no relevant relationships.
Abbreviations:
- AI
- artificial intelligence
- CNN
- convolutional neural network
- DL
- deep learning
- ML
- machine learning
References
- 1.Louis DN, Perry A, Reifenberger G, et al. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol 2016;131(6):803–820. [DOI] [PubMed] [Google Scholar]
- 2.Geraldo LHM, Garcia C, da Fonseca ACC, et al. Glioblastoma therapy in the age of molecular medicine. Trends Cancer 2019;5(1):46–65. [DOI] [PubMed] [Google Scholar]
- 3.Mabray MC, Cha S. Advanced MR imaging techniques in daily practice. Neuroimaging Clin N Am 2016;26(4):647–666. [DOI] [PubMed] [Google Scholar]
- 4.Therasse P, Arbuck SG, Eisenhauer EA, et al. New guidelines to evaluate the response to treatment in solid tumors. European Organization for Research and Treatment of Cancer, National Cancer Institute of the United States, National Cancer Institute of Canada. J Natl Cancer Inst 2000;92(3):205–216. [DOI] [PubMed] [Google Scholar]
- 5.Sharma M, Juthani RG, Vogelbaum MA. Updated response assessment criteria for high-grade glioma: beyond the MacDonald criteria. Chin Clin Oncol 2017;6(4):37. [DOI] [PubMed] [Google Scholar]
- 6.Karu K, Jain AK, Bolle RM. Is there any texture in the image? Pattern Recognit 1996;29(9):1437–1446. [Google Scholar]
- 7.Chen DR, Chang RF, Huang YL. Computer-aided diagnosis applied to US of solid breast nodules by using neural networks. Radiology 1999;213(2):407–412. [DOI] [PubMed] [Google Scholar]
- 8.Tourassi GD. Journey toward computer-aided diagnosis: role of image texture analysis. Radiology 1999;213(2):317–320. [DOI] [PubMed] [Google Scholar]
- 9.Castellano G, Bonilha L, Li LM, Cendes F. Texture analysis of medical images. Clin Radiol 2004;59(12):1061–1069. [DOI] [PubMed] [Google Scholar]
- 10.Davnall F, Yip CS, Ljungqvist G, et al. Assessment of tumor heterogeneity: an emerging imaging tool for clinical practice? Insights Imaging 2012;3(6):573–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ganeshan B, Miles KA. Quantifying tumour heterogeneity with CT. Cancer Imaging 2013;13(1):140–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lubner MG, Smith AD, Sandrasegaran K, Sahani DV, Pickhardt PJ. CT texture analysis: definitions, applications, biologic correlates, and challenges. RadioGraphics 2017;37(5):1483–1503. [DOI] [PubMed] [Google Scholar]
- 13.Julesz B, Gilbert EN, Shepp LA, Frisch HL. Inability of humans to discriminate between visual textures that agree in second-order statistics-revisited. Perception 1973;2(4):391–405. [DOI] [PubMed] [Google Scholar]
- 14.Haralick RM, Shanmugam K, Dinstein I. Textural features for image classification. IEEE Trans Syst Man Cybern 1973;SMC-3(6):610–621. [Google Scholar]
- 15.Gillies RJ, Kinahan PE, Hricak H. Radiomics: images are more than pictures, they are data. Radiology 2016;278(2):563–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zwanenburg A, Leger S, Vallières M, Löck S. Image biomarker standardisation initiative. ArXiv 1612.07003 [preprint] https://arxiv.org/abs/1612.07003. Posted October 23, 2019. Accessed December 1, 2019.
