Summary
A few studies have holistically examined successive changes in coral holobionts in response to increased temperatures. Here, responses of the coral host Pocillopora damicornis, its Symbiodiniaceae symbionts, and associated bacteria to increased water temperatures were investigated. High temperatures induced bleaching, but no coral mortality was observed. Transcriptome analyses showed that P. damicornis responded more quickly to elevated temperatures than its algal symbionts. Numerous genes putatively associated with apoptosis, exocytosis, and autophagy were upregulated in P. damicornis, suggesting that Symbiodiniaceae can be eliminated or expelled through these mechanisms when P. damicornis experiences heat stress. Furthermore, apoptosis in P. damicornis is presumably induced through tumour necrosis factor and p53 signalling and caspase pathways. The relative abundances of several coral disease‐associated bacteria increased at 32°C, which may affect immune responses in heat‐stressed corals and potentially accelerates the loss of algal symbionts. Additionally, consistency of Symbiodiniaceae community structures under heat stress suggests non‐selective loss of Symbiodiniaceae. We propose that heat stress elicits interrelated response mechanisms in all parts of the coral holobiont.
Introduction
Coral reefs are threatened by global climate change, increasing sea surface temperatures and ocean acidification (Anthony, 2016). It is increasingly recognized that higher sea surface temperatures are the most immediate threat to coral reef survival (Hoegh‐Guldberg et al., 2007; Hughes et al., 2017) as they cause coral bleaching which can lead to coral morbidity and mortality and has substantially decimated coral reefs over the past two decades (Heron et al., 2016; Oliver et al., 2018). An additional adverse effect is the global increase in the prevalence of various coral diseases due to environmental changes (e.g., sea surface temperature increases) caused by anthropogenic activities (Sokolow, 2009).
Corals exhibit a range of cellular responses to heat stress, and the underlying molecular mechanisms have been studied in depth (Pinzón et al., 2015; Rosic et al., 2015; Brener‐Raffalli et al., 2018; Hou et al., 2018; van de Water et al., 2018). In particular, corals subjected to heat stress are known to upregulate expression of heat shock proteins (Lesser, 2011; Brener‐Raffalli et al., 2018; van de Water et al., 2018). Furthermore, over‐expression of p53 proteins in Montastraea faveolata after exposure to heat stress was shown to be associated with DNA damage and coral bleaching (Lesser and Farrell, 2004). Caspase activity, which is involved in apoptosis, was also found to increase in corals subjected to heat stress (Yu et al., 2017; Brener‐Raffalli et al., 2018; van de Water et al., 2018). In addition to apoptosis, cell necrosis was also observed to be induced by oxidative stress, which may have been caused by increased temperatures (Lesser, 2011). Moreover, multiple genes associated with immune responses in corals are known to be upregulated in response to heat stress (Rodriguez‐Lanetty et al., 2009; Barshis et al., 2013; Pinzón et al., 2015; Anderson et al., 2016; Davies et al., 2016; van de Water et al., 2018). These cellular responses to environmental stress are directly associated with coral health and viability.
Corals are associated with diverse microbiota comprising photosynthetic algae of the family Symbiodiniaceae, as well as bacteria, fungi, archaea, protists and viruses, which together with their coral host comprise the holobiont (Rohwer et al., 2002). Many coral‐associated microorganisms play important roles in a range of biological processes, and they are sensitive to environmental perturbation (Muscatine and Porter, 1977; Raina et al., 2009; Lema et al., 2012). The microbiome is critical to host fitness and viability (Bourne et al., 2016); for example, a symbiosis of corals and Symbiodiniaceae may be disrupted by heat stress, resulting in the expulsion of algal endosymbionts, leading to coral bleaching (Weis, 2008; Baird et al., 2009; Davy et al., 2012). Coral bleaching thus eliminates a major source of energy for the coral holobiont (Davy et al., 2012); however, coral bleaching may also exert protective or adaptive functions for the coral–algal symbiosis. Expulsion of algal endosymbionts may reduce oxidative stress in corals and may allow establishing new symbiotic relationships between coral and other Symbiodiniaceae species with a higher tolerance for increased temperatures (Jones et al., 2008; Sampayo et al., 2008; Baird et al., 2009; Obura, 2009; Cunning and Baker, 2013; Fujise et al., 2014). This concept is termed ‘adaptive bleaching hypothesis’ (Buddemeier and Fautin, 1993; Baker, 2003). Moreover, when exposed to increased temperatures, Symbiodiniaceae show substantial transcriptomic alterations, particularly regarding genes associated with photosynthesis, metabolism, antioxidant activity and immune responses, which may contribute to the disruption of symbiosis (Lesser, 2011; Rosic et al., 2015; Gierz et al., 2017) and may be exacerbated by heat stress‐induced pathogen invasion or viral infection (Levin et al., 2017; van de Water et al., 2018).
Structural changes in communities of coral‐associated bacteria due to heat stress have been observed frequently in previous studies (Ainsworth and Hoegh‐Guldberg, 2009; Littman et al., 2010, 2011; Lee et al., 2015; Nguyen‐Kim et al., 2015; Tout et al., 2015; Ziegler et al., 2017; Grottoli et al., 2018), and these changes in the coral‐associated bacteria may increase the abundance of potential pathogens (Littman et al., 2010, 2011; Nguyen‐Kim et al., 2015; Tout et al., 2015). Hence, a considerable abundance of pathogenic agents may help explain the collapse of a coral holobiont. In addition, coral‐associated bacterial communities seem to affect tolerance of the coral host to temperature stress (Littman et al., 2010; Ziegler et al., 2017; Grottoli et al., 2018; Epstein et al., 2019), suggesting that adaptation through changes in bacterial communities may constitute another possible mechanism to counteract adverse environmental effects (as described by the coral probiotic hypothesis; Reshef et al., 2006; Ziegler et al., 2017). Furthermore, putatively beneficial bacteria isolated from corals were shown in inoculating experiments to act as probiotics that can partially mitigate coral bleaching (Rosado et al., 2019). Taken together, these insights highlight the complexity and importance of comprehensive responses of the coral host, algal endosymbionts and coral‐associated bacteria to heat stress (Littman et al., 2010; Ziegler et al., 2017; van de Water et al., 2018).
Recent increases in frequency and severity of coral bleaching and other heat‐stress‐associated coral diseases prompted many studies to help understand the intricate mechanisms underlying these events (Bourne et al., 2016). It is crucial, however, to understand how each compartment of the coral holobiont responds to increased temperatures and which response interactions may occur between them. So far, most studies investigated the responses of coral host, algal symbionts and bacteria to environmental changes in isolation, and only a few studies attempted a holistic approach to assess effects of heat stress on the microbiome and gene expression patterns in corals (Leggat et al., 2011; Rosic et al., 2015; Lin et al., 2017; Grottoli et al., 2018; van de Water et al., 2018). In addition, successive changes in coral microbiome communities due to increasing temperature have rarely been studied (Lee et al., 2015). We therefore investigated the effects of increased temperatures on cell densities, community structures and transcriptome profiles of the Pocillopora damicornis coral holobiont, which is widely distributed in the Indo‐Pacific region (Veron, 1993). Uncontrolled environmental factors in in situ studies may confound results (Lee et al., 2015); therefore, responses of the coral holobiont to gradually increasing temperatures were examined under controlled laboratory conditions in the present study. We hypothesized that the coral host, Symbiodiniaceae and coral‐associated bacteria would respond in different ways to increasing seawater temperatures, and that their sequential responses would be interdependent. Investigating responses of coral holobionts to gradually increasing temperatures will help understand the dynamic relationships between corals and their associated microorganisms.
Results
Abundance of Symbiodiniaceae algae
Water temperature was increased from 25 to 32°C over a period of 16 days and was subsequently maintained at 32°C for 7 days. At the beginning of the experiment (T0; 25°C), the density of Symbiodiniaceae algae in P. damicornis was 3.13 ± 0.93 × 106 (mean ± SD) cells cm−2 and was decreased to 2.88 ± 1.47 × 105 cells cm−2 at the end of the experiment (T7; corals maintained at 32°C for 7 days; Fig. 1A). A slight decrease was observed in corals maintained at 25°C for 23 days (group Cf) with a final density of 1.68 ± 0.25 × 106 cells cm−2 (P = 0.198, Dunn–Bonferroni post hoc test after a Kruskal–Wallis test). Dunn–Bonferroni post hoc analyses executed after significant Kruskal–Wallis tests revealed that the strongest effect of temperature occurred at 31°C (T4, day 16; T0 vs. T4: P = 0.027) and at 32°C (T0 vs. T5 [corals maintained at 32°C for 2 days; day 18]: P = 0.014; T0 vs. T6 [corals maintained at 32°C for 5 days; day 21]: P = 0.002; T0 vs. T7 [corals maintained at 32°C for 7 days; day 23], P < 0.001). Although Symbiodiniaceae cell density was significantly reduced in response to heat stress, coral mortality was not observed during the experiment.
Fig 1.

(A) Density of Symbiodiniaceae cells in coral tissue. Numbers are shown as means ± standard deviations, which were calculated based on triplicate coral nubbins. Asterisks indicate significant differences at P < 0.05, compared with the initial 25°C treatment. B. Durusdinium subtype composition of each replicate. Bars indicate relative abundance of each subtype. Subtypes with relative abundance <1% are combined as ‘others’.
Symbiodiniaceae community changes
The SymTyper pipeline (Edmunds et al., 2014) was used to assign Symbiodiniaceae sequences of groups T0–T7 to the genus Durusdinium. Of these, 2 789 749 sequences were assigned to 14 sub‐clade types, and 10 393 sequences were considered ‘putatively novel’. These novel sequences clustered in eight de novo operational taxonomic units ([OTUs]; 97% similarity; Table S1). Durusdinium subtypes D110, D112 and D_I:0 were predominant across all samples and accounted for 94.85% of the Symbiodiniaceae sequences (Fig. 1B). Subtype D1a.3 was relatively abundant in one sample cultured at 27°C (relative abundance: 37.38%) and in one sample cultured at 29°C (relative abundance: 32.64%). In all other samples, the relative abundance of subtype D1a.3 was below 5.64% (Fig. 1B). All other previously described and novel subtypes constituted the ‘Symbiodiniaceae rare biosphere’ (Boulotte et al., 2016). Symbiodiniaceae community structure was relatively stable despite increasing temperatures (permutational multivariate analysis of variance [PERMANOVA]: F = 0.976, R 2 = 0.299; P = 0.517). Beta diversity dispersion did not differ significantly between groups (F = 1.288; P = 0.305).
