Abstract
Living cells exploit bistable and oscillatory behaviors as memory mechanisms, facilitating the integration of transient stimuli into sustained molecular responses that control downstream functions. Synthetic bistable networks have also been studied as memory entities, but have rarely been utilized to control orthogonal functions in coupled dynamic systems. We herein present a new cascade pathway, for which we have exploited a well‐characterized switchable peptide‐based replicating network, operating far from equilibrium, that yields two alternative steady‐state outputs, which in turn serve as the input signals for consecutive processes that regulate various features of Au nanoparticle shape and assembly. This study further sheds light on how bridging together the fields of systems chemistry and nanotechnology may open up new opportunities for the dynamically controlled design of functional materials.
Keywords: dynamic networks, molecular evolution, nanotechnology, nonequilibrium processes, supramolecular chemistry
A peptide‐based bistable reaction network was programmed to control a cascade reaction influencing Au nanoparticle morphology, self‐assembly kinetics and shape. This study offers a clear demonstration of how bridging the fields of systems chemistry and nanotechnology can expand our repertoire of functional materials.
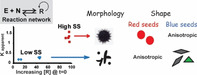
Life on earth has probably evolved from simple precursors interacting through multiple prebiotic chemical reactions, which were later on wired together into feedback‐controlled autocatalytic networks. [1] Therefore, taking the key steps towards realizing life in entirely synthetic systems would demand obtaining a clear picture of how feedback processes, such as bistability and oscillations, can yield the elaborate non‐linear functions usually observed in cellular (signaling) systems. [2] Intriguingly, several existing synthetic bistable networks—possessing two successive steady states (SSs)—have been successfully applied for engineering chemical memory.[ 2d , 3 ] However, none of these networks have yet been implemented as a pre‐encoded molecular programming tool for downstream design of nano‐scale materials. To that aim, in the current study we have combined together two fundamentally different reaction systems, namely a peptide‐based bistable replication network, and a set of nanoparticle formation reaction and growth (NRG) processes. The success of this assay relies on a cascade pathway, in which the outputs of the bistable network serve as input signals for the consecutive NRG processes, which in turn, depending on the applied experimental configuration and reaction conditions, regulate various features of Au nanoparticle (NP) shape and assembly (Figure 1 A).
Figure 1.
A) A schematic description of a cascade mechanism in which the bistable network that reaches either high or low steady states (Hss and Lss, respectively) regulates nanoparticle formation reaction and growth (NRG) processes and consequently yields variable output in Au NP morphology, self‐assembly, and shape. B) Representative results highlighting bistability in the SS product distribution (manifested by Kapp; Table S1), obtained in the reaction shown in (A) for different initial concentration combinations of the peptides (E, N and R; Table 1) and small molecule (S). In all cases, the network reactions were carried out at pH 7, with [E] and [N] kept in equal amounts, and total peptide concentration [N+R] remains constant at 100 μm. Inset: the equation used to calculate the SS constant Kapp. [3c]
The new cascade reaction provides a clear demonstration bridging systems chemistry of replicating networks with nanotechnology. Since life is an inherently out‐of‐equilibrium process, the investigation of replicating systems and emerging replicative functions under far from equilibrium conditions holds particular interest for Systems Chemistry research aiming to develop synthetic entities with life‐like behavior. [4] Metal nanoparticles were chosen for this study because they present a wide substrate scope for synthetic design, as they can respond to tiny variations in molecular compositions, thereby capable of producing signaling anisotropy. Inspired in part by previous studies with protein networks that mediated the synthesis of magnetite nanoparticle chains in magnetotactic bacteria, [5] we aim here to gain precise control over nanomaterial shape and assembly properties using the programmable steady states (SSs) in the bistable peptide network (Figure 1 A). Initial effort in this direction recently demonstrated control over the self‐assembly of micrometer size beads, using a significantly different system that relies on a robust enzyme‐DNA bistable network. [6] In other related studies, self‐assembly of nanoparticles was controlled by ordinary (non‐bistable) networks, applying either fuel‐assisted or autonomous processes far from equilibrium. [7] We herein propose that regulation of the various aspects of nanomaterial structures (i.e., morphology, shape, and self‐assembly) using the bistable network can help in the design and understanding of related memory‐like systems, providing them with primitive decision making capabilities, which are frequently found to be operative in nature.
