ABSTRACT
Objective
Identifying molecular changes that contribute to the onset and progression of Huntington's disease (HD) is of importance for the development and evaluation of potential therapies.
Methods
We conducted an unbiased mass‐spectrometry proteomic analysis on the cerebrospinal fluid of 12 manifest HD patients (ManHD), 13 pre‐manifest (preHD), and 38 controls. A biologically plausible and significant possible biomarker was validated in samples from a separate cohort of patients and controls consisting of 23 ManHD patients and 23 controls.
Results
In ManHD compared to preHD, 10 proteins were downregulated and 43 upregulated. Decreased levels of proenkephalin (PENK) and transthyretin were closely linked to HD symptom severity, whereas levels of 15 upregulated proteins were associated with symptom severity. The decreased PENK levels were replicated in the separate cohort where absolute quantitation was performed.
Conclusions
We hypothesize that declining PENK levels reflect the degeneration of medium spiny neurons (MSNs) that produce PENK and that assays for PENK may serve as a surrogate marker for the state of MSNs in HD. © 2020 The Authors. Movement Disorders published by Wiley Periodicals LLC on behalf of International Parkinson and Movement Disorder Society
Keywords: Huntington's disease; biomarkers; proenkephalin; proteomics
Huntington's disease (HD) is a severe monogenic neurodegenerative disorder with a prevalence of 5.7 of 100,000 in populations of European ancestry. 1 Although the ultimate cause for HD, a CAG‐expansion in the HTT‐gene, has been well known for almost 30 years, we still lack understanding of the underlying pathophysiological process, and there are no proven disease‐modifying therapies.
Biomarkers for HD have been investigated and developed, including magnetic resonance imaging (MRI) protocols and biochemical markers as neurofilament light chain protein (NFL) 2 , 3 , 4 and mutant huntingtin protein (mHTT). 5 These allow for disease progression monitoring and have been introduced as surrogate endpoints in clinical trials. 6
HD is characterized by motor symptoms (initially usually choreatic movements), psychiatric symptoms (that may manifest as a wide variety of symptoms), and cognitive decline. Initial symptoms may vary, but clinically the motor symptoms are generally used to establish disease onset. 7 This may not reflect fully the neurodegenerative process, because many patients experience severe psychiatric and/or cognitive symptoms before onset of motor symptoms.
Several ongoing and planned trials are targeting disease‐specific processes such as mRNA coding for huntingtin (HTT). 8 This approach has recently shown promising results with a decrease of mHTT in cerebrospinal fluid (CSF) in HD patients. 6
Still, there is insufficient understanding of the processes underlying symptom development in HD, and data suggest roles for mitochondrial dysfunction, intracellular transportation systems, and neuroinflammation, as well as other processes. 7 With the possible advent of disease‐modifying therapies, biomarkers for disease onset—more specific and objective than motor symptom scoring—are needed.
Unbiased (or “hypothesis‐free”) proteomics has the potential to identify new protein markers tied to pathophysiological processes in the brains of HD patients reflected in CSF. Two previous studies investigated potential markers in HD. 9 , 10 It remains to conduct a study including pre‐manifest HD patients (preHD) combined with capability to identify all proteins detected. In the light of this, we conducted a liquid chromatography‐mass spectrometry (LC‐MS) proteomics study of CSF from a clinically well‐characterized HD cohort of different disease stages (including preHD) to identify proteins linked to disease progression.
Methods
Enrollment of Study Participants
The cohort included 12 manifest HD patients (5 women, mean age 51.5), 13 preHD (6 women, mean age 36.2), and 38 matched controls (26 women, mean age 46.2), enrolled at Uppsala University Hospital. preHD was defined as individuals with an HTT gene expansion (>36 repeats) and a diagnostic confidence level below four, whereas manifest HD was defined by an HTT gene expansion (>36 repeats) and diagnostic confidence level of four. 11 Additionally, a validation cohort was included in the absolute quantitation of PENK, comprising 23 manifest HD patients and 23 controls enrolled from Sahlgrenska University Hospital in Gothenburg and Karolinska University Hospital in Stockholm.
Standard Protocol Approvals, Registrations, and Patient Consents
The study was approved by the regional ethics review board in Uppsala (No. 2012/274) and all participants provided their written informed consent.
Clinical Assessment
All HD gene carriers were assessed with a battery of clinical tests including unified Huntington's disease rating scale (UHDRS) total motor score (TMS), 12 Symbol digit modality test (SDMT), Stroop interference (S‐I), Stroop word reading (S‐W), Stroop color naming (S‐C), category verbal fluency (CVF), letter verbal fluency (LVF), and total functional capacity (TFC). 13 Composite UHDRS (cUHDRS) scores were calculated by combining TFC, TMS, SDMT, and S‐W according to the formula published by Schobel et al. 14
The CAG repeat number and age were used to calculate the extent of exposure to expanded CAG repeat, (CAGn‐35.5)*age (CAP score), a statistic that reflects the level of accumulated genetic toxicity. CAP score is a robust predictor of the clinical HD phenotype. 15 Five‐year probability of disease onset was estimated using age and repeat number. 16
Sample Collection
CSF was collected at the same visit as the clinical assessment. The procedure was standardized as previously described. 17 CSF from matched controls was obtained from a biobank at the same clinic consisting of patients who had undergone lumbar puncture for investigation purposes and in whom neurological disease had been ruled out. The procedure was similar to that of the HD sample collection and has been published before. 18 Controls were selected according to the following exclusion criteria: previously known disorder of the central nervous system (CNS); investigation that resulted in a CNS disorder (inflammatory, degenerative, or infectious) or other significant medical diagnosis (eg, cancer); significantly abnormal standard analyses of CSF (eg, cells, albumin, glucose, and lactate) or other CSF analyses (where available); and significant neuroradiological abnormalities (where available).
Experimental Procedures
Chemicals and Reagents
Acetonitrile (ACN), methanol (MeOH), acetic acid (HAc), formic acid (FA), ammonium bicarbonate (NH4HCO3) were obtained from Merck (Darmstadt, Germany). Acetone, protease inhibitor cocktail, phosphate buffered saline (PBS), and trifluoroacetic acid (TFA) were purchased from Sigma Aldrich (St. Louis, MO). For tryptic digestion iodoacetamide (IAA), urea and dithiothreitol (DTT) were obtained from Sigma Aldrich and trypsin/Lys‐C mixture (mass spectrometry grade) was obtained from Promega (Mannheim, Germany). Ultrapure water was prepared by Milli‐Q water purification system (Millipore, Bedford, MA).
Sample Preparation
Before nanoLC‐MS/MS analysis, the low abundant proteins were enriched through depletion of the seven most highly abundant proteins (albumin, IgG, α‐1‐antitrypsin, IgA, haptoglobin, transferrin, and fibrinogen) using a human multiple affinity removal (MARS) spin cartridge‐Hu‐7 (Agilent Technologies, Palo Alto, CA). Immunodepletion using the MARS spin cartridge was performed according to the manufacturer's instruction. Before depletion, 19 an aliquot of 260 μL of each CSF sample was filtered through a 0.22 μm cellulose acetate spin filters (Agilent Technologies) by centrifugation at 15,000 g for 2 minutes. To minimize any proteolytic degradation during the depletion, the samples were kept at 6°C whenever possible. Next, an aliquot of 250 μL of each CSF sample was loaded onto the spin cartridge and the flow‐through (FT) fraction was collected by centrifugation for 2 minutes at 100 g. Two successive wash steps with 400 μL of MARS‐7 Buffer A were carried out to obtain maximum yield. The flow‐through and wash (W) fractions were combined. The spin cartridge was washed with 2 mL of MARS‐7 Buffer B to remove bound proteins and was then re‐equilibrated with Buffer A. The remaining fractions (FT+W) were dried using a SpeedVac (Thermo Fisher Scientific, Waltham, MA).
