Abstract
Objectives
Due to the maternally‐inherited nature of mitochondrial DNA (mtDNA), there is a lack of information regarding fetal mtDNA in the plasma of pregnant women. We aim to explore the presence and topologic forms of circulating fetal and maternal mtDNA molecules in surrogate pregnancies.
Methods
Genotypic differences between fetal and surrogate maternal mtDNA were used to identify the fetal and maternal mtDNA molecules in plasma. Plasma samples were obtained from the surrogate pregnant mothers. Using cleavage‐end signatures of BfaI restriction enzyme, linear and circular mtDNA molecules in maternal plasma could be differentiated.
Results
Fetal‐derived mtDNA molecules were mainly linear (median: 88%; range: 80%–96%), whereas approximately half of the maternal‐derived mtDNA molecules were circular (median: 51%; range: 42%–60%). The fetal DNA fraction of linear mtDNA was lower (median absolute difference: 9.8%; range: 1.1%–27%) than that of nuclear DNA (median: 20%; range: 9.7%–35%). The fetal‐derived linear mtDNA molecules were shorter than the maternal‐derived ones.
Conclusion
Fetal mtDNA is present in maternal plasma, and consists mainly of linear molecules. Surrogate pregnancies represent a valuable clinical scenario for exploring the biology and potential clinical applications of circulating mtDNA, for example, for pregnancies conceived following mitochondrial replacement therapy.
What is already known about this topic?
In natural, non‐surrogate pregnancies, fetal nuclear DNA in maternal plasma consists of short linear DNA fragments that have a shorter size distribution than the maternal background nuclear DNA.
Both linear and circular mitochondrial DNA (mtDNA) molecules co‐exist in plasma of liver and bone marrow transplant patients.
Liver tissues contribute a considerable amount of mtDNA predominantly in a linear form to the plasma DNA pool.
mtDNA molecules of hematopoietic origin are mainly in a circular form in plasma.
What does this study add?
Fetal and maternal mtDNA molecules could be distinguished in the plasma of surrogate pregnant mothers.
Linear and circular mtDNA molecules co‐existed in the plasma of surrogate pregnant women.
The fetal mtDNA molecules in surrogate maternal plasma appeared to be predominantly in a linear form, while approximately half of the surrogate maternal mtDNA molecules were in a circular form.
The fetal mtDNA fraction was less abundant than the fetal nuclear DNA fraction in surrogate pregnant women.
Circulating fetal mtDNA molecules consisted of more short molecules compared with the surrogate maternal mtDNA molecules.
1. INTRODUCTION
Most studies of cell‐free DNA in plasma focus on DNA molecules in a linear form derived from the nucleus. These linear plasma DNA molecules display characteristic fragmentation patterns, and are associated with nucleosomal structures. 1 For instance, the modal frequency of fragment sizes is approximately 166 bp, accompanied by multiple 10‐bp periodic peaks of small sizes. Such characteristic patterns have inspired recent interest in plasma DNA fragmentomics, encompassing areas such as plasma DNA preferred ends, 2 , 3 fragment sizes, 1 , 4 , 5 end motifs, 6 , 7 nucleosome footprints, 8 , 9 , 10 plasma DNA end orientations 11 as well as biological links between fragmentation processes and nucleases. 6 , 12 , 13
In addition to studying the properties of nuclear DNA molecules in plasma, more recently, on the study of subjects following liver and bone marrow transplantation, we have demonstrated that there are two different topologic forms of mitochondrial DNA (mtDNA), namely linear and circular forms, present in plasma DNA. 14 Of note, mtDNA molecules of hematopoietic origin appear to be mainly circular, in sharp contrast to the liver‐derived mtDNA molecules that mainly display a linear form. 14 These results suggest that plasma mtDNA topology might vary depending of the tissues of origin of the detected mtDNA.
Topologic study of plasma DNA can also be extended to autosomal DNA molecules. 15 For example, the presence of extrachromosomal circular DNA (eccDNA) was observed in both maternal and fetal DNA molecules in pregnant women, and displaying distinct size patterns. 15 While circular mtDNA and eccDNA in plasma are governed by different biologic mechanisms, our work has highlighted their commonality in their accessibility to being studied using similar methodologies.