- 17.Forghani R, Savadjiev P, Chatterjee A, Muthukrishnan N, Reinhold C, Forghani B. Radiomics and artificial intelligence for biomarker and prediction model development in oncology. Comput Struct Biotechnol J 2019;17:995–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kumar V, Gu Y, Basu S, et al. Radiomics: the process and the challenges. Magn Reson Imaging 2012;30(9):1234–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lambin P, Rios-Velazquez E, Leijenaar R, et al. Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer 2012;48(4):441–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chang P, Grinband J, Weinberg BD, et al. Deep-learning convolutional neural networks accurately classify genetic mutations in gliomas. AJNR Am J Neuroradiol 2018;39(7):1201–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee H, Yune S, Mansouri M, et al. An explainable deep-learning algorithm for the detection of acute intracranial haemorrhage from small datasets. Nat Biomed Eng 2019;3(3):173–182. [DOI] [PubMed] [Google Scholar]
- 22.Zhou Z, Chen L, Sher D, et al. Predicting lymph node metastasis in head and neck cancer by combining many-objective radiomics and 3-dimensioal convolutional neural network through evidential reasoning. ArXiv e-prints. https://ui.adsabs.harvard.edu/abs/2018arXiv180507021Z. Posted 2018. Accessed May 1, 2018. [DOI] [PMC free article] [PubMed]
- 23.Breiman L. Random forests. Mach Learn 2001;45(1):5–32. [Google Scholar]
- 24.Hinton GE, Osindero S, Teh YW. A fast learning algorithm for deep belief nets. Neural Comput 2006;18(7):1527–1554. [DOI] [PubMed] [Google Scholar]
- 25.Hastie T, Tibshirani R, Friedman J. The elements of statistical learning: data mining, inference, and prediction. 2nd ed. Berlin, Germany: Springer Verlag, 2009. [Google Scholar]
- 26.Lam HK, Ling S, Nguyen HT. Computational intelligence and its applications: evolutionary computation, fuzzy logic, neural network and support vector machine techniques. London, England: Imperial College Press, 2012. [Google Scholar]
- 27.Du KL, Swamy MNS. Neural networks and statistical learning. London, England: Springer, 2014. [Google Scholar]
- 28.Russell SJ, Norvig P. Artificial intelligence: a modern approach. Malaysia: Pearson Education Limited, 2016. [Google Scholar]
- 29.Kubát M. An introduction to machine learning. 2nd ed. Cham, Switzerland: Springer International Publishing, 2017. [Google Scholar]
- 30.Erickson BJ, Korfiatis P, Akkus Z, Kline TL. Machine learning for medical imaging. RadioGraphics 2017;37(2):505–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chartrand G, Cheng PM, Vorontsov E, et al. Deep learning: a primer for radiologists. RadioGraphics 2017;37(7):2113–2131. [DOI] [PubMed] [Google Scholar]
- 32.Nie D, Zhang H, Adeli E, Liu L, Shen D. 3D deep learning for multi-modal imaging-guided survival time prediction of brain tumor patients. Med Image Comput Comput Assist Interv 2016;9901:212–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nie D, Lu J, Zhang H, et al. Multi-channel 3D deep feature learning for survival time prediction of brain tumor patients using multi-modal neuroimages. Sci Rep 2019;9(1):1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li ZC, Bai H, Sun Q, et al. Multiregional radiomics profiling from multiparametric MRI: identifying an imaging predictor of IDH1 mutation status in glioblastoma. Cancer Med 2018;7(12):5999–6009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Laukamp KR, Thiele F, Shakirin G, et al. Fully automated detection and segmentation of meningiomas using deep learning on routine multiparametric MRI. Eur Radiol 2019;29(1):124–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cha YJ, Jang WI, Kim MS, et al. Prediction of response to stereotactic radiosurgery for brain metastases using convolutional neural networks. Anticancer Res 2018;38(9):5437–5445. [DOI] [PubMed] [Google Scholar]
- 37.Papp L, Pötsch N, Grahovac M, et al. Glioma survival prediction with combined analysis of in vivo 11C-MET PET features, ex vivo features, and patient features by supervised machine learning. J Nucl Med 2018;59(6):892–899. [DOI] [PubMed] [Google Scholar]
