Abstract
Pseudomonas aeruginosa causes chronic airway infections, a major determinant of lung inflammation and damage in cystic fibrosis (CF). Loss-of-function lasR mutants commonly arise during chronic CF infections, are associated with accelerated lung function decline in CF patients and induce exaggerated neutrophilic inflammation in model systems. In this study, we investigated how lasR mutants modulate airway epithelial membrane bound ICAM-1 (mICAM-1), a surface adhesion molecule, and determined its impact on neutrophilic inflammation in vitro and in vivo. We demonstrated that LasR-deficient strains induce increased mICAM-1 levels in airway epithelial cells compared to wild-type strains, an effect attributable to the loss of mICAM-1 degradation by LasR-regulated proteases and associated with enhanced neutrophil adhesion. In a subacute airway infection model, we also observed that lasR mutant-infected mice displayed greater airway epithelial ICAM-1 expression and increased neutrophilic pulmonary inflammation. Our findings provide new insights into the intricate interplay between lasR mutants, LasR-regulated proteases and airway epithelial ICAM-1 expression, and reveal a new mechanism involved in the exaggerated inflammatory response induced by lasR mutants.
Author summary
Cystic fibrosis (CF) patients develop progressive lung disease characterized by chronic airway infections, commonly caused by the opportunistic pathogen Pseudomonas aeruginosa, and excessive non-resolving neutrophilic inflammation. Loss of function mutations of the lasR quorum sensing transcription regulator gene commonly arise during chronic P. aeruginosa infections and are associated with increased lung inflammation. In this study, we demonstrated that loss-of-function lasR mutants induced increased mICAM-1 levels on airway epithelial cells compared to wild-type P. aeruginosa strains in cell culture and in murine infection models. This effect was caused by the loss of ICAM-1 degradation by LasR-regulated secreted protease, facilitated neutrophil adhesion, and was associated with increased neutrophilic lung inflammation. Our study provides novel insights into the P. aeruginosa—airway epithelial–neutrophil interactions, and demonstrates how a common pathoadaptation of P. aeruginosa may drive lung disease progression by exacerbating inflammation.
Introduction
Individuals with the genetic disease Cystic Fibrosis (CF) develop progressive lung disease characterized by chronic airway infections and exuberant neutrophilic inflammation. Mutations in the Cystic Fibrosis Transmembrane Conductance Regulator (CFTR) gene lead to impaired chloride secretion and poor mucociliary clearance, making CF patients prone to lung infections. Chronic airway infections are major drivers of persistent inflammation in CF lung disease [1]. The majority of adult CF patients are infected with Pseudomonas aeruginosa for decades, which is associated with increased morbidity and mortality [2–4]. Within the airways, P. aeruginosa interacts with airway epithelial cells (AEC) to induce expression of pro-inflammatory mediators, modulate inflammatory pathways and further exacerbate lung inflammation [5]. Adding to a hyper-inflammatory state associated with the intrinsic CFTR defects [6,7], neutrophil responses to P. aeruginosa in the CF lung are both ineffective and excessive, further contributing to lung damage [8,9].
During chronic infection, P. aeruginosa evolves and genetically adapts to the host lung environment [10]. P. aeruginosa isolates recovered from chronic “late” stage CF infections are genotypically and phenotypically distinct from those found in “early” stage infections, and commonly share phenotypes such as mucoidy, loss of motility or protease production [11,12]. A notable example of this convergent evolution is the loss of LasR function, typically attributable to mutations in lasR, the gene encoding the major quorum sensing transcriptional regulator in P. aeruginosa [13–17]. Loss of function lasR variants are found in at least one third of CF patients chronically infected with P. aeruginosa [15]. Quorum sensing is a bacterial communication system that allows the coordinated expression of hundreds of P. aeruginosa genes, including many that encode exoproducts and virulence factors [18–20].
Although loss of LasR function leads to the attenuation of bacterial virulence in models of acute infection [12,21,22], chronic infections with lasR mutants in CF patients have been associated with accelerated decline in lung function [15] and increased markers of inflammation [23]. To understand the impact of P. aeruginosa lasR mutants on host-pathogen interactions and on the inflammatory responses relevant to CF lung disease, our group previously demonstrated that lasR variants elicited an enhanced pro-inflammatory cytokine response in AEC due to a loss of cytokine degradation by LasR-regulated proteases, leading to greater neutrophil recruitment in vitro. lasR variants also caused exaggerated neutrophilic inflammation and immunopathology in a murine model of sub-acute airway infection [23].
Adhesion molecules, such as cell surface intercellular adhesion molecule 1 (ICAM-1), play an important role in lung inflammation [24]. Membrane bound ICAM-1, expressed on endothelial and epithelial cells, is a ligand to β2 integrins on leukocytes and is involved in neutrophil migration, adhesion, and other functions [25–28]. Although the role of epithelial ICAM-1 is less well established than endothelial ICAM-1, which functions as the major adhesion receptor for leukocyte rolling-adhesion and transendothelial migration [28], studies suggest that airway epithelial ICAM-1 likely plays an important role in lung inflammation. Epithelial ICAM-1 mediates neutrophil adhesion and retention in the respiratory compartment, and contributes to lung inflammation in the setting of pulmonary infection and endotoxin-induced injury [26,29–33]. In fact, epithelial cells express low levels of ICAM-1 unless infected or stimulated by inflammatory cytokines [34,35] or bacterial products including P. aeruginosa secreted products [31,36,37]. Hubeau et al have also reported that AEC in the human CF lung over-express ICAM-1 compared to non-CF tissues, and ICAM-1 surface epithelial expression is associated with spatially adjacent neutrophil accumulation [38].
Since airway epithelial ICAM-1 expression is upregulated in P. aeruginosa infection, is a feature of CF lung disease and mediates neutrophilic inflammation in the lung, this led us to investigate the effect of lasR mutants on membrane bound ICAM-1 (mICAM-1) in AEC and its contribution to neutrophilic inflammation in vitro and in vivo.
Material and methods
Ethics statement
All experiments using human neutrophils were approved by the Research Ethics Board of the McGill University Health Centre (protocol 14–169), with informed written consent obtained from all subjects. All animal experiments were carried out with approval from the Animal Care Committee of the RI MUHC (AUP #2015–7586).
Bacterial strains, plasmids and growth conditions
All bacterial strains used in this study are described in detail in S1 Table. Primer sequences and plasmids are listed in S2 and S3 Tables respectively. To generate the constructs for inducible protease expression, the lasA, aprA and prpL coding sequences were amplified by PCR from the PAO1 genome using primers lasA-GWB5-RBS, lasA-GWB2, aprA-GWB5-RBS, aprA-GWB2, prpL-GWB5-RBS and prpL-GWB2 pairs respectively. The PCR products were recombined into pDONR221P5P2 using BP clonase II to generate the entry vectors pENTR-lasA, pENTR-prpL and pENTR-aprA. The constructed entry vectors pENTR-lasA, pENTR-prpL and pENTR-aprA were each recombined with the vector pJJH187 and the destination vector miniCTX2.1-Tc-GW using LR Clonase II Plus (Invitrogen) to generate the arabinose inducible constructs pEXP-lasA, pEXP-prpL and pEXP-aprA. The individual expression vectors were subsequently integrated into the genome of the Late strain as previously described [23]. Complemented strains were selected on LB agar containing 50 μg/ml tetracycline. Protease expression was induced with 1% (w/v) L-arabinose (ACROS Organics) added to the growth media.
P. aeruginosa filtrate preparation
Bacteria were grown overnight in 5 mL LB medium (BD Difco), washed twice in sterile phosphate-buffered saline (PBS) and the cell pellets were resuspended at an OD600 of 0.05 in synthetic CF medium (SCFM), which was developed to resemble the nutrient composition in CF sputum [39]. 5 mL planktonic cultures were incubated at 37°C for 24h, with shaking at 250 rpm, then centrifuged for 5 min at 5000 rpm to pellet cells. The supernatants were filtered with 0.22 μm cellulose acetate filters (Fisher Scientific) to generate sterile cell-free filtrates. The remaining cell pellets were resuspended in PBS to measure the OD600 for estimation of the cell biomass. Filtrates were first normalized to the OD600 of the pellet by dilution in SCFM and then stored at -20°C until use. As control, we note that the normalization of each filtrate to the OD600 or total protein content of the pellet were equivalent. Each filtrate aliquot was discarded after one freeze-thaw cycle. Where indicated, filtrates were heat-inactivated for 10 min at 95°C.
Airway epithelial cell culture conditions and stimulation with P. aeruginosa filtrates
Immortalized human airway epithelial cells (BEAS-2B) were cultured in cell culture-grade plates in DMEM (Wisent) supplemented with 10% heat-inactivated fetal bovine serum (FBS, Wisent), penicillin and streptomycin (Wisent) at 37°C with 5% CO2. Once 80–90% confluent, cells were seeded with 150,000 cells/well into 12- well tissue culture plates (Sarstedt) for flow cytometry experiments, or with 37,500 cells per well in 48-well tissue culture plates (Sarstedt) for neutrophil adhesion assays. Cells were seeded 48 h prior to stimulation with filtrates, and 24 h before stimulation, the culture medium was changed to starvation media (DMEM containing penicillin, streptomycin and 0.5% heat-inactivated FBS). BEAS-2Bs were stimulated with 30 μL P. aeruginosa filtrate (in 1 mL total volume) for flow cytometry experiments or 7.5 μL filtrate (in 250 μL total volume) for neutrophil adhesion assays for a duration of 24 h in fresh starvation media at 37°C with 5% CO2. Equal volumes of SCFM were used as a negative control and a final concentration of 20 ng/mL TNF-α (BioLegend) was used as a positive control for ICAM-1 induction and neutrophil binding.
mICAM-1 measurement
Following stimulation with P. aeruginosa filtrates, the supernatants of BEAS-2B cultures were collected and 500 μL cold Accutase (STEMCELL Technologies) was added to each well and incubated at room temperature for 10 min to detach cells. The BEAS-2B cell suspensions were then added to their respective supernatants of the same well and centrifuged at 2000 rpm. The cells were resuspended in FACS buffer (PBS+1% heat-inactivated FBS), stained with 1:1000 Fixable Viability Dye eFluor 780 (eBioscience) for 25 min on ice, then with 1:50 FITC-conjugated anti-human ICAM-1 antibody or IgG1-FITC isotype control (R&D Systems) for 30 min on ice, followed by fixation with 0.5% PFA for 10 min at room temperature. Cells were analyzed with a LSR II flow cytometer (BD Biosciences) and results were analyzed using FlowJo (BD Biosciences). For our analysis, debris (low FSC-A/SSC-A), doublets (higher SSC-A than SSC-H) and dead cells (high eFluor 780) were excluded. For each condition, the results were calculated by subtracting the median fluorescence intensity (MFI) of the isotype control from the corresponding sample’s ICAM-1 MFI.
