Abstract
Background and Aim
Magnifying endoscopy with narrow‐band imaging (ME‐NBI) has made a huge contribution to clinical practice. However, acquiring skill at ME‐NBI diagnosis of early gastric cancer (EGC) requires considerable expertise and experience. Recently, artificial intelligence (AI), using deep learning and a convolutional neural network (CNN), has made remarkable progress in various medical fields. Here, we constructed an AI‐assisted CNN computer‐aided diagnosis (CAD) system, based on ME‐NBI images, to diagnose EGC and evaluated the diagnostic accuracy of the AI‐assisted CNN‐CAD system.
Methods
The AI‐assisted CNN‐CAD system (ResNet50) was trained and validated on a dataset of 5574 ME‐NBI images (3797 EGCs, 1777 non‐cancerous mucosa and lesions). To evaluate the diagnostic accuracy, a separate test dataset of 2300 ME‐NBI images (1430 EGCs, 870 non‐cancerous mucosa and lesions) was assessed using the AI‐assisted CNN‐CAD system.
Results
The AI‐assisted CNN‐CAD system required 60 s to analyze 2300 test images. The overall accuracy, sensitivity, specificity, positive predictive value, and negative predictive value of the CNN were 98.7%, 98%, 100%, 100%, and 96.8%, respectively. All misdiagnosed images of EGCs were of low‐quality or of superficially depressed and intestinal‐type intramucosal cancers that were difficult to distinguish from gastritis, even by experienced endoscopists.
Conclusions
The AI‐assisted CNN‐CAD system for ME‐NBI diagnosis of EGC could process many stored ME‐NBI images in a short period of time and had a high diagnostic ability. This system may have great potential for future application to real clinical settings, which could facilitate ME‐NBI diagnosis of EGC in practice.
Keywords: artificial intelligence, convolutional neural network, early gastric cancer, magnifying endoscopy, narrow‐band imaging
Introduction
White light imaging in esophagogastroduodenoscopy is the most sensitive method of early detection of gastric cancer. 1 However, accurate diagnosis of early gastric cancer (EGC) is sometimes difficult with white light imaging alone, especially for small lesions. Magnifying endoscopy with narrow‐band imaging (ME‐NBI) is a recently developed, image‐enhanced endoscopic technique. 2 For ME‐NBI diagnosis of EGC, the vessel plus surface classification system (VSCS) proposed by Yao et al. 3 has proven prospectively to be very useful in distinguishing between EGC and non‐cancer 4 , 5 , 6 , 7 , 8 , 9 and could require fewer biopsies to diagnose each cancer. 7 Recently, the magnifying endoscopy simple diagnostic algorithm for EGC (MESDA‐G) has been proposed as a unified system of ME‐NBI diagnosis for EGC and has become widely known. 9 The VSCS is an important diagnostic algorithm, serving as the base algorithm for the MESDA‐G and has been used in our hospital for ME‐NBI diagnosis. However, these advantages have been reported from the results obtained by expert endoscopists. Although ME‐NBI is thought to make a huge contribution to clinical practice, acquiring skill at ME‐NBI diagnosis using VSCS requires substantial expertise and experience. While there are reports claiming that an e‐learning system is an effective tool for training ME‐NBI diagnosis efficiently, a further development of diagnostic technology is still required. 10
As deep learning (DL) technology has rapidly gained attention as an optimal machine learning method, the application of artificial intelligence (AI) in medicine has been explored enthusiastically. 11 , 12 In the field of upper gastrointestinal endoscopy, AI is expected to provide the solution to clinical hurdles that are believed difficult to overcome with currently available imaging technology. There have been various reports of AI‐assisted endoscopic diagnostic systems of the upper gastrointestinal tract and many of those in the gastric region are related to the detection of gastric cancer and Helicobacter pylori infection status. 13 , 14 , 15 , 16 , 17 , 18 , 19 Recently, a few reports have assessed the usefulness of computer‐aided diagnosis (CAD) systems for ME‐NBI diagnosis of EGC using AI. 20 , 21 These results are almost equivalent to or slightly lower than the previously reported diagnostic accuracies of ME‐NBI (90.4% 6 and 96.1% 7 ) performed by endoscopists in Japan. Moreover, although maximal magnification was stated as the method used in these two articles, the water immersion technique was not used. We have been using water immersion technique with maximal magnification in our hospital and reported excellent diagnostic accuracies in gastric cancer cases of H. pylori post‐eradication status that were thought to be difficult to diagnose. 22 However, so far, there are no research articles published evaluating the diagnostic accuracy of AI‐assisted convolutional neural network (CNN)‐CAD systems applied to imaging data captured using the water immersion technique with maximal magnification. For this reason, and in order to solve the various issues mentioned above, we constructed an AI‐assisted CNN‐CAD system based on ME‐NBI images to diagnose EGC and evaluated the diagnostic accuracy of the AI‐assisted CNN‐CAD system.