The proteins in the depleted CSF sample were digested using a trypsin/Lys‐C mixture. Briefly, the proteins were re‐dissolved in 50 μL of digestion buffer (6 M urea, 50 mM NH4HCO3). Ten microliters of 45 mM aqueous DTT was then added to all samples and the mixtures were incubated at 50°C for 150 minutes to reduce the disulfide bridges. The samples were cooled to room temperature (RT) and 10 μL of 100 mM aqueous IAA was added before incubating the mixtures for an additional 15 minutes at RT in darkness to carabamidomethylate the cysteines. Finally, a volume of 50 μL of 100 mM NH4HCO3 was added to all samples followed by the trypsin/Lys‐C mixture dissolved in 500 mM NH4HCO3, yielding a final trypsin/protein concentration of 5% (w/w). The tryptic digestion was performed overnight at 37°C. Before MS analysis, the peptides were purified and desalted by Pierce C18 Spin Columns (Thermo Fisher Scientific). These columns were activated by 2 × 200 μL of 50% ACN and equilibrated with 2 × 200 μL of 0.5% TFA. The tryptic peptides were adsorbed to the media using two repeated cycles of 40 μL sample loading and the column was washed using 3 × 200 μL of 0.5% TFA. Finally, the peptides were eluted in 3 × 50 μL of 70% ACN and dried.
LC‐MS/MS
The samples were analyzed using a QExactive Plus Orbitrap mass spectrometer (Thermo Fisher Scientific, Bremen, Germany) equipped with a nano electrospray ion source. The peptides were separated by reversed phase liquid chromatography using an EASY‐nLC 1000 system (Thermo Fisher Scientific). A set‐up of pre‐column and analytical column was used. The pre‐column was a 2 cm EASY‐column (1D 100 μm, 5 μm C18) (Thermo Fisher Scientific) while the analytical column was a 10 cm EASY‐column (ID 75 μm, 3 μm, C18; Thermo Fisher Scientific). Peptides were eluted with a 150 minutes linear gradient from 4% to 100% ACN at 250 nL min/L. The mass spectrometer was operated in positive ion mode acquiring a survey mass spectrum with resolving power 70,000 (full width half maximum), m/z = 400–1750 using an automatic gain control (AGC) target of 3 × 106. The 10 most intense ions were selected for higher‐energy collisional dissociation (HCD) fragmentation (25% normalized collision energy) and MS/MS spectra were generated with an AGC target of 5 × 105 at a resolution of 17,500. The mass spectrometer worked in data‐dependent mode. Quantitation is relative between the samples run together. The instrument is subject to “drift” during an experiment, which can cause false differences, if for instance samples belonging to the same group are run in a sequence. The sample run order was planned to avoid this type of bias.
Mass Spectrometry Data Handling
The acquired data (.RAW‐files) were processed in MaxQuant 1.5.1.2,20 and database searches were performed using the implemented Andromeda search engine. MS/MS spectra were correlated to a FASTA database containing proteins from Homo sapiens extracted from the Uniprot database (release May 2018). A decoy search database, including common contaminants and a reverse database, was used to estimate the identification false discovery rate (FDR). An FDR of 1% was accepted. The search parameters included: maximum 10 ppm and 0.6 Da error tolerances for the survey scan and MS/MS analysis, respectively; enzyme specificity was trypsin; maximum one missed cleavage site allowed; cysteine carbamidomethylation was set as static modification and oxidation (M) was set as variable modification. The search criteria for protein identification were set to at least two matching peptides. Label free quantification was applied for comparative proteomics.
Absolute Quantification of Proenkephalin
The MS experiment was conducted as described above except that the heavy‐labeled synthetic peptide ELLETGDNR of proenkephalin (6.25 pmol/250 μL CSF or 0.025 nmol/L CSF) was spiked into the digests. Thereafter, the samples were desalted using the SPE Pierce C18 Spin Columns (Thermo Fisher Scientific) as described above. Dried peptides were resolved in 50 μL of 0.1% formic acid and further diluted 4 times before nano‐LC‐MS/MS. For data analysis, the SkyLine 3.7.0 software preloaded with peptide sequence ELLETGDNR of proenkephalin was used to identify fragment ions and to extract peak area for the peptide of interest.
Statistics
A pre‐specified data analysis was applied to curate proteins of interest as markers of disease progression. Only proteins that were identified in at least 80% of samples were subjected to statistical testing. Proteins with either up‐ or downregulated levels were identified (ManHD vs preHD, using t tests). Proteins with age‐dependent levels were identified (Spearman rank correlations) and in such cases the age‐dependent contribution estimated from the control group was removed in the comparison between ManHD and preHD, using linear regression. Finally, the proteins that also had moderate‐ to strong correlations (Spearman ρ > 0.5) with cUHDRS were considered as of interest. The level of significance was set at P < 0.05.
Data Availability Statement
Raw data sets including all quantified protein levels can be made available on request.
Results
A total of 1,045 unique proteins (protein groups) were identified by LC‐MS. Potential effects of differing storage time on protein levels were tested in the control group. Only phosphatidylcholine sterol acyltransferase (LCAT) was significantly correlated with days stored (r = 0.342, P = 0.041). The number of proteins identified did not differ between groups (Kruskal‐Wallis test, P = 0.358).
The levels of two proteins were higher in men compared with women in the control group (ACTG1, P = 0.021; COL1A1, P = 0.047).
Proteomic Differences Between HD Gene Expansion Carriers and Controls
Altered protein levels in all gene expansion carriers (ManHD and preHD) versus controls were identified (Tables 1 and 2). Of the 371 proteins that were identified in at least 80% of samples, the levels of 38 proteins significantly differed between groups (26 proteins were upregulated and 12 downregulated; Table 1). The most significantly upregulated proteins were Adipocyte enhancer‐binding protein 1 (AEBP1, P = <0.0001) and latent‐transforming growth factor β‐binding protein 4 (LTBP4, P = 0.0106), with a ratio of over 1.3 between gene expansion carriers versus controls. Proenkephalin‐A (PENK, P = 0.0104) was most significantly downregulated with a ratio of 0.72. Adjustment for age was not performed as the two groups were age matched.
TABLE 1.