Protease activity measurements of P. aeruginosa filtrates
Total protease activity in filtrates was measured using the Hide-Remazol Brilliant Blue assay as previously described [40]. Caseinolytic activity in filtrates was assessed by spotting 30 μL of filtrates on sterile 6mm paper disk placed on skim milk 1.5% agar plates as previously described [41] and the zone of clearance (diameter minus the 6mm filter) was measured after 16 h incubation at 37°C. Elastolytic activity in filtrates was measured by Elastin-Congo Red (ECR) assay, as previously described in detail [23].
Recombinant human ICAM-1 (rhICAM-1) degradation assay
To measure the ICAM-1 degradation by P. aeruginosa secreted proteases in vitro, 25 μL of 10 μg/mL (250 ng) rhICAM-1 (Peprotech) was incubated with 5 μL P. aeruginosa filtrate or PBS (negative control) for 24 h at 37°C, shaking at 200 rpm. The samples were then diluted in 4X loading buffer containing DTT, incubated at 95°C for 5 min, then stored at -20°C until quantification of rhICAM-1 by Western Blotting. 20 μL of each rhICAM-1 degradation sample was loaded onto 4–20% Mini-PROTEAN gels (Bio-Rad), separated by SDS-PAGE and then transferred onto PVDF membranes. The blots were blocked in 5% (w/v) skim milk prior to incubation with polyclonal anti-rhICAM-1 antibody (500-P287, Peprotech) overnight at 4°C. The protein bands were detected with anti-rabbit IgG DyLight 800 4X PEG conjugated secondary antibody (Cell Signaling Technology), imaged with the Odyssey imaging system (Li-Cor Biosciences) and quantified using the Odyssey V3.0 software (Li-Cor Biosciences). Results are shown as % band density compared to the negative control (rhICAM-1 incubated with PBS control) on the respective blot. Full blots are provided in S1 Spreadsheet.
Neutrophil adhesion assay
Primary human neutrophils were isolated from 5 mL whole blood collected from healthy volunteer donors using the EasySep Direct Human Neutrophil Isolation Kit (Stemcell) according to the manufacturer’s instructions in phenol red-free RPMI-1640 with 1% heat-inactivated donor serum, and included incubation in RBC lysis buffer (Stemcell) for 10 min. Neutrophils were stained with 1 μM calcein-AM for 30 min in the dark at room temperature, then washed twice with media and filtered through a 40 μm nylon cell strainer cap (Fisherbrand). BEAS-2B cells were first stimulated with 30 μL P. aeruginosa filtrates for 24h, and after removal of the filtrate-containing media, BEAS-2B cells were co-incubated with 4x105 neutrophils in phenol red-free DMEM supplemented with 1% heat-inactivated donor serum for 2 h at 37°C with 5% CO2. Neutrophils were then removed and BEAS-2B cells were washed three times with media to remove non-adherent neutrophils. For control experiments, 4x105 neutrophils were stained and co-incubated with AEC as described above or incubated in wells containing media without AEC for 2h, then washed as described above. Adherent neutrophils were counted in a blinded manner using three representative fields of view per well obtained by laser scanning confocal microscopy (10X objective, Zeiss LSM700). Statistical analysis was performed using the average number of neutrophils per field of view (1.64 mm2) for each individual well.
Subacute murine P. aeruginosa airway infection
Mice were infected with P. aeruginosa embedded in agar beads to generate a subacute non-lethal airway infections, as previously described [23]. Briefly, bacterial suspensions were mixed at 1:1 (v/v) in 2% LB and 3% molten agar with continuous stirring into mineral oil to generate bacteria embedded agar beads. Sterile PBS LB agar beads were used as a negative control. Adult male C57BL/6 mice (6 to 9 weeks old, Charles River) were infected by non-surgical intratracheal injection of 50 μL bead suspension containing ~5*105 CFU/mouse. The mice were sacrificed at 2 or 4 days post infection (p.i). After cardiac puncture, the lungs were perfused by PBS injection into the vena cava to remove blood leukocytes prior to harvest. Perfused lungs were then lavaged with 4 x 0.5 mL ice-cold PBS through an intra-tracheal catheter for collection of the bronchoalveolar fluid (BALF). To measure the pulmonary bacterial load, lungs were harvested, homogenized and serially diluted for viable CFU counts on LB agar plates.
Immune cell counts in lung homogenates and bronchoalveolar fluid (BALF)
For lung homogenates, perfused and lavaged lungs were placed in RPMI, minced, digested with 150 U/mL collagenase (Sigma-Aldrich) for 1 h at 37°C, then homogenized through a 16G needle. Cell suspensions were then filtered through a 100 μm cell strainer (BD Biosciences) and red blood cells were lysed with 0.2% (w/v) NaCl. The remaining cells were washed, resuspended in RPMI and enumerated using a Z1 cell counter (Beckmann-Coulter). Single cell suspensions were stained with Fixable Viability Dye eFluor780 (1:1000, eBioscience), blocked with anti-murine CD16/CD32 (1:100, eBioscience) then surface stained with eFluor610-conjugated anti-murine CD45 (1:40, 30-F11, eBioscience), FITC-conjugated anti-mouse CD11c (1:200, N418, eBioscience) and eFluor710-conjugated anti-murine Ly6G (1:160, 1A8, eBioscience). Cells were fixed with Cytofix (BD) and analyzed by flow cytometry using a LSR II Fortessa X-20 (BD Biosciences) and FlowJo 10.0.7 software (BD Biosciences). Neutrophils were defined as single, live, CD45+, Ly6G high and CD11c- cells, and total neutrophil counts were calculated by multiplying the proportion of neutrophils by the total number of live cells.
For the BALF, cells were spun down at 1000 rpm for 10 min, resuspended in PBS and counted by hemocytometer. For immune cell counts, cells collected from the BALF were loaded onto Shandon Cytoslides (Thermo Fisher), air-dried, stained with Shandon Kwik-Diff (Thermo Fisher) and analyzed by light microscopy to obtain average neutrophil, monocyte/macrophage and lymphocyte counts.
Lung histopathology and ICAM-1 immunofluorescence
Perfused and lavaged lungs were inflated, fixed overnight with 10% formalin phosphate solution and sectioned in 5 μM thick slices. For histopathology, paraffin-embedded lung sections were stained with H&E and images were acquired using an Olympus BX51 microscope fitted with an Olympus DP70 CCD camera. For immunofluorescence, lung sections were stained with an anti-mouse ICAM-1 primary antibody (1:1000, eBioscience, YN1/1.7.4) and OmniMap anti-rat HRP (Ventana). All slides were processed using a Ventana DISCOVERY ULTRA automated slide preparation system (Roche) at the same time. The lung sections were imaged by laser scanning confocal microscopy (20x objective, Zeiss LSM700) at Ex 488 nm/Em 518 nm to visualize the autofluorescence of the lung tissues, and Ex 542 nm/Em 568 nm to visualize the ICAM-1 signal. All lung sections were imaged using identical confocal microscopy settings. Seven representative fields of view containing at least one airway cross-section were randomly selected per lung and imaged in a blinded manner.
Image analyses to quantify airway epithelial ICAM-1 expression were performed in two independent manners. First, the airway epithelial ICAM-1 expression of each airway was scored by visual assessment from 1 (no signal) to 10 (very strong) in a blinded manner by two independent reviewers. Second, ICAM-1 fluorescence intensity was quantified within the airway epithelium based on a manually designated region of interest (ROI) using the Icy software (V2.0.3.0, Institut Pasteur) [42]. The total fluorescence intensity of each ROI above a set threshold (defined by ICAM-1 negative regions in PBS lungs) was measured and normalized to the ROI surface area (in pixels).
P. aeruginosa colony morphology
Bacteria were grown on LB agar for 16h. The presence of a metallic sheen caused by the accumulation of 4-hydroxy-2-heptylquinoline, which has previously been linked to loss of LasR function [16], was assessed visually.
N-3-oxo-dodecanoyl-homoserine lactone (3-oxo-C12-HSL) measurement
Production of the LasR autoinducer 3-oxo-C12-HSL was measured as previously described [43,44]. Briefly, bacterial strains were grown in LB medium + 50 mM MOPS for 16h. 3-oxo-C12-HSL levels were quantified using the bioassay strain E. coli DH5α expressing pJN105L and pSC11.
Statistical analyses
All results are shown as mean ± SD or SEM (as indicated), unless otherwise stated in the figure legends. Statistical analyses between two groups were performed using an unpaired two-tailed t-test or Mann-Whitney test as appropriate. Analyses between three or more groups were performed using a one-way ANOVA and Sidak’s multiple comparisons test. Correlation between two measurements were estimated by linear regression and statistical analysis was conducted using Pearson’s correlation analysis. Proportions within two groups were compared by Chi-Square test. Statistical analyses were done in Graphpad Prism 8.3.0 (GraphPad Software).
Results
Stimulation with LasR-deficient variants induces higher mICAM-1 levels in AECs than wild-type P. aeruginosa
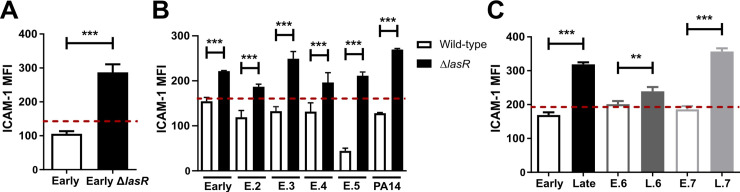
In order to compare the AEC mICAM-1 responses to secreted factors from loss-of-function lasR mutant and wild-type strains, we first stimulated BEAS-2B cells with filtrates of a P. aeruginosa clinical isolate isolated from early CF infection (“Early”) and its isogenic lasR mutant, and measured cell surface mICAM-1 levels by flow cytometry. We observed a 2.7-fold increase in mICAM-1 levels in cells stimulated with the lasR mutant filtrates compared to the wild-type filtrates (Fig 1A). Of note, stimulation with TNF-α was used as a positive control for mICAM-1 induction in every experiment (S1A Fig) and stimulation with SCFM medium, which was used as a negative control, did not induce significantly greater mICAM-1 expression than starvation media alone (S1B Fig).
Fig 1. LasR deficient variants induce increased levels of mICAM-1 in AEC.
BEAS-2B cells were stimulated for 24h with 30 μL sterile filtrates of (A) the Early clinical isolate and its isogenic lasR mutant, (B) five “early” clinical isolates or PA14 wild-type and their isogenic lasR mutants or (C) three “early” clinical isolates paired with their clonally-related “late” isolates (Early/Late, E6/L6, E7/L7). mICAM-1 levels (MFI) were measured by flow cytometry, with SCFM (media) serving as negative control (—dashed line). Results are shown as the mean ± SD of one representative experiment (from ≥ 2 independent experiments, each with biological triplicates). *P < 0.05; **P < 0.01; ***P < 0.001.