Changes in bacterial community structure
After quality filtering, the remaining 870 582 sequences from 27 sequencing libraries were classified in 26 bacterial phyla. The most abundant coral‐associated bacterial phyla or classes (relative abundance >5% in at least one coral sample) were Actinobacteria, Bacteroidetes, Cyanobacteria, Firmicutes, Halanaerobiaeota, Alphaproteobacteria, Deltaproteobacteria, Gammaproteobacteria and Verrucomicrobia (Fig. 2A). Furthermore, we selected OTUs with significantly enriched (i.e., more abundant) or reduced relative abundance using a corrected threshold of significance (P < 0.05; Fig. 2B). When temperatures were increased from 25°C to 27–30°C, we found no enrichment or reduction of any OTU, compared with original communities; however, at 31°C, the relative abundance of one Ruegeria OTU increased 44‐fold and that of an Endozoicomonas OTU was reduced by half. After 5 days at 32°C, an OTU assigned to the taxon Terasakiellaceae was enriched. After 7 days at 32°C, more pronounced OTU changes were observed, including a reduction in relative abundance by more than 70% in five OTUs belonging to the taxa Clostridium (sensu stricto), Endozoicomonas (two OTUs), Nitrincolaceae and Terasakiellaceae, as well as over 28‐fold enrichment of OTUs including Alphaproteobacteria (Altererythrobacter, Ruegeria, Micavibrionaceae [two OTUs]), Dadabacteriales, Flavobacteriaceae (two OTUs), Gammaproteobacteria, Haliangium (Deltaproteobacteria), Phycisphaeraceae (two OTUs) and phylum WPS‐2.
Fig 2.
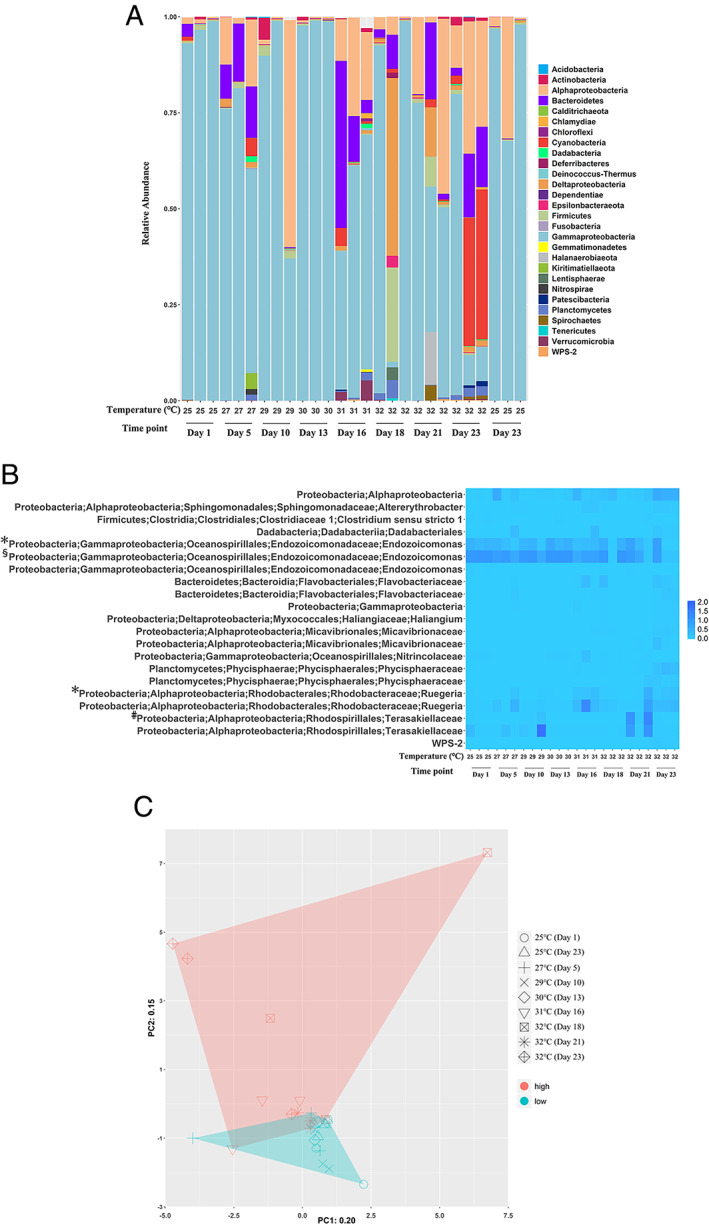
(A) Bacterial composition of each replicate. B. Bacterial OTUs with significant alterations in relative abundance due to the higher temperatures (≥31°C) in comparison with the initial 25°C treatment. Each cell represents a log10‐transformed relative abundance of each OTU. Before log10‐transformation, relative abundance was multiplied by one hundred, and then 1 was added to cope with zero in the data. OTUs without symbols appeared to be reduced/enriched after 7 days at 32°C; symbols indicate following results: *OTU was reduced/enriched at 31°C; #OTU was enriched after 5 days at 32°C; §For this OTU, before log10‐transformation, the relative abundance was multiplied by 10, and then 1 was added. C. Principal component analysis plot of bacterial community structures in each sample. Symbols indicate bacterial populations in each sample. Samples collected at 25°C, 27°C, 29°C and 30°C are shown in turquoise; samples collected at 31°C and 32°C are shown in red.
Overall, coral‐associated bacterial communities differed significantly between treatments at <31°C and ≥31°C, as revealed by a principal component analysis (PC1 and PC2 explained 20% and 15% of the variance respectively; Fig. 2C) and PERMANOVA (F = 4.290, R 2 = 0.146; P = 0.001). No significant differences in bacterial communities were observed among samples cultured at <31°C (F = 0.739, R 2 = 0.228; P = 0.877; PERMANOVA) or at ≥31°C (F = 1.454, R 2 = 0.353; P = 0.057; PERMANOVA). Beta diversity dispersion did not differ significantly between low‐temperature (<31°C) and high‐temperature (31–32°C) groups (F = 2.953; P = 0.101) or among high‐temperature (F = 0.334; P = 0.810) and low‐temperature groups (F = 1.426; P = 0.298). The results showed that coral‐associated bacterial communities were stable at 25–30°C, but not at temperatures of 31°C or higher.
Transcriptomic responses of the coral holobiont to heat stress
Samples collected from specimens cultured at 25°C (day 1), 29°C (day 10), 31°C (day 16) and 32°C for 7 days (day 23) were used for transcriptomic analyses. In total, 12 188 coral unigenes (Table S2) and 4460 Symbiodiniaceae unigenes (Table S3) were identified. In addition, 78 unigenes were considered to belong to both corals and Symbiodiniaceae. We observed 12.94%–19.12% dissimilarity in coral gene expression profiles between samples collected at each time point (Fig. S1a). Significant differences in coral gene expression levels between temperature treatments were observed using non‐metric multidimensional scaling and PERMANOVA analyses (Fig. S1b; F = 7.389, R 2 = 0.425; P = 0.001; PERMANOVA). Compared with culturing at 25°C (samples collected on the first day), differential expression was observed in 267 coral host genes (2.19%) at 29°C, in 1380 genes (11.32%) at 31°C and in 2911 genes (23.88%) at 32°C (T7, day 23; Fig. 3). Most differentially expressed genes (DEGs) were upregulated (Figs 3 and 4).
Fig 3.
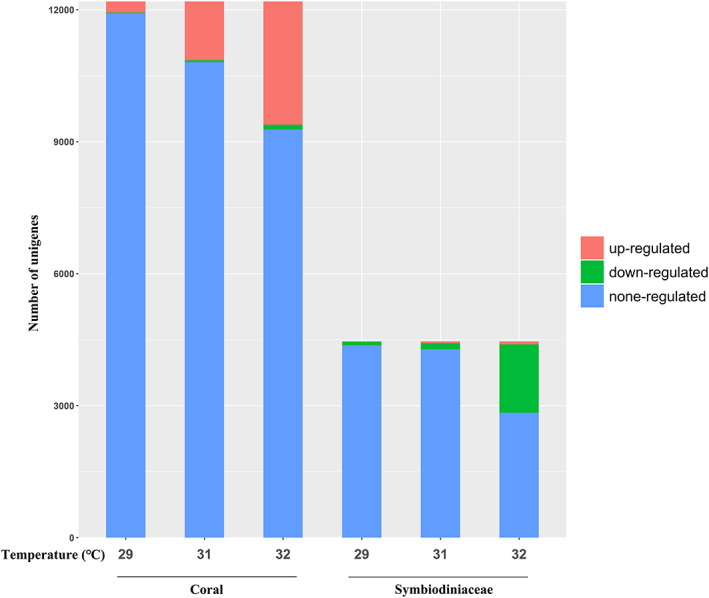
Differentially expressed coral and Symbiodiniaceae genes in response to elevated water temperatures, compared with the initial 25°C treatment.
Fig 4.
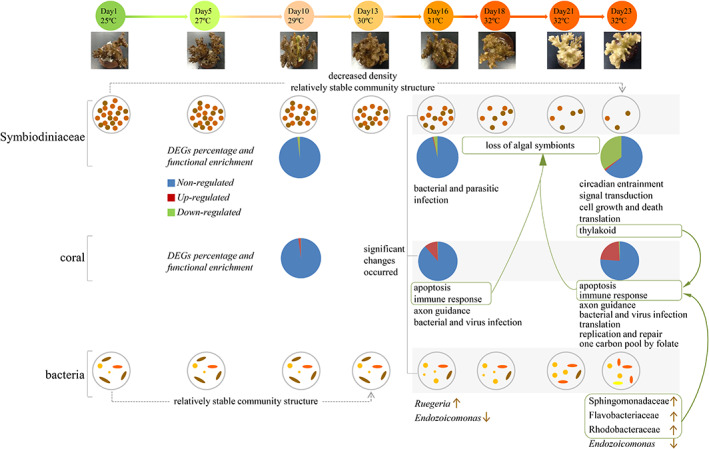
Timeline showing changes in the coral host, Symbiodiniaceae algae, and bacteria at different temperatures and their potential interactions. Symbiodiniaceae cell density was significantly decreased after water temperature was increased to 31°C, and community structures were stable. Only a few genes were differentially expressed in Symbiodiniaceae at 29°C and 31°C, compared with the initial 25°C treatment; in contrast, a higher number of DEGs was observed at 32°C. Most DEGs were downregulated in Symbiodiniaceae, and functions putatively associated with circadian entrainment, signal transduction, cell growth and death, translation, and photosynthesis were affected at 32°C. Damage to Symbiodiniaceae thylakoid caused by heat stress was likely involved in coral apoptosis (indicated by green wireframe and curved arrows). In total, 267 DEGs were observed in corals at 29°C, 1380 genes at 31°C, and 2911 genes at 32°C, compared coral genes in the initial 25°C treatment, and most were upregulated. Many over‐expressed genes at 31 and 32°C were associated with apoptosis and immune responses; they presumably contributed to the loss of algal symbionts (indicated by green wireframe and curved arrows). OTUs that were significantly enriched (more abundant) or reduced (P < 0.05) due to increasing water temperatures are indicated by ↑ (enriched) and ↓ (reduced). Enrichment of coral disease‐associated bacteria at 32°C, such as OTUs assigned to Sphingomonadaceae, Flavobacteriaceae and Rhodobacteraceae may elicit immune responses in heat‐stressed corals, which may then accelerate the loss of algal symbionts (indicated by green wireframe and curved arrows).
Symbiodiniaceae gene expression profiles showed 10.17%–17.02% dissimilarity between samples collected at each time point (Fig. S2a). Symbiodiniaceae genes were also significantly differentially expressed among groups (Fig. S2b; F = 14.591, R 2 = 0.593; P = 0.001; PERMANOVA). A small number of genes were differentially expressed at 29°C (88 genes; 1.97%) and at 31°C (179 genes; 4.01%), compared with the initial 25°C treatments, and 1620 genes (36.32%) were differentially expressed in Symbiodiniaceae algae at 32°C. In contrast to coral DEGs, most algal DEGs were downregulated (Figs 3 and 4).
