Summary
Organogenesis of legume root nodules begins with the nodulation factor‐dependent stimulation of compatible root cells to initiate divisions, signifying an early nodule primordium formation event. This is followed by cellular differentiation, including cell expansion and vascular bundle formation, and we previously showed that Lotus japonicus NF‐YA1 is essential for this process, presumably by regulating three members of the SHORT INTERNODES/STYLISH (STY) transcription factor gene family.
In this study, we used combined genetics, genomics and cell biology approaches to characterize the role of STY genes during root nodule formation and to test a hypothesis that they mediate nodule development by stimulating auxin signalling.
We show here that L. japonicus STYs are required for nodule emergence. This is attributed to the NF‐YA1‐dependent regulatory cascade, comprising STY genes and their downstream targets, YUCCA1 and YUCCA11, involved in a local auxin biosynthesis at the post‐initial cell division stage. An analogous NF‐YA1/STY regulatory module seems to operate in Medicago truncatula in association with the indeterminate nodule patterning.
Our data define L. japonicus and M. truncatula NF‐YA1 genes as important nodule emergence stage‐specific regulators of auxin signalling while indicating that the inductive stage and subsequent formation of early nodule primordia are mediated through an independent mechanism(s).
Keywords: auxin, legume, Lotus, Medicago, NF‐YA1, root nodule, symbiosis
Introduction
Symbiotic nodules develop as lateral organs on legume and some nonleguminous plant roots (Sprent & James, 2007; Doyle, 2011; Werner et al., 2014), usually following induction by nodulation factors (NFs), morphogenic lipo‐chitooligosaccharides synthesized by symbiotic rhizobia (Lerouge et al., 1990). Their perception by a compatible LysM‐type receptor kinase (Limpens et al., 2003; Madsen et al., 2003; Radutoiu et al., 2003, 2007; Arrighi et al., 2006; Broghammer et al., 2012; Liang et al., 2014; Kelly et al., 2017) induces local reprograming of root cells towards symbiotic development (Geurts et al., 2016; Wong et al., 2019). Despite commonalities with lateral root formation in both ontogeny (Herrbach et al., 2014; Xiao et al., 2019) and molecular mechanism (Schiessl et al., 2019; Soyano et al., 2019), nitrogen‐fixing root nodules are anatomically and functionally distinct organs (Oldroyd et al., 2009; Madsen et al., 2010).
Downstream from NF perception, cytokinins are endogenous inducers of root nodule formation (Cooper & Long, 1994; Gonzalez‐Rizzo et al., 2006; Murray et al., 2007; Tirichine et al., 2007; Plet et al., 2011; Ariel et al., 2012; Boivin et al., 2016; Miri et al., 2016; Gamas et al., 2017; Reid et al., 2017; H. Liu et al., 2018), distinguishing nodulation from lateral root evelopment, where auxin priming is required (Du & Scheres, 2018). A local increase in sensitivity to cytokinin in the root cortex was proposed as essential for the initiation of nodule primordia (NP) formation (Held et al., 2014). Given more recent data showing that legumes differ from nonlegumes in this respect, the acquisition of unique responsiveness to cytokinin by the root cortex might have contributed to legume‐specific diversification, perhaps underpinning the evolution of nodulation (Gauthier‐Coles et al., 2019).
Absence of nodulation in several plant lineages in the N2‐fixing clade, including legumes and nonlegumes, coincided with independent losses or pseudogenization of a limited number of symbiotic loci, including NODULE INCEPTION (NIN) (Griesmann et al., 2018; Van Velzen et al., 2018), which responds to cytokinin signalling in the root cortex (Vernie et al., 2015; Gamas et al., 2017; Murray, 2017). NIN, which encodes an RWP‐RK domain‐containing transcription regulator belonging to the NIN‐like protein (NLP) family (Chardin et al., 2014; Nishida et al., 2018), plays a complex role during N2‐fixing symbiosis. Essential in the epidermis for rhizobial entry via a root hair infection thread (IT)‐dependent mechanism (Schauser et al., 1999; Marsh et al., 2007; Kosuta et al., 2011; C. W. Liu, et al., 2019; Soyano et al., 2019), NIN also mediates the initiation of NP formation in the subtending root cortex and pericycle. Responsiveness of NIN to cytokinin is critical in the latter context (Heckmann et al., 2011; Yoro et al., 2014; J. Liu et al., 2019) and its overexpression in both L. japonicus and M. truncatula roots was sufficient for pseudonodule formation, mimicking the effect of NFs and cytokinins (Soyano et al., 2013; Vernie et al., 2015).
Several direct NIN targets, including LOB‐Domain Protein 16/Asymmetric Leaves 2‐Like 18 (LBD16/ASL18), Nuclear Factor‐YA1 (NF‐YA1) and NF‐YB1, have been identified as relevant to NP formation (Soyano et al., 2013, 2019; Schiessl et al., 2019). NIN‐dependent activation of the cytokinin responsive LBD16/ASL18 promoted cell divisions during early NP formation (Schiessl et al., 2019) and L. japonicus LBD16/ASL18 was shown to interact with NF‐YA1 in vitro and also in vivo when ectopically expressed in planta (Soyano et al., 2019).
NF‐Ys are heterotrimeric transcription factors comprising NF‐YA, NF‐YB, and NF‐YC subunits (Mantovani 1999; Laloum et al., 2013) that are essential during rhizobial infection and nodule formation (Combier et al., 2006, 2008; Zanetti et al., 2010; Soyano et al., 2013; Battaglia et al., 2014; Laloum et al., 2014; Laporte et al., 2014; Baudin et al., 2015; Hossain et al., 2016; Zanetti et al., 2017; Ripodas et al., 2019; Bu et al., 2020). Ectopic expression of NF‐YA1 and NF‐YB1, along with LBD16/ASL18, coordinately stimulated cortical cell divisions and partially rescued the defective nodulation phenotype of L. japonicus daphne, which carries a mutant NIN allele, suggesting that their interaction is important during nodule initiation (Soyano et al., 2013, 2019; Yoro et al., 2014). However, we showed that L. japonicus NF‐YA1 was dispensable at this early developmental phase yet essential for nodule differentiation, including cell expansion and vascular bundle formation, presumably through regulation of three SHORT INTERNODES/STYLISH (STY) transcription factor genes, STY1, STY2 and STY3 (Hossain et al., 2016). STYs are known to regulate auxin homeostasis (Sohlberg et al., 2006; Eklund et al., 2010a,b; Baylis et al., 2013; Estornell et al., 2018) and we postulated a similar function during nodule differentiation (Hossain et al., 2016).
We demonstrate here that upon inoculation with Mesorhizobium loti, L. japonicus NF‐YA1 facilitates the expression of seven STYs that regulate at least two YUCCA genes involved in auxin biosynthesis. The STY genes, like NF‐YA1, proved to be essential for nodule emergence but were also crucial for M. loti infection. A partial, functional redundancy was found between NF‐YA1 and NF‐YA4. The nf‐ya1 nf‐ya4 double mutant, unable to regulate the STY and YUCCA gene expression in response to M. loti infection, developed many small primordia that did not differentiate into nodules, a phenotype that persisted in nf‐ya triple and quadruple mutant lines. These results indicate that initiation of cell divisions for NP does not require NF‐YA1 and other partially redundantly acting NF‐YAs. However, NF‐YA1 and STYs are indispensable during the nodule emergence stage. An equivalent M. truncatula NF‐YA1/STY regulatory circuit probably partakes in mediation of the patterning of indeterminate nodules.
Materials and Methods
Plant materials, growth conditions and assessment of phenotypes
Mutants were identified using the L. japonicus LORE1 retrotransposon insertion line collection at Aarhus University in Denmark (https://lotus.au.dk/; Mun et al., 2016). Plants were genotyped using the gene‐ and LORE1‐specific primers, following established procedures (Urbanski et al., 2012).
For the STY3 locus, additional mutant alleles, called sty3‐1 to sty3‐9, were identified using the L. japonicus Targeting Induced Local Lesions In Genomes (TILLING) resource at the John Innes Centre (RevGenUK; https://www.jic.ac.uk/technologies/genomic‐services/revgenuk‐tilling‐reverse‐genetics/; Perry et al., 2009). Two homozygous lines, sty3‐1 and sty3‐9, with predicted premature termination codons were genotyped using derived cleavage amplified polymorphic sequence markers, followed by enzymatic digest with either AgeI (sty3‐1) or PvuII (sty3‐9) (Table S1). Higher‐order mutant lines were developed by performing genetic crosses between selected homozygous single and double mutants. The F3 progeny derived from homozygous higher‐order mutant lines were utilized for subsequent phenotypic analyses.
Germination, growth conditions and assessment of symbiotic phenotypes were as previously described (Wopereis et al., 2000). For nonsymbiotic phenotypic analyses, vermiculite : sand (6 : 1) growth medium was supplemented with Broughton & Dilworth solution containing 1 mM KNO3.
For gene expression analysis (quantitative reverse transcription polymerase chain reaction (qRT‐PCR)), L. japonicus seedlings were grown in sterile conditions for 7 d and inoculated with M. loti. Control uninoculated roots were harvested at this time, while inoculated roots were harvested 4, 7, 12 or 21 d later. At least three independent biological replicates per treatment were collected. For gene expression analysis in the primary transformants, T0 plants, STY3::SRDX5 and STY3::SRDX6 (see below) were propagated by cuttings, as follows: shoot tip cuttings of c. 4 cm in length from branches of T0 plants, along with control wild‐type cuttings, were set into a vermiculite : sand (6 : 1) mixture soaked in B&D solution containing 0.5 mM KNO3, where they were grown for 14 d under sterile conditions and then inoculated with M. loti.
STY3::SRDX dominant negative constructs
The NF‐YA1Pro:STY3::SRDX:NF‐YA1‐3′UTR binary vector (Hossain et al., 2016) was transferred in parallel with an empty pKGWD,0 vector, used as a negative control, to Agrobacterium tumefaciens strain LBA4404. Standard transformation protocols were employed to generate fully transgenic L. japonicus plants (Lombari et al., 2005) The primary transgenic T0 plants were allowed to self‐fertilize and the resulting T1 populations were genotyped for the NF‐YA1Pro:STY3::SRDX transgene (Table S1) and evaluated for nodulation phenotypes.
Gene expression analysis using qRT‐PCR
Total RNA was extracted from roots using the Plant/Fungi Total RNA Purification Kit (Norgen Biotek Corp., Thorold, ON, Canada), treated with TURBO DNase I (Invitrogen) and assessed for purity and quality. cDNA was prepared from 500 ng of RNA using the SuperScript IV VILO Master Mix (Invitrogen). qRT‐PCR was performed using three to five biological and three technical replicates, on a CFX384 Real‐Time PCR Detection System (Bio‐Rad, Mississauga, ON, Canada) using the SensiFAST SYBR No‐ROX kit (Bioline, Memphis, TN, USA) under the following conditions: 95°C for 3 min followed by 40 cycles of 95°C for 5 s, 60°C for 15 s and 72°C for 15s. Expression levels were normalized against three reference genes (UBQ, PP2A and ATP‐s) as previously described (Held et al., 2014). Primer sequences used for the qRT‐PCR expression analyses are listed in Table S1.
Gene promoter activity localization using GUS histochemical assay
To develop the localization constructs, fragments encompassing the STY1 to STY9 promoter regions, were prepared by gene synthesis (Bio Basic Inc., Markham, ON, Canada). The promoter sequences were synthesized as such, for STY1 (position −3422 to −1), STY2 (−4000 to −1), STY3 (−4000 to −1), STY4 (−4000 to −1), STY5 (−3000 to −1), STY6 (−2177 to −1), STY7 (−4000 to −1), STY8 (−4000 to −1), and STY9 (−2487 to −1), where −1 denotes the first base upstream from predicted ATG initiation codons. These were recombined in the pKGWFS7 destination vector containing an in‐frame fusion between the regions coding for green fluorescent protein (GFP)/β‐glucuronidase (GUS) (Karimi et al., 2002; https://gateway.psb.ugent.be/), using the Gateway technology (Invitrogen).
The TAC/BAC clones, LjT09B24 (TM2451) and LjB04P03 (BM2658) (http://www.kazusa.or.jp/lotus/), were used to amplify the promoter regions of YUCCA1 (position −2466 to −1) and YUCCA11 (−2249 to −1), respectively. The resulting promoter fragments were cloned into the pENTR/D‐TOPO vector (Invitrogen) and subsequently recombined in the pKGWFS7 destination vector.
After validation by sequencing, constructs were transformed into Agrobacterium rhizogenes strains AR1193 and ARquA1 for Lotus and Medicago hairy root transformation, respectively. At least 10 independent hairy root systems were analysed for GUS reporter activity, following an established protocol (Held et al., 2014). Histochemical assays of M. truncatula hairy roots were performed as previously described (Cerri et al., 2012). Magenta‐Gal (5‐bromo‐6‐chloro‐3‐indolyl‐β‐d‐galactopyranoside; Biosynth B7200) and X‐Gluc (5‐bromo‐4‐chloro‐3‐indolyl‐β‐d‐glucuronic acid, cyclohexylammonium salt; Thermo Scientific, Waltham, MA, USA) substrates were used for histochemical staining of Sinorhizobium meliloti (β‐galactosidase; purple color) and GUS (blue color) activity, respectively, within ITs and nodules.
YUCCA1 and YUCCA11 overexpression experiments
The YUCCA1 and YUCCA11 overexpression constructs were prepared by synthesizing a 769 bp long DNA fragment encompassing the sequence of the 2 × Cauliflower mosaic virus 35S promoter with attL1 and attR5 recombination sites at its 5′ and 3′ ends, respectively (Bio Basic Inc.). The YUCCA1 cDNA and YUCCA11 genomic sequences were likewise synthesized. The former encompassed the entire 1251bplong coding region, predicted 5′‐ (201bp) and 3′‐ (585bp) UTRs, and attL5 and attL2 at 5′ and 3′ ends, respectively. The YUCCA11 genomic region included all exons and introns (1896 bp long, from ATG to the predicted stop codon), 500 bp 3′UTR and attL5 and attL2 recombination sites.
