Summary
A LuxI/R‐like quorum sensing (QS) system (AfeI/R) has been reported in the acidophilic and chemoautotrophic Acidithiobacillus spp. However, the function of AfeI/R remains unclear because of the difficulties in the genetic manipulation of these bacteria. Here, we constructed different afeI mutants of the sulfur‐ and iron‐oxidizer A. ferrooxidans, identified the N‐acyl homoserine lactones (acyl‐HSLs) synthesized by AfeI, and determined the regulatory effects of AfeI/R on genes expression, extracellular polymeric substance synthesis, energy metabolism, cell growth and population density of A. ferrooxidans in different energy substrates. Acyl‐HSLs‐mediated distinct regulation strategies were employed to influence bacterial metabolism and cell growth of A. ferrooxidans cultivated in either sulfur or ferrous iron. Based on these findings, an energy‐substrate‐dependent regulation mode of AfeI/R in A. ferrooxidans was illuminated that AfeI/R could produce different types of acyl‐HSLs and employ specific acyl‐HSLs to regulate specific genes in response to different energy substrates. The discovery of the AfeI/R‐mediated substrate‐dependent regulatory mode expands our knowledge on the function of QS system in the chemoautotrophic sulfur‐ and ferrous iron‐oxidizing bacteria, and provides new insights in understanding energy metabolism modulation, population control, bacteria‐driven bioleaching process, and the coevolution between the acidophiles and their acidic habitats.
Introduction
Acidophiles, a class of important extremophiles and geo‐microbes, are widely distributed in the hot springs and acid mines. In these natural habitats, the chemoautotrophic acidophiles participate in the global element cycles of sulfur and iron via the oxidation of reduced inorganic sulfur compounds (RISCs) to sulfate and the conversions between ferrous and ferric ions (Menzel et al., 2015; Quatrini and Johnson, 2016). The acidophile‐driven bioleaching process has given rise to a worldwide problem of water and solid contaminations in natural and man‐made mine environments (Chen et al., 2014). On the other hand, this process has been advantageously utilized in the biomining industry for the recovery of valuable metals from sulfide ores, such as copper or gold (Rawlings, 1998; Chen et al., 2014). Thus, the researches on the bioleaching microbes and their metabolism and regulation mechanisms are of significance for the treatments of acid mine contaminations and the development of high‐efficient biological metallurgy technology. Acidithiobacillus spp., a group of acidophilic chemolithoautotrophic Gram‐negative bacteria, are prevalent in the sulfur‐ and ferrous iron‐ contained acidic ecosystems, and are the predominant player in acidophile communities structures in acid mine drainages (AMD) and terrestrial hot springs (Rawlings, 1998; Liljeqvist et al., 2015; Quatrini and Johnson, 2018). They are the most active and wide‐used bioleaching bacteria in the biomining industry (Olson et al., 2003; Rohwerder et al., 2003). All Acidithiobacillus strains are capable of oxidizing various RISCs for autotrophic growth, and some of them can use ferrous iron as an energy substrate (Bosecker, 1997; Rohwerder et al., 2003). Seven species have been identified in the Acidithiobacillus genus, including four sulfur‐ and ferrous iron‐oxidizing species (A. ferrooxidans, A. ferridurans, A. ferriphilus and A. ferrivorans), and three sulfur‐oxidizing‐only species (A. thiooxidans, A. caldus and A. albertensis).
Acidithiobacillus. ferrooxidans has become an important model bacterium for the researches of the acidophilic bacteria on physiological biochemistry, molecular biology, microbial mineralogy and so on (Sugio et al., 1987; Rawlings, 2002). It can gain energy by the oxidation of ferrous iron and reduced sulfur compounds at the aerobic condition, and can also obtain energy via anaerobic metabolisms including the oxidation of sulfur and hydrogen by using ferric iron as an electron acceptor and the oxidation of hydrogen by using sulfur as electron acceptor (Ohmura et al., 2002). For the variety of RISCs, the sulfur metabolism is achieved by different kinds of enzymes located in different cellular compartments in A. ferrooxidans, such as thiosulfate dehydrogenase, thiosulfate quinone oxidoreductase, tetrathionate hydrolase (TetH) in periplasmic space; persulfide dioxygenase (formerly named as sulfur dioxygenase, SDO), HDR, Hdr‐like complex in the cytoplasm; and sulfide:quinone oxidoreductase (SQR) located in the inner membrane (Ng et al., 2000; Sugio et al., 2009; Wang et al., 2019). Ferrous iron oxidation in A. ferrooxidans involves the petI and rus operons, two transcriptional units that mediate downhill and uphill electron pathways to generate ATP and NADH respectively (Quatrini et al., 2009). Therefore, A. ferrooxidans exhibits distinct physiological characteristics and gene expression profiles depending on the availability of these two energy substrates.
Quorum‐sensing (QS) is a cell‐to‐cell communication mechanism that enables bacteria to control gene expression in response to changes in cell density (Parsek and Greenberg, 2000; Wackett, 2008). QS regulation depends on the production, release, accumulation and detection of signaling autoinducers, and this process is generally mediated by the autoinducer synthase and cognate autoinducer receptor (An et al., 2006; Schaefer et al., 2008). QS has been widely identified in Gram‐negative and Gram‐positive bacteria and is fundamental for cell‐to‐cell communication (Juhas et al., 2005; Kai and Bassler, 2016). Hundreds of traits can be regulated via QS in both pathogenic and environmental bacteria, such as EPS synthesis, biofilm formation, cell colonization, bioluminescence and the secretion of virulence factors (Goo et al., 2015; Ben‐Yaakov and Salomon, 2019). QS can also regulate metabolic processes in some bacteria, such as sugar and phosphate metabolism, as well as secondary metabolites (Goo et al., 2015; Certner and Vollmer, 2018; Ha et al., 2018). Moreover, QS has been also observed in many extremophiles; however, such studies are limited by the difficulty of genetic manipulation in these bacteria (Inaba et al., 2018).
A LuxI/R‐like QS system (AfeI/R), encoded by the afeI‐orf3‐afeR operon, was discovered in A. ferrooxidans (Farah et al., 2005; Rivas et al., 2005). Similar to the prototypical LuxI/R‐like system in many Gram‐negative bacteria, the QS system in A. ferrooxidans consists of a LuxI‐type autoinducer synthase (AfeI) and a LuxR‐type receptor (AfeR) that mediates the production of N‐acyl homoserine lactones (acyl‐HSLs) and controls genes expression by binding signaling molecules respectively (An et al., 2006; Schaefer et al., 2008). Additionally, another potential acyl‐HSL synthetase (Act) in A. ferrooxidans was discovered in an operon. This operon encompasses four co‐transcribed genes (glyQ, glysS, gph and act), which encode for the α and β subunits of glycine tRNA synthetase, a phosphatase and an acyltransferase respectively (Rivas et al., 2007). Given that Act has only been previously confirmed to produce C14‐HSL in Escherichia coli in the absence of a corresponding signal molecule receptor gene in the operon, the role of the Act‐like QS system in A. ferrooxidans remains largely unclear (Rivas et al., 2007; Gonzalez et al., 2013).
Nine acyl‐HSLs have been identified from A. ferrooxidans cultures, including C12‐HSL, C14‐HSL, 3‐OH‐C8‐HSL, 3‐OH‐C10‐HSL, 3‐OH‐C12‐HSL, 3‐OH‐C14‐HSL, 3‐OH‐C16‐HSL, 3‐O‐C12‐HSL and 3‐O‐C14‐HSL (Farah et al., 2005). The addition of exogenous acyl‐HSLs led to notable phenotypic changes. For instance, the addition of C12/C14‐HSLs mixtures or acyl‐HSL analogs promoted biofilm formation on the surfaces of elemental sulfur and pyrite (Gonzalez et al., 2013; Bellenberg et al., 2014). Moreover, the addition of a C14‐HSLs mixture also improved A. ferrooxidans electroactivity on an inert carbon electrode (Chabert et al., 2017). Furthermore, more than 100 genes were differentially expressed in A. ferrooxidans exposed to a tetrazolic acyl‐HSL analog (tetrazole 9c), of which 60 were involved in biofilm formation (Mamani et al., 2016). Exposure to a synthetic QS blocker also revealed that AfeI/R mediates Cu2+ resistance in A. ferrooxidans (Wenbin et al., 2011). In addition, the overexpression of afeI/R operon suggested the important roles of Afe/R in the growth of A. ferrooxidans in S0‐enriched media and in improving the bioleaching efficiency of A. ferrooxidans to ores (Gao et al., 2020). However, although some functions of AfeI/R have been identified via the addition assays of exogenous signal molecules, the understanding of the roles of AfeI/R in A. ferrooxidans has not been fully achieved. For example, the acyl‐HSLs synthesized by AfeI were not determined due to the interference of the Act system. Meanwhile, the roles of AfeI‐produced acyl‐HSLs were also unclear.
The question that whether AfeI/R has a regulatory function in Fe2+‐cultivating A. ferrooxidans remains to be answered. Unlike elemental sulfur, ferrous iron exists as an ion in bacterial cultures (Quatrini et al., 2009). The utilization of elemental sulfur by A. ferrooxidans requires EPS‐mediated attachment, while ferrous iron metabolism employs very different pathways (Gehrke et al., 1998; Harneit et al., 2006; Quatrini et al., 2009). Notably, a lower transcriptional level of the afeI/R operon has been observed in Fe2+‐enriched media compared with S0‐enriched media (Farah et al., 2005). However, the role of AfeI/R in A. ferrooxidans when Fe2+ is used as the energy source is not clear.