Gene ontology and pathway functional enrichment in corals
Gene ontology (GO) enrichment results showed that hydrolase activity was significantly enriched, mainly due to upregulation of the respective coral genes at 29, 31 and 32°C (Table S4). In addition, corals cultured at 32°C showed significant differential responses in the GO categories ‘guanyl nucleotide binding’, ‘guanyl ribonucleotide binding’, ‘GTP binding’, ‘ion transmembrane transporter activity’, ‘regulation of RNA splicing’, ‘mitotic checkpoint complex’ and ‘bub1‐bub3 complex’. While not enriched in any GO term, genes related to heat shock proteins (Hsp40, Hsp70 and αB‐crystallin) were upregulated after the temperature increased to 29°C; however, the expression of Hsp70 was seen to reduce at 32°C.
No significant Kyoto Encyclopaedia of Genes and Genomes (KEGG) pathway enrichment was observed in coral DEGs at 29°C, compared with the samples collected at 25°C (day 1). At 31 and 32°C, several KEGG pathways were significantly enriched, predominantly in upregulated coral genes, including those assigned to the pathways ‘apoptosis’, ‘cytokine‐cytokine receptor interaction’, ‘bacterial and viral infection’, ‘p53 signalling pathway’, ‘transcriptional misregulation in cancer’ and ‘axon guidance’ (Table 1, Fig. S3a). In addition, DEGs associated with ‘one carbon pool by folate’, ‘central carbon metabolism in cancer’, ‘aminoacyl‐tRNA biosynthesis’, ‘non‐homologous end‐joining’ and ‘NOD‐like receptor signalling’ pathways were significantly enriched at 32°C, which occurred primarily in upregulated genes (Tables 1 and S5; Fig. S3b). DEGs annotated as NLRP3, APAF1, CASP3, CASP8, hspa2, ROCK1, ROCK2, TNFRSF16, TRAF1, ANKRD17, SAPK/JNK, NOD1, NOD2 and NLRC4 were involved in more than one pathway which were presumably associated with immune and stress responses (Fig. S3). Moreover, in pathways associated with transcriptional misregulation and central carbon metabolism in cancer, most DEGs were associated with immune responses or apoptosis (NLRP3, TNFRSF16 and TRAF1), oncogenesis (FGFR and RET) and regulation of cell proliferation (GTPase KRas; Table S5; Fig. S3).
Table 1.
Pathways significantly enriched in the DEGs (corrected P‐value ≤0.01).
| Pathway | Pathway ID | No. of DEGs at 31°C | No. of DEGs at 32°C | |
|---|---|---|---|---|
| Coral | ||||
| Downregulated | Axon guidance | ko04360 | 1 | – |
| Legionellosis | ko05134 | – | 8 | |
| Salmonella infection | ko05132 | – | 1 | |
| Virus infection | ko05164 | – | 5 a | |
| Transcriptional misregulation in cancer | ko05202 | – | 2 | |
| Central carbon metabolism in cancer | ko05230 | – | 2 | |
| NOD‐like receptor signalling pathway | ko04621 | – | 1 | |
| One carbon pool by folate | ko00670 | – | 2 | |
| Aminoacyl‐tRNA biosynthesis | ko00970 | – | 4 | |
| Non‐homologous end‐joining | ko03450 | – | 1 | |
| Upregulated | Axon guidance | ko04360 | 30 | 58 |
| Apoptosis ‐ multiple species | ko04215 | 20 | 27 | |
| p53 signalling pathway | ko04115 | 30 | 36 b | |
| Cytokine–cytokine receptor interaction | ko04060 | 18 | 25 | |
| Legionellosis | ko05134 | 25 | 38 | |
| Salmonella infection | ko05132 | 33 | 59 | |
| Virus infection | ko05164 | 40 | 60 a | |
| Transcriptional misregulation in cancer | ko05202 | 33 | 52 | |
| Central carbon metabolism in cancer | ko05230 | ‐ | 45 | |
| NOD‐like receptor signalling pathway | ko04621 | – | 92 | |
| One carbon pool by folate | ko00670 | – | 17 | |
| Aminoacyl‐tRNA biosynthesis | ko00970 | – | 16 | |
| Non‐homologous end‐joining | ko03450 | – | 11 | |
| Symbiodiniaceae | ||||
| Downregulated | Parasitic infection | ko05144 | 9 | 11 |
| Legionellosis | ko05134 | 3 | – | |
| Circadian entrainment | ko04713 | – | 42 | |
| Calcium signalling pathway | ko04020 | – | 56 | |
| cAMP signalling pathway | ko04024 | – | 45 | |
| Meiosis | ko04114 | – | 21 | |
| Cellular senescence | ko04218 | – | 20 | |
| RNA transport | ko03013 | – | 82 | |
| mRNA surveillance pathway | ko03015 | – | 66 | |
| Upregulated | Parasitic infection | ko05144 | 1 | – |
| Legionellosis | ko05134 | 6 | – | |
| Calcium signalling pathway | ko04020 | – | 1 | |
| cAMP signalling pathway | ko04024 | – | 8 | |
| Meiosis | ko04114 | – | 8 | |
| Cellular senescence | ko04218 | – | 6 | |
| RNA transport | ko03013 | – | 2 | |
| mRNA surveillance pathway | ko03015 | – | 6 |
This pathway was enriched in the DEGs at 32°C with corrected P‐value 0.02.
This pathway was enriched in the DEGs at 32°C with corrected P‐value 0.34.
GO and pathway functional enrichment in Symbiodiniaceae
Gene enrichment analysis based on GO annotations showed that catalytic activity, binding molecular functions and protein folding were affected by a temperature increase to 29°C (Table S4). Both upregulated Hsp70 and downregulated Hsp60 were involved in binding functions, and Hsp60 was also involved in protein folding. Several molecular functions associated with transferase, ligase and hydrolase activities were enriched in upregulated Symbiodiniaceae genes at 32°C, whereas translation, protein folding, and amide and peptide biosynthetic processes were primarily enriched in downregulated genes. Regarding cellular components, phosphatase complex, endoplasmic reticulum exit site, cytoplasm, thylakoid and ribosome were also significantly affected by the 32°C treatment (Table S4). Although the GO term ‘thylakoid’ was not enriched, the genes putatively annotated as thylakoid formation protein were significantly downregulated after water temperature increased to 29°C.
As in corals, no KEGG pathways were significantly enriched in Symbiodiniaceae DEGs at 29°C, compared with the samples collected at 25°C (day 1). While only pathways associated with bacterial and parasitic diseases were significantly enriched in DEGs at 31°C (Table 1; Fig. S4a), several pathways associated with circadian entrainment, calcium signalling pathway, cAMP signalling, cellular senescence, meiosis, RNA transport and messenger RNA (mRNA) surveillance were enriched at 32°C, predominantly in downregulated genes (Table 1; Fig. S4b). DEGs annotated as calmodulin‐like protein (CALML12), calcium channel subunit, calcium/calmodulin‐dependent protein kinase (CAMK2) and protein phosphatase 1 subunit were involved in multiple pathways associated with signal transduction, cell growth and death, and environmental adaptation (Table S6; Fig. S4).
Discussion
Individual responses of coral host, Symbiodiniaceae and bacteria to increased temperatures are crucial for the holobiont regarding effects of heat stress; however, also interactions between these responses are likely critical for acclimation and survival of corals exposed to temperature changes. In the current study, tripartite dynamic responses to increased temperatures were examined, and interactions between them were explored.
Deep sequencing of Symbiodiniaceae showed that only species in the genus Durusdinium were associated with P. damicornis corals collected from the Luhuitou fringing reef at Sanya. This was consistent with the results of a previous study that used PCR‐Restriction Fragment Length Polymorphism analysis of the 28S rRNA gene (Dong et al., 2008). In contrast, on Lord Howe Island, Taiwan, and in New Caledonia, P. damicornis was predominantly associated with the Symbiodiniaceae genus Cladocopium (formerly known as Clade C; Boulotte et al., 2016; Brener‐Raffalli et al., 2018). Pocillopora damicornis has been found to harbour more than one Symbiodiniaceae clade, and typically, one clade is dominant (Cunning et al., 2015; Boulotte et al., 2016; Brener‐Raffalli et al., 2018). These results support Pocillopora can establish endosymbiotic relationships with different Symbiodiniaceae species (Putnam et al., 2012).
The density of symbiotic algae cells decreased with increasing temperatures, however, no changes in Symbiodiniaceae community were observed. It should be noted that although Symbiodiniaceae densities did not differ significantly between the initial and the final 25°C treatments, a comparison of control (25°C) and heat‐stressed corals at the same time points is relevant. The results of the present study suggest that elimination or expulsion of algal symbionts from P. damicornis is a non‐selective process with respect to the taxon of symbiotic algae. However, further studies are required to investigate whether such selection depends on the physiological status of algal cells, for instance, the preferential expulsion of dividing, photosynthetically damaged or competent symbiotic algal cells from P. damicornis (Baghdasarian and Muscatine, 2000; Ladriere et al., 2008; Fujise et al., 2014). In previous studies, no changes in the dominance of Symbiodiniaceae taxa during bleaching were observed in experimental or natural settings (Bellantuono et al., 2012; Boulotte et al., 2016 respectively), whereas several switching events have been known to occur in the rare biosphere (Boulotte et al., 2016). No switching in species composition was detected in the present study, likely due to the experiment being conducted in a laboratory, which prevented the acquisition of new exogenous Symbiodiniaceae taxa.
In the present study, substantial changes in Symbiodiniaceae gene expression occurred at 32°C, whereas in corals, gradual changes were already observed at 29°C. These results suggest that at a transcriptional level, corals responded more quickly to increased seawater temperatures than Symbiodiniaceae (Fig. 4). Moreover, as the water temperature increased, Symbiodiniaceae DEGs predominantly showed downregulation, whereas DEGs of coral hosts were mostly upregulated at 29, 31 and 32°C. In Symbiodiniaceae, several genes associated with protein folding and thylakoid formation were downregulated when the temperature increased to 29°C, which suggested misfolding of proteins and damage to proteins and thylakoid caused by increased temperature. Meanwhile, hydrolase activity was increased in the coral host. These results suggest that photosystem and nutrient availability were affected by increased water temperature (Morris et al., 2019). Following this, many genes associated with translation, meiosis, signal transduction and circadian entrainment of Symbiodiniaceae were downregulated, whereas most genes associated with apoptosis, immunity, translation, and replication and repair were upregulated in corals. Growth and physiological activity appeared to be negatively affected in Symbiodiniaceae, whereas immune and stress responses in corals were enhanced in response to increasing water temperature. The functionally intact coral‐zooxanthella symbiosis is a controlled homeostatic state (Weis, 2008; Obura, 2009), and responses in Symbiodiniaceae and coral hosts suggest impaired homeostatic symbiosis under heat stress. The design of the current study may have introduced bias due to non‐synchronized sampling of treatments and controls; potential effects of such bias should be assessed in future studies to test comparability at different time points.
Previous studies proposed a model based on the stress response syndrome or the general adaptive mechanism (Obura, 2009). It was hypothesized that before bleaching, there is a controlled state in the coral‐zooxanthellae symbiosis (i.e., functioning symbiosis modulated by healthy host‐symbiont interactions) under low levels of stress maintained by various stress responses (Obura, 2009). Coral and Symbiodiniaceae algae genes that were putatively annotated, such as Hsp70 and αB‐crystallin which were upregulated at 29°C, may be involved in first‐order stress responses so as to keep the internal physiological stress below a threshold for bleaching (Downs et al., 2000; Obura, 2009); however, bleaching occurred under increased stress in the current study.