Our bistable system underlies a non‐symmetric reversible trans‐thioesterification reaction between an electrophile peptide E and a nucleophile peptide N to produce the replicator R and a thiol leaving group S (Figure 1 A; see sequences in Table 1). This reaction network comprises a positive feedback, generated by efficient self‐replication of R as a coiled coil template. [8] The resulted disparity in forward and backward reactions is a network response that leads to bistability, when two distinct steady states (SSs) [3c] are obtained depending on the reaction's starting compositions. The replication reaction is carried out in a closed tube but, as we have recently analyzed experimentally, theoretically and via simulation, it is nevertheless active far from equilibrium (as in the open environment), since it contains an excess of the small thiol molecule (S) and a continuous supply of chemical “fuel” in the form of tris (2‐carboxyethyl) phosphine hydrochloride (TCEP).[ 2c , 2k , 9 ] Multiple concentration combinations of the network components are taken as the 1st inputs to initiate the bistable reaction, which generates only two types of output steady state product distribution signals, named high steady state (Hss) and low steady state (Lss) and characterized by the apparent reaction constant values (Kapp; Figure 1 B). The current experiments, conducted with constant total peptide concentration ([N]+[R]) of 100 μm, yielded 6±2 % of (each) E and N (Kapp=1310±180) and 20±3 % of (each) E and N (Kapp=243±54), for the Hss and Lss, respectively (Table s1). As elaborated below in detail, these two distinct SSs store different types of molecular information that can control the NRG processes and produce signaling anisotropy. We analyze the origin of the effects of the two SSs on this anisotropy phenomenon, and furthermore demonstrate that, as in biological bistable systems, it is possible to actively switch between the SSs and, therefore, to reverse the effects on the coupled (NRG) processes.
Table 1.
Peptide names and sequences.
|
Peptide[a] |
Sequence[b] |
|---|---|
|
E |
Ar‐ RVARLEKKVSALEKKVA‐COSR |
|
Es |
Ar‐EKKVA‐COSR |
|
N |
H‐ZLEXEVARLKKLVGE‐CONH2 |
|
Nβ |
H‐ZβLEXEVARLKKLVGE‐CONH2 |
|
Ns |
H‐ZLEXE‐CONH2 |
|
R |
Ar‐RVARLEKK‐VSALEKKVAZLEXEVARLKKLVGE‐CONH2 |
|
Rβ |
Ar‐RVARLEKKVSALEKKVAZβLEXEVA‐RLKKLVGE‐CONH2 |
|
Rs |
Ar‐EKKVAZLEXE‐CONH2 |
[a] All peptides were synthesized and purified in house. [b] Ar=4‐acetamidobenzoate, X=Lys‐Ar, SR=S=2‐mercapto‐ethane sulfonate, Z=‐SCH2CO and Zβ=‐SCH2CH2CO.
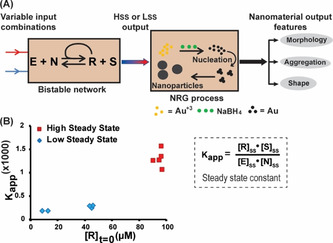
Au NPs were chosen as the working model due to their diversity in shape, ranging from isotropic spherical [10] to anisotropic nanostar, [11] bipyramid, [12] prism, [13] rods, [14] and others. [15] This large variability has been used to control applications in different fields, including nanomedicine, sensing, signal generation, non‐linear optics and catalysis.[ 7e , 16 ] The interactions of Au NPs with free thiol, thiosulphate and/or carboxylic acid functional groups, as well as the effects of these interactions on NP morphologies, have been documented in many studies.[ 16a , 17 ] Therefore, in order to control the NP morphology using the dynamic bistable network, we have first allowed the network reaction to reach the respective Hss and Lss, and then introduced each one of these mixtures separately for in situ synthesis of Au NPs in the presence of the reducing agent NaBH4. The high SS resulted from the bistable reaction network initiated with the input signal of only R (100 μm), whereas the low SS mixtures were generated from two different sets of input signals, 10 μm of R (+ 90 μm of each E and N) or 50 μm of R (+ 50 μm of each E and N), respectively. At the outset, we started these experiments with 15 μm of Au salt and 500 μm NaBH4. Specific regulation of NP morphology was evidenced when two different patterns, nano‐urchin and hyperbranched nanorods, were obtained as the final outputs after employing the Hss and Lss mixtures, respectively (Figure 2 and Figures S1–S3 in SI). The synthesized Au NPs that induced the nucleation towards different morphology from the two different SS mixtures were ultra‐small, having their diameters below 2.5 nm. Such small NPs are nonmetallic, they do not display the surface plasmon resonance (Figure S4), and their synthesis could not be monitored using the UV/Vis spectroscopy. [18] Nevertheless, transmission electron microscope (TEM) images provided valuable structural analysis, revealing that the urchin nanoaggregates formed in the Hss varied in diameter from 50 to 90 nm, and were thus larger than the hyperbranched nanorod aggregates generated in the Lss which had their lengths ranging from 10 to 30 nm (Figure 2 and Figures S1 and S2). Similar experiments with a different Au salt concentration (5 μm) reproduced similar morphologies from these concomitant SSs (Figure S3).