Regulated proteins in HD gene expansion carriers vs controls
| Gene names | Protein names | ↓ | ↑ | Ratio HD/C | P value |
| AEBP1 | Adipocyte enhancer‐binding protein 1 | ↑ | 1.43 | <0.0001 | |
| LTBP4 | Latent‐transforming growth factor β‐binding protein 4 | ↑ | 1.34 | 0.0106 | |
| APOA4 | Apolipoprotein A‐IV | ↑ | 1.27 | 0.0227 | |
| PRELP;MST161 | Prolargin | ↑ | 1.24 | 0.0006 | |
| SORCS3 | VPS10 domain‐containing receptor SorCS3 | ↑ | 1.24 | 0.0002 | |
| RAB1;RNASE4 | Ribonuclease 4 | ↑ | 1.22 | 0.0001 | |
| PLG | Plasminogen | ↑ | 1.22 | 0.0436 | |
| RARRES2 | Retinoic acid receptor responder protein 2 | ↑ | 1.20 | 0.0006 | |
| ECM2 | Extracellular matrix protein 2 | ↑ | 1.20 | 0.0103 | |
| CLU | Clusterin | ↑ | 1.20 | <0.0001 | |
| PCSK1N | ProSAAS | ↑ | 1.19 | 0.0017 | |
| C4A | Complement C4‐A | ↑ | 1.17 | 0.0305 | |
| IGFBP2 | Insulin‐like growth factor‐binding protein 2 | ↑ | 1.17 | <0.0001 | |
| PROC | Vitamin K‐dependent protein C | ↑ | 1.17 | 0.0488 | |
| GNPTG;RJD9 | N‐acetylglucosamine‐1‐phosphotransferase subunit gamma | ↑ | 1.15 | 0.0058 | |
| LDHB | L‐lactate dehydrogenase | ↑ | 1.15 | 0.0061 | |
| ITIH5 | Inter‐α‐trypsin inhibitor heavy chain H5 | ↑ | 1.15 | 0.0355 | |
| ACTG1 | Actin, cytoplasmic 2 | ↑ | 1.14 | 0.0371 | |
| COL18A1 | Collagen α‐1(XVIII) chain; endostatin | ↑ | 1.13 | 0.0069 | |
| THBS2 | Thrombospondin‐2 | ↑ | 1.13 | 0.0179 | |
| PPIA | Peptidyl‐prolyl cis‐trans isomerase | ↑ | 1.12 | 0.0058 | |
| IGFBP4 | Insulin‐like growth factor‐binding protein 4 | ↑ | 1.12 | 0.0460 | |
| CLEC3B | Tetranectin | ↑ | 1.10 | 0.0064 | |
| GSN | Gelsolin | ↑ | 1.09 | 0.0285 | |
| PCOLCE | Procollagen C‐endopeptidase enhancer 1 | ↑ | 1.07 | 0.0306 | |
| QSOX1 | Sulfhydryl oxidase 1 | ↑ | 1.06 | 0.0428 | |
| SPARCL1 | SPARC‐like protein 1 | ↓ | 0.92 | 0.0483 | |
| SEZ6 | Seizure protein 6 homolog | ↓ | 0.91 | 0.0385 | |
| CLSTN1 | Calsyntenin‐1; soluble Alc‐α; CTF1‐α | ↓ | 0.89 | 0.0267 | |
| ALCAM | CD166 antigen | ↓ | 0.89 | 0.0141 | |
| NELL2 | Protein kinase C‐binding protein NELL2 | ↓ | 0.88 | 0.0352 | |
| L1CAM | Neural cell adhesion molecule L1 | ↓ | 0.88 | 0.0200 | |
| CSPG3 | Neurocan core protein | ↓ | 0.88 | 0.0255 | |
| C4orf48 | Neuropeptide‐like protein C4orf48 | ↓ | 0.83 | 0.0093 | |
| LGALS3BP | Galectin‐3‐binding protein | ↓ | 0.79 | 0.0001 | |
| WFDC2 | WAP four‐disulfide core domain protein 2 | ↓ | 0.79 | 0.0008 | |
| UBB | Ubiquitin‐60S ribosomal protein L40 | ↓ | 0.77 | 0.0253 | |
| PENK | Proenkephalin‐A | ↓ | 0.72 | 0.0104 |
TABLE 2.
Top canonical pathways altered in HD gene expansion carriers vs controls
| Name | P value | Overlap |
| Clathrin‐mediated endocytosis signaling | 4.19E−04 | 1.9% 4/207 |
| LXR/RXR activation | 1.15E −03 | 2.5% 3/121 |
| FXR/RXR activation | 1.29E −03 | 2.4% 3/126 |
| Atherosclerosis signaling | 1.32E −03 | 2.4% 3/127 |
| Coagulation system | 1.63E −03 | 5.7% 2/35 |
Proteomic Differences Between Manifest and Pre‐Manifest HD
Altered protein levels in the ManHD group versus the preHD group were identified. Of the 376 proteins that were identified in at least 80% of samples, the levels of 68 proteins significantly differed between the manifest group and the preHD group.
Twenty‐one proteins (31%) of the 68 investigated were associated with ageing. The age‐dependent contribution was estimated in the control group and removed in the comparison between the manifest patients and preHD, using linear regression. PENK (P = 0.005), transthyretin (TTR, P = 0.011), carboxypeptidase E (CPE, P = 0.044), protocadherin FAT2 (FAT2, P = 0.001), and vasorin (VASN, P = 0.034) remained significantly altered, whereas clusterin was borderline significant (CLU, P = 0.05). The association for the remaining 15 proteins of the 21 age‐dependent proteins did not survive age‐adjustment, leaving a total of 53 changed proteins (Table 3). Biological functions of protein functions were curated manually. The four proteins PENK, CLU, PPIA, and ACTG1 were significantly altered in both comparisons (HD vs controls and ManHD vs preHD).
TABLE 3.