These DNA fragments were recombined into the pKGWD,0 gateway destination vector to generate 2X35SPro:YUCCA1 and 2X35SPro:YUCCA11. They were transformed into A. rhizogenes strain AR1193, which was used to generate hairy roots on transgenic, L. japonicus shoots carrying the DR5:GUS reporter.
Development of DR5:GUS reporter line in nf‐ya1‐2 nf‐ya4 background
The transgenic L. japonicus line carrying the DR5:GUS reporter was crossed with homozygous nf‐ya1‐2 nf‐ya4. The F1 plants were allowed to self‐fertilize to produce segregating F2 populations, and homozygous nf‐ya1‐2 nf‐ya4 plants carrying DR5:GUS were selected for propagation.
Next‐generation RNA sequencing
Mesorhizobium loti‐inoculated root samples were harvested 4 d after inoculation (dai), along with corresponding uninoculated roots of the same age. Total RNA was extracted using the Plant/Fungi Total RNA Purification Kit (Norgen Biotek Corp., Thorold, ON, Canada). RNA quality was verified using an Agilent Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA, USA). The RNA library was constructed and sequenced at the Center for Applied Genomics at Sick Kids Hospital (Toronto, Canada) using Illumina Hi‐Seq 2500 paired‐end reads. Mapping of reads to the L. japonicus MG20 v.3.0 genome was performed using clc genomics workbench with the following parameters: mapping type: map to gene regions only; mismatch cost: 2; insertion cost: 3; deletion cost: 3; length fraction: 0.8; similarity fraction: 0.8; global alignment: no; auto‐detect paired distances: yes; strand‐specific: both; maximum number of hits for a read: 10; count paired reads as two: no; expression value: reads per kilobase of transcript, per million mapped reads (RPKM) and transcripts per kilobase million (TPM); calculate RPKM and TPM for genes without transcripts: no; use EM estimation: yes; create fusion gene table: no. After the expression analysis, Gaussian statistical analysis was performed on all the tables using the t‐test function, and the P‐values were false discovery rate (FDR)‐corrected.
Phylogenetic analyses and microscopy
Protein sequences were aligned with clustalw and the corresponding phylogenetic trees were generated using Mega 7 software and the neighbour‐joining method with bootstrap replicates of 1000. All microscopic observations were performed using either a Nikon Eclipse Ni upright or Nikon SMZ25 stereo microscope integrated with a DsRi2 digital camera and epifluorescence capability (Nikon, Tokyo, Japan). All images were captured in a TIFF format and subsequently processed using Adobe photoshop cs6.
Results
Activities of L. japonicus and M. truncatula SHI/STY genes associate with root and nodule development
Using L. japonicus STY1, STY2 and STY3 and A. thaliana SHI/STY protein sequences as queries, nonredundant sequence collections at the NCBI server (https://www.ncbi.nlm.nih.gov/) and the L. japonicus (www.kazusa.or.jp/lotus/; https://lotus.au.dk/) and M. truncatula (https://medicago.toulouse.inra.fr/MtrunA17r5.0‐ANR/) databases were analysed. Predicted L. japonicus and M. truncatula SHI/STY gene families of nine and eight members, respectively, were identified (Fig. 1). All encoded proteins contained the evolutionarily conserved RING‐type zinc finger (IPR001841) and IGGH domains, the latter probably specific to the SHI/STY protein family (Fridborg et al., 2001; Eklund et al., 2010b; Gomariz‐Fernandez et al., 2017) (Figs S1–S3).
Fig. 1.
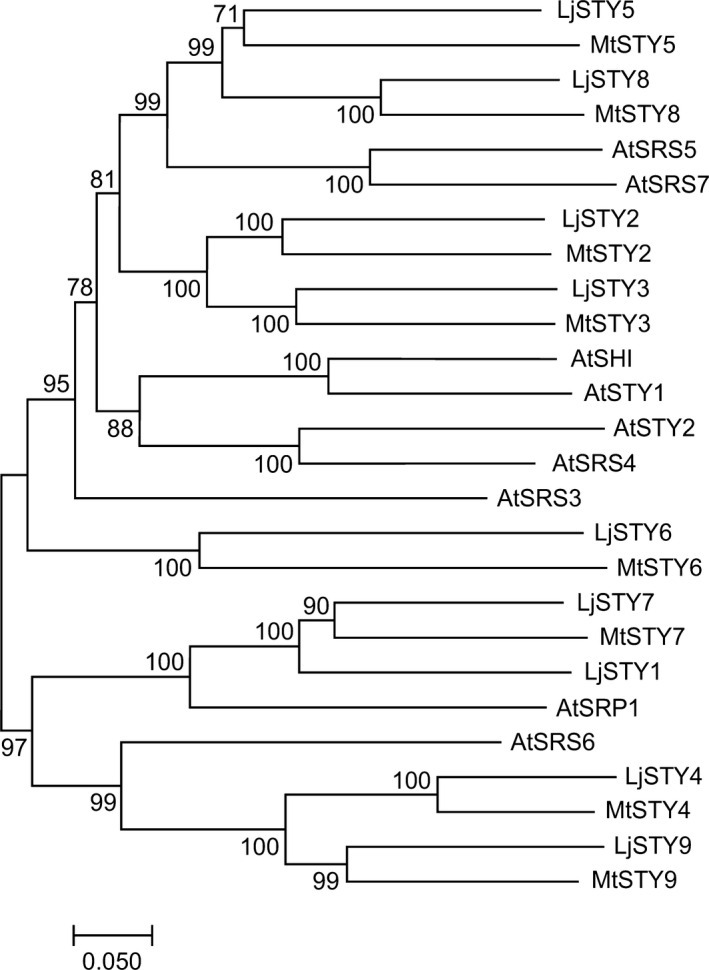
Phylogenetic relationship between predicted STY proteins from Lotus japonicus, Medicago truncatula and Arabidopsis thaliana. The unrooted tree was constructed using full‐length protein sequences. For the sake of coherence the same number was given to Medicago and Lotus STY genes when a clear homologue was present in both species. The following accession numbers refer to the protein sequences used: L. japonicus – LjSTY1 (Lj6g3v0959410), LjSTY2 (Lj0g3v0059359), LjSTY3 (Lj2g3v1728900), LjSTY4 (Lj3g3v0766120), LjSTY5 (Lj1g3v2140900), LjSTY6 (Lj3g3v3376040), LjSTY7 (Lj2g3v3044220), LjSTY8 (Lj5g3v0155490)¸ and LjSTY9 (Lj0g3v0258549); M. truncatula – MtSTY2 (MtrunA17Chr8g0372461), MtSTY3 (MtrunA17Chr5g0404781), MtSTY4 (MtrunA17Chr3g0082511), MtSTY5 (MtrunA17Chr3g0142171), MtSTY6 (MtrunA17Chr4g0035591), MTSTY7 (MtrunA17Chr5g0441921), MtSTY8 (MtrunA17Chr1g0155791) MtSTY9 (MtrunA17Chr8g0353111); and A. thaliana SHI (At5g66350), STY1 (NM_114966), STY2 (At4g36260), LRP1 (At5g12330), SRS3 (At2g21400), SRS4 (At2g18120), SRS5 (At1g75520), SRS6 (At3g54430), SRS7 (At1g19790), SRS8 (At5g33210).
Follow‐up experiments were directed toward functional characterization of the L. japonicus genes but limited comparative studies with M. truncatula STYs were also performed. Lotus japonicus and M. truncatula form determinate and indeterminate (i.e. meristem containing) nodules, respectively (Szczyglowski et al., 1998; Sprent & James, 2007; Xiao et al., 2014), which both require NF‐YA1 (Combier et al., 2006, 2008; Soyano et al., 2013), and we showed previously that MtNF‐YA1 can functionally replace LjNF‐YA1 during nodule formation (Hossain et al., 2016). We have postulated, therefore, that an equivalent NF‐YA1/STY regulatory module must operate and be relevant during the indeterminate nodule development.
Promoter‐GUS reporter constructs were developed for each LjSTY gene and their activities were tested in A. rhizogenes‐induced hairy roots (Fig. 2). Similar to Arabidopsis (Kuusk et al., 2002, 2006; Eklund et al., 2011), the GUS reporter activity was associated with early lateral root primordia, including the initial pericycle cell divisions and the central vasculature and root apices of emerged lateral roots. These activity patterns were shared by all LjSTY promoters, except for LjSTY7, for which GUS staining was only weakly detectable at the base of emerging lateral roots. Remarkably, all STY promoters were active in dividing cortical cells of young NP and later, at the base and in the vasculature of mature nodules (Fig. 2), resembling expression domains of NF‐YA1 (Hossain et al., 2016).
Fig. 2.
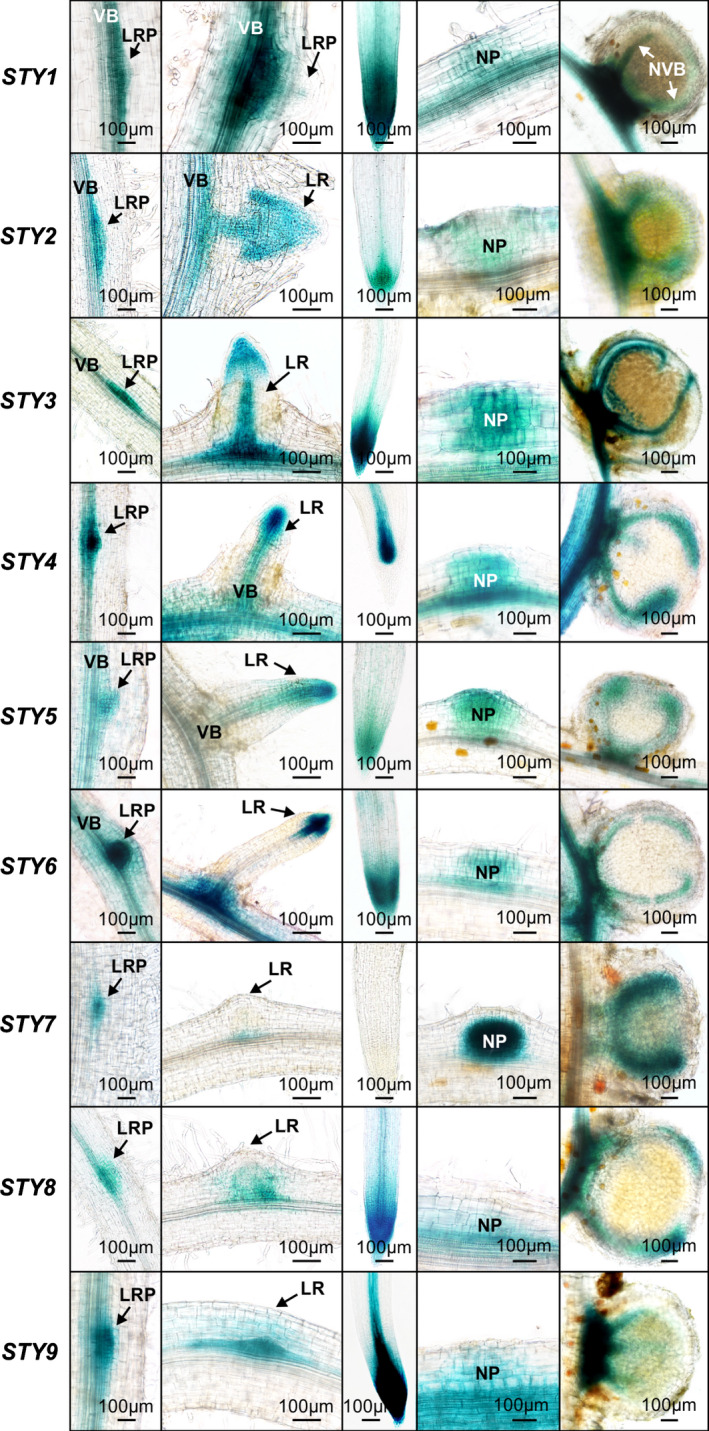
The activity of Lotus japonicus STY promoters during root and nodule development. Representative images of hairy root segments are shown. Columns from left to right depict the β‐glucuronidase (GUS) activity (in blue) as associated with small lateral root primordia (LRP), emerging or already emerged lateral roots (LR), root apical regions, nodule primordia (NP) and fully mature nodules. All specimens were collected 10–14 d after inoculation with Mesorhizobium loti. For each promoter:GUS construct, 10–15 individual hairy roots were analysed. VB, root vascular bundle; NVB, nodule vascular bundle.
We also explored the expression pattern of M. truncatula STY2 (MtSTY2) using a 3089 bp promoter fragment fused to the GUS reporter gene. MtSTY2 was chosen because its mRNA had one of the highest steady‐state levels in nodules (Roux et al., 2014). Similar to L. japonicus STYs, the MtSTY2 promoter activity was localized primarily in the apical region of growing lateral root primordia and main roots (Fig. 3a,b). Upon S. meliloti infection, GUS activity was detected concurrent with early cell divisions associated with penetrating ITs in the inner root cortex (Fig. 3c,d). In young primordia and emerging nodules, GUS staining was apparent in most dividing cells (Fig. 3e), which was reminiscent of L. japonicus nodules (e.g. LjSTY7; Fig. 2). However, with further development, the reporter gene expression rapidly became restricted to uninfected cells of the nodule parenchyma and vascular cambium (Fig. 3f) and, later, to the nodule meristematic zone (Fig. 3g), often developing a patchy pattern as nodules grew older (Fig. 3h).
Fig. 3.