Transcriptional responses to heat stress offered some insight regarding mechanisms associated with bleaching in P. damicornis due to heat stress. We observed the effects on the thylakoid when the water temperature reached 29°C; as the temperature increased further, these effects became stronger. This probably led to the generation of reactive oxygen species (Weis, 2008). The Symbiodiniaceae DEGs identified in the present study did not include superoxide dismutase or ascorbate peroxidase; however, superoxide dismutase was over‐expressed in corals, suggesting oxidative stress due to increased temperatures which could cause P. damicornis bleaching through activation of immunity in corals (Weis, 2008; Lesser, 2011). Apoptosis and p53 signalling pathway‐associated genes were enriched in corals at 31 and 32°C, which contributed to the expulsion of algal symbionts (Gates et al., 1992; Douglas, 2003; Weis, 2008; Lesser, 2011); this is one possible explanation for the observed significant reduction in algal symbiont density after the water temperature increased to 31°C. In addition to oxidative stress induced by increased temperature, photosynthate may also be reduced due to damaged photosystems in Symbiodiniaceae; therefore, a nutrient reduction may also lead to coral bleaching through the photo‐oxidative pathway (Morris et al., 2019). Moreover, coral disease‐associated bacteria enriched at 32°C are possibly other potential elicitors of coral innate immunity, which may contribute to the elimination of symbiont algae (as discussed in detail below). Notably, genes putatively annotated as TNF receptors and TNF receptor‐associated factors showed significantly increased expression under heat stress. The respective gene products initiate signal transduction, which can result in caspase activation and apoptosis and eventually induce coral bleaching (Barshis et al., 2013; Quistad et al., 2014; Yuan et al., 2017). These results suggest that these genes are important regulators of apoptosis in P. damicornis under heat stress. In addition to apoptosis over‐expression of putative autophagy and exocytosis‐related proteins mainly was observed at 32°C, which provides evidence of the in situ degradation and exocytosis of symbionts from heat‐stressed P. damicornis (Weis, 2008). Previous studies showed that CAMK is expressed in various Symbiodiniaceae clades, and it may play an important role in aiding communication between eukaryotic cells and establishment of coral–algal symbiosis (Nagamune and Sibley, 2006; Rosic et al., 2015). Thus, downregulation of the genes annotated as CAMK in Symbiodiniaceae due to heat stress may contribute to the disintegration of coral–algal symbiosis, which may be involved in the bleaching cascade.
Algal symbiont density in corals decreased considerably when water temperature increased to ≥31°C, which also produced significant changes in bacterial community structure (Fig. 4). No significant changes in the relative abundance of OTUs were observed at lower temperatures (<31°C). Similar phenomena were previously observed in Acropora muricata (Lee et al., 2015). These results suggest complex interactions between coral bleaching and changes in coral‐associated bacterial communities. During heat stress, photosynthate translocation and metabolite profiles of corals and their symbionts are substantially altered (Sogin et al., 2016; Hillyer et al., 2017; Morris et al., 2019). In the current study, numerous genes presumably associated with hydrolase activity and proteolysis were up‐regulated in corals after water temperature increased to 29°C, which may contribute to increased catabolism of carbohydrates and amino acids (Hillyer et al., 2017). We suggest that the changes in bacterial community structure resulted from the physicochemical environmental perturbations in the coral microhabitats during bleaching. A large diversity of metabolic capabilities and regulatory pathways likely contribute to the enrichment of Ruegeria spp. when nutrients are limited during coral bleaching (Luo and Moran, 2014). Loss of Endozoicomonas from the P. damicornis bacterial community during heat stress may be associated with the depletion of products such as dimethylsulfopropionate (DMSP; Bourne et al., 2013; Correa et al., 2013) that are derived from photosymbionts, the abundance of which was substantially reduced. Moreover, Endozoicomonas is commonly associated with corals, likely as beneficial symbionts, which can regulate further bacterial colonization through the production of bioactive secondary metabolites or by competitive exclusion (Rua et al., 2014; Neave et al., 2016, 2017). Reduced abundance of this group would therefore also diminish its regulatory effect on the microbial community associated with the coral. Apart from nutrients, light and temperature in coral microenvironments are also likely to vary during coral bleaching (Fabricius, 2006; Wangpraseurt et al., 2017), which may also affect bacterial community structures. However, comprehensive understanding of bacterial responses to altered physicochemical environmental factors in coral microhabitats during bleaching requires comprehensive investigations of microenvironmental properties, metabolic networks and microbial consortia, particularly the abundance and activity of microbes, which are sensitive to oxygen content or which take part in carbon and nitrogen cycling and DMSP degradation (e.g., Endozoicomonas which were correlated with the presence of photosymbionts) (Bourne et al., 2013; Bourne and Webster, 2013; Morris et al., 2019).
Several bacterial taxa such as Sphingomonadaceae, Flavobacteriaceae and Rhodobacteraceae, which have been associated with coral diseases (Gignoux‐Wolfsohn and Vollmer, 2015; Rosales et al., 2019), were found to occur at relatively high abundance in corals at 32°C. This was probably related to the reduction of Endozoicomonas, the regulatory effect of which on coral‐associated microbes was diminished. Here, it should be mentioned that the reduction/augmentation of the relative abundance does not necessarily imply a decrease/increase in the number of bacterial cells within the coral holobiont. In addition, genes presumably associated with bacterial and viral infections were upregulated in corals, suggesting that increased temperatures cause pathogenic threats to corals. This may explain the higher prevalence of many coral diseases in the context of global warming (Willis et al., 2004; Bruno et al., 2007; Maynard et al., 2015). Furthermore, these alterations were probably associated with immune responses in P. damicornis as we observed that a large number of upregulated genes were presumably associated with the NOD‐like receptor signalling pathway at 32°C, which may perceive the presence of pathogens and induce apoptosis. Besides playing a crucial role in host innate immune responses to pathogens, apoptosis is also one of the mechanisms involved in algal symbiont elimination (Weis, 2008; Lesser, 2011). Therefore, our results suggest that immune responses elicited by microbiome alterations following heat stress, which may increase pathogen pressure, accelerate the loss of algal symbionts.
Among indicator bacteria that have been identified in heat‐tolerant corals by Ziegler et al. (2017), Phycisphaerales and Myxococcales were also found to be enriched in P. damicornis under heat stress in the present study. According to the previous report, these bacteria may contribute to coral resilience under heat stress (Ziegler et al., 2017). No mortality was observed in corals in the present study, despite bleaching induced by heat stress. Therefore, we suggest that the Phycisphaerales and Myxococcales bacteria in heat‐stressed corals may have contributed to the individual fitness of the bleached P. damicornis. Studying the transcriptional characteristics of bacteria in the future will facilitate a better understanding of their role in coral holobiont faced with heat stress.
Transcriptional misregulation and central carbon metabolism in cancer pathways were enriched in coral DEGs at 31 and 32°C, most of which were related to immune responses, apoptosis, oncogenesis and regulation of cell proliferation. Ten overexpressed genes were annotated as fibroblast growth factor receptor (FGFRs), which are involved in various biological processes, including cell proliferation, differentiation, survival, migration and apoptosis (Haugsten et al., 2010), and they play a critical role in cnidarian development (Matus et al., 2007; Turwankar and Ghaskadbi, 2019). Deregulation of FGFR signalling is implicated in several developmental syndromes and in many types of cancer (Haugsten et al., 2010), and it is associated with growth anomalies in Platygyra carnosa (Zhang et al., 2017). Additionally, the RET gene, which is associated with oncogenesis in mammals (Takahashi et al., 1985), was over‐expressed in P. damicornis under heat stress. Expression of RET protooncogenes is also significantly increased in heat‐stressed Acropora hyacinthus, presumably as a regulative function of apoptosis or other forms of cell death (Barshis et al., 2013). Changes in FGFR and RET expression were observed in heat‐stressed P. damicornis; however, their roles in thermal bleaching remain to be elucidated.
In response to elevated temperatures, corals showed earlier transcriptional responses than Symbiodiniaceae algae. During heat stress, algal symbionts of P. damicornis were probably eliminated by the coral host through mechanisms including apoptosis, autophagy and exocytosis. Moreover, apoptosis may be induced by thermal stress through the tumour necrosis factor and p53 signalling and caspase pathways in P. damicornis. Bacterial community alterations seemed to accompany substantial reductions in Symbiodiniaceae algae densities. In bacterial communities, a drastic reduction in the abundance of the potential symbiont Endozoicomonas might produce niches for other bacteria including coral disease‐associated taxa. Potential pathogens may cause immune responses in the coral host, which may also contribute to algal symbiont elimination. Consistent Symbiodiniaceae community structure suggests non‐selective elimination or expulsion of Symbiodiniaceae algae during P. damicornis bleaching. As no mortality was observed in the present study, we suggest that P. damicornis bleach to resist heat stress and that bleaching is likely regulated by responses of all three compartments of the holobiont. Based on the results of the present study, heat stress likely threatens coral health due to the interconnected responses of the coral host and its associated microbiome.
Experimental procedures
Heat stress experiment
Four P. damicornis colonies were collected from the Luhuitou fringing reef at Sanya, China (109°28′ E, 18°13′ N) in December 2015 by SCUBA diving at a depth of 3–5 m while maintaining a distance of 0.5 m between colonies. Seawater temperature was approximately 25–26°C, the average pH was 8.18 ± 0.01 and salinity was approximately 34‰. Coral samples were kept in seawater and were transported to the South China Sea Institute of Oceanology, Guangzhou, China. The four donor corals were divided into 72 nubbins (approximately 6–7 cm) which were then attached to ceramic reef discs. The nubbins were transferred to an indoor aquarium with circulating artificial seawater (25°C), which was prepared by mixing deionized water with Reef Crystals sea salt (Aquarium Systems). Corals were then allowed to acclimate for 15 days. After this, coral nubbins were removed from the acclimation tank and were randomly assigned to 12 10‐L tanks, with six nubbins per tank (Fig. 5). The tanks were supplied with artificial seawater at a flow rate of 300 ml min−1 from the same circulation pump and were illuminated with approximately 150 μmol photons m−2 s−1 in a 12:12‐h light/dark cycle.
Fig 5.
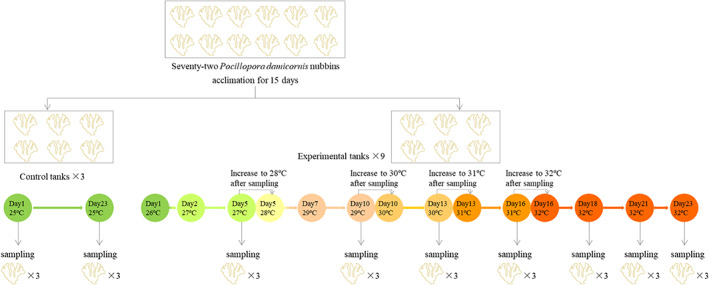
Heat‐stress experiment set‐up. Tanks contained artificial seawater that was circulated using the same pump.