In this study, we explore whether tissue‐associated topologic forms of mtDNA might be present in the plasma of pregnant women. Pregnancy offers an attractive model for studying the biology of tissue‐specific plasma DNA molecules because there is the co‐existence of fetal‐ and maternal‐derived DNA in maternal plasma. 16 However, the study of fetal mtDNA in maternal plasma is complicated by the fact that mitochondria are inherited from the mother. Hence, it would not normally be possible to differentiate fetal and maternal mtDNA molecules in natural pregnancies. In the context of this study, a surrogate mother is a woman engaged by a couple to become pregnant with an embryo fertilized using gametes of the couple. Thus, in this study, surrogate pregnancy was chosen as a model to explore the topologic forms of circulating fetal mtDNA.
2. METHODS AND MATERIALS
2.1. Subjects
The subject recruitment and sample collection were conducted in Altravita IVF‐clinic, ECO Center, Moscow, Russia. Five surrogate pregnant cases were recruited. Biological parents involved in the five surrogate pregnancies gave informed written consent. Informed verbal consent was obtained from the five surrogate pregnant women. Plasma and buffy coat samples of surrogate pregnant subjects were collected during the second or third trimester (21–31 weeks). Buffy coat or saliva samples from the oocyte donor subjects were collected. Review of the clinical research protocol was conducted and approved by the ethics committee of the ECO Center, Moscow, Russia (reference number: Protocol No. 3). The Joint Chinese University of Hong Kong – New Territories East Cluster Clinical Research Ethics Committee approved (reference number: 2019.568) the data generation and data analysis of the study.
2.2. DNA extraction and sequencing library preparation
Peripheral blood samples of the surrogate pregnant women were collected into EDTA‐containing tubes and centrifuged at 1600 g at 4°C for 10 min. The upper portion of plasma was recentrifuged at 16,000 g at 4°C for 10 min to obtain cell‐free plasma. Plasma DNA was extracted from 3–4 ml of plasma using the QIAamp Circulating Nucleic Acid Kit (Qiagen). Half of the plasma DNA was digested by the restriction enzyme BfaI (New England Biolabs) while the other half of the plasma DNA was processed without the restriction enzyme digestion step. Ten units of BfaI (restriction recognition site: C^TAG; ^ denoting the cutting site) were used in the cleavage of 20 ng plasma DNA at 37°C for 120 min. Tissue DNA samples of buffy coat or saliva from the oocyte donors and buffy coat from the surrogate mothers were extracted using the QIAamp DNA Mini and Blood Mini Kit. The S220 Focused‐ultrasonicator (Covaris) was used to shear tissue DNA (from both surrogate mothers and oocyte donor mothers) into sizes of around 200 bp.
DNA libraries were generated by the TruSeq Nano DNA Library Prep Kit (Illumina) and ligated with xGEN Dual Index UMI adapters (IDT). A unique dual‐index sequence is a sequence tag that recognizes a sample identity for sample multiplexing purpose. Unique molecular identifier (UMI) is a technology for ascertaining molecular identity by adding a unique sequence tag together with an adaptor to the original DNA template before PCR amplification and sequencing. The size and concentration of DNA libraries were then analyzed respectively by 2100 TapeStation (Agilent) and Qubit 3.0 Fluorometer (Thermo Fisher Scientific).
2.3. Hybridization‐based target enrichment
SeqCap EZ Prime Developer Probes (Roche) targeting human mtDNA were used in the target‐capture enrichment procedures after the DNA library preparation. The NimbleGen SeqCap Hybridization and Wash Kit (Roche) was used during capture procedures. The 4200 TapeStation (Agilent) and the Qubit 3.0 Fluorometer (Thermo Fisher Scientific) were used to evaluate the quality of the target‐captured DNA libraries. Targeted regions of interest included 1000 SNP (single nucleotide polymorphism) sites and human mtDNA regions for probe design and synthesis (Roche NimbleGen Inc.) as previously described. 14
2.4. Sequencing and alignment
Target‐captured DNA libraries were sequenced in a 2 × 70 bp paired‐end mode on a NextSeq 500 system (Illumina) using the NextSeq 500 High Output Reagent Cartridge v2 Kit (Illumina). Sequencing reads were attributed to multiple samples based on unique dual‐index sequences using Picard tools (https://broadinstitute.github.io/picard/). The demultiplexed reads were aligned to the reference genome database including the human reference genome (NCBI37/hg19) and the mitochondrial genome using BWA. 17 Reads with UMIs information were used to generate consensus reads using fgbio tools. 18 SOAP2 19 was utilized to realign the consensus reads to determine the genomic origins for each paired‐end sequenced reads. During the alignment based on SOAP2, 19 we allowed up to two mismatches to make those reads with the single nucleotide variants mappable for the downstream analysis. The fetal and maternal DNA molecules were differentiated from the mapped reads based on informative single nucleotide variants. 1
2.5. Determination of linear and circular mtDNA
The restriction enzyme BfaI was used for differentiating linear and circular mtDNA as previously described. 14 This enzyme cleaves C^TAG sites and is expected to cut DNA once every 256 bp (1/44). Briefly, after plasma DNA was treated with BfaI, the linear mtDNA molecules in plasma would mainly either remain uncleaved or be cleaved into two fragments both with one BfaI‐cleaved end signature. On the contrary, an intact circular mtDNA (~16.5 kb), containing a total of 98 BfaI cutting sites, would be cleaved into linear mtDNA fragments each bearing two cleavage ends. The principle of the topologic analysis of plasma mtDNA for surrogate pregnant subjects is illustrated in Figure 1.