Methods
Preparation of training and test image sets
In this single‐center retrospective study, from the 745 lesions of differentiated type EGC resected by endoscopy at our hospital between April 2013 and March 2018, a total of 349 lesions were selected. These provided 5227 ME‐NBI images using the water immersion technique at the maximal magnification that had sufficient quality to permit diagnosis by the MESDA‐G (Fig. 1a–c). ME‐NBI images of gastric adenocarcinoma of fundic gland‐type (GAFG) 23 , 24 and diffuse‐type EGC, as well as unanalyzable and low‐quality images, were excluded, because it is unlikely that the ME‐NBI findings of these cases could be accurately diagnosed as cancer. In all EGC patients, histopathological diagnosis was made by a pathologist specialized in gastrointestinal pathology (T.Y.). The main histological type was determined according to the Japanese Classification of Gastric Carcinoma, third English edition. 25 In addition, 2647 ME‐NBI images of non‐cancerous mucosa or non‐cancerous lesions that were obtained under the same conditions were collected. These ME‐NBI images included fundic gland mucosa, pyloric gland mucosa, patchy redness, adenoma, xanthoma, focal atrophy, and ulcer scars (Fig. 1d–o) and were subjected to endoscopic diagnosis but not to pathological examination of the biopsy.
Figure 1.
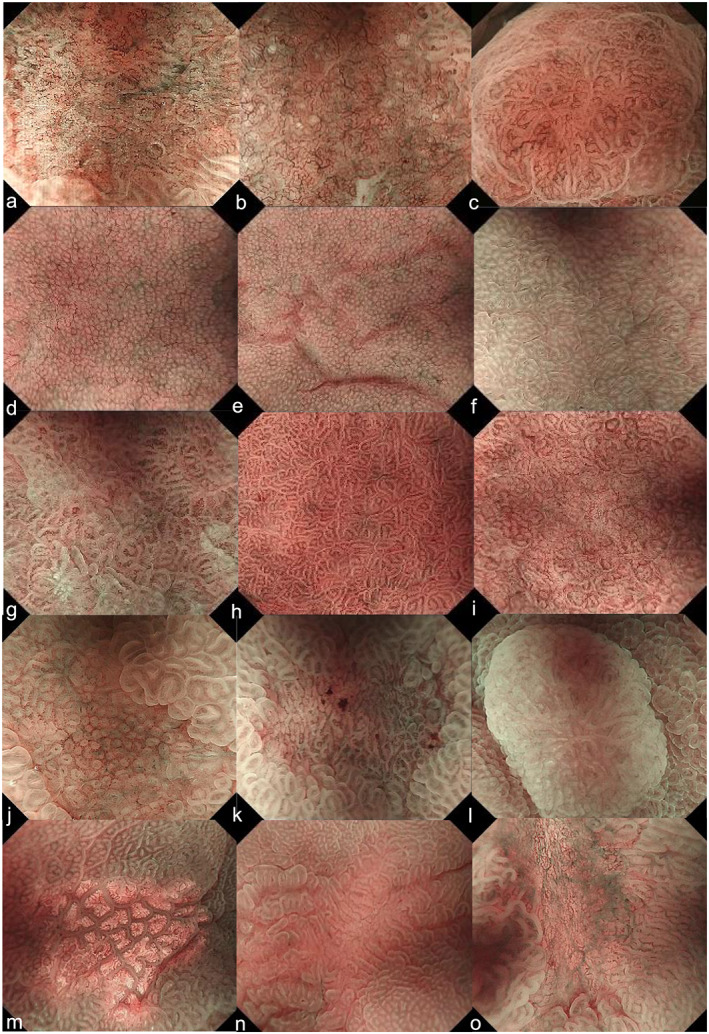
Educational endoscopic images used for the CNN. (a) Differentiated‐type cancer (0‐IIc, tub1), (b) differentiated‐type cancer (0‐IIc, tub2), (c) differentiated‐type cancer (0‐IIa, tub1), (d–f) fundic gland mucosa, (g–i) pyloric gland mucosa, (j, k) patchy redness, (l) adenoma, (m) xanthoma, (n) focal atrophy, and (o) ulcer scar. 0‐IIa, flatly elevated; 0‐IIc, flatly depressed; CNN, convolution neural network; tub1, well differentiated adenocarcinoma; tub2, moderately differentiated adenocarcinoma.
Constructing a convolutional neural network algorithm
The AI‐assisted CNN‐CAD system was developed through transfer learning leveraging a CNN architecture, Deep Residual Network 26 (ResNet‐50), which was pretrained on the ImageNet database containing over 14 million images. A Caffe DL framework, one of the most widely used frameworks originally developed at the Berkeley Vision and Learning Center, was used to train, validate, and test the CNN. Using transfer learning, we replaced the final classification layer with another fully connected layer, retrained it using our training dataset, and fine‐tuned the parameters of all layers. Images were resized to 224 × 224 pixels to suit the original dimensions of the models. A rotation augmentation was also applied to images. Data augmentation was strictly performed only on the training dataset to improve the system's classification performance. All layers of the CNN were fine‐tuned using stochastic gradient descent with a global learning rate of 0.001. These values were set up by trial and error to ensure all data were compatible with ResNet‐50.