To test effects of gradual heat stress on corals, the water temperature was increased gradually from 25 to 32°C in nine tanks and was maintained at 25°C in three control tanks (Fig. 5). We used 32°C as the maximum temperature because this was identified as the bleaching threshold temperature of corals on the Luhuitou reef (Li et al., 2008, 2012). In each of the nine experimental tanks, the temperature was increased to 26°C on the first day of the experiment, and to 27°C on the second day which was maintained for 3 days; it was increased to 28°C on the fifth day which was maintained for 2 days, and then increased to 29°C on the seventh day, and thereafter increased by 1°C every 3 days to 32°C. The temperature in experimental tanks was then maintained at 32°C for 7 days. The total experimental period was 23 days. The water temperature of each tank was regulated using a 25‐W submersible aquarium heater (EHEIM, GmbH & KG, Deizisau, Germany).
For collecting control samples, we selected a random nubbin from each control tank according to case randomization that was executed using SPSS, and nubbins were collected using sterile forceps. Samples were collected at the beginning (day 1, group T0; 25°C) and at the end (day 23, group Cf; 25°C) of the experiment. For sample collecting from experimental tanks, three tank numbers were randomly selected, and one random coral nubbin per tank was collected from treatments cultured at 27°C (group T1, day 5), 29°C (group T2, day 10), 30°C (group T3, day 13), 31°C (group T4, day 16), after 2 days at 32°C (group T5, day 18), after 5 days at 32°C (group T6, day 21) and after 7 days at 32°C (group T7, day 23) respectively. Corals from temperature treatments lasting 3 days (27, 29, 30 and 31°C) were sampled on the respective third day. Selected nubbins were removed from ceramic reef discs and were gently washed using filtered autoclaved artificial seawater, which was produced using Sea salts (Sigma‐Aldrich, USA). Subsequently, each nubbin was cut into three fragments (approximately 2 cm) using a sterilized bone cutter. One fragment of each nubbin was placed in a falcon tube on ice and was used to determine Symbiodiniaceae cell abundance within 30 min after sampling. The remaining fragments were immediately placed in 15‐ml falcon tubes containing sufficient RNAlater solution (QIAGEN GmbH, Hilden, Germany) to completely immerse the fragment. Samples were then stored at −80°C until DNA or RNA extraction.
Symbiodiniaceae density
Tissue was removed from the coral skeleton using 0.2‐μm‐filtered autoclaved artificial seawater that was sprayed using a syringe. Samples were vortexed to maintain micro‐algae in suspension, and Symbiodiniaceae cells in droplets of the resulting homogenized tissue suspension were counted using a Neubauer improved haemocytometer (Paul Marienfeld GmbH & KG, Lauda‐Königshofen, Germany). Ten replicate counts using 10 droplets of 10 μl each were performed per sample. The coral surface area was measured using the aluminium foil coating method (Marsh Jr., 1970), and the density of Symbiodiniaceae algae was calculated as an average number of cells cm−2 coral surface area. Normality and homogeneity of variance of Symbiodiniaceae algal density data were tested using SPSS Statistics Version 21. Variances were non‐equal, thus densities were compared using a Kruskal–Wallis test.
DNA and RNA extraction
Tubes containing coral fragments were placed on ice, samples were removed from RNAlater solution, and excess solution was removed using kimwipes. Coral tissue was removed from skeletons in conical flasks using a water pick and 0.2‐μm‐filtered autoclaved artificial seawater. Tissue suspensions were then centrifuged at 13 000g for 5 min, and DNA was extracted from the cell pellet using the MO BIO Power Soil DNA Kit (Mo Bio, Carlsbad, CA, USA), according to the manufacturer's instructions. Two replicates of blank extractions (i.e., without coral DNA) were produced simultaneously.
The remaining coral fragments were placed in bead tubes (RNA PowerSoil Total RNA Isolation Kit; Mo Bio) containing 5 ml QIAzol Lysis Reagent (QIAGEN Sciences, Valencia, CA, USA). The sample was then homogenized for 6 min using a Vortex‐Genie 2 (MO BIO), and RNA was extracted using the RNeasy Plus Universal Midi Kit (QIAGEN GmbH). Isolated RNA was dissolved in RNase‐free water and was stored at −80°C. RNA quality and quantity were assessed using electrophoretic profiling, performed on an Agilent 2100 BioAnalyzer (Agilent Technologies, Santa Clara, CA, USA).
16S rRNA gene amplification, sequencing and analysis
PCR was performed using two universal bacterial primers, 338F (5′‐ACTCCTACGGGAGGCAGC‐3′) and 806R (5′‐GGACTACVSGGGTATCTAAT‐3′) incorporated a 12‐bp barcode on the 5′ end unique to each sample. These primers were used to amplify the hypervariable V3–V4 region of the bacterial 16S rRNA gene. The 50‐μl PCR reaction mixture contained 25 μl TaKaRa Ex Taq Premix (TaKaRa Biotechnology [Dalian], China), 1 μl of each primer (10 μM) and 5 μl (10–20 ng) template DNA. Extraction blanks and sterilized deionized water were used as negative controls. The PCR cycling conditions were 95°C for 5 min, followed by 30 cycles of 94°C for 30 s, 53°C for 1 min, and 72°C for 1 min, and a final extension step at 72°C for 10 min. Each PCR was performed using three replicates. Product replicates of each sample were pooled, and amplicons were purified using the QIAquick Gel Extraction Kits (QIAGEN GmbH). Product quality was assessed using a NanoDrop spectrophotometer (Thermo Scientific, Vantaa, Finland), and sequencing was performed using an Illumina MiSeq platform with 2 × 300 bp paired‐end reads.
Raw reads were quality‐filtered using Trimmomatic (V0.33, Bolger et al., 2014), and filtered paired‐end reads were merged using FLASH v1.2.11, (Magoč and Salzberg, 2011) with a minimum overlap of 10 nucleotides. Maximum allowed error rate in the overlap region was 0.2. The resulting sequences were grouped by barcode using QIIME v1.9.1 (Caporaso et al., 2010) and were clustered as OTUs at 97% sequence identity using UCLUST (Edgar, 2010). After clustering, singleton OTUs were removed using usearch v10.0.240 (Edgar, 2010), and chimeras were detected and removed using the UCHIME v4.2.40 de novo algorithm (Edgar et al., 2011). OTUs were taxonomically identified using the SILVA_132 database and VSEARCH global alignments (Rognes et al., 2016). Non‐target OTUs such as mitochondria, chloroplasts, archaea, eukaryotes, and unidentified sequences, and OTUs with abundances below 0.005% (minimum number of representative sequences, Bokulich et al., 2013) were removed.
Internal transcribed spacer 2 fragment amplification, sequencing, and analysis
The internal transcribed spacer 2 region of Symbiodiniaceae nuclear ribosomal DNA was PCR‐amplified using specific primers (forward: 5′‐GTGAATTGCAGAACTCCGTG‐3′; reverse: 5′‐CCTCCGCTTACTTATATGCTT‐3′ contained a 12‐bp barcode on the 5′ end unique to each sample; Boulotte et al., 2016). The 50‐μl PCR mixtures contained 25 μl TaKaRa Ex Taq Premix, 1 μl of each primer (10 μM) and 5 μl (10–20 ng) template DNA. PCR cycling conditions were 95°C for 5 min, followed by 30 cycles of 95°C for 30 s, 52°C for 40 s, and 72°C for 45 s and a final extension step of 72°C for 5 min. PCRs were performed in triplicates, and products were pooled and purified using the QIAquick Gel Extraction Kits (QIAGEN). Quality of purified PCR products was assessed using a NanoDrop spectrophotometer (Thermo Scientific). Sequencing was performed on an Illumina HiSeq 2500 platform with 2 × 250 bp paired‐end reads.
Raw sequencing reads were quality‐filtered, merged and demultiplexed as described above. For taxonomic classification, the Symbiodiniaceae‐specific bioinformatics pipeline SymTyper was used with a reference database comprising 719 Symbiodiniaceae sequences (Dryad data archive, doi: 10.5061/dryad.32md8). Sequences were assigned to Symbiodiniaceae clades using a Hidden Markov Model in SymTyper, with a threshold of e‐value 10−20 and a ratio of 10−5 relative to the next best clade hit. Sequences classified in clades were then assigned to subtypes according to Cunning et al. (2015). Sequences not assigned to a subtype were clustered at 97% similarity (Arif et al., 2014) using the ‘uclust’ algorithm of QIIME. Symbiodiniaceae subtypes with abundances below 0.005% were removed (Bokulich et al., 2013). Leaf taxa closest to internal nodes and BLAST hits closest to the de novo clusters are shown in Table S1. Read counts of both known Symbiodiniaceae taxa and newly designated clusters were used for diversity analyses.
RNA sequencing and data analyses
According to observed changes in Symbiodiniaceae density and in the bacterial community, samples collected at 25°C (day 1), 29°C (day 10), 31°C (day 16) and 32°C for 7 days (day 23) were used for transcriptomic analyses. Eukaryotic mRNA was isolated from total RNA, which had been treated with DNase I, using magnetic bead‐labelled oligo(dT)s. RNA sequencing libraries were prepared and were sequenced on an Illumina HiSeq 4000 platform using a commercial service (BGI, Shenzhen, China). Approximately 120.57 Gb sequencing data were generated, and after quality filtration, Trinity v2.0.6 (Grabherr et al., 2011) was used to de novo assemble clean reads. Tgicl v2.0.6 (Pertea et al., 2003) was used to cluster transcripts in unigenes.
Unigenes were annotated using databases including the nucleotide sequence database, the non‐redundant protein sequence database (NR), clusters of orthologous groups, KEGG and SWISSPROT using Blast v2.2.23 (Altschul et al., 1990). Blast2GO v2.5.0 (Conesa et al., 2005) was used to produce GO annotations based on NR annotation. To identify coral and Symbiodiniaceae sequences, unigenes were aligned with the mRNA sequences of Acropora digitifera (Shinzato et al., 2011) and Breviolum minutum (formerly Symbiodinium minutum; Shoguchi et al., 2013) using BLAST, with an e‐value threshold of 1−5.
Clean reads were mapped to unigenes using Bowtie2 v2.2.5 (Langmead and Salzberg, 2012), and gene expression levels in each sample were calculated using RSEM v1.2.12 (Li and Dewey, 2011). Based on unigene expression results, differential gene expression between groups was assessed using DEseq2 (Love et al., 2014) with a negative binomial distribution. Genes were considered significantly differentially expressed when the fold change was ≥2, and the adjusted P‐value was ≤0.05. GO and KEGG pathway functional enrichments of DEGs were determined using phyper in R; P‐values were corrected using the false discovery rate. DEG functions were considered significantly enriched when corrected P‐values were ≤0.01.
Statistical analyses
To all read counts, 1 was added to cope with zero in the data for the centred log‐ratio (clr) transformation (Gloor et al., 2017). Read counts were transformed using the clr transformation by using the R package compositions (van den Boogaart and Tolosana‐Delgado, 2008), and then Aitchison distances were calculated using the robCompositions package (Templ et al., 2011). A principal component analysis of Aitchison distances was performed using the R package vegan (Dixon, 2003). Symbiodiniaceae cell density was significantly reduced after water temperature was increased to 31°C, providing a physiological sign of coral bleaching (Siebeck et al., 2006). Using this threshold, samples were grouped in ‘low temperature’ (<31°C) and ‘high temperature’ (≥31°C) groups. Differences between microbial community structures were tested using PERMANOVA with 999 permutations (Clarke, 1993) in vegan, based on Aitchison distances. Group dispersions were calculated based on Aitchison distances using the betadisper function of the vegan package. Differential taxa between temperatures were determined using clr‐transformed data and the aldex.glm function of ALDEx2 (Gloor et al., 2017).