FIGURE 1.
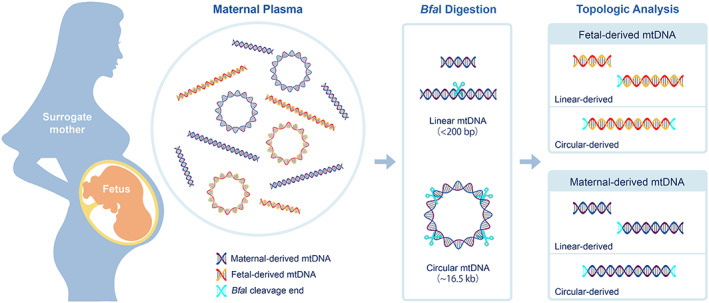
Schematic illustration of the topological assessment of mtDNA in surrogate pregnancies. Fetal mtDNA may be released into surrogate maternal plasma through the placenta during pregnancy. BfaI cleavage signatures were used for differentiating the intact circular mtDNA molecules (~16.5 kb) from pre‐existing linear mtDNA molecules (generally <200 bp). BfaI has a 4‐bp recognition sequence which by random chance should occur once every 256 bp (1/44). Therefore, fragments originated from intact circular mtDNA molecules would carry two BfaI‐cleavage ends (Cyan DNA) after BfaI digestion (Cyan scissor), whereas the majority of the fragments derived from pre‐existing linear mtDNA molecules would have no more than one cleavage end (Cyan DNA). Single nucleotide variants in mitochondrial genomes were used to determine the physical forms of fetal and maternal mtDNA in plasma of surrogate pregnant women. Combined with topological assessment of mtDNA, we could identify if the fetal‐derived (red‐yellow DNA) or maternal‐derived (purple‐blue DNA) mtDNA molecules were in a circular or linear form according to the patterns of BfaI‐cleavage signatures [Colour figure can be viewed at wileyonlinelibrary.com]
2.6. Determination of informative variants in mitochondrial genomes and nuclear genomes
The genotypes of fetal and surrogate mother's mitochondrial genomes were determined by sequencing relevant tissue DNA including buffy coat or saliva DNA from oocyte donors or surrogate mother's buffy coat DNA. A cell can contain numerous copies of mtDNA among which some may carry different variants in certain locations of the mtDNA genome, but some may not, which is referred to as mtDNA heteroplasmy. To minimize the influence of the mtDNA heteroplasmy in a tissue, we focused on the informative variants in the mitochondrial genome for which the alleles of a single nucleotide variant site were identical (i.e., homoplasmic) but different between the surrogate mother and oocyte donor. Due to the maternal inheritance of mtDNA, fetal‐specific mtDNA variants were represented by oocyte donor specific mtDNA variants. Fetal mtDNA fraction (F m) was deduced by the allelic ratio between fetal‐specific variants (e.g., A) and surrogate maternal specific variants (e.g., B) present in the mitochondrial genome:
In our present study, the placental tissues were not available and the fetal genotypes were determined non‐invasively. Thus, we could not directly obtain the fetal genotype information. In our analysis, we utilized the nuclear genotypes from tissue genomic DNA from the surrogate mother and the oocyte donor mother, in conjugation with sequencing result from one plasma aliquot, to deduce the informative SNPs allowing the fetal nuclear DNA fraction (F n) estimation in the second plasma aliquot. For the sake of simplicity, we first focused on the SNPs which were homozygous for the same allele both in the surrogate mother (AA) and the oocyte donor mother (AA). Among those SNPs, if a locus showed a different allele in one plasma aliquot, such allele would likely be derived from the fetus. Due to our bioinformatics algorithm, the fetal SNPs used in the fetal nuclear DNA fraction calculation would be heterozygous (AB) in the fetus:
2.7. Statistical analysis
Sequencing data analysis was performed using in‐house bioinformatics programs written in Perl and R languages. A p‐value of <0.05 was considered as statistically significant with all two‐tailed probabilities.