An AI‐assisted CNN‐CAD system was trained and validated on a dataset of 5574 ME‐NBI images (EGCs: 3797 images of 267 cases, non‐cancerous images: 1777 images). During training, we randomly divided the training dataset into a training dataset (4460 images) and a validation dataset (1114 images), with a ratio of 8:2, to refine the model during training (Fig. S1).
Outcome measures of artificial intelligence diagnosis
To evaluate the diagnostic accuracy (diagnosis of cancer or non‐cancer), a separate test dataset of 2300 ME‐NBI images (EGCs: 1430 images of 82 cases, non‐cancerous images: 870 images) was applied to the resultant CNN (Fig. S1). The test dataset was not augmented. In the test, the trained neural network generated a continuous number between 0 and 1 for cancer or non‐cancer images, corresponding to the probability of that condition being represented by the image. The cut‐off value for final diagnosis of each condition (cancerous/non‐cancerous) was set at 0.5.
We present the definition of the evaluation criteria in Table S1. Moreover, we calculated the area under the receiver‐operating characteristic curve (AUC) to evaluate the accuracy of the AI‐assisted CNN‐CAD system as a device. The overall test speed was defined as from the start to the end of the analysis of the test images by time measurement built into the AI‐assisted CNN‐CAD system.
We also attempted to understand how the AI‐assisted CNN‐CAD system recognized the input images by applying a Gradient‐weighted Class Activation Map (Grad‐CAM 27 ) to determine which area of the images was most essential to the classification result. Grad‐CAM produces a coarse localization map highlighting important regions in the image for predicting the target concept, in our case, gastric cancer. We created the heatmap image from the localization map data.
Endoscopic assessment
All images were captured using high‐resolution endoscopes (GIF‐H260Z or GIF‐H290Z; Olympus, Tokyo, Japan) and standard endoscope video systems (EVIS LUCERA or EVIS LUCERA ELITE; Olympus). The video processor was always used with the structure enhancement function set at the B8 level for ME‐NBI and with the NBI color mode fixed at level 1. In all study patients, ME‐NBI with the maximal magnification using the water immersion technique was performed by three experienced endoscopists (K.M., H.U., and K.M.).
Statistical analysis
All statistical analyses were performed with EZR (Easy R; Saitama Medical Center, Jichi Medical University, Saitama, Japan), 28 which is a graphical user interface for R (The R Foundation for Statistical Computing, Vienna, Austria). Continuous data were compared using the Mann–Whitney U‐test. Categorical analysis of variables was performed using Fisher's exact test. A P value < 0.05 was considered to indicate a statistically significant difference.
Ethics
This study was reviewed and approved by the Institutional Review Board of Juntendo University School of Medicine (approval number: #18‐229). Patients were not required to give consent for the study because the analysis used anonymous clinical data that were obtained after each patient had agreed to treatment by verbal and documental consent. Individuals cannot be identified from the data presented.
Results
Patient and lesion characteristics in the training and test datasets
A total of 349 EGCs were enrolled. According to the time frame of the ME‐NBI procedure, the data obtained from these EGCs were divided into two sets, the training dataset (n = 267) and the test dataset (n = 82). The test dataset included data collected at a later time. Table 1 presents the patient and lesion characteristics used in the training and test datasets. Compared with the training dataset, the test dataset included significantly more cases of H. pylori‐negative gastric cancer (including uninfected and post‐eradication cases), those with smaller tumor diameters, and lesions in the lower (L) part of the stomach.
Table 1.
Patient and lesion characteristics in the training and test datasets
| Characteristics | Training dataset (n = 267) | Test dataset (n = 82) | P value |
|---|---|---|---|
| Male/female, n | 205/62 | 58/24 | 0.27 |
| Age, average (range), years | 72.3 (35–92) | 72.2 (52–87) | 0.99 |
| Tumor location, n, upper third/middle third/lower third | 30/114/123 | 11/17/54 | < 0.01 |
| Tumor size, average (range), mm | 15.8 (2–70) | 13.5 (4–50) | < 0.05 |
| Macroscopic types, n, 0‐I/0‐IIa/0‐IIb/0‐IIc/Type 0 mixed | 6/88/10/144/19 | 0/19/3/52/8 | 0.46 |
| Depth of tumor, n, T1a (mucosa)/T1b (submucosa) | 230/37 | 69/13 | 0.65 |
| Helicobacter pylori infection, n, positive/negative (eradication/uninfected) | 102/143 (135/8) | 21/53 (50/3) | < 0.05 |
0‐I, protruded; 0‐IIa, flatly elevated; 0‐IIb, flat; 0‐IIc, flatly depressed.
Vessel plus surface classification system findings of early gastric cancers in the training and test datasets
Table 2 presents the VSCS findings of EGC in the training and test datasets. There are no significant differences in diagnosis between the training and test datasets. There were 18 lesions that were not diagnosed endoscopically as cancer. Among them, 8 lesions, 7/267 (2.6%) in the training dataset and 1/82 (1.2%) in the test dataset, were falsely diagnosed as non‐cancer because a demarcation line was absent. The remaining 10 lesions, 6/267 (2.2%) in the training dataset and 4/82 (4.9%) in the test dataset, were falsely diagnosed as non‐cancer due to presence of regular microvascular pattern (MVP) and microsurface pattern (MSP), although a demarcation line was present.