To compare gene expression profiles of corals and Symbiodiniaceae, a Bray–Curtis distance matrix was produced from the gene expression matrix (in fragments per kilobase of transcript per million mapped reads), and a non‐metric multidimensional scaling profile was generated. Differences in transcriptomes of corals and Symbiodiniaceae among temperature treatments were tested using PERMANOVA with 999 permutations (Clarke, 1993).
Author Contributions
J.L., L.L., and S.Z. designed this study. J.L. and L.L. acquired and analysed the data. Y.Z. analysed the data. J.L. and Y.Z. drew graphs. J.L. drafted the manuscript, and all authors commented on the manuscript and made suggestions.
Supporting information
Figure S1. (a) Dissimilarity in coral gene expression profiles between samples collected at each time point. (b) Non‐metric multidimensional scaling (nMDS) plot representing the differences in the overall gene expression patterns in corals exposed to elevated water temperatures.
Figure S2. (a) Dissimilarity in Symbiodiniaceae gene expression profiles between samples collected at each time point. (b) Non‐metric multidimensional scaling (nMDS) plot representing the differences in the overall gene expression patterns in Symbiodiniaceae endosymbionts exposed to elevated water temperatures.
Figure S3. Heatmap of differentially expressed genes involved in the enriched pathways in the coral Pocillopora damicornis following exposure to 31°C (a) and 32°C (b). Each cell represents the log2‐fold change value of each gene. The dot shows the gene involved in a certain pathway.
Figure S4. Heatmap of differentially expressed genes involved in the enriched pathways in Symbiodiniaceae following exposure to 31°C (a) and 32°C (b). Each cell represents the log2‐fold change value of each gene. The dot shows the gene involved in a certain pathway.
Table S1. Internal node and de novo taxa among reference sequences of Symbiodiniaceae. The closest leaf taxon and distance to that taxon are given for internal nodes (‘D_I:X’). The closest BLAST hit from the reference database and associated e‐value are given for de novo taxa (‘D_d:X’).
Table S2. Coral gene expression profiles under different temperatures. Data in columns 7 to 18 are FPKM (Fragments per kilobase Million) for each gene at 25, 29, 31 and 32°C.
Table S3. Algal symbiont gene expression profiles under different temperatures. Data in columns 7 to 18 are FPKM (Fragments per Kilobase Million) for each gene at 25, 29, 31 and 32°C.
Table S4. Gene ontology terms significantly (with corrected P‐value ≤0.01) enriched in the differentially expressed genes in the coral Pocillopora damicornis and Symbiodiniaceae following exposure to elevated seawater temperatures.
Table S5. Kyoto Encyclopaedia of Genes and Genomes (KEGG) pathways significantly (with corrected P‐value ≤0.01) enriched in the differentially expressed genes in the coral Pocillopora damicornis following exposure to elevated seawater temperatures.
Table S6. Kyoto Encyclopaedia of Genes and Genomes (KEGG) pathways significantly (with corrected P‐value ≤0.01) enriched in the differentially expressed genes in Symbiodiniaceae following exposure to elevated seawater temperatures.
Acknowledgements
We thank the anonymous reviewers for their insightful comments that improved our manuscript. Prof. Junde Dong is gratefully thanked for his help in the coral sampling. We are grateful to Luping Zhang for providing help in aquarium system maintenance. We acknowledge Dr. Yanyan Zhou for the fruitful discussions to improve this manuscript. This work was supported by the National Natural Science Foundation of China (Nos. 41676155 and 41890853), Guangdong Natural Science Funds for Distinguished Young Scholars (2017A030306025), the National Key R&D Program of China (2017YFC0506303), and the Youth Innovation Promotion Association of the Chinese Academy of Sciences (No. 2017396).
Data Availability Statement
The datasets generated during the current study have been deposited and are publicly available in the Sequence Read Archive repository under BioProject ID PRJNA478260 and PRJNA559028.
References
- Ainsworth, T.D. , and Hoegh‐Guldberg, O. (2009) Bacterial communities closely associated with coral tissues vary under experimental and natural reef conditions and thermal stress. Aquat Biol 4: 289–296. [Google Scholar]
- Altschul, S.F. , Gish, W. , Miller, W. , Myers, E.W. , and Lipman, D.J. (1990) Basic local alignment search tool. J Mol Biol 215: 403–410. [DOI] [PubMed] [Google Scholar]
- Anderson, D.A. , Walz, M.E. , Weil, E. , Tonellato, P. , and Smith, M.C. (2016) RNA‐Seq of the Caribbean reef‐building coral Orbicella faveolata (Scleractinia‐Merulinidae) under bleaching and disease stress expands models of coral innate immunity. Peer J 4: e1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthony, K.R.N. (2016) Coral reefs under climate change and ocean acidification: challenges and opportunities for management and policy. Annu Rev Env Resour 41: 59–81. [Google Scholar]
- Arif, C. , Daniels, C. , Bayer, T. , Banguera‐Hinestroza, E. , Barbrook, A. , Howe, C.J. , et al. (2014) Assessing Symbiodinium diversity in scleractinian corals via next‐generation sequencing‐based genotyping of the ITS2 rDNA region. Mol Ecol 23: 4418–4433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baghdasarian, G. , and Muscatine, L. (2000) Preferential expulsion of dividing algal cells as a mechanism for regulating algal‐cnidarian symbiosis. Biol Bull 199: 278–286. [DOI] [PubMed] [Google Scholar]
- Baird, A.H. , Bhagooli, R. , Ralph, P.J. , and Takahashi, S. (2009) Coral bleaching: the role of the host. Trends Ecol Evol 24: 16–20. [DOI] [PubMed] [Google Scholar]
- Baker, A.C. (2003) Flexibility and specificity in coral‐algal symbiosis: diversity, ecology and biogeography. Annu Rev Ecol Evol Syst 34: 661–689. [Google Scholar]
- Barshis, D.J. , Ladner, J.T. , Oliver, T.A. , Seneca, F.O. , Traylor‐Knowles, N. , and Palumbi, S.R. (2013) Genomic basis for coral resilience to climate change. Proc Natl Acad Sci USA 110: 1387–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellantuono, A.J. , Hoegh‐Guldberg, O. , and Rodriguez‐Lanetty, M. (2012) Resistance to thermal stress in corals without changes in symbiont composition. P R Soc Lond B Biol Sci 279: 1100–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokulich, N.A. , Subramanian, S. , Faith, J.J. , Gevers, D. , Gordon, J.I. , Knight, R. , et al. (2013) Quality‐filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat Methods 10: 57–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger, A.M. , Lohse, M. , and Usadel, B. (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulotte, N.M. , Dalton, S.J. , Carroll, A.G. , Harrison, P.L. , Putnam, H.M. , Peplow, L.M. , and van Oppen, M.J. (2016) Exploring the Symbiodinium rare biosphere provides evidence for symbiont switching in reef‐building corals. ISME J 10: 2693–2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourne, D.G. , Dennis, P.G. , Uthicke, S. , Soo, R.M. , Tyson, G.W. , and Webster, N. (2013) Coral reef invertebrate microbiomes correlate with the presence of photosymbionts. ISME J 7: 1452–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourne, D.G. , Morrow, K.M. , and Webster, N.S. (2016) Insights into the coral microbiome: underpinning the health and resilience of reef ecosystems. Annu Rev Microbiol 70: 317–340. [DOI] [PubMed] [Google Scholar]
- Bourne, D.G. , and Webster, N.S. (2013) Coral reef bacterial communities. In The Prokaryotes–Prokaryotic Communities and Ecophysiology, Rosenberg, E. , DeLong, E.F. , Lory, S. , Stackebrandt, E. , and Thompson, F. (eds). Heidelberg: Springer, pp. 163–187. [Google Scholar]
- Brener‐Raffalli, K. , Clerissi, C. , Vidal‐Dupiol, J. , Adjeroud, M. , Bonhomme, F. , Pratlong, M. , et al. (2018) Thermal regime and host clade, rather than geography, drive Symbiodinium and bacterial assemblages in the scleractinian coral Pocillopora damicornis sensu lato . Microbiome 6: 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruno, J.F. , Selig, E.R. , Casey, K.S. , Page, C.A. , Willis, B.L. , Harvell, C.D. , et al. (2007) Thermal stress and coral cover as drivers of coral disease outbreaks. PLoS Biol 5: e124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buddemeier, R.W. , and Fautin, D.G. (1993) Coral bleaching as an adaptive mechanism–a testable hypothesis. Bioscience 43: 320–326. [Google Scholar]
- Caporaso, J.G. , Kuczynski, J. , Stombaugh, J. , Bittinger, K. , Bushman, F.D. , Costello, E.K. , et al. (2010) QIIME allows analysis of high‐throughput community sequencing data. Nat Methods 7: 335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke, K.R. (1993) Non‐parametric multivariate analyses of changes in community structure. Aust J Ecol 18: 117–143. [Google Scholar]
- Conesa, A. , Götz, S. , García‐Gómez, J.M. , Terol, J. , Talón, M. , and Robles, M. (2005) Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21: 3674–3676. [DOI] [PubMed] [Google Scholar]
- Correa, H. , Haltli, B. , Duque, C. , and Kerr, R. (2013) Bacterial communities of the gorgonian octocoral Pseudopterogorgia elisabethae . Microb Ecol 66: 972–985. [DOI] [PubMed] [Google Scholar]
- Cunning, R. , and Baker, A.C. (2013) Excess algal symbionts increase the susceptibility of reef corals to bleaching. Nat Clim Change 3: 259–262. [Google Scholar]
- Cunning, R. , Yost, D.M. , Guarinello, M.L. , Putnam, H.M. , and Gates, R.D. (2015) Variability of Symbiodinium communities in waters, sediments, and corals of thermally distinct reef pools in American Samoa. PLoS One 10: e0145099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies, S.W. , Marchetti, A. , Ries, J.B. , and Castillo, K.D. (2016) Thermal and pCO2 stress elicit divergent transcriptomic responses in a resilient coral. Front Mar Sci 3: 112. [Google Scholar]
- Davy, S.K. , Allemand, D. , and Weis, V.M. (2012) Cell biology of cnidarian‐dinoflagellate symbiosis. Microbiol Mol Biol Rev 76: 229–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon, P. (2003) VEGAN, a package of R functions for community ecology. J Veg Sci 14: 927–930. [Google Scholar]
- Dong, Z. , Huang, H. , Huang, L. , Li, Y. , and Li, X. (2008) PCR‐RFLP analysis of large subunit rDNA of symbiotic dinoflagellates in scleractinian corals from Luhuitou fringing reef of Sanya, Hainan. Biodivers Sci 16: 498–502 (In Chinese). [Google Scholar]
- Douglas, A.E. (2003) Coral bleaching–how and why? Mar Pollut Bull 46: 385–392. [DOI] [PubMed] [Google Scholar]