Mutagenesis of the QS genes has become a powerful and effective approach to study the biological functions and regulation mechanisms of QS in many bacteria. In this study, we explored the distribution of AfeI/R‐like QS system in Acidithiobacillus and other acidophiles. Several mutants of the acyl‐HSLs synthetase genes were successfully constructed and used to study the effect of gene knockout and overexpression on acyl‐HSLs synthesize, energy metabolism, cell growth, EPS secretion and gene transcript profile in A. ferrooxidans cultivated with different energy substrates. Moreover, the acyl‐HSLs produced by AfeI were identified, and two key acyl‐HSLs were found to influence A. ferrooxidans growth. Therefore, our results revealed that AfeI/R‐mediated regulation effects in A. ferrooxidans were versatile and substrate‐dependent. The results in this study provide new insights in understanding the QS‐mediated regulation in acidophilic sulfur‐oxidizing and/or ferrous iron‐oxidizing bacteria.
Results
Distribution of AfeI/R‐like QS system in Acidithiobacillus and other acidophiles
The protein sequences of AfeI, AfeR and Orf3 from A. ferrooxidans ATCC 23270 were used to explore the homologous proteins in Acidithiobacillus and other acidophiles based on the reported acidophilic species and their published genome sequences on the NCBI database (Quatrini and Johnson, 2016). As shown in Fig. 1, AfeI/R‐like QS system could be identified from A. ferrooxidans, A. ferridurans, A. ferrivorans and A. thiooxidans in the genus of Acidithiobacillus. AfeI/R system was found in all the sulfur‐ and ferrous iron‐oxidizing species except A. ferriphilus that did not have the published genomic information, while only A. thiooxidans possesses the QS system in the three reported sulfur‐oxidizing‐only species of Acidithiobacillus. The result indicated the distribution of AfeI/R in the sulfur‐ and ferrous iron‐oxidizing species is more pervasive than that in sulfur‐oxidizing‐only species of Acidithiobacillus. AfeI/R‐like QS system also found in the genus of Thiomonas that is a group of chemoautotrophic sulfur‐oxidizing‐only bacteria. AfeI/R system showed some variations at the gene arrangement and protein sequence in Acidithiobacillus. The afeR‐orf3‐afeI operon could be identified from the species of A. ferrooxidans, A. ferridurans and A. thiooxidans, while A. ferrivorans only possesses two separated genes afeI and afeR encoding the proteins with low identities to that from A. ferrooxidans ATCC 23270. Although almost all of the strains in A. thiooxidans have the conserved afeI/R operon, afeR gene and the afeI‐orf3 are separated on the genome of A. thiooxidans ATCC 19377. A truncated AfeR and the low‐identity AfeI and orf3 are found in A. thiooxidans ATCC 19377, in contrast with those in other A. thiooxidans strains. Therefore, the gene arrangement and protein sequence of AfeI/R system in Acidithiobacillus would be variant in different species or strains.
Fig 1.

Distribution of AfeI/R‐like QS system in Acidithiobacillus and other acidophiles. [Color figure can be viewed at wileyonlinelibrary.com]
Generation of A. ferrooxidans afeI mutants
To characterize the biological function of the AfeI/R QS system, the acyl‐HSL synthase gene (afeI, AFE_1999) was targeted to generate afeI deletion and overexpression mutant strains. The afeI knockout strains were screened and identified by PCR using different primer sets (Fig. 2A and B). The ΔafeI strain was a markerless in‐frame AfeI mutation with a 552‐bp deletion from the start site (ATG) to the stop codon (TAA). The afeI expression plasmid was constructed using a tac promoter to initiate gene transcription, as well as the autonomously replicating plasmid pJRD215 as the backbone of the expression plasmid (Fig. 2C). The constructed afeI expression plasmid and the backbone plasmid were respectively conjugated into wild type A. ferrooxidans strain WT, generating the afeI overexpression strain OEafeI and the wild‐type control strain WT(pJRD215) (Fig. 2D).
Fig 2.

Verification of different types of engineered bacteria during the generation of the afeI deletion and overexpression strains. A. Diagram of the afeI gene cluster and verification primers. UHA and DHA represent upstream and downstream homologous arms respectively. B. Electrophoretic analysis of PCR products to verify ΔafeI. Lanes 1, 3 and 5, PCR products from ΔafeI using the primer pair P1F/R, P2F/R and P3F/R respectively; lanes 2, 4 and 6, PCR products from wild type using the primer pair P1F/R, P2F/R and P3F/R respectively. C. Illustration of the afeI expression vector pJRD215‐Ptac‐afeI and verification primers. D. Electrophoretic analysis of PCR products to verify the afeI‐overexpression strains. Lanes 1, 2, 3 and 4, PCR products from the afeI overexpression strain, the wild‐type strain, pJRD215 and pJRD215‐Ptac‐afeI respectively, using the primer pair P4F/R. E. Diagram of the act gene cluster and verification primers. F. Electrophoretic analysis of PCR products to verify Δact(afeI). Lanes 1, 3 and 5, PCR products from Δact(afeI) using the primer pair PA1F/R, PA2F/R and PA3F/R respectively; lanes 2, 4 and 6, PCR products from wild type using the primer pair PA1F/R, PA2F/R and PA3F/R respectively; lanes 7, 8, 9 and 10, PCR products from the Δact(afeI) strain, the wild‐type strain, pJRD215 and pJRD215‐Ptac‐afeI respectively, using the primer pair P4F/R. G. Diagram of the ΔafeI&act gene cluster and verification primers. H. Electrophoretic analysis of PCR products to verify ΔafeI&act. Lanes 1, 3, 5, 7, 9 and 11, PCR products from ΔafeI&act using the primer pair P1F/R, P2F/R, P3F/R, PA1F/R, PA2F/R and PA3F/R respectively; lanes 2, 4, 6, 8, 10 and 12, PCR products from wild type using the primer pair P1F/R, P2F/R, P3F/R, PA1F/R, PA2F/R and PA3F/R respectively. [Color figure can be viewed at wileyonlinelibrary.com]
To identify the molecules synthesized by AfeI, a 639‐bp sequence of the acyltransferase gene (act) was deleted from strains of WT and ΔafeI, generating Δact and ΔafeI&act respectively (Fig. 2E and G). Furthermore, the afeI expression plasmid (pJRD215‐Ptac‐afeI) and the empty plasmid (pJRD215) were conjugated into Δact and ΔafeI&act, resulting in the afeI‐expression‐only strain Δact(afeI) and the afeI and act deletion strain ΔafeI&act(215) respectively.
Substrate‐dependent regulatory effects of the AfeI/R on A. ferrooxidans growth and energy metabolism
When cells were grown in Fe2+‐enriched media, the cell growth rate, maximum cell density and ferrous iron oxidation rate of the afeI overexpression strain were dramatically decreased, and the maximum cell density of the afeI overexpression strain reached only approximately 70% of that of the control strain (Fig. 3A and C). In contrast, almost no difference was observed between the afeI knockout and the WT strain in terms of cell growth and ferrous iron oxidation (Fig. 3B and D). These results indicate that overexpression of afeI could inhibit A. ferrooxidans ferrous iron oxidation and cell population in Fe2+‐enriched media.
Fig 3.

Analyses of the growth and metabolism of engineered A. ferrooxidans strains. Growth (A, B) and ferrous oxidation (C, D) of the afeI overexpression (OEafeI) and knockout (ΔafeI) strains in Fe2+‐enriched media. Growth (E, F) and sulfate production (G, H) curves for the afeI overexpression (OEafeI) and knockout (ΔafeI) strains S0‐enriched media. CK indicates control. NS indicates no significant difference. [Color figure can be viewed at wileyonlinelibrary.com]
When elemental sulfur was used as an energy substrate, overexpression of afeI significantly increased the cell density in the lag and exponential growth phases of the afeI overexpression strain compared with the wild‐type control strain WT (pJRD215) (Fig. 3E), and simultaneously increased sulfate production on days 3, 6, 8 and 10 (Fig. 3G). However, the enhanced growth caused by afeI overexpression decreased and ultimately disappeared when the cells entered the late exponential and stationary growth phases (Fig. 3E and G). Deletion of afeI did not distinctly affect cell density, although a slight decrease in sulfate production was observed on days 8 and 10 for the ΔafeI mutant (Fig. 3F and H). These results indicated that a high level of AfeI expression could enhance sulfur metabolism and cell growth in A. ferrooxidans, and this AfeI/R‐mediated regulation was dependent on bacterial growth stage in the S0‐enriched media.
Substrate‐dependent influences of AfeI/R on EPS synthesis in A. ferrooxidans
In Fe2+‐enriched media, the main EPS components (proteins and carbohydrates) of afeI knockout and overexpression strains had no significant difference compared with that of the control strains (Fig. 4A). The results indicated that the regulatory effect of AfeI/R on A. ferrooxidans EPS synthesis did not occur in the Fe2+‐enriched media.
Fig 4.