Table 2.
Vessel plus surface classification system findings of early gastric cancer in the training and test datasets
| Training dataset (n = 267) | Test dataset (n = 82) | P value | |
|---|---|---|---|
| Demarcation line (positive/negative, %) | 260/7, 97.4% | 81/1, 98.8 | 0.75 |
| Microvascular pattern (regular/irregular/absent) | 13/238/16 | 5/73/4 | 0.98 |
| Microsurface pattern (regular/irregular/absent) | 14/205/48 | 8/60/14 | 0.49 |
| Diagnosis (cancer/non‐cancer, %) | 254/13, 95.1% | 77/5, 93.9% | 0.88 |
Performance of the artificial intelligence‐assisted convolutional neural network computer‐aided diagnosis system
The overall test speed was 38.3 images/s (0.026 s/image). The performance of the AI‐assisted CNN‐CAD system is shown in Table 3. The accuracy was 98.7%, with 2271 of the 2300 images being correctly diagnosed. The sensitivity, specificity, positive predictive value, negative predictive value, false‐positive rate, and false‐negative rate were 98% (1401/1430), 100% (870/870), 100% (1401/1401), 96.8% (870/899), 0% (0/870), and 2% (29/1430), respectively. When we evaluated the receiver‐operating characteristic curves for CNN trained by development images, the AUC was 99% (Fig. S2).
Table 3.
Diagnostic accuracy of the artificial intelligence‐assisted convolutional neural network computer‐aided diagnosis system
| Number of endoscopic images | Accuracy | Sensitivity | Specificity | PPV | NPV |
|---|---|---|---|---|---|
| EGC: 1430 | 98.7% | 98% | 100% | 100% | 96.8% |
| Non‐cancerous images: 870 | 2271/2300 | 1401/1430 | 870/870 | 1401/1401 | 870/899 |
EGC, early gastric cancer; NPV, negative predictive value; PPV, positive predictive value.
A total of 12 EGCs, represented by 29 ME‐NBI images, were misdiagnosed as non‐cancer. Six cases were difficult to distinguish from intestinal metaplasia or gastritis, even by experienced endoscopists (Table 4, Fig. 2a–c). Moreover, it appeared that lesions located in the middle or lower part of the stomach, of relatively small size and presenting as 0‐IIc, were found more frequently in the test dataset. In terms of the invasion depth, all cases were diagnosed as intramucosal carcinoma and most of them were H. pylori negative. Images from the other six cases were of insufficient quality to permit diagnosis because of bleeding, low‐power view, and being out of focus (Fig. 2d,e). The false‐positive images were not shown in the test dataset.
Table 4.
Characteristics of false‐negative cases
| Characteristics | 6 EGCs (18 ME‐NBI images) |
|---|---|
| Male/female, n | 2/4 |
| Age, average (range), years | 65.3 (44–75) |
| Tumor location, n, upper third/middle third/lower third | 1/3/2 |
| Tumor size, average (range), mm | 10.2 (3–32) |
| Macroscopic types, n, 0‐I/0‐IIa/0‐IIb/0‐IIc/Type 0 mixed | 0/0/0/5/1 (0‐IIb + IIc) |
| Depth of tumor, n, T1a (mucosa)/T1b (submucosa) | 6/0 |
| Helicobacter pylori infection, n, positive/negative (eradication/uninfected) | 1/5 (4/1) |
0‐I, protruded; 0‐IIa, flatly elevated; 0‐IIb, flat; 0‐IIc, flatly depressed.
EGC, early gastric cancer; ME‐NBI, magnifying endoscopy with narrow‐band imaging.
Figure 2.
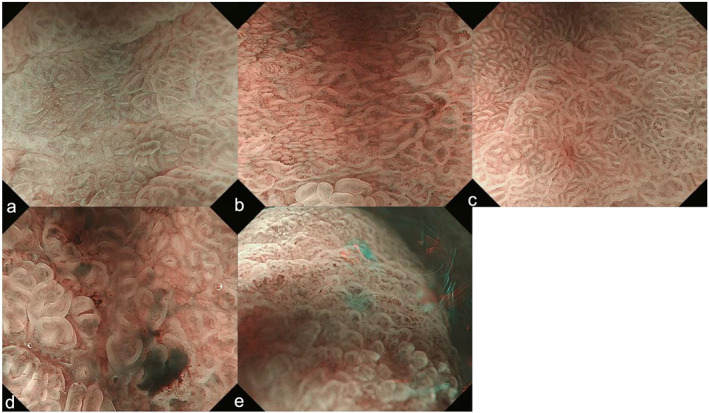
Misdiagnosed endoscopic images shown by the CNN. (a–c) False negatives: lesions appear as intestinal metaplasia or gastritis. (a) Differentiated‐type cancer (0‐IIc, tub1, after Hp eradication): regular MVP + regular MSP with a DL. (b) Differentiated‐type cancer (0‐IIc, tub2, after Hp eradication): regular MVP + regular MSP with a DL. (c) Differentiated‐type cancer (0‐IIa, tub1, after Hp eradication): regular MVP + regular MSP with a DL. (d) False negative: bleeding. (e) False negative: low‐power view and out of focus. 0‐IIc, flatly depressed; CNN, convolution neural network; DL, demarcation line; Hp, Helicobacter pylori; MSP, microsurface pattern; MVP, microvascular pattern; tub1, well differentiated adenocarcinoma; tub2, moderately differentiated adenocarcinoma.
