Abstract
Association studies have linked microbiome alterations with many human diseases, but not always reported consistent results, which necessitates cross-study comparisons. Here, a meta-analysis of eight geographically and technically diverse fecal shotgun metagenomic studies of colorectal cancer (CRC, N = 768), which was controlled for several confounders, identified a core set of 29 species significantly enriched in CRC metagenomes (FDR < 1E-5). CRC signatures derived from single studies maintained accuracy in other studies. By training on multiple studies we improved detection accuracy and disease specificity for CRC. Functional analysis of CRC metagenomes revealed enriched protein and mucin catabolism genes and depleted carbohydrate degradation genes. Moreover we inferred elevated production of secondary bile acids from CRC metagenomes suggesting a metabolic link between cancer-associated gut microbes and a fat- and meat-rich diet. Through extensive validations, this meta-analysis firmly establishes globally generalizable, predictive taxonomic and functional microbiome CRC signatures as a basis for future diagnostics.
INTRODUCTION
Studying microbial communities colonizing the human body in a culture-independent manner has been enabled by metagenomic sequencing technologies [1]. These have yielded glimpses into the complex yet incompletely understood interactions between the gut microbiome – the microbial ecosystem residing primarily in the large intestine – and its host [2]. To explore microbiome-host interactions in a disease context, metagenome-wide association studies (MWAS) have begun to map gut microbiome alterations in diabetes, inflammatory bowel disease, colorectal cancer and many other conditions [3–12]. However, due to the many biological factors possibly influencing gut microbiome composition in addition to the condition studied, a current challenge for MWAS is confounding, which can cause false associations [13, 14]. This issue is further aggravated by a lack of standards in metagenomic data generation and processing, making it difficult to disentangle technical from biological effects [15].
Robustness of microbiome-disease associations can be assessed through comparisons across multiple metagenomic case-control studies, i.e. meta-analyses. These aim at identifying associations that are consistent across studies and thus less likely attributable to biological or technical confounders. Most informative are meta-analyses of populations from diverse geographic and cultural regions. Previous microbiome meta-analyses based on 16S rRNA gene amplicon data found stark technical differences between studies and the reported taxonomic disease associations were either of low effect size or not well resolved [16–18]. In contrast, shotgun metagenomics enables analyses with higher taxonomic resolution and of gene functions to improve statistical power for fine-mapping disease-associated strains and aid in the interpretation of host-microbial co-metabolism. Thus far however, meta-analyses of shotgun metagenomic data have either reported on features of general dysbiosis in comparisons across multiple diseases [19], or have left it unclear how well microbiome signatures generalize across studies of the same disease when data are rigorously separated to avoid over-optimistic evaluations of their prediction accuracy [20].
Here, we present a meta-analysis of a total of eight studies of CRC including fecal metagenomic data from 386 cancer cases and 392 tumor-free controls. After consistent data reprocessing, we examined an initial set of five studies for CRC-associated changes in the gut microbiome. Firstly, we investigated potential confounders, followed by identifying (univariate) microbial species associations, and inferring species co-occurrence patterns in CRC. Secondly, we trained multivariable classification models for recognition of CRC status, from both taxonomic and functional microbiome profiles and tested how accurately these models generalized to data from studies not used for training. Moreover, we evaluated performance improvements achieved by pooling data across studies and the disease-specificity of the resulting classification models. Thirdly, targeted investigation of virulence and toxicity genes as candidate functional biomarkers for CRC revealed several of these to be enriched in CRC metagenomes indicative of their prevalence and potential relevance in CRC patients. Three additional, more recent studies were finally used to independently validate these taxonomic and functional CRC signatures.
RESULTS
Consistent processing of published and new data for meta-analysis of CRC metagenomes
In this meta-analysis we included four published studies which used fecal shotgun metagenomics to characterize CRC patients compared to healthy controls (referred to by the country codes FR, AT, CN, and US, corresponding to the respective main study population; see Table 1, Supplementary Table S1, and Methods for inclusion criteria). For an additional fifth study population, we generated new fecal metagenomic data from samples collected in Germany (herein abbreviated as DE); a subset of samples from this patient collective were published previously (Table 1, Methods, [8]). These five studies were conducted on three continents and differed in sampling procedures, sample storage, and DNA extraction protocols. Notably, the fecal specimen of the US study were freeze-dried and stored at −80°C for more than 25 years before DNA extraction and sequencing [10]. In all studies, however, samples were collected prior to treatment, thus excluding cancer therapy as a potential confounding effect [14, 21]. Most samples were even taken before bowel preparation for colonoscopy, with some exceptions in the DE, CN and US studies (Supplementary Table S2). To ensure consistency in bioinformatic analyses, all raw sequencing data were (re-)processed using mOTUs2 for taxonomic profiling [22] and MOCAT2 for functional profiling [23].
Table 1. Fecal metagenomic studies of colorectal cancer included in this meta-analysis.
See Methods for inclusion criteria and Supplementary Table S2 for extended meta-data. For a detailed description of patient recruitment and data generation for the DE study, see Methods. The data for 38 samples from the DE study had been published previously as part of an independent validation cohort in [8].
Univariate meta-analysis of species associated with CRC
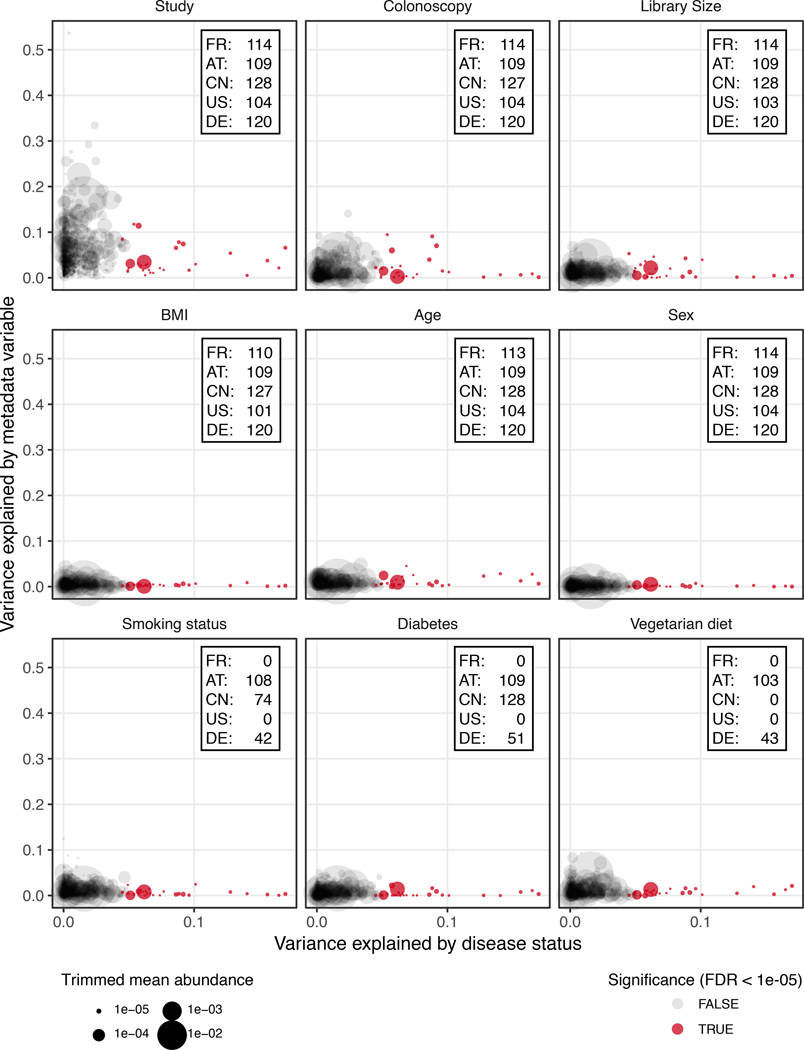
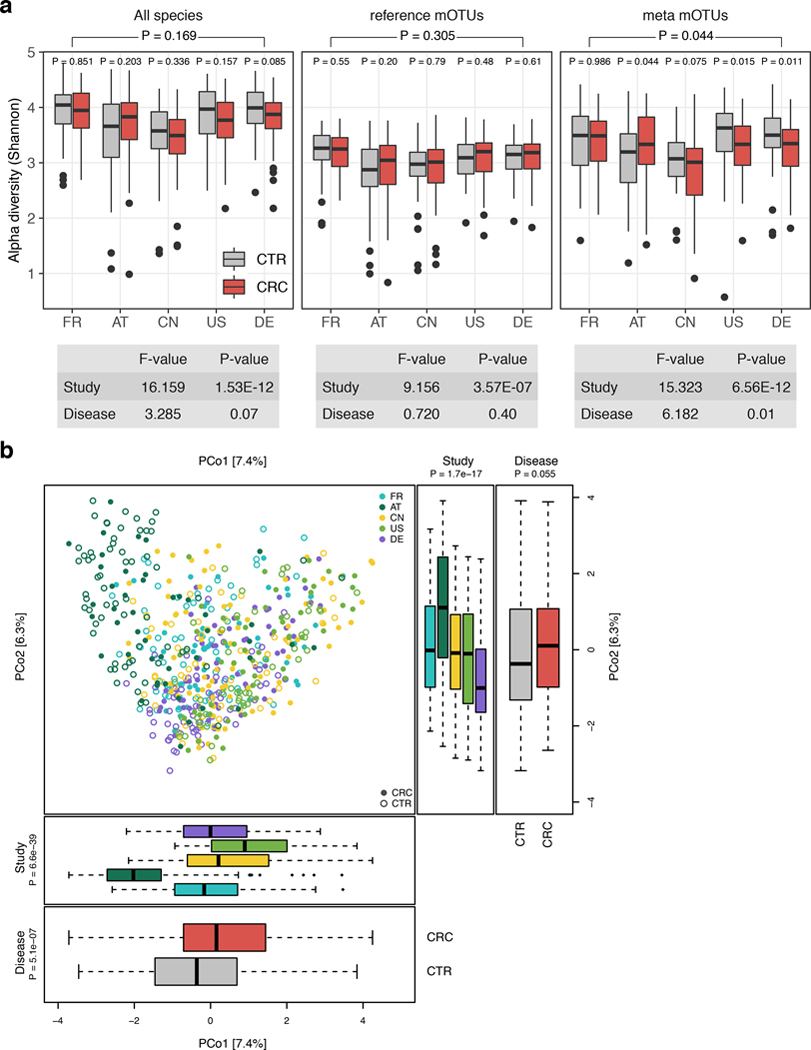
The first aim of the meta-analysis was to determine gut microbial species that are enriched or depleted in CRC metagenomes in a consistent manner across the five study populations. However, as these studies differed from one another in many biological and technical aspects, we first quantified the effect of study-associated heterogeneity on microbiome composition. We contrasted this with other potential confounders (‘patient age’, ‘BMI’, ‘sex’, ‘sampling after colonoscopy’, and ‘library size’; additionally, ‘smoking status’, ‘type II diabetes comorbidity’, and ‘vegetarian diet’ where available Extended Data 1, Supplementary Table S3). This analysis revealed the factor ‘study’ to have a predominant impact on species composition, which is supported by a recent comparison of DNA extraction protocols, as these typically differ between studies [15]. An analysis of microbial alpha and beta diversity showed study heterogeneity to also have a larger effect on overall microbiome composition than CRC in our data (Extended Data 2).
For the identification of microbial taxa significantly differing in abundance in CRC, parametric effect size measures are not well established, because microbiome data is characterized by non-Gaussian distributions with extreme dispersion; we thus used a generalisation of the fold change (Extended Data 3) and non-parametric significance testing. In this permutation test framework [24] (herein referred to as blocked univariate Wilcoxon tests) differential abundance in CRC can be assessed while accounting for ‘study’ as a nuisance effect that is treated as a blocking factor; additionally, motivated by our confounder analysis, we also blocked for ‘colonoscopy’ in all analyses (Methods, Extended Data 1). To rule out spurious associations due to the compositional nature of microbial relative abundance data, we additionally compared the results of this test with a method [25] employing log-ratio transformation (and found highly correlated results, Supplementary Fig. 1, Supplementary Table S4).