- 38.Bae S, Choi YS, Ahn SS, et al. Radiomic MRI phenotyping of glioblastoma: improving survival prediction. Radiology 2018;289(3):797–806. [DOI] [PubMed] [Google Scholar]
- 39.Artzi M, Bressler I, Ben Bashat D. Differentiation between glioblastoma, brain metastasis and subtypes using radiomics analysis. J Magn Reson Imaging 2019;50(2):519–528. [DOI] [PubMed] [Google Scholar]
- 40.Qian Z, Li Y, Wang Y, et al. Differentiation of glioblastoma from solitary brain metastases using radiomic machine-learning classifiers. Cancer Lett 2019;451:128–135. [DOI] [PubMed] [Google Scholar]
- 41.Shrot S, Salhov M, Dvorski N, Konen E, Averbuch A, Hoffmann C. Application of MR morphologic, diffusion tensor, and perfusion imaging in the classification of brain tumors using machine learning scheme. Neuroradiology 2019;61(7):757–765. [DOI] [PubMed] [Google Scholar]
- 42.Kim Y, Cho HH, Kim ST, Park H, Nam D, Kong DS. Radiomics features to distinguish glioblastoma from primary central nervous system lymphoma on multi-parametric MRI. Neuroradiology 2018;60(12):1297–1305. [DOI] [PubMed] [Google Scholar]
- 43.Nakagawa M, Nakaura T, Namimoto T, et al. Machine learning based on multi-parametric magnetic resonance imaging to differentiate glioblastoma multiforme from primary cerebral nervous system lymphoma. Eur J Radiol 2018;108:147–154. [DOI] [PubMed] [Google Scholar]
- 44.Kunimatsu A, Kunimatsu N, Yasaka K, et al. Machine learning-based texture analysis of contrast-enhanced MR imaging to differentiate between glioblastoma and primary central nervous system lymphoma. Magn Reson Med Sci 2019;18(1):44–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dong F, Li Q, Xu D, et al. Differentiation between pilocytic astrocytoma and glioblastoma: a decision tree model using contrast-enhanced magnetic resonance imaging-derived quantitative radiomic features. Eur Radiol 2019;29(8):3968–3975. [DOI] [PubMed] [Google Scholar]
- 46.Chen X, Tong Y, Shi Z, et al. Noninvasive molecular diagnosis of craniopharyngioma with MRI-based radiomics approach. BMC Neurol 2019;19(1):6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Niu L, Zhou X, Duan C, et al. Differentiation researches on the meningioma subtypes by radiomics from contrast-enhanced magnetic resonance imaging: a preliminary study. World Neurosurg 2019;126:e646–e652. [DOI] [PubMed] [Google Scholar]
- 48.Zhang S, Song G, Zang Y, et al. Non-invasive radiomics approach potentially predicts non-functioning pituitary adenomas subtypes before surgery. Eur Radiol 2018;28(9):3692–3701. [DOI] [PubMed] [Google Scholar]
- 49.Kniep HC, Madesta F, Schneider T, et al. Radiomics of brain MRI: utility in prediction of metastatic tumor type. Radiology 2019;290(2):479–487. [DOI] [PubMed] [Google Scholar]
- 50.Hu LS, Ning S, Eschbacher JM, et al. Multi-parametric MRI and texture analysis to visualize spatial histologic heterogeneity and tumor extent in glioblastoma. PLoS One 2015;10(11):e0141506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ranjith G, Parvathy R, Vikas V, Chandrasekharan K, Nair S. Machine learning methods for the classification of gliomas: initial results using features extracted from MR spectroscopy. Neuroradiol J 2015;28(2):106–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang G, Nawaz T, Barrick TR, Howe FA, Slabaugh G. Discrete wavelet transform-based whole-spectral and subspectral analysis for improved brain tumor clustering using single voxel MR spectroscopy. IEEE Trans Biomed Eng 2015;62(12):2860–2866. [DOI] [PubMed] [Google Scholar]
- 53.Li-Chun Hsieh K, Chen CY, Lo CM. Quantitative glioma grading using transformed gray-scale invariant textures of MRI. Comput Biol Med 2017;83:102–108. [DOI] [PubMed] [Google Scholar]
- 54.Zhang X, Yan LF, Hu YC, et al. Optimizing a machine learning based glioma grading system using multi-parametric MRI histogram and texture features. Oncotarget 2017;8(29):47816–47830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cho HH, Lee SH, Kim J, Park H. Classification of the glioma grading using radiomics analysis. PeerJ 2018;6:e5982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ditmer A, Zhang B, Shujaat T, et al. Diagnostic accuracy of MRI texture analysis for grading gliomas. J Neurooncol 2018;140(3):583–589. [DOI] [PubMed] [Google Scholar]