Heatmaps are shown in Figure 3. In the EGC images, the regions that were determined as cancer by the AI‐assisted CNN‐CAD system are displayed in red and were consistent with those determined as cancer by the endoscopists. The images of gastric adenoma and those showing patchy redness were determined as non‐cancer by the AI‐assisted CNN‐CAD system and are not displayed in red on the heatmap. Again, these non‐cancer regions were consistent with those determined as non‐cancer by the endoscopists.
Figure 3.
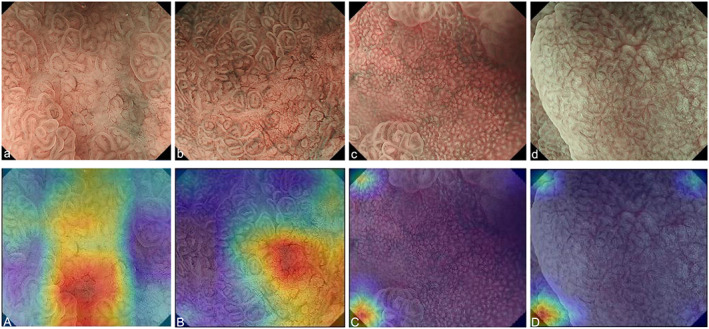
Heatmap of occlusion analysis. (a) Differentiated‐type cancer (0‐IIc, tub1). (b) Differentiated‐type cancer (0‐IIc, tub1). (c) Patchy redness. (d) Adenoma. (A–D) Heatmaps of each lesion. 0‐IIc, flatly depressed; tub1, well differentiated adenocarcinoma.
Discussion
Here, we developed an AI‐assisted CNN‐CAD system, based on a large number of ME‐NBI images, to diagnose EGC. The accuracy, sensitivity, specificity, positive predictive value, and negative predictive value of the AI‐assisted CNN‐CAD system were 98.7%, 98%, 100%, 100%, and 96.8%, respectively, and were especially high (Table 3). In addition, the AUC of our result was 99% (Fig. S2). To the best of our knowledge, the system developed in our study achieved the highest diagnostic accuracy among the AI‐assisted CNN‐CAD systems that diagnose EGCs, based on ME‐NBI images.
In the test dataset of 1430 images of gastric cancer cases, only 29 images from 12 cases were falsely diagnosed as non‐cancer by the AI‐assisted system, resulting in a false‐negative rate of 2%. There were no non‐cancer images falsely diagnosed as cancer by the AI‐assisted system, resulting in a false‐positive rate of 0%. Half of the false negatives were caused by the image quality being insufficient to permit diagnosis because of bleeding, low‐power view, and being out of focus, in which unanalyzable areas were sometimes misdiagnosed as non‐cancer (Fig. 2d,e). This can be corrected by DL about unanalyzable/low‐quality images and by introducing a new system applicable only to certain images suitable for AI diagnosis. Although most of the previously reported studies of AI diagnosis were retrospective, using only high‐quality still images, 29 , 30 , 31 , 32 DL of low‐quality images is expected to improve the accuracy achieved by AI diagnostic systems. It is certainly reasonable to expect an improvement in diagnostic accuracy using this approach. Nevertheless, because the AI‐assisted system developed here has already achieved an extremely high level of diagnostic accuracy, it seems more effective and practical to develop a method in which only high‐quality images are used, to obtain even greater diagnostic accuracy. Thus, it is considered necessary to introduce a new diagnostic system to use only high‐quality images suitable for diagnosis. The remaining false negatives were lesions difficult to distinguish from intestinal metaplasia or gastritis (Table 4, Fig. 2a–c). Of these six lesions, five were H. pylori negative, including four post‐H. pylori eradication and one uninfected gastric cancer. They were depressed lesions with a small tumor diameter and all were intramucosal carcinoma with features of gastric cancer of post‐H. pylori eradication status. All lesions falsely diagnosed as non‐cancer by the AI‐assisted system were also difficult for the endoscopists to diagnose correctly, and indeed, three of these six lesions were falsely diagnosed as non‐cancer by the endoscopists (Table 4, Fig. 2a–c). Thus, it is reasonable to speculate that the AI‐assisted system developed in our study had a diagnostic ability of virtually the same level as endoscopists (Table 2). Previously, we reported some cases that were diagnosed as non‐cancer according to the VSCS because of a low level of irregularity in internal MVP and MSP, despite the presence of a demarcation line. 22 The training dataset and the test dataset were divided according to the time frame in which they were collected and the test dataset included data collected at a later time. This difference in the data collection time might have contributed to the finding that H. pylori‐negative gastric cancer was significantly more frequent in the test dataset than the training dataset as the transition of H. pylori infection status in Japan, likely leading to a higher incidence of lesions diagnosed as non‐cancer by endoscopists, based on ME‐NBI data. In the future, this can be corrected by DL about cases that are difficult to diagnose and diagnostic limitations of EGCs, which will surely reduce false negatives and improve the negative predictive value significantly. Furthermore, we excluded ME‐NBI images of GAFG and diffuse‐type EGC to keep the quality of the AI‐assisted CNN‐CAD system, because some previous studies have reported that they should be dealt as the diagnostic limitations of ME‐NBI by VSCS. 3 , 