To validate that the increased mICAM-1 response was attributable to the loss of LasR function in multiple P. aeruginosa strain backgrounds, we then stimulated AEC with filtrates from five other pairs of wild-type and isogenic lasR mutant strains, including four “early” clinical isolates (E.2 to E.5) and the common P. aeruginosa reference strain PA14. Stimulation with all lasR mutant filtrates induced mICAM-1 levels to a greater degree than their wild-type parental filtrates, ranging from a 1.4- to a 4.7-fold increase in mICAM-1 levels (Fig 1B). Furthermore, all mICAM-1 flow cytometry measurements were gated on live cells, and the cell viability ranged from 86.4% to 96.4% in different conditions (S1C Fig). We therefore conclude that these modest differences in cell viability were unlikely to be sufficient to explain the magnitude of mICAM-1 reduction observed in wild-type filtrate-stimulated AEC.
Since loss-of-function lasR mutations are commonly found in P. aeruginosa clinical isolates recovered during late stage CF infections, we next compared several “early” infection LasR-competent clinical isolates to their clonally related, “late” infection LasR-deficient isolates (characterized by low protease production, low 3-oxo-C12 HSL autoinducer levels and metallic colony sheen) [16] recovered from the same patients, namely the Early/Late, E.6/L.6 and E.7/L.7 paired isolates as characterized in S4 Table. We observed a similar pattern in mICAM-1 response, with stimulation by all “late” isolates eliciting a higher mICAM-1 response (Fig 1C). Both the Late and L.7 filtrate resulted in 1.9-fold greater mICAM-1 levels compared to their clonally related “early” isolates, but the difference in mICAM-1 levels was more modest (1.2-fold) with the E.6 - L.6 pair. The variability observed across the different “early-late” pairs was not surprising since “late” isolates harbour numerous genetic and phenotypic differences compared to their clonally related “early” isolates. Furthermore, different lasR mutations have varying effects on LasR function, and the LasR regulon can vary across different strain backgrounds [45]. These results thus indicated that loss-of-function lasR variants, both genetically engineered and naturally occurring ones, elicited higher mICAM-1 responses in human AEC than their respective wild-type counterparts, but that the magnitude of this phenotype varied depending on the P. aeruginosa strain background. Since the P. aeruginosa blue-green pigment pyocyanin can stimulate ICAM-1 expression [36] and is typically a LasR-controlled secreted secondary metabolite, we also noted that pyocyanin production was significantly decreased in the Early ΔlasR and Late strains compared to the Early strain (S1D Fig), thus indicating that the mICAM-1 expression in lasR mutant-stimulated AEC was unlikely to be attributable to increased pyocyanin levels.
Increased mICAM-1 levels on lasR mutant-stimulated AEC correlate with decreased caseinolytic and elastolytic activity in lasR mutant filtrates
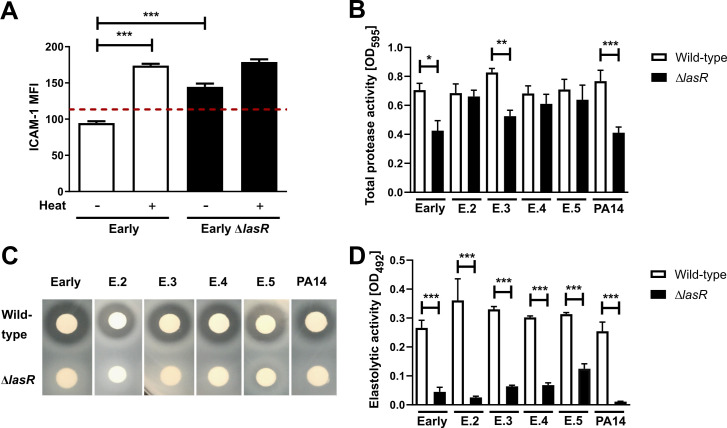
In nearly all wild-type stimulation conditions, we also noted that mICAM-1 levels were below those observed in media control conditions (Figs 1A–1C and 2A), raising the possibility that mICAM-1 might be degraded or downregulated by wild-type filtrates. To start investigating this hypothesis, we heat-treated wild-type and lasR mutant filtrates prior to stimulation to inactivate heat labile bacterial exoproducts, which include secreted proteases. As shown in Fig 2A, heat inactivation of wild-type filtrates restored mICAM-1 to levels above those seen with non-heat treated lasR mutant filtrates, indicating that LasR-regulated heat-labile compound(s) present in wild-type filtrates degraded or down-regulated mICAM-1. We also noted that heat inactivation in both wild-type and lasR mutant filtrates led to a 1.8- and 1.2-fold increase in mICAM-1 levels compared to their respective non-heat-treated filtrates (Fig 2A), suggesting the concurrent presence of heat-stable compound(s) that either induced ICAM-1 production or prevented its degradation in both wild-type and lasR mutant filtrates.
Fig 2. Induction of mICAM-1 is associated with reduced caseinolytic and elastolytic activity in lasR mutant filtrates.
BEAS-2B cells were stimulated for 24h with 30 μL filtrates of (A) the Early and isogenic Early ΔlasR mutant (+/- heat inactivation). mICAM-1 levels (MFI) were measured by flow cytometry, with SCFM (media) serving as negative control (—dashed line). Protease activity in wild-type and lasR mutant filtrates was characterized for (B) total protease activity using the Hide-Remazol Brilliant Blue assay, (C) for caseinolytic activity using the clearance zone diameter on skim milk agar and (D) for elastolytic activity using the Elastin-Congo Red assay. Results for (A) are shown as mean ± SD of one representative experiment (from ≥ 2 independent experiments, each with biological triplicates). Results for (B) and (D) are shown as mean ± SEM, with pooled data (n ≥ 4 biological replicates from two independent experiments). Results for (C) are representative of ≥ 2 independent experiments in biological duplicates. *P < 0.05; **P < 0.01; ***P < 0.001.
LasR regulates the expression of several secreted proteases such as AprA, LasA, LasB and type IV protease (T4P) that can degrade components of host defenses and immunity [23,46–48], some of which have been shown to be heat-labile [23,49]. Therefore, we hypothesized that the loss of mICAM-1 protein signal upon stimulation with wild-type filtrates was due to the activity of these proteases, and sought to characterize the proteolytic activity in wild-type and lasR mutant filtrates. We measured protease activity in several manners, with a Hide Blue degradation assay for total protease activity, skim milk plates for caseinolytic activity and Elastin-Congo Red assay for elastolytic activity. Although the total protease activity only showed differences in three (Early, E.3, PA14) out of the six pairs (Fig 2B), caseinolytic (Fig 2C) and elastolytic activity (Fig 2D) were highly reduced (at least by 68%) or undetectable in all lasR mutants compared to parental wild-type strains. We also confirmed both caseinolytic and elastolytic activities to be heat-labile (S2A and S2B Fig). These results therefore indicated that there was a significant loss in caseinolytic and elastolytic activities among all lasR mutants tested. We further observed a negative correlation between the filtrates’ protease activity and ICAM-1 induction on AEC, with caseinolytic protease activity showing a stronger correlation (R2 = 0.76) than elastolytic protease activity (R2 = 0.66) (S2C and S2D Fig).
LasR-regulated proteases degrade mICAM-1
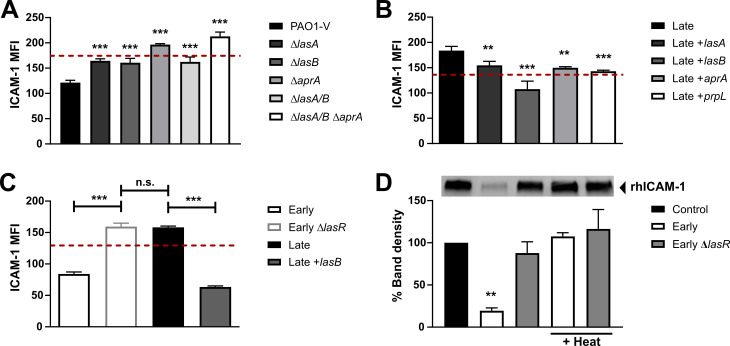
To test the contribution of the different LasR-regulated proteases on mICAM-1, we measured the effect of PAO1-V, an invasive P. aeruginosa isolate, and its single (lasA, lasB, aprA), double (lasA/lasB) and triple (lasA/lasB/aprA) protease mutants on mICAM-1 levels by flow cytometry. We observed a modest increase (1.3- to 1.4-fold) in mICAM-1 levels upon stimulation with filtrates from the ΔlasA, ΔlasB and ΔlasA ΔlasB strains (compared to wild-type) as well as a more pronounced increase (1.6- and 1.8-fold) upon stimulation with the ΔaprA and triple mutant, respectively (Fig 3A), suggesting that AprA might have the greatest effect on ICAM-1 levels in this strain background. The caseinolytic activity of the protease mutants further confirmed the relative contribution of each proteases in the PAO1-V strain background, as loss of AprA caused the greatest reduction in caseinolytic activity among the three proteases, while loss of all three LasB/LasB/AprA completely abrogated all caseinolytic activity (S3A Fig). Not surprisingly, elastolytic activity was nearly undetectable in the lasB and triple protease mutant (S3B Fig).
Fig 3. LasR-regulated proteases degrade ICAM-1.
BEAS-2B cells were stimulated for 24h with 30 μL filtrates of (A) PAO1-V and its isogenic protease mutants of lasA, lasB and aprA, (B) the Late isolate and complemented strains Late +lasA, Late +lasB, Late +aprA or Late +prpL (T4P) or (C) Early, Early ΔlasR, Late, Late +lasB. mICAM-1 levels were measured by flow cytometry, with SCFM (media) serving as negative control (—dashed line). (D) in vitro degradation of recombinant human ICAM-1 (rhICAM-1) by P. aeruginosa filtrate. In each sample, 250 ng rhICAM-1 was incubated with 5 μL filtrates of the Early and Early ΔlasR strains (+/- heat inactivation) or PBS control for 24h, and the remaining intact rhICAM-1 following degradation was quantified by Western Blotting. Results in (A), (B) and (C) are shown as mean ± SD from one representative experiment (from ≥ 2 independent experiments, each with biological triplicates). Results in (D) are displayed as a representative Western Blot and quantification of the % band density compared to the PBS control condition (n ≥ 3 biological replicates from two independent experiments). Different lanes were cropped from the same blot and imaged at the same exposure. Full blots can be found in the S1 Spreadsheet. *P < 0.05; **P < 0.01; ***P < 0.001.