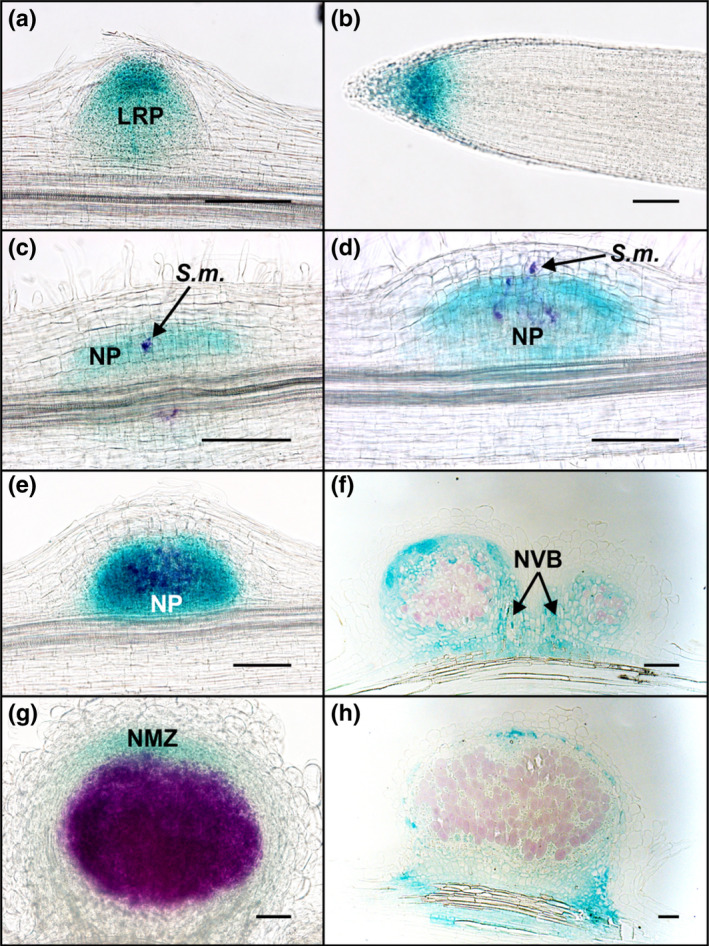
Expression analysis of MtSTY2 during the symbiotic interaction between Medicago truncatula and Sinorhizobium meliloti. Roots expressing a pMtSTY2:GUS fusion were analysed at different stages after inoculation with S. meliloti. Double staining using Magenta‐Gal and X‐Gluc was performed to allow the visualization of the infecting S. meliloti in purple and pMtSTY2 expression in blue. (a) pMtSTY2:GUS expression in a lateral root primordium (LRP) just before its emergence. (b) Expression in the meristematic zone of a main root. (c) pMtSTY2:GUS expression in dividing cells of the inner and central cortex of a 2‐d‐old nodule primordium. (d) pMtSTY2:GUS expression in dividing cells of a 3‐d‐old nodule primordium (NP). (e) pMtSTY2:GUS expression in the central zone of a 5‐d‐old nodule. (f) Thin section through a 5‐d‐old nodule. (g) pMtSTY2:GUS expression in a 10‐d‐old nodule. (h) Thin section through a 12‐d‐old nodule. Bars, 100 μm. S.m., Sinorhizobium meliloti; NVB, nodule vascular bundle; NMZ, nodule meristematic zone.
The level of STY mRNAs is regulated during nodule development
STY gene expression was further characterized through in silico analysis of our next‐generation RNA sequencing (RNA‐seq) data (BioProject ID PRJNA630938; http://www.ncbi.nlm.nih.gov/bioproject/630938). At 4 dai with M. loti, the steady‐state levels of LjSTY1, 2, 3 and 7 mRNAs were significantly upregulated (Table S2). As the histochemical data indicated activity of all LjSTY promoters during nodule formation, the steady‐state level of the nine LjSTY mRNAs was further evaluated across four developmental stages (Fig. 4). Equivalent RNA samples derived from roots of nf‐ya1‐2 (Hossain et al., 2016) were analysed in parallel in order to determine whether STY genes are regulated by NF‐YA1 upon M. loti infection.
Fig. 4.
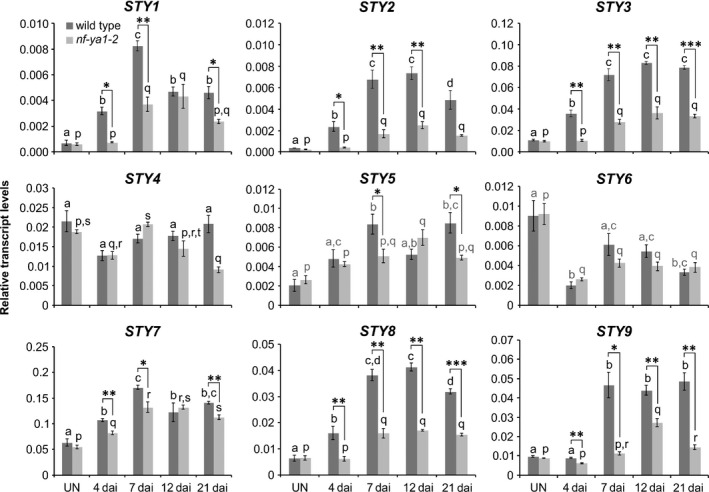
Expression of Lotus japonicus STY genes is regulated upon Mesorhizobium loti infection. Quantitative reverse transcription polymerase chain reaction data showing steady‐state levels of Lotus japonicus STY mRNAs in control, uninoculated (UN) roots of L. japonicus wild‐type (dark grey) and the nf‐ya1‐2 mutant (light grey) and in corresponding root samples collected at various time points (d) after inoculation (dai) with M. loti. The means ± SE are given for three biological replicates. Statistical groupings across different time points, reflected by the same letters, have been determined separately for each genotype using one‐way ANOVA with the Tukey’s honestly significant difference post hoc test (P < 0.05). Student’s t‐test was used to carry out pairwise comparisons between the wild‐type and nf‐ya1‐2 at each time point (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
Consistent with the RNA sequencing results, at 4 dai, STY1, 2, 3, 7 and additionally STY8 mRNAs were significantly upregulated in wild‐type roots, above control levels. This upregulation was lost in nf‐ya1‐2, except for STY7, which showed only a partial dependency on NF‐YA1 (Fig. 4).
At 7 dai, levels of these five STY mRNAs were further enhanced and two additional STY mRNAs, STY5 and STY9, were also significantly upregulated (Fig. 4). Interestingly, the initially upregulated STY mRNAs (i.e. STY1, 2, 3, 7 and 8) displayed only a partial dependency on NF‐YA1 at this particular stage after M. loti infection, indicated by the somewhat attenuated response in nf‐ya1‐2 (Fig. 4). These observations predicted that in addition to NF‐YA1, other regulators are likely to partake in mediating STY gene expression, which was later confirmed (see below).
In comparison to uninoculated roots, levels of all seven STY mRNAs described were also significantly upregulated at subsequent 12 and 21 dai time points. Conversely, STY4 did not respond significantly and STY6 mRNA showed decreased levels upon M. loti infection (Fig. 4).
These observations indicated that at least seven L. japonicus STY genes were regulated during nodule development, although all nine genes could be pertinent, based on histochemical data. Similarly, in silico analysis of M. truncatula laser caption microscopy/RNA sequencing (LCM‐RNA‐seq) expression data (Roux et al., 2014) showed that seven out of eight MtSTY genes had significantly higher mRNA levels in mature nodules than in uninoculated roots, primarily in the meristematic zone, with MtSTY2 and MtSTY9 displaying the highest fold upregulation (Table S3a). Importantly, upregulated levels of at least six MtSTY mRNAs (MtSTY2, 3, 4, 7, 8 and 9), expressed early on upon S. meliloti infection (Larrainzar et al., 2015), were dependent on MtNF‐YA1 (Table S3b), indicating that the relevant NF‐YA1/STY regulatory modules operate in both determinate and indeterminate nodules.
Lotus japonicus sty mutants
To assess the functional relevance of STY genes during root nodule development, mutant L. japonicus lines carrying LORE1 retrotransposon insertions in exonic regions were identified for all but the STY3 gene (Table S4) (https://lotus.au.dk/) (Malolepszy et al., 2016; Mun et al., 2016). A TILLING approach (Perry et al., 2009) was therefore used to identify deleterious mutations at the STY3 locus. Two identified alleles, sty3‐1 and sty3‐9, carried independent single nucleotide substitutions (C187 to T and C493 to T, respectively) that were predicted to result in premature translation termination (STOP) codons (Fig. S4). The line homozygous for the sty3‐9 allele was used in subsequent analyses.
Efforts to characterize the impact of individual sty mutations on nodulation were focused on STY1, 2, 3, 7 and 8, which had upregulated expression 4 d after M. loti infection (Fig. 4).
sty mutants show weak symbiotic phenotypes
At 7 dai, there were no significant differences in epidermal IT (eIT) formation between wild‐type and the sty mutants (Fig. S5a). However, while forming a near wild‐type number of nodules, several sty lines had significantly fewer primordia, suggesting gene relevance, particularly where two alleles show this characteristic (e.g. sty1 and sty7 loci) (Fig. S5b). Shoot and root weight, used as measures of plant performance, were diminished in most sty mutants (Fig. S6a,b), but a simple correlation between plant growth under nonsymbiotic conditions and the number of NP could not be fully supported.
To assess the redundancy of STY gene function, a triple sty mutant was developed. When evaluated at 21 dai, sty1‐2 sty2‐1 sty3‐9 had significantly fewer NP, nodules (Fig. 5a) and eITs, while microcolonies (MCs) were unaffected (Fig. 5b). Under nonsymbiotic conditions, 28 d after sowing (das), other developmental failings, including diminished shoot and root growth were apparent (Fig. 5c,d). Given these effects, which confounded the interpretation of the nodulation results, a dominant negative approach, targeting STY genes in nodules, was implemented in fully transgenic L. japonicus plants, extending our earlier observations in the hairy root system (Hossain et al., 2016).
Fig. 5.
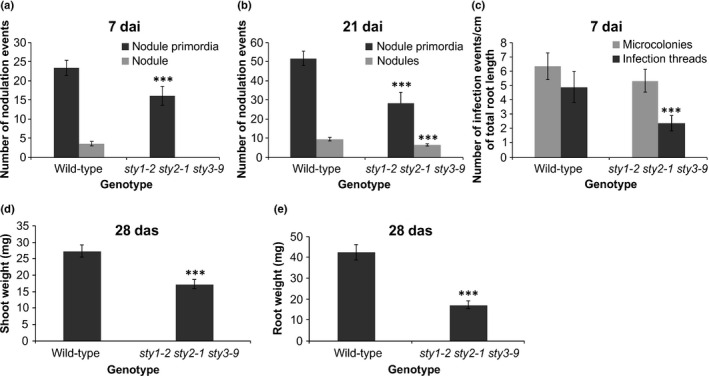
Lotus japonicus sty1‐2 sty2‐1 sty3‐9 triple mutant has significant symbiotic and nonsymbiotic defects. (a, b) Nodulation events (nodule primordia and nodules) were scored in wild‐type and the triple mutant at 7 (a) and 21 d after inoculation (dai) (b) with Mesorhizobium loti. (c) The frequency of infection events (microcolonies and epidermal infection threads) is given. (d, e) Nonsymbiotic phenotypes, including shoot weight (d) and root weight (e) of plants grown under sterile conditions, in the absence of M. loti, were evaluated 28 d after sowing (das). Twenty and 30 individuals per genotype were analysed for symbiotic and nonsymbiotic phenotypes, respectively. The scores represent means ± 95% confidence intervals for three biological replicates. Asterisks represent a significant difference from wild‐type for a given category (Student’s t‐test: ***, P < 0.001).
STY3::SRDX blocks infection thread and nodule formation
As STY3 expression was relatively high in comparison to other STY genes and entirely dependent on NF‐YA1 at 4 dai with M. loti (Fig. 4), the LjNF‐YA1Pro:STY3::SRDX chimeric construct (Hossain et al., 2016) was used in the dominant negative approach. As documented for several transcription factors (Hiratsu et al., 2003), the 12‐amino‐acid‐long Ethylene‐Responsive Element‐Binding Factor (ERF)‐associated amphiphilic repression (SRDX) domain was expected to convert STY3 into a dominant transcriptional repressor.
In contrast to profusely nodulated control plants that lacked the construct, 10 independent T0 plants carrying LjNF‐YA1Pro:STY3::SRDX (STY3::SRDX +) formed no visible nodules (Fig. 6a). The remaining, STY3::SRDX‐negative (STY3::SRDX ‐) plants formed wild‐type nodules (Table S5). Detailed evaluation of STY3::SRDX + T1 progeny, at 7 and 21 dai, showed that while unable to develop mature nodules, NP were initiated (Fig. 6b,c), albeit significantly less frequently than in control, STY3::SRDX ‐ roots (Fig. 6b–f). Despite the STY3::SRDX + plants having many MCs, more than the wild‐type in the case of STY3::SRDX5, very few eITs formed (Fig. 6g) and most terminated prematurely within root hairs (Fig. 6h–j). Together, these data indicated an essential role for STYs during both eIT formation and nodule organogenesis.
Fig. 6.