33 However, we should evaluate the diagnostic ability of the AI‐assisted CNN‐CAD system for diagnostic limitation cases such as GAFG and diffuse‐type EGC for application to real clinical settings in future. In the test dataset, each EGC had more than 10 images (82 lesions, 1430 NBI‐ME images). The consistency of the AI diagnosis for different images related to one lesion was almost maintained. The average of inconsistency rate of the AI diagnosis was 10.9% (range 3–32.3%) in 12 false‐negative EGCs. We speculated that the inconsistency of the AI diagnosis was caused by the image quality and condition of focus or heterogeneity of MV and MS irregularity in one lesion according to the results of analyzing 12 false‐negative EGCs.
It was not clear what properties of images the AI‐assisted CNN‐CAD system developed in this study specifically evaluates; however, by using heatmaps, we were able to confirm which parts of the images the AI system focuses on (Fig. 3). We did not investigate heatmaps of all images; however, the regions evaluated by the AI‐assisted CNN‐CAD system appeared to be those with a high density of blood vessels with uneven diameters. Thus, it is speculated that the AI‐assisted system might put greater focus on regions presenting blood vessels of irregular shape. Because the VSCS also makes diagnoses focusing on MVP in principle, if the AI‐assisted CNN‐CAD system places importance on MVP, it is possible to infer that the AI‐assisted CNN‐CAD system may make diagnoses in a similar way to endoscopists.
As mentioned above, there are two reports about ME‐NBI diagnosis of gastric cancer using AI 20 , 21 and the AI‐assisted diagnostic system developed in our study demonstrated a higher diagnostic accuracy than was obtained in those studies. The most important difference was the ME‐NBI observation method employed in those studies. The water immersion technique with maximal magnification used in our study can eliminate halation so as to generate perfectly focused, clear, and sharp images of uniform quality, which are suitable for endoscopic diagnosis, and thus, these images are optimal for AI‐assisted diagnosis. Therefore, the AI‐assisted CNN‐CAD system demonstrated lower false‐negative detection rate and lower levels of variation in information volume and categories of imaging data, than the other available AI diagnostic systems. Moreover, the accuracy of analysis is believed to be high, because this study provided analysis of the diagnostic findings of EGC by the VSCS, comparison on heatmaps, evaluation of diagnosis using the VSCS by endoscopists, and evaluation of diagnostic accuracy.
This study has several limitations. First, this was an exploratory research for evaluating feasibility of the AI‐assisted CNN‐CAD system for ME‐NBI diagnosis of EGC. We will compare the diagnostic ability between the AI‐assisted CNN‐CAD system and endoscopists to assess the clinical usefulness as the next step. In addition, we will analyze the final AI diagnosis for each lesion instead of each image, because the endoscopic diagnosis of each lesion is usually made based on the interpretation of all images. Second, this was a single‐center retrospective study. However, we believe the results are reliable because verification was made prospectively. Third, we used only high‐quality endoscopic images for the training and test images. Fourth, the number of images used in this study was limited and more images should be collected. In particular, it is necessary to collect more images from which it is difficult to make a diagnosis by the VSCS, such as those of gastric cancer not infected with or post‐eradication of H. pylori. Furthermore, even though the false‐positive rate was 0%, non‐cancer images were scarce and more images from non‐cancer cases should be included. Fifth, non‐cancerous mucosa and lesions were subjected only to endoscopic diagnosis, not to biopsy. However, because the endoscopists in charge of image capturing are experienced physicians, any lesions suspected of cancer, even in the slightest, would have been subjected to biopsy examination. Because no such images were included in this study, there was considered to be little chance that images of cancer were mixed in by mistake. Thus, we believe the diagnostic quality of this study.
In conclusion, the AI‐assisted CNN‐CAD system demonstrated high diagnostic accuracy for ME‐NBI diagnosis of EGC with surprising efficiency. The AI‐assisted CNN‐CAD system may have great potential for future application to real clinical settings. For the first step, we analyzed only still ME‐NBI images. Therefore, we plan to demonstrate the usefulness of AI‐assisted CNN‐CAD system for medical videos, which could help to diagnose EGC during ME‐NBI in real time. We hope the AI‐assisted CNN‐CAD system will support us in ME‐NBI diagnosis of EGC in daily clinical practice in near future.