Regulated proteins manifest HD vs pre‐manifest gene expansion carriers after age‐adjustment
| Main biological function | Gene names | Protein names | ↓ | ↑ | Ratio ManHD/pGEC | P value | |
| Un‐adjusted | Age‐adjusted | ||||||
| Immune response | C1S | Complement C1s subcomponent | ↑ | 1.094 | 0.0413 | N/A | |
| C7 | Complement component C7 | ↑ | 1.292 | 0.0023 | N/A | ||
| CFHR1 | Complement factor H‐related protein 1 | ↓ | 0.705 | 0.0121 | N/A | ||
| CD44 | CD44 antigen | ↑ | 1.436 | 0.0045 | N/A | ||
| IL6ST | Interleukin‐6 receptor subunit β | ↑ | 1.262 | 0.0121 | N/A | ||
| ORM2 | α‐1‐acid glycoprotein 2 | ↓ | 0.649 | 0.0034 | N/A | ||
| Lipid metabolism | GM2A | Ganglioside GM2 activator | ↑ | 1.328 | 0.0028 | N/A | |
| LCAT | Phosphatidylcholine‐sterol acyltransferase | ↑ | 1.327 | 0.0033 | N/A | ||
| Oxidative stress | SOD3 | Extracellular superoxide dismutase [Cu‐Zn] | ↓ | 0.776 | 0.0156 | N/A | |
| HPX | Hemopexin | ↓ | 0.684 | 0.0038 | N/A | ||
| Hormone | TTR | Transthyretin | ↓ | 0.615 | 0.0045 | 0.011 | |
| RBP4 | Retinol‐binding protein 4 | ↓ | 0.758 | 0.0118 | N/A | ||
| PENK | Proenkephalin‐A | ↓ | 0.693 | 0.0045 | 0.005 | ||
| CPE | Carboxypeptidase E | ↑ | 1.294 | 0.0051 | 0.044 | ||
| STC2 | Stanniocalcin‐2 | ↑ | 1.282 | 0.0064 | N/A | ||
| AGT | Angiotensinogen | ↑ | 1.290 | 0.0086 | N/A | ||
| AZGP1 | Zinc‐α‐2‐glycoprotein | ↓ | 0.763 | 0.0444 | N/A | ||
| Cell death/survival | CLU | Clusterin | ↑ | 1.184 | 0.0147 | 0.05 | |
| VASN | Vasorin | ↑ | 1.293 | 0.0090 | 0.034 | ||
| TYRO3 | Receptor protein‐tyrosine kinase | ↑ | 1.307 | 0.0395 | N/A | ||
| PLXDC2 | Plexin domain‐containing protein 2 | ↑ | 1.277 | 0.0160 | N/A | ||
| Energy | MDH1 | Malate dehydrogenase | ↑ | 1.280 | 0.0367 | N/A | |
| ENO2 | γ‐enolase, “NSE” | ↑ | 1.272 | 0.0292 | N/A | ||
| LDHA | L‐lactate dehydrogenase | ↑ | 1.275 | 0.0131 | N/A | ||
| PKM2 | Pyruvate kinase | ↑ | 1.266 | 0.0151 | N/A | ||
| Coagulation, angiogenesis | F12 | Coagulation factor XII | ↓ | 0.643 | 0.0187 | N/A | |
| Protein modification | MAN2A2 | α‐mannosidase 2x | ↑ | 1.379 | 0.0104 | N/A | |
| ART3 | NAD(P)(+)—arginine ADP‐ribosyltransferase; ecto‐ADP‐ribosyltransferase 3 | ↑ | 1.299 | 0.0029 | N/A | ||
| Neuronal outgrowth | CNTN1 | Contactin‐1 | ↑ | 1.167 | 0.0070 | N/A | |
| SEMA4B | Semaphorin‐4B | ↑ | 1.306 | 0.0203 | N/A | ||
| SEMA3G | Semaphorin‐3G | ↑ | 1.290 | 0.0322 | N/A | ||
| NEO1 | Neogenin | ↑ | 1.135 | 0.0325 | N/A | ||
| EFEMP1 | EGF‐containing fibulin‐like extracellular matrix protein 1 | ↑ | 1.185 | 0.0328 | N/A | ||
| EPHA4 | Receptor protein‐tyrosine kinase; ephrin type‐A receptor 4 | ↑ | 1.189 | 0.0463 | N/A | ||
| NDRG2 | Protein NDRG2 | ↑ | 1.289 | 0.0203 | N/A | ||
| B4GAT1 | β‐1,4‐glucuronyltransferase 1 | ↑ | 1.277 | 0.0184 | N/A | ||
| Cell adhesion, signaling, extra‐cellular matrix | SGCE | Epsilon‐sarcoglycan | ↑ | 1.304 | 0.0147 | N/A | |
| NCAM2 | Neural cell adhesion molecule 2 | ↑ | 1.198 | 0.0410 | N/A | ||
| FAT2 | Protocadherin fat 2 | ↑ | 1.499 | 0.0226 | 0.001 | ||
| OMG | Oligodendrocyte‐myelin glycoprotein | ↑ | 1.295 | 0.0414 | N/A | ||
| VTN | Vitronectin | ↓ | 0.661 | 0.0077 | N/A | ||
| ADAM22 | Disintegrin and metalloproteinase domain‐containing protein 22 | ↑ | 1.280 | 0.0276 | N/A | ||
| NRXN2 | Neurexin‐2 | ↑ | 1.214 | 0.0463 | N/A | ||
| NRXN3 | Neurexin‐3 | ↑ | 1.234 | 0.0471 | N/A | ||
| CASPR4 | Contactin‐associated protein 4 | ↑ | 1.318 | 0.0427 | N/A | ||
| SEZ6L2 | Seizure 6‐like protein 2 | ↑ | 1.231 | 0.0340 | N/A | ||
| LRRC4B | Leucine‐rich repeat‐containing protein 4B | ↑ | 1.325 | 0.0165 | N/A | ||
| ACTG1 | Actin, cytoplasmic 2 | ↑ | 1.244 | 0.0030 | N/A | ||
| NFASC | Neurofascin | ↑ | 1.247 | 0.0262 | N/A | ||
| ISLR | Immunoglobulin superfamily containing leucine‐rich repeat protein | ↑ | 1.209 | 0.0132 | N/A | ||
| COL6A1 | Collagen α‐1(VI) chain | ↑ | 1.261 | 0.0069 | N/A | ||
| Miscellaneous | PPIA | Peptidyl‐prolyl cis‐trans isomerase | ↑ | 1.173 | 0.0111 | N/A | |
| GPC1 | Glypican‐1; secreted glypican‐1 | ↑ | 1.267 | 0.0254 | N/A | ||
Age‐adjusted P‐values are derived from comparison between ManHD and preHD where age‐contribution estimated from controls has been removed using linear regression. Age‐adjustment was performed only if a significant age‐association was found among controls, otherwise marked “N/A.”
Protein Level Associations to the Composite UHDRS
A correlation matrix of regulated proteins and disease progression parameters is available below (Fig. 1). Only two (20%) of the down‐regulated proteins—PENK and TTR—displayed moderate to strong correlations with cUHDRS (Spearman R > 0.5). PENK and TTR also correlated with CAP score in all HTT gene expansion carriers (GECs) (PENK R = 0.696, P < 0.01, TTR R = −0.409, P = 0.042) and 5‐year risk of onset in the preHD participants (PENK R = −0.712, P < 0.01) TTR (R = −0.561, P = 0.046).
FIG. 1.
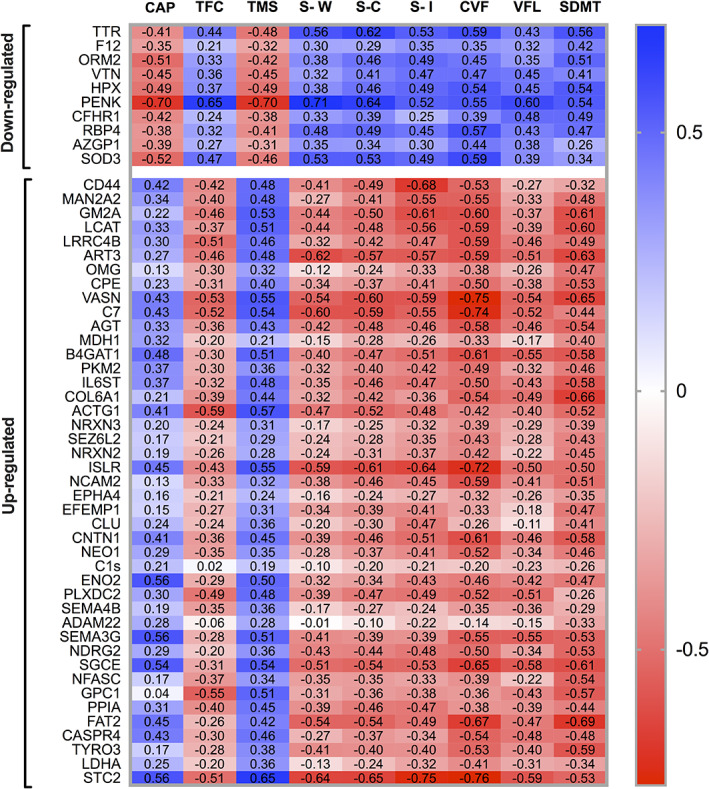
Significantly up‐ or downregulated proteins. Significantly regulated proteins (manifest HD vs pre‐manifest HD) and their correlations with measures of symptom severity. Unadjusted Spearman correlations (ρ values). CAP, CAG age product; TFC, total functional capacity; TMS, total motor score; S‐W, Stroop word; S‐C, Stroop color; S‐I, Stroop interference; CVF, category verbal fluency (animals); VFL, verbal fluency letters; SDMT symbol digit modalities test. [Color figure can be viewed at wileyonlinelibrary.com]
Correlations of PENK are displayed in Fig. 2.