3. RESULTS
3.1. BfaI enzymatic digestion for maternal plasma DNA of surrogate pregnant women
We obtained a median on‐target depth of 1005× (range: 529–1467×) and 1081× (range: 693–2106×) for samples with and without BfaI digestion, respectively. There was a 7.9‐fold increase in the median proportion of mtDNA among all plasma DNA increased in plasma DNA samples with BfaI digestion (median: 4.0%; range: 3.0%–24%), compared with those without digestion (median: 0.51%; range: 0.2–1.4%) (p‐value: 0.004, Mann–Whitney U test) (Figure 2(A)). The median proportion of circular mtDNA fragments (i.e., for those carrying two enzymatic cleavage ends) among all mtDNA molecules in BfaI digested samples was 50% (range: 36–55%), which was a 76‐fold increase compared with those without digestion (median: 0.66%; range: 0.41–1.2%) (p‐value = 0.004, Mann–Whitney U test) (Figure 2(B)). Overall, these results suggested the existence of circular mtDNA in plasma DNA of surrogate pregnant subjects.
FIGURE 2.

Effects of BfaI cleavage on plasma mtDNA. (A) Comparison of the proportion of mtDNA among all DNA in plasma of surrogate pregnant women with and without BfaI digestion. (B) Comparison of the proportion of mtDNA molecules with two BfaI‐cleavage ends (circular‐derived mtDNA) among all mtDNA in plasma of surrogate pregnant women with and without BfaI digestion [Colour figure can be viewed at wileyonlinelibrary.com]
As shown in Figure S1A, in contrast to plasma DNA of surrogate pregnant women without BfaI treatment exhibiting a relatively smooth size distribution of mtDNA peaked at around 60 bp, plasma DNA with BfaI digestion displayed an irregular distribution with a series of sporadic spikes up to 550 bp. The results were largely consistent with what was previously observed in plasma DNA of liver transplant patients. 14 However, we did not observe such differences in the size distribution of nuclear DNA molecules between before and after BfaI digestion (Figure S1B). The data provided further evidence that there may exist circular mtDNA in plasma of surrogate pregnant women.
3.2. Topologic analysis of fetal DNA and maternal mtDNA
We identified single nucleotide variants in the mitochondrial genome whose sequences differed between oocyte donor and paired surrogate maternal tissues, termed informative variants. To get fetal genotype information, buffy coat or saliva DNA samples from oocyte donors were sequenced to a median on‐target depth of 1636× (939–2283×). The matched buffy coat samples of surrogate mothers (5 pairs) were sequenced to a median on‐target depth of 1575× (interquartile range: 618–2361×). We obtained a median of 29 informative variants (range: 13–46) in the mitochondrial genome and 674 informative SNPs in the nuclear genome (range: 469–836, out of 1000 SNPs designed for target capture). The molecules carrying fetal‐specific mitochondrial variants (fetal‐derived mtDNA) and surrogate maternal‐specific mitochondrial variants were determined. We identified a median of 1176 (range: 260–2864) fetal‐specific mtDNA molecules and a median of 49,623 (range: 13,227–214,898) surrogate maternal‐specific mtDNA molecules among five maternal plasma samples.
For the maternal plasma DNA samples without BfaI digestion, the median fetal nuclear DNA fraction was 20% (range: 9.7%–35%), whereas the fetal mtDNA fraction was found to be much lower (median: 7.8%; range: 1.4%–11%) (p‐value = 0.008, Mann–Whitney U test) (Figure 3). No significant correlation was observed between the fetal fractions of circulating nuclear DNA and mtDNA (r = 0.23; p‐value = 0.71).
FIGURE 3.

Fetal DNA fraction in nuclear DNA and mtDNA among surrogate maternal plasma DNA samples without Bfa I treatment [Colour figure can be viewed at wileyonlinelibrary.com]
On the basis of BfaI cleavage end signature analysis, we found that the proportion of linear mtDNA among fetal‐derived mtDNA molecules (i.e., placenta‐derived; median: 88%; range: 80%–96%) was much higher than that among maternal‐derived mtDNA molecules (i.e., mainly of hematopoietic and liver origin; median: 49%; range: 40%–58%) (Figure 4).