- Downs, C.A. , Mueller, E. , Phillips, S. , Fauth, J.E. , and Woodley, C.M. (2000) A molecular biomarker system for assessing the health of coral (Montastraea faveolata) during heat stress. Marine Biotechnol 2: 533–544. [DOI] [PubMed] [Google Scholar]
- Edgar, R.C. (2010) Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26: 2460–2461. [DOI] [PubMed] [Google Scholar]
- Edgar, R.C. , Haas, B.J. , Clemente, J.C. , Quince, C. , and Knight, R. (2011) UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27: 2194–2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmunds, P.J. , Pochon, X. , Levitan, D.R. , Yost, D.M. , Belcaid, M. , Putnam, H.M. , and Gates, R.D. (2014) Long‐term changes in Symbiodinium communities in Orbicella annularis in St. John, US Virgin Islands. Mar Ecol‐Prog Ser 506: 129–144. [Google Scholar]
- Epstein, H.E. , Torda, G. , and van Oppen, M.J.H. (2019) Relative stability of the Pocillopora acuta microbiome throughout a thermal stress event. Coral Reefs 38: 373–386. [Google Scholar]
- Fabricius, K.E. (2006) Effects of irradiance, flow, and colony pigmentation on the temperature microenvironment around corals: implications for coral bleaching? Limnol Oceanogr 51: 30–37. [Google Scholar]
- Fujise, L. , Yamashita, H. , Suzuki, G. , Sasaki, K. , Liao, L.M. , and Koike, K. (2014) Moderate thermal stress causes active and immediate expulsion of photosynthetically damaged zooxanthellae (Symbiodinium) from corals. PLoS One 9: e114321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gates, R.D. , Baghdasarian, G. , and Muscatine, L. (1992) Temperature stress causes host cell detachment in symbiotic cnidarians: implications for coral bleaching. Biol Bull 182: 324–332. [DOI] [PubMed] [Google Scholar]
- Gierz, S.L. , Forêt, S. , and Leggat, W. (2017) Transcriptomic analysis of thermally stressed symbiodinium reveals differential expression of stress and metabolism genes. Front Plant Sci 8: 271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gignoux‐Wolfsohn, S.A. , and Vollmer, S.V. (2015) Identification of candidate coral pathogens on White band disease‐infected staghorn coral. PLoS One 10: e0134416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gloor, G.B. , Macklaim, J.M. , Pawlowsky‐Glahn, V. , and Egozcue, J.J. (2017) Microbiome datasets are compositional: and this is not optional. Front Microbiol 8: 2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr, M.G. , Haas, B.J. , Yassour, M. , Levin, J.Z. , Thompson, D.A. , Amit, I. , et al. (2011) Full‐length transcriptome assembly from RNA‐Seq data without a reference genome. Nat Biotechnol 29: 644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grottoli, A.G. , Martins, P.D. , Wilkins, M.J. , Johnston, M.D. , Warner, M.E. , Cai, W.J. , et al. (2018) Coral physiology and microbiome dynamics under combined warming and ocean acidification. PLoS One 13: e0191156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haugsten, E.M. , Wiedlocha, A. , Olsnes, S. , and Wesche, J. (2010) Roles of fibroblast growth factor receptors in carcinogenesis. Mol Cancer Res 8: 1439–1452. [DOI] [PubMed] [Google Scholar]
- Heron, S.F. , Maynard, J.A. , van Hooidonk, R. , and Eakin, C.M. (2016) Warming trends and bleaching stress of the World's coral reefs 1985–2012. Sci Rep 6: 38402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillyer, K.E. , Dias, D.A. , Lutz, A. , Wilkinson, S.P. , Roessner, U. , and Davy, S.K. (2017) Metabolite profiling of symbiont and host during thermal stress and bleaching in the coral Acropora aspera . Coral Reefs 36: 105–118. [Google Scholar]
- Hoegh‐Guldberg, O. , Mumby, P.J. , Hooten, A.J. , Steneck, R.S. , Greenfield, P. , Gomez, E. , et al. (2007) Coral reefs under rapid climate change and ocean acidification. Science 318: 1737–1742. [DOI] [PubMed] [Google Scholar]
- Hou, J. , Xu, T. , Su, D. , Wu, Y. , Cheng, L. , Wang, J. , et al. (2018) RNA‐Seq reveals extensive transcriptional response to heat stress in the stony coral Galaxea fascicularis . Front Genet 9: 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes, T.P. , Kerry, J.T. , Álvarez‐Noriega, M. , Álvarez‐Romero, J.G. , Anderson, K.D. , Baird, A.H. , et al. (2017) Global warming and recurrent mass bleaching of corals. Nature 543: 373–377. [DOI] [PubMed] [Google Scholar]
- Jones, A.M. , Berkelmans, R. , van Oppen, M.J.H. , Mieog, J.C. , and Sinclair, W. (2008) A community change in algal endosymbionts of scleractinian coral following a natural bleaching event: field evidence of acclimatization. P R Soc Lond B Biol Sci 275: 1359–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladriere, O. , Compere, P. , Decloux, N. , Vandewalle, P. , and Poulicek, M. (2008) Morphological alterations of zooxanthellae in bleached cnidarian hosts. Cah Biol Mar 49: 215–227. [Google Scholar]
- Langmead, B. , and Salzberg, S.L. (2012) Fast gapped‐read alignment with Bowtie 2. Nat Methods 9: 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, S.T.M. , Davy, S.K. , Tang, S.L. , Fan, T.Y. , and Kench, P.S. (2015) Successive shifts in the microbial community of the surfacemucus layer and tissues of the coral Acropora muricata under thermal stress. FEMS Microbiol Ecol 91: fiv142. [DOI] [PubMed] [Google Scholar]
- Leggat, W. , Seneca, F. , Wasmund, K. , Ukani, L. , Yellowlees, D. , and Ainsworth, T.D. (2011) Differential responses of the coral host and their algal symbiont to thermal stress. PLoS One 6: e26687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lema, K.A. , Willis, B.L. , and Bourne, D.G. (2012) Corals form characteristic associations with symbiotic nitrogen‐fixing bacteria. Appl Environ Microbiol 78: 3136–3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesser, M.P. (2011) Coral bleaching:causes and mechanisms. In Coral Reefs: An Ecosystem in Transition, Dubinsky, Z. , and Stambler, N. (eds). Dordrecht: Springer, pp. 405–419. [Google Scholar]
- Lesser, M.P. , and Farrell, J. (2004) Solar radiation increases the damage to both host tissues and algal symbionts of corals exposed to thermal stress. Coral Reefs 23: 367–377. [Google Scholar]
- Levin, R.A. , Voolstra, C.R. , Weynberg, K.D. , and van Oppen, M.J.H. (2017) Evidence for a role of viruses in the thermal sensitivity of coral photosymbionts. ISME J 11: 808–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, B. , and Dewey, C.N. (2011) RSEM: accurate transcript quantification from RNA‐Seq data with or without a reference genome. BMC Bioinformatics 12: 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, S. , Yu, K. , Shi, Q. , Chen, T. , Zhao, M. , and Yan, H. (2008) Experimental study of stony coral response to the high temperature in Luhuitou of Hainan Island. Trop Geogr 28: 534–539 In Chinese. [Google Scholar]
- Li, X. , Liu, S. , Huang, H. , Huang, L. , Jing, Z. , and Zhang, C. (2012) Coral bleaching caused by an abnormal water temperature rise at Luhuitou fringing reef, Sanya Bay, China. Aquat Ecosyst Health 15: 227–233. [Google Scholar]
- Lin, Z. , Chen, M. , Dong, X. , Zheng, X. , Huang, H. , Xu, X. , and Chen, J. (2017) Transcriptome profiling of Galaxea fascicularis and its endosymbiont Symbiodinium reveals chronic eutrophication tolerance pathways and metabolic mutualism between partners. Sci Rep 7: 42100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Littman, R. , Willis, B.L. , and Bourne, D.G. (2011) Metagenomic analysis of the coral holobiont during a natural bleaching event on the Great Barrier Reef. Environ Microbiol Rep 3: 651–660. [DOI] [PubMed] [Google Scholar]
- Littman, R.A. , Bourne, D.G. , and Willis, B.L. (2010) Responses of coral‐associated bacterial communities to heat stress differ with Symbiodinium type on the same coral host. Mol Ecol 19: 1978–1990. [DOI] [PubMed] [Google Scholar]
- Love, M.I. , Huber, W. , and Anders, S. (2014) Moderated estimation of fold change and dispersion for RNA‐seq data with DESeq2. Genome Biol 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, H. , and Moran, M.A. (2014) Evolutionary ecology of the marine Roseobacter clade. Microbiol Mol Biol Rev 78: 573–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magoč, T. , and Salzberg, S.L. (2011) FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27: 2957–2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh, J.A., Jr. (1970) Primary productivity of reef‐building calcareous red algae. Ecology 51: 255–263. [Google Scholar]
- Matus, D.Q. , Thomsen, G.H. , and Martindale, M.Q. (2007) FGF signaling in gastrulation and neural development in Nematostella vectensis, an anthozoan cnidarian. Dev Genes Evol 217: 137–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maynard, J. , van Hooidonk, R. , Eakin, C.M. , Puotinen, M. , Garren, M. , Williams, G. , et al. (2015) Projections of climate conditions that increase coral disease susceptibility and pathogen abundance and virulence. Nat Clim Change 5: 688–694. [Google Scholar]
- Morris, L.A. , Voolstra, C.R. , Quigley, K.M. , Bourne, D.G. , and Bay, L.K. (2019) Nutrient availability and metabolism affect the stability of coral–symbiodiniaceae symbioses. Trends Microbiol 27: 678–689. [DOI] [PubMed] [Google Scholar]
- Muscatine, L. , and Porter, J.W. (1977) Reef corals: mutualistic symbioses adapted to nutrient‐poor environments. Bioscience 27: 454–460. [Google Scholar]
- Nagamune, K. , and Sibley, L.D. (2006) Comparative genomic and phylogenetic analyses of calcium ATPases and calcium‐regulated proteins in the apicomplexa. Mol Biol Evol 23: 1613–1627. [DOI] [PubMed] [Google Scholar]
- Neave, M.J. , Apprill, A. , Ferrier‐Pagès, C. , and Voolstra, C.R. (2016) Diversity and function of prevalent symbiotic marine bacteria in the genus Endozoicomonas . Appl Microbiol Biotechnol 100: 8315–8324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neave, M.J. , Michell, C.T. , Apprill, A. , and Voolstra, C.R. (2017) Endozoicomonas genomes reveal functional adaptation and plasticity in bacterial strains symbiotically associated with diverse marine hosts. Sci Rep 7: 40579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen‐Kim, H. , Bouvier, T. , Bouvier, C. , Bui, V.N. , Le‐Lan, H. , and Bettarel, Y. (2015) Viral and bacterial epibionts in thermally‐stressed corals. J Mar Sci Eng 3: 1272–1286. [Google Scholar]
- Obura, D.O. (2009) Reef corals bleach to resist stress. Mar Pollut Bull 58: 206–212. [DOI] [PubMed] [Google Scholar]
- Oliver, J.K. , Berkelmans, R. , and Eakin, C.M. (2018) Coral bleaching in space and time. In Coral Bleaching: patterns, Processes, Causes and Consequences. van Oppen, M.J.H. and Lough, J.M. (eds). Cham: Springer, pp. 21–39. [Google Scholar]