Figure 2.
Regulating nanoparticle morphology using the bistable network Hss and Lss. A) General experimental layout of the cascade reaction configuration. B,C) TEM images of nanoparticles grown for 3 hours and 2 days in the Hss and Lss mixtures. The NRG processes were carried out with 15 μm of AuCl3 in the presence of 500 μm of NaBH4 at pH 7 and 22 °C.
The general propensity of bistable systems to induce alternative NP morphologies was further demonstrated by employing a network made of another replicating thiodepsipeptide mutant and its precursors (Rβ, Nβ and E; Table 1). Rβ possesses lower replication capacity [19] and the respective bistable network featured a smaller difference between the two SSs (Kapp=71±14 and 3.6±0.6 for the Hss and Lss, respectively; Figure S5 and Table S1). Nevertheless, distinctly different NP morphology patterns, nano‐urchin and small aggregates, were obtained as the main features after employing these new Hss and Lss mixtures, respectively (Figure S5). As a target for future experiments, we intend to study additional peptide sequences, in order to correlate sequence changes with the propensity to form bistable systems, and consequently to control the NRG processes. A negative control experiment with a network of shorter peptides that cannot replicate (Rs, Ns and Es; Table 1) yielded only one SS product distribution for all studied initial concentration combinations (Table S1), and consequently did not induce anisotropy in NP formation morphology (Figure S6)
We propose that the two assembly manifestations appear due to different degrees of nanoparticle stabilization with the different SS mixtures, since each one of the latter possess significantly different sets of interacting components (i.e., thiol residues, carboxylic acids and backbone amides). This assumption was first supported by the results of two control experiments with only the thiol‐containing peptide N, or the small molecule thiol S, which were studied at two different concentrations corresponding to their abundances in the Hss and Lss and did not produce remarkable differences in nanomorphology (Figures S7 and S8). Then, although it is difficult to dictate the exact amounts of Au‐NP‐bound components (E, N or R) in the SS mixtures, we performed a set of experiments to assess their relative preferences to functionalize the NP surfaces. We therefore synthesized the Au NPs in buffered water (in absence of peptides), immediately mixed them with a single network component (E, N or R) or with one of the SS mixtures (Hss or Lss, separately), and investigated the NPs aggregation over time by UV/Vis spectroscopy. In all three cases studied with the individual peptide mixtures, the NPs aggregation was slowed down versus aggregation of the peptide‐free NPs, and we further noticed that the rate of aggregation in the presence of the replicator R was slower than in the presence of E or N (Figure S9), suggesting a tighter NP binding or higher coverage by R. We propose that the higher extent of multivalency provided by R, versus E or N, makes it more effective in surface functionalization. This finding is also supported by the lower rate of aggregation in the presence of the Hss versus Lss (Figure s10), as the Hss mixture contains higher amount of R.