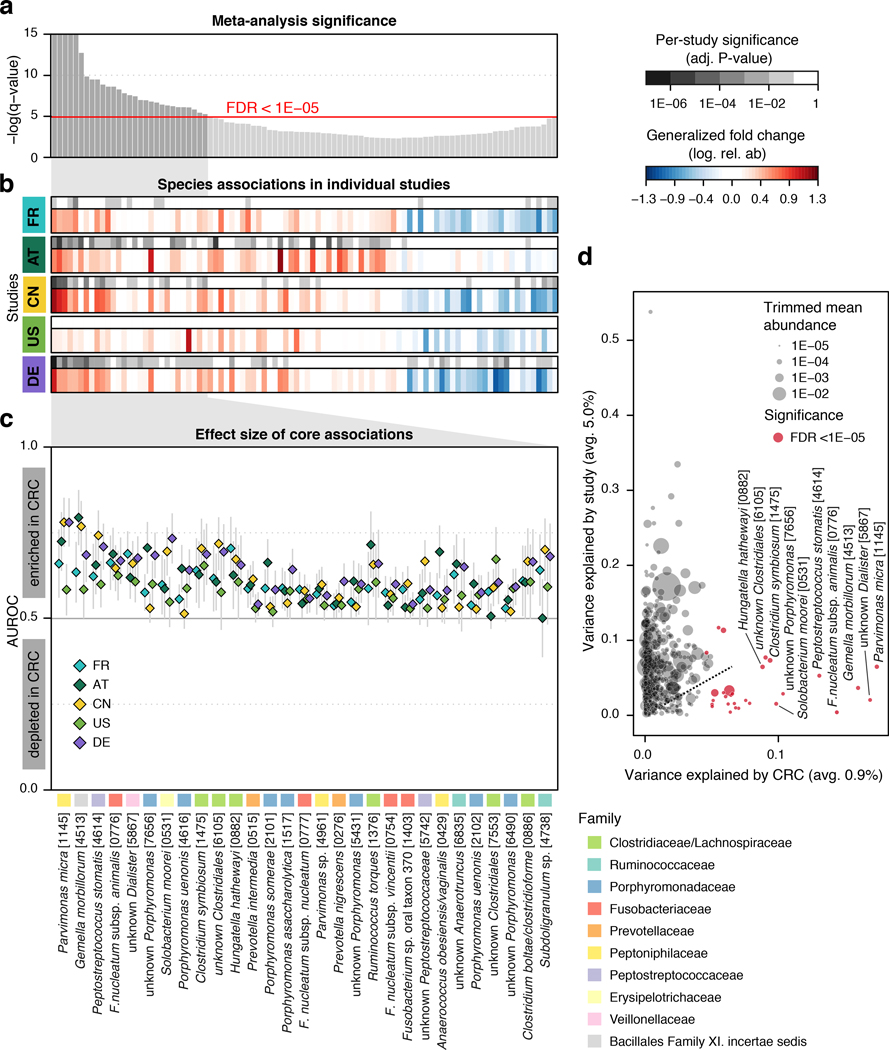
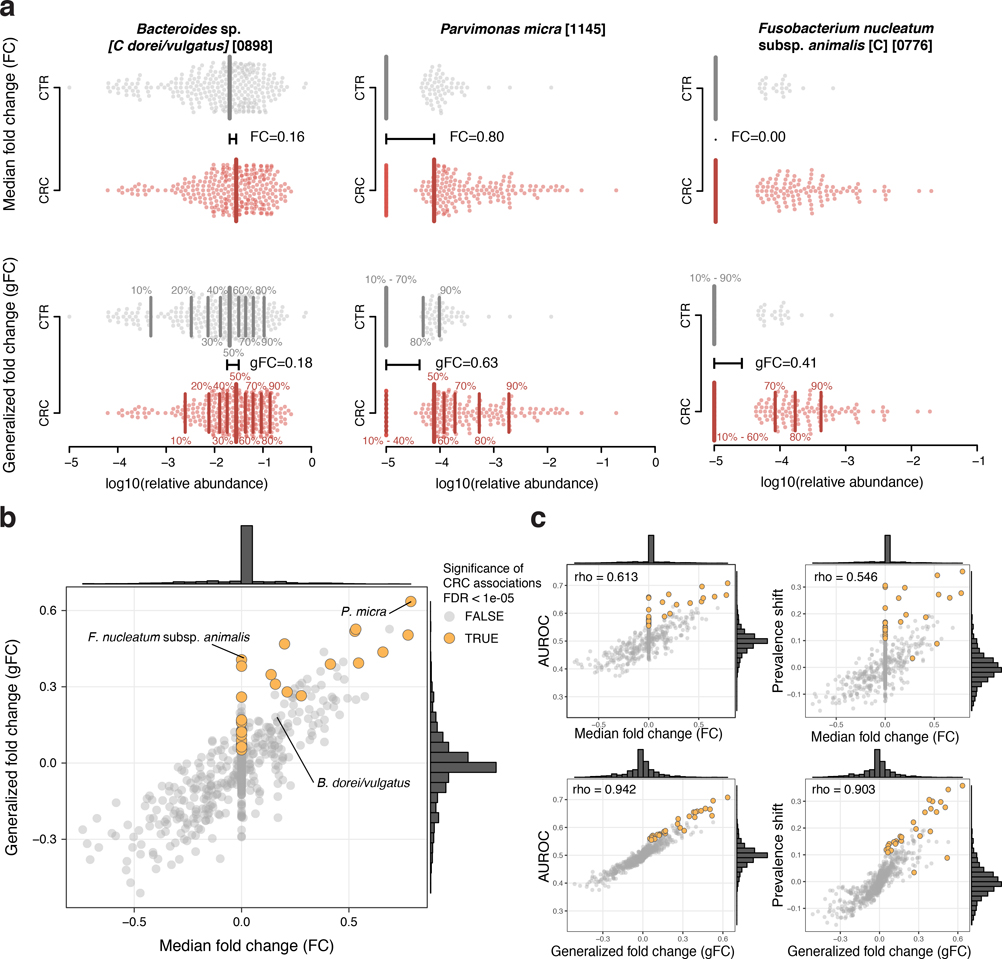
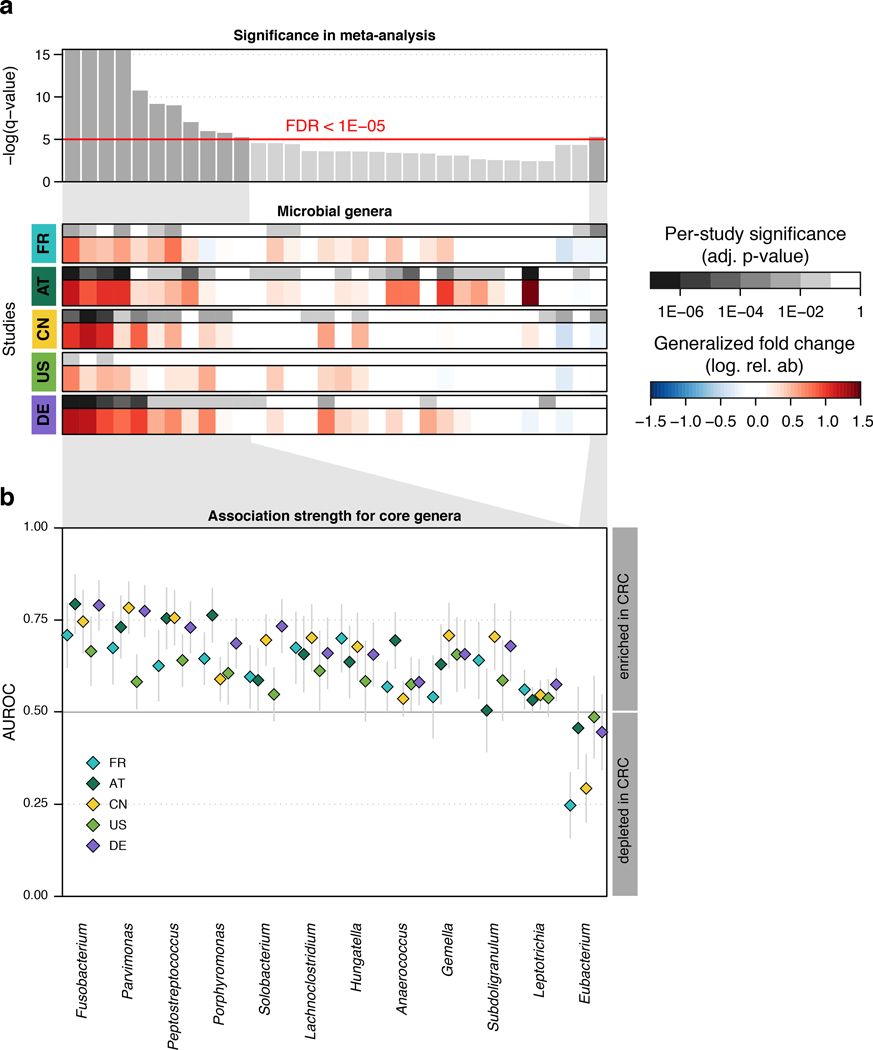
At a meta-analysis false discovery rate (FDR) of 0.005, we identified 94 microbial species to be differentially abundant in the CRC microbiome, out of 849 species consistently detected across studies (Supplementary Table S4, Methods). Among these, we focused on a core set of the 29 most significant markers (FDR < 1E-5, Fig. 1a) for further analysis. The latter included members of several genera previously associated with CRC, such as Fusobacterium, Porphyromonas, Parvimonas, Peptostreptococcus, Gemella, Prevotella, and Solobacterium (Fig. 1b, [8–11]), and 8 additional species without genomic reference sequences (meta-mOTUs, Methods, [22]) mostly from the Porphyromonas and Dialister genera and the Clostridiales order (see Extended Data 4 and Supplementary Table S4 for genus-level associations). Collectively, these 29 core CRC-associated species show a previously underappreciated diversity of 11 Clostridiales species to be enriched in CRC (Fig. 1b). In contrast to the majority of species that are more strongly affected by study heterogeneity than by CRC status, 26 out of the 29 CRC-associated species varied more by disease status (Fig. 1d).
Figure 1. Despite study differences, meta-analysis identifies a core set of gut microbes strongly associated with CRC.
(a) Meta-analysis significance of gut microbial species derived from blocked Wilcoxon tests (n=574 independent observations) is given by bar height (false discovery rate, FDR, of 0.05). (b) Underneath, species-level significance as computed by two-sided Wilcoxon test (FDR-corrected P-value) and generalized fold change (Methods) within individual studies are displayed as heatmaps in gray and color, respectively (see color bars and Table 1 for details on studies included). Species are ordered by meta-analysis significance and direction of change. (c) For a core of highly significant species (meta-analysis FDR 1E-5), association strength is quantified by the area under the Receiver Operating Characteristics curve (AUROC) across individual studies (color coded diamonds) and 95% confidence intervals are indicated by gray lines. Family-level taxonomic information is color-coded above species names (numbers in brackets are mOTU species identifiers, see Methods). (d) Variance explained by disease status (CRC vs controls) is plotted against variance explained by study effects for individual microbial species with dot size proportional to abundance (Methods); core microbial markers are highlighted in red. F. nucleatum – Fusobacterium nucleatum.
All of the core CRC-associated species were enriched in patients and were often undetectable in metagenomes from non-neoplastic controls. While previous studies were contradictory in the reported proportion of positive versus negative associations [8, 9, 17, 20], our meta-analysis results are more easily reconciled with a model in which – potentially many – gut microbes contribute to or benefit from tumorigenesis than with the opposing model in which a lack of protective microbes contributes to CRC development (Fig. 1b). Although these core taxonomic CRC associations were highly significant and consistent, individual studies showed marked discrepancies in the species identified as significant (Fig. 1a). Retrospective examination of the precision and sensitivity with which individual studies detected this core of CRC-associated species showed relatively low sensitivity for the US study (consistent with the original report [10]) and low precision of the AT study due to associations that were not replicated in other studies (Supplementary Fig. 2).
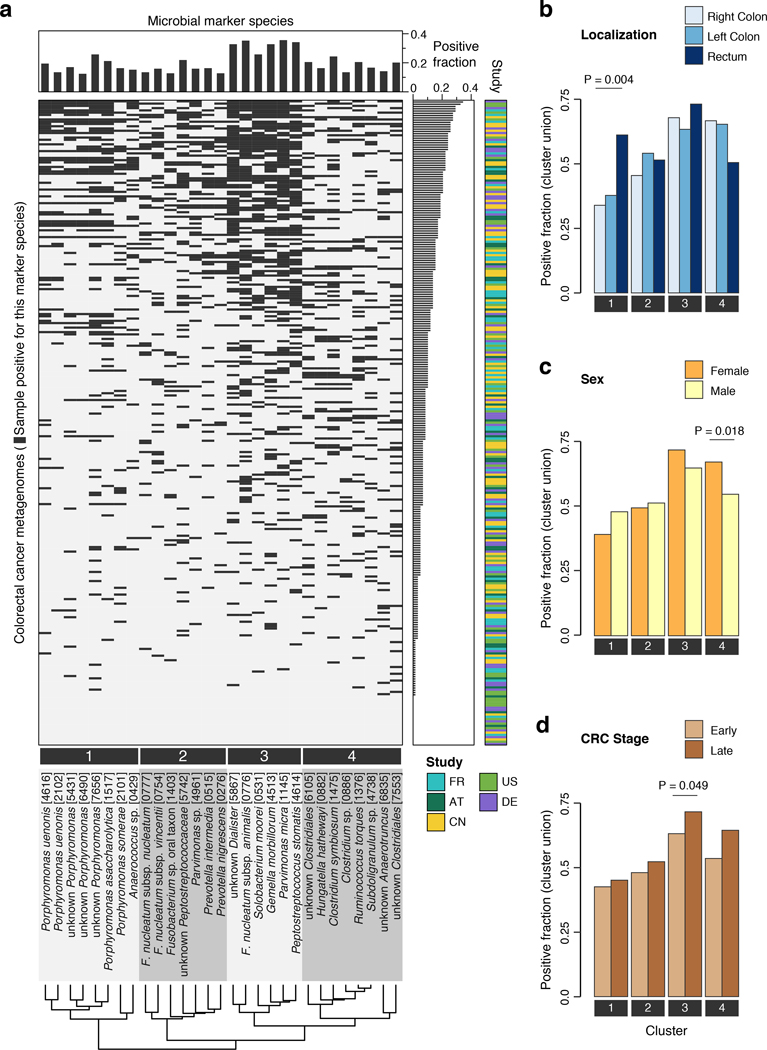
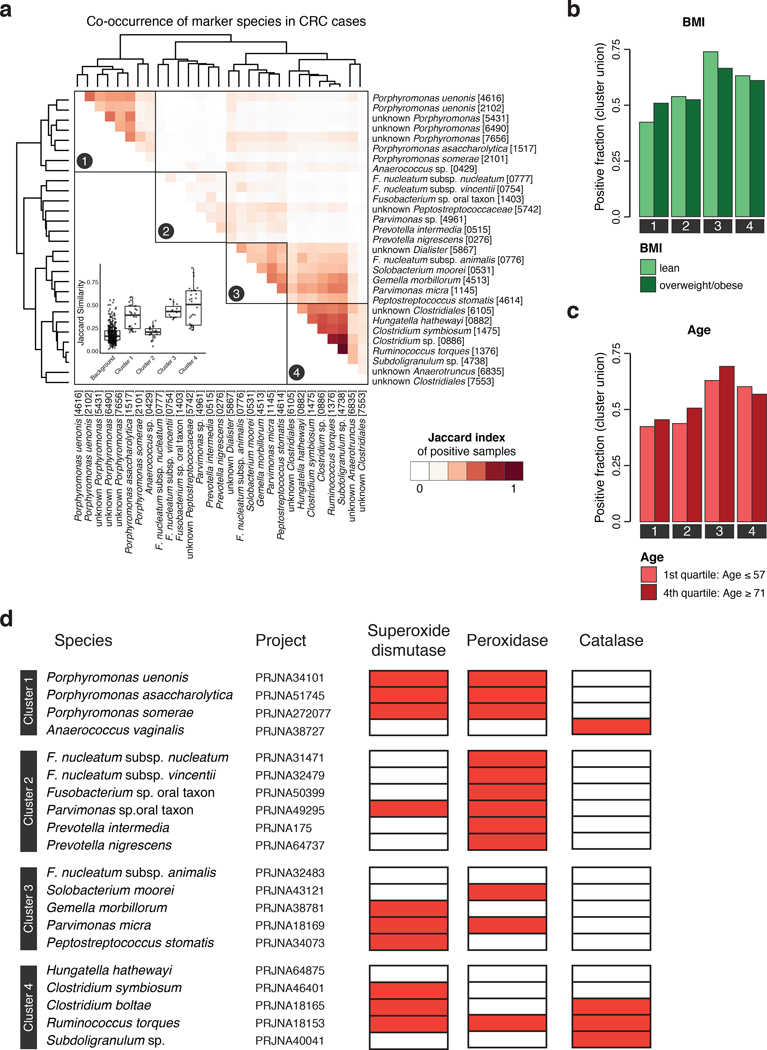
Analyzing patient metagenomes for co-occurrences among the core set of 29 species that are strongly enriched in the CRC microbiome revealed four species clusters with distinct taxonomic composition (Fig. 2a, Extended Data 5, Methods). Two of them showed strong taxonomic consistency: Cluster 1 exclusively comprised Porphyromonas spp., and cluster 4 only contained members of the Clostridiales order. In contrast, the other two clusters were taxonomically more heterogeneous with cluster 3 grouping together the species with highest prevalence in CRC cases (all among the ten most highly significant markers), consistent with a co-occurrence analysis of one of the data sets included here [11]. Cluster 2 contained species with intermediate prevalence.
Figure 2. Co-occurrence analysis of CRC-associated gut microbial species reveals four clusters preferentially linked to specific patient subgroups.