FIGURE 4.
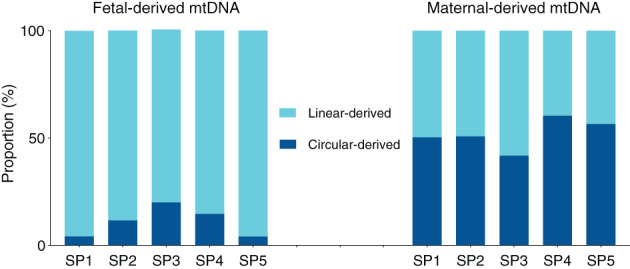
The proportions of linear and circular forms in fetal‐ and maternal‐derived mtDNA in plasma across five surrogate pregnancies (SP1 to SP5) [Colour figure can be viewed at wileyonlinelibrary.com]
3.3. Size distribution of fetal‐ and maternal‐derived mtDNA
We next analyzed the size profiles of the linear mtDNA molecules in plasma DNA of surrogate pregnant women without BfaI digestion. The fetal‐derived mtDNA molecules were much shorter than the maternal counterparts (Figure 5(A) and Figure S2). The fetal‐derived nuclear DNA fragments in all five surrogate maternal plasma DNA were found to be shorter than the maternal nuclear DNA (Figure S3), showing a 166‐bp peak with a series of 10‐bp periodic peaks in small sizes that were not observable in circulating fetal and maternal mtDNA. Such characteristic sizes of fetal and maternal nuclear DNA results were highly consistent with the previous reports. 1 , 2 Interestingly, the size difference between fetal and maternal mtDNA appeared to be much sharper than counterparts of nuclear DNA, as evinced by the fact that there was a 44% increase in fetal mtDNA size <100 bp but only a 1.1% increase in fetal nuclear DNA compared with corresponding maternal results, respectively.
FIGURE 5.
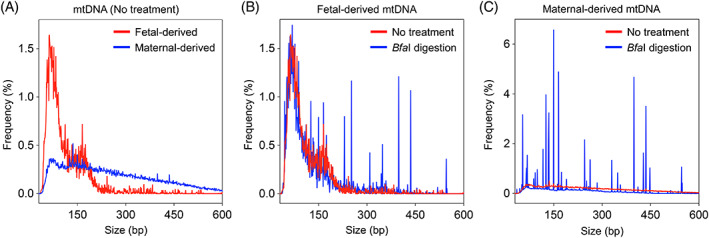
Size profiles for mtDNA molecules of fetal‐ or maternal‐derived in plasma of surrogate pregnant women. (A) Size distributions for fetal‐ and maternal‐derived mtDNA molecules in the plasma of surrogate pregnant women without enzymatic cleavage. (B) Size distributions for fetal‐derived mtDNA molecules with and without Bfa I cleavage in the plasma of surrogate pregnant women. (C) Size distributions for maternal‐derived mtDNA molecules with and without Bfa I cleavage in the plasma of surrogate pregnant women [Colour figure can be viewed at wileyonlinelibrary.com]
Intriguingly, by looking at plasma mtDNA results pooled from five surrogate subjects after BfaI digestion (Figure 5(B)), we observed that there were multiple discrete sporadic spikes present in the fetal mtDNA size profile, but no such spiked peaks in those samples without BfaI digestion. These results suggested that the BfaI cutting acted on the fetal circular mitochondrial genome in plasma DNA of surrogate pregnant women. As expected, we observed much more and more prominent spikes in the maternal mtDNA size profile, which was in line with the fact that a higher abundance of circular mtDNA was present in the maternal background DNA (Figure 5(C)).
4. DISCUSSION
In the present study, we showed for the first time the presence of fetal mtDNA in surrogate maternal plasma. We further investigated the linear and circular forms of mtDNA between fetal and maternal DNA, by taking advantage of the genotypic differences between fetal and maternal mitochondrial genomes in the surrogate pregnant women. Our data revealed that the linear and circular mtDNA coexisted in the plasma DNA of surrogate pregnant subjects. The fetal‐derived mtDNA in plasma was shown to be predominantly of a linear form (88%), which is reminiscent of the preponderance of linear liver‐derived mtDNA in the plasma of liver transplant recipients. 14 In contrast, half of surrogate maternal background mtDNA (mainly of hematopoietic and liver origin) appeared to be of circular origin. The abundance of circular mtDNA in the surrogate maternal background mtDNA (51%) was lower than that reported in hematopoietically derived mtDNA (86%) in plasma of liver transplant patients. 14 One possible explanation is that the hematopoietic cells mainly contribute circular mtDNA into plasma, whereas the liver tissues containing thousands of copies of mitochondrial genome per cell 20 are considered as one major contributor of linear mtDNA in plasma DNA pool (74%). 14 Therefore, the liver tissues are believed to release linear mtDNA into plasma DNA of surrogate pregnant women, thus diluting the circular mtDNA in the maternal background DNA.