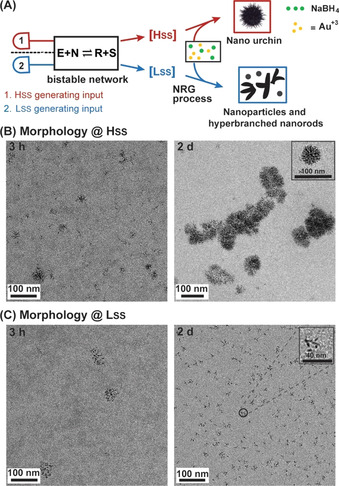
Since monitoring the progress of Au NP synthesis by UV/Vis spectroscopy proved to be challenging, we ran the following experiments with relatively larger UV/Vis active Au NP seeds. This was accomplished by mixing in a 1:1 ratio (v/v) of the Good buffer MOPS (3‐(N‐morpholino) propanesulfonic acid; 500 mm, pH 7) with an Au salt solution (4.5 mm). Transient red colored seeds were obtained in this mixture after 30 s vortex, while during continuous vortex for additional 30 s, the red seeds were spontaneously transformed into blue colored seeds. Both seed types were then characterized using UV/Vis and TEM measurements. The red seeds contained ≈94 % of spherical nanoparticles having diameter of ca. 13 nm, while the blue seeds were found as a mixture of several anisotropic nanoparticles, including ≈6 % (each) of the nanoprism and bipyramid structures, and other less well‐defined architectures (Figure S11). Then, in order to control the self‐assembly and shape of NP seeds by the bistable reaction networks, we cultivated the pre‐synthesized seeds into the two SS solutions, separately (Figure 3 A), and followed the outcome features by additional UV/Vis spectroscopy and TEM measurements.
Figure 3.
Bistability inducing disparity in nanoparticle self‐assembly kinetics. A) General layout of the experiments performed to regulate the Au NPs self‐assembly and shape by the bistable reaction network SSs. B,C) UV/Vis spectra that enabled us to follow the real‐time aggregation of bluish Au NP seeds within the Hss and Lss mixtures, respectively. D) Au NP plasmon resonance absorption (603 nm) as a function of time, extracted from the data shown in (B) and (C). E,F) TEM images of Au NP aggregates obtained after 1 h and 3 h incubation times in the Hss and Lss mixtures, respectively. G) Layout of the switching experiments, in which the Hss was triggered to convert into the Lss, and vice versa, prior to their reactions with the bluish Au NP seeds. H,I) TEM images of Au NP aggregates obtained after 1 h and 3 h incubation times in the Lss‐to‐Hss and Hss‐to‐Lss switch mixtures, respectively. The NRG processes were carried out in the respective SS mixtures with 30 μm of Au blue seeds (anisotropic NP seeds) at pH 7 and 22 °C.
The self‐assembly induced aggregation of Au NPs was analyzed by transferring the bluish seeds into the high and low SS mixtures in separate vessels, yielding final Au concentration of 30 μm. The aggregation kinetics of the NPs were investigated by following the reduction over time of the surface plasmon resonance signal at 603 nm. This assay clearly indicated slower aggregation in the Hss mixture versus the Lss mixture (Figure 3 B–D). Similar experiments with a higher concentration of the bluish Au NP seeds (100 μm) exhibited the same self‐assembly trend (Figure S12). Furthermore, when the Au NP aggregates obtained after one and three hours within both SS mixtures were visualized by TEM measurements, we observed fewer and less dense aggregates after equilibration in the Hss versus Lss (Figure 3 E,F). Using complementary ligand exchange experiments (SI Section 3.3) and LCMS measurements (Figure S13), we have also confirmed that all three peptide network components (E, N and R) were involved in functionalizing the Au blue seeds during the self‐assembly process. Experiments with Au NPs red seeds (100 μm) also followed similar self‐assembly kinetics, as they reacted through faster aggregation in the Lss relative to Hss mixtures (Figure S14).
Previous studies have shown that biological bistable networks are switchable, namely that they can be flipped from one state to the other by application of specific stimuli. Similarly, we performed active switching between the Hss and Lss, in both directions (Figure 3 G; experimental details in SI Section 2), and analyzed the potency of the switched mixtures to control the bluish NP seeds self‐assembly. The Lss‐to‐Hss and Hss‐to‐Lss switch reactions indeed resulted in product distributions similar to that observed in the original Hss and Lss mixtures, respectively, as deduced from the calculated Kapp values (Table S1). The aggregation kinetics of the NPs were again investigated in the presence of the SS mixtures obtained after the switch experiments, Both the UV/Vis experiments that followed the decrease in the surface plasmon resonance signal (Figure S15), and TEM measurements at 1 h and 3 h after mixing (Figure 3 H,I), have clearly indicated slower aggregation in the Lss‐to‐Hss mixture versus the Hss‐to‐Lss mixture. This experiment demonstrates the utility of the bistable system to control downstream functions through fairly complex pathways.