(a) The heatmap shows for all CRC patients (n=285 independent samples) if the respective sample is positive for each of the core set of microbial marker species (see Methods for adjustment of positivity threshold). Samples are ordered according to the sum of positive markers and marker species are clustered based on Jaccard similarity of positive samples, resulting in four clusters (Methods). Barplots in (b), (c), and (d) show the fraction of CRC samples that are positive for marker species clusters (defined as the union of positive marker species) broken down by patient subgroups based on differences in tumor location, sex, or CRC stage, respectively. Statistically significant associations between CRC subgroups and marker species clusters were identified using the Cochran–Mantel–Haenszel test blocked for study effects and are indicated above bars (P < 0.1).
Investigating whether these four clusters were associated with different tumor characteristics, we found the Porphyromonas cluster 1 to be significantly enriched in rectal tumors (Fig. 2b), consistent with the presence of superoxide dismutase genes in Porphyromonas genomes possibly conferring tolerance to a more aerobic milieu in the rectum (Extended Data 5). The Clostridiales cluster 4 was significantly more prevalent in female CRC patients. All species clusters showed a slight tendency towards latestage CRC (i.e. AJCC stages III and IV), but this was only significant for cluster 3. Associations with patient age and BMI were weaker and not significant (Extended Data 5). To rule out secondary effects due to differences in patient composition among studies, all of these tests were corrected for study effects (by blocking for ‘study’ and ‘colonoscopy’, see Methods). At the level of individual species, significant stage-specific enrichments could not be detected suggesting CRC-associated microbiome changes to be less dynamic during cancer progression than previously postulated [26], although fecal material may be less suitable to address this question than tissue samples.
Metagenomic CRC classification models
To establish metagenomic signatures for CRC detection across studies in face of geographic and technical heterogeneity, we developed multivariable statistical modeling workflows with rigorous external validation to avoid prevailing issues of overfitting and over-optimistic reports of model accuracy [19]. As a precaution against over-optimistic evaluation, these workflows are independent of the above-described differential abundance analysis. Instead, LASSO (Least Absolute Shrinkage and Selection Operator) logistic regression classifiers were employed to select predictive microbial features and eliminated uninformative ones (Methods).
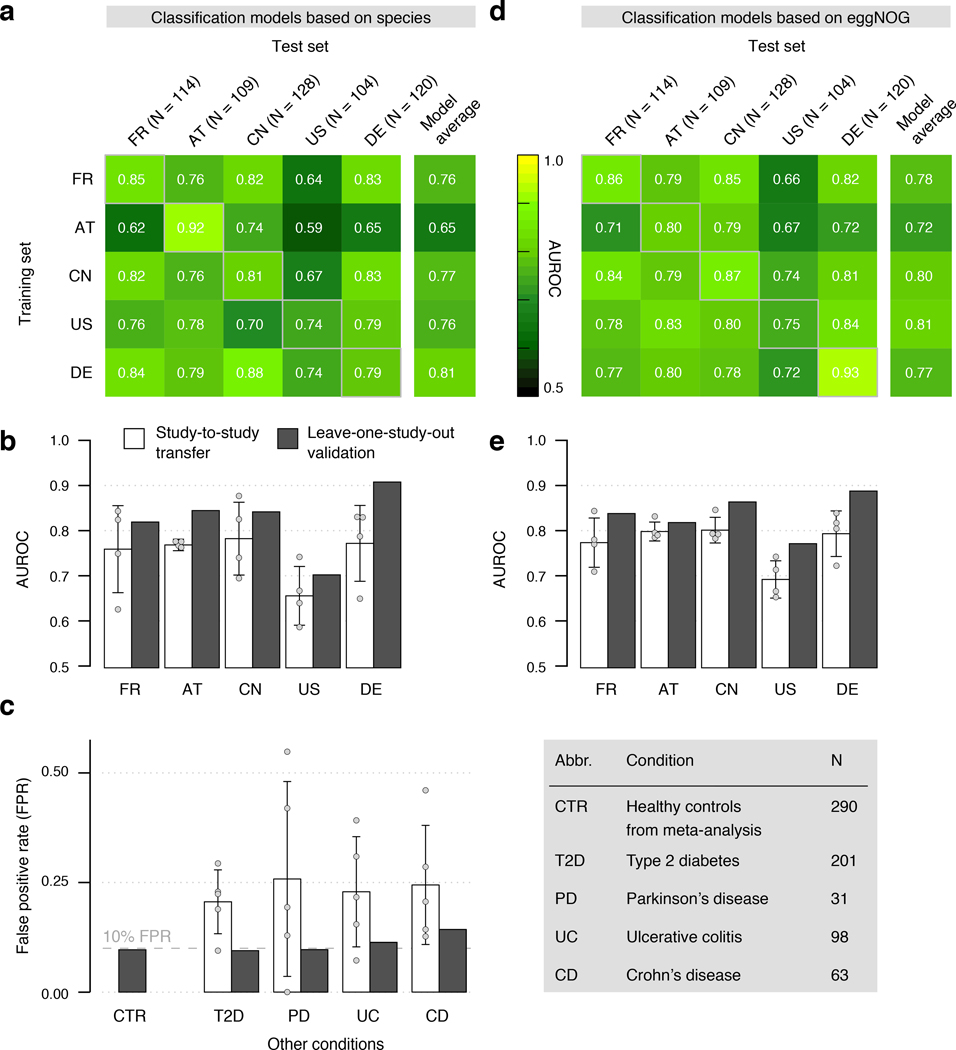
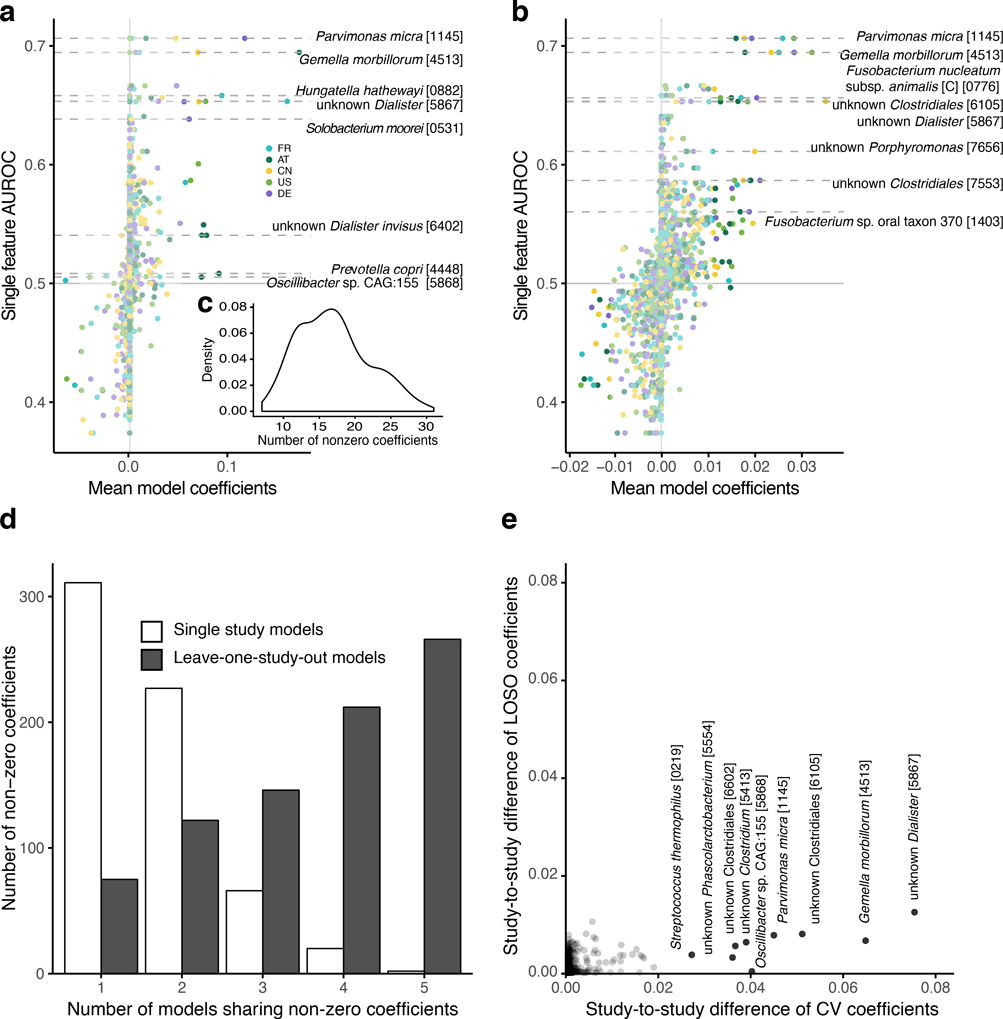
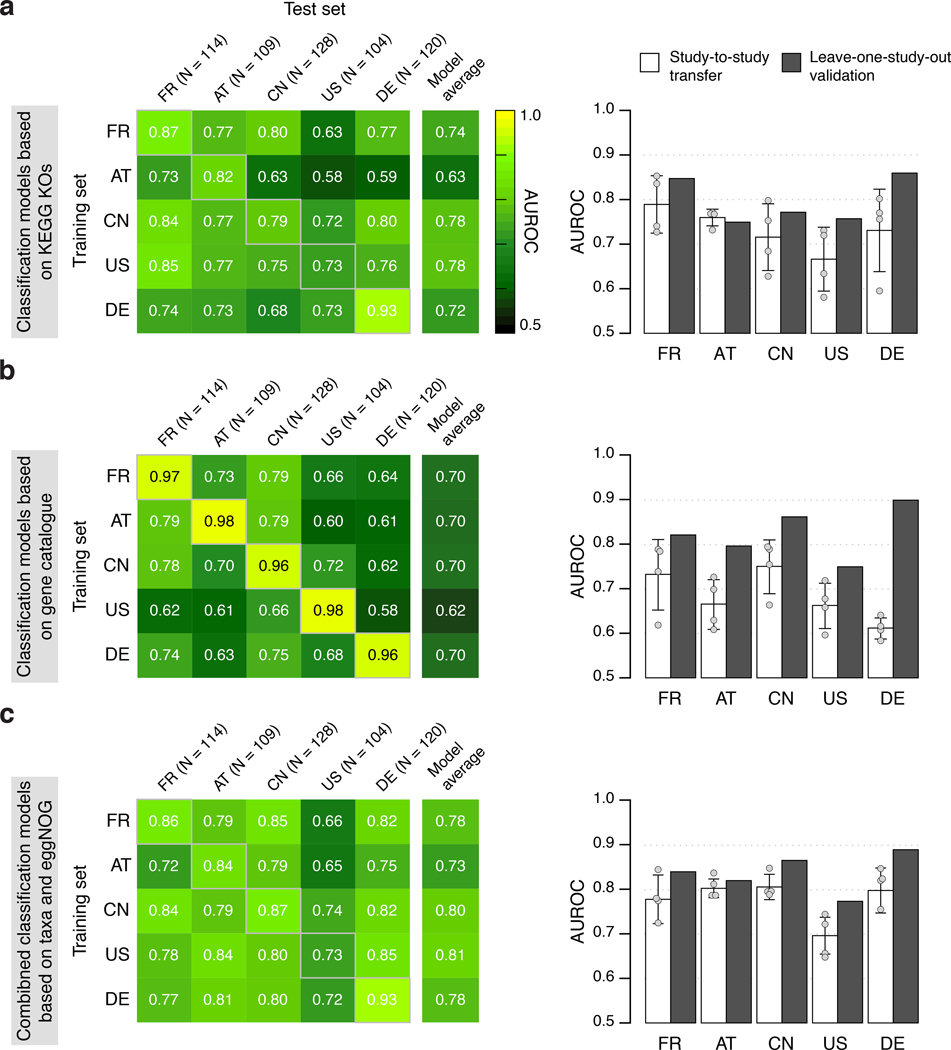
In a first step, we used abundance profiles from five studies including the 849 most abundant microbial species and assessed how well classifiers trained in cross validation (CV) on one study generalize in evaluations on the other four studies (study-to-study transfer of classifiers) (Fig. 3a). Within-study cross-validation performance, as quantified by the Area Under the Receiver Operating Characteristics (AUROC) curve, ranged between 0.69 and 0.92 and was generally maintained in study-to-study transfer (AUROC dropping by 0.07±0.12 on average) with two notable exceptions. First, in line with the univariate analysis of species associations, CRC detection accuracy on the US study was lower than for the other studies, both in cross-validation and in study-to-study transfer. This could potentially be explained by the US fecal specimen, unlike in the other studies, being freeze-archived for >25 years before metagenomic sequencing [10]. Second, classifiers trained on the AT study did not generalize as well to the other studies, consistent with low study precision seen in univariate meta-analysis (Supplementary Fig. 2). Given the microbial co-occurrence clusters described above, we wondered whether species-species interactions would provide additional information relevant for CRC recognition that is not contained in species abundance profiles. However, nonlinear classifiers able to exploit such interactions did not yield significantly better accuracies (Supplementary Fig. 3, see also [27]), suggesting that the linear model based on few biomarkers (on average 17 species account for more than 80% of the classifier weight, Extended Data 6) is near optimal for CRC prediction.
Figure 3. Both taxonomic and functional metagenomic classification models generalize across studies in particular when trained on data from multiple studies.
CRC classification accuracy resulting from cross validation within each study (gray boxes along diagonal) and study-to-study model transfer (external validations off diagonal) as measured by AUROC for classifiers trained on (a) species and (d) eggNOG gene family abundance profiles. The last column depicts the average AUROC across external validations. Classification accuracy, as evaluated by AUROC on a held-out study, improves if taxonomic (b) or functional (e) data from all other studies are combined for training (leave-one-study-out, LOSO validation) relative to models trained on data from a single study (study-to-study transfer, average and standard deviation shown). Bar height for study-to-study transfer corresponds to the average of four classifiers (error bars indicate standard deviation, n=4). (c) Combining training data across studies substantially improves CRC specificity of the (LOSO) classification models relative to models trained on data from a single study (depicted by bar color, as in (c) and (d)) as assessed by the false positive rate (FPR) on fecal samples from patients with other conditions (see legend). Bar height for study-to-study transfer corresponds to the average FPR across classifiers (n=5) with error bars indicating the standard deviation of FPR values observed.