However, we could not completely rule out the possibility that fragments carrying two BfaI sites were originated from long linear mtDNA. According to our previous study, there was a median of 18% of pre‐existing linear mtDNA carrying at least two BfaI‐cleavage sites. 14 However, 50% of mtDNA molecules in plasma mtDNA were found to carry two BfaI sites after digestion, suggesting that the circular mtDNA molecules were present in the surrogate maternal plasma DNA.
In plasma DNA without BfaI digestion, the mtDNA being analyzed are likely to be spontaneously‐fragmented linear molecules, as sequencing adaptors could not be ligated to circular DNA molecules. Among the linear mtDNA molecules, the fetal mtDNA fraction was found to be relatively lower in surrogate maternal plasma compared with the fetal contribution in nuclear DNA fragments, which was opposite to the observation that liver DNA contribution was greatly enriched in linear mtDNA molecules. 14 One possible reason would be attributed to the difference between the fetal mtDNA fraction and fetal nuclear DNA fraction. The placental cells contain lower mtDNA content 21 than the liver cells. 20 Thus, more linear mtDNA shed from liver tissues would dilute the placental‐derived linear mtDNA contribution.
It is surprising to see that BfaI cutting patterns, originating from complete circular mitochondrial genomes, was present in fetal mtDNA size profile. These suggested the potential existence of complete fetal mitochondrial genomes in plasma. As the clearance of fetal DNA was reported to be very fast (a mean half‐life in the order of tens of minutes 22 , 23 ), the complete circular fetal mtDNA molecules might be protected from clearance by mtDNA binding proteins like mitochondrial transcription factor A (TFAM), 24 or organelles like mitochondria and platelet. 25 The mechanism of tissue‐specific complete circular mtDNA resistant to degradation would require further investigation.
A recent study revealed the presence of fetal and maternal extrachromosomal circular DNA (eccDNA, mainly <600 bp) in the plasma of pregnant women, showing a bimodal size distribution peaking at ~202 and ~338 bp with sharp 10‐bp periodicities. 15 These results suggest that restriction enzyme‐based topologic analysis is not only suited for eccDNA identification but can also be used to infer the large circular DNA such as an entire circular mitochondrial genome. It would be interesting to use this approach to study whether circular DNA molecules might be associated with diseases such as cancer in the future study.
We have shown that placental mtDNA molecules are present at readily detectable amounts in maternal plasma and that they are mainly linear in nature. We believe that these realizations may catalyze the development of placental‐specific mtDNA assays even in non‐surrogate pregnancies. For example, one may focus on enriching the linear mtDNA molecules. Our research group is currently studying many other fragmentation features of cell‐free fetal DNA. 2 , 26 , 27 Some of these features are more pronounced among the fetal nuclear DNA than the maternal nuclear DNA. If such additional features are identified for placental mtDNA, even more specific assays could be developed in conjunction with the findings of the present study. It needs to be noted that, in the present study, we tested surrogate pregnancies during the second and third trimester (21–31 weeks) of pregnancy. It would be desirable to analyze the plasma DNA collected from subjects during earlier pregnancy in the future. By understanding the physical features of placental mtDNA we may one day be able to detect and confirm the success, or otherwise, of mitochondrial replacement therapy (MRT) 28 , 29 through the analysis of placental mtDNA in the plasma samples of pregnant women whose embryos has undergone MRT.
In summary, studying surrogate pregnancies, we have demonstrated fetal mtDNA is present in the plasma of pregnant women. This observation may have clinical applications for noninvasive prenatal testing in the follow‐up of pregnancies conceived following MRT. Topologic analysis of mtDNA represents an emerging direction in plasma DNA research and may have applications beyond pregnancies to fields including cancer liquid biopsy and transplantation monitoring.