We further investigated the contribution of individual proteases by genetically complementing the Late (LasR-deficient) strain with the lasA, lasB, aprA or prpL (T4P) genes under control of an arabinose-inducible promoter to generate the Late +lasA, Late +lasB, Late +aprA and Late +prpL constructs (S1 Table). We confirmed that caseinolytic activity was increased upon complementation with the +lasB, +aprA and +prpL constructs (S3C Fig) and that elastolytic activity, which is mostly attributed to LasB, was only restored upon complementation with the +lasB construct (S3D Fig). Complementation with any of the four proteases resulted in decreased mICAM-1 levels compared to stimulation with the parental Late strain filtrate, with the greatest reduction in mICAM-1 levels (1.7-fold) observed with the Late +lasB strain (Fig 3B). Notably, lasB complementation of the Late isolate was sufficient to restore mICAM-1 levels to those observed in AECs stimulated with wild-type filtrates (Fig 3C).
To examine whether the decreased ICAM-1 levels induced by wild-type filtrates were due to direct degradation of ICAM-1, we incubated recombinant human ICAM-1 (rhICAM-1) with Early and Early ΔlasR filtrates and measured the in vitro rhICAM-1 degradation by Western blotting. Incubation with the Early, but not the Early ΔlasR filtrate, resulted in a significant reduction (81% decrease) in detectable and thus intact rhICAM compared to the PBS control (Fig 3D). We also confirmed that in vitro rhICAM-1 degradation was abrogated upon heat-inactivation of filtrates (Fig 3D), and reduced upon loss of one or more LasR-regulated proteases (S3E Fig). These results suggested that several LasR-regulated proteases present in wild-type filtrates can degrade rhICAM-1, the individual contribution of which might be strain-dependent.
Neutrophil binding to AEC is increased upon stimulation with lasR mutant filtrates
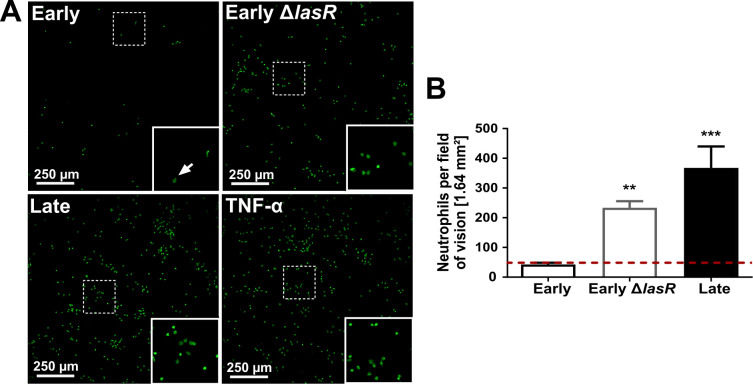
Next, we sought to characterize the functional consequences of the increased mICAM-1 responses in AEC stimulated with mutant lasR filtrates. Since mICAM-1 is involved in neutrophil adhesion to endothelial and epithelial cells [28,34,50], we hypothesized that AEC stimulated with lasR mutant filtrates will bind more neutrophils than AEC stimulated with wild-type filtrates. To test this, we quantified the adhesion of primary human neutrophils to AECs pre-stimulated with bacterial filtrates by confocal microscopy, and observed that significantly higher numbers of neutrophils adhered to AEC stimulated with lasR mutant filtrates (both Early ΔlasR and Late) compared to wild-type filtrate (5.5- and 8.6-fold increase respectively) or media control (Fig 4A and 4B). We also confirmed that neutrophil binding was specific to AEC, as control experiments using wells without AEC showed no adherent neutrophils (S3F Fig).
Fig 4. Neutrophil adhesion is increased in AEC stimulated with lasR mutant filtrates.
BEAS-2B cells were pre-stimulated with 30 μL filtrates of Early, Early ΔlasR and Late strains, then co-incubated with calcein-stained primary neutrophils (green). 20 ng/mL TNF-α served as positive control and SCFM served as negative control (—dashed line). Adherent neutrophils were visualized by confocal microscopy (10X objective) in (A) and quantified in (B). Results are shown as mean ± SD from one representative experiment (from 2 independent experiments, each with 3 biological replicates). *P < 0.05; **P < 0.01; ***P < 0.001.
The lasR mutant induces greater bronchial ICAM-1 levels and neutrophilic lung infiltration in a subacute murine lung infection model
We previously reported that lasR mutants induced a hyperinflammatory phenotype with higher levels of pro-inflammatory cytokines IL-6 and IL-8, greater neutrophilic inflammation and increased immunopathology compared to wild-type strains in a murine model of subacute lung infection [23]. In this well-established model, bacteria are embedded in agar beads and inoculated endotracheally, causing a non-lethal airway-centric infection that persist and is associated with neutrophilic inflammation. Our observations of ICAM-1 modulation in vitro therefore led us to ask whether lasR mutant infections were also associated with increased airway ICAM-1 expression in vivo. We infected C57BL/6 mice with wild-type or lasR mutant bacteria and analyzed the airway epithelial ICAM-1 expression by immunofluorescence, with confocal microscopy imaging of airway cross section on whole lung thin sections (S4A Fig). We analyzed mice at 2 and 4 days post-infection (p.i) and confirmed that both infection groups harboured equivalent bacterial burden at all time points (S4C Fig).
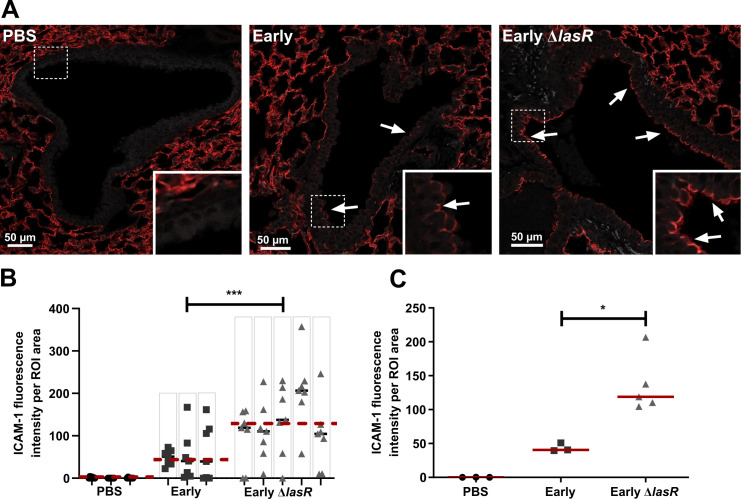
Although ICAM-1 can be expressed by multiple cell types in the lung, including endothelial and alveolar epithelial cells, we focused on the ICAM-1 expression of the bronchial epithelium where we observed a strong induction in expression with P. aeruginosa infection, whereas ICAM-1 expression in the alveolar compartment remained constant in all conditions and time points (Fig 5A, area surrounding airways). To quantify ICAM-1 expression, we first developed an automated image analysis method to measure ICAM-1 signals across multiple airway cross-sections per lung section, and to normalize the total ICAM-1 fluorescence of each airway epithelial cross- section to its surface area (as outlined in S4A Fig). As highlighted in Figs 5A and S4B, bronchial ICAM-1 expression was largely restricted to the apical (luminal) side of the bronchial epithelium. We validated our automated ICAM-1 quantification method using a conventional semi-quantitative scoring system, and given the very good correlation between the two methods (R2 = 0.84, S4D Fig), we proceeded with the automated approach. The median ICAM-1 expression of all airways was significantly (3.2-fold) higher in lasR mutant infected mice compared to wild-type infected mice (Fig 5B). Analysis of pooled lung sections demonstrated a 2.9-fold higher median ICAM-1 airway signal in lasR mutant infected mice compared to wild-type infected mice at 2 days p.i, and consistently low (or undetectable) levels of ICAM-1 in PBS control mice (Fig 5C).
Fig 5. lasR mutant infected mice display increased airway epithelial ICAM-1 expression.
Mice were infected with Early, Early ΔlasR or PBS embedded agar beads and lungs were harvested at 2 days p.i. for ICAM-1 immunofluorescence (shown as red). (A) Images shown are representative of airway cross-sections displaying the median ICAM fluorescence scores of each infection group. Images were taken with a 20X objective, inset boxes are digital magnification of the bronchial epithelium and the white arrows point to regions of high ICAM-1 expression localized to the apical epithelial surface (luminal side). The green autofluorescence (Ex 488/ Em 518, shown as grey) was imaged to visualize the tissue structure. (B) ICAM-1 total fluorescence normalized to the airway surface area (ROI) in every image, with results shown as median within each lung (black line) and median within each group (—dashed line), each dot corresponding to one distinct airway, and columns corresponding to one distinct mouse lung. (C) Median normalized ICAM-1 fluorescence, with results shown as medians using data pooled from 7 randomly selected airways per lung, and each dot corresponding to one mouse (n ≥ 3 mice per condition). *P < 0.05; **P < 0.01; ***P < 0.001.
We also noted considerable heterogeneity in airway mICAM-1 expression within the same mouse lung (each mouse with its individual airway data points is represented in columns, Fig 5B) in both infection groups. This anatomically heterogeneous pattern was not surprising given that we used an airway infection model where agar bead embedded bacteria are entrapped within airway lumens and cause foci of infection rather than a diffuse infection throughout the lung [51]. As a result, lung regions in closest proximity to bacteria-containing beads displayed the greatest host responses to infection while more distant regions remained relatively normal, a pattern observed in other studies using a similar infection model [26,51]. By 4 days p.i, the majority of airways showed little to no mICAM-1 expression, and no significant differences were detected in the two infection groups (S4E Fig). While the proportion of ICAM-1 negative airways (i.e. airways with ICAM-1 signal comparable to PBS control) was not significantly different across both infection groups at 2 days p.i. (81% vs 91%, p = 0.251), we observed a trend towards a higher percentage of ICAM-1-positive airways in lasR mutant-infected mice at 4 days p.i. compared to wild-type-infected mice (40% vs 18%, p = 0.057, S4F Fig). Together, these results thus suggested that lasR mutants were associated with increased airway ICAM-1 induction both in vitro and in vivo.