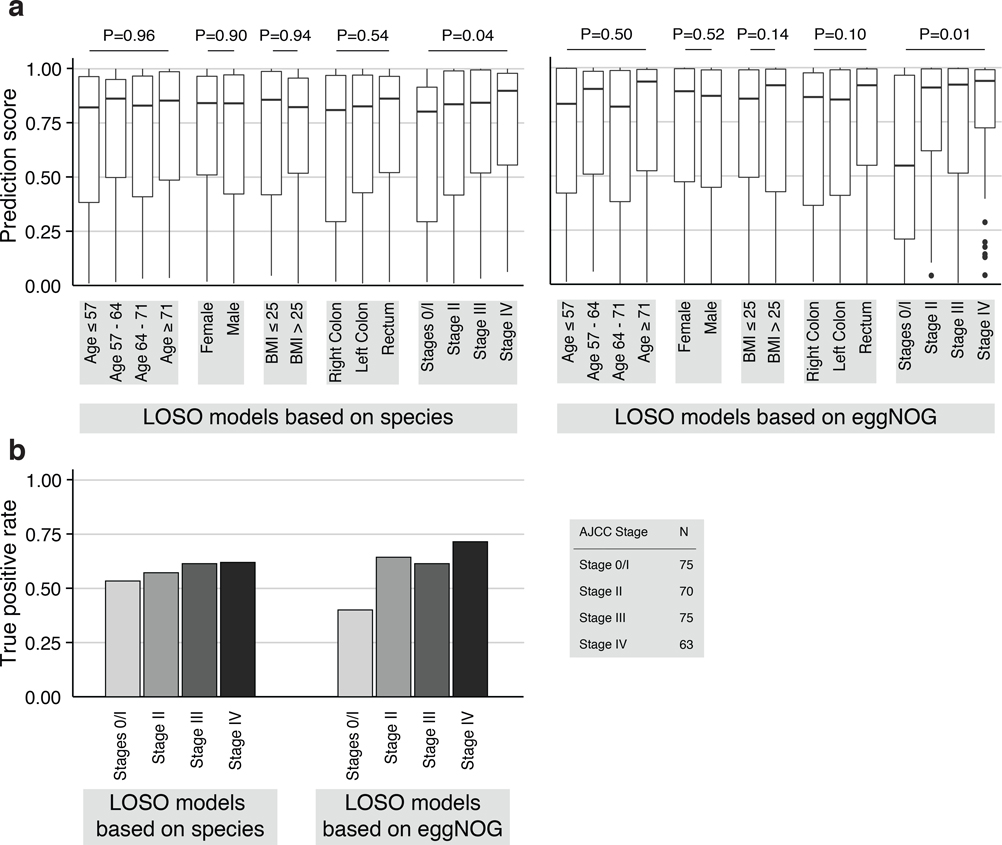
We further assessed if including data from all but one study in model training improves prediction on the remaining held-out study (leave-one-study-out validation, LOSO). LOSO performance of species-level models ranged between 0.71 and 0.91, and when disregarding the US study as an outlier was ≥0.83 (Fig. 3b). This corresponds to a LOSO accuracy increase of 0.076±0.03 compared to study-to-study transfer. These results suggest that one can expect a CRC detection accuracy ≥0.8 (AUROC) for any new CRC study using similarly generated metagenomic data. We moreover verified that metagenomic CRC classification models trained on species composition were not biased for clinical subgroups. With the exception of slightly more sensitive detection of late stage CRC (P = 0.03, mostly originating from the US study, Extended Data 7), we did not observe any classification bias by patient age, sex, BMI, or localization. Together this suggests that these metagenomic classifiers are unlikely to be strongly confounded by the clinical parameters recorded.
Several previous studies comparing microbiome changes across multiple diseases reported primarily general dysbiotic alterations and highlighted the need to examine the disease specificity of microbiome signatures [17, 19]. Therefore, we assessed false positive (FP) predictions of our metagenomic CRC classifiers on fecal metagenomes of type 2 diabetes [4, 5], Parkinson’s disease [12], ulcerative colitis and Crohn’s disease [6, 7] patients, reasoning that classifiers relying on biomarkers for general dysbiosis would yield an excess of FPs on these cohorts. However, our LOSO classification models calibrated to have a false-positive rate (FPR) of 0.1 on CRC datasets in fact maintained similarly low FPRs on other disease datasets ranging from 0.09 to 0.13 (Fig. 3c). Interestingly, disease specificity of LOSO models was significantly improved over that observed for classifiers trained on a single study, indicating that inclusion of multiple studies in the training set of a classifier can substantially improve its specificity for a given disease.
Functional metagenomic signatures for CRC
As shotgun metagenomics data, in contrast to 16S rRNA gene amplicon data, allow for a direct analysis of the functional potential of the gut microbiome, we examined how predictive metabolic pathways and orthologous gene families differing in abundance between CRC patients and controls would be of CRC status. When applying the same classification workflow as above to eggNOG orthologous gene family abundances [28], CRC detection accuracy was very similar to that observed for taxonomic models (Fig. 3de). AUROC values ranged from 0.70 to 0.81 for study-to-study transfer (per-study averages, Fig. 3e) and from 0.78 to 0.89 in LOSO validation with a pattern of generalization across studies resembling that for taxonomic classifiers. The accuracy of functional signatures did not strongly depend on eggNOG as an annotation source, but was similar when based on other comprehensive functional databases, such as KEGG [29] (Extended Data 8). When using individual gene abundances from metagenomic gene catalogues as a classifier input [30], we observed higher within-study cross-validation AUROC values of ≥0.96 in all studies, but lower generalization to other studies (AUROC between 0.60 and 0.79) (Extended Data 8).
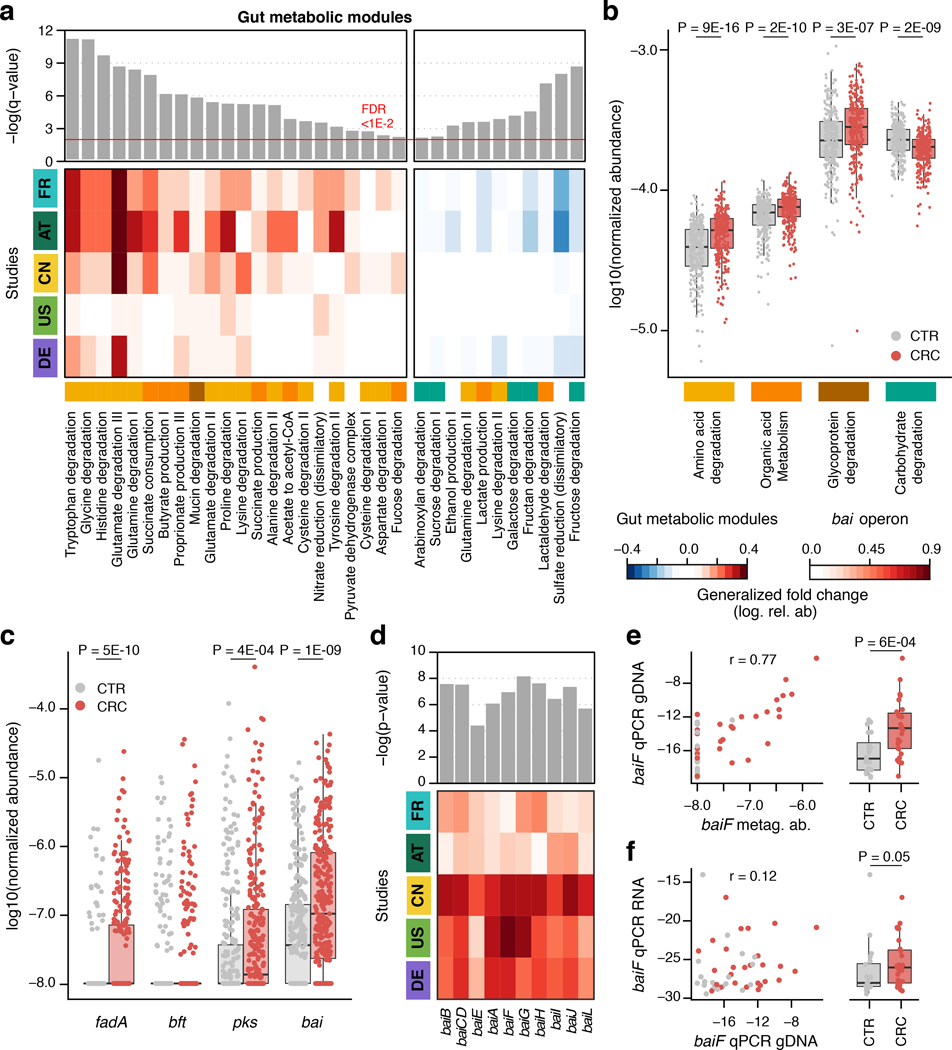
To explore changes in metabolic capacity of gut microbiomes from CRC patients more broadly, we quantified gut metabolic modules (defined in [31]) and subjected these to the same differential abundance analysis developed for microbial species. Gut metabolic modules with significantly higher abundance (FDR < 0.01, Wilcoxon test blocked for study and colonoscopy) in CRC metagenomes predominantly belonged to pathways for the degradation of amino acids, mucins (glycoproteins) and organic acids. This clear trend was accompanied by a depletion of genes from carbohydrate degradation modules (Fig. 4ab). Differences in all four high-level categories were highly significant (P < 1E-6 in all cases, blocked Wilcoxon tests) and consistent across studies (Fig. 4b). Overall these results establish a clear shift from dietary carbohydrate utilization in a healthy gut microbiome to amino acid degradation in CRC consistent with an earlier report based on a subset of the data [8]. Correlation analysis suggests that increased capacity for amino acid degradation is mostly contributed by CRC-associated Clostridiales (cf. cluster 4 in Fig. 2, Supplementary Fig. 4). About one half of these metagenomic pathway enrichments are also in agreement with independent metabolomics data suggesting increased availability of amino acids in epithelial cells or feces of CRC patients (Supplementary Table S5, [32–36]). While the observed pathway enrichments could potentially result from many factors, including unmeasured ones [13], they are consistent with established dietary risk factors for CRC, which include red and processed meat consumption [37] and low fiber intake [38].
Figure 4. Meta-analysis identifies consistent functional changes in CRC metagenomes.
(a) Meta-analysis significance of gut metabolic modules derived from blocked Wilcoxon tests (n=574 independent samples) is indicated by bar height (top panel, FDR of 0.01). Underneath, the generalized fold change (Methods) for gut metabolic modules [31] within individual studies is displayed as heatmap (see color key below (b)). Metabolic modules are ordered by significance and direction of change. A higher-level classification of the modules is color-coded below the heatmap for the four most common categories (colors as in (b), white indicating other classes). (b) Normalized log abundances for these selected functional categories is compared between controls (CTR) and colorectal cancer cases (CRC). Abundances are summarized as geometric mean of all modules in the respective category and statistical significance determined using blocked Wilcoxon tests (n=574 independent samples, see Methods). (c) Normalized log abundances for virulence factors and toxins compared between metagenomes of controls (CTR) and colorectal cancer cases (CRC) (significant differences P < 0.05 were determined by blocked Wilcoxon test, n=574 independent samples, see Methods for gene identification and quantification in metagenomes; fadA: gene encoding Fusobacterium nucleatum adhesion protein A, bft: gene encoding Bacteroides fragilis enterotoxin, pks: genomic island in Escherichia coli encoding enzymes for the production of genotoxic colibactin, and bai: bile acid inducible operon present in some Clostridiales species encoding bile acid converting enzymes). (d) Meta-analysis significance (uncorrected P-value) as determined by blocked Wilcoxon tests (n=574 independent samples) and generalized fold change within individual studies are displayed as bars and heatmap, respectively, for the genes contained in the bai operon. Due to high sequence similarity to baiF, baiK was not independently detectable with our approach. (e) Metagenomic quantification of baiF (metag. ab. – normalized relative abundance) is plotted against qPCR quantification in genomic DNA (gDNA) extracted from a subset of DE samples (n=47), with Pearson correlation (r) indicated (see Methods). (f) Expression of baiF determined via qPCR on reverse-transcribed RNA from the same samples in contrast to genomic DNA (as in e). The boxplots on the side of (e), (f) show the difference between cancer (CRC) and control (CTR) samples in the respective qPCR quantification (P-values on top were computed using a one-sided Wilcoxon test). All boxplots show interquartile ranges (IQR) as boxes with the median as a black horizontal line and whiskers extending up to the most extreme points within 1.5-fold IQR.
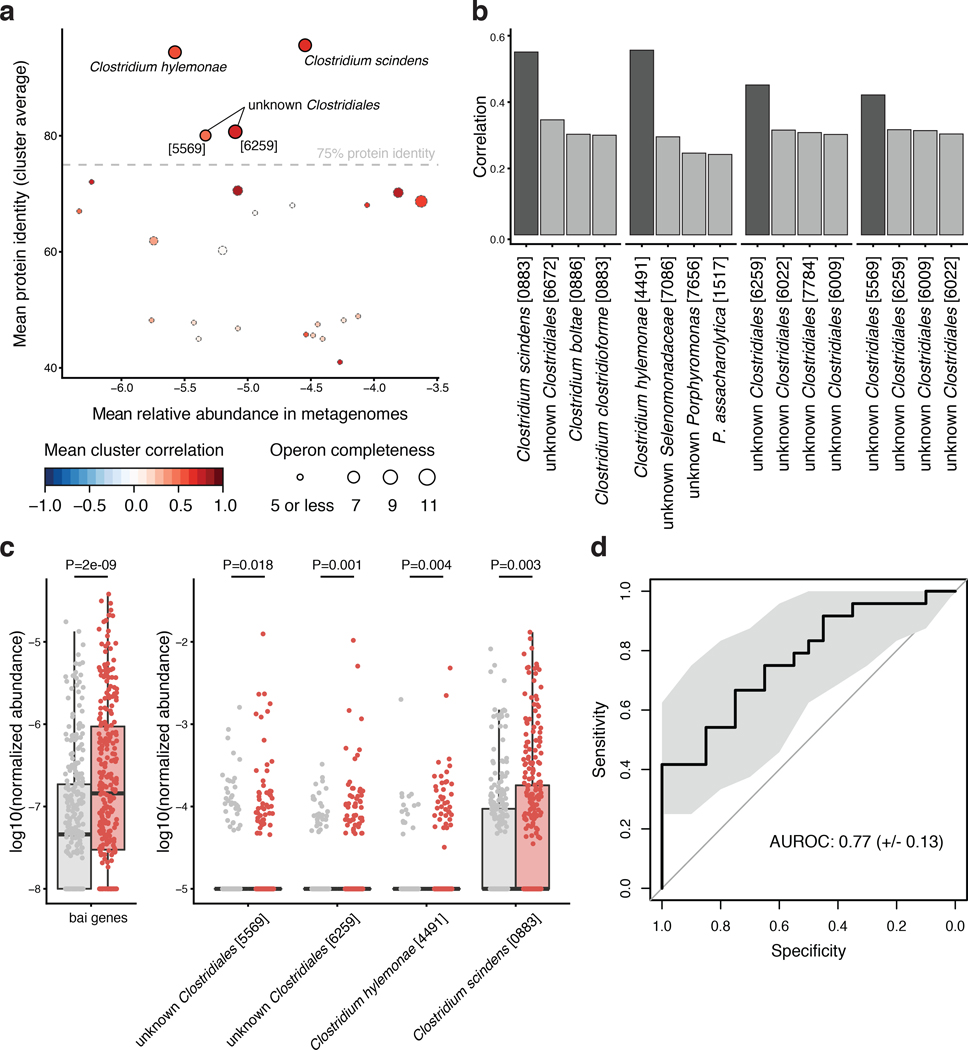
The large metagenomic data set analyzed here allowed us to quantify the prevalence of gut microbial virulence and toxicity mechanisms thought to play a role in colorectal carcinogenesis. Prominent examples include the Fusobacterium nucleatum adhesion protein A (encoded by the fadA gene), the Bacteroides fragilis enterotoxin (bft gene) and colibactin produced by some Escherichia coli strains (pks genomic island) [39, 40]. Moreover, intestinal Clostridium spp. are known to contribute to the conversion of primary to secondary bile acids using several metabolic pathways including 7α-dehydroxylation, encoded in the bai operon [41]. The products of this 7α-dehydroxylation pathway, deoxycholate and lithocholate, are known hepatotoxins associated with liver cancer [42] and hypothesized to also promote CRC [43]. Although intensely studied at a mechanistic level, these factors are not (well) represented in general databases that can be used for metagenome annotation (Supplementary Fig. 5). Thus, we built a targeted metagenome annotation workflow based on Hidden Markov Models to identify and quantify virulence factors and toxicity pathways of interest in CRC. Additionally, we used co-abundance clustering to infer operon completeness for factors encoded by multiple genes (Methods, Extended Data 9, Supplementary Fig. 5). While fadA, bft, the pks island and the bai operon were clearly detectable in deeply sequenced fecal metagenomes, they varied broadly with respect to abundance, significance and cross-study consistency of enrichment (Fig. 4c): fadA and pks were significantly enriched in CRC metagenomes (P = 5.3E-10 and 4.1E-4 respectively), whereas no significant abundance difference could be detected for bft in fecal metagenomes, despite reports on its enrichment in the mucosa of CRC patients [44], its carcinogenic effect in mouse models [45], and synergistic action with pks [46]. Our quantification of the bai operon showed a highly significant enrichment in CRC metagenomes (P = 1.6E-9) observed across all five studies (Fig. 4d) at an average abundance that exceeded fadA and pks copy numbers (Fig. 4c). Metagenome analysis indicated that at least four Clostridiales species (including the well characterized C. scindens and C. hylemonae [47, 48]) have a (near) complete 7α-dehydroxylation pathway contributing to the observed enrichment of bai operon copies (Extended Data 9). To validate this finding and further explore its value towards diagnostic application, we developed a targeted quantification assay for the baiF gene based on quantitative PCR (qPCR, see Methods). Quantification of baiF by qPCR using genomic DNA from 47 fecal samples of the DE study population was found to be similar to, yet more sensitive than by metagenomics (Fig. 4e). Gut microbial baiF copy numbers clearly distinguished CRC patients from controls (P = 0.001) at an AUROC of 0.77, which in this subset of samples is surpassed by only a single species marker for CRC (Extended Data 9). Although consistent with increased deoxycholate metabolite levels reported for serum and stool samples of CRC patients [49], this finding does not imply 7α-dehydroxylation pathway activity. We therefore quantified baiF expression using RNA extracts from the same set of fecal samples, and found also transcript levels to be elevated in CRC patients (Fig. 4f). The observed weak correlation of baiF expression with genomic abundance (Fig. 4f) might be explained by dynamic transcriptional regulation [47] and bai expression in feces might not accurately reflect the tumor microenvironment. Taken together, these data suggest gut microbial metabolic markers to be meaningful and highly predictive of CRC status.