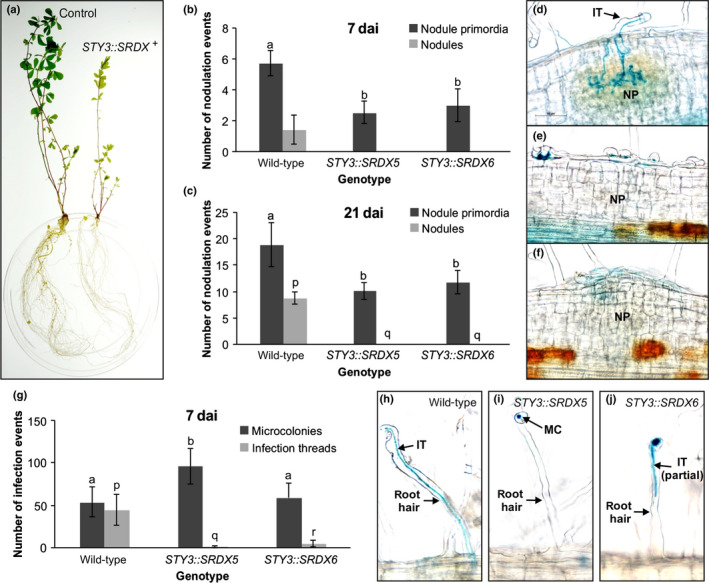
Transgenic Lotus japonicus plants carrying the LjNF‐YA1Pro:STY3::SRDX construct do not form nodules. (a) Images, taken 28 d after inoculation (dai) with Mesorhizobium loti, of representative, fully transgenic T0 L. japonicus plants carrying either empty vector (control), or the same vector containing the LjNF‐YA1Pro:STY3::SRDX transgene (STY3::SRDX +). (b, c) Scores of nodulation events at 7 (b) and 21 dai (c) with M. loti, from two independent L. japonicus T1 populations (STY3::SRDX5 and STY3::SRDX6) segregating the LjNF‐YA1Pro:STY3::SRDX transgene. Note that ‘wild‐type’ denotes T1 segregants without the transgene. (d–f) Representative images of wild‐type nodule primordia (NP) (d) and those formed by transgenic plants carrying the LjNF‐YA1Pro:STY3::SRDX transgene (e, f). The image in (e) shows a typical, small NP event while panel (f) represents an infrequent, larger NP that forms a visible bump at the root surface. (g) Scores of infection events (i.e. microcolonies and epidermal infection threads) in L. japonicus T1 plants that either lack (wild‐type) or carry the LjNF‐YA1Pro:STY3::SRDX transgene. The scores represent means ± 95% confidence intervals. Statistical groupings, reflected by the same letters, have been determined separately for each of the two infection event categories using one‐way ANOVA with Tukey’s honestly significant difference post hoc test. (h–j) Representative images of epidermal infection events in the wild‐type (h) and the transgene‐containing (STY3::SRDX5) T1 plants (i, j) are shown. MC, microcolony; IT, epidermal infection thread; IT (partial), IT that was terminated within the root hair shaft.
STYs are required to regulate YUCCA1 and YUCCA11 expression during symbiosis
Downstream targets of L. japonicus NF‐YA1/STY‐dependent regulation were identified via a targeted approach. Focusing on YUCCA genes was deemed relevant given the importance of auxin signalling for both symbiotic infection and nodule formation (Suzaki et al., 2012, 2013; Breakspear et al., 2014; Kohlen et al., 2018) and the postulated role of NF‐YA1 in regulating the relevant auxin maxima during symbiosis (Hossain et al., 2016). YUCCA genes encode flavin monooxygenase‐like enzymes and some, including Arabidopsis YUCCA4 and YUCCA8, were shown to mediate the rate‐limiting step in tryptophan‐dependent auxin biosynthesis and to be regulated by STY1 (Eklund et al., 2010a).
Using Arabidopsis YUCCA protein sequences as blast queries (Cheng et al., 2006; Zhao, 2018), 21 YUCCA‐like proteins were predicted to be encoded by the L. japonicus genome, including those represented by only partial sequences (Fig. S7).
An in silico analysis of RNA‐seq data (BioProject ID PRJNA630938) showed that at 4 dai with M. loti the steady‐state level of only Lj4g3v3081700 (YUCCA11) mRNA was significantly (FDR < 0.05) upregulated in L. japonicus roots (Table S6). A phylogenetic tree constructed with the corresponding protein sequences revealed that L. japonicus YUCCA11 (LjYUCCA11) had the highest primary sequence homology to M. truncatula MtrunA17Chr6g0485621/MtYUC2 (Fig. S8), shown previously to respond to S. meliloti infection (Schiessl et al., 2019). Interestingly, three other Medicago YUCCA genes, namely MtrunA17Chr7g0262591 (Medtr7g099330/MtYUC8), MtrunA17Chr7g0262471 (Medtr7g099160) and MtrunA17Chr3g0139441 (Medtr3g109520/MtYUC1), were upregulated in response to rhizobial inoculation or NF application (Larrainzar et al., 2015; Schiessl et al., 2019). The predicted protein products of the first two M. truncatula genes had the highest homology to Lj1g3v4528740.1 (LjYUCCA1) while the third had the highest homology to Lj1g3v2036560.1 (Fig. S8). Considering both RNA‐seq data and phylogenic relationships, LjYUCCA1 and LjYUCCA11 genes were chosen for subsequent analyses.
YUCCA1 mRNA, while detectable in uninoculated L. japonicus roots, was significantly increased upon M. loti infection (Fig. 7a). By contrast, YUCCA11 mRNA could be detected only in M. loti‐inoculated roots (Fig. 7b). Importantly, the responsiveness of the two genes was significantly attenuated in STY3::SRDX5 and STY3::SRDX6 roots (Fig. 7a,b).
Fig. 7.
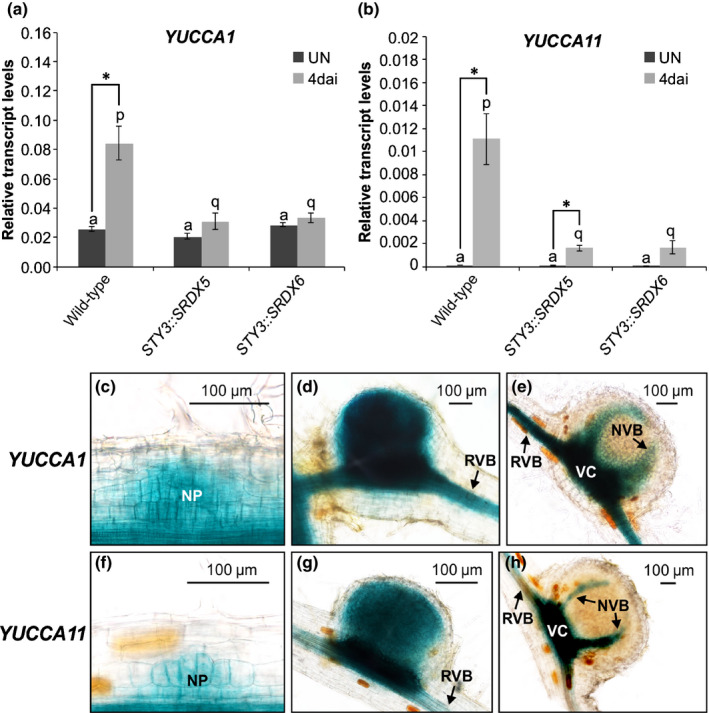
Lotus japonicus YUCCA1 and YUCCA11 expression associates with nodule development and is dependent upon STYs. (a, b) Steady‐state levels of YUCCA1 (a) and YUCCA11 (b) mRNAs were determined by quantitative reverse transcription polymerase chain reaction in uninoculated (UN) L. japonicus roots of the wild‐type genotype and the corresponding roots from T0 transgenic plants, propagated from cuttings of two independent T0 lines (STY3::SRDX5 and STY3::SRDX6) carrying the STY3‐SRDX transgene. These were compared with equivalent root samples collected 4 d after inoculation (dai) with Mesorhizobium loti. The mean ± SE is given for three biological replicates. Statistical groupings across different genotypes, reflected by the same letters, have been determined separately for M. loti inoculated and uninoculated samples using one‐way ANOVA with Tukey’s honestly significant difference post hoc test. Asterisks (*) denote significant differences between uninoculated and inoculated samples (Student’s t‐test: *, P < 0.05). (c–h) The β‐glucuronidase (GUS) reporter activity (in blue) as driven by YUCCA1 (c–e) and YUCCA11 (f–h) promoters. Representative images of nodule primordia (c, f) and small (d, g) and fully mature nodules (e, h) are shown. The images were captured at 10 dai with M. loti. NP, nodule primordium; NVB, nodule vascular bundle; RVB, root vascular bundle; VC, vascular cambium.
The YUCCA1 promoter showed activity along the entire root vasculature and also in dividing cortical cells of NP (Fig. 7c,e). In developing nodules, this activity appeared to be present in all centrally located cells, only to be confined later to the nodule vasculature in mature nodules (Fig. 7d,e). The activity of the YUCCA11 promoter, though similar, was confined to regions of the root associated with nodules, including vasculature at the place of nodule emergence (Fig. 7f,h).
NF‐YA1 regulates expression of YUCCA1 and YUCCA11
Because responsiveness of YUCCA1 and YUCCA11 upon M. loti infection required STY‐dependent functions and STYs were regulated by NF‐YA1, the two YUCCA genes should also be subjected to the NF‐YA1‐dependent regulation.
For YUCCA1 this was most apparent at 4 dai, where the mRNA level was significantly upregulated above control, uninoculated roots and this was entirely NF‐YA1‐dependent (Fig. 8a). By contrast, levels of YUCCA11 mRNA remained high in M. loti‐inoculated roots at all time points analysed. Interestingly, reaching peak levels at 4 and 12 dai required NF‐YA1, while maintaining moderately upregulated levels at 7 and 21 dai time points was NF‐YA1‐independent (Fig. 8b).
Fig. 8.
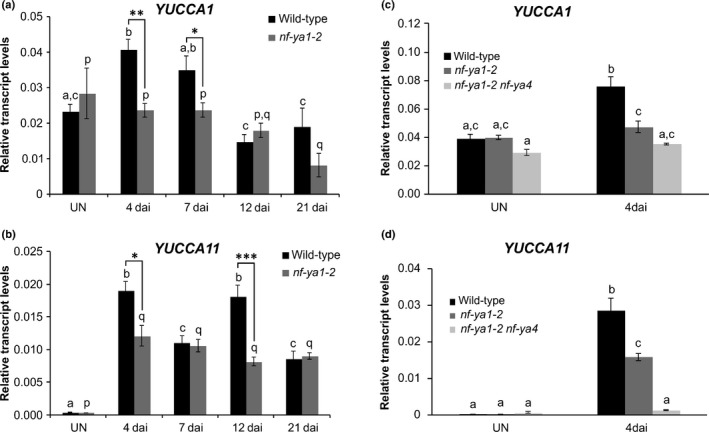
Lotus japonicus NF‐YA1 and NF‐YA4 work partially redundantly to regulate YUCCA11 gene expression. (a–d) Quantitative reverse transcription polymerase chain reaction data showing steady‐state levels of YUCCA1 (a, c) and YUCCA11 (b, d) mRNAs in roots of L. japonicus wild‐type (black), nf‐ya1‐2 (dark grey) and the nf‐ya1‐2 nf‐ya4 double mutant (light grey). Root samples from the uninoculated (UN) control and those collected at various time points (d) after inoculation (dai) with Mesorhizobium loti were analysed. In all graphs, the mean ± SE is given for three biological replicates. Small letters denote significant differences in transcript abundance between time points, as determined separately for each genotype by one‐way ANOVA with Tukey’s honestly significant difference post hoc test. Student’s t‐test was used to carry out pairwise comparisons between wild‐type and nf‐ya1‐2 in (a) and (b). Asterisks denote significant differences in pairwise comparisons (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
NF‐YA1 and NF‐YA4 function partially redundantly to regulate YUCCA11 and STYs
It was surmised that another, partially redundantly operating NF‐YA could be responsible for the observed incomplete NF‐YA1 dependency of the YUCCA11 gene expression. As L. japonicus NF‐YA4 is predicted to be the closest paralogue of NF‐YA1 (Hossain et al., 2016), we tested whether it is pertinent to YUCCA11 and perhaps also to YUCCA1 expression.
The nf‐ya4 mutant allele was derived from a L. japonicus line carrying the LORE1 insertion in the first exon of the NF‐YA4 locus (Table S7). At 4 dai with M. loti, the steady‐state level of YUCCA1 mRNA was significantly upregulated in wild‐type but not in nf‐ya1‐2 mutant roots, and absence of functional NF‐YA4 in nf‐ya1‐2 nf‐ya4 had no additional significant impact (Fig. 8c). By contrast, strong upregulation of YUCCA11 mRNA, in the M. loti‐inoculated wild‐type, was significantly attenuated in nf‐ya1‐2 and further diminished to the background, uninoculated level, in nf‐ya1‐2 nf‐ya4 roots, (Fig. 8d).
We then asked whether this apparent functional redundancy between NF‐YA1 and NF‐YA4 could also explain the NF‐YA1‐independent regulation of STY gene expression, as observed at 7 dai with M. loti (see Fig. 4) and we found this to be the case (Fig. 9).
Fig. 9.