CONFLICT OF INTEREST
K. C. Allen Chan, Rossa W.K. Chiu, and Y.M. Dennis Lo hold equities in DRA, Take2 Holdings and Grail. K.C. Allen Chan, Rossa W.K. Chiu, and Y.M. Dennis Lo are consultants to Grail. Y.M. Dennis Lo is a scientific co‐founder of Grail and serves on the scientific advisory board of Grail. Peiyong Jiang hold equities in Grail. Rossa W.K. Chiu is a consultant to Illumina. Peiyong Jiang is a director of KingMed Future. Haiqiang Zhang, Suk Hang Cheng, Peiyong Jiang, K.C. Allen Chan, Rossa W.K. Chiu, Y.M. Dennis Lo have filed a number of patent applications on cell‐free DNA based molecular diagnostics, including the technology present in this work. Patent royalties are received from Grail, Illumina, Sequenom, DRA, Take2 Health and Xcelom.
Supporting information
Figure S1. Size profiles for nuclear DNA and mtDNA molecules with and without BfaI digestion in plasma of surrogate pregnant women. (A) Size profiles of mtDNA fragments in surrogate maternal plasma DNA with and without BfaI digestion. (B) Size profiles of nuclear DNA fragments in surrogate maternal plasma DNA with and without BfaI digestion.
Figure S2. Plots of size profiles for fetal‐ and maternal‐derived mtDNA molecules in plasma of five surrogate pregnant women (SP1‐5).
Figure S3. Plots of size profiles for fetal‐ and maternal‐derived nuclear DNA molecules in plasma of five surrogate pregnant women (SP1‐5).
ACKNOWLEDGMENTS
We thank Ms. Lee Wing Shan and Ms. Jin Yongjie for their technical assistance in DNA sequencing and data analysis, and Landon Chan for early exploratory work. This work was supported by the Hong Kong Research Grants Council Theme‐Based Research Scheme (T12‐403/15‐N). Y.M. Dennis Lo is supported by an endowed chair from the Li Ka Shing Foundation.
DATA AVAILABILITY STATEMENT
Research methods are available by contacting the corresponding author. Participants have not consented for sequence data sharing.
REFERENCES
- 1. Lo YM, Chan KC, Sun H, et al. Maternal plasma DNA sequencing reveals the genome‐wide genetic and mutational profile of the fetus. Sci Transl Med. 2010;2(61):61‐91. [DOI] [PubMed] [Google Scholar]
- 2. Chan KC, Jiang P, Sun K, et al. Second generation noninvasive fetal genome analysis reveals de novo mutations, single‐base parental inheritance, and preferred DNA ends. Proc Natl Acad Sci U S A. 2016;113(50):E8159‐E8168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Jiang P, Sun K, Tong YK, et al. Preferred end coordinates and somatic variants as signatures of circulating tumor DNA associated with hepatocellular carcinoma. Proc Natl Acad Sci U S A. 2018;115(46):E10925‐E10933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Underhill HR, Kitzman JO, Hellwig S, et al. Fragment length of circulating tumor DNA. PLoS Genet. 2016;12(7):e1006162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cristiano S, Leal A, Phallen J, et al. Genome‐wide cell‐free DNA fragmentation in patients with cancer. Nature. 2019;570(7761):385‐389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Serpas L, Chan RWY, Jiang P, et al. Dnase1l3 deletion causes aberrations in length and end‐motif frequencies in plasma DNA. Proc Natl Acad Sci U S A. 2019;116(2):641‐649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chandrananda D, Thorne NP, Bahlo M. High‐resolution characterization of sequence signatures due to non‐random cleavage of cell‐free DNA. BMC Med Genet. 2015;8:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Snyder MW, Kircher M, Hill AJ, Daza RM, Shendure J. Cell‐free DNA comprises an in vivo nucleosome footprint that informs its tissues‐of‐origin. Cell. 2016;164(1–2):57‐68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Straver R, Oudejans CB, Sistermans EA, Reinders MJ. Calculating the fetal fraction for noninvasive prenatal testing based on genome‐wide nucleosome profiles. Prenat Diagn. 2016;36(7):614‐621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ulz P, Thallinger GG, Auer M, et al. Inferring expressed genes by whole‐genome sequencing of plasma DNA. Nat Genet. 2016;48(10):1273‐1278. [DOI] [PubMed] [Google Scholar]
- 11. Sun K, Jiang P, Cheng SH, et al. Orientation‐aware plasma cell‐free DNA fragmentation analysis in open chromatin regions informs tissue of origin. Genome Res. 2019;29(3):418‐427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cheng THT, Lui KO, Peng XL, et al. DNase1 does not appear to play a major role in the fragmentation of plasma DNA in a knockout mouse model. Clin Chem. 2018;64(2):406‐408. [DOI] [PubMed] [Google Scholar]