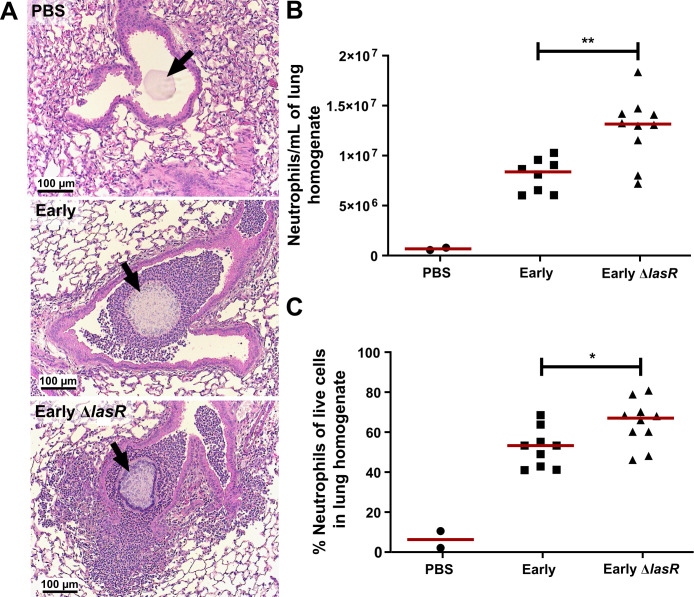
Next, we measured neutrophil counts in the lung homogenates and bronchoalveolar lavage fluid (BALF) to determine whether the increased ICAM-1 response to lasR mutant infections at 2 days p.i was also associated with increased lung neutrophilic infiltration. H&E staining of Early and Early ΔlasR-infected mouse lungs revealed that peri-bronchial and parenchymal inflammation in Early ΔlasR-infected mice was more extensive than in Early-infected or PBS control mice (Fig 6A). We note that the H&E lung sections could not be accurately assessed for intraluminal airway inflammation because the mouse airways were flushed with PBS for BALF collection prior to fixation. Both total lung neutrophil counts (1.6-fold increase, p = 0.006) and percentage of neutrophils (68% vs. 53%, p = 0.033) were significantly elevated in lasR mutant compared to wild-type infected mice (Fig 6B and 6C). We also noted no significant differences in the BALF analyses at day 2 p.i. (S5A and S5B Fig), although our previous studies using the same agar bead infection model showed that, by day 4 p.i, Early ΔlasR-infected mice displayed 25-fold greater BALF neutrophil counts compared to Early-infected mice (8.6x105 vs 3.4x104) [23]. Furthermore, our previous studies also reported greater BALF protein and more severe lung histopathology scores in Early ΔlasR-infected mice at day 4 p.i, confirming the presence of immunopathology in association with increased lung inflammation [23]. These results suggested that lasR mutants induced an early enhanced airway epithelial mICAM-1 response in vivo at 2 days p.i. which subsides by 4 days p.i. and are associated with increased neutrophil lung inflammation. Given that the bacterial burden in both Early and Early ΔlasR strains remained equivalent from the time of infection until day 4 p.i, the differences in ICAM-1 and inflammatory responses most likely resulted from differences in pathogen-host interactions rather than bacterial burden.
Fig 6. lasR mutant induces greater pulmonary neutrophilic inflammation than wild-type infection.
Mice were infected with Early, Early ΔlasR or PBS embedded agar beads and lungs were harvested at 2 days p.i. for lung histopathology and immune cell counts. (A) Representative images of H&E stained lung sections (10X objective), demonstrating airway cross-sections and surrounding peri-bronchial tissues. Lung homogenates were analyzed by flow cytometry for enumeration of (B) total neutrophils (CD45+, Ly6G high, CD11c-) and (C) the percentage of neutrophils among all live single cells. Results are shown as medians, with each dot representing one mouse. *P < 0.05; **P < 0.01.
Discussion
Loss-of-function lasR mutants are common in chronic CF infections [13,52], and are associated with more severe lung disease [15] and increased markers of inflammation [23] in CF patients. They cause dysregulated bacterial-host interactions through multiple mechanisms [23,46,53–55] that likely contribute to their propensity to cause greater pathology in chronic infection. Our lab has previously shown that lasR mutant infections caused an increased pro-inflammatory cytokine and chemokine response in airway epithelial cells, and an exaggerated neutrophilic inflammation and lung immunopathology in vivo [23]. In this study, we showed that loss-of-function lasR mutants also induced increased mICAM-1 levels on AEC compared to wild-type strains in cell culture models, and this effect facilitated neutrophil adhesion. We also found that lasR mutant infected mice showed increased airway epithelial ICAM-1 expression and neutrophilic lung inflammation compared to wild-type infected mice in a model of subacute airway infection. Our findings thus provide new insights into the intricate interplay between LasR-regulated proteases and airway ICAM-1 expression. As P. aeruginosa adapts to the CF lung and lasR variants emerge, these bacterial-host interactions and their effects in modulating lung inflammation change over the course of chronic infections.
Loss-of-function lasR mutations can emerge under laboratory conditions [16,56,57] and in human infections [58,59]. They are highly prevalent in chronic CF infections, as previously reported by our group and others [13,15,23,60], and numerous genomic studies of longitudinally collected P. aeruginosa clinical strains indicate that the lasR gene is under strong positive selection, with evidence of convergent evolution and pathoadaptation to the CF host [10,13,14,61–63]. Several studies have reported that loss-of-function lasR mutants have increased fitness in conditions such as low oxygen [64,65], denitrification [66], high cell density [67] and growth on certain amino acids [16,68]. It is therefore plausible that loss of LasR function confers a growth or survival advantage in such conditions relevant to the CF lung environment. We thus surmise that the exaggerated neutrophilic inflammation is a consequence rather than the selection pressure that drives the emergence of loss-of-function lasR variants in the CF lung.
Our results also demonstrated that the enhanced mICAM-1 response was primarily attributable to the loss of LasR-regulated proteases that degraded ICAM-1, findings which are consistent with previous reports by our group and others that LasR-regulated proteases can degrade mediators of the innate immune system and the complement system [23,46–48,54,69]. Furthermore, proteolytic cleavage of mICAM-1 by host-proteases such as neutrophil elastase and cathepsin G has been described [70,71], and degradation by bacterial proteases has been considered a potential mechanism of bacterial virulence [72]. While our previous findings suggested a key role for LasB in the degradation of IL-6 and IL-8 [23], we now observed that several LasR-regulated proteases, primarily LasB, AprA and T4P can degrade mICAM-1, and that their contributions likely vary depending on strain specific expression and secretion levels of each protease. The proteolytic activity of distinct proteases can also show interdependence, as LasA requires activation through proteolytic cleavage by LasB, and T4P function is significantly increased upon proteolytic cleavage by LasB and AprA [73,74].
P. aeruginosa can modulate ICAM-1 expression through other mechanisms. P. aeruginosa and other gram-negative bacterial lipopolysaccharide (LPS) can induce ICAM-1 expression in epithelial and other cell types [37,75,76]. Although LPS expression or structures may vary across P. aeruginosa clinical strains, we doubt that this variable had a major effect on the ICAM-1 induction by the Early and Early ΔlasR strains since heat-inactivated filtrates of both strains, which contain heat-stable compounds such as LPS, stimulated mICAM-1 to comparable levels (Fig 2A). The P. aeruginosa type III effector ExoU also induces cleavage of mICAM-1 to soluble ICAM-1, but through a host cyclooxygenase-dependent pathway [77,78]. We note that the effect of P. aeruginosa ExoU on mICAM-1 does not explain our results since ExoU secretion requires an active type III secretion system [79] and is likely negligible in the cell-free P. aeruginosa filtrates used in our in vitro experimental system. Finally, Look et al. have reported that phenazines (such as pyocyanin) secreted by P. aeruginosa also induce ICAM-1 expression in AEC [36]. However, these secondary metabolites are unlikely to account for the increased ICAM-1 response to our lasR mutant strains which are pyocyanin deficient (S1D Fig).
Our results support an important role for airway epithelial mICAM-1 responses in neutrophil adhesion. As an adhesion molecule expressed on the apical surface of epithelial cells [27,37], mICAM-1 allows immune cells, notably neutrophils, to bind to the airway epithelial surface, an interaction that promotes transepithelial migration and thus recruitment of inflammatory cells [26,30]. It also facilitates neutrophil-mediated clearance of pulmonary pathogens through yet unclear mechanisms [25,26]. We recognize that other adhesion molecules such as vascular cell adhesion molecule 1 (VCAM-1) also contribute to neutrophil adhesion to AEC. For example, VCAM-1 basal expression in AEC is low but upregulated in response to pro-inflammatory cytokine and respiratory syncytial virus [80–82]. This is consistent with our observation that neutrophil adhesion to AEC stimulated with Late filtrates is greater than Early ΔlasR filtrates despite similar mICAM-1 levels in both conditions, indicating that mICAM-1 is not the sole adhesion factor involved.
LasR regulates the expression of many secreted proteins and small molecules. We thus recognize that loss-of-function lasR variants likely modulate host inflammatory responses through additional pathways. For example, LasR-regulated secreted molecules such as pyocyanin and rhamnolipids can cause cell death, dampen immune cell function and trigger inflammation [83–85]. LasR-regulated proteases can degrade a broad range of host proteins that mediate lung inflammation, from cytokines such as IL-6, IL-8, MCP-1 and IFN-γ [23,48,69,86] to protease-activated receptors such as PAR2 [87,88], as well as flagellin monomers, a well established pathogen-associated molecular pattern [89,90]. Although the ability of LasR-regulated proteases to target host immune and inflammatory mediators has typically been considered as a mechanism of immune evasion in the context of acute infections, we propose that the loss of such activity contributes in fact to the hyper-inflammation and immunopathology observed in the setting of subacute or chronic infections.
In our in vivo infection model, we observed that lasR mutant infections were associated with higher pulmonary neutrophil counts by flow cytometry and peri-bronchial inflammation by immunohistopathology. While BALF neutrophil counts did not differ significantly at 2 days p.i., we have previously reported that they were significantly increased at 4 days p.i. in Early ΔlasR-infected mice [23]. This suggests that neutrophil recruitment into the airways likely lags and manifests later than in the peri-bronchial compartments. Although we cannot directly infer the causal contribution of airway ICAM-1 to the pulmonary neutrophilic inflammation in our in vivo model, the association between the two is consistent with our in vitro neutrophil adhesion data.
The role of airway epithelial mICAM-1 in lung inflammation during infection is emerging but complex and incompletely understood. Previous studies have also suggested that airway epithelial mICAM-1 expression significantly affects neutrophil recruitment to the lungs in vivo and in vitro [26,34]. For example, in a model of subacute H. influenzae lung infection, Humlicek et al demonstrated a marked reduction of leukocyte recruitment upon intratracheal instillation of anti-ICAM-1 blocking antibody which predominantly targets epithelial cells of the lungs [26]. Studies have reported that basal mICAM-1 expression is minimal in the airway epithelium in human and rodent lung tissues and cell cultures, but induced in states of inflammation and infection [29,38,82]. This stands in contrast to alveolar epithelial cells which show high constitutive expression levels of mICAM-1 in the absence of infection, as we and others observed in mice, rats and humans [33,91]. Induction of mICAM-1 expression in AEC has been observed with infection by H. influenzae, C. pneumoniae, Pneumocystis carinii [26,29–31] or P. aeruginosa [36] as seen in our study, either directly in response to bacterial products, or indirectly in response to inflammatory cytokines such as TNF-α [82,92]. Epithelial ICAM-1 thus promotes neutrophil recruitment and retention to the airway compartment [24,26], a response that may aid in pathogen clearance during infection but also exacerbates inflammation-mediated pathology. Our study thus demonstrates that loss-of-function lasR mutants induce greater mICAM-1 expression and neutrophil adhesion to human airway epithelial cells in vitro. We also observed that lasR mutant infections elicit greater ICAM-1 expression in the bronchial epithelium and greater lung inflammation in murine infections, but this association remains to be mechanistically proven in vivo.