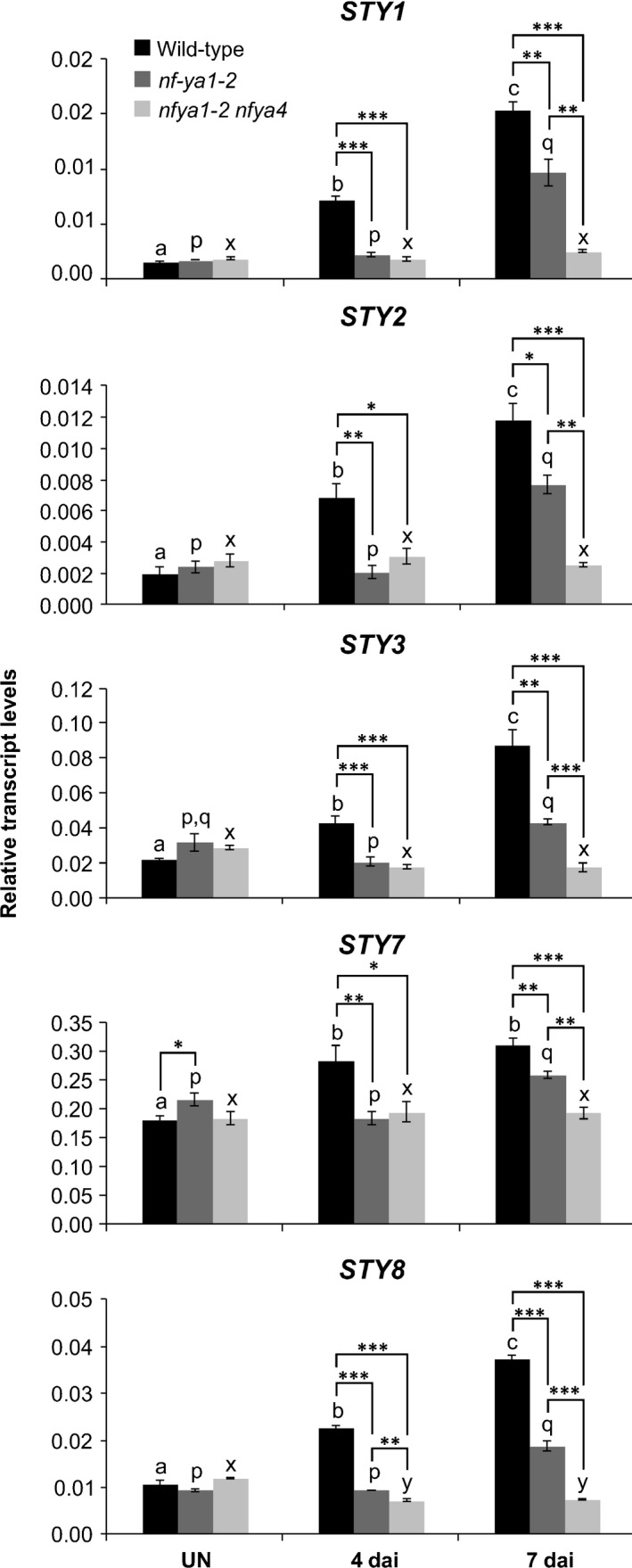
Lotus japonicus NF‐YA1 and NF‐YA4 work partially redundantly to regulate expression of STY genes. The steady‐state levels of L. japonicus STY mRNAs in control, uninoculated (UN) roots of L. japonicus wild‐type (black), nf‐ya1‐2 (dark grey) and nf‐ya1‐2 nf‐ya4 mutants (light grey) and in corresponding root samples collected at 4 and 7 d after inoculation (dai) with Mesorhizobium loti. The mean ± SE is given for four to five biological replicates. Statistical groupings across different time points, reflected by the same letters, have been determined separately for each genotype using one‐way ANOVA with Tukey’s honestly significant difference post hoc test (P < 0.05). Dunnett’s test was used to carry out pairwise comparisons between wild‐type, nf‐ya1‐2 and nf‐ya1‐2 nf‐ya4 mutants at each time point (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
NF‐YA4 acts partially redundantly with NF‐YA1 to regulate symbiosis
Consistent with its impact on STY and YUCCA gene expression, nf‐ya4 potentiated the symbiotic defect of nf‐ya1‐2. At 21 dai with M. loti, nf‐ya1‐2 roots developed a few nodules (Fig. 10a), while the nf‐ya1‐2 nf‐ya4 double mutant formed only small NP, developmentally arrested at a stage comparable to those formed by LjNF‐YA1Pro:STY3::SRDX transgenic plants (Fig. 6e,f). However, unlike the transgenic plants, the number of NP in nf‐ya1‐2 nf‐ya4 was elevated compared with wild‐type and nf‐ya1‐2 single mutant plants (Fig. 10a). At 7 dai with M. loti, nf‐ya1‐2 nf‐ya4 also showed a weak but significant infection defect, forming fewer eITs and more MCs than the wild‐type (Fig. 10b).
Fig. 10.
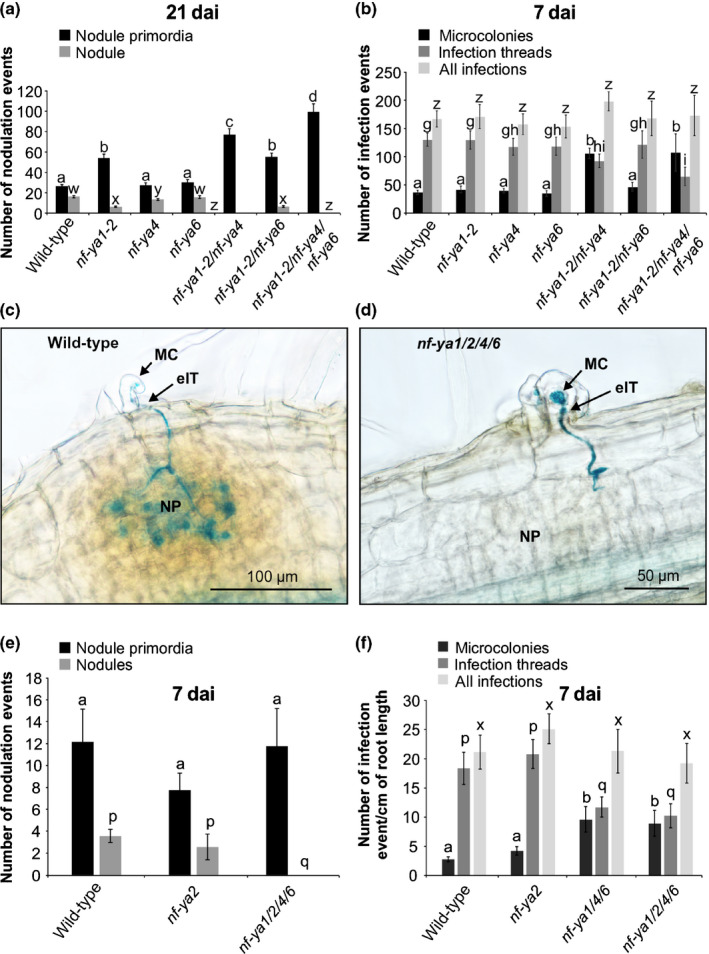
Lotus japonicus NF‐YAs function partially redundantly to regulate nodule organogenesis and Mesorhizobium loti infection. (a, b, e, f) Scores of nodulation events (nodule primordia and nodules) at 21 d after inoculation (dai) (a) and the number of nodulation (e) and infection events (b, f) (at 7 dai with M. loti strain NZP2235 carrying the hemA:LacZ reporter cassette for the indicated genotypes. Means ± 95% confidence intervals are presented for 15–54 individuals per genotype and statistical groupings, reflected by the same letters, have been determined separately for each category using one‐way ANOVA with Tukey’s honestly significant difference post hoc test (P < 0.05). (c, d) Typical wild‐type (c) and nf‐ya1/2/4/6 mutant (d) nodule primordia (NP). MC, microcolony; eIT, epidermal infection thread.
Our RNA‐seq data (BioProject ID PRJNA630938) from uninoculated L. japonicus roots collected 11 das showed that the NF‐YA4 mRNA was the most abundant among all root NF‐YA mRNA species, followed by NF‐YA2 > 6 > 5 > 7 > 8 > 3 and > 1 (Table S8). We therefore tested symbiotic phenotypes of higher‐order mutants, nf‐ya1‐2 nf‐ya4 nf‐ya6 and nf‐ya1‐2 nf‐ya2 nf‐ya4 nf‐ya6. Lotus japonicus lines carrying exonic LORE1 insertions at the NF‐YA2 and NF‐YA6 loci were used to develop these mutant lines (Table S7). Like nf‐ya1‐2 nf‐ya4, the triple and quadruple nf‐ya mutants did not develop nodules but retained the capacity to make small NP (Fig. 10c,e). At 21 dai, these were significantly more abundant than in corresponding wild‐type roots (Figs 10a, S9) and this was in spite of somewhat diminished root growth of the mutant lines. By contrast, the number of eITs was reduced by c. 50%, while the number of MCs was significantly increased in both triple and quadruple mutants (Fig. 10b,f). The direct comparison between the nf‐ya1/4/6 and nf‐ya1/2/4/6 mutant lines showed no significant difference with respect to number and category of infection events (Fig. 10f).
Early auxin signalling remains unaffected in the nf‐ya1 nf‐ya4 double mutant
Having determined that NF‐YA/STY module‐dependent signalling regulates YUCCA1 and YUCCA11 in the context of symbiosis, and that the relevant members of the three gene families shared nodule expression domains, we asked whether this contributes to auxin signalling. The corresponding YUCCA1 cDNA and YUCCA11 genomic sequences were therefore expressed under the control of the CaMV 2x35S constitutive promoter in hairy roots induced on transgenic L. japonicus shoots carrying the DR5:GUS auxin reporter (Ulmasov et al., 1997; Liao et al., 2015). Control hairy roots had typical elongated morphology, with the reporter activity confined to lateral root initiation sites and root apical regions (Fig. 11a,e). By contrast, 2x35SPro:YUCCA1 and 2x35SPro:YUCCA11‐transformed hairy roots had a super‐root‐like morphology (Boerjan et al., 1995), with 2x35SPro:YUCCA11 having the most pronounced impact on phenotype (Fig. 11b,d). Their short, thick and highly branched architecture was associated with ectopic DR5:GUS activity, which extended well beyond apical root regions, indicative of enhanced auxin activity (Fig. 11f,g). These observations suggested that YUCCA1 and YUCCA11 participate in regulation of auxin homeostasis, which was further tested.
Fig. 11.
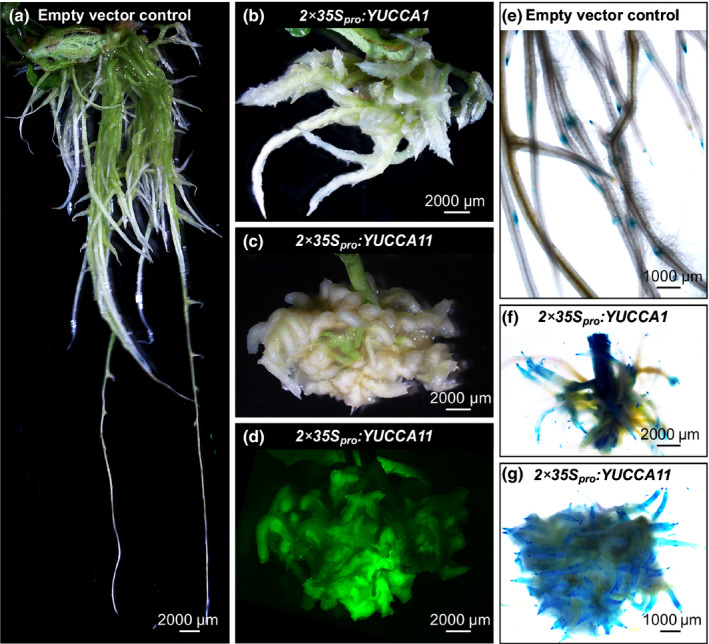
Ectopic expression of YUCCA1 and YUCCA11 results in auxin overproduction root phenotypes. (a–c) Representative images of hairy roots generated on transgenic Lotus japonicus shoots, carrying the DR5:GUS auxin reporter, using Agrobacterium rhizogenes carrying empty pKGWD,0 vector (a), or the same vector containing either the YUCCA1 mRNA sequence (b) or the YUCCA11 genomic fragment (including all exons, introns and the 3′UTR) (c) under the control of 2x CaMV 35S promoter (2x35Spro). (d) Example of the green fluorescent protein (GFP) fluorescence for the specimen shown in (c), used as proof of successful transformation. Panels (e–g) show activity of the DR5:GUS reporter, indicated in blue, in the corresponding hairy roots. Note that independent hairy root specimens were used for live imaging (a–d) and the histochemical analysis of β‐glucuronidase (GUS) activity (e–g).
The DR5:GUS reporter activity in wild‐type M. loti‐inoculated L. japonicus plants was associated with both early and late symbiotic events, initially observed in conjunction with early cell divisions in subepidermal and two subtending cortical cell layers (Fig. 12a) and slightly later during development, across all cell layers of small NP (Fig. 12b). Notably, similar early DR5:GUS activity patterns were also present in the nf‐ya1‐2 nf‐ya4 double mutant (Fig. 12d,e). At 21 dai with M. loti, wild‐type plants had mature nodules with DR5:GUS activity confined to the base and vasculature (Fig. 12c), reminiscent of the L. japonicus NF‐YA1 and STY gene expression (Fig. 2; Hossain et al., 2016). By contrast, in the double mutant, which is unable to make nodules, the reporter gene activity continued to associate with small NP (Fig. 12f). We attempted to rescue the nf‐ya1 nf‐ya4 mutant nodulation phenotype in transgenic hairy root experiments using a chimeric NF‐YA1 Pro:YUCCA11 expression module, but this was unsuccessful as no emerged nodules were present on transformed hairy roots. It is likely that in spite of a similar spatiotemporal expression patterns during nodule development, the strong NF‐YA1 promoter cannot functionally mimic the weak YUCCA11 promoter, resulting in lack of complementation. NF‐YA1 may also regulate other necessary processes in parallel to YUCCA genes.
Fig. 12.
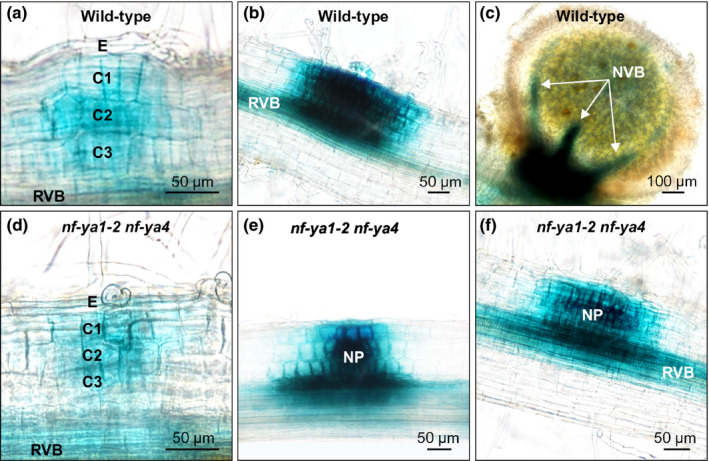
Localization of auxin responses during nodule development in Lotus japonicus wild‐type and nf‐ya1‐2 nf‐ya4. Representative images of the DR5:GUS reporter activity, indicated in blue, during different stages of nodule formation in L. japonicus wild‐type (upper row) and the nf‐ya1‐2 nf‐ya4 double mutant (lower row). (a–f) The specimens were collected and imaged at 14 (a, b, d, e) and 21 (c, f) d after inoculation (dai) with Mesorhizobium loti. Note that nodule development is blocked in nf‐ya1‐2 nf‐ya4 such that at 21 dai only nodule primordia (NP) are present (f). RVB, root vascular bundle; NVB, nodule vascular bundle; E, epidermis; C1, C2 and C3, consecutive root cortical cell layers.