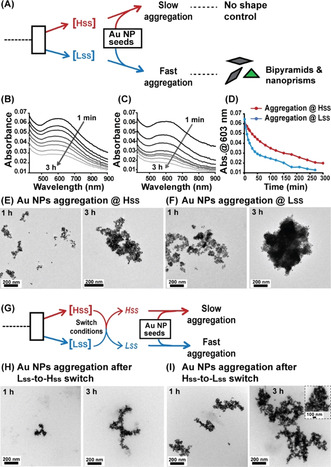
Finally, in order to probe the shape‐directing capability, Au NP seeds were first allowed to cultivate in the respective SS solutions separately, where their final concentrations were maintained at 100 μm. TEM images of aged red seeds samples (after 2 days) displayed that incubation in the Hss solution retained 90 % of the seeds as spherical NPs. Interestingly, incubation in the Lss mixture retained only 55 % as spherical NPs, while 45 % of the parent NP seeds were converted into various anisotropic NPs (Figure 4 A,B,E and Figure S17). A similar experiment with a lower concentration of the spherical red seeds (30 μm) also yielded the same results (Figure S18).
Figure 4.
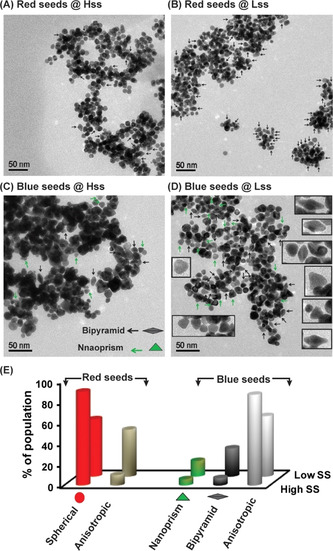
Bistability inducing disparity in nanoparticle shapes. A,B) TEM images of Au NPs obtained after 2 days of incubation of the red seeds in the Hss and Lss mixtures. Arrows depicting anisotropic NPs. C,D) TEM images of Au NPs obtained after 30 min of incubation of the bluish seeds in the Hss and Lss mixtures. Arrows depicting selected bipyramid and nanoprism objects; higher magnification images of such objects are given as inset in panel (D). E) Relative population of the various shaped NPs in the Hss and Lss, calculated by analysis of the above TEM images. More than 150 NP images in each sample were used to calculate the percentage of the different shapes; note additional images in Figures S17–S19. The NRG processes were carried out in the respective SS mixtures with 100 μm of Au red or blue seeds at pH 7 and 22 °C.
Exciting results were also obtained in similar experiments when the bluish seeds (100 μm) of anisotropic Au NPs were cultivated separately in concomitant Hss and Lss solutions. TEM analysis after incubation for 30 minutes again demonstrated significant shape retention in the Hss mixture, which contained similar shapes distribution as in parent blue seeds, namely ≈6 % of (each) bipyramid and nanoprism NPs, and 88 % of others anisotropic structures. On the other hand, incubation in the Lss mixture yielded 25 % and 15 % of bipyramid and nanoprism Au architectures, respectively. The rest 60 % population included with other anisotropic NPs (Figures 4 C–E and Figure S19). It has been shown earlier that Au NP shape transformation can be driven under certain circumstances by interacting ligands in the environment. [20] Our experiments with the bistable networks clearly display a shape‐retention capability for the Hss, while the Lss mixture can more actively direct the evolution of new architectures, i.e., the bipyramids and nanoprisms.
In conclusion, we have disclosed a new concept methodology, by which non‐enzymatic switchable bistable reaction networks can be coupled for downstream functions, particularly for the regulation of NP morphology, shape and self‐assembly structures. The analysis of this feedback‐controlled cascade system is fairly general, as it was performed with different peptide sequences, and by applying variable experimental conditions (e.g., different concentrations of Au salts and Au NP seeds). It can hence contribute to advance several contemporary scientific goals in systems chemistry. Firstly, it features a new type of biological mimicry, where a synthetic bioorganic circuit operates out‐of‐equilibrium to control the behavior of inorganic materials. This may open the door for exploiting dynamic molecular programming of more complex nanomaterials, valuable for various applications in (bio)nanotechnology. Furthermore, the construction of such artificial memory‐like systems will improve our understanding of other bioengineered networks and signal transduction apparatus. Altogether, we propose that research in systems chemistry reaching out to the nanomaterials field would expand the chance for success in the design of life‐like systems and may in the future pave the way towards materialization of artificial cells.
Conflict of interest
The authors declare no conflict of interest.