FIG. 2.
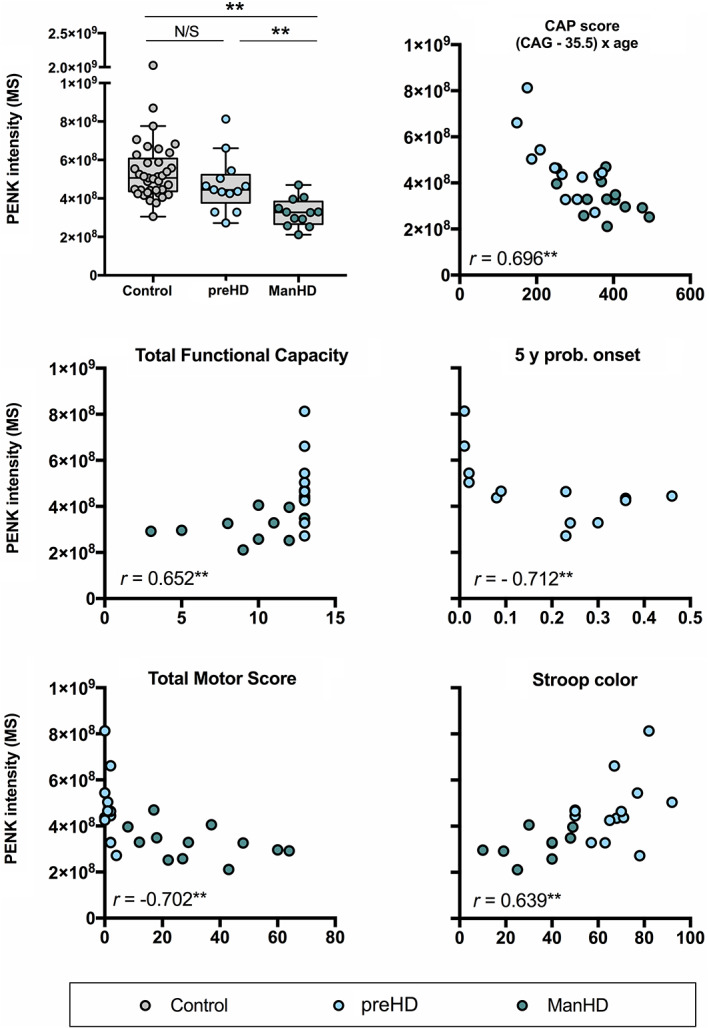
Proenkephalin (PENK) intensity levels. PENK relative intensity levels from the original study cohort plotted unadjusted on all scatter plots above. Significance tests in the top left intergroup comparisons were age‐adjusted by removing the age‐related effect estimated from the control group. Correlations with total motor score, total functional capacity, and Stroop color are shown in the respective graphs and have not been adjusted for age. *P < 0.05, **P < 0.01. [Color figure can be viewed at wileyonlinelibrary.com]
Out of up‐regulated proteins, 15 (35%) had moderate to strong correlations with cUHDRS (Spearman R > 0.5), GM2A, LCAT, ART3, CPE, VASN, B4GAT1, COL1A6, ACTG1, ISLR, SGCE, GPC1, PPIA, FAT2, TYRO3, and STC2. Of these, VASN, B4GAT1, ACTG1, ISLR, SGCE, FAT2, and STC2 also correlated with disease burden score in all GECs.
Absolute Quantitation of PENK in a Separate Cohort
A cohort with ManHD patient samples (n = 23) as well as controls (n = 23) from Stockholm and Gothenburg showed a lower concentration of PENK in ManHD compared with controls after direct age‐adjustment (median 2.75 and 9.38 nmol/mL, respectively, P = 0.004; Fig. 3). The mean age of the HD patients was 54.6 years, and 48.5 years for the controls. The mean CAG repeat number was 43.65. There was no significant interaction between age and PENK among controls (Spearman ρ −0.151, P = 0.492). Fifteen of 23 controls and 10 of 23 ManHD were women.
FIG. 3.
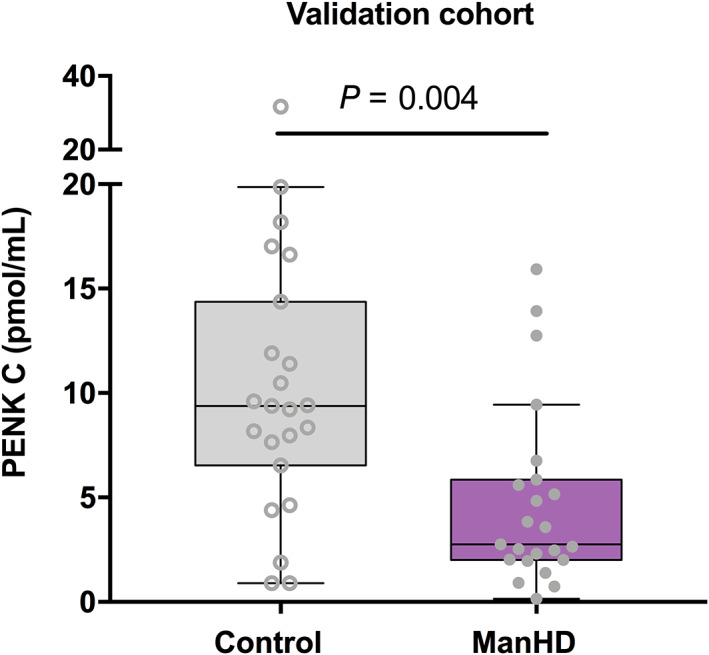
PENK intergroup comparisons. The validation cohort consisting of controls (n = 23) and manifest HD patient samples (n = 23) from Stockholm and Gothenburg. Unadjusted concentrations are plotted whereas the P value is derived from age‐adjusted comparison. [Color figure can be viewed at wileyonlinelibrary.com]
Pathway Analysis
Impact of altered pathways between groups were investigated in the software Ingenuity pathway analysis (IPA) (Tables 1 and 4). In comparisons both between preHD and ManHD patients as well as between all HD gene carriers versus controls LXR/RXR and FXR/RXR activation were identified as top canonical pathways. Atherosclerosis signaling was also identified as a top canonical pathway in both comparisons. Further pathways significantly differing between ManHD and preHD were acute phase response signaling and the intrinsic prothrombin activation pathway. Top upstream regulators were APP, dihydrotestosterone, beta‐estradiol, IL‐6, and high‐mobility group at‐hook 1 (HMGA1).
TABLE 4.