Discussion
We have demonstrated previously that NF‐YA1 regulates expression of three members of the L. japonicus SHI/STY gene family (Hossain et al., 2016). This was extended by showing that the activity of all nine L. japonicus STY genes is associated with nodule development, and that seven are NF‐YA1‐dependent. Similarly, expression of seven M. truncatula STY genes was upregulated in nodules and at least six of them showed MtNF‐YA1 dependency. Arabidopsis LRP1 (Smith & Fedoroff, 1995), STY2 (Kuusk et al., 2002), SRS5 (Kuusk et al., 2006) and STY1 (Eklund et al., 2011) shared identical or highly overlapping expression domains. This was also true for promoters of the L. japonicus STY genes, implying their functional redundancy in mediating root and nodule development. Importantly, STY gene activities in mature nodules associated with vascular bundles and meristematic regions in L. japonicus and M. truncatula nodules, respectively, probably reflecting areas of persisting cell cycle activities in these two developmentally distinct organs.
A remarkable feature of SHI/STY genes is their functional synergism and redundancy. Phenotypic features of single and higher‐order Arabidopsis mutants indicate that members of the SHI/STY family act in a dosage‐dependent manner to regulate carpel and leaf formation (Kuusk et al., 2006). Unlike leaves and carpels, however, nodules are dispensable plant organs. Therefore, finding that expression of nine L. japonicus and at least six M. truncatula STY genes associates with nodule development was surprising. As the STYs were expressed at a relatively low level in L. japonicus roots, a dosage‐dependency requirement for accurate SHI/STY functioning during nodule formation could be a relevant factor. On the other hand, selective constraints for maintaining such a large number of SHI/STY genes in the symbiotic programme could be owing to its initial, extensive overlap with the lateral root formation programme (Schiessl et al., 2019; Soyano et al., 2019).
STYs regulate rhizobial infection and early nodule primordia formation events
Both the sty1/2/3 triple mutant and the LjNF‐YA1Pro:STY3::SRDX transgenic plants had significantly fewer eITs than did the wild‐type, despite an unchanged or even increased number of MCs, as in the STY3::SRDX5 background (Figs 5, 6). Auxin has been shown to play an important role in mediating symbiotic infection in root hairs (Breakspear et al., 2014; Nadzieja et al., 2018). Hence, STYs and YUCCAs may contribute to the mechanism that regulates auxin biosynthesis during IT formation. This notion is consistent with the observation that the soybean GmYUC2a gene, while contributing to local auxin biosynthesis, was essential for both Bradyrhizobium diazoefficiens infection and nodule organogenesis (Wang et al., 2019).
STYs may also partake in the regulation of early NP formation. Transgenic LjNF‐YA1Pro:STY3::SRDX plants formed significantly fewer (c. 50%) early NP compared with the wild‐type. A similar negative effect was observed in sty1/2/3, suggesting that STYs are indeed required in this developmental context. If so, this is most likely independent of NF‐YA1/NF‐YA4.
Lotus japonicus NF‐YAs, unlikely participants of early nodule primordia formation events
Both nf‐ya1‐2 and nf‐ya1‐2 nf‐ya4 formed significantly more early NP than did the wild‐type, yet the additive effect of the nf‐ya4 mutation totally blocked nodule emergence in the double mutant (Fig. 10). Formation of eITs was also affected to some degree in nf‐ya1 nf‐ya4. However, the total number of infections (i.e. number of MCs and eITs) was similar to the wild‐type, indicating that root susceptibilty to M. loti infection was unchanged in the absence of NF‐YA1 and NF‐YA4. Higher‐order nf‐ya mutants were tested but no additional significant effects were found, and at 21 dai the capacity of nf‐ya1/2/4/6 to form early NP was enhanced relative to the wild‐type. Additional NF‐YA genes are expressed at relatively low levels in L. japonicus roots (Table S8). We cannot, therefore, entirely rule out the possibility that higher redundancy, beyond the NF‐YA1/2/4/6 level, accounts for the sustained early NP formation in the quadruple mutant. However, we consider this rather unlikely because in the presence of nf‐ya1‐2, the nf‐ya4 mutation totally blocked the responsiveness of STY and YUCCA genes to M. loti infection, but NP were still formed. We therefore favour a model wherein L. japonicus NF‐YA1 is essential only downstream of the initial cortical cell divisions. Together with other partially redundantly operating NF‐YAs, such as NF‐YA4, NF‐YA1 probably mediates the nodule emergence stage by regulating STYs and YUCCAs involved in auxin biosynthesis (Fig. 13). Consistent with this model, early auxin activity, required for or associated with the initial cell divisions (Suzaki et al., 2012; Ng & Mathesius, 2018; Kohlen et al., 2018), appeared to be wild‐type in nf‐ya1‐2 nf‐ya4, indicating that an independent mechanism must mediate this process. Future experiments should test the impact of YUCCA1 and YUCCA11 silencing on the nodulation phenotype.
Fig. 13.
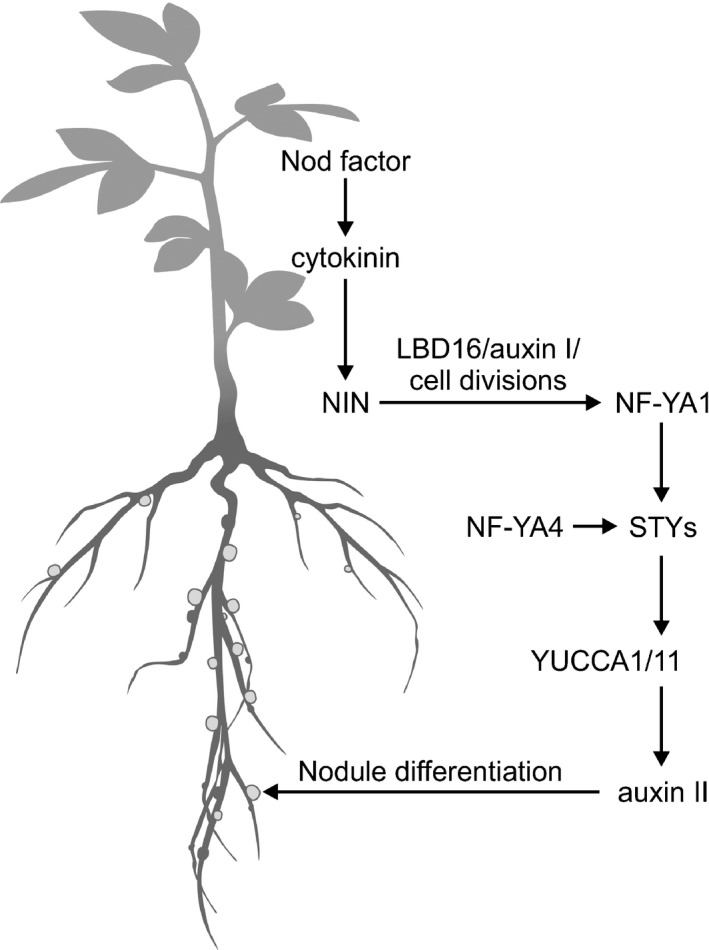
A working model for the NF‐YA‐dependent regulation of nodule organogenesis in Lotus japonicus. The Mesorhizobium loti‐derived nodulation (Nod) factor regulates expression of NF‐YA1 in a cytokinin‐ and NODULE INCEPTION (NIN)‐dependent manner. Acting as a presumed subunit of a heterotrimeric NF‐Y complex, NF‐YA1, along with NF‐YA4, partakes in the activation of STY gene expression in young nodule primordia, which in turn regulates the activity of YUCCA1 and YUCCA11 genes. The latter two genes are predicted to encode flavin monooxygenases, which contribute to local auxin signalling (auxin II) that mediates differentiation of mature nodules, downstream from the initial cell divisions that form nodule primordium in association with the first auxin peak (auxin I).
Lotus japonicus NF‐YA1 mediates local auxin signalling required for nodule emergence
A rapid increase in acropetal auxin transport below the M. loti infection sites has been documented in L. japonicus roots, but how this relates to early nodule primodium formation remains unclear (Pacios‐Bras et al., 2003; Ng & Mathesius, 2018). An increase in the cellular sensitivity to auxin was also considered in this developmental context, but this assertion awaits further confirmation (Ng & Mathesius, 2018). Finding that LBD16/ASL1, a member of a plant‐specific transcription factor gene family (Shuai et al., 2002), mediates local auxin biosynthesis preceding activation of the cell cycle for NP therefore provides an important insight into at least one of the contributing mechanisms (Schiessl et al., 2019).
LOB‐domain proteins regulate developmental and metabolic processes (Shuai et al., 2002; Xu et al., 2016) and Arabidopsis LBD16/ASL18 was shown to participate in the initiation of various root and root‐like organs by serving as a point of convergence for different priming events (W. Liu et al., 2018). This is probably also the role of legume LBD16/ASL18 during nodule formation (Schiessl et al., 2019; Soyano et al., 2019). Consistent with our results in L. japonicus and also previous observations (Combier et al., 2006; Laloum et al., 2014; Laporte et al., 2014; Baudin et al., 2015), the M. truncatula nf‐ya1‐1 mutation had little impact in this context (Schiessl et al., 2019). A fate mapping approach has shown that the apical meristem, typical of indeterminate nodules, specifically originates from divisions in the third root cortical layer (C3). In the M. truncatula nf‐ya1‐1 knockout mutant, NP development was unaffected, while cell divisions in the C3 layer and consequently nodule meristem formation were strongly impaired (Xiao et al., 2014).
Clearly, both LBD16 and NF‐YA1 are positive regulators of cell divisions (Schiessl et al., 2019; Soyano et al., 2019). However, their interaction in L. japonicus, under innate conditions, is likely to be pertinent only downstream of the initial cell divisions, perhaps during the subsequent cell division maintenance phase (Suzaki et al., 2012). Interestingly, different developmental processes apparently require a gradual attenuation of the LBD16 expression at the patterning stage (W. Liu et al., 2018) and based on the published expression data (Schiessl et al., 2019), this also seems to be the case during root nodule formation. By contrast, NF‐YA1s remain highly expressed and are indispensable for nodule differentiation (Combier et al., 2006; Laloum et al., 2014; Hossain et al., 2016).
We propose that the initial cell divisions and subsequent nodule emergence, encompassing further cell divisions and nodule patterning, are regulated by NF‐YA1‐independent and dependent mechanisms, respectively (Fig. 13). Our data lend experimental support for the previously postulated, developmental stage‐specific regulation of determinate and indeterminate nodule formation (Suzaki et al., 2012; Xiao et al., 2014) while depicting L. japonicus and M. truncatula NF‐YA1 genes as important nodule emergence stage‐specific regulators of auxin signalling.
Author contributions
KS, AS and AN conceived the idea and designed the experiments. AS, SZ, JT, TH, AL and LR performed the experiments. SS, AS and AN analysed the gene families. TM, SUA and JS analysed RNA‐seq data and provided bioinformatics and LORE1 mutant support. KS, AS and LR wrote the article.
Supporting information
Fig. S1 Lotus japonicus STY proteins.
Fig. S2 The putative RING zinc‐finger domain.
Fig. S3 Predicted IGGH domain.
Fig. S4 Lotus japonicus mutant sty alleles.
Fig. S5 Lotus japonicus single sty mutants have only very subtle symbiotic defects.
Fig. S6 Mutations at most STY loci affect nonsymbiotic plant growth.
Fig. S7 Primary sequence conservation between predicted Lotus japonicus YUCCA proteins.
Fig. S8 Relationship tree between predicted Lotus japonicus and Medicago truncatula YUCCA proteins.
Fig. S9 Lotus japonicus NF‐YAs function partially redundantly.
Table S1 Primers used in this study.
Table S2 Expression of four Lotus japonicus STY genes is significantly upregulated during the early stages of symbiosis.
Table S3 Analysis of Medicago truncatula STY gene expression.
Table S4 List of sty alleles carrying a LORE1 insertion, as identified from the Lotus Base information portal (https://lotus.au.dk/).
Table S5 Segregation of the NF‐YA1Pro:STY3::SRDX transgene in T1 populations, STY3::SRDX5 and STY3::SRDX6, derived from two independent T0 plants.
Table S6 YUCCA11 is regulated upon Mesorhizobium loti inoculation.
Table S7 A list of mutant nf‐ya alleles used in this study.
Table S8 Levels of different NF‐YA mRNAs in uninoculated L. japonicus roots.
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Acknowledgements
We thank A. Molnar for his expert help in preparation of figures. This work was supported by grants from the Agriculture and Agri‐Food Canada Crop Genomics Initiative and the National Science and Engineering Research Council of Canada (NSERC grant no. 3277A01) to KS, the Danish National Research Foundation grant DNRF79 to JS and the Agence Nationale de la Recherche (ANR) grant CE20‐012 ‘NODCCAAT’ and the ‘Laboratoire d’Excellence (LABEX)’‐‘TULIP’ (ANR‐10‐LABX‐41) to AN.