Supporting information
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supplementary
Acknowledgements
We acknowledge the financial support by the Israeli Science Foundation (ISF 707/16), and IM is grateful to PBC post‐doctoral fellowship program. We thank Dr. R. Mukherjee and Dr. R. Cohen‐Luria for help in early stages of the project, and Dr. A. Upcher (IKI BGU) for performing the TEM measurements. Open access funding enabled and organized by Projekt DEAL.
I. Maity, D. Dev, K. Basu, N. Wagner, G. Ashkenasy, Angew. Chem. Int. Ed. 2021, 60, 4512.
Contributor Information
Dr. Indrajit Maity, Email: maityindrajitchem@gmail.com.
Prof. Dr. Gonen Ashkenasy, Email: gonenash@bgu.ac.il.
References
- 1.
- 1a. Eigen M., Schuster P., The hypercycle. A principle of natural self-organization, Springer, Berlin, 1979; [DOI] [PubMed] [Google Scholar]
- 1b. Kauffman S. A., J. Theor. Biol. 1986, 119, 1–24; [DOI] [PubMed] [Google Scholar]
- 1c. Orgel L. E., Nature 1992, 358, 203–209. [DOI] [PubMed] [Google Scholar]
- 2.
- 2a. An Introduction to Nonlinear Chemical Dynamics: Oscillations, Waves, Patterns, and Chaos (Eds.: Epstein I. R., Pojman J. A.), Oxford University Press, New York, 1998; [Google Scholar]
- 2b. T. Bánsági, Jr. , Taylor A. F., Isr. J. Chem. 2018, 58, 706–713; [Google Scholar]
- 2c. Wagner N., Hochberg D., Peacock-Lopez E., Maity I., Ashkenasy G., Life 2019, 9, 45; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2d. Kim J., White K. S., Winfree E., Mol. Syst. Biol. 2006, 2, 68; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2e. Padirac A., Fujii T., Rondelez Y., Proc. Natl. Acad. Sci. USA 2012, 109, E3212–E3220; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2f. Wagner N., Alasibi S., Peacock-Lopez E., Ashkenasy G., J. Phys. Chem. Lett. 2015, 6, 60–65; [DOI] [PubMed] [Google Scholar]
- 2g. Semenov S. N., Wong A. S. Y., van der Made R. M., Postma S. G. J., Groen J., van Roekel H. W. H., de Greef T. F. A., Huck W. T. S., Nat. Chem. 2015, 7, 160–165; [DOI] [PubMed] [Google Scholar]
- 2h. Kundu P. K., Samanta D., Leizrowice R., Margulis B., Zhao H., Borner M., Udayabhaskararao T., Manna D., Klajn R., Nat. Chem. 2015, 7, 646–652; [DOI] [PubMed] [Google Scholar]
- 2i. Semenov S. N., Kraft L. J., Ainla A., Zhao M., Baghbanzadeh M., Campbell V. E., Kang K., Fox J. M., Whitesides G. M., Nature 2016, 537, 656; [DOI] [PubMed] [Google Scholar]
- 2j. Leira-Iglesias J., Tassoni A., Adachi T., Stich M., Hermans T. M., Nat. Nanotechnol. 2018, 13, 1021–1027; [DOI] [PubMed] [Google Scholar]
- 2k. Maity I., Wagner N., Mukherjee R., Dev D., Peacock-Lopez E., Cohen-Luria R., Ashkenasy G., Nat. Commun. 2019, 10, 4636; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2l. Joesaar A., Yang S., Bögels B., van der Linden A., Pieters P., Kumar B. V. V. S. P., Dalchau N., Phillips A., Mann S., de Greef T. F. A., Nat. Nanotechnol. 2019, 14, 369–378; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2m. Cafferty B. J., Wong A. S. Y., Semenov S. N., Belding L., Gmur S., Huck W. T. S., Whitesides G. M., J. Am. Chem. Soc. 2019, 141, 8289–8295; [DOI] [PubMed] [Google Scholar]
- 2n. Schaffter S. W., Schulman R., Nat. Chem. 2019, 11, 829–838; [DOI] [PubMed] [Google Scholar]
- 2o. Bohner B., T. Bansagi, Jr. , Toth A., Horvath D., Taylor A. F., Angew. Chem. Int. Ed. 2020, 59, 2823–2828; [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem. 2020, 132, 2845–2850. [Google Scholar]
- 3.