- 13. Han DSC, Ni M, Chan RWY, et al. The biology of cell‐free DNA fragmentation and the roles of DNASE1, DNASE1L3, and DFFB. Am J Hum Genet. 2020;106(2):202‐214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ma ML, Zhang H, Jiang P, et al. Topologic analysis of plasma mitochondrial DNA reveals the coexistence of both linear and circular molecules. Clin Chem. 2019;65(9):1161‐1170. [DOI] [PubMed] [Google Scholar]
- 15. Sin STK, Jiang P, Deng J, et al. Identification and characterization of extrachromosomal circular DNA in maternal plasma. Proc Natl Acad Sci U S A. 2020;117(3):1658‐1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lo YM, Corbetta N, Chamberlain PF, et al. Presence of fetal DNA in maternal plasma and serum. Lancet. 1997;350(9076):485‐487. [DOI] [PubMed] [Google Scholar]
- 17. Li H, Durbin R. Fast and accurate long‐read alignment with burrows‐wheeler transform. Bioinformatics. 2010;26(5):589‐595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. MacConaill LE, Burns RT, Nag A, et al. Unique, dual‐indexed sequencing adapters with UMIs effectively eliminate index cross‐talk and significantly improve sensitivity of massively parallel sequencing. BMC Genomics. 2018;19(1):30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Li R, Yu C, Li Y, et al. SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics. 2009;25(15):1966‐1967. [DOI] [PubMed] [Google Scholar]
- 20. Yin PH, Lee HC, Chau GY, et al. Alteration of the copy number and deletion of mitochondrial DNA in human hepatocellular carcinoma. Br J Cancer. 2004;90(12):2390‐2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Qiu C, Hevner K, Abetew D, et al. Mitochondrial DNA copy number and oxidative DNA damage in placental tissues from gestational diabetes and control pregnancies: a pilot study. Clin Lab. 2013;59(5–6):655‐660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lo YM, Zhang J, Leung TN, Lau TK, Chang AM, Hjelm NM. Rapid clearance of fetal DNA from maternal plasma. Am J Hum Genet. 1999;64(1):218‐224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yu SC, Lee SW, Jiang P, et al. High‐resolution profiling of fetal DNA clearance from maternal plasma by massively parallel sequencing. Clin Chem. 2013;59(8):1228‐1237. [DOI] [PubMed] [Google Scholar]
- 24. Gilkerson R, Bravo L, Garcia I, et al. The mitochondrial nucleoid: integrating mitochondrial DNA into cellular homeostasis. Cold Spring Harb Perspect Biol. 2013;5(5):a011080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Shuster RC, Rubenstein AJ, Wallace DC. Mitochondrial DNA in anucleate human blood cells. Biochem Biophys Res Commun. 1988;155(3):1360‐1365. [DOI] [PubMed] [Google Scholar]
- 26. Jiang P, Xie T, Ding SC, et al. Detection and characterization of jagged ends of double‐stranded DNA in plasma. Genome Res. 2020;30(8):1144‐1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sun K, Jiang P, Wong AIC, et al. Size‐tagged preferred ends in maternal plasma DNA shed light on the production mechanism and show utility in noninvasive prenatal testing. Proc Natl Acad Sci U S A. 2018;115(22):E5106‐E5114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Slone J, Huang T. The special considerations of gene therapy for mitochondrial diseases. NPJ Genom Med. 2020;5:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tachibana M, Amato P, Sparman M, et al. Towards germline gene therapy of inherited mitochondrial diseases. Nature. 2013;493(7434):627‐631. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Size profiles for nuclear DNA and mtDNA molecules with and without BfaI digestion in plasma of surrogate pregnant women. (A) Size profiles of mtDNA fragments in surrogate maternal plasma DNA with and without BfaI digestion. (B) Size profiles of nuclear DNA fragments in surrogate maternal plasma DNA with and without BfaI digestion.
Figure S2. Plots of size profiles for fetal‐ and maternal‐derived mtDNA molecules in plasma of five surrogate pregnant women (SP1‐5).
Figure S3. Plots of size profiles for fetal‐ and maternal‐derived nuclear DNA molecules in plasma of five surrogate pregnant women (SP1‐5).
Data Availability Statement
Research methods are available by contacting the corresponding author. Participants have not consented for sequence data sharing.