- 57.Su C, Jiang J, Zhang S, et al. Radiomics based on multicontrast MRI can precisely differentiate among glioma subtypes and predict tumour-proliferative behaviour. Eur Radiol 2019;29(4):1986–1996. [DOI] [PubMed] [Google Scholar]
- 58.Vamvakas A, Williams SC, Theodorou K, et al. Imaging biomarker analysis of advanced multiparametric MRI for glioma grading. Phys Med 2019;60:188–198. [DOI] [PubMed] [Google Scholar]
- 59.Sengupta A, Agarwal S, Gupta PK, et al. On differentiation between vasogenic edema and non-enhancing tumor in high-grade glioma patients using a support vector machine classifier based upon pre and post-surgery MRI images. Eur J Radiol 2018;106:199–208. [DOI] [PubMed] [Google Scholar]
- 60.Zhang B, Chang K, Ramkissoon S, et al. Multimodal MRI features predict isocitrate dehydrogenase genotype in high-grade gliomas. Neuro Oncol 2017;19(1):109–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Arita H, Kinoshita M, Kawaguchi A, et al. Lesion location implemented magnetic resonance imaging radiomics for predicting IDH and TERT promoter mutations in grade II/III gliomas. Sci Rep 2018;8(1):11773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Park YW, Han K, Ahn SS, et al. Whole-tumor histogram and texture analyses of DTI for evaluation of IDH1-mutation and 1p/19q-codeletion status in World Health Organization grade II gliomas. AJNR Am J Neuroradiol 2018;39(4):693–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Park YW, Han K, Ahn SS, et al. Prediction of IDH1-mutation and 1p/19q-codeletion status using preoperative MR imaging phenotypes in lower grade gliomas. AJNR Am J Neuroradiol 2018;39(1):37–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kuthuru S, Deaderick W, Bai H, et al. A visually interpretable, dictionary-based approach to imaging-genomic modeling, with low-grade glioma as a case study. Cancer Inform 2018;17:1176935118802796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lee MH, Kim J, Kim ST, et al. Prediction of IDH1 mutation status in glioblastoma using machine learning technique based on quantitative radiomic data. World Neurosurg 2019;125:e688–e696. [DOI] [PubMed] [Google Scholar]
- 66.Wu S, Meng J, Yu Q, Li P, Fu S. Radiomics-based machine learning methods for isocitrate dehydrogenase genotype prediction of diffuse gliomas. J Cancer Res Clin Oncol 2019;145(3):543–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Akkus Z, Ali I, Sedlář J, et al. Predicting deletion of chromosomal arms 1p/19q in low-grade gliomas from MR images using machine intelligence. J Digit Imaging 2017;30(4):469–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shofty B, Artzi M, Ben Bashat D, et al. MRI radiomics analysis of molecular alterations in low-grade gliomas. Int J Comput Assist Radiol Surg 2018;13(4):563–571. [DOI] [PubMed] [Google Scholar]
- 69.Korfiatis P, Kline TL, Coufalova L, et al. MRI texture features as biomarkers to predict MGMT methylation status in glioblastomas. Med Phys 2016;43(6):2835–2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Macyszyn L, Akbari H, Pisapia JM, et al. Imaging patterns predict patient survival and molecular subtype in glioblastoma via machine learning techniques. Neuro Oncol 2016;18(3):417–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li Y, Qian Z, Xu K, et al. MRI features predict p53 status in lower-grade gliomas via a machine-learning approach. Neuroimage Clin 2017;17:306–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Larroza A, Moratal D, Paredes-Sánchez A, et al. Support vector machine classification of brain metastasis and radiation necrosis based on texture analysis in MRI. J Magn Reson Imaging 2015;42(5):1362–1368. [DOI] [PubMed] [Google Scholar]