Validation of CRC signatures in independent study populations
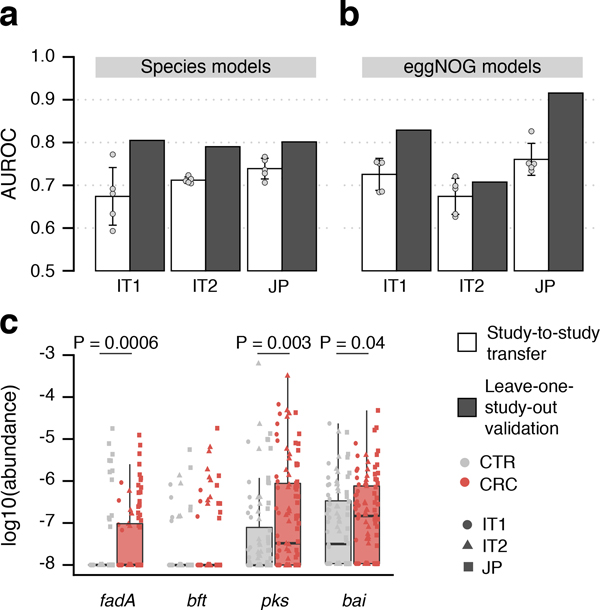
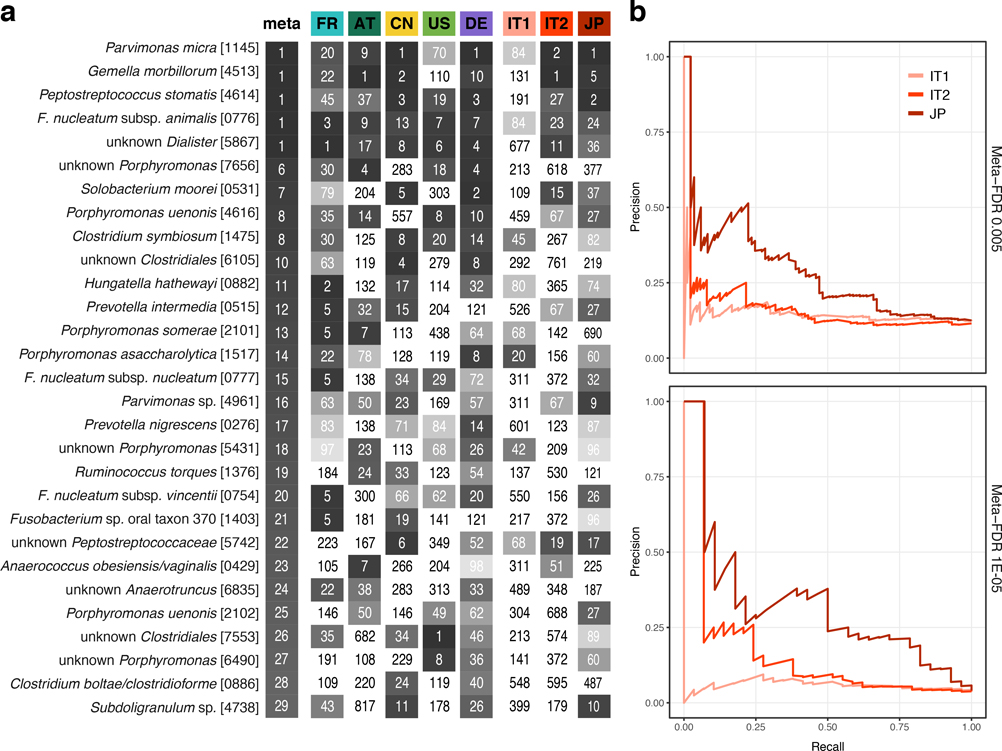
Even though CRC classification accuracy for both species and functions were evaluated on independent data, we nonetheless sought to confirm it using two additional study populations from Italy (IT1 and IT2, combined N = 61 CRC, N = 62 CTR, [27], see Methods, Table 1) and one from Japan (JP, N = 40 CRC, N = 40 CTR, see Methods, Table 1). The overlap of single species associations detected in the IT2 study and those from the meta-analysis was found to vary within the range seen for the other studies, whereas for IT1 and JP the overlap was slightly lower (cf. study precision in Supplementary Fig. 2, Extended Data 10). Nonetheless, the AUROC of LOSO classification models based on species ranged between 0.79 and 0.81 and that for the classifiers based on eggNOG from 0.71 to 0.92 (Fig. 5ab). We also validated CRC enrichment of fadA, pks and bai genes in these three study populations (Fig. 5c). Altogether these results highlight consistent alterations in the gut microbiome of CRC patients across eight study populations from seven countries in three continents.
Figure 5. Meta-analysis results are validated in three independent study populations.
CRC classification accuracy for independent datasets, two from Italy and one from Japan (see Supplementary Table S2), is indicated by bar height for single study (white) and leave-one-study-out (grey) models using either (a) species or (b) eggNOG gene family abundance profiles (cf. Fig. 3). Bar height for single study models corresponds to the average of five classifiers (error bars indicate standard deviation, n=5). (c) Normalized log abundances for virulence factors and toxins (cf. Figure 4c) compared between controls (CTR) and colorectal cancer cases (CRC). P-values were determined by blocked, one-sided Wilcoxon tests (n=193 independent samples). Boxes represent interquartile ranges (IQR) with the median as a black horizontal line and whiskers extending up to the most extreme points within 1.5-fold IQR.
DISCUSSION
Through extensive and statistically rigorous validation, in which data from studies used for training is strictly separated from that for testing, our meta-analysis firmly establishes that gut microbial signatures are highly predictive of CRC (see also [27]). In particular metagenomic classifiers trained on species profiles from multiple studies maintained an AUROC of at least 0.8 in seven out of eight data sets and achieved an accuracy similar to the fecal occult blood test, a standard non-invasive clinical test for CRC (Supplementary Fig. 6, cf. [8]). These results thus suggest that polymicrobial CRC classifiers are globally applicable and can overcome technical and geographical study differences, which we found to generally impact observed microbiome composition more than the disease itself (Fig. 1c, Extended Data 1, 2). The generalization accuracy of classifiers across studies seen here is higher than that reported in 16S rRNA gene amplicon sequencing studies, which are characterized by even larger heterogeneity across studies [16, 18] (Supplementary Fig. 7).
Previous microbiome meta-analyses suggested that the majority of gut microbial taxa differing in any given case-control study reflect general dysbiosis rather than disease-specific alterations illustrating the difficulty of establishing disease-specific microbiome signatures [17, 19]. Here, by combining data across studies for training (LOSO), we were able to develop disease-specific signatures that maintained false positive control on diabetes and IBD metagenomes at a very similar level as for CRC (Fig. 3c) despite these diseases having shared effects on the gut microbiome [17, 50] and an increased comorbidity risk [51].
Although for diagnostic purposes, unresolved causality between microbial and host processes during CRC development are not a central concern, elucidating the underlying mechanisms would greatly enhance our understanding of colorectal tumorigenesis. Towards this goal, we developed both broad and targeted annotation workflows for functional metagenome analysis. First, we found functional signatures based on the abundances of orthologous groups of microbial genes to yield accuracies as high as taxonomic signatures (Fig. 3), which raises the hope for future improvements in metagenome annotation to translate into microbiome signature refinements. Second, by investigating potentially carcinogenic bacterial virulence and toxicity mechanisms taking a targeted metagenome annotation approach, we confirmed highly significant enrichments of the colibactin-producing pks gene cluster and the Fusobacterium nucleatum adhesin FadA in CRC metagenomes (Fig. 4c). Our results support the clinical relevance of these factors adding to the experimental evidence for their carcinogenic potential [46, 52–54]. We further examined the bai operon, encoding enzymes that produce secondary bile acids via 7α-dehydroxylation, as an example of toxic host-microbial co-metabolism (see [27] for another intriguing example). While α-dehydroxylated bile acids are established liver carcinogens [42], their contribution to CRC is less clear [43]. Here, we have, for the first time, shown bai to be highly enriched in stool from CRC patients (Fig. 4cd) and confirmed this finding at both the genomic and the transcriptomic level using qPCR (Fig. 4ef). As bai enrichment (and expression) is likely a consequence of a diet rich in fat and meat [55], it is intriguing to explore whether bai could be used as a surrogate microbiome marker for such difficult-to-measure dietary CRC risk factors. To further unravel the molecular underpinning of these dietary CRC risk factors, molecular pathological epidemiology studies that investigate the mucosal microbiome as part of the tumor microenvironment, hold great potential [56, 57]. However, they will require more comprehensive diet questionnaires, medical records, and molecular tumor characterizations than are available for the study populations analyzed here. In this context, carcinogens possibly contained in the virome also warrant further investigation [58, 59], but for this goal, metagenomic data needs to be generated with protocols optimized for virus enrichment [60].
Taken together, our results and those by Thomas, Manghi et al. [27], strongly support the promise of microbiome-based CRC diagnostics. Both taxonomic and metabolic gut microbial marker genes established in these meta-analyses could form the basis of future diagnostic assays that are sufficiently robust, sensitive, and cost-effective for clinical application. The targeted qPCR-based quantification of the baiF gene is a first step in this direction. Our metagenomic analysis of this and other virulence and toxicity markers bridge to existing mechanistic work in preclinical models and could enable future work aiming to precisely determine the contribution of gut microbiota to CRC development.
Data and Code Availability
The raw sequencing data for the samples in the DE study that had not been published before (see Methods), are made available in the European Nucleotide Archive (ENA) under the study identifier PRJEB27928. Metadata for these samples are available as Supplementary Table S6.
For the other studies included here, the raw sequencing data can be found under the following ENA identifiers: PRJEB10878 for [11], PRJEB12449 for [10], ERP008729 for [9], and ERP005534 for [8]. The independent validation cohorts can be found in SRA under the identifier SRP136711 for [27] and in the DDBJ database under the ID DRA006684.
Filtered taxonomic and functional profiles used as input for the statistical modeling pipeline are available in Supplementary Data 1.
The code and all analysis results can be found under https://github.com/zellerlab/crc_meta.
Methods
Study inclusion and data acquisition
We used PubMed to search for studies that published fecal shotgun metagenomic data of human colorectal cancer patients and healthy controls. The search term, all hits, and the justification for exclusion or inclusion are available in Supplementary Table S1. Raw fastq files were downloaded for the four included studies from the European Nucleotide Archive, using the following ENA identifiers: PRJEB10878 for [11], PRJEB12449 for [10], ERP008729 for [9], and ERP005534 for [8].
DE study recruitment and sequencing
The German (DE) study population data consist of 60 fecal CRC metagenomes, 38 of which were sequenced and published in [8] under ENA accession ERP005534. The fecal metagenomes from additional 22 CRC patients recruited for the same ColoCare study (DKFZ, Heidelberg, [61, 62]) were sequenced later as part of this work. All fecal samples were collected after colonoscopy. Sixty gender- and age-matched participants of the PRÄVENT study run by the same clinical investigators were included as healthy controls; as these were not subjected to colonoscopy, the presence of undiagnosed colorectal carcinomas cannot be completely ruled out but is expected to be unlikely due to low prevalence of preclinical CRC in the general population [63].
Written informed consent was obtained from all additional 22 CRC patients and 60 controls. The study protocol was approved by the institutional review board (EMBL Bioethics Internal Advisory Board) and the ethics committee of the Medical Faculty at the University of Heidelberg. The study is in agreement with the WMA Declaration of Helsinki and the Department of Health and Human Services Belmont Report.
Genomic DNA was extracted from the fecal samples (preserved in RNALater) and libraries were prepared as previously described [8]. Whole-genome shotgun sequencing was performed by using Illumina HiSeq 2000 / 2500 / 4000 (Illumina, San Diego, USA) platforms at the Genomics Core Facility, European Molecular Biology Laboratory, Heidelberg.
Independent validation cohorts
During the revision of this manuscript, we included three independent study populations for external validation. Two of them were recruited in Italy (IT1 and IT2) with informed consent from all participants and ethical approval by the Ethics committee of Azienda Ospedaliera of Alessandria and that of the European Institute of Oncology of Milan. Shotgun fecal metagenomic data was generated as described in [27].