Analysis of EPS synthesis and cell attachment. The protein and carbohydrate levels of EPS in Fe2+‐enriched media (A) and S0‐enriched media (B and C). Observation of cell attachment on sulfur coupons by SEM (D). OEafeI indicates afeI overexpression strains.
In S0‐enriched media, the levels of the EPS components of the afeI overexpression strain were more than threefold higher than those of the control strain in the exponential growth phase (Fig. 4B), whereas the difference disappeared in the stationary phase (Fig. 4C). The deletion of afeI did not result in a significant change in EPS protein and carbohydrate content in S0‐enriched media (Fig. 4B and C). Scanning electron microscopy (SEM) results showed that the surfaces of sulfur coupons cultivated with the afeI overexpression strain were uneven, bumpy and gully‐like, and the cells tended to aggregate and form biofilms (Fig. 4D). In contrast, the results for the ΔafeI and WT strains were considerably different, with smooth‐surface sulfur coupons and scattered cells (Fig. 4D).
These results suggested that the regulation of AfeI/R on the EPS synthesize was dependent on the energy substrates.
Substrate‐dependent regulatory role of acyl‐HSLs synthesized by AfeI on the growth of A. ferrooxidans
The significant influence of afeI overexpression on A. ferrooxidans growth in Fe2+‐ or S0‐enriched media implied that the acyl‐HSLs synthesized by AfeI could influence cell growth. To further validate this speculation, add‐back assays with A. ferrooxidans WT strain were carried out using acyl‐HSLs extracted from the culture broth of the afeI overexpression or deletion strains. When Fe2+ was used as the sole energy substrate, the addition of acyl‐HSLs extracted from S0‐ or Fe2+‐enriched afeI overexpression strain cultures suppressed cell growth and final bacterial population (Fig. 5A and B).
Fig 5.

Growth of the A. ferrooxidans wild‐type strain supplemented with the extracted acyl‐HSLs. A, B. The addition of acyl‐HSLs into A. ferrooxidans cultures grown in Fe2+‐enriched media at the beginning of cultivation; D–F, addition of acyl‐HSLs into A. ferrooxidans cultures grown in S0‐enriched media at different cultivation stages. HSLs/S‐OEafeI and HSLs/S‐ΔafeI indicate acyl‐HSLs extracted from culture broths of the afeI overexpression and deletion strains respectively, of A. ferrooxidans grown in S0‐enriched media; HSLs/F‐OEafeI and HSLs/F‐ΔafeI indicate acyl‐HSLs extracted from culture broths of the afeI overexpression and deletion strains respectively, of A. ferrooxidans grown in Fe2+‐enriched media. CK indicates the blank control. [Color figure can be viewed at wileyonlinelibrary.com]
Cell growth was enhanced in A. ferrooxidans cultivated in S0‐enriched media upon the exogenous addition of acyl‐HSLs from S0‐enriched afeI overexpression strain cultures at the initial or mid‐growth phase (Fig. 5C and D). However, no growth advantage occurred when the acyl‐HSLs were added in the stationary growth phase (Fig. 5E). No apparent change in cell growth was observed upon the addition of extracts from ΔafeI cultures (Fig. 5).
Thus, the regulatory effects of the extracted acyl‐HSLs were consistent with the effects obtained by overexpressing afeI, indicating the influence of the AfeI‐synthesized acyl‐HSLs on cell growth of A. ferrooxidans and showing the different growth effects depending on the different energy substrates.
Identification of AfeI‐synthesizing acyl‐HSLs in different energy substrates
To avoid interference from another acyl‐HSLs synthetase (Act) in A. ferrooxidans (Rivas et al., 2007), the afeI‐expression‐only strain Δact(afeI) and the control strain ΔafeI&act(215) were constructed to identify the AfeI‐synthetized acyl‐HSLs by LC‐MS‐MS (Table 1 and Fig. S1). In S0‐enriched media, five types of acyl‐HSLs were verified, with [MS+H] + values of 284.22, 312.25, 272.18, 300.21 and 328.24, which were similar to the [MS+H] + values of C12, C14, 3‐OH‐C10, 3‐OH‐C12 and 3‐OH‐C14 respectively. In addition, these five acyl‐HSLs exhibited the characteristic protonation of homoserine lactone (m/z 102.05) (Morin et al., 2003), and the MS2 spectra of these five acyl‐HSLs were the same as those of the standard compounds. Because 3‐OH‐C16 is not commercially available, it was identified using a previously reported method (Morin et al., 2003; Farah et al., 2005). The [M+H]+ value of 356.2801 observed in the extracts was almost identical to the theoretical [M+H]+ number of 3‐OH‐C16 (356.2790; Fig. S1F and I). And the LC‐MS‐MS experiments detected a value of m/z 102.0541 in the MS2 results (356.2801), which is characteristic of acyl‐HSLs protonation. Thus, we concluded that afeI can synthesize six types of acyl‐HSLs (C12, C14, 3‐OH‐C10, 3‐OH‐C12, 3‐OH‐C14 and 3‐OH‐C16) in A. ferrooxidans cells grown in S0‐enriched media. Furthermore, the relative content of these six acyl‐HSLs in the extracts was detected via the peak area normalization method (Ni et al., 2019); the acyl‐HSL concentrations were found to occur in the following order: 3‐OH‐C14 > 3‐OH‐C12 > 3‐OH‐C16 > C12 > C14 > 3‐OH‐C10. In Fe2+‐enriched media, three acyl‐HSLs were identified with the following relative concentration order: 3‐OH‐C14 > 3‐OH‐C12 > 3‐OH‐C16. Thus, different signal molecules were synthesized by afeI depending on the presence of different energy substrates, among which 3‐OH‐C14 was the most abundant and may play important roles in growth and metabolism regulation. Moreover, the relative quantification results showed that the contents of acyl‐HSLs detected in afeI overexpression strain cultures were 2.97 and 2.47 times higher than those of the control strain in S0‐ and Fe2+‐enriched media respectively.
Table 1.
Identification of the acyl‐HSLs in S0‐ or Fe2+‐containing medium by LC‐MS/MS.
| Acyl‐HSLs | Chemical formula | [M+H]+ ion detected (m/z) | Structural formula | S0 | Fe2+ |
|---|---|---|---|---|---|
| 3‐OH‐C14 | C18H33O4N | 328.2400 |

|
****** | *** |
| 3‐OH‐C12 | C16H29O4N | 300.2100 |
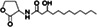
|
***** | ** |
| 3‐OH‐C16 | C20H37O4N | 356.2790 |

|
**** | * |
| C12 | C16H29O3N | 284.2220 |

|
*** | ND |
| C14 | C18H33O3N | 312.2530 |

|
** | ND |
| 3‐OH‐C10 | C14H25O4N | 272.1850 |

|
* | ND |
Asterisk indicates relative concentration in the extracts; ND indicates not detected.
Identification of functional acyl‐HSLs involved in A. ferrooxidans growth modulation
To identify the key acyl‐HSLs involved in A. ferrooxidans growth regulation, five acyl‐HSL standards, including C12, C14, 3‐OH‐C10, 3‐OH‐C12 and 3‐OH‐C14 were purchased to perform add‐back assays. C12‐HSL inhibited A. ferrooxidans growth in Fe2+‐enriched media (Fig. 6A), while other acyl‐HSLs had no obvious effect (Fig. 6C). Moreover, the addition of 3‐OH‐C14‐HSL on the 4th day stimulated cell growth at the log growth phase (Fig. 6B) and promoted EPS synthesis in A. ferrooxidans grown in S0‐enriched media (Fig. S2). However, adding other acyl‐HSLs to S0‐enriched A. ferrooxidans media did not have any statistically significant effect on cell growth (Fig. 6D). Therefore, two key signal molecules with the function of influencing A. ferrooxidans growth were discovered, including the regulation of C12‐HSL on A. ferrooxidans in Fe2+‐enriched media and the modulation of 3‐OH‐C14‐HSL on A. ferrooxidans in S0‐enriched media.
Fig 6.

Growth curves of the A. ferrooxidans wild‐type strain with the addition of different kinds of standard acyl‐HSLs products. HSLs indicate N‐acyl homoserine lactones. [Color figure can be viewed at wileyonlinelibrary.com]
Metabolic pathways regulated by AfeI/R in different energy substrate
The differentially expressed genes (DEGs) were detected by RNA‐seq (Table S1), and the DEGs of interest were verified by Real‐time quantitative PCR (RT‐qPCR).
Genes associated with energy metabolism were differentially expressed in the afeI overexpression strains in different energy substrates. In Fe2+‐enriched media, overexpression of afeI led to downregulation of genes in the rus, cyo, pet, doxDA, and hdr operons as well as the dsrE, tusA and sqr genes (AFE_1792), and upregulation of petB1 in the petI operon as well as cydA, tetH and sdo (AFE_2644), suggesting inhibitory effects on ferrous iron‐ and sulfur‐oxidizing pathways (Fig. 7). Notably, a hydrogenase gene cluster (AFE_0700‐AFE_0719, AFE_0700 encodes a sigma54‐dependent regulator (SDR), Fig. S3) exhibited significant downregulation in this condition (Fig. 7), implying the significance of AfeI/R mediated hydrogen metabolism on the growth of A. ferrooxidans in Fe2+‐media. When S0 was used as the energy substrate, overexpression of afeI resulted in the obvious upregulation of genes in the doxDA, and hdr clusters, and downregulation of the sqr (AFE_0267) and sdo (AFE_0269) genes (Fig. 7). The high expression levels of the majority of sulfur‐oxidizing genes indicated the sulfur‐oxidizing system was stimulated by overexpression of afeI in S0‐culture. Simultaneously, the iron‐oxidizing system, rus operon and pet operon, was significantly inhibited in the S0‐cultivated afeI overexpression strain (Fig. 7). Therefore, AfeI/R could effectively modulate the metabolisms of sulfur, iron and hydrogen to control cell growth and population size of A. ferrooxidans in different energy‐substrates.