- 73.Zhang J, Yu H, Qian X, et al. Pseudo progression identification of glioblastoma with dictionary learning. Comput Biol Med 2016;73:94–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tiwari P, Prasanna P, Wolansky L, et al. Computer-extracted texture features to distinguish cerebral radionecrosis from recurrent brain tumors on multiparametric MRI: a feasibility study. AJNR Am J Neuroradiol 2016;37(12):2231–2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Peng L, Parekh V, Huang P, et al. Distinguishing true progression from radionecrosis after stereotactic radiation therapy for brain metastases with machine learning and radiomics. Int J Radiat Oncol Biol Phys 2018;102(4):1236–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chang K, Zhang B, Guo X, et al. Multimodal imaging patterns predict survival in recurrent glioblastoma patients treated with bevacizumab. Neuro Oncol 2016;18(12):1680–1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Liu L, Zhang H, Rekik I, Chen X, Wang Q, Shen D. Outcome prediction for patient with high-grade gliomas from brain functional and structural networks. Med Image Comput Comput Assist Interv 2016;9901:26–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Liu Y, Xu X, Yin L, Zhang X, Li L, Lu H. Relationship between glioblastoma heterogeneity and survival time: an MR imaging texture analysis. AJNR Am J Neuroradiol 2017;38(9):1695–1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhou M, Chaudhury B, Hall LO, Goldgof DB, Gillies RJ, Gatenby RA. Identifying spatial imaging biomarkers of glioblastoma multiforme for survival group prediction. J Magn Reson Imaging 2017;46(1):115–123. [DOI] [PubMed] [Google Scholar]
- 80.Liu Y, Zhang X, Feng N, et al. The effect of glioblastoma heterogeneity on survival stratification: a multimodal MR imaging texture analysis. Acta Radiol 2018;59(10):1239–1246. [DOI] [PubMed] [Google Scholar]
- 81.Liu X, Li Y, Qian Z, et al. A radiomic signature as a non-invasive predictor of progression-free survival in patients with lower-grade gliomas. Neuroimage Clin 2018;20:1070–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sanghani P, Ang BT, King NKK, Ren H. Overall survival prediction in glioblastoma multiforme patients from volumetric, shape and texture features using machine learning. Surg Oncol 2018;27(4):709–714. [DOI] [PubMed] [Google Scholar]
- 83.Chen X, Fang M, Dong D, et al. Development and validation of a MRI-based radiomics prognostic classifier in patients with primary glioblastoma multiforme. Acad Radiol 2019;26(10):1292–1300. [DOI] [PubMed] [Google Scholar]
- 84.Li C, Wang S, Serra A, et al. Multi-parametric and multi-regional histogram analysis of MRI: modality integration reveals imaging phenotypes of glioblastoma. Eur Radiol 2019;29(9):4718–4729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kim BS, Kim ST, Kim JH, et al. Apparent diffusion coefficient as a predictive biomarker for survival in patients with treatment-naive glioblastoma using quantitative multiparametric magnetic resonance profiling. World Neurosurg 2019;122:e812– e820. [DOI] [PubMed] [Google Scholar]
- 86.Wu G, Shi Z, Chen Y, et al. A sparse representation-based radiomics for outcome prediction of higher grade gliomas. Med Phys 2019;46(1):250–261. [DOI] [PubMed] [Google Scholar]
- 87.Zhang X, Lu H, Tian Q, et al. A radiomics nomogram based on multiparametric MRI might stratify glioblastoma patients according to survival. Eur Radiol 2019;29(10):5528–5538. [DOI] [PubMed] [Google Scholar]
- 88.Niu J, Zhang S, Ma S, et al. Preoperative prediction of cavernous sinus invasion by pituitary adenomas using a radiomics method based on magnetic resonance images. Eur Radiol 2019;29(3):1625–1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lipkova J, Angelikopoulos P, Wu S, et al. Personalized radiotherapy design for glioblastoma: integrating mathematical tumor models, multimodal scans, and Bayesian inference. IEEE Trans Med Imaging 2019;38(8):1875–1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gates EDH, Lin JS, Weinberg JS, et al. Guiding the first biopsy in glioma patients using estimated Ki-67 maps derived from MRI: conventional versus advanced imaging. Neuro Oncol 2019;21(4):527–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Fathi Kazerooni A, Mohseni M, Rezaei S, Bakhshandehpour G, Saligheh Rad H. Multi-parametric (ADC/PWI/T2-w) image fusion approach for accurate semi-automatic segmentation of tumorous regions in glioblastoma multiforme. MAGMA 2015;28(1):13–22. [DOI] [PubMed] [Google Scholar]