In conclusion, we report on a novel mechanism through which P. aeruginosa modulates innate immune and inflammatory responses in the host lung, and how loss of LasR function, a common patho-adaptation during chronic CF infections, enhances neutrophilic inflammation. Additionally, modulation of airway epithelial mICAM-1 expression may also have other implications through its function as the major entry receptor of human rhinoviruses (HRV) [93]. For example, stimulation of AEC with H. influenzae, which also induces mICAM-1 expression, leads to increased susceptibility to HRV-infection [94]. Whether AEC stimulated with P. aeruginosa lasR mutants are more susceptible to HRV infection remains to be determined. HRV infections are common in CF patients and may be linked to pulmonary exacerbations [95], an important determinant of lung function decline in CF patients. Whether CF patients chronically infected with P. aeruginosa lasR mutants are more susceptible to HRV infection as a result of enhanced airway mICAM-1 levels is an intriguing hypothesis to be explored.
Supporting information
BEAS-2B cells were stimulated for 24h with (A) 30 μL filtrate of the Early or Early ΔlasR strain or 20 ng/mL TNF-α; (B) starvation media (SM) +/- 30 μL SCFM medium; (C) 30 μL filtrates from six pairs of wild-type clinical isolates and isogenic lasR mutant. In (A) and (C), SCFM served as negative control (—dashed line) and 20 ng/mL TNF-α as positive control. (A+B) mICAM-1 levels were measured by flow cytometry and (B) AEC viability following filtrate stimulation was measured as the percentage of live cells (low eFluor 780) among all single cells by flow cytometry. The results are shown as mean ± SD of one representative experiment (from ≥ 2 independent experiments, each with biological triplicates). (D) Representative bacterial cultures of the Early, Early ΔlasR and Late strains, with pyocyanin (blue-green pigment) production only evident with the Early strain. Cultures were grown in SCFM, as used for filtrate production. *P < 0.05; **P < 0.01; ***P < 0.001.
(TIF)
(A) Caseinolytic activity in Early and Early ΔlasR filtrates (+/- heat treatment) was measured on skim milk agar plates. (B) Elastolytic activity in Early and Early ΔlasR filtrates (+/- heat treatment) was measured by Elastin-Congo Red assay. Correlation between (C) caseinolytic or (D) elastolytic activity of different P. aeruginosa filtrates and mICAM-1 levels on AEC stimulated with the respective filtrates. Results in (A) and (B) are shown as mean +SEM and are representative of ≥ 2 independent experiments, each with biological duplicates. In (C) and (D), each data point represents one wild-type or lasR mutant strain, with the X value displaying the mean ±SD ICAM-1 induction in one representative experiment (in biological triplicates) and the Y value displaying the mean ± SEM (C) caseinolytic or (D) elastolytic activity (two independent experiments, each with biological duplicates). The trendlines in (C) and (D) were calculated by linear regression. ND = not detectable. *P < 0.05; **P < 0.01; ***P < 0.001.
(TIF)
Caseinolytic (A+C) and elastolytic (B+D) activity of (A+B) PAO1-V and its isogenic protease mutants of lasA, lasB and aprA or (C+D) the Late strain complemented with lasA, lasB, aprA or prpL (T4P) was measured on skim milk agar plates and by Elastin-Congo Red assay, respectively. (E) rhICAM-1 was quantified by Western Blotting with a polyclonal anti rhICAM-1 antibody, following incubation for 24h with PBS (- control) or filtrates of PAO1-V and its isogenic protease mutants as indicated. (F) Adhesion of calcein-stained human primary neutrophils (green) after 2h of incubation in wells with or without AEC was analyzed by confocal imaging. Results in (A-D) are shown as mean ± SEM, with pooled data (n ≥ 3 biological replicates from ≥ 2 independent experiments). Results in (G) are representative of 2 independent experiments. Results in (F) are representative of 2 independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001.
(TIF)
(A) Representative image of the region of interest (ROI, green area) manually drawn to define the airway epithelium on mouse lung sections. (B) Representative image of an airway cross-section (20X objective, with digital magnification in the inset box) displaying high bronchial ICAM-1 expression (red) localized to the apical side the bronchial epithelium facing the lumen (arrows). The autofluorescence of the tissue was imaged in the green channel (Ex 488/ Em 518) and is shown both in grey (to better highlight the ICAM-1 signal) or green. (C) Total lung bacterial burden at 2 and 4 days p.i, in mice infected with the Early or Early ΔlasR strain. Results are pooled from two independent experiments. (D) Correlation between manual and automated scoring of the airway epithelial mICAM-1 fluorescence intensity. Each dot represents one distinct airway section analyzed. (E) ICAM-1 fluorescence intensity per ROI area in mice infected with the Early, Early ΔlasR or PBS control. Each dot represents one airway section. (F) Percentage of ICAM-1 positive airways in mice infected with the Early or Early ΔlasR strain at 2 and 4 days p.i. Results are shown as mean ± SEM (C), median ± IQR (E) or percentages (F). *P < 0.05; **P < 0.01; ***P < 0.001; n.s. P ≥ 0.05.
(TIF)
C57BL/6 mice were infected with the Early, Early ΔlasR or PBS control and sacrificed at 2 days p.i. (A) In BALF, the proportion of neutrophils was determined by Kwik-Diff staining. (B) Total BALF neutrophil counts, calculated by multiplying the proportion of neutrophils by the total number of live cells. Results are shown as medians (n = 2 mice in the control group, n ≥ 8 mice in infected groups). *P < 0.05; **P < 0.01; ***P < 0.001.
(TIF)
(DOCX)
(DOCX)
(DOCX)
Low protease production, the presence of a metallic sheen and low 3-oxo-C12-HSL signal level are characteristic phenotypes of LasR-deficient strains. NA not available.
(DOCX)
(XLSX)
Acknowledgments
We would like to thank S. Fleiszig and J. A. Hobden for providing the PAO1-V, PAO1-V ΔlasA, PAO1-V ΔaprA, PAO1-V ΔlasA/B and PAO1-V ΔlasA/B ΔaprA strains. The Gateway miniCTX2.1 vector was provided by J. J. Harrison and the E. coli DH5α (pJN105L, pSC11) bioassay strain was obtained from A. A. Dandekar. We acknowledge D. P. Speert, James Slosnik and J. L. Burns for providing the P. aeruginosa clinical isolates. We would also like to thank C. Hupperetz for his technical support with the filtrates.
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
L.H has been supported by scholarships from the German Academic Exchange Service (DAAD), and D.N is supported by a FRQS salary award. This work was supported by funds from Cystic Fibrosis Canada #2469 (D.N), CIHR PJT-148827 (D.N) and the Vertex Cystic Fibrosis Research Innovation Award (D.N). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Ratjen F, Bell SC, Rowe SM, Goss CH, Quittner AL, Bush A. Cystic fibrosis. Nature reviews Disease primers. 2015;1:15010. 10.1038/nrdp.2015.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Folkesson A, Jelsbak L, Yang L, Johansen HK, Ciofu O, Hoiby N, et al. Adaptation of Pseudomonas aeruginosa to the cystic fibrosis airway: an evolutionary perspective. Nature reviews Microbiology. 2012;10(12):841–51. 10.1038/nrmicro2907 [DOI] [PubMed] [Google Scholar]
- 3.Lynch SV, Bruce KD. The cystic fibrosis airway microbiome. Cold Spring Harbor perspectives in medicine. 2013;3(3):a009738. 10.1101/cshperspect.a009738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kosorok MR, Zeng L, West SE, Rock MJ, Splaingard ML, Laxova A, et al. Acceleration of lung disease in children with cystic fibrosis after Pseudomonas aeruginosa acquisition. Pediatric pulmonology. 2001;32(4):277–87. 10.1002/ppul.2009.abs [DOI] [PubMed] [Google Scholar]
- 5.Lin CK, Kazmierczak BI. Inflammation: A Double-Edged Sword in the Response to Pseudomonas aeruginosa Infection. Journal of innate immunity. 2017;9(3):250–61. 10.1159/000455857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rosen BH, Evans TIA, Moll SR, Gray JS, Liang B, Sun X, et al. Infection Is Not Required for Mucoinflammatory Lung Disease in CFTR-Knockout Ferrets. American journal of respiratory and critical care medicine. 2018;197(10):1308–18. 10.1164/rccm.201708-1616OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Paemka L, McCullagh BN, Abou Alaiwa MH, Stoltz DA, Dong Q, Randak CO, et al. Monocyte derived macrophages from CF pigs exhibit increased inflammatory responses at birth. Journal of cystic fibrosis: official journal of the European Cystic Fibrosis Society. 2017;16(4):471–4. 10.1016/j.jcf.2017.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Konstan MW, Berger M. Current understanding of the inflammatory process in cystic fibrosis: onset and etiology. Pediatric pulmonology. 1997;24(2):137–42; discussion 59–61. [DOI] [PubMed] [Google Scholar]
- 9.Cantin AM, Hartl D, Konstan MW, Chmiel JF. Inflammation in cystic fibrosis lung disease: Pathogenesis and therapy. Journal of cystic fibrosis: official journal of the European Cystic Fibrosis Society. 2015;14(4):419–30. 10.1016/j.jcf.2015.03.003 [DOI] [PubMed] [Google Scholar]