Fig 7.

Effect of the AfeI/R QS system on the transcriptional profiles of A. ferrooxidans. This is the valid mean value of fold changes (FC) determined by RT‐qPCR. S‐OEafeI and Fe‐OEafeI represent the afeI overexpression in S0‐ and Fe2+‐enriched media respectively. FC ≥ 2, P ≤ 0.05, upregulated; FC ≤ 0.5, P ≤ 0.05, downregulated; 0.5 ≤ FC ≤ 2, P ≥ 0.05, no change (data are not shown in the figure). [Color figure can be viewed at wileyonlinelibrary.com]
The transcription of eight SDRs was clearly influenced by the afeI (Fig. 7). Upon growth on Fe2+‐containing media, there were four SDR genes (AFE_0693, 0700, 0957 and 2597) as well as the sigma 54 gene (AFE_3025) downregulated in the afeI overexpression strain (Fig. 7). Upon growth on S0‐containing media, there were three SDR genes (AFE_0693, 0957 and 2696) downregulated in the afeI overexpression strain. Thus, the differential expression of SDR genes indicated the strong and extensive impacts of the AfeI/R QS system on the sigma54‐regulated pathways in different energy‐substrates.
Genes associated with membrane permeability were differentially expressed in the afeI overexpression A. ferrooxidans strains (Fig. 7). The oprD (AFE_1497) and tonB (AFE_1991) genes involved in EPS transport (Abbas et al., 2007; Zhang et al., 2018) were markedly downregulated in the afeI overexpression strain grown on Fe2+‐media, while both of these genes were upregulated in the afeI overexpression strain grown on S0‐media (Fig. 7). Conjugal transfer related genes were markedly downregulated in the afeI overexpression strain grown on S0‐media, while both of these genes were no changed in the afeI overexpression strain grown on Fe2+‐media (Fig. 7)
The transcription levels caused by afeI overexpression were different under different energy substrates. This indicated that the versatile regulation of AfeI/R QS system in A. ferrooxidans was dependent on energy substrates.
Discussion
In this study, we revealed that AfeI/R‐like QS system could not only function in the S0‐cultivating process through the use of key acyl‐HSLs but also play an important regulatory role in bacterial ferrous iron oxidation, cell growth and quorum size in Fe2+‐enriched media. Sulfur and ferrous iron are the two crucial energy substrates for acidophiles, which could affect population development and community formation in the natural habitats of AMD sites and terrestrial hot springs and in the industrial bioleaching processes (Rawlings, 2002; Rohwerder et al., 2003). The prevalence of AfeI/R system in the sulfur‐ and ferrous iron‐oxidizing species of Acidithiobacillus (Fig. 1), together with the inhibiting effect caused by the overexpression of afeI or the addition of exogenous acyl‐HSLs (Figs 3A, C, 5A, B and 6A), implied that the AfeI/R system in Acidithiobacillus have evolved the regulatory capacity on bacterial ferrous metabolism. To our knowledge, this is the first report that QS system is involved in the regulation of bacterial ferrous metabolism, cell growth and population density in Fe2+ cultivation. Moreover, our results manifested that the key acyl‐HSL‐inducible EPS synthesis could influence the sulfur oxidation and cell growth of A. ferrooxidans in the S0‐enriched media (Figs 3E, G and 4B, D, Fig. S2).
An AfeI/R‐mediated energy‐substrate‐dependent regulation model in A. ferrooxidans was proposed on the basis of the extensive influences of energy substrates on the synthesis of acyl‐HSLs, the regulatory effects of signal molecules and the AfeI/R‐regulated pathways/systems (Fig. 8). According to the energy substrates, AfeI could synthesize different kinds of acyl‐HSLs, and some of the signal molecules could function as the ‘stimulator’ or ‘inhibitor’ with the prerequisite of specific energy substrate to regulate the expression of genes involved in metabolic pathways and regulatory systems. Thus, the AfeI/R‐mediated versatile regulation could offer varied strategies for A. ferrooxidans to modulate its genes expression and phenotypes in sulfur‐ and ferrous iron‐contained extremely acidic environments.
Fig 8.

Model of the substrate‐dependent AfeI/R‐regulated network in A. ferrooxidans. CTP indicates conjugal transfer protein; hyp indicates hypothetical protein. [Color figure can be viewed at wileyonlinelibrary.com]
The regulatory effects of AfeI/R on sulfur metabolism and cell growth of A. ferrooxidans in S0‐enriched media could be attributed to its regulation of EPS synthesis. EPS can enhance the adhesion of cells to solid energy substrates, and provide an active reaction space between the cells and the surface of the substrates (Gehrke et al., 1998; Harneit et al., 2006). Overexpression of afeI stimulated the EPS synthesis (Fig. 4B), which in turn enhanced the attachment and bioerosion of cells on elemental sulfur (Fig. 4D). This process could accelerate the activation and oxidation of extracellular elemental sulfur (Gehrke et al., 1998; Harneit et al., 2006), resulting in the increase of sulfur‐oxidizing capacity (upregulation of sulfur‐oxidizing genes in Fig. 7). Thus, we confirmed that the AfeI/R‐mediated regulation on EPS synthesis could influence the sulfur metabolism and cell growth of A. ferrooxidans in S0‐enriched media. The add‐back assays suggested that 3‐OH‐C14‐HSL could function as a ‘stimulator’ to regulate EPS synthesis and cell growth of A. ferrooxidans in S0‐enriched media (Fig. 6B and Fig. S2). Therefore, the 3‐OH‐C14‐HSL‐inducible EPS synthesis could be the regulatory strategy of AfeI/R for A. ferrooxidans to modulate its sulfur metabolism and cell growth in S0‐enriched media. The increase in the levels of signal molecules upon overexpression of afeI or addition of exogenous acyl‐HSLs could promote sulfur metabolism and cell growth in favorable growth environments (lag and log phases) but did not change the final population density in the S0‐enriched media (Figs 3E, G, 5C–E and 6B). Thus, the role of AfeI/R could be defined as an ‘accelerator’ for the development of the population, but not a ‘quorum maker’ for A. ferrooxidans in S0‐enriched media.
The strong inhibitory effect of either overexpression of afeI or addition of acyl‐HSLs on the ferrous iron oxidation, cell growth and quorum size of A. ferrooxidans in Fe2+‐enriched media (Figs 3A, C, 5A, B and 6A) suggested that AfeI/R could be considered as an efficient ‘inhibitor’ for A. ferrooxidans cultivated in Fe2+‐enriched media. Neither overexpression nor deletion of afeI was observed to have an effect on EPS synthesis for cells grown in Fe2+‐enriched media (Fig. 4A). These results provide two evidence: first, AfeI/R could not regulate the synthesis of EPS in the Fe2+‐enriched media; second, the QS‐mediated regulation was not caused by EPS in this condition. The obvious downregulation of genes encoding electron transporter and respiratory chain (rus, pet and cyo operon) may be the reason for the decrease of ferrous oxidation capacity and cell density of the afeI overexpression stain in Fe2+‐enriched media (Figs 3A, C and 7). The striking downregulation of a hydrogenase gene cluster in afeI overexpression strain provides another clue for understanding the AfeI/R‐mediated regulation in Fe2+‐media (Fig. 7 and Fig. S3). The HupR‐containing hydrogenase genes cluster was suggested to catalyze the conversion of dihydrogen to protons and electrons in A. ferrooxidans (Schröder et al., 2007; Kalms et al., 2018). The significant downregulation of the hydrogenase cluster probably reduced the generation of intracellular protons, which likely altered the pH homeostasis that is important for ATP biosynthesis (Lubitz et al., 2014; Hansen and Perner, 2016; Kalms et al., 2018). Thus, AfeI/R may participate in the regulation of hydrogen metabolism, which could be another reason for the decrease of cell growth and population density of the afeI overexpression stain in Fe2+‐enriched media.