References
- Ariel F, Brault‐Hernandez M, Laffont C, Huault E, Brault M, Plet J, Moison M, Blanchet S, Ichante JL, Chabaud M et al. 2012. Two direct targets of cytokinin signaling regulate symbiotic nodulation in Medicago truncatula . Plant Cell 24: 3838–3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrighi JF, Barre A, Ben Amor B, Bersoult A, Soriano LC, Mirabella R, de Carvalho‐Niebel F, Journet EP, Gherardi M, Huguet T et al. 2006. The Medicago truncatula lysin motif‐receptor‐like kinase gene family includes NFP and new nodule‐expressed genes. Plant Physiology 142: 265–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battaglia M, Ripodas C, Clua J, Baudin M, Aguilar OM, Niebel A, Zanetti ME, Blanco FA. 2014. A nuclear factor Y interacting protein of the GRAS family is required for nodule organogenesis, infection thread progression, and lateral root growth. Plant Physiology 164: 1430–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudin M, Laloum T, Lepage A, Ripodas C, Ariel F, Frances L, Crespi M, Gamas P, Blanco FA, Zanetti ME et al. 2015. A phylogenetically conserved group of Nuclear Factor‐Y transcription factors interact to control nodulation in legumes. Plant Physiology 169: 2761–2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baylis T, Cierlik I, Sundberg E, Mattsson J. 2013. SHORT INTERNODES/STYLISH genes, regulators of auxin biosynthesis, are involved in leaf vein development in Arabidopsis thaliana . New Phytologist 197: 737–750. [DOI] [PubMed] [Google Scholar]
- Boerjan W, Cervera MT, Delarue M, Beeckman T, Dewitte W, Bellini C, Caboche M, Van Onckelen H, Van Montagu M, Inze D. 1995. Superroot, a recessive mutation in Arabidopsis, confers auxin overproduction. Plant Cell 7: 1405–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boivin S, Kazmierczak T, Brault M, Wen J, Gamas P, Mysore KS, Frugier F. 2016. Different cytokinin histidine kinase receptors regulate nodule initiation as well as later nodule developmental stages in Medicago truncatula . Plant, Cell & Environment 39: 2198–2209. [DOI] [PubMed] [Google Scholar]
- Breakspear A, Liu C, Roy S, Stacey N, Rogers C, Trick M, Morieri G, Mysore KS, Wen J, Oldroyd GE et al. 2014. The root hair "infectome" of Medicago truncatula uncovers changes in cell cycle genes and reveals a requirement for auxin signaling in rhizobial infection. Plant Cell 26: 4680–4701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broghammer A, Krusell L, Blaise M, Sauer J, Sullivan JT, Maolanon N, Vinther M, Lorentzen A, Madsen EB, Jensen KJ et al. 2012. Legume receptors perceive the rhizobial lipochitin oligosaccharide signal molecules by direct binding. Proceedings of the National Academy of Sciences, USA 109: 13859–13864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bu F, Rutten L, Roswanjaya YP, Kulikova O, Rodriguez‐Franco M, Ott T, Bisseling T, van Zeijl A, Geurts R. 2020. Mutant analysis in the nonlegume Parasponia andersonii identifies NIN and NF‐YA1 transcription factors as a core genetic network in nitrogen‐fixing nodule symbioses. New Phytologist 226: 541–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerri MR, Frances L, Laloum T, Auriac MC, Niebel A, Oldroyd GE, Barker DG, Fournier J, de Carvalho‐Niebel F. 2012. Medicago truncatula ERN transcription factors: regulatory interplay with NSP1/NSP2 GRAS factors and expression dynamics throughout rhizobial infection. Plant Physiology 160: 2155–2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chardin C, Girin T, Roudier F, Meyer C, Krapp A. 2014. The plant RWP‐RK transcription factors: key regulators of nitrogen responses and of gametophyte development. Journal of Experimental Botany 65: 5577–5587. [DOI] [PubMed] [Google Scholar]
- Cheng Y, Dai X, Zhao Y. 2006. Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis . Genes and Development 20: 1790–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Combier JP, de Billy F, Gamas P, Niebel A, Rivas S. 2008. Trans‐regulation of the expression of the transcription factor MtHAP2‐1 by a uORF controls root nodule development. Genes and Development 22: 1549–1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Combier JP, Frugier F, de Billy F, Boualem A, El‐Yahyaoui F, Moreau S, Vernie T, Ott T, Gamas P, Crespi M et al. 2006. MtHAP2‐1 is a key transcriptional regulator of symbiotic nodule development regulated by microRNA169 in Medicago truncatula . Genes and Development 20: 3084–3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper JB, Long SR. 1994. Morphogenetic rescue of Rhizobium meliloti nodulation mutants by trans‐zeatin secretion. Plant Cell 6: 215–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle JJ. 2011. Phylogenetic perspectives on the origins of nodulation. Molecular Plant–Microbe Interactions 24: 1289–1295. [DOI] [PubMed] [Google Scholar]
- Du Y, Scheres B. 2018. Lateral root formation and the multiple roles of auxin. Journal of Experimental Botany 69: 155–167. [DOI] [PubMed] [Google Scholar]
- Eklund DM, Cierlik I, Staldal V, Claes AR, Vestman D, Chandler J, Sundberg E. 2011. Expression of Arabidopsis SHORT INTERNODES/STYLISH family genes in auxin biosynthesis zones of aerial organs is dependent on a GCC box‐like regulatory element. Plant Physiology 157: 2069–2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eklund DM, Staldal V, Valsecchi I, Cierlik I, Eriksson C, Hiratsu K, Ohme‐Takagi M, Sundstrom JF, Thelander M, Ezcurra I et al. 2010a. The Arabidopsis thaliana STYLISH1 protein acts as a transcriptional activator regulating auxin biosynthesis. Plant Cell 22: 349–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eklund DM, Thelander M, Landberg K, Staldal V, Nilsson A, Johansson M, Valsecchi I, Pederson ER, Kowalczyk M, Ljung K et al. 2010b. Homologues of the Arabidopsis thaliana SHI/STY/LRP1 genes control auxin biosynthesis and affect growth and development in the moss Physcomitrella patens . Development 137: 1275–1284. [DOI] [PubMed] [Google Scholar]
- Estornell LH, Landberg K, Cierlik I, Sundberg E. 2018. SHI/STY genes affect pre‐ and post‐meiotic anther processes in auxin sensing domains in Arabidopsis . Frontiers in Plant Science 9: 150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridborg I, Kuusk S, Robertson M, Sundberg E. 2001. The Arabidopsis protein SHI represses gibberellin responses in Arabidopsis and barley. Plant Physiology 127: 937–948. [PMC free article] [PubMed] [Google Scholar]
- Gamas P, Brault M, Jardinaud MF, Frugier F. 2017. Cytokinins in symbiotic nodulation: when, where, what for? Trends in Plant Science 22: 792–802. [DOI] [PubMed] [Google Scholar]
- Gauthier‐Coles C, White RG, Mathesius U. 2019. Nodulating legumes are distinguished by a sensitivity to cytokinin in the root cortex leading to pseudonodule development. Frontiers in Plant Science 9: 1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geurts R, Xiao TT, Reinhold‐Hurek B. 2016. What does it take to evolve a nitrogen‐fixing endosymbiosis? Trends in Plant Science 21: 199–208. [DOI] [PubMed] [Google Scholar]
- Gomariz‐Fernandez A, Sanchez‐Gerschon V, Fourquin C, Ferrandiz C. 2017. The role of SHI/STY/SRS genes in organ growth and carpel development is conserved in the distant eudicot species Arabidopsis thaliana and Nicotiana benthamiana . Frontiers in Plant Science 8: 814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez‐Rizzo S, Crespi M, Frugier F. 2006. The Medicago truncatula CRE1 cytokinin receptor regulates lateral root development and early symbiotic interaction with Sinorhizobium meliloti . Plant Cell 18: 2680–2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griesmann M, Chang Y, Liu X, Song Y, Haberer G, Crook MB, Billault‐Penneteau B, Lauressergues D, Keller J, Imanishi L et al. 2018. Phylogenomics reveals multiple losses of nitrogen‐fixing root nodule symbiosis. Science 361: eaat1743. [DOI] [PubMed] [Google Scholar]
- Heckmann AB, Sandal N, Bek AS, Madsen LH, Jurkiewicz A, Nielsen MW, Tirichine L, Stougaard J. 2011. Cytokinin induction of root nodule primordia in Lotus japonicus is regulated by a mechanism operating in the root cortex. Molecular Plant–Microbe Interactions 24: 1385–1395. [DOI] [PubMed] [Google Scholar]
- Held M, Hou H, Miri M, Huynh C, Ross L, Hossain MS, Sato S, Tabata S, Perry J, Wang TL et al. 2014. Lotus japonicus cytokinin receptors work partially redundantly to mediate nodule formation. Plant Cell 26: 678–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrbach V, Rembliere C, Gough C, Bensmihen S. 2014. Lateral root formation and patterning in Medicago truncatula . Journal of Plant Physiology 171: 301–310. [DOI] [PubMed] [Google Scholar]
- Hiratsu K, Matsui K, Koyama T, Ohme‐Takagi M. 2003. Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis . The Plant Journal 34: 733–739. [DOI] [PubMed] [Google Scholar]
- Hossain MS, Shrestha A, Zhong S, Miri M, Austin RS, Sato S, Ross L, Huebert T, Tromas A, Torres‐Jerez I et al. 2016. Lotus japonicus NF‐YA1 plays an essential role during nodule differentiation and targets members of the SHI/STY gene family. Molecular Plant–Microbe Interactions 29: 950–964. [DOI] [PubMed] [Google Scholar]
- Karimi M, Inze D, Depicker A. 2002. GATEWAY vectors for Agrobacterium‐mediated plant transformation. Trends in Plant Science 7: 193–195. [DOI] [PubMed] [Google Scholar]
- Kelly S, Radutoiu S, Stougaard J. 2017. Legume LysM receptors mediate symbiotic and pathogenic signalling. Current Opinion in Plant Biology 39: 152–158. [DOI] [PubMed] [Google Scholar]
- Kohlen W, Ng JLP, Deinum EE, Mathesius U. 2018. Auxin transport, metabolism, and signalling during nodule initiation: indeterminate and determinate nodules. Journal of Experimental Botany 69: 229–244. [DOI] [PubMed] [Google Scholar]
- Kosuta S, Held M, Hossain MS, Morieri G, Macgillivary A, Johansen C, Antolin‐Llovera M, Parniske M, Oldroyd GE, Downie AJ et al. 2011. Lotus japonicus symRK‐14 uncouples the cortical and epidermal symbiotic program. The Plant Journal 67: 929–940. [DOI] [PubMed] [Google Scholar]
- Kuusk S, Sohlberg JJ, Long JA, Fridborg I, Sundberg E. 2002. STY1 and STY2 promote the formation of apical tissues during Arabidopsis gynoecium development. Development 129: 4707–4717. [DOI] [PubMed] [Google Scholar]
- Kuusk S, Sohlberg JJ, Magnus Eklund D, Sundberg E. 2006. Functionally redundant SHI family genes regulate Arabidopsis gynoecium development in a dose‐dependent manner. The Plant Journal 47: 99–111. [DOI] [PubMed] [Google Scholar]
- Laloum T, De Mita S, Gamas P, Baudin M, Niebel A. 2013. CCAAT‐box binding transcription factors in plants: Y so many?. Trends in Plant Science 18: 157–166. [DOI] [PubMed] [Google Scholar]
- Laloum T, Baudin M, Frances L, Lepage A, Billault‐Penneteau B, Cerri MR, Ariel F, Jardinaud MF, Gamas P, de Carvalho‐Niebel F et al. 2014. Two CCAAT‐box‐binding transcription factors redundantly regulate early steps of the legume‐rhizobia endosymbiosis. The Plant Journal 79: 757–768. [DOI] [PubMed] [Google Scholar]
- Laporte P, Lepage A, Fournier J, Catrice O, Moreau S, Jardinaud MF, Mun JH, Larrainzar E, Cook DR, Gamas P et al. 2014. The CCAAT box‐binding transcription factor NF‐YA1 controls rhizobial infection. Journal of Experimental Botany 65: 481–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larrainzar E, Riely BK, Kim SC, Carrasquilla‐Garcia N, Yu HJ, Hwang HJ, Oh M, Kim GB, Surendrarao AK, Chasman D et al. 2015. Deep sequencing of the Medicago truncatula root transcriptome reveals a massive and early interaction between nodulation factor and ethylene signals. Plant Physiology 169: 233–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerouge P, Roche P, Faucher C, Maillet F, Truchet G, Promé JC, Dénarié J. 1990. Symbiotic host‐specificity of Rhizobium meliloti is determined by a sulphated and acylated glucosamine oligosaccharide signal. Nature 344: 781–784. [DOI] [PubMed] [Google Scholar]
- Liang Y, Toth K, Cao Y, Tanaka K, Espinoza C, Stacey G. 2014. Lipochitooligosaccharide recognition: an ancient story. New Phytologist 204: 289–296. [DOI] [PubMed] [Google Scholar]
- Liao CY, Smet W, Brunoud G, Yoshida S, Vernoux T, Weijers D. 2015. Reporters for sensitive and quantitative measurement of auxin response. Nature Methods 12: 207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limpens E, Franken C, Smit P, Willemse J, Bisseling T, Geurts R. 2003. LysM domain receptor kinases regulating rhizobial Nod factor‐induced infection. Science 302: 630–633. [DOI] [PubMed] [Google Scholar]
- Liu CW, Breakspear A, Guan D, Cerri MR, Jackson K, Jiang S, Robson F, Radhakrishnan GV, Roy S, Bone C et al. 2019. NIN acts as a network hub controlling a growth module required for rhizobial infection. Plant Physiology 179: 1704–1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Zhang C, Yang J, Yu N, Wang E. 2018. Hormone modulation of legume‐rhizobial symbiosis. Journal of Integrative Plant Biology 60: 632–648. [DOI] [PubMed] [Google Scholar]
- Liu J, Rutten L, Limpens E, van der Molen T, van Velzen R, Chen R, Chen Y, Geurts R, Kohlen W, Kulikova O et al. 2019. A remote cis‐regulatory region is required for NIN expression in the pericycle to initiate nodule primordium formation in Medicago truncatula . Plant Cell 31: 68–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Yu J, Ge Y, Qin P, Xu L. 2018. Pivotal role of LBD16 in root and root‐like organ initiation. Cellular and Molecular Life Sciences 75: 3329–3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombari P, Ercolano E, El Alaouri E, Chiurazzi M. 2005. Agrobacterium‐mediated in vitro transformation. In: Márquez AJ, Stougaard J, Udvardi M, Parniske M, Spaink H, Saalbach G, Webb J, Chiurazzi M, Márquez AJ, eds. Lotus japonicus Handbook. Berlin, Germany: Springer, 87–95. [Google Scholar]