- 10.Winstanley C O ’Brien S, Brockhurst MA. Pseudomonas aeruginosa Evolutionary Adaptation and Diversification in Cystic Fibrosis Chronic Lung Infections. Trends in microbiology. 2016;24(5):327–37. 10.1016/j.tim.2016.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Faure E, Kwong K, Nguyen D. Pseudomonas aeruginosa in Chronic Lung Infections: How to Adapt Within the Host? Frontiers in immunology. 2018;9:2416. 10.3389/fimmu.2018.02416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lorè NI, Cigana C, De Fino I, Riva C, Juhas M, Schwager S, et al. Cystic fibrosis-niche adaptation of Pseudomonas aeruginosa reduces virulence in multiple infection hosts. PloS one. 2012;7(4):e35648. 10.1371/journal.pone.0035648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith EE, Buckley DG, Wu Z, Saenphimmachak C, Hoffman LR, D’Argenio DA, et al. Genetic adaptation by Pseudomonas aeruginosa to the airways of cystic fibrosis patients. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(22):8487–92. 10.1073/pnas.0602138103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marvig RL, Sommer LM, Molin S, Johansen HK. Convergent evolution and adaptation of Pseudomonas aeruginosa within patients with cystic fibrosis. Nature genetics. 2015;47(1):57–64. 10.1038/ng.3148 [DOI] [PubMed] [Google Scholar]
- 15.Hoffman LR, Kulasekara HD, Emerson J, Houston LS, Burns JL, Ramsey BW, et al. Pseudomonas aeruginosa lasR mutants are associated with cystic fibrosis lung disease progression. Journal of cystic fibrosis: official journal of the European Cystic Fibrosis Society. 2009;8(1):66–70. 10.1016/j.jcf.2008.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.D’Argenio DA, Wu M, Hoffman LR, Kulasekara HD, Déziel E, Smith EE, et al. Growth phenotypes of Pseudomonas aeruginosa lasR mutants adapted to the airways of cystic fibrosis patients. Molecular microbiology. 2007;64(2):512–33. 10.1111/j.1365-2958.2007.05678.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bjarnsholt T, Jensen P, Jakobsen TH, Phipps R, Nielsen AK, Rybtke MT, et al. Quorum sensing and virulence of Pseudomonas aeruginosa during lung infection of cystic fibrosis patients. PloS one. 2010;5(4):e10115. 10.1371/journal.pone.0010115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schuster M, Lostroh CP, Ogi T, Greenberg EP. Identification, timing, and signal specificity of Pseudomonas aeruginosa quorum-controlled genes: a transcriptome analysis. Journal of bacteriology. 2003;185(7):2066–79. 10.1128/jb.185.7.2066-2079.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wagner VE, Bushnell D, Passador L, Brooks AI, Iglewski BH. Microarray analysis of Pseudomonas aeruginosa quorum-sensing regulons: effects of growth phase and environment. Journal of bacteriology. 2003;185(7):2080–95. 10.1128/jb.185.7.2080-2095.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schuster M, Greenberg EP. A network of networks: quorum-sensing gene regulation in Pseudomonas aeruginosa. International journal of medical microbiology: IJMM. 2006;296(2–3):73–81. 10.1016/j.ijmm.2006.01.036 [DOI] [PubMed] [Google Scholar]
- 21.Lelong E, Marchetti A, Simon M, Burns JL, van Delden C, Köhler T, et al. Evolution of Pseudomonas aeruginosa virulence in infected patients revealed in a Dictyostelium discoideum host model. Clinical microbiology and infection: the official publication of the European Society of Clinical Microbiology and Infectious Diseases. 2011;17(9):1415–20. [DOI] [PubMed] [Google Scholar]
- 22.Lesprit P, Faurisson F, Join-Lambert O, Roudot-Thoraval F, Foglino M, Vissuzaine C, et al. Role of the quorum-sensing system in experimental pneumonia due to Pseudomonas aeruginosa in rats. American journal of respiratory and critical care medicine. 2003;167(11):1478–82. 10.1164/rccm.200207-736BC [DOI] [PubMed] [Google Scholar]
- 23.LaFayette SL, Houle D, Beaudoin T, Wojewodka G, Radzioch D, Hoffman LR, et al. Cystic fibrosis-adapted Pseudomonas aeruginosa quorum sensing lasR mutants cause hyperinflammatory responses. Science advances. 2015;1(6). 10.1126/sciadv.1500199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beck-Schimmer B, Schimmer RC, Pasch T. The airway compartment: chambers of secrets. News in physiological sciences: an international journal of physiology produced jointly by the International Union of Physiological Sciences and the American Physiological Society. 2004;19:129–32. [DOI] [PubMed] [Google Scholar]
- 25.Schnitzler N, Haase G, Podbielski A, Lütticken R, Schweizer KG. A co-stimulatory signal through ICAM-beta2 integrin-binding potentiates neutrophil phagocytosis. Nature medicine. 1999;5(2):231–5. 10.1038/5597 [DOI] [PubMed] [Google Scholar]
- 26.Humlicek AL, Pang L, Look DC. Modulation of airway inflammation and bacterial clearance by epithelial cell ICAM-1. American journal of physiology Lung cellular and molecular physiology. 2004;287(3):L598–607. 10.1152/ajplung.00073.2004 [DOI] [PubMed] [Google Scholar]
- 27.Sumagin R, Robin AZ, Nusrat A, Parkos CA. Transmigrated neutrophils in the intestinal lumen engage ICAM-1 to regulate the epithelial barrier and neutrophil recruitment. Mucosal immunology. 2014;7(4):905–15. 10.1038/mi.2013.106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lawson C, Wolf S. ICAM-1 signaling in endothelial cells. Pharmacological reports: PR. 2009;61(1):22–32. 10.1016/s1734-1140(09)70004-0 [DOI] [PubMed] [Google Scholar]
- 29.Yu ML, Limper AH. Pneumocystis carinii induces ICAM-1 expression in lung epithelial cells through a TNF-alpha-mediated mechanism. The American journal of physiology. 1997;273(6):L1103–11. 10.1152/ajplung.1997.273.6.L1103 [DOI] [PubMed] [Google Scholar]
- 30.Jahn HU, Krüll M, Wuppermann FN, Klucken AC, Rosseau S, Seybold J, et al. Infection and activation of airway epithelial cells by Chlamydia pneumoniae. The Journal of infectious diseases. 2000;182(6):1678–87. 10.1086/317608 [DOI] [PubMed] [Google Scholar]
- 31.Frick AG, Joseph TD, Pang L, Rabe AM, St Geme JW 3rd, Look DC. Haemophilus influenzae stimulates ICAM-1 expression on respiratory epithelial cells. Journal of immunology (Baltimore, Md: 1950). 2000;164(8):4185–96. 10.4049/jimmunol.164.8.4185 [DOI] [PubMed] [Google Scholar]
- 32.Beck-Schimmer B, Madjdpour C, Kneller S, Ziegler U, Pasch T, Wüthrich RP, et al. Role of alveolar epithelial ICAM-1 in lipopolysaccharide-induced lung inflammation. The European respiratory journal. 2002;19(6):1142–50. 10.1183/09031936.02.00236602 [DOI] [PubMed] [Google Scholar]
- 33.Burns AR, Takei F, Doerschuk CM. Quantitation of ICAM-1 expression in mouse lung during pneumonia. Journal of immunology (Baltimore, Md: 1950). 1994;153(7):3189–98. [PubMed] [Google Scholar]
- 34.Tosi MF, Stark JM, Hamedani A, Smith CW, Gruenert DC, Huang YT. Intercellular adhesion molecule-1 (ICAM-1)-dependent and ICAM-1-independent adhesive interactions between polymorphonuclear leukocytes and human airway epithelial cells infected with parainfluenza virus type 2. Journal of immunology (Baltimore, Md: 1950). 1992;149(10):3345–9. [PubMed] [Google Scholar]
- 35.Look DC, Rapp SR, Keller BT, Holtzman MJ. Selective induction of intercellular adhesion molecule-1 by interferon-gamma in human airway epithelial cells. The American journal of physiology. 1992;263(1 Pt 1):L79–87. 10.1152/ajplung.1992.263.1.L79 [DOI] [PubMed] [Google Scholar]
- 36.Look DC, Stoll LL, Romig SA, Humlicek A, Britigan BE, Denning GM. Pyocyanin and its precursor phenazine-1-carboxylic acid increase IL-8 and intercellular adhesion molecule-1 expression in human airway epithelial cells by oxidant-dependent mechanisms. Journal of immunology (Baltimore, Md: 1950). 2005;175(6):4017–23. 10.4049/jimmunol.175.6.4017 [DOI] [PubMed] [Google Scholar]
- 37.Madjdpour C, Oertli B, Ziegler U, Bonvini JM, Pasch T, Beck-Schimmer B. Lipopolysaccharide induces functional ICAM-1 expression in rat alveolar epithelial cells in vitro. American journal of physiology Lung cellular and molecular physiology. 2000;278(3):L572–9. 10.1152/ajplung.2000.278.3.L572 [DOI] [PubMed] [Google Scholar]
- 38.Hubeau C, Lorenzato M, Couetil JP, Hubert D, Dusser D, Puchelle E, et al. Quantitative analysis of inflammatory cells infiltrating the cystic fibrosis airway mucosa. Clinical & Experimental Immunology. 2001;124(1):69–76. 10.1046/j.1365-2249.2001.01456.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Palmer KL, Aye LM, Whiteley M. Nutritional cues control Pseudomonas aeruginosa multicellular behavior in cystic fibrosis sputum. Journal of bacteriology. 2007;189(22):8079–87. 10.1128/JB.01138-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Woods DE, Schaffer MS, Rabin HR, Campbell GD, Sokol PA. Phenotypic comparison of Pseudomonas aeruginosa strains isolated from a variety of clinical sites. Journal of clinical microbiology. 1986;24(2):260–4. 10.1128/JCM.24.2.260-264.1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bjorn MJ, Sokol PA, Iglewski BH. Influence of iron on yields of extracellular products in Pseudomonas aeruginosa cultures. Journal of bacteriology. 1979;138(1):193–200. 10.1128/JB.138.1.193-200.1979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.de Chaumont F, Dallongeville S, Chenouard N, Hervé N, Pop S, Provoost T, et al. Icy: an open bioimage informatics platform for extended reproducible research. Nature methods. 2012;9(7):690–6. 10.1038/nmeth.2075 [DOI] [PubMed] [Google Scholar]
- 43.Sappington KJ, Dandekar AA, Oinuma K, Greenberg EP. Reversible signal binding by the Pseudomonas aeruginosa quorum-sensing signal receptor LasR. mBio. 2011;2(1):e00011–11. 10.1128/mBio.00011-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee JH, Lequette Y, Greenberg EP. Activity of purified QscR, a Pseudomonas aeruginosa orphan quorum-sensing transcription factor. Molecular microbiology. 2006;59(2):602–9. 10.1111/j.1365-2958.2005.04960.x [DOI] [PubMed] [Google Scholar]
- 45.Chugani S, Kim BS, Phattarasukol S, Brittnacher MJ, Choi SH, Harwood CS, et al. Strain-dependent diversity in the Pseudomonas aeruginosa quorum-sensing regulon. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(41):E2823–31. 10.1073/pnas.1214128109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sorensen M, Kantorek J, Byrnes L, Boutin S, Mall MA, Lasitschka F, et al. Pseudomonas aeruginosa Modulates the Antiviral Response of Bronchial Epithelial Cells. Frontiers in immunology. 2020;11:96. 10.3389/fimmu.2020.00096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Laarman AJ, Bardoel BW, Ruyken M, Fernie J, Milder FJ, van Strijp JA, et al. Pseudomonas aeruginosa alkaline protease blocks complement activation via the classical and lectin pathways. Journal of immunology (Baltimore, Md: 1950). 2012;188(1):386–93. 10.4049/jimmunol.1102162 [DOI] [PubMed] [Google Scholar]