- 3a. Lebar T., Bezeljak U., Golob A., Jerala M., Kadunc L., Pirš B., Stražar M., Vučko D., Zupančič U., Benčina M., Forstnerič V., Gaber R., Lonzarić J., Majerle A., Oblak A., Smole A., Jerala R., Nat. Commun. 2014, 5, 5007; [DOI] [PubMed] [Google Scholar]
- 3b. Muzika F., Bansagi T., Schreiber I., Schreiberova L., Taylor A. F., Chem. Commun. 2014, 50, 11107–11109; [DOI] [PubMed] [Google Scholar]
- 3c. Mukherjee R., Cohen-Luria R., Wagner N., Ashkenasy G., Angew. Chem. Int. Ed. 2015, 54, 12452–12456; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2015, 127, 12629–12633; [Google Scholar]
- 3d. Meijer L. H. H., Joesaar A., Steur E., Engelen W., van Santen R. A., Merkx M., de Greef T. F. A., Nat. Commun. 2017, 8, 1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.
- 4a. Ruiz-Mirazo K., Briones C., de la Escosura A., Chem. Rev. 2014, 114, 285–366; [DOI] [PubMed] [Google Scholar]
- 4b. Mattia E., Otto S., Nat. Nanotechnol. 2015, 10, 111–119; [DOI] [PubMed] [Google Scholar]
- 4c. Alfonso I., Chem. Commun. 2016, 52, 239–250; [DOI] [PubMed] [Google Scholar]
- 4d. Ashkenasy G., Hermans T. M., Otto S., Taylor A. F., Chem. Soc. Rev. 2017, 46, 2543–2554; [DOI] [PubMed] [Google Scholar]
- 4e. Li J., Nowak P., Otto S., J. Am. Chem. Soc. 2013, 135, 9222–9239; [DOI] [PubMed] [Google Scholar]
- 4f. Bissette A. J., Fletcher S. P., Angew. Chem. Int. Ed. 2013, 52, 12800–12826; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2013, 125, 13034–13061; [Google Scholar]
- 4g. Kosikova T., Philp D., Chem. Soc. Rev. 2017, 46, 7274–7305; [DOI] [PubMed] [Google Scholar]
- 4h. Ragazzon G., Prins L. J., Nat. Nanotechnol. 2018, 13, 882–889; [DOI] [PubMed] [Google Scholar]
- 4i. Bai Y., Chotera A., Taran O., Liang C., Ashkenasy G., Lynn D. G., Chem. Soc. Rev. 2018, 47, 5444–5456; [DOI] [PubMed] [Google Scholar]
- 4j. Kroiss D., Ashkenasy G., Braunschweig A. B., Tuttle T., Ulijn R. V., Chem 2019, 5, 1917–1920. [Google Scholar]
- 5. Mirabello G., Lenders J. J. M., Sommerdijk N. A. J. M., Chem. Soc. Rev. 2016, 45, 5085–5106. [DOI] [PubMed] [Google Scholar]
- 6. Zadorin A. S., Rondelez Y., Gines G., Dilhas V., Urtel G., Zambrano A., Galas J.-C., Estevez-Torres A., Nat. Chem. 2017, 9, 990. [DOI] [PubMed] [Google Scholar]
- 7.
- 7a. Grötsch R. K., Wanzke C., Speckbacher M., Angı A., Rieger B., Boekhoven J., J. Am. Chem. Soc. 2019, 141, 9872–9878; [DOI] [PubMed] [Google Scholar]
- 7b. Sahoo J. K., Roy S., Javid N., Duncan K., Aitken L., Ulijn R. V., Nanoscale 2017, 9, 12330–12334; [DOI] [PubMed] [Google Scholar]
- 7c. Heinen L., Walther A., Chem. Sci. 2017, 8, 4100–4107; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7d. Sahoo J. K., Pappas C. G., Sasselli I. R., Abul-Haija Y. M., Ulijn R. V., Angew. Chem. Int. Ed. 2017, 56, 6828–6832; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2017, 129, 6932–6936; [Google Scholar]
- 7e. della Sala F., Maiti S., Bonanni A., Scrimin P., Prins L. J., Angew. Chem. Int. Ed. 2018, 57, 1611–1615; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2018, 130, 1627–1631; [Google Scholar]
- 7f. van Ravensteijn B. G. P., Voets I. K., Kegel W. K., Eelkema R., Langmuir 2020, 36, 10639–10656; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7g. Kruse J., Merkens S., Chuvilin A., Grzelczak M., ACS Appl. Nano Mater. 2020, 3, 9520–9527. [Google Scholar]
- 8. Dadon Z., Wagner N., Alasibi S., Samiappan M., Mukherjee R., Ashkenasy G., Chem. Eur. J. 2015, 21, 648–654. [DOI] [PubMed] [Google Scholar]
- 9. Wagner N., Mukherjee R., Maity I., Peacock-Lopez E., Ashkenasy G., ChemPhysChem 2017, 18, 1842–1850. [DOI] [PubMed] [Google Scholar]
- 10. Lloyd J. A., Liu Y., Ng S. H., Thai T., Gómez D. E., Widmer-Cooper A., Bach U., Nanoscale, 2019, 11, 22841–22848. [DOI] [PubMed] [Google Scholar]
- 11.