Supporting information
Table S1. Definition of evaluation criteria.
Figure S1. Training and test image sets.
Figure S2. Receiver operating characteristic curve for the test dataset.
Ueyama, H. , Kato, Y. , Akazawa, Y. , Yatagai, N. , Komori, H. , Takeda, T. , Matsumoto, K. , Ueda, K. , Matsumoto, K. , Hojo, M. , Yao, T. , Nagahara, A. , and Tada, T. (2021) Application of artificial intelligence using a convolutional neural network for diagnosis of early gastric cancer based on magnifying endoscopy with narrow‐band imaging. Journal of Gastroenterology and Hepatology, 36: 482–489. 10.1111/jgh.15190.
Declaration of conflict of interest: No author has a financial relationship relevant to this publication. Aoyama K and Kato Y are technical staff of AI Medical Service. Tada T is a shareholder of AI Medical Service.
Author contribution: Ueyama H, Nagahara A, and Tada T conceived and designed the study and wrote, edited, and reviewed the manuscript. Yao T performed all histopathological diagnosis. Akazawa Y, Yatagai N, Komori H, Takeda T, Matsumoto K, Ueda K, Matsumoto K, and Hojo M gathered ME‐NBI images and patients' clinical information. Kato Y provided valuable advice regarding the technical information and managed the AI‐assisted CNN‐CAD system and analyzed the data in this manuscript. All authors gave final approval for publication. Ueyama H, Nagahara A, and Tada T take full responsibility for the work as a whole, including the study design, access to data, and the decision to submit and publish the manuscript.
References
- 1. Hamashima C, Okamoto M, Shabana M et al. Sensitivity ofendoscopic screening for gastric cancer by the incidence method. Int. J. Cancer 2013; 133: 653–659. [DOI] [PubMed] [Google Scholar]
- 2. Gono K, Yamazaki K, Douguchi N et al. Endoscopic observation of tissue by narrow band illumination. Optical Review 2003; 10: 211–215. [Google Scholar]
- 3. Yao K, Anagnostopoulos GK, Ragunath K. Magnifying endoscopy for diagnosing and delineating early gastric cancer. Endoscopy 2009; 41: 462–467. [DOI] [PubMed] [Google Scholar]
- 4. Yao K, Iwashita A, Tanabe H et al. Novel zoom endoscopy technique for diagnosis of small flat gastric cancer: a prospective, blind study. Clin. Gastroenterol. Hepatol. 2007; 5: 869–878. [DOI] [PubMed] [Google Scholar]
- 5. Kato M, Kaise M, Yonezawa J et al. Magnifying endoscopy with narrow‐band imaging achieves superior accuracy in the differential diagnosis of superficial gastric lesions identified with white‐light endoscopy: a prospective study. Gastrointest. Endosc. 2010; 72: 523–529. [DOI] [PubMed] [Google Scholar]
- 6. Ezoe Y, Muto M, Uedo N et al. Magnifying narrowband imaging is more accurate than conventional white‐light imaging in diagnosis of gastric mucosal cancer. Gastroenterology 2011; 141: 2017–2025. [DOI] [PubMed] [Google Scholar]
- 7. Yao K, Doyama H, Gotoda T et al. Diagnostic performance and limitations of magnifying narrow‐band imaging in screening endoscopy of early gastric cancer: a prospective multicenter feasibility study. Gastric Cancer 2014; 17: 669–679. [DOI] [PubMed] [Google Scholar]
- 8. Yamada S, Doyama H, Yao K et al. An efficient diagnostic strategy for small, depressed early gastric cancer with magnifying narrow‐band imaging: a post‐hoc analysis of a prospective randomized controlled trial. Gastrointest. Endosc. 2014; 79: 55–63. [DOI] [PubMed] [Google Scholar]
- 9. Muto M, Yao K, Kaise M et al. Magnifying endoscopy simplediagnostic algorithm for early gastric cancer (MESDA‐G). Dig. Endosc. 2016; 28: 379–393. [DOI] [PubMed] [Google Scholar]
- 10. Nakanishi H, Doyama H, Ishikawa H et al. Evaluation of an e‐learning system for diagnosis of gastric lesions using magnifying narrow‐band imaging: a multicenter randomized controlled study. Endoscopy 2017; 49: 957–967. [DOI] [PubMed] [Google Scholar]
- 11. Esteva A, Kuprel B, Novoa RA et al. Dermatologist‐level classification of skin cancer with deep neural networks. Nature 2017; 542: 115–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ehteshami Bejnordi B, Veta M, Johannes van Diest P et al. Diagnostic assessment of deep learning algorithms for detection of lymph node metastases in women with breast cancer. JAMA 2017; 318: 2199–2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kodashima S, Fujishiro M, Takubo K et al. Ex vivo pilot study using computed analysis of endo‐cytoscopic images to differentiate normal and malignant squamous cell epithelia in the oesophagus. Dig. Liver Dis. 2007; 39: 762–766. [DOI] [PubMed] [Google Scholar]