Top canonical pathways altered in manifest HD vs pre‐manifest gene expansion carriers
| Name | P value | Overlap |
| LXR/RXR activation | 8.66E−14 | 9.1% 11/121 |
| FXR/RXR activation | 4.97E −12 | 7.9% 10/126 |
| Acute phase response signaling | 3.14E −09 | 5.1% 9/176 |
| Atherosclerosis signaling | 2.46E −06 | 4.7% 6/127 |
| Intrinsic prothrombin activation pathway | 8.08E −06 | 9.5% 4/42 |
Comparing HD gene carriers versus controls, other pathways significantly differing were clathrin‐mediated endocytosis signaling and coagulation system. Top upstream regulators were VCAN, mibolerone, TWIST1, inosine, and β‐estradiol.
Discussion
In the present study, we identified CSF proteomic differences between ManHD patients and preHD. Because the goal was to identify markers of disease progression, we highlight proteins that also displayed correlations with symptom severity. Below, we will discuss the potential markers identified and their connection to HD pathophysiology.
Decreasing Levels of Proenkephalin May Reflect the Function of Medium Spiny Neurons of the Striatum
CSF PENK was decreased in ManHD patients compared with preHD. There was also a trend toward lower levels in preHD compared to controls. Although this trend did not reach significance, there was an inverse correlation between PENK and the 5‐year risk of onset among the preHD group. PENK levels also decreased along with the progression of HD symptoms. PENK is expressed highly in a subset of medium spiny neurons (MSNs) of the putamen/caudate nucleus, 21 and has, to our knowledge, not previously been measured in the CSF of HD gene carriers. Because loss of MSNs is thought to be an important part of the degenerative process in HD, validation of this finding in a separate cohort was deemed important. A PENK‐related peptide (Met‐enkephalin‐Arg6‐Gly7‐Leu8 [MERGL]) was reported to be down‐regulated in HD (and progressive supranuclear palsy) compared to controls. 22 MERGL levels seemed to be affected by antipsychotics in the former study, but we could not find differing levels of PENK in the participants using antipsychotics. Contrary from the GABA‐ and enkephalinergic MSNs, neuropeptide Y (NPY)‐containing interneurons are spared in HD and higher levels of NPY have been reported in CSF of HD patients. 23 The authors suggested that a ratio between NPY and dynorphin‐A or enkephalin peptides could serve as a biomarker for HD onset or disease progression. Furthermore, PENK is a stabile surrogate marker for the enkephalins that, because of their instability (T1/2 15 minutes), are less likely to serve as meaningful HD biomarkers. Development of PENK as a blood marker for HD may be challenging as it is also expressed in non‐neuronal tissues 24 and increases in renal and cardiac dysfunction. 25 PENK, as part of the endogenous opioid system, modulates central pain perception, which has been reported to be abnormal in HD patients. 26 Immunoassays for PENK quantification are commercially available.
Possible Implications of Reduced Transthyretin in HD
Strikingly low levels of TTR (fold change 0.61) were found in the ManHD group compared with preHD, and TTR levels correlated moderately with HD symptoms. Decreased TTR has indeed been reported in manifest HD before, 9 , 10 although associations to symptoms were not reported.
TTR is a transporter of thyroxine, and in a recent study 27 in the same HD cohort, we found that the tyrosine metabolism was the most impacted pathway, and CSF thyroxine was markedly reduced in manifest HD patients compared with preHD. The combined finding of reduced TTR and thyroxine are remarkable. Thyroxine have profound and important roles in the development of the CNS, with lack of thyroxine causing the neurological deficits of cretinism. 28
TTR is also an amyloidigenic protein and point mutations in the TTR gene facilitates protein misfolding and amyloidogenesis resulting in familiar amyloid polyneuropathy and/or cardiomyopathy. TTR has also been implicated as a therapeutic target in Alzheimer's disease (AD), 29 where patients have reduced CSF TTR. 30 The reduction of TTR could, according to the hypothesis from AD, reflect a failing protective mechanism against aggregation of toxic β‐amyloid. Because mutant huntingtin forms amyloid‐like aggregates, 31 this mechanism warrants further study in HD.
Inflammatory Mechanisms in HD Reflected in the CSF Proteome
In agreement with previous studies in HD, 32 , 33 , 34 we found evidence of complement activation. C7, a component of the membrane attack complex, was elevated and correlated highly with symptoms. Elevated plasma C7 in HD patients has also been reported. 33 C7 is regulated by CLU, which was also upregulated in our study, but hardly correlated significantly with any HD symptoms. C4A and C4B both increased with severity of symptoms, but neither differed between ManHD and preHD after age‐adjustment. C1s was on the contrary elevated but did not correlate with HD symptoms. Both IL‐6ST and CD44 were increased but displayed only weak to moderate correlations with symptoms. Elevated IL‐6 levels in plasma and CSF in HD have been reported before. 35
VASN, which was strongly associated to symptom severity, is an hypoxia‐induced protein that attenuates transforming growth factor‐β by binding. 36 Other proteins that were closely linked to HD symptoms include STC2, a regulator of insulin‐like growth factor bioavailability, 37 and the brain enriched protein SGCE, which is encoded by the gene linked to myoclonus dystonia syndrome (DYT11). 38
Relating to the top canonical pathway liver X receptor/retinoid X receptor (LXR/RXR) activation, wild‐type huntingtin has previously been shown to interact with this group of nuclear receptors. In fact, it has been suggested that the polyglutamine expansion impairs the normal function where huntingtin activates LXR‐mediated transcription 39 —a central mechanism regulating the homeostasis of cholesterol in the brain. In plasma from HD patients, brain‐derived 24‐OH cholesterol deficiency has been found, 40 and impaired cholesterol synthesis in the CNS has also been observed in a HD mouse model. 41 Further, evidence suggests that polyglutamine toxicity may be countered by high mobility group box proteins HMGB1/2 42 —homologs of the HMGA1, 43 a pathway that was also disrupted in our study.
Limitations
The limited HD sample in the exploratory part of the study, a common restraint of single center studies, reduces statistical power. Detailed clinical data nevertheless allowed us to strengthen the findings by selecting proteins of interest as biomarkers based on associations to disease progression parameters. Current progression parameters may not reflect disease in a perfect manner, and findings such as these may of course reflect other, hitherto unknown processes influencing the pathology.
Natural age differences between the manifest HD patients and the preHD group poses another statistical challenge. Direct age‐adjustment between the groups could potentially remove true differences produced by a combination of age and neurodegeneration in HD. Instead we estimated normal age‐effects on proteins levels from the relatively large number of controls that matched the ages of both groups and removed this effect in the comparison.
Although the explorative MS proteomics design is aimed at and in theory the best suited for biomarker discovery, it offers several challenges, especially for the reproducibility of findings. Part of the reason for this is instrument “drift,” which was addressed by adjusting sample run order.
Conclusion
To our knowledge, this study is the first advanced CSF proteomics study to include a clinically well‐characterized HD cohort spanning a wide range of disease stages. The pathways correlated to disease progression strengthen previously suggested pathophysiological mechanisms such as overactivation of LXR‐activation and brain cholesterol metabolism. Biological functions of altered proteins suggest derangement of several systems including immune system activity, synaptic function, neuronal outgrowth, and hormonal homeostasis.