- 11a. Chandra K., Culver K. S. B., Werner S. E., Lee R. C., Odom T. W., Chem. Mater. 2016, 28, 6763–6769; [Google Scholar]
- 11b. Xi W., Haes A. J., J. Am. Chem. Soc. 2019, 141, 4034–4042. [DOI] [PubMed] [Google Scholar]
- 12. Lee J.-H., Gibson K. J., Chen G., Weizmann Y., Nat. Commun. 2015, 6, 7571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhai Y., DuChene J. S., Wang Y.-C., Qiu J., Johnston-Peck A. C., You B., Guo W., DiCiaccio B., Qian K., Zhao E. W., Ooi F., Hu D., Su D., Stach E. A., Zhu Z., Wei W. D., Nat. Mater. 2016, 15, 889. [DOI] [PubMed] [Google Scholar]
- 14.
- 14a. Hill H. D., Millstone J. E., Banholzer M. J., Mirkin C. A., ACS Nano 2009, 3, 418–424; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14b. Kim J.-Y., Han M.-G., Lien M.-B., Magonov S., Zhu Y., George H., Norris T. B., Kotov N. A., Sci. Adv. 2018, 4, e1700682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.
- 15a. Bahng J. H., Yeom B., Wang Y., Tung S. O., Hoff J. D., Kotov N., Nature 2015, 517, 596; [DOI] [PubMed] [Google Scholar]
- 15b. Yan F., Liu L., Walsh T. R., Gong Y., El-Khoury P. Z., Zhang Y., Zhu Z., De Yoreo J. J., Engelhard M. H., Zhang X., Chen C.-L., Nat. Commun. 2018, 9, 2327; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15c. O′Brien M. N., Jones M. R., Lee B., Mirkin C. A., Nat. Mater. 2015, 14, 833. [DOI] [PubMed] [Google Scholar]
- 16.
- 16a. Daniel M.-C., Astruc D., Chem. Rev. 2004, 104, 293–346; [DOI] [PubMed] [Google Scholar]
- 16b. Maiti S., Pezzato C., Garcia Martin S., Prins L. J., J. Am. Chem. Soc. 2014, 136, 11288–11291; [DOI] [PubMed] [Google Scholar]
- 16c. Zhao H., Sen S., Udayabhaskararao T., Sawczyk M., Kučanda K., Manna D., Kundu P. K., Lee J.-W., Král P., Klajn R., Nat. Nanotechnol. 2016, 11, 82. [DOI] [PubMed] [Google Scholar]
- 17. Chen Y., Xianyu Y., Jiang X., Acc. Chem. Res. 2017, 50, 310–319. [DOI] [PubMed] [Google Scholar]
- 18. Zhou M., Zeng C., Chen Y., Zhao S., Sfeir M. Y., Zhu M., Jin R., Nat. Commun. 2016, 7, 13240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dadon Z., Samiappan M., Shahar A., Zarivach R., Ashkenasy G., Angew. Chem. Int. Ed. 2013, 52, 9944–9947; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2013, 125, 10128–10131. [Google Scholar]
- 20.
- 20a. Rambukwella M., Sakthivel N. A., Delcamp J. H., Sementa L., Fortunelli A., Dass A., Front. Chem. 2018, 6, 331–317; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20b. Stoeva S. I., Zaikovski V., Prasad B. L. V., Stoimenov P. K., Sorensen C. M., Klabunde K. J., Langmuir 2005, 21, 10280–10283. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supplementary