- 14. Horie Y, Yoshio T, Aoyama K et al. Diagnostic outcomes of esophageal cancer by artificial intelligence using convolutional neural networks. Gastrointest. Endosc. 2019; 89: 25–32. [DOI] [PubMed] [Google Scholar]
- 15. Miyaki R, Yoshida S, Tanaka S et al. Quantitative identification of mucosal gastric cancer under magnifying endoscopy with flexible spectral imaging color enhancement. J. Gastroenterol. Hepatol. 2013; 28: 841–847. [DOI] [PubMed] [Google Scholar]
- 16. Kanesaka T, Lee TC, Uedo N et al. Computer‐aided diagnosis for identifying and delineating early gastric cancers in magnifying narrow‐band imaging. Gastrointest. Endosc. 2018; 87: 1339–1344. [DOI] [PubMed] [Google Scholar]
- 17. Hirasawa T, Aoyama K, Tanimoto T et al. Application of artificial intelligence using a convolutional neural network for detecting gastric cancer in endoscopic images. Gastric Cancer 2018; 21: 653–660. [DOI] [PubMed] [Google Scholar]
- 18. Shichijo S, Nomura S, Aoyama K et al. Application of convolutional neural networks in the diagnosis of Helicobacter pylori infection based on endoscopic images. EBioMedicine 2017; 25: 106–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nakashima H, Kawahira H, Kawachi H, Sakaki N. Artificial intelligence diagnosis of Helicobacter pylori infection using blue laser imaging‐bright and linked color imaging: a singlecenter prospective study. Ann. Gastroenterol. 2018; 31: 462–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Horiuchi Y, Aoyama K, Tokai Y et al. Convolutional neural network for differentiating gastric cancer from gastritis using magnified endoscopy with narrow band imaging. Dig. Dis. Sci. 2020; 65: 1355–1363. [DOI] [PubMed] [Google Scholar]
- 21. Li L, Chen Y, Shen Z et al. Convolutional neural network for the diagnosis of early gastric cancer based on magnifying narrow band imaging. Gastric Cancer 2020; 23: 126–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Akazawa Y, Ueyama H, Yao T et al. Usefulness of demarcation of differentiated‐type early gastric cancers after Helicobacter pylori eradication by magnifying endoscopy with narrow‐band imaging. Digestion 2018; 98: 175–184. [DOI] [PubMed] [Google Scholar]
- 23. Ueyama H, Yao T, Nakashima Y et al. Gastric adenocarcinoma of fundic gland type (chief cell predominant type) proposal for a new entity of gastric adenocarcinoma. Am. J. Surg. Pathol. 2010; 34: 609–619. [DOI] [PubMed] [Google Scholar]
- 24. Ueyama H, Matsumoto K, Nagahara A et al. Gastric adenocarcinoma of the fundic gland type (chief cell predominant type). Endoscopy 2014; 46: 153–157. [DOI] [PubMed] [Google Scholar]
- 25. Japanese Gastric Cancer Association . Japaneses classification of gastric carcinoma: 3rd English edition. Gastric Cancer 2011; 14: 101–112. [DOI] [PubMed] [Google Scholar]
- 26. He K, Zhang X, Ren S, Sun J, Deep residual learning for image recognition. arXiv:1512.03385.
- 27. Selvaraju RR, Cogswell M, Das A, Vedantam R, Parikh D, Batra D. Grad‐CAM: visual explanations from deep networks via gradient‐based localization. arXiv:1610.02391. 2017.
- 28. Kanda Y. Investigation of the freely available easy‐to‐use software “EZR” for medical statistics. Bone Marrow Transplant. 2013; 48: 452–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhu Y, Wang QC, Xu MD et al. Application of convolutional neural network in the diagnosis of the invasion depth of gastric cancer based on conventional endoscopy. Gastrointest. Endosc. 2019; 89: 806–815. [DOI] [PubMed] [Google Scholar]
- 30. Shichijo S, Endo Y, Aoyama K et al. Application of convolutional neural networks for evaluating Helicobacter pylori infection status on the basis of endoscopic images. Scand. J. Gastroenterol. 2018; 53: 940–946. 10.1080/00365521.2019.1577486. [DOI] [PubMed] [Google Scholar]
- 31. Takiyama H, Ozawa T, Ishihara S et al. Automatic anatomical classification of esophagogastroduodenoscopy images using deep convolutional neural networks. Scientific Reports 2018; 8: 7497. 10.1038/s41598-018-25842-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mori Y, Kudo SE, Wakamura K et al. Novel computer‐aided diagnostic system for colorectal lesions by using endocytoscopy (with videos). Gastrointest. Endosc. 2015; 81: 621–629. [DOI] [PubMed] [Google Scholar]
- 33. Matumoto K, Ueyama H, Yao K et al. Diagnostic limitations of magnifying endoscopy with narrow band imaging in early gastric cancer. Endosc Int Open. 2020. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Definition of evaluation criteria.
Figure S1. Training and test image sets.
Figure S2. Receiver operating characteristic curve for the test dataset.