The third study population was recruited in Japan (JP) with informed consent and ethical approval of the institutional review boards of the National Cancer Center Japan - Research Institute and the Tokyo Institute of Technology. DNA was extracted from frozen fecal samples using a GNOME DNA Isolation Kit (MP Biomedicals, Santa Ana, CA) with an additional bead-beating step as previously described [64]. DNA quality was assessed with an Agilent 4200 TapeStation (Agilent Technologies, Santa Clara CA). After final precipitation, the DNA samples were resuspended in TE buffer and stored at −80°C before further analysis. Sequencing libraries were generated with the Nextera XT DNA Sample Preparation Kit (Illumina, San Diego, CA). Library quality was confirmed with an Agilent 4200 TapeStation. Whole-genome shotgun sequencing was carried out on the HiSeq2500 platform (Illumina). All samples were paired-end sequenced with a 150-bp read length to a targeted data set size of 5.0 Gb.
Taxonomic profiling and data preprocessing
The metagenomic samples were quality controlled using MOCAT2’s -rtf procedure, which is based on the ‘solexaqa’ algorithm [23]. In particular, reads that map with at least 95% sequence identity and alignment length of at least 45 bp to the human genome hg19 were removed. In a second step, taxonomic profiles were generated with the mOTU profiler version 2.0.0 ([22, 65, 66] – see motu-tool.org and GitHub version tag 2.0.0) using the following parameters: -l 75, -g 2 and -c. Briefly, this profiler is based on ten universal single-copy marker-gene families (COG0012, COG0016, COG0018, COG0172, COG0215, COG0495, COG0525, COG0533, COG0541 and COG0552) [66]. These marker-genes were extracted from >25,000 reference genomes and >3,000 metagenomic samples allowing to profile prokaryotic species with a sequenced reference genome (ref-mOTUs) and ones without (meta-mOTUs). The read count for a mOTU was calculated as median of the read count of the genes that belonged to that mOTU.
mOTU profiles were first converted to relative abundances to account for library size. Then, profiles were filtered to focus on a set of species that are confidently detectable in multiple studies. Specifically, microbial species that did not exceed a maximum relative abundance of 1E-03 in at least 3 of the studies were excluded from further analysis, together with the fraction of unmapped metagenomic reads.
Functional metagenome profiling and data preprocessing
High-quality reads (same quality filtering as for taxonomic profiling) were aligned against a combined database (IGChg38 hereafter) consisting of the hg38 release of the human reference genome and the integrated gene catalog (IGC) containing 9.9 million non-redundant microbial genes [30] using BWA mem [67] (Version: 0.7.15-r1140) with default parameters. The purpose of adding the human genome to the reference database was to filter out reads that mapped as well or better to some human sequence than to any bacterial gene. Alignments were computed separately for paired-end and single read libraries (single reads could result from read pairs where one read was filtered out in the quality filtering procedure described above). Alignments were then filtered to only retain those longer than 50bp with >95% sequence identity. Then the highest scoring alignment(s) was/were kept for each read. As IGChg38 is a database of predominantly genes and not genomes, there will be a substantial proportion of read-pairs where one end maps within the gene while the other end does not – it either maps to an adjacent gene or remains unmapped due to intergenic regions not contained in the database. Therefore, we counted a whole read-pair aligning to a gene when (i) both ends from a read pair map to the same gene, (ii) only one end from a read-pair maps to the gene, or (iii) a read from the single read library maps to the gene. We then counted only the read-pairs that map uniquely to one gene in the IGC, thus excluding ambiguous read pairs mapping with similarly high scores to multiple genes in the database. For a given metagenomic sample, we further normalized the abundance of each IGC gene by the length of that gene. We then estimated relative abundance of IGC genes by dividing gene abundances by the total abundance of all genes in IGC (excluding the human chromosomes).
Because metagenomes from CRC patients were not included when the IGC was constructed, we analyzed how well CRC-associated species as identified in this meta-analysis were represented in the IGC. Using a phylogenetic marker gene (COG0533), which is also used by the species profiling workflow on which the meta-analysis is based, for 24 out of the 29 core CRC-associated species we found a match in the IGC with at least 90% nucleotide identity, indicating that a sequence from the same species (above 93.1% identity) or a slightly more distant relative is present in the IGC (Supplementary Fig. 8). The relative abundance of eggNOG orthologous groups [28] was estimated by summing relative abundances of genes annotated to belong to the same eggNOG orthologous group as of the most recent annotations provided by MOCAT2 [23]. To obtain KEGG orthologous groups (KO) and pathway abundances, we applied the same procedure, but using KEGG annotations for IGC provided by MOCAT2 [29].
Overview over statistical analyses
For univariate association testing between the abundances of microbial taxa or gene functions we used nonparametric tests throughout; all of these were two-sided Wilcoxon tests except were otherwise noted. To account for potential confounding and heterogeneity between data sets we employed a stratified version of the Wilcoxon test [24] (see below for details). ANOVA was conducted on rank-transformed data. Significance of binary co-occurrence patterns was assessed using (stratified) Cochrane-Mantel-Haenszel tests.
Multivariable analysis was done with strict separation between training and test data. This importantly also pertained to feature selection, which was either done via the LASSO [68] or by nested cross-validation procedures to avoid overoptimistic performance assessment [69] (see below for details). All samples included in this meta-analysis came from distinct individuals to ensure that generalization across subjects – rather than across timepoints within a given subject – is assessed.
Confounder analysis
To quantify the effect of potential confounding factors relative to that of CRC on single microbial species, we used an ANOVA-type analysis. The total variance within the abundance of a given microbial species was compared to the variance explained by disease status and the variance explained by the confounding factor akin to a linear model including both CRC status and confounding factor as explanatory variables for species abundance. Variance calculations were performed on ranks in order to account for non-Gaussian distribution of microbiome abundance data. Potential confounders with continuous values were transformed into categorical data either as quartiles or for the case of body mass index (BMI) into lean/obese/overweight according to conventional cutoffs (lean: < 25, obese: 25 – 30, overweight: > 30).
Univariate meta-analysis for the identification of CRC-associated gut microbial species
Significance of differential abundance was tested on a per-species basis using a blocked Wilcoxon test implemented in the R coin package [24]. Informed by the results of the preceding confounder analysis, we blocked for `study` and additionally `colonoscopy` in the CN study. Within this framework, significance is tested against a conditional null distribution derived from permutations of the observed data. Notably, permutations are performed within each block in order to control for variations in block size and composition. To adjust for multiple hypothesis testing, P-values were adjusted using the false-discovery rate (FDR) method [70].
As nonparametric effect size measures we used the area under the ROC curve (AUROC) with permutation-based confidence intervals computed using the pROC package in R [71]. We further developed a generalization of the (logarithmic) fold change that is widely used for other types of read abundance data. This generalization is designed to have better resolution for sparse microbiome profiles (where 0 entries can render median-based fold change estimates uninformative for the large portion of species with a prevalence below 0.5). The generalized fold change (gFC) is computed as mean difference in a set of pre-defined quantiles of the logarithmic CTR and CRC distributions (see Extended Data 3 for further details; we used quantiles ranging from 0.1 to 0.9 in increments of 0.1).
For the retrospective analysis of study precision and recall for detecting microbial species associations from the meta-analysis, the true set was defined as the species which were associated at a given FDR in the meta-analysis. Then, we checked how well this set of species would be recovered using the single-study significance as determined by the Wilcoxon test. Study precision corresponds to the proportion of meta-analysis significant species among those detected as significant in a single study. Similarly, recall (or sensitivity) corresponds to the proportion of species out of the true set of meta-analysis significant species that were recovered in a given study.
Species co-occurrence and cluster analysis in CRC metagenomes
For the analysis of gut bacterial species co-occurring in CRC microbiomes, relative abundances of the core set of associated species (excluding the CRC-depleted Clostridiales meta-mOTU [1296]) were discretized into binary values to determine whether a CRC (metagenomic) sample is “positive” or “negative” for a given microbial marker. To normalize for differences in prevalence (and therefore specificity) of these markers we adjusted the threshold value, above which a sample is labeled “positive” based on the abundance in healthy controls. For each microbial species, the 95th percentile in healthy controls was used as threshold, which effectively results in adjusting the per-marker false positive rate to 0.05. Based on the binarized species-by-sample matrix, species were then clustered using the Jaccard dissimilarity as implemented in the vegan package in R [72]. Associations between species clusters and meta-variables were tested as 2-by-n (where n is the number of categories in the meta-variable tested) contingency tables using a Cochrane-Mantel-Haenszel test with study as blocking factor as implemented in the coin package [24].
Multivariable statistical modeling workflow and model evaluation
As a main goal of our work is to assess the generalization accuracy of microbiome-based CRC classifiers across technical and geographic differences in patient populations, we extensively validated classification models across studies taking the following two approaches.
In study-to-study transfer validation, metagenomic classifiers were trained on a single study and their performance externally assessed on all other studies (off-diagonal cells in Fig. 3ac). Effectively we implemented a nested cross validation procedure on the training study to compute within-study accuracy (cells on the diagonal in Fig. 3ac) and tune the model hyperparameters.
In leave-one-study-out (LOSO) validation, data from one study was set aside as an external validation set, while the data from the remaining 4 studies was pooled as a training set on which we implemented the same nested cross validation procedure as for study-to-study transfer (see [19] for a more detailed description of LOSO).
Data preprocessing, model building, and model evaluation was performed using the SIAMCAT R package (https://bioconductor.org/packages/SIAMCAT, version 1.1.0).
Preprocessing of taxonomic abundance profiles for statistical modeling
Relative abundances were first filtered to remove markers with low overall abundance and no variance (an artifact for single-study data arising from the joint data filtering described above), log-transformed (after adding a pseudo-count of 1E-05 to avoid non-finite values resulting from log(0), [73]) and finally standardized as z-scores. Data were split into training and test set for 10 times repeated 10-fold stratified cross validation (balancing class proportions across folds). For each split, a L1-regularized (LASSO) logistic regression model [68] was trained on the training set, which was then used to predict the test set. The lambda parameter, i.e. regularization strength was selected for each model to maximize the area under the precision recall curve under the constraint that the model contained at least 5 non-zero coefficients. Models were then evaluated by calculating the area under the Receiver Operating Characteristics curve (AUROC) based on the posterior probability for the CRC class.
In model transfer to a hold-out study, the holdout data were normalized for comparability in the same way as the training dataset by using the frozen normalization function in SIAMCAT, which retains the same features and re-uses the same normalization parameters (e.g. the mean of a feature for z-score standardization). Then, all 100 models derived from the cross validation on the training dataset (10 times repeated 10-fold CV) were applied to the holdout dataset and predictions were averaged across all models.
In the LOSO setting, data from the four training studies were jointly processed as a single dataset in the same way as described above using 10 times repeated 10-fold stratified cross validation.
Preprocessing of functional abundance profiles
Functional profiles, such as eggNOG gene family or KEGG module abundance profiles were preprocessed as described above for species profiles, but using 1E-06 as maximum abundance cutoff and 1E-09 as a pseudo-count during log transformation. Since these abundance tables contained several thousand input features we implemented an additional feature selection step, which was nested properly into the cross-validation procedures as described above. This nested approach is crucial to avoid over-optimistically biased performance estimates ([74], Chapter 7.10). Specifically, features were filtered inside each training fold (without using any information from the test fold) by selecting the 1600 features with highest single-feature AUROC values (for features depleted in CRC, 1 - AUROC was used for feature selection).
Preprocessing of gene abundance profiles
To ascertain the predictive power of a classifiers based on IGC gene abundances [30] we applied a series of filters to the abundance tables to reduce the number of genes that would be the input of the LASSO modelling. These filters where applied once on a per-study level and once in a leave-one-study-out (LOSO) mode, where they were applied jointly to all studies in the training set, with the remaining one being held out for external validation.
The following filters were applied in this order:
All genes with 0 abundance in ≥15% of samples (regardless of CRC status) were discarded.
The remaining data was discretized using the equal frequencies method implemented in the ‘discretize’ function of the sideChannelAttack R package (version 1.0–6) as a preparation to the minimal-redundancy-maximal-relevance (mRMR) algorithm [75].
As a feature selection procedure, mRMR (code version from 20 April 2009 downloaded from http://home.penglab.com/proj/mRMR/ on 3 Dec 2016) was run on the gene abundance table to retain the 100 top genes as output.
LASSO models were then built on log10-transformed abundances (pseudo-count of 10E-09, centered and scaled) of the sets of 100 top genes returned by mRMR. The whole process was repeated 10 times in a 5-fold stratified cross-validation scheme to allow for an estimation of the confidence of the AUROCs of the resulting models. We used the LiblineaR package (version 2.10–8) to build the LASSO models in R and tested a sequence of 20 cost parameters (equivalent or the lambda parameter controlling regularization strength) evenly spaced from 0.0012 to 0.22. The cost parameter was selected to maximize the AUROC within the training set.
External evaluation of disease-specificity of the metagenomic classifiers
To assess how disease-specific the predictions of the CRC models are, we applied these to data from case-control studies investigating other human diseases. Fecal metagenomic data of patients with Parkinson’s disease [12], type 2 diabetes [4, 5], and inflammatory bowel disease [6, 7] were taxonomically profiled as described above. The parameters for quality control with MOCAT2 and for the mOTU profiler were the same as described above, except for the data from [6], where we used -l 50 (to set the threshold for minimum alignment length to 50) as the read length is shorter (average read length 71) compared to the other more recently generated Illumina shotgun metagenomic data.
Relative abundance data were treated exactly as another holdout dataset for each model, i.e. applying the frozen normalization prediction routines as described above. For each CRC model applied to the external datasets, a cutoff on its prediction output was adjusted to yield a false positive rate (FPR) of 0.1 on the controls of its respective (CRC) training set. Subsequently its FPR on metagenomes from patients suffering from the above-mentioned (non-CRC) conditions was assessed to evaluate its disease specificity. The rationale behind this is that a metagenomic classifier recognizing general features of dysbiosis would be expected to predict CRC patients and those suffering from other conditions at a similar rate; such a classifier would thus in the above-described evaluation display a much higher FPR than on the controls of its training set. In contrast maintaining a low FPR in this evaluation indicates that the classification model is based on CRC-specific features rather than hallmarks of general dysbiosis or nonspecific inflammation.
Functional profiling of gut metabolic modules (GMMs)
Gut metabolic modules were computed as originally proposed [31], using the KEGG KO profiles based on the IGC (see Functional metagenome profiling above) as input. Statistical analysis and generalized fold change calculations were performed analogously to species profiles (see above). Gut metabolic modules were summarized across functional groups (e.g. amino acid degradation) as geometric mean of all modules within the respective group.
Targeted functional analysis of virulence and toxicity pathways of potential relevance in CRC
To investigate toxins and virulence mechanisms that have previously been implicated with CRC [40], we constructed for each gene belonging to the respective virulence or toxicity pathway a hidden Markov model (HMM). Each HMM was built from a multiple sequence alignment generated by MUSCLE [76], containing the respective reference sequences and close homologs identified using PSI-Blast [77]. Multiple sequence alignments are available together with the code for this paper (https://github.com/zellerlab/crc_meta). Then, we screened the IGC metagenomic gene catalogue [30] with each HMM using the HMMER software (version 3.1b2) [78]. Genes with an E-value below 1E-10 were filtered for uniqueness, since in some cases the HMMs would call different regions in the same gene. For single gene virulence factors (i.e. fadA and bft), potential IGC hits were aligned against the reference sequence using the Needleman-Wunsch algorithm in the EMBOSS package [79]. Hits were then filtered based on percentage of sequence identity (cutoff: 40%) and sequence similarly to the species relative abundance profiles based on maximum relative abundance (cutoff: 1E-07) in order to exclude genes with limited relevance. Statistical analysis was performed on the sum of all genes.