It appears that the levels of PENK begin to decrease before disease onset and follow disease progression. Clearly decreased PENK levels were validated in a separate cohort of manifest HD patients. We hypothesize that declining PENK levels reflect the degeneration or dysfunction of MSNs that produce PENK, and assays for PENK may serve as a surrogate marker for the state of MSNs in HD.
Author Roles
(1) Research Project: A. Conception, B. Organization, C. Execution; (2). Statistical Analysis: A. Design, B. Execution, C. Review and Critique; (3) Manuscript Preparation: A. Writing of the First Draft, B. Review and Critique.
V.M.: 1A, 2A, 3A
A.M.L: 3B
D.N.: 3B
M.K.: 3B
R.C.: 3B
M.P.A.: 3B
P.S.: 3B
S.A.: 3B
J.B.: 3B
G.S.: 3B
J.B.: 1A, 2A, 3B
J.S.: 1A, 2A, 3A
| Name | Contribution |
| Valter Niemelä | Design and conceptualization; manuscript writing; data aquisition |
| Anne‐Marie Landtblom | Interpretation; revised manuscript for intellectual content |
| Dag Nyholm | Interpretation; revised manuscript for intellectual content |
| Maria Kneider | Sample acquisition; revised manuscript for intellectual content |
| Radu Constantinescu | Sample acquisition; revised manuscript for intellectual content |
| Martin Paucar Arce | Sample acquisition; revised manuscript for intellectual content |
| Per Svenningson | Sample acquisition; revised manuscript for intellectual content |
| Sandy Abujrais | Sample analysis; data acquisition; revised manuscript for intellectual content |
| Joachim Burman | Sample acquisition; revised manuscript for intellectual content |
| Ganna Shevchenko | Sample analysis; data acquisition; revised manuscript for intellectual content |
| Jonas Bergquist | Design and conceptualization; ample analysis; data acquisition; revised manuscript for intellectual content |
| Jimmy Sundblom | Design and conceptualization; funding acquisition; sample acquisition; drafted manuscript for intellectual content |
Financial Disclosures
V.N., A.M.L., D.N., and J.B. Uppsala University Hospital and The Swedish agreement concerning research and education of doctors (ALF‐medel). M.K. and R.C. Sahlgrenska University Hospital and The Swedish agreement concerning research and education of doctors (ALF‐medel). M.P.A. and P.S. Karolinska university hospital and The Swedish agreement concerning research and education of doctors (ALF‐medel). S.A. and G.S. Uppsala university. J.B. Uppsala University and SciLife lavoratory. J.S. Uppsala University Hospital and the Swedish agreement concerning research and education of doctors (ALF‐medel): Foundation for medical research, Aland islands (Stiftelsen för Åländsk medicinsk forskning).
| Valter Niemelä | Uppsala university hospital; The swedish agreement concerning research and education of doctors (ALF‐medel). |
| Anne‐Marie Landtblom | Uppsala university hospital; The swedish agreement concerning research and education of doctors (ALF‐medel). |
| Dag Nyholm | Uppsala university hospital; The swedish agreement concerning research and education of doctors (ALF‐medel). |
| Maria Kneider | Sahlgrenska university hospital; The swedish agreement concerning research and education of doctors (ALF‐medel). |
| Radu Constantinescu | Sahlgrenska university hospital; The swedish agreement concerning research and education of doctors (ALF‐medel). |
| Martin Paucar Arce | Karolinska university hospital;The swedish agreement concerning research and education of doctors (ALF‐medel). |
| Per Svenningson | Karolinska university hospital; The swedish agreement concerning research and education of doctors (ALF‐medel). |
| Sandy Abujrais | Uppsala university |
| Joachim Burman | Uppsala university hospital; The swedish agreement concerning research and education of doctors (ALF‐medel). |
| Ganna Shevchenko | Uppsala university |
| Jonas Bergquist | Uppsala university; SciLife lavoratory |
| Jimmy Sundblom | Uppsala university hospital;The swedish agreement concerning research and education of doctors (ALF‐medel): Foundation for medical research, Aland islands (Stiftelsen för Åländsk medicinsk forskning) |
Funding agencies: The study was funded by the Foundation for medical research, Aland islands (Stiftelsen för Åländsk medicinsk forskning), and by the Swedish agreement concerning research and education of doctors (ALF‐medel).
References
- 1. Pringsheim T, Wiltshire K, Day L, Dykeman J, Steeves T, Jette N. The incidence and prevalence of Huntington's disease: a systematic review and meta‐analysis. Mov Disord 2012;27(9):1083–1091. [DOI] [PubMed] [Google Scholar]
- 2. Constantinescu R, Romer M, Oakes D, Rosengren L, Kieburtz K. Levels of the light subunit of neurofilament triplet protein in cerebrospinal fluid in Huntington's disease. Parkinsonism Relat Disord 2009;15(3):245–248. [DOI] [PubMed] [Google Scholar]
- 3. Vinther‐Jensen T, Bornsen L, Budtz‐Jorgensen E, et al. Selected CSF biomarkers indicate no evidence of early neuroinflammation in Huntington disease. Neurol Neuroimmunol Neuroinflamm 2016;3(6):e287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Niemela V, Landtblom AM, Blennow K, Sundblom J. Tau or neurofilament light‐which is the more suitable biomarker for Huntington's disease? PLoS One 2017;12(2):e0172762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wild EJ, Boggio R, Langbehn D, et al. Quantification of mutant huntingtin protein in cerebrospinal fluid from Huntington's disease patients. J Clin Invest 2015;125(5):1979–1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tabrizi SJ, Leavitt BR, Landwehrmeyer GB, et al. Targeting Huntingtin expression in patients with Huntington's disease. N Engl J Med 2019;380(24):2307–2316. [DOI] [PubMed] [Google Scholar]
- 7. Ross CA, Aylward EH, Wild EJ, et al. Huntington disease: natural history, biomarkers and prospects for therapeutics. Nat Rev Neurol 2014;10(4):204–216. [DOI] [PubMed] [Google Scholar]
- 8. Caron NS, Dorsey ER, Hayden MR. Therapeutic approaches to Huntington disease: from the bench to the clinic. Nat Rev Drug Discov 2018;17(10):729–750. [DOI] [PubMed] [Google Scholar]
- 9. Fang Q, Strand A, Law W, et al. Brain‐specific proteins decline in the cerebrospinal fluid of humans with Huntington disease. Mol Cell Proteomics 2009;8(3):451–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Vinther‐Jensen T, Simonsen AH, Budtz‐Jorgensen E, Hjermind LE, Nielsen JE. Ubiquitin: a potential cerebrospinal fluid progression marker in Huntington's disease. Eur J Neurol 2015;22(10):1378–1384. [DOI] [PubMed] [Google Scholar]
- 11. Reilmann R, Leavitt BR, Ross CA. Diagnostic criteria for Huntington's disease based on natural history. Mov Disord 2014;29(11):1335–1341. [DOI] [PubMed] [Google Scholar]
- 12. Huntington Study Group CI . Unified Huntington's disease rating scale: reliability and consistency. Mov Disord 1996;11(2):136–142. [DOI] [PubMed] [Google Scholar]