- 92.Fathi Kazerooni A, Nabil M, Zeinali Zadeh M, et al. Characterization of active and infiltrative tumorous subregions from normal tissue in brain gliomas using multiparametric MRI. J Magn Reson Imaging 2018;48(4):938–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kanas VG, Zacharaki EI, Thomas GA, Zinn PO, Megalooikonomou V, Colen RR. Learning MRI-based classification models for MGMT methylation status prediction in glioblastoma. Comput Methods Programs Biomed 2017;140:249–257. [DOI] [PubMed] [Google Scholar]
- 94.Kickingereder P, Bonekamp D, Nowosielski M, et al. Radiogenomics of glioblastoma: machine learning-based classification of molecular characteristics by using multiparametric and multiregional MR imaging features. Radiology 2016;281(3):907–918. [DOI] [PubMed] [Google Scholar]
- 95.Emblem KE, Due-Tonnessen P, Hald JK, et al. Machine learning in preoperative glioma MRI: survival associations by perfusion-based support vector machine outperforms traditional MRI. J Magn Reson Imaging 2014;40(1):47–54. [DOI] [PubMed] [Google Scholar]
- 96.Emblem KE, Pinho MC, Zöllner FG, et al. A generic support vector machine model for preoperative glioma survival associations. Radiology 2015;275(1):228–234. [DOI] [PubMed] [Google Scholar]
- 97.Artzi M, Liberman G, Nadav G, et al. Differentiation between treatment-related changes and progressive disease in patients with high grade brain tumors using support vector machine classification based on DCE MRI. J Neurooncol 2016;127(3):515–524. [DOI] [PubMed] [Google Scholar]
- 98.Gurbani SS, Sheriff S, Maudsley AA, Shim H, Cooper LAD. Incorporation of a spectral model in a convolutional neural network for accelerated spectral fitting. Magn Reson Med 2019;81(5):3346–3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Traverso A, Wee L, Dekker A, Gillies R. Repeatability and reproducibility of radiomic features: a systematic review. Int J Radiat Oncol Biol Phys 2018;102(4):1143–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Drozdzal M, Chartrand G, Vorontsov E, et al. Learning normalized inputs for iterative estimation in medical image segmentation. Med Image Anal 2018;44:1–13. [DOI] [PubMed] [Google Scholar]
- 101.Choe J, Lee SM, Do KH, et al. Deep learning-based image conversion of CT reconstruction kernels improves radiomics reproducibility for pulmonary nodules or masses. Radiology 2019;292(2):365–373. [DOI] [PubMed] [Google Scholar]
- 102.Shen C, Liu Z, Guan M, et al. 2D and 3D CT radiomics features prognostic performance comparison in non-small cell lung cancer. Transl Oncol 2017;10(6):886–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Yang L, Yang J, Zhou X, et al. Development of a radiomics nomogram based on the 2D and 3D CT features to predict the survival of non-small cell lung cancer patients. Eur Radiol 2019;29(5):2196–2206. [DOI] [PubMed] [Google Scholar]
- 104.Zhao B, Tan Y, Tsai WY, Schwartz LH, Lu L. Exploring variability in CT characterization of tumors: a preliminary phantom study. Transl Oncol 2014;7(1):88–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Mackin D, Fave X, Zhang L, et al. Measuring computed tomography scanner variability of radiomics features. Invest Radiol 2015;50(11):757–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Nyflot MJ, Yang F, Byrd D, Bowen SR, Sandison GA, Kinahan PE. Quantitative radiomics: impact of stochastic effects on textural feature analysis implies the need for standards. J Med Imaging (Bellingham) 2015;2(4):041002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zhao B, Tan Y, Tsai WY, et al. Reproducibility of radiomics for deciphering tumor phenotype with imaging. Sci Rep 2016;6(1):23428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Shafiq-Ul-Hassan M, Zhang GG, Latifi K, et al. Intrinsic dependencies of CT radiomic features on voxel size and number of gray levels. Med Phys 2017;44(3):1050–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Baeßler B, Weiss K, Pinto Dos Santos D. Robustness and reproducibility of radiomics in magnetic resonance imaging: a phantom study. Invest Radiol 2019;54(4):221–228. [DOI] [PubMed] [Google Scholar]
- 110.Tixier F, Um H, Bermudez D, et al. Preoperative MRI-radiomics features improve prediction of survival in glioblastoma patients over MGMT methylation status alone. Oncotarget 2019;10(6):660–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.