The energy substrates could influence both the types of acyl‐HSLs produced by AfeI and the regulatory function of these acyl‐HSLs. Due to the presence of other potential acyl‐HSL‐synthase genes (act) in A. ferrooxidans, the specific acyl‐HSLs produced by AfeI remained unclear. Therefore, we constructed the afeI‐expression‐only strain Δact(afeI) and the afeI and act double knockout strain ΔafeI&act(215) to determine which acyl‐HSLs were generated by AfeI. Our results demonstrated that acyl‐HSLs with C‐3 hydrogen and hydroxyl substituents were synthesized by AfeI in S0‐enriched media, whereas only 3‐hydroxyl‐HSLs were found in the Fe2+‐enriched media (Table 1). Interestingly, previously reported acyl‐HSLs in A. ferrooxidans (i.e., 3‐oxo‐HSLs and 3‐hydroxy‐C8‐HSL) were not detected in this study (Farah et al., 2005). Synthesis of acyl‐HSLs requires S‐adenosylmethionine substrates and an acylated acyl carrier protein (acyl‐ACP) from the fatty acid synthesis pathway, and growth conditions could influence the availability of acyl‐ACP substrates (Parsek and Greenberg, 2000; Teplitski et al., 2003). Thus, the differences between the acyl‐HSLs observed herein and in previous reports could be due to the influence of other potential QS systems (Act), as well as differences in cultivation methods and environments. Abundant 3‐OH‐C14‐HSL was detected in both Fe2+‐ and S0‐enriched media (Table 1), but this compound was functional only in the S0‐enriched media (Fig. 6B and C). Although C12‐HSL showed an inhibitory effect on the growth of A. ferrooxidans in Fe2+‐enriched media (Fig. 6A), it was detected only in the S0‐media but not in Fe2+‐media (Table 1). Due to the lack of a 3‐OH‐C16 standard, an add‐back assay for this acyl‐HSL was not performed in this study. The role of 3‐OH‐C16‐HSL and other unidentified acyl‐HSLs produced by AfeI in the regulation of A. ferrooxidans in Fe2+‐enriched media remains an open question for future studies. These results indicated that different signal molecules were used by AfeI/R to modulate specific regulation pathways in different substrates. Therefore, the substrate‐dependent synthesis and regulation of the acyl‐HSLs is a key characteristic of the AfeI/R QS system, which probably allow A. ferrooxidans to effectively cope with the different energy substrates in the growth environments.
The significant changes in the transcriptomes of afeI knockout and overexpression strains both in S0‐ and Fe2+‐enriched media (Fig. 7 and Table S1) suggested that AfeI/R is an important means for A. ferrooxidans regulating its genes transcription in different energy substrates. In the QS‐mediated gene regulation system, the receptor reacts to a signal molecule and then binds to the lux‐box sequence to control genes transcription (An et al., 2006; Schaefer et al., 2008). The lux‐box sequences in A. ferrooxidans were predicted via the bioinformatic approach (Banderas and Guiliani, 2013), and the lux‐box region upstream of the afeI gene was determined via gel mobility shift assays (Mamani et al., 2016). Based on these results, DEGs containing lux‐box sequences were found in this study, including sulfur metabolism gene (AFE_0269), pet operon (AFE_3107‐3111), sigma‐54‐dependent transcriptional regulator gene (AFE_0693 and 0957) and conjugal transfer gene (AFE_1694). These results implied the direct regulation of AfeR on these genes, highlighting the control of the AfeI/R QS system on these pathways in A. ferrooxidans.
The AfeI/R‐mediated substrate‐dependent versatile regulation could be a noteworthy characteristic of QS regulation in these chemoautotrophic sulfur‐ and ferrous iron‐oxidizing bacteria, differentiating to other reported LuxI/R‐like QS regulation in bacteria. AfeR, for its important roles in discriminating different acyl‐HSLs and modulating genes transcription, maybe a key factor for the formation of the AfeI/R‐mediated versatile regulation in different energy‐substrates. The homology model suggested that AfeR has a receptor domain that binds to signal molecules and a regulatory domain that interacts with DNA to regulate gene transcription (Zhang et al., 2002; Soulère et al., 2008). The receptor domain of the LuxR family has evolved differently to suit its hosts (Bottomley et al., 2007). Alignment of AfeR and LuxR family protein sequences revealed that the acyl‐HSLs receptor domain has highly conserved key amino acid residues (Tyr58, Trp63, Asp75, Trp90, Ala105 and Gly113), as well as some amino acid residues that are less conservative than other strains (amino acids colored in cyan in Fig. S4). Therefore, the receptor domain of the AfeR may evolve some unique features, which leads to the different recognition ability of AfeR on acyl‐HSLs in different energy substrates. In addition, it was reported that the conformations of an acyl‐HSL are diverse, such as the linear and curved‐shape alkyl chain (Soulère et al., 2008). Acyl‐HSLs with different conformations showed different affinities to the AfeR receptor (Soulère et al., 2008). Besides, it has been reported that the pattern of acyl‐HSLs produced by a single strain depends largely on the media (Teplitski et al., 2003). Therefore, it is speculated that different energy substrates may affect the conformations of the acyl‐HSLs, which in turn affects the binding of the acyl‐HSLs to the AfeR receptor, and ultimately leads to the differences in signal recognition and gene regulation of AfeI/R system in different energy substrates. Therefore, the structural differences between AfeR and other LuxR family proteins as well as the change of the conformations of signal molecules in different substrates could contribute to the formation of AfeI/R‐mediated substrate‐dependent versatile regulation in the sulfur‐ and ferrous iron‐oxidizing bacteria.
In summary, AfeI/R have evolved distinct regulatory strategies specific to the energy substrates, and the AfeI/R‐mediated substrate‐dependent regulation could be an important mechanism employed by these sulfur‐ and ferrous iron‐oxidizing bacteria to maintain the balance between their energy metabolisms and population development in the sulfur‐ and ferrous iron‐containing extremely acidic environments. This study would be a basis for further studies on the ecological functions of AfeI/R‐like QS systems in the natural habitats and provide new insights in the synthetic biological research of the chemoautotrophic bacteria.
Experimental procedures
Bacteria and growth conditions
The bacteria and plasmids used in this study are listed in Table 2. Escherichia coli was cultivated at 37 °C in LB media (Sambrook et al., 1982). The A. ferrooxidans ATCC 23270 strain was grown in 9K inorganic salt media with Fe2+ (10 g/L) or S0 (0.8% w/v) as energy sources, and the pH was adjusted to 2.0 using H2SO4. Starkey‐Na2S2O3 agar media was used for A. ferrooxidans plate cultures (Wang et al., 2012). Cell growth in the 9K‐S0 and 9K‐Fe2+ media was monitored via OD600 nm measurements and the microscopic counting method respectively.
Table 2.
Bacteria and plasmids used in this study.
| Strain and plasmids | Description | Source |
|---|---|---|
| Strain | ||
|
Acidithiobacillus ferrooxidans ATCC 23270 |
Type strain | ATCC |
| WT | Wild type | ATCC |
| WT (pJRD215) | Wild type including the plasmid of pJRD215 | This study |
| ΔafeI | afeI gene deletion | This study |
| OEafeI | overexpress afeI gene, including the plasmid of pJRD215‐Ptac‐afeI | This study |
| Δact (afeI) | act gene deletion and afeI gene overexpress | This study |
| ΔafeI&act | Both afeI and act genes deletion | This study |
| Escherichia coli | ||
| DH5α | F‐φ80dlacZΔM15Δ(lacZYA‐argF)U169 end A1 recA1 hsdR17(rk ‐,mk + ) supE44λ‐thi‐1 gyr96 rela1 phoA | TransGen Biotech |
| S17‐1λpir | Tpr Smr recAthi pro r k ‐ m k ‐RP4:2‐Tc:MuKmTn7λpir | Bilecen and Yildiz (2009) |
| SM10 | Kmr thi‐1 thr leu tonA lacY supE recARP4‐2‐Tc::Mu | Simon et al. (1983) |
| Plasmids | ||
| pSDUDI | Suicide plasmid; Apr Kmr oiTRP4 multi‐cloning sites | Wang et al. (2016) |
| pSDUDI::afeI(UHA + DHA) | Suicide plasmid for ΔafeI construction | This study |
| pSDUDI::act(UHA + DHA) | Suicide plasmid for Δact construction | This study |
| pMSD1‐I‐SecI | pMSD containing the I‐SecI gene | Wang et al. (2012) |
| pJRD215 | Smr Kmr IncQ Mob+ | Davison et al. (1987) |
| pJRD215‐Ptac‐afeI | Smr Kmr IncQ Mob+ Ptac afeI gene | This study |
Mutant strain construction
The sequences for all primers used in this section are listed in Table S2. The markerless deletion of the afeI gene (AFE_1999) in the A. ferrooxidans ATCC 23270 was performed as described previously (Wang et al., 2012). Two homologous arms were first amplified using the IUPF/R and IDWF/R primer pairs and ligated to the pSDUDI plasmid. The generated suicide pSDUDI‐HomafeI plasmid was then transferred into A. ferrooxidans via conjugation (Peng et al., 1994). Single‐crossover recombinant strains were then selected. Then, the pMSD1‐I‐SecI plasmid was conjugated into the single‐crossover recombinants, resulting in a second homologous recombination to generate the gene knockout and wild‐type (WT) reversion. The ΔafeI was then identified via PCR using three primer pairs: P1F/P1R, P2F/P2R and P3F/P3R; the purified P1F/P1R‐amplified PCR fragments were sequenced to confirm the mutation.
The afeI gene and the tac promoter were amplified via PCR using the PtacF/PtacR and PIF/PIR primer pairs respectively. The two fragments were digested and ligated into the pJRD215 plasmid. The pJRD215‐Ptac‐afeI plasmid was conjugated into the A. ferrooxidans ATCC 23270 strain to construct the afeI overexpression strain. PCR amplification using plasmid‐specific P4F/R primers was performed to confirm the overexpression strain.