For virulence pathways containing more than one gene, the IGC hits of each functional group within the pathway were aligned against the respective reference sequence and filtered for percentage of sequence identity and maximum abundance. Then, all hits were clustered based on the Pearson correlation of the log-abundances across all samples using the Ward algorithm as implemented in the hclust function in R. The gene clusters were filtered based on operon completeness (how many genes of the operon were present in the cluster) and average correlation within the cluster (Extended Data 9). For statistical analysis, the genes in the selected gene clusters were summed up within each group or all together for the overall analysis.
Quantitative PCR for baiF
Real-time quantitative PCR to quantify the abundance and expression of baiF was performed on a subset of samples in the DE cohort (20 control and 24 colorectal cancer samples, see Supplementary Table S6). For these samples, DNA and RNA extraction was done with the Allprep PowerFecal DNA/RNA kit (Qiagen, Cat No: 80244) with additional RNAse and DNAse digestion steps, respectively, as described by the manufacturer. DNA and RNA concentrations were determined by Qubit Fluorometer (Invitrogen) and quality control of all RNA samples was done using an Agilent 2100 Bioanalyzer in combination with RNA 6000 Nano and Pico LabChip kits.
First-strand cDNA was synthesized by SuperScript IV VILO Master Mix with ezDNAse enzyme and random hexamer primers (Invitrogen, catalogue number 11766500) as recommended by the manufacturer. Reaction were performed as described in the protocol with one minor change of temperature (incubation for the reverse transcription step at 55°C).
To quantify baiF relative to the total bacterial RNA/DNA in a sample, qPCR was performed in triplicates for 16S rRNA and the baiF genes, using both cDNA and genomic DNA (gDNA) as template. We used the following primers for baiF: TTCAGYTTCTACACCTG (forward), GGTTRTCCATRCCGAACAGCG (reverse), and standard primers F515 and R806 for 16S [80]. RT-PCR reactions were prepared with a final primer concentration of 0.5 μM, including 5 ng of genomic DNA or 10 ng of cDNA in 20 μl final reaction volume, and reactions were performed with SYBR Green qPCR mix on StepOne Real-Time PCR system (Thermo Fisler Scientific). Cycling conditions were as follows; initial denaturation of 95°C for 10 min, then 40 cycles of denaturing at 95°C for 15 s, annealing at 60°C for 60 s followed by melt curve analysis.
Delta-Ct values were calculated as difference between baiF and 16S Ct values. Significance of the comparison between control and colorectal cancer samples was tested on the delta-Ct values using a one-sided Wilcoxon test as a confirmation of metagenomic enrichment.
Extended Data
Extended Data Figure 1: Potential confounder of individual microbial species associations by patient demographics and technical factors.
Variance explained by disease status (CRC vs control) is plotted against variance explained by different putative confounding factors for individual microbial species. Each species is represented by a dot proportional in size to its abundance (see legend and Methods); core microbial markers identified in meta-analysis (tested by two-sided blocked Wilcoxon test, n=574 independent observations) are highlighted in red. For the confounder analysis, factors with continuous values were discretized into quartiles and BMI was split into lean/overweight/obese according to conventional cutoffs. The variance explained by disease status was computed all data; accordingly, the x-values are the same in all panels and also in Fig. 1d. Variance explained by different confounding factors was computed using all samples for which data were available (indicated by insets).
Extended Data Figure 2: Study shows a strong influence on alpha and beta diversity.
(a) Alpha diversity as measured by the Shannon index was computed for all gut microbial species (n=849), reference mOTUs (n=246), and meta mOTUs (n=603) separately. P-values were computed using two-sided Wilcoxon test, while the overall p-value (on top) was calculated using a two-sided blocked Wilcoxon test (n=575 independent observations, see Methods). Anova F statistics below the panel were computed using the R function aov. (b) Principal coordinate analysis of samples from all five included studies based on Bray-Curtis distance; study is color-coded and disease status (CRC vs control) indicated by filled/unfilled circles. The boxplots on the side and below show samples projected onto the first two principle coordinates broken down by study and disease status, respectively. P-values were computed using a two-sided Wilcoxon test for disease status and a Kruskal-Wallis test for study, (n=575 independent observations). For all boxplots, boxes denote the interquartile ranges (IQR) with the median as thick black line and whiskers extending up to the most extreme points within 1.5-fold IQR.
Extended Data Figure 3: The generalized fold change extends the established (median-based) fold change to provide higher resolution in sparse microbiome data.
(a) In the top row, the logarithmic relative abundances for Bacteroides dorei/vulgatus, Parvimonas micra, and Fusobacterium nucleatum subsp. animalis -examples for a highly prevalent and two low-prevalence species- are shown as swarmplot for the control (CTR) and colorectal cancer (CRC) groups. The thick vertical lines indicate the medians in the different groups and the black horizontal line shows the difference between the two medians, which corresponds to the classical (median-based) fold change. Since Fusobacterium nucleatum subsp. animalis is not detectable in more than 50% of the cancer cases, there is no difference between the CTR and CRC median and thus the fold change is 0. The lower row shows the same data, but instead of only the median (or 50th percentile), 9 quantiles ranging from 10% to 90% are shown by thinner vertical lines. The generalized fold change is indicated by the horizontal black line again, computed as mean of the differences between the corresponding quantiles in both groups. In the case of the sparse data (e.g. Fusobacterium), the differences in the 70%, 80% and 90% quantiles cause the generalized fold change to be higher than 0. (b) The median fold change is plotted against the newly developed generalized fold change (gFC) for all microbial species (core set of microbial CRC marker species highlighted in orange). Marginal histograms visualize the distribution for both FC and gFC. (c) Scatter plots showing the relationship between FC and gFC and area under the Receiver Operating Characteristics (AUROC) or shift in prevalence between CRC and CTR, with Spearman correlations added in the top-left corners; gFC provides higher resolution (wider distribution around 0) and better correlation with the nonparametric AUROC effect size measure as well as prevalence shift, which captures the difference in prevalence of a species in CRC metagenomes relative to control metagenomes.
Extended Data Figure 4: Microbial genera identified in meta-analysis to be associated with CRC.
(a) Meta-analysis significance of microbial genera, computed using univariate two-sided Wilcoxon test blocked for study and colonoscopy (n=574 independent observations) is given by bar height (FDR 0.005). Underneath, significance (FDR-corrected p-value computed from two-sided Wilcoxon test) and generalized fold changes (see Methods) within individual studies are displayed as heatmaps in gray and color, respectively (see keys). Genera are ordered by meta-analysis significance and direction of change. (b) For highly significant genera (meta-analysis FDR 1E-05), association strength is quantified by the area under the Receiver Operating Characteristics (AUROC) across individual studies (color-coded diamonds) and 95% confidence interval are depicted by gray lines.
Extended Data Figure 5: The core set of CRC-enriched microbial species can be stratified into four clusters based on co-occurrence in CRC metagenomes.
(a) The heatmap shows the Jaccard index (computed by comparing marker-positive samples, see Methods) for the core set of microbial marker species, compute on CRC cases only. Clustering was performed using the Ward algorithm as implemented in the R function hclust. The inset shows the distribution of Jaccard similarities within each cluster and for the background (all similarities between species not in the same cluster, n=841). Boxes denote the interquartile ranges (IQR) with the median as thick black line and whiskers extending up to the most extreme points within 1.5-fold IQR. (b) Barplots show the fraction of CRC samples that are positive for a marker species clusters (defined as the union of positive marker species) broken down by patient subgroups based on BMI and (c) age (see Fig. 2bcd for other patient subgroups). Significance of the associations between CRC subgroups and marker species clusters were tested using the Cochran-Mantel-Haenszel test blocked for study (but no significant associations were detected). (d) For the core set of microbial species with a genomic reference, the presence (red) or absence (white) of superoxide dismutase, peroxidase, and catalase are shown as heatmap (see Methods).
Extended Data Figure 6: Coefficients of leave-one-study-out LASSO logistic regression models compared to models trained on individual studies.
(a) Mean coefficients (feature weights) from LASSO cross-validation models traind on single studies (color-coded) are plotted against the single feature AUROC for each species feature. Horizontal lines highlight microbial species that are -for at least one study- selected in more than 50% of the models in cross-validation and account for more than 10% of the absolute model weight in at least 10% of the cross-validation models. Similarly, (b) shows the same for models trained in the leave-one-study-out (LOSO) setting (see Methods). Colors indicate which study has been left out of the the training set (and is used for validation). Since the weights of the LOSO models are spread across more species and thus generally lower, species are highlighted by horizontal lines if their weights explain more than 2.5% of the absolute model in at least 10% of cross-validation models and they have been selected in more than 50% of models in cross-validation. (c) Inset shows the distribution of the number of non-zero coefficients across all cross-validation models. (d) Bar height indicates the number of non-zero coefficients that are shared between the mean models for each study or left-out study, respectively. (e) The study-to-study difference (computed as median of all pairwise differences between model weights for a single species across the mean models) for cross-validation (CV) single-study models are plotted against the same measure for the LOSO models. Species with a study-to-study difference of more than 0.02 in the cross-validation models are highlighted and annotated, showing much larger variability between models trained on single studies compared to LOSO models.
Extended Data Figure 7: Analysis of leave-one-study-out models for prediction bias.
(a) To examine whether species and gene-family-level classification models are confounded, i.e. biased towards certain patient subgroups, prediction scores from leave-one-study-out models are plotted broken down into strata for each clinical parameter (e.g. female and male for sex). Prediction bias for each variable was tested by two-sided Wilcoxon (for sex and BMI) or Kruskal-Wallis (all others) tests while blocking for study as confounder (n=575 independent observations). Boxes denote interquartile ranges (IQR) with the median as horizontal black line and whiskers extending up to the most extreme point within 1.5-fold IQR. A significant difference in prediction score was detected only for CRC stage. This stage-bias is more pronounced for gene-family then for species models. (b) To examine CRC stage bias further the barplots show the true positive rate (TPR) corresponding to an overall 10% false positive rate (see also Fig. 3c) for the different CRC stages displaying slightly higher classification sensitivity for late stage CRC for both species and gene-family models.
Extended Data Figure 8: Cross-study performance of statistical models based on KEGG KO abundances, single-gene abundances from the metagenomic gene catalogue (IGC), and the combination of taxonomic and eggNOG abundance profiles.
CRC classification accuracy resulting from cross validation within each study (gray boxed along diagonal) and study-to-study model transfer (external validations off diagonal) as measured by AUROC for classification models trained on KEGG KO (a), models based on the gene catalogue (b), and models based on the combination of taxonomic and eggNOG abundance profiles (c) (see Methods for details on statistical modeling workflows). The last column depicts the average AUROC across external validations. The barplots on the right show that the classification accuracy on a held-out study improves if data from all other studies are combined for training (leave-one-study-out, LOSO validation) relative to the mean of models trained on data from a single study (study-to-study transfer, n=4, error bars show standard deviation) consistently across different types of input data.
Extended Data Figure 9: Identification of bai genes in metagenomes.
Putative bai genes identified in the metagenomic gene catalogue (IGC) were clustered by co-abundance in metagenomes to infer genomic linkage (see Methods) to be able to infer operon completeness and species of origin. (a) For each resulting cluster of putative bile acid converting genes, the mean relative abundance is plotted against the mean percentage of protein identity derived from global alignment against the know bile acid converting genes from C. scindens and C. hylemonae (see Methods). Completeness, i.e. how many of the 11 different bai gene functions are represented in each cluster, and mean gene-to-gene Pearson correlation of log-relative abundance within each cluster are encoded by dot size and color, respectively (see legend). The four clusters with mean protein identity above 75% to know bai operon containing genomes are included in the subsequent analysis and labeled with the most highly correlated mOTU (see (b)). (b) Pearson correlation between gene cluster abundances and most highly correlated species relative abundance (in logarithmic space) is given by bar height for the four gene clusters identified in (a). The most highly correlating species is highlighted in darker grey (see labeling of gene clusters in (a)). (c) The log-transformed abundances of all bai genes and the four species identified in (b) are shown as boxplots for controls (grey) and CRC cases (red). Assessing the significance of differences between CRC and controls (by a two-sided Wilcoxon test blocked for study and colonoscopy, n=574 independent observations) demonstrates a much more significant CRC enrichment of aggregated metagenomic bai gene abundance than of the individual clostridial species to which these belong. Boxes denote the interquartile ranges (IQR) with the median as thick black line and whiskers extending up to the most extreme points within 1.5-fold IQR. (d) Receiver operating characteristic (ROC) curve for the qPCR quantification of the baiF gene in genomic DNA of a subset of samples in the DE study (n=47, see Methods and Fig. 4e) is shown as black line. Shaded grey area depicts the 95% confidence interval.
Extended Data Figure 10: Validation of meta-analysis single species associations in three independent cohorts.
(a) Heatmap showing for the core set of CRC-associated species (see Fig. 1) the rank of the respective species within the associations of each study (tested by two-sided Wilcoxon test), including the three independent validation cohorts (see Table 1), compared to the rank in the meta-analysis (meta, tested by two-sided blocked Wilcoxon test) on the left. (b) Precision-recall curves for the different independent validation cohorts using the meta-analysis set of associated species at FDR 0.005 (n=94, top) and 1E-05 (n=29, bottom) as “true” set (tested by two-sided blocked Wilcoxon test, see Methods) and the naïve (uncorrected) within-cohort significance (tested by two-sided Wilcoxon test) as predictor (see Supplementary Figure X).
Supplementary Material
Acknowledgements
We are thankful to members of the Zeller, Bork and Arumugam groups for inspiring discussions. Additionally, we thank Yan Ping Yuan and the EMBL Information Technology Core Facility for support with high-performance computing as well as the EMBL Genomics Core Facility for sequencing support. We are also grateful for advice from Bern Klaus, EMBL Centre for Statistical Data Analysis. We acknowledge funding from EMBL, DKFZ, the Huntsman Cancer Foundation, the Intramural Research Program of the National Cancer Institute, ETH Zürich, and the following external sources: the European Research Council (CancerBiome ERC-2010-AdG_20100317 to P.B., Microbios ERC-AdG-669830 to P.B., Meta-PG ERC-2016-STG-716575 to N.S.), the Novo Nordisk Foundation (grant NNF10CC1016515 to M.A.), the Danish Diabetes Academy supported by the Novo Nordisk Foundation and TARGET research initiative (Danish Strategic Research Council [0603-00484B] to M.A.), the Matthias-Lackas Foundation (to C.M.U), the National Cancer Institute (grants R01 CA189184, R01 CA207371, U01 CA206110, P30 CA042014 all to C.M.U), the BMBF (the de.NBI network #031A537B to P.B. and the ERA-NET TRANSCAN project 01KT1503 to C.M.U.), the Helmut Horten Foundation (to S.Sunagawa), and the Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP – 16/23527-2 to A.M.T.).
For the IT Validation Cohorts, funding was provided by Lega Italiana per La Lotta contro i Tumori.