- Pertea, G. , Huang, X. , Liang, F. , Antonescu, V. , Sultana, R. , Karamycheva, S. , et al. (2003) TIGR gene indices clustering tools (TGICL): a software system for fast clustering of large EST datasets. Bioinformatics 19: 651–652. [DOI] [PubMed] [Google Scholar]
- Pinzón, J.H. , Kamel, B. , Burge, C.A. , Harvell, C.D. , Medina, M. , Weil, E. , and Mydlarz, L.D. (2015) Whole transcriptome analysis reveals changes in expression of immune‐related genes during and after bleaching in a reef‐building coral. R Soc Open Sci 2: 140214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam, H.M. , Stat, M. , Pochon, X. , and Gates, R.D. (2012) Endosymbiotic flexibility associates with environmental sensitivity in scleractinian corals. P R Soc Lond B Biol Sci 279: 4352–4361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quistad, S.D. , Stotland, A. , Barott, K.L. , Smurthwaite, C.A. , Hilton, B.J. , Grasis, J.A. , et al. (2014) Evolution of TNF‐induced apoptosis reveals 550 My of functional conservation. Proc Natl Acad Sci U S A 111: 9567–9572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raina, J.B. , Tapiolas, D. , Willis, B.L. , and Bourne, D.G. (2009) Coral‐associated bacteria and their role in the biogeochemical cycling of sulfur. Appl Environ Microbiol 75: 3492–3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reshef, L. , Koren, O. , Loya, Y. , Zilber‐Rosenberg, I. , and Rosenberg, E. (2006) The coral probiotic hypothesis. Environ Microbiol 8: 2068–2073. [DOI] [PubMed] [Google Scholar]
- Rodriguez‐Lanetty, M. , Harii, S. , and Hoegh‐Guldberg, O.V.E. (2009) Early molecular responses of coral larvae to hyperthermal stress. Mol Ecol 18: 5101–5114. [DOI] [PubMed] [Google Scholar]
- Rognes, T. , Flouri, T. , Nichols, B. , Quince, C. , and Mahé, F. (2016) VSEARCH: a versatile open source tool for metagenomics. PeerJ 4: e2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohwer, F. , Seguritan, V. , Azam, F. , and Knowlton, N. (2002) Diversity and distribution of coral‐associated bacteria. Mar Ecol Prog Ser 243: 1–10. [Google Scholar]
- Rosado, P.M. , Leite, D.C.A. , Duarte, G.A.S. , Chaloub, R.M. , Jospin, G. , da Rocha, U.N. , et al. (2019) Marine probiotics: increasing coral resistance to bleaching through microbiome manipulation. ISME J 13: 921–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosales, S.M. , Miller, M.W. , Williams, D.E. , Traylor‐Knowles, N. , Young, B. , and Serrano, X.M. (2019) Microbiome differences in disease‐resistant vs. susceptible Acropora corals subjected to disease challenge assays. Sci Rep 9: 18279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosic, N. , Ling, E.Y.S. , Chan, C.K.K. , Lee, H.C. , Kaniewska, P. , Edwards, D. , et al. (2015) Unfolding the secrets of coral–algal symbiosis. ISME J 9: 844–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rua, C.P.J. , Trindade‐Silva, A.E. , Appolinario, L.R. , Venas, T.M. , Garcia, G.D. , Carvalho, L.S. , et al. (2014) Diversity and antimicrobial potential of culturable heterotrophic bacteria associated with the endemic marine sponge Arenosclera brasiliensis . PeerJ 2: e419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampayo, E. , Ridgway, T. , Bongaerts, P. , and Hoegh‐Guldberg, O. (2008) Bleaching susceptibility and mortality of corals are determined by fine‐scale differences in symbiont type. Proc Natl Acad Sci U S A 105: 10444–10449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinzato, C. , Shoguchi, E. , Kawashima, T. , Hamada, M. , Hisata, K. , Tanaka, M. , et al. (2011) Using the Acropora digitifera genome to understand coral responses to environmental change. Nature 476: 320–323. [DOI] [PubMed] [Google Scholar]
- Shoguchi, E. , Shinzato, C. , Kawashima, T. , Gyoja, F. , Mungpakdee, S. , Koyanagi, R. , et al. (2013) Draft assembly of the Symbiodinium minutum nuclear genome reveals dinoflagellate gene structure. Curr Biol 23: 1399–1408. [DOI] [PubMed] [Google Scholar]
- Siebeck, U.E. , Marshall, N.J. , Klüter, A. , and Hoegh‐Guldberg, O. (2006) Monitoring coral bleaching using a colour reference card. Coral Reefs 25: 453–460. [Google Scholar]
- Sogin, E.M. , Putnam, H.M. , Anderson, P.E. , and Gates, R.D. (2016) Metabolomic signatures of increases in temperature and ocean acidification from the reef‐building coral, Pocillopora damicornis . Metabolomics 12: 71. [Google Scholar]
- Sokolow, S. (2009) Effects of a changing climate on the dynamics of coral infectious disease: a review of the evidence. Dis Aquat Organ 87: 5–18. [DOI] [PubMed] [Google Scholar]
- Takahashi, M. , Ritz, J. , and Cooper, G.M. (1985) Activation of a novel human transforming gene, ret, by DNA rearrangement. Cell 42: 581–588. [DOI] [PubMed] [Google Scholar]
- Templ, M. , Hron, K. , and Filzmoser, P. (2011) robCompositions: an R‐package for robust statistical analysis of compositional data. In Compositional Data Analysis: Theory and Applications, Pawlowsky‐Glahn, V. , and Buccianti, A. (eds). United Kingdom: John Wiley & Sons Ltd, pp. 341–355. [Google Scholar]
- Tout, J. , Siboni, N. , Messer, L.F. , Garren, M. , Stocker, R. , Webster, N.S. , et al. (2015) Increased seawater temperature increases the abundance and alters the structure of natural Vibrio populations associated with the coral Pocillopora damicornis . Front Microbiol 6: 432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turwankar, A. , and Ghaskadbi, S. (2019) VEGF and FGF signaling during head regeneration in hydra. Gene 717: 144047. [DOI] [PubMed] [Google Scholar]
- van de Water, J.A.J.M. , Mares, M.C.D. , Dixon, G.B. , Raina, J.B. , Willis, B.L. , Bourne, D.G. , and van Oppen, M.J.H. (2018) Antimicrobial and stress responses to increased temperature and bacterial pathogen challenge in the holobiont of a reef building coral. Mol Ecol 27: 1065–1080. [DOI] [PubMed] [Google Scholar]
- van den Boogaart, K.G. , and Tolosana‐Delgado, R. (2008) “Compositions”: a unified R package to analyze compositional data. Comput Geosci‐UK 34: 320–338. [Google Scholar]
- Veron, J.E.N. (1993) Corals of Australia and the Indo‐Pacific. Honolulu: University of Hawaii Press. [Google Scholar]
- Wangpraseurt, D. , Holm, J.B. , Larkum, A.W.D. , Pernice, M. , Ralph, P.J. , Suggett, D.J. , et al. (2017) In vivo microscale measurements of light and photosynthesis during coral bleaching: evidence for the optical feedback loop? Front Microbiol 8: 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weis, V.M. (2008) Cellular mechanisms of Cnidarian bleaching: stress causes the collapse of symbiosis. J Exp Biol 211: 3059–3066. [DOI] [PubMed] [Google Scholar]
- Willis, B.L. , Page, C.A. , and Dinsdale, E.A. (2004) Coral disease on the Great Barrier Reef. In Coral Health and Disease, Rosenberg, E. , and Loya, Y. (eds). Berlin, Heidelberg: Springer, pp. 69–104. [Google Scholar]
- Yu, X. , Huang, B. , Zhou, Z. , Tang, J. , and Yu, Y. (2017) Involvement of caspase3 in the acute stress response to high temperature and elevated ammonium in stony coral Pocillopora damicornis . Gene 637: 108–114. [DOI] [PubMed] [Google Scholar]
- Yuan, C. , Zhou, Z. , Zhang, Y. , Chen, G. , Yu, X. , Ni, X. , et al. (2017) Effects of elevated ammonium on the transcriptome of the stony coral Pocillopora damicornis . Mar Pollut Bull 114: 46–52. [DOI] [PubMed] [Google Scholar]
- Zhang, Y. , Sun, J. , Mu, H. , Lun, J.C.Y. , and Qiu, J. (2017) Molecular pathology of skeletal growth anomalies in the brain coral Platygyra carnosa: a meta‐transcriptomic analysis. Mar Pollut Bull 124: 660–667. [DOI] [PubMed] [Google Scholar]
- Ziegler, M. , Seneca, F.O. , Yum, L.K. , Palumbi, S.R. , and Voolstra, C.R. (2017) Bacterial community dynamics are linked to patterns of coral heat tolerance. Nat Commun 8: 14213. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. (a) Dissimilarity in coral gene expression profiles between samples collected at each time point. (b) Non‐metric multidimensional scaling (nMDS) plot representing the differences in the overall gene expression patterns in corals exposed to elevated water temperatures.
Figure S2. (a) Dissimilarity in Symbiodiniaceae gene expression profiles between samples collected at each time point. (b) Non‐metric multidimensional scaling (nMDS) plot representing the differences in the overall gene expression patterns in Symbiodiniaceae endosymbionts exposed to elevated water temperatures.
Figure S3. Heatmap of differentially expressed genes involved in the enriched pathways in the coral Pocillopora damicornis following exposure to 31°C (a) and 32°C (b). Each cell represents the log2‐fold change value of each gene. The dot shows the gene involved in a certain pathway.
Figure S4. Heatmap of differentially expressed genes involved in the enriched pathways in Symbiodiniaceae following exposure to 31°C (a) and 32°C (b). Each cell represents the log2‐fold change value of each gene. The dot shows the gene involved in a certain pathway.
Table S1. Internal node and de novo taxa among reference sequences of Symbiodiniaceae. The closest leaf taxon and distance to that taxon are given for internal nodes (‘D_I:X’). The closest BLAST hit from the reference database and associated e‐value are given for de novo taxa (‘D_d:X’).
Table S2. Coral gene expression profiles under different temperatures. Data in columns 7 to 18 are FPKM (Fragments per kilobase Million) for each gene at 25, 29, 31 and 32°C.
Table S3. Algal symbiont gene expression profiles under different temperatures. Data in columns 7 to 18 are FPKM (Fragments per Kilobase Million) for each gene at 25, 29, 31 and 32°C.
Table S4. Gene ontology terms significantly (with corrected P‐value ≤0.01) enriched in the differentially expressed genes in the coral Pocillopora damicornis and Symbiodiniaceae following exposure to elevated seawater temperatures.
Table S5. Kyoto Encyclopaedia of Genes and Genomes (KEGG) pathways significantly (with corrected P‐value ≤0.01) enriched in the differentially expressed genes in the coral Pocillopora damicornis following exposure to elevated seawater temperatures.
Table S6. Kyoto Encyclopaedia of Genes and Genomes (KEGG) pathways significantly (with corrected P‐value ≤0.01) enriched in the differentially expressed genes in Symbiodiniaceae following exposure to elevated seawater temperatures.
Data Availability Statement
The datasets generated during the current study have been deposited and are publicly available in the Sequence Read Archive repository under BioProject ID PRJNA478260 and PRJNA559028.