An act gene‐specific suicide plasmid was produced with the ACTUPF/R, ACTDWF/R primer pairs. The generated plasmid was then conjugated into WT and ΔafeI. Δact was identified using primers PA1F/R, PA2F/R and PA3F/R. ΔafeI&act was identified using primers P1F/R, P2F/R, P3F/R, PA1F/R, PA2F/R and PA3F/R. Then, the pJRD215‐Ptac‐afeI and pJRD215 plasmids were conjugatively transferred into Δact and ΔafeI&act generating strains Δact(afeI) and ΔafeI&act (pJRD215) respectively.
Determination of Fe2+ and SO42 ‐ concentrations in culture media
The concentration of Fe2+ in the liquid media was determined via the o‐phenanthroline method as described previously (Herrera et al., 1989), whereas the concentration of SO4 2‐ was measured via ion chromatography (ICS‐1100AR, DIONEX, USA) (Miura and Kawaoi, 2000).
EPS extraction and analysis
EPS extraction was performed as described previously (More et al., 2014; Xiao et al., 2017). Cells were collected by centrifugation and adjusted to their final concentration (OD600 nm = 1). Then, 1 ml of these cell suspensions were centrifuged at 12 000g for 1 min at 4 °C. The cells were then resuspended in 4 ml of TNE buffer (10 mM Tris, 100 mM NaCl, 5 mM EDTA, pH = 7.5) and centrifuged at 12 000g for 10 min. The pellet was then resuspended in 4 ml TNE + SDS (0.1%). After a 5‐min reaction period at room temperature, the samples were centrifuged at 12 000g for 10 min to obtain EPS extracts. Afterward, the extracts were washed three times with TNE buffer and eluted in 50 mM Tris (pH 7.5). The total carbohydrate content in the EPS extracts was determined using the anthrone‐sulfuric acid method (Ding et al., 2019). The protein concentration in the EPS extracts was measured using the Modified Bradford Protein Assay Kit (Sangon Biotech). The experiments were performed three times and each sample was set three biological replications. Statistical analysis was conducted via Student's t‐test using the GraphPad Prism software (version 7.0; GraphPad).
Sulfur coupon preparation and SEM
Sulfur coupons were prepared by melting sulfur powder and then pouring the liquid sulfur on a glass coverslip to cool and solidify (Bellenberg et al., 2014). The cell solutions were added to the sulfur‐coupon‐contained medium, and the cell density was adjusted to OD600 = 0.1. After 8 days of cultivation, the sulfur coupons were taken out, fixed with 2.5% glutaraldehyde, dehydrated in a series of graded ethanol solutions and critical point‐dried. After gold sputtering, the sulfur coupons were observed by SEM (Quanta 250 FEG, FEI) (Liu et al., 2003).
Crude acyl‐HSL extraction and identification
The cells were cultivated until they reached the stationary growth phase (OD600 = 0.30–0.32), after which 5 L of culture was collected and centrifuged at 12 000g for 5 min. The supernatant from the culture broth was extracted twice using an equal volume of HPLC‐grade dichloromethane (Rivas et al., 2005; Ruiz et al., 2008). The residues were dissolved in 1 mL of HPLC‐grade methanol and stored at −20 °C. Acyl‐HSLs extraction and identification were performed three times. The acyl‐HSL extracts were identified via liquid chromatography with tandem mass spectrometry (LC‐MS‐MS; Altimate3000 (LC) Thermo Fisher, USA; ImpactHD (MS), Bruker, Germany) as described previously (Morin et al., 2003). C12, C14, 3‐OH‐C10, 3‐OH‐C12 and 3‐OH‐C14 standards were purchased from Sigma (USA). Relative acyl‐HSL quantification in the extracts was achieved by calculating the peak area from the LC‐MS‐MS results (Ni et al., 2019).
Add‐back experiments
300 μl and 600 μl of the acyl‐HSLs extracts from the S0‐ and Fe2+‐enriched media were respectively added to 150 ml of media. The extract add‐back assays in S0‐enriched media were performed at 4th, 9th and 12th day. When Fe2+ was used as an energy substrate, the extracts were only added at the beginning of cultivation. The concentration used for each acyl‐HSL standard was 10 μM. The acyl‐HSLs were added on the 4th day in S0‐enriched media and on the 0th day in Fe2+‐enriched media. The experiments were performed three times with three biological replications.
RNA extraction, real‐time quantitative PCR and RNA sequencing
RNA was extracted from cell samples in the mid‐log growth phase. The RNAprep Pure Cell/Bacteria Kit (Tiangen, China) was used for all RNA extractions according to the manufacturer's instructions. The extracted RNA was then visualized via formaldehyde degeneration electrophoresis (Rivas et al., 2005). The A260 value and A260/A280 ratio were measured to determine RNA concentration and purity respectively. Reverse transcription was performed using the PrimeScript™ RT Reagent Kit (TaKaRa, China). RT‐qPCR reactions were performed in a Roche LightCycler480 (Roche, USA) using the SYBR®Premix Ex Taq™ (TaKaRa) enzyme; alaS was used as a reference gene (Nieto et al., 2009). The 2‐ΔΔCt method was used to analyse relative changes in gene expression (Livak and Schmittgen, 2001). RT‐qPCR primers are listed in Table S2.
RNA sequences were supplied by Novogene, China. The raw data of RNA‐seq were deposited in NCBI with accession numbers SRR9208397, SRR9208398, SRR9208395, SRR9208396, SRR9208401, SRR9208402, SRR9208399 and SRR9208400. The 179 genes were randomly selected to verify the DEGs obtained by RNA‐seq. The original data of DEGs and verification results of RT‐qPCR were listed in supplementary Table S1. The primers used in the RT‐qPCR were listed in Table S2.
Statistical analysis
All experiments were performed three times with three biological replications. One‐way analysis of variance coupled with Bonferroni's multiple comparison test was used to compare. Statistical analysis was conducted via the Student's t‐test. All statistical analyses were performed using the GraphPad Prism software (version 7.0). Statistical significance is indicated with asterisks (**** indicates P < 0.0001, *** indicates P < 0.001, ** indicates P < 0.01 and * indicates P < 0.05) in the results section.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supporting information
Appendix S1: Supporting Information
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (31570036, 31872621, 31570041, 41877400), the Startup Funding of the Chinese Academy of Sciences (2017‐020), the National Key Research and Development Project of China (2018YFC1802601), the Project of Taishan Industry Leading Talent in Shandong province (LJNY201603), the State Key Laboratory of Microbial Technology Foundation (M2017‐01), and People's Republic of China. We are grateful to Prof. Yue‐Zhong Li and associate Prof. Hai‐Nan Su from Shandong University for suggestions and supports regarding the study. We thank the supports from Core Facilities Sharing Platform for Life Sciences of Shandong University, including Sen Wang for SEM technical assistance, Jing‐Yao Qu for LC‐MS analysis, Zhi‐Feng Li for RT‐qPCR instruction and Nan‐nan Dong for providing bacteriological incubators.
Contributor Information
Jian‐Qun Lin, Email: jianqunlin@sdu.edu.cn.
Lin‐Xu Chen, Email: linxuchen@sdu.edu.cn.