For the JP Validation Cohort, funding was provided by the National Cancer Center Research and Development Fund (25-A-4,28-A-4, and 29-A-6), Practical Research Project for Rare/Intractable Diseases from the Japan Agency for Medical Research and Development (AMED) (JP18ek0109187), JST (Japan Science and Technology Agency)-PRESTO (JPMJPR1507), JSPS (Japan Society for the Promotion of Science) KAKENHI (16J10135, 142558 and 221S0002), Joint Research Project of the Institute of Medical Science, the University of Tokyo, and the Takeda Science Foundation and Suzuken Memorial Foundation.
Footnotes
Competing Interest
P. Bork, G. Zeller, A.Y. Voigt, and S. Sunagawa are named inventors on a patent (EP2955232A1: Method for diagnosing colorectal cancer based on analyzing the gut microbiome).
References
- 1.Tringe SG and Rubin EM, Metagenomics: DNA sequencing of environmental samples. Nat. Rev. Genet, 2005. 6(11): p. 805–814. [DOI] [PubMed] [Google Scholar]
- 2.Tremaroli V and Bäckhed F, Functional interactions between the gut microbiota and host metabolism. Nature, 2012. 489(7415): p. 242–249. [DOI] [PubMed] [Google Scholar]
- 3.Lynch SV and Pedersen O, The Human Intestinal Microbiome in Health and Disease. N. Engl. J. Med, 2016. 375(24): p. 2369–2379. [DOI] [PubMed] [Google Scholar]
- 4.Qin J, et al. , A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature, 2012. 490(7418): p. 55–60. [DOI] [PubMed] [Google Scholar]
- 5.Karlsson FH, et al. , Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature, 2013. 498(7452): p. 99–103. [DOI] [PubMed] [Google Scholar]
- 6.Qin J, et al. , A human gut microbial gene catalogue established by metagenomic sequencing. Nature, 2010. 464(7285): p. 59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schirmer M, et al. , Dynamics of metatranscription in the inflammatory bowel disease gut microbiome. Nat Microbiol, 2018. 3(3): p. 337–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zeller G, et al. , Potential of fecal microbiota for early-stage detection of colorectal cancer. Mol. Syst. Biol, 2014. 10(11): p. 766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feng Q, et al. , Gut microbiome development along the colorectal adenoma-carcinoma sequence. Nat. Commun, 2015. 6: p. 6528. [DOI] [PubMed] [Google Scholar]
- 10.Vogtmann E, et al. , Colorectal Cancer and the Human Gut Microbiome: Reproducibility with Whole-Genome Shotgun Sequencing. PLoS One, 2016. 11(5): p. e0155362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yu J, et al. , Metagenomic analysis of faecal microbiome as a tool towards targeted non-invasive biomarkers for colorectal cancer. Gut, 2017. 66(1): p. 70–78. [DOI] [PubMed] [Google Scholar]
- 12.Bedarf JR, et al. , Functional implications of microbial and viral gut metagenome changes in early stage L-DOPA-naïve Parkinson’s disease patients. Genome Med, 2017. 9(1): p. 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schmidt TSB, Raes J, and Bork P, The Human Gut Microbiome: From Association to Modulation. Cell, 2018. 172(6): p. 1198–1215. [DOI] [PubMed] [Google Scholar]
- 14.Forslund K, et al. , Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature, 2015. 528(7581): p. 262–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Costea PI, et al. , Towards standards for human fecal sample processing in metagenomic studies. Nat. Biotechnol, 2017. 35(11): p. 1069–1076. [DOI] [PubMed] [Google Scholar]
- 16.Lozupone CA, et al. , Meta-analyses of studies of the human microbiota. Genome Res, 2013. 23(10): p. 1704–1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Duvallet C, et al. , Meta Analysis Of Microbiome Studies Identifies Shared And Disease-Specific Patterns. 2017. [DOI] [PMC free article] [PubMed]
- 18.Shah MS, et al. , Leveraging sequence-based faecal microbial community survey data to identify a composite biomarker for colorectal cancer. Gut, 2018. 67(5): p. 882–891. [DOI] [PubMed] [Google Scholar]
- 19.Pasolli E, et al. , Machine Learning Meta-analysis of Large Metagenomic Datasets: Tools and Biological Insights. PLoS Comput. Biol, 2016. 12(7): p. e1004977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dai Z, et al. , Multi-cohort analysis of colorectal cancer metagenome identified altered bacteria across populations and universal bacterial markers. Microbiome, 2018. 6(1): p. 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maier L, et al. , Extensive impact of non-antibiotic drugs on human gut bacteria. Nature, 2018. 555(7698): p. 623–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Milanese A, et al. , Microbial abundance, activity, and population genomic profiling with mOTUs. Nature Communications, 2019. formally accepted for publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kultima JR, et al. , MOCAT2: a metagenomic assembly, annotation and profiling framework. Bioinformatics, 2016. 32(16): p. 2520–2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hothorn T, et al. , A Lego System for Conditional Inference. Am. Stat, 2006. 60(3): p. 257–263. [Google Scholar]
- 25.Mandal S, et al. , Analysis of composition of microbiomes: a novel method for studying microbial composition. Microb Ecol Health Dis, 2015. 26: p. 27663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tjalsma H, et al. , A bacterial driver-passenger model for colorectal cancer: beyond the usual suspects. Nat Rev Microbiol, 2012. 10(8): p. 575–82. [DOI] [PubMed] [Google Scholar]
- 27.Thomas AM, et al. , Metagenomic analysis of colorectal cancer datasets identifies cross-cohort microbial diagnostic signatures and a link with choline degradation. co-submitted to Nature Medicine, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huerta-Cepas J, et al. , eggNOG 4.5: a hierarchical orthology framework with improved functional annotations for eukaryotic, prokaryotic and viral sequences. Nucleic Acids Res, 2016. 44(D1): p. D286–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kanehisa M, et al. , Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res, 2014. 42(Database issue): p. D199–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li J, et al. , An integrated catalog of reference genes in the human gut microbiome. Nat. Biotechnol, 2014. 32(8): p. 834–841. [DOI] [PubMed] [Google Scholar]
- 31.Vieira-Silva S, et al. , Species-function relationships shape ecological properties of the human gut microbiome. Nat Microbiol, 2016. 1(8): p. 16088. [DOI] [PubMed] [Google Scholar]
- 32.Hirayama A, et al. , Quantitative metabolome profiling of colon and stomach cancer microenvironment by capillary electrophoresis time-of-flight mass spectrometry. Cancer Res, 2009. 69(11): p. 4918–25. [DOI] [PubMed] [Google Scholar]
- 33.Denkert C, et al. , Metabolite profiling of human colon carcinoma--deregulation of TCA cycle and amino acid turnover. Mol Cancer, 2008. 7: p. 72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mal M, et al. , Metabotyping of human colorectal cancer using two-dimensional gas chromatography mass spectrometry. Anal Bioanal Chem, 2012. 403(2): p. 483–93. [DOI] [PubMed] [Google Scholar]
- 35.Weir TL, et al. , Stool microbiome and metabolome differences between colorectal cancer patients and healthy adults. PLoS One, 2013. 8(8): p. e70803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goedert JJ, et al. , Fecal metabolomics: assay performance and association with colorectal cancer. Carcinogenesis, 2014. 35(9): p. 2089–2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aykan NF, Red meat and colorectal cancer. Oncology Reviews, 2015. 9(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.World Cancer Research Fund / American Institute for Cancer Research, Diet, Nutrition, Physical Activity and Cancer: a Global Perspective, in Continuous Update Project Expert Report. 2018.
- 39.Dutilh BE, et al. , Screening metatranscriptomes for toxin genes as functional drivers of human colorectal cancer. Best Pract Res Clin Gastroenterol, 2013. 27(1): p. 85–99. [DOI] [PubMed] [Google Scholar]
- 40.Sears CL and Garrett WS, Microbes, Microbiota, and Colon Cancer. Cell Host Microbe, 2014. 15(3): p. 317–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ridlon JM, et al. , Consequences of bile salt biotransformations by intestinal bacteria. Gut Microbes, 2016. 7(1): p. 22–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yoshimoto S, et al. , Obesity-induced gut microbial metabolite promotes liver cancer through senescence secretome. Nature, 2013. 499(7456): p. 97–101. [DOI] [PubMed] [Google Scholar]
- 43.Ajouz H, Mukherji D, and Shamseddine A, Secondary bile acids: an underrecognized cause of colon cancer. World Journal of Surgical Oncology, 2014. 12(1): p. 164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Boleij A, et al. , The Bacteroides fragilis toxin gene is prevalent in the colon mucosa of colorectal cancer patients. Clin. Infect. Dis, 2015. 60(2): p. 208–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu S, et al. , A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nat. Med, 2009. 15(9): p. 1016–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dejea CM, et al. , Patients with familial adenomatous polyposis harbor colonic biofilms containing tumorigenic bacteria. Science, 2018. 359(6375): p. 592–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ridlon JM, Kang DJ, and Hylemon PB, Isolation and characterization of a bile acid inducible 7alpha-dehydroxylating operon in Clostridium hylemonae TN271. Anaerobe, 2010. 16(2): p. 137–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mallonee DH, White WB, and Hylemon PB, Cloning and sequencing of a bile acid-inducible operon from Eubacterium sp. strain VPI 12708. Journal of Bacteriology, 1990. 172(12): p. 7011–7019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ocvirk S and O’Keefe SJD, Influence of Bile Acids on Colorectal Cancer Risk: Potential Mechanisms Mediated by Diet-Gut Microbiota Interactions. Curr. Nutr. Rep, 2017. 6(4): p. 315–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gevers D, et al. , The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe, 2014. 15(3): p. 382–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Viennot S, et al. , Colon cancer in inflammatory bowel disease: recent trends, questions and answers. Gastroenterol. Clin. Biol, 2009. 33 Suppl 3: p. S190–201. [DOI] [PubMed] [Google Scholar]
- 52.Rubinstein MR, et al. , Fusobacterium nucleatum Promotes Colorectal Carcinogenesis by Modulating E-Cadherin/β-Catenin Signaling via its FadA Adhesin. Cell Host Microbe, 2013. 14(2): p. 195–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kostic AD, et al. , Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe, 2013. 14(2): p. 207–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Arthur JC, et al. , Intestinal inflammation targets cancer-inducing activity of the microbiota. Science, 2012. 338(6103): p. 120–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Reddy BS, Diet and excretion of bile acids. Cancer Res, 1981. 41(9 Pt 2): p. 3766–8. [PubMed] [Google Scholar]
- 56.Ogino S, et al. , Integrative analysis of exogenous, endogenous, tumour and immune factors for precision medicine. Gut, 2018. 67(6): p. 1168–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ogino S, et al. , Molecular pathological epidemiology of colorectal neoplasia: an emerging transdisciplinary and interdisciplinary field. Gut, 2011. 60(3): p. 397–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hannigan GD, et al. , Diagnostic Potential and Interactive Dynamics of the Colorectal Cancer Virome. MBio, 2018. 9(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.zur Hausen H, Red meat consumption and cancer: reasons to suspect involvement of bovine infectious factors in colorectal cancer. Int J Cancer, 2012. 130(11): p. 2475–83. [DOI] [PubMed] [Google Scholar]
- 60.Shkoporov AN, et al. , Reproducible protocols for metagenomic analysis of human faecal phageomes. Microbiome, 2018. 6(1): p. 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
Additional References
- 61.Bohm J, et al. , Discovery of novel plasma proteins as biomarkers for the development of incisional hernias after midline incision in patients with colorectal cancer: The ColoCare study. Surgery, 2017. 161(3): p. 808–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liesenfeld DB, et al. , Metabolomics and transcriptomics identify pathway differences between visceral and subcutaneous adipose tissue in colorectal cancer patients: the ColoCare study. Am J Clin Nutr, 2015. 102(2): p. 433–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pox CP, et al. , Efficacy of a nationwide screening colonoscopy program for colorectal cancer. Gastroenterology, 2012. 142(7): p. 1460–7 e2. [DOI] [PubMed] [Google Scholar]
- 64.Furet JP, et al. , Comparative assessment of human and farm animal faecal microbiota using real-time quantitative PCR. FEMS Microbiol Ecol, 2009. 68(3): p. 351–62. [DOI] [PubMed] [Google Scholar]
- 65.Mende DR, et al. , Accurate and universal delineation of prokaryotic species. Nat. Methods, 2013. 10(9): p. 881–884. [DOI] [PubMed] [Google Scholar]
- 66.Sunagawa S, et al. , Metagenomic species profiling using universal phylogenetic marker genes. Nat. Methods, 2013. 10(12): p. 1196–1199. [DOI] [PubMed] [Google Scholar]
- 67.Li H and Durbin R, Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics, 2009. 25(14): p. 1754–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tibshirani R, Regression Shrinkage and Selection via the Lasso. J.R. Stat. Soc. Series B Stat. Methodol, 1996. 58(1): p. 267–288. [Google Scholar]
- 69.Smialowski P, Frishman D, and Kramer S, Pitfalls of supervised feature selection. Bioinformatics, 2010. 26(3): p. 440–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Benjamini Y and Hochberg Y, Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Series B Stat. Methodol, 1995. 57(1): p. 289–300. [Google Scholar]
- 71.Robin X, et al. , pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics, 2011. 12(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Oksanen J, et al. , vegan: Community Ecology Package. 2018.
- 73.Costea PI, et al. , A fair comparison. Nat. Methods, 2014. 11(4): p. 359–359. [DOI] [PubMed] [Google Scholar]
- 74.Hastie T, Tibshirani R, and Friedman J, The Elements of Statistical Learning: Data Mining, Inference, and Prediction. 2013: Springer Science & Business Media. 536. [Google Scholar]
- 75.Peng H, Long F, and Ding C, Feature selection based on mutual information: criteria of max-dependency, max-relevance, and min-redundancy. IEEE Trans Pattern Anal Mach Intell, 2005. 27(8): p. 1226–38. [DOI] [PubMed] [Google Scholar]
- 76.Edgar RC, MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res, 2004. 32(5): p. 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Altschul SF, et al. , Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res, 1997. 25(17): p. 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Eddy SR, Accelerated Profile HMM Searches. PLoS Comput. Biol, 2011. 7(10): p. e1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rice P, Longden I, and Bleasby A, EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet, 2000. 16(6): p. 276–277. [DOI] [PubMed] [Google Scholar]
- 80.Caporaso JG, et al. , Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci U S A, 2011. 108 Suppl 1: p. 4516–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw sequencing data for the samples in the DE study that had not been published before (see Methods), are made available in the European Nucleotide Archive (ENA) under the study identifier PRJEB27928. Metadata for these samples are available as Supplementary Table S6.
For the other studies included here, the raw sequencing data can be found under the following ENA identifiers: PRJEB10878 for [11], PRJEB12449 for [10], ERP008729 for [9], and ERP005534 for [8]. The independent validation cohorts can be found in SRA under the identifier SRP136711 for [27] and in the DDBJ database under the ID DRA006684.
Filtered taxonomic and functional profiles used as input for the statistical modeling pipeline are available in Supplementary Data 1.
The code and all analysis results can be found under https://github.com/zellerlab/crc_meta.