- 13. Shoulson I, Fahn S. Huntington disease: clinical care and evaluation. Neurology 1979;29(1):1–3. [DOI] [PubMed] [Google Scholar]
- 14. Schobel SA, Palermo G, Auinger P, et al. Motor, cognitive, and functional declines contribute to a single progressive factor in early HD. Neurology 2017;89(24):2495–2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Penney JB Jr, Vonsattel JP, MacDonald ME, Gusella JF, Myers RH. CAG repeat number governs the development rate of pathology in Huntington's disease. Ann Neurol 1997;41(5):689–692. [DOI] [PubMed] [Google Scholar]
- 16. Langbehn DR, Brinkman RR, Falush D, Paulsen JS, Hayden MR, nternational Huntington's Disease Collaborative Group . A new model for prediction of the age of onset and penetrance for Huntington's disease based on CAG length. Clin Genet 2004;65(4):267–277. [DOI] [PubMed] [Google Scholar]
- 17. Niemela V, Burman J, Blennow K, Zetterberg H, Larsson A, Sundblom J. Cerebrospinal fluid sCD27 levels indicate active T cell‐mediated inflammation in premanifest Huntington's disease. PLoS One 2018;13(2):e0193492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Feresiadou A, Nilsson K, Ingelsson M, et al. Measurement of sCD27 in the cerebrospinal fluid identifies patients with neuroinflammatory disease. J Neuroimmunol 2019;332:31–36. [DOI] [PubMed] [Google Scholar]
- 19. Khoonsari PE, Haggmark A, Lonnberg M, et al. Analysis of the cerebrospinal fluid proteome in Alzheimer's disease. PLoS One 2016;11(3):e0150672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.‐range mass accuracies and proteome‐wide protein quantification. Nat Biotechnol 2008;26(12):1367–1372. [DOI] [PubMed] [Google Scholar]
- 21. Gerfen CR, Young WS 3rd. Distribution of striatonigral and striatopallidal peptidergic neurons in both patch and matrix compartments: an in situ hybridization histochemistry and fluorescent retrograde tracing study. Brain Res 1988;460(1):161–167. [DOI] [PubMed] [Google Scholar]
- 22. Iadarola MJ, Mouradian MM. Decrease in a proenkephalin peptide in cerebrospinal fluid in Huntington's disease and progressive supranuclear palsy. Brain Res 1989;479(2):397–401. [DOI] [PubMed] [Google Scholar]
- 23. Wagner L, Bjorkqvist M, Lundh SH, et al. Neuropeptide Y (NPY) in cerebrospinal fluid from patients with Huntington's disease: increased NPY levels and differential degradation of the NPY1‐30 fragment. J Neurochem 2016;137(5):820–837. [DOI] [PubMed] [Google Scholar]
- 24. Denning GM, Ackermann LW, Barna TJ, et al. Proenkephalin expression and enkephalin release are widely observed in non‐neuronal tissues. Peptides 2008;29(1):83–92. [DOI] [PubMed] [Google Scholar]
- 25. Emmens JE, Ter Maaten JM, Damman K, et al. Proenkephalin, an opioid system surrogate, as a novel comprehensive renal marker in heart failure. Circ Heart Fail 2019;12(5):e005544. [DOI] [PubMed] [Google Scholar]
- 26. Sprenger GP, van der Zwaan KF, Roos RAC, Achterberg WP. The prevalence and the burden of pain in patients with Huntington disease: a systematic review and meta‐analysis. Pain 2019;160(4):773–783. [DOI] [PubMed] [Google Scholar]
- 27. Herman S, Niemela V, Emami Khoonsari P, et al. Alterations in the tyrosine and phenylalanine pathways revealed by biochemical profiling in cerebrospinal fluid of Huntington's disease subjects. Sci Rep 2019;9(1):4129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Di Liegro I. Thyroid hormones and the central nervous system of mammals (review). Mol Med Rep 2008;1(3):279–295. [PubMed] [Google Scholar]
- 29. Buxbaum JN, Reixach N. Transthyretin: the servant of many masters. Cell Mol Life Sci 2009;66(19):3095–3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Serot JM, Christmann D, Dubost T, Couturier M. Cerebrospinal fluid transthyretin: aging and late onset Alzheimer's disease. J Neurol Neurosurg Psychiatry 1997;63(4):506–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Scherzinger E, Lurz R, Turmaine M, et al. Huntingtin‐encoded polyglutamine expansions form amyloid‐like protein aggregates in vitro and in vivo. Cell 1997;90(3):549–558. [DOI] [PubMed] [Google Scholar]
- 32. Leblhuber F, Walli J, Jellinger K, et al. Activated immune system in patients with Huntington's disease. Clin Chem Lab Med 1998;36(10):747–750. [DOI] [PubMed] [Google Scholar]
- 33. Dalrymple AWE, Joubert R, Sathasivam K, et al. Proteomic profiling of plasma in Huntington's disease reveals neuroinflammatory activation and biomarker candidates. J Proteome Res 2007;6(7):2833–2840. [DOI] [PubMed] [Google Scholar]
- 34. Singhrao SK, Neal JW, Morgan BP, Gasque P. Increased complement biosynthesis by microglia and complement activation on neurons in Huntington's disease. Exp Neurol 1999;159(2):362–376. [DOI] [PubMed] [Google Scholar]
- 35. Bjorkqvist M, Wild EJ, Thiele J, et al. A novel pathogenic pathway of immune activation detectable before clinical onset in Huntington's disease. J Exp Med 2008;205(8):1869–1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ikeda Y, Imai Y, Kumagai H, et al. Vasorin, a transforming growth factor ‐binding protein expressed in vascular smooth muscle cells, modulates the arterial response to injury in vivo. Proc Natl Acad Sci USA 2004;101(29):10732–10737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Fujimoto M, Hwa V, Dauber A. Novel modulators of the growth hormone ‐ insulin‐like growth factor axis: pregnancy‐associated plasma protein‐A2 and stanniocalcin‐2. J Clin Res Pediatr Endocrinol 2017;9(Suppl 2):1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Xiao J. Cloning, developmental regulation and neural localization of rat ɛ‐sarcoglycan. Brain Res Mol Brain Res 2003;119(2):132–143. [DOI] [PubMed] [Google Scholar]
- 39. Futter M, Diekmann H, Schoenmakers E, Sadiq O, Chatterjee K, Rubinsztein DC. Wild‐type but not mutant huntingtin modulates the transcriptional activity of liver X receptors. J Med Genet 2009;46(7):438–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Leoni V, Mariotti C, Tabrizi SJ, et al. Plasma 24S‐hydroxycholesterol and caudate MRI in pre‐manifest and early Huntington's disease. Brain 2008;131(Pt 11):2851–2859. [DOI] [PubMed] [Google Scholar]
- 41. Leoni V, Caccia C. The impairment of cholesterol metabolism in Huntington disease. Biochim Biophys Acta 2015;1851(8):1095–1105. [DOI] [PubMed] [Google Scholar]
- 42. Qi ML, Tagawa K, Enokido Y, et al. Proteome analysis of soluble nuclear proteins reveals that HMGB1/2 suppress genotoxic stress in polyglutamine diseases. Nat Cell Biol 2007;9(4):402–414. [DOI] [PubMed] [Google Scholar]
- 43. Stroedicke M, Bounab Y, Strempel N, et al. Systematic interaction network filtering identifies CRMP1 as a novel suppressor of huntingtin misfolding and neurotoxicity. Genome Res 2015;25(5):701–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Raw data sets including all quantified protein levels can be made available on request.