References
- Abbas, A. , Adams, C. , Scully, N. , Glennon, J. , and O'Gara, F. (2007) A role for TonB1 in biofilm formation and quorum sensing in Pseudomonas aeruginosa . FEMS Microbiol Lett 274: 269–278. 10.1111/j.1574-6968.2007.00845.x. [DOI] [PubMed] [Google Scholar]
- An, D. , Danhorn, T. , Fuqua, C. , and Parsek, M.R. (2006) Quorum sensing and motility mediate interactions between Pseudomonas aeruginosa and Agrobacterium tumefaciens in biofilm cocultures. Proc Natl Acad Sci U S A 103: 3828–3833. 10.1073/pnas.0511323103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banderas, A. , and Guiliani, N. (2013) Bioinformatic prediction of gene functions regulated by quorum sensing in the bioleaching bacterium Acidithiobacillus ferrooxidans . Int J Mol Sci 14: 16901–16916. 10.3390/ijms140816901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellenberg, S. , Diaz, M. , Noel, N. , Sand, W. , Poetsch, A. , Guiliani, N. , and Vera, M. (2014) Biofilm formation, communication and interactions of leaching bacteria during colonization of pyrite and sulfur surfaces. Res Microbiol 165: 773–781. 10.1016/j.resmic.2014.08.006. [DOI] [PubMed] [Google Scholar]
- Ben‐Yaakov, R. , and Salomon, D. (2019) The regulatory network of Vibrio parahaemolyticus type VI secretion system 1. Environ Microbiol 21: 2248–2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilecen, K. , and Yildiz, F.H. (2009) Identification of a calcium‐controlled negative regulatory system affecting Vibrio cholerae biofilm formation. Environ Microbiol 11: 2015–2029. 10.1111/j.1462-2920.2009.01923.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosecker, K. (1997) Bioleaching: metal solubilization by microorganisms. FEMS Microbiol Rev 20: 591–604. 10.1016/s0168-6445(97)00036-3. [DOI] [Google Scholar]
- Bottomley, M.J. , Muraglia, E. , Bazzo, R. , and Carfì, A. (2007) Molecular insights into quorum sensing in the human pathogen Pseudomonas aeruginosa from the structure of the virulence regulator LasR bound to its autoinducer. J Biol Chem 282: 13592–13600. 10.1074/jbc.M700556200. [DOI] [PubMed] [Google Scholar]
- Certner, R.H. , and Vollmer, S.V. (2018) Inhibiting bacterial quorum sensing arrests coral disease development and disease‐associated microbes. Environ Microbiol 20: 645–657. [DOI] [PubMed] [Google Scholar]
- Chabert, N. , Bonnefoy, V. , and Achouak, W. (2017) Quorum sensing improves current output with Acidithiobacillus ferrooxidans . Microb Biotechnol 11: 136. 10.1111/1751-7915.12797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y.T. , Li, J.T. , Chen, L.X. , Hua, Z.S. , Huang, L.N. , Liu, J. , et al. (2014) Biogeochemical processes governing natural pyrite oxidation and release of acid metalliferous drainage. Environ Sci Technol 48: 5537–5545. 10.1021/es500154z. [DOI] [PubMed] [Google Scholar]
- Davison, J. , Heusterspreute, M. , Chevalier, N. , Ha‐Thi, V. , and Brunel, F. (1987) Vectors with restriction site banks. V. pJRD215, a wide‐host‐range cosmid vector with multiple cloning sites. Gene 51: 275–280. 10.1016/0378-1119(87)90316-7. [DOI] [PubMed] [Google Scholar]
- Ding, X.S. , Zhao, B. , An, Q. , Tian, M. , and Guo, J.S. (2019) Role of extracellular polymeric substances in biofilm formation by Pseudomonas stutzeri strain XL‐2. Appl Microbiol Biotechnol 103: 9169–9180. 10.1007/s00253-019-10188-4. [DOI] [PubMed] [Google Scholar]
- Farah, C. , Vera, M. , Morin, D. , Haras, D. , Jerez, C.A. , and Guiliani, N. (2005) Evidence for a functional quorum‐sensing type AI‐1 system in the extremophilic bacterium Acidithiobacillus ferrooxidans . Appl Environ Microbiol 71: 7033–7040. 10.1128/aem.71.11.7033-7040.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, X.‐Y. , Liu, X.‐J. , Fu, C.‐A. , Gu, X.‐F. , Lin, J.‐Q. , Liu, X.‐M. , et al. (2020) Novel Strategy for Improvement of the Bioleaching Efficiency of Acidithiobacillus ferrooxidans Based on the AfeI/R Quorum Sensing System. Minerals 10: 222. 10.3390/min10030222. [DOI] [Google Scholar]
- Gehrke, T. , Telegdi, J. , Thierry, D. , and Sand, W. (1998) Importance of extracellular polymeric substances from Thiobacillus ferrooxidans for bioleaching. Appl Environ Microbiol 64: 2743–2747. 10.0000/PMID9647862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez, A. , Bellenberg, S. , Mamani, S. , Ruiz, L. , Echeverria, A. , Soulere, L. , et al. (2013) AHL signaling molecules with a large acyl chain enhance biofilm formation on sulfur and metal sulfides by the bioleaching bacterium Acidithiobacillus ferrooxidans . Appl Microbiol Biotechnol 97: 3729–3737. 10.1007/s00253-012-4229-3. [DOI] [PubMed] [Google Scholar]
- Goo, E. , An, J.H. , Kang, Y. , and Hwang, I. (2015) Control of bacterial metabolism by quorum sensing. Trends Microbiol 23: 567–576. 10.1016/j.tim.2015.05.007. [DOI] [PubMed] [Google Scholar]
- Ha, J.‐H. , Hauk, P. , Cho, K. , Eo, Y. , Ma, X. , Stephens, K. , et al. (2018) Evidence of link between quorum sensing and sugar metabolism in Escherichia coli revealed via cocrystal structures of LsrK and HPr. Sci Adv 4: eaar7063. 10.1126/sciadv.aar7063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen, M. , and Perner, M. (2016) Hydrogenase gene distribution and H2 consumption ability within the Thiomicrospira Lineage. Front Microbiol 7: 99. 10.3389/fmicb.2016.00099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harneit, K. , Göksel, A. , Kock, D. , Klock, J.H. , Gehrke, T. , and Sand, W. (2006) Adhesion to metal sulfide surfaces by cells of Acidithiobacillus ferrooxidans, Acidithiobacillus thiooxidans and Leptospirillum ferrooxidans . Hydrometallurgy 83: 245–254. 10.1016/j.hydromet.2006.03.044. [DOI] [Google Scholar]
- Herrera, L. , Ruiz, P. , Aguillon, J.C. , and Fehrmann, A. (1989) A new spectrophotometric method for the determination of ferrous iron in the presence of ferric iron. J Chem Technol Biotechnol 44: 171–181. 10.1002/jctb.280440302. [DOI] [Google Scholar]
- Inaba, Y. , Banerjee, I. , Kernan, T. , and Banta, S. (2018) Transposase‐mediated chromosomal integration of exogenous genes in Acidithiobacillus ferrooxidans . Appl Environ Microbiol 84: e01381–e01318. 10.1128/aem.01381-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juhas, M. , Eberl, L. , and Tümmler, B. (2005) Quorum sensing: the power of cooperation in the world of Pseudomonas . Environ Microbiol 7: 459–471. [DOI] [PubMed] [Google Scholar]
- Kai, P. , and Bassler, B.L. (2016) Quorum sensing signal‐response systems in Gram‐negative bacteria. Nat Rev Microbiol 14: 576. 10.1038/nrmicro.2016.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalms, J. , Schmidt, A. , Frielingsdorf, S. , Utesch, T. , Gotthard, G. , Von, S.D. , et al. (2018) Tracking the route of molecular oxygen in O2‐tolerant membrane‐bound [NiFe] hydrogenase. Proc Natl Acad Sci U S A 115: 201712267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liljeqvist, M. , Ossandon, F.J. , Gonzalez, C. , Rajan, S. , Stell, A. , Valdes, J. , et al. (2015) Metagenomic analysis reveals adaptations to a cold‐adapted lifestyle in a low‐temperature acid mine drainage stream. FEMS Microbiol Ecol 91: fiv011. 10.1093/femsec/fiv011. [DOI] [PubMed] [Google Scholar]
- Liu, H.L. , Chen, B.Y. , Lan, Y.W. , and Cheng, Y.C. (2003) SEM and AFM images of pyrite surfaces after bioleaching by the indigenous Thiobacillus thiooxidans . Appl Microbiol Biotechnol 62: 414–420. 10.1007/s00253-003-1280-0. [DOI] [PubMed] [Google Scholar]
- Livak, K.J. , and Schmittgen, T.D. (2001) Analysis of relative gene expression data using real‐time quantitative PCR and the 2−ΔΔCT method. Methods 25: 402–408. 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lubitz, W. , Ogata, H. , Rüdiger, O. , and Reijerse, E. (2014) Hydrogenases. Chem Rev 114: 4081–4148. 10.1021/cr4005814. [DOI] [PubMed] [Google Scholar]
- Mamani, S. , Moinier, D. , Denis, Y. , Soulere, L. , Queneau, Y. , Talla, E. , et al. (2016) Insights into the quorum sensing regulon of the acidophilic Acidithiobacillus ferrooxidans revealed by transcriptomic in the presence of an acyl homoserine lactone superagonist analog. Front Microbiol 7: 1365. 10.3389/fmicb.2016.01365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menzel, P. , Gudbergsdottir, S.R. , Rike, A.G. , Lin, L. , Zhang, Q. , Contursi, P. , et al. (2015) Comparative Metagenomics of Eight Geographically Remote Terrestrial Hot Springs. Microb Ecol 70: 411–424. 10.1007/s00248-015-0576-9. [DOI] [PubMed] [Google Scholar]
- Miura, Y. , and Kawaoi, A. (2000) Determination of thiosulfate, thiocyanate and polythionates in a mixture by ion‐pair chromatography with ultraviolet absorbance detection. J Chromatogr A 884: 81–87. 10.1016/S0021-9673(00)00221-1. [DOI] [PubMed] [Google Scholar]
- More, T.T. , Yadav, J.S.S. , Yan, S. , Tyagi, R.D. , and Surampalli, R.Y. (2014) Extracellular polymeric substances of bacteria and their potential environmental applications. J Environ Manage 144: 1–25. 10.1016/j.jenvman.2014.05.010. [DOI] [PubMed] [Google Scholar]