- Madsen EB, Madsen LH, Radutoiu S, Olbryt M, Rakwalska M, Szczyglowski K, Sato S, Kaneko T, Tabata S, Sandal N et al. 2003. A receptor kinase gene of the LysM type is involved in legume perception of rhizobial signals. Nature 425: 637–640. [DOI] [PubMed] [Google Scholar]
- Madsen LH, Tirichine L, Jurkiewicz A, Sullivan JT, Heckmann AB, Bek AS, Ronson CW, James EK, Stougaard J. 2010. The molecular network governing nodule organogenesis and infection in the model legume Lotus japonicus . Nature Communications 1: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malolepszy A, Mun T, Sandal N, Gupta V, Dubin M, Urbanski D, Shah N, Bachmann A, Fukai E, Hirakawa H et al. 2016. The LORE1 insertion mutant resource. The Plant Journal 88: 306–317. [DOI] [PubMed] [Google Scholar]
- Mantovani R. 1999. The molecular biology of the CCAAT‐binding factor NF‐Y. Gene 239: 15–27. [DOI] [PubMed] [Google Scholar]
- Marsh JF, Rakocevic A, Mitra RM, Brocard L, Sun J, Eschstruth A, Long SR, Schultze M, Ratet P, Oldroyd GE. 2007. Medicago truncatula NIN is essential for rhizobial‐independent nodule organogenesis induced by autoactive calcium/calmodulin‐dependent protein kinase. Plant Physiology 144: 324–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miri M, Janakirama P, Held M, Ross L, Szczyglowski K. 2016. Into the root: how cytokinin controls rhizobial infection. Trends in Plant Science 21: 178–186. [DOI] [PubMed] [Google Scholar]
- Mun T, Bachmann A, Gupta V, Stougaard J, Andersen SU. 2016. Lotus Base: an integrated information portal for the model legume Lotus japonicus . Scientific Reports 6: 39447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray JD. 2017. The cell cycle in nodulation. In: Rose RJ, ed. Molecular cell biology of the growth and differentiation of plant cells. Boca Raton, FL, USA: CRC Press, 220–235. [Google Scholar]
- Murray JD, Karas BJ, Sato S, Tabata S, Amyot L, Szczyglowski K. 2007. A cytokinin perception mutant colonized by Rhizobium in the absence of nodule organogenesis. Science 315: 101–104. [DOI] [PubMed] [Google Scholar]
- Nadzieja M, Kelly S, Stougaard J, Reid D. 2018. Epidermal auxin biosynthesis facilitates rhizobial infection in Lotus japonicus . The Plant Journal 95: 101–111. [DOI] [PubMed] [Google Scholar]
- Ng JLP, Mathesius U. 2018. Acropetal auxin transport inhibition is involved in indeterminate but not determinate nodule formation. Frontiers in Plant Science 9: 169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida H, Tanaka S, Handa Y, Ito M, Sakamoto Y, Matsunaga S, Betsuyaku S, Miura K, Soyano T, Kawaguchi M et al. 2018. A NIN‐LIKE PROTEIN mediates nitrate‐induced control of root nodule symbiosis in Lotus japonicus . Nature Communications 9: 499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldroyd GE, Harrison MJ, Paszkowski U. 2009. Reprogramming plant cells for endosymbiosis. Science 324: 753–754. [DOI] [PubMed] [Google Scholar]
- Pacios‐Bras C, Schlaman HR, Boot K, Admiraal P, Langerak JM, Stougaard J, Spaink HP. 2003. Auxin distribution in Lotus japonicus during root nodule development. Plant Molecular Biology 52: 1169–1180. [DOI] [PubMed] [Google Scholar]
- Perry J, Brachmann A, Welham T, Binder A, Charpentier M, Groth M, Haage K, Markmann K, Wang TL, Parniske M. 2009. TILLING in Lotus japonicus identified large allelic series for symbiosis genes and revealed a bias in functionally defective ethyl methanesulfonate alleles toward glycine replacements. Plant Physiology 151: 1281–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plet J, Wasson A, Ariel F, Le Signor C, Baker D, Mathesius U, Crespi M, Frugier F. 2011. MtCRE1‐dependent cytokinin signaling integrates bacterial and plant cues to coordinate symbiotic nodule organogenesis in Medicago truncatula . The Plant Journal 65: 622–633. [DOI] [PubMed] [Google Scholar]
- Radutoiu S, Madsen LH, Madsen EB, Felle HH, Umehara Y, Gronlund M, Sato S, Nakamura Y, Tabata S, Sandal N et al. 2003. Plant recognition of symbiotic bacteria requires two LysM receptor‐like kinases. Nature 425: 585–592. [DOI] [PubMed] [Google Scholar]
- Radutoiu S, Madsen LH, Madsen EB, Jurkiewicz A, Fukai E, Quistgaard EM, Albrektsen AS, James EK, Thirup S, Stougaard J. 2007. LysM domains mediate lipochitin‐oligosaccharide recognition and Nfr genes extend the symbiotic host range. European Molecular Biology Organization Journal 26: 3923–3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid D, Nadzieja M, Novak O, Heckmann AB, Sandal N, Stougaard J. 2017. Cytokinin biosynthesis promotes cortical cell responses during nodule development. Plant Physiology 175: 361–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripodas C, Castaingts M, Clua J, Villafane J, Blanco FA, Zanetti ME. 2019. The PvNF‐YA1 and PvNF‐YB7 subunits of the heterotrimeric NF‐Y Transcription factor influence strain preference in the Phaseolus vulgaris‐Rhizobium etli symbiosis. Frontiers in Plant Science 10: 221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux B, Rodde N, Jardinaud MF, Timmers T, Sauviac L, Cottret L, Carrere S, Sallet E, Courcelle E, Moreau S et al. 2014. An integrated analysis of plant and bacterial gene expression in symbiotic root nodules using laser‐capture microdissection coupled to RNA sequencing. The Plant Journal 77: 817–837. [DOI] [PubMed] [Google Scholar]
- Schauser L, Roussis A, Stiller J, Stougaard J. 1999. A plant regulator controlling development of symbiotic root nodules. Nature 402: 191–195. [DOI] [PubMed] [Google Scholar]
- Schiessl K, Lilley JLS, Lee T, Tamvakis I, Kohlen W, Bailey PC, Thomas A, Luptak J, Ramakrishnan K, Carpenter MD et al. 2019. NODULE INCEPTION recruits the lateral root developmental program for symbiotic nodule organogenesis in Medicago truncatula . Current Biology 29: e3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuai B, Reynaga‐Pena CG, Springer PS. 2002. The LATERAL ORGAN BOUNDARIES gene defines a novel, plant‐specific gene family. Plant Physiology 129: 747–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DL, Fedoroff NV. 1995. LRP1, a gene expressed in lateral and adventitious root primordia of Arabidopsis . Plant Cell 7: 735–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohlberg JJ, Myrenas M, Kuusk S, Lagercrantz U, Kowalczyk M, Sandberg G, Sundberg E. 2006. STY1 regulates auxin homeostasis and affects apical‐basal patterning of the Arabidopsis gynoecium. The Plant Journal 47: 112–123. [DOI] [PubMed] [Google Scholar]
- Soyano T, Kouchi H, Hirota A, Hayashi M. 2013. NODULE INCEPTION directly targets NF‐Y subunit genes to regulate essential processes of root nodule development in Lotus japonicus . PLoS Genetics 9: e1003352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soyano T, Shimoda Y, Kawaguchi M, Hayashi M. 2019. A shared gene drives lateral root development and root nodule symbiosis pathways in Lotus . Science 366: 1021–1023. [DOI] [PubMed] [Google Scholar]
- Sprent JI, James EK. 2007. Legume evolution: where do nodules and mycorrhizas fit in? Plant Physiology 144: 575–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzaki T, Ito M, Kawaguchi M. 2013. Genetic basis of cytokinin and auxin functions during root nodule development. Frontiers in Plant Science 4: 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzaki T, Yano K, Ito M, Umehara Y, Suganuma N, Kawaguchi M. 2012. Positive and negative regulation of cortical cell division during root nodule development in Lotus japonicus is accompanied by auxin response. Development 139: 3997–4006. [DOI] [PubMed] [Google Scholar]
- Szczyglowski K, Shaw RS, Wopereis J, Copeland S, Hamburger D, Kasiborski B, Dazzo FB, De Bruijn FJ. 1998. Nodule organogenesis and symbiotic mutants of the model legume Lotus japonicus . Molecular Plant–Microbe Interactions 11: 684–697. [Google Scholar]
- Tirichine L, Sandal N, Madsen LH, Radutoiu S, Albrektsen AS, Sato S, Asamizu E, Tabata S, Stougaard J. 2007. A gain‐of‐function mutation in a cytokinin receptor triggers spontaneous root nodule organogenesis. Science 315: 104–107. [DOI] [PubMed] [Google Scholar]
- Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ. 1997. Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9: 1963–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbanski DF, Malolepszy A, Stougaard J, Andersen SU. 2012. Genome‐wide LORE1 retrotransposon mutagenesis and high‐throughput insertion detection in Lotus japonicus . The Plant Journal 69: 731–741. [DOI] [PubMed] [Google Scholar]
- van Velzen R, Holmer R, Bu F, Rutten L, van Zeijl A, Liu W, Santuari L, Cao Q, Sharma T, Shen D et al. 2018. Comparative genomics of the nonlegume Parasponia reveals insights into evolution of nitrogen‐fixing rhizobium symbioses. Proceedings of the National Academy of Sciences, USA 115: E4700–E4709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernie T, Kim J, Frances L, Ding Y, Sun J, Guan D, Niebel A, Gifford ML, de Carvalho‐Niebel F, Oldroyd GE. 2015. The NIN transcription factor coordinates diverse nodulation programs in different tissues of the Medicago truncatula root. Plant Cell 27: 3410–3424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Yang W, Zuo Y, Zhu L, Hastwell AH, Chen L, Tian Y, Su C, Ferguson BJ, Li X. 2019. GmYUC2a mediates auxin biosynthesis during root development and nodulation in soybean. Journal of Experimental Botany 70: 3165–3176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner GD, Cornwell WK, Sprent JI, Kattge J, Kiers ET. 2014. A single evolutionary innovation drives the deep evolution of symbiotic N2‐fixation in angiosperms. Nature Communications 5: 4087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong JEMM, Nadzieja M, Madsen LH, Bücherl CA, Dam S, Sandal NN, Couto D, Derbyshire P, Uldum‐Berentsen M, Schroeder S et al. 2019. A Lotus japonicus cytoplasmic kinase connects Nod factor perception by the NFR5 LysM receptor to nodulation. Proceedings of the National Academy of Sciences, USA 116: 14339–14348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wopereis J, Pajuelo E, Dazzo FB, Jiang Q, Gresshoff PM, De Bruijn FJ, Stougaard J, Szczyglowski K. 2000. Short root mutant of Lotus japonicus with a dramatically altered symbiotic phenotype. The Plant Journal 23: 97–114. [DOI] [PubMed] [Google Scholar]
- Xiao TT, Schilderink S, Moling S, Deinum EE, Kondorosi E, Franssen H, Kulikova O, Niebel A, Bisseling T. 2014. Fate map of Medicago truncatula root nodules. Development 141: 3517–3528. [DOI] [PubMed] [Google Scholar]
- Xiao TT, van Velzen R, Kulikova O, Franken C, Bisseling T. 2019. Lateral root formation involving cell division in both pericycle, cortex and endodermis is a common and ancestral trait in seed plants. Development 146: dev182592. [DOI] [PubMed] [Google Scholar]
- Xu C, Luo F, Hochholdinger F. 2016. LOB domain proteins: beyond lateral organ boundaries. Trends in Plant Science 21: 159–167. [DOI] [PubMed] [Google Scholar]
- Yoro E, Suzaki T, Toyokura K, Miyazawa H, Fukaki H, Kawaguchi M. 2014. A positive regulator of nodule organogenesis, NODULE INCEPTION, Acts as a negative regulator of rhizobial infection in Lotus japonicus . Plant Physiology 165: 747–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanetti ME, Blanco FA, Beker MP, Battaglia M, Aguilar OM. 2010. A C subunit of the plant nuclear factor NF‐Y required for rhizobial infection and nodule development affects partner selection in the common bean–Rhizobium etli symbiosis. Plant Cell 22: 4142–4157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanetti ME, Ripodas C, Niebel A. 2017. Plant NF‐Y transcription factors: key players in plant‐microbe interactions, root development and adaptation to stress. Biochimica et Biophysica Acta. Gene Regulatory Mechanisms 1860: 645–654. [DOI] [PubMed] [Google Scholar]
- Zhao Y. 2018. Essential roles of local auxin biosynthesis in plant development and in adaptation to environmental changes. Annual Review of Plant Biology 69: 417–435. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Lotus japonicus STY proteins.
Fig. S2 The putative RING zinc‐finger domain.
Fig. S3 Predicted IGGH domain.
Fig. S4 Lotus japonicus mutant sty alleles.
Fig. S5 Lotus japonicus single sty mutants have only very subtle symbiotic defects.
Fig. S6 Mutations at most STY loci affect nonsymbiotic plant growth.
Fig. S7 Primary sequence conservation between predicted Lotus japonicus YUCCA proteins.
Fig. S8 Relationship tree between predicted Lotus japonicus and Medicago truncatula YUCCA proteins.
Fig. S9 Lotus japonicus NF‐YAs function partially redundantly.
Table S1 Primers used in this study.
Table S2 Expression of four Lotus japonicus STY genes is significantly upregulated during the early stages of symbiosis.
Table S3 Analysis of Medicago truncatula STY gene expression.
Table S4 List of sty alleles carrying a LORE1 insertion, as identified from the Lotus Base information portal (https://lotus.au.dk/).
Table S5 Segregation of the NF‐YA1Pro:STY3::SRDX transgene in T1 populations, STY3::SRDX5 and STY3::SRDX6, derived from two independent T0 plants.
Table S6 YUCCA11 is regulated upon Mesorhizobium loti inoculation.
Table S7 A list of mutant nf‐ya alleles used in this study.
Table S8 Levels of different NF‐YA mRNAs in uninoculated L. japonicus roots.
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.