- Morin, D. , Grasland, B. , Vallee‐Rehel, K. , Dufau, C. , and Haras, D. (2003) On‐line high‐performance liquid chromatography‐mass spectrometric detection and quantification of N‐acylhomoserine lactones, quorum sensing signal molecules, in the presence of biological matrices. J Chromatogr A 1002: 79–92. 10.1016/s0021-9673(03)00730-1. [DOI] [PubMed] [Google Scholar]
- Ng, K.Y. , Kamimura, K. , and Sugio, T. (2000) Production of hydrogen sulfide from tetrathionate by the iron‐oxidizing bacterium Thiobacillus ferrooxidans NASF‐1. J Biosci Bioeng 90: 193–198. 10.1016/s1389-1723(00)80109-7. [DOI] [PubMed] [Google Scholar]
- Ni, Z. , Sousa, B.C. , Colombo, S. , Afonso, C.B. , Melo, T. , Pitt, A.R. , et al. (2019) Evaluation of air oxidized PAPC: a multi laboratory study by LC‐MS/MS. Free Radic Biol Med 144: 156–166. 10.1016/j.freeradbiomed.2019.06.013. [DOI] [PubMed] [Google Scholar]
- Nieto, P.A. , Covarrubias, P.C. , Jedlicki, E. , Holmes, D.S. , and Quatrini, R. (2009) Selection and evaluation of reference genes for improved interrogation of microbial transcriptomes: case study with the extremophile Acidithiobacillus ferrooxidans . BMC Mol Biol 10: 63. 10.1186/1471-2199-10-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohmura, N. , Sasaki, K. , Matsumoto, N. , and Saiki, H. (2002) Anaerobic respiration using Fe(3+), S(0), and H(2) in the chemolithoautotrophic bacterium Acidithiobacillus ferrooxidans . J Bacteriol 184: 2081–2087. 10.1128/jb.184.8.2081-2087.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson, G.J. , Brierley, J.A. , and Brierley, C.L. (2003) Bioleaching review part B: progress in bioleaching: applications of microbial processes by the minerals industries. Appl Microbiol Biotechnol 63: 249–257. 10.1007/s00253-003-1404-6. [DOI] [PubMed] [Google Scholar]
- Parsek, M.R. , and Greenberg, E.P. (2000) Acyl‐homoserine lactone quorum sensing in Gram‐negative bacteria: A signaling mechanism involved in associations with higher organisms. Proc Natl Acad Sci U S A 97: 8789–8793. 10.1073/pnas.97.16.8789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, J.B. , Yan, W.M. , and Bao, X.Z. (1994) Plasmid and transposon transfer to Thiobacillus ferrooxidans . J Bacteriol 176: 2892–2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quatrini, R. , Appia‐Ayme, C. , Denis, Y. , Jedlicki, E. , Holmes, D.S. , and Bonnefoy, V. (2009) Extending the models for iron and sulfur oxidation in the extreme Acidophile Acidithiobacillus ferrooxidans . BMC Genomics 10: 394. 10.1186/1471-2164-10-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quatrini, R. , and Johnson, D.B. (2016) Acidophiles: Life in Extremely Acidic Environments. Norfolk, UK: Caister Academic Press. [Google Scholar]
- Quatrini, R. , and Johnson, D.B. (2018) Microbiomes in extremely acidic environments: functionalities and interactions that allow survival and growth of prokaryotes at low pH. Curr Opin Microbiol 43: 139–147. 10.1016/j.mib.2018.01.011. [DOI] [PubMed] [Google Scholar]
- Rawlings, D.E. (1998) Industrial practice and the biology of leaching of metals from ores The 1997 Pan Labs Lecture. J Ind Microbiol Biotechnol 20: 268–274. 10.1038/sj.jim.2900522. [DOI] [Google Scholar]
- Rawlings, D.E. (2002) Heavy metal mining using microbes. Annu Rev Microbiol 56: 65. 10.1146/annurev.micro.56.012302.161052. [DOI] [PubMed] [Google Scholar]
- Rivas, M. , Seeger, M. , Holmes, D.S. , and Jedlicki, E. (2005) A Lux‐like quorum sensing system in the extreme acidophile Acidithiobacillus ferrooxidans . Biol Res 38: 283. 10.4067/S0716-97602005000200018. [DOI] [PubMed] [Google Scholar]
- Rivas, M. , Seeger, M. , Jedlicki, E. , and Holmes, D.S. (2007) Second acyl homoserine lactone production system in the extreme acidophile Acidithiobacillus ferrooxidans . Appl Environ Microbiol 73: 3225. 10.1128/aem.02948-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohwerder, T. , Gehrke, T. , Kinzler, K. , and Sand, W. (2003) Bioleaching review part A: progress in bioleaching: fundamentals and mechanisms of bacterial metal sulfide oxidation. Appl Microbiol Biotechnol 63: 239. 10.1007/s00253-003-1448-7. [DOI] [PubMed] [Google Scholar]
- Ruiz, L.M. , Valenzuela, S. , Castro, M. , Gonzalez, A. , Frezza, M. , Soulère, L. , et al. (2008) AHL communication is a widespread phenomenon in biomining bacteria and seems to be involved in mineral‐adhesion efficiency. Hydrometallurgy 94: 133–137. 10.1016/j.hydromet.2008.05.028. [DOI] [Google Scholar]
- Sambrook, J. , Fritsch, E.F. , and Maniatis, T. (1982) Molecular Cloning: A Laboratory Manual. New York, USA: Cold Spring Harbor Laboratory. [Google Scholar]
- Schaefer, A.L. , Greenberg, E.P. , Oliver, C.M. , Oda, Y. , Huang, J.J. , Bittan‐Banin, G. , et al. (2008) A new class of homoserine lactone quorum‐sensing signals. Nature 454: 595–599. 10.1038/nature07088. [DOI] [PubMed] [Google Scholar]
- Schröder, O. , Bleijlevens, B. , Jongh, T.E.D. , Chen, Z. , Li, T. , Fischer, J. , et al. (2007) Characterization of a cyanobacterial‐like uptake [NiFe] hydrogenase: EPR and FTIR spectroscopic studies of the enzyme from Acidithiobacillus ferrooxidans . J Biol Inorg Chem 12: 212. 10.1007/s00775-006-0185-7. [DOI] [PubMed] [Google Scholar]
- Simon, R. , Priefer, U. , and Pühler, A. (1983) A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Nat Biotechnol 1: 784–791. [Google Scholar]
- Soulère, L. , Guiliani, N. , Queneau, Y. , Jerez, C.A. , and Doutheau, A. (2008) Molecular insights into quorum sensing in Acidithiobacillus ferrooxidans bacteria via molecular modelling of the transcriptional regulator AfeR and of the binding mode of long‐chain acyl homoserine lactones. J Mol Model 14: 599. 10.1007/s00894-008-0315-y. [DOI] [PubMed] [Google Scholar]
- Sugio, T. , Mizunashi, W. , Inagaki, K. , and Tano, T. (1987) Purification and some properties of sulfur:ferric ion oxidoreductase from Thiobacillus ferrooxidans . J Bacteriol 169: 4916–4922. 10.1128/jb.169.11.4916-4922.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugio, T. , Taha, T.M. , and Takeuchi, F. (2009) Ferrous iron production mediated by tetrathionate hydrolase in tetrathionate‐, sulfur‐, and iron‐Grown Acidithiobacillus ferrooxidans ATCC 23270 Cells. Biosci Biotechnol Biochem 73: 1381–1386. 10.1271/bbb.90036. [DOI] [PubMed] [Google Scholar]
- Teplitski, M. , Eberhard, A. , Gronquist, M.R. , Gao, M. , Robinson, J.B. , and Bauer, W.D. (2003) Chemical identification of N‐acyl homoserine lactone quorum‐sensing signals produced by Sinorhizobium meliloti strains in defined medium. Arch Microbiol 180: 494–497. 10.1007/s00203-003-0612-x. [DOI] [PubMed] [Google Scholar]
- Wackett, L.P. (2008) Quorum sensing. Environ Microbiol 10: 2899–2900. [Google Scholar]
- Wang, H. , Liu, X. , Liu, S. , Yu, Y. , Lin, J. , Lin, J. , et al. (2012) Development of a markerless gene replacement system for Acidithiobacillus ferrooxidans and construction of a pfkB mutant. Appl Environ Microbiol 78: 1826–1835. 10.1128/aem.07230-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, R. , Lin, J.‐Q. , Liu, X.‐M. , Pang, X. , Zhang, C.‐J. , Yang, C.‐L. , et al. (2019) Sulfur oxidation in the acidophilic autotrophic Acidithiobacillus spp . Front Microbiol 9: 3290. 10.3389/fmicb.2018.03290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Z.B. , Li, Y.Q. , Lin, J.Q. , Pang, X. , Liu, X.M. , Liu, B.Q. , et al. (2016) The two‐component system RsrS‐RsrR regulates the tetrathionate intermediate pathway for thiosulfate oxidation in Acidithiobacillus caldus . Front Microbiol 7: 1755. 10.3389/fmicb.2016.01755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenbin, N. , Dejuan, Z. , Feifan, L. , Lei, Y. , Peng, C. , Xiaoxuan, Y. , and Hongyu, L. (2011) Quorum‐sensing system in Acidithiobacillus ferrooxidans involved in its resistance to Cu(2)(+). Lett Appl Microbiol 53: 84–91. 10.1111/j.1472-765X.2011.03066.x. [DOI] [PubMed] [Google Scholar]
- Xiao, Y. , Zhang, E. , Zhang, J. , Dai, Y. , Yang, Z. , Christensen, H.E.M. , et al. (2017) Extracellular polymeric substances are transient media for microbial extracellular electron transfer. Sci Adv 3: e1700623. 10.1126/sciadv.1700623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, R.‐G. , Pappas, K.M. , Brace, J.L. , Miller, P.C. , Oulmassov, T. , Molyneaux, J.M. , et al. (2002) Structure of a bacterial quorum‐sensing transcription factor complexed with pheromone and DNA. Nature 417: 971–974. 10.1038/nature00833. [DOI] [PubMed] [Google Scholar]
- Zhang, Y. , Gao, J. , Wang, L. , Liu, S. , Bai, Z. , Zhuang, X. , and Zhuang, G. (2018) Environmental adaptability and quorum sensing: iron uptake regulation during biofilm formation by Paracoccus denitrificans . Appl Environ Microbiol 84: e00865‐18. 10.1128/aem.00865-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1: Supporting Information