- 48.Leidal KG, Munson KL, Johnson MC, Denning GM. Metalloproteases from Pseudomonas aeruginosa degrade human RANTES, MCP-1, and ENA-78. Journal of interferon & cytokine research: the official journal of the International Society for Interferon and Cytokine Research. 2003;23(6):307–18. [DOI] [PubMed] [Google Scholar]
- 49.Park SJ, Kim SK, So YI, Park HY, Li XH, Yeom DH, et al. Protease IV, a quorum sensing-dependent protease of Pseudomonas aeruginosa modulates insect innate immunity. Molecular microbiology. 2014;94(6):1298–314. 10.1111/mmi.12830 [DOI] [PubMed] [Google Scholar]
- 50.Sumagin R, Brazil JC, Nava P, Nishio H, Alam A, Luissint AC, et al. Neutrophil interactions with epithelial-expressed ICAM-1 enhances intestinal mucosal wound healing. Mucosal immunology. 2016;9(5):1151–62. 10.1038/mi.2015.135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Facchini M, De Fino I, Riva C, Bragonzi A. Long term chronic Pseudomonas aeruginosa airway infection in mice. Journal of visualized experiments: JoVE. 2014(85). 10.3791/51019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jiricny N, Molin S, Foster K, Diggle SP, Scanlan PD, Ghoul M, et al. Loss of social behaviours in populations of Pseudomonas aeruginosa infecting lungs of patients with cystic fibrosis. PloS one. 2014;9(1):e83124. 10.1371/journal.pone.0083124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Skopelja-Gardner S, Theprungsirikul J, Lewis KA, Hammond JH, Carlson KM, Hazlett HF, et al. Regulation of Pseudomonas aeruginosa-Mediated Neutrophil Extracellular Traps. Frontiers in immunology. 2019;10(1670). 10.3389/fimmu.2019.01670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Alcorn JF, Wright JR. Degradation of pulmonary surfactant protein D by Pseudomonas aeruginosa elastase abrogates innate immune function. The Journal of biological chemistry. 2004;279(29):30871–9. 10.1074/jbc.M400796200 [DOI] [PubMed] [Google Scholar]
- 55.Maillé É, Ruffin M, Adam D, Messaoud H, Lafayette SL, McKay G, et al. Quorum Sensing Down-Regulation Counteracts the Negative Impact of Pseudomonas aeruginosa on CFTR Channel Expression, Function and Rescue in Human Airway Epithelial Cells. Frontiers in cellular and infection microbiology. 2017;7:470. 10.3389/fcimb.2017.00470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Köhler T, Buckling A, van Delden C. Cooperation and virulence of clinical Pseudomonas aeruginosa populations. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(15):6339–44. 10.1073/pnas.0811741106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sandoz KM, Mitzimberg SM, Schuster M. Social cheating in Pseudomonas aeruginosa quorum sensing. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(40):15876–81. 10.1073/pnas.0705653104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hammond JH, Hebert WP, Naimie A, Ray K, Van Gelder RD, DiGiandomenico A, et al. Environmentally Endemic Pseudomonas aeruginosa Strains with Mutations in lasR Are Associated with Increased Disease Severity in Corneal Ulcers. mSphere. 2016;1(5). 10.1128/mSphere.00140-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dénervaud V, TuQuoc P, Blanc D, Favre-Bonté S, Krishnapillai V, Reimmann C, et al. Characterization of cell-to-cell signaling-deficient Pseudomonas aeruginosa strains colonizing intubated patients. Journal of clinical microbiology. 2004;42(2):554–62. 10.1128/jcm.42.2.554-562.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ciofu O, Johansen HK, Aanaes K, Wassermann T, Alhede M, von Buchwald C, et al. P. aeruginosa in the paranasal sinuses and transplanted lungs have similar adaptive mutations as isolates from chronically infected CF lungs. Journal of cystic fibrosis: official journal of the European Cystic Fibrosis Society. 2013;12(6):729–36. 10.1016/j.jcf.2013.02.004 [DOI] [PubMed] [Google Scholar]
- 61.Nguyen D, Singh PK. Evolving stealth: genetic adaptation of Pseudomonas aeruginosa during cystic fibrosis infections. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(22):8305–6. 10.1073/pnas.0602526103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Damkiær S, Yang L, Molin S, Jelsbak L. Evolutionary remodeling of global regulatory networks during long-term bacterial adaptation to human hosts. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(19):7766–71. 10.1073/pnas.1221466110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dettman JR, Kassen R. Evolutionary Genomics of Niche-Specific Adaptation to the Cystic Fibrosis Lung in Pseudomonas aeruginosa. Molecular biology and evolution. 2021;38(2):663–75. 10.1093/molbev/msaa226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Clay ME, Hammond JH, Zhong F, Chen X, Kowalski CH, Lee AJ, et al. Pseudomonas aeruginosa lasR mutant fitness in microoxia is supported by an Anr-regulated oxygen-binding hemerythrin. Proceedings of the National Academy of Sciences of the United States of America. 2020;117(6):3167–73. 10.1073/pnas.1917576117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hammond JH, Dolben EF, Smith TJ, Bhuju S, Hogan DA. Links between Anr and Quorum Sensing in Pseudomonas aeruginosa Biofilms. Journal of bacteriology. 2015;197(17):2810–20. 10.1128/JB.00182-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Toyofuku M, Nomura N, Fujii T, Takaya N, Maseda H, Sawada I, et al. Quorum sensing regulates denitrification in Pseudomonas aeruginosa PAO1. Journal of bacteriology. 2007;189(13):4969–72. 10.1128/JB.00289-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Heurlier K, Dénervaud V, Haenni M, Guy L, Krishnapillai V, Haas D. Quorum-sensing-negative (lasR) mutants of Pseudomonas aeruginosa avoid cell lysis and death. Journal of bacteriology. 2005;187(14):4875–83. 10.1128/JB.187.14.4875-4883.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hoffman LR, Richardson AR, Houston LS, Kulasekara HD, Martens-Habbena W, Klausen M, et al. Nutrient availability as a mechanism for selection of antibiotic tolerant Pseudomonas aeruginosa within the CF airway. PLoS pathogens. 2010;6(1):e1000712. 10.1371/journal.ppat.1000712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Horvat RT, Parmely MJ. Pseudomonas aeruginosa alkaline protease degrades human gamma interferon and inhibits its bioactivity. Infection and immunity. 1988;56(11):2925–32. 10.1128/IAI.56.11.2925-2932.1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Champagne B, Tremblay P, Cantin A, St Pierre Y. Proteolytic cleavage of ICAM-1 by human neutrophil elastase. Journal of immunology (Baltimore, Md: 1950). 1998;161(11):6398–405. [PubMed] [Google Scholar]
- 71.Robledo O, Papaioannou A, Ochietti B, Beauchemin C, Legault D, Cantin A, et al. ICAM-1 isoforms: specific activity and sensitivity to cleavage by leukocyte elastase and cathepsin G. European journal of immunology. 2003;33(5):1351–60. 10.1002/eji.200323195 [DOI] [PubMed] [Google Scholar]
- 72.Tada H, Sugawara S, Nemoto E, Imamura T, Potempa J, Travis J, et al. Proteolysis of ICAM-1 on human oral epithelial cells by gingipains. Journal of dental research. 2003;82(10):796–801. 10.1177/154405910308201007 [DOI] [PubMed] [Google Scholar]
- 73.Kessler E, Safrin M, Gustin JK, Ohman DE. Elastase and the LasA protease of Pseudomonas aeruginosa are secreted with their propeptides. The Journal of biological chemistry. 1998;273(46):30225–31. 10.1074/jbc.273.46.30225 [DOI] [PubMed] [Google Scholar]
- 74.Oh J, Li XH, Kim SK, Lee JH. Post-secretional activation of Protease IV by quorum sensing in Pseudomonas aeruginosa. Scientific reports. 2017;7(1):4416. 10.1038/s41598-017-03733-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Perfetto B, Donnarumma G, Criscuolo D, Paoletti I, Grimaldi E, Tufano MA, et al. Bacterial components induce cytokine and intercellular adhesion molecules-1 and activate transcription factors in dermal fibroblasts. Research in microbiology. 2003;154(5):337–44. 10.1016/S0923-2508(03)00084-6 [DOI] [PubMed] [Google Scholar]
- 76.Beck-Schimmer B, Schimmer RC, Warner RL, Schmal H, Nordblom G, Flory CM, et al. Expression of lung vascular and airway ICAM-1 after exposure to bacterial lipopolysaccharide. American journal of respiratory cell and molecular biology. 1997;17(3):344–52. 10.1165/ajrcmb.17.3.2861 [DOI] [PubMed] [Google Scholar]
- 77.Lins RX, de Assis MC, Mallet de Lima CD, Freitas C, Maciel Plotkowski MC, Saliba AM. ExoU modulates soluble and membrane-bound ICAM-1 in Pseudomonas aeruginosa-infected endothelial cells. Microbes and infection. 2010;12(2):154–61. 10.1016/j.micinf.2009.11.005 [DOI] [PubMed] [Google Scholar]
- 78.Grenier D, Bodet C. Streptococcus suis stimulates ICAM-1 shedding from microvascular endothelial cells. FEMS immunology and medical microbiology. 2008;54(2):271–6. 10.1111/j.1574-695X.2008.00476.x [DOI] [PubMed] [Google Scholar]
- 79.Galle M, Carpentier I, Beyaert R. Structure and function of the Type III secretion system of Pseudomonas aeruginosa. Current protein & peptide science. 2012;13(8):831–42. 10.2174/138920312804871210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wang SZ, Hallsworth PG, Dowling KD, Alpers JH, Bowden JJ, Forsyth KD. Adhesion molecule expression on epithelial cells infected with respiratory syncytial virus. The European respiratory journal. 2000;15(2):358–66. 10.1034/j.1399-3003.2000.15b23.x [DOI] [PubMed] [Google Scholar]
- 81.Sabatini F, Silvestri M, Sale R, Serpero L, Di Blasi P, Rossi GA. Cytokine release and adhesion molecule expression by stimulated human bronchial epithelial cells are downregulated by salmeterol. Respiratory medicine. 2003;97(9):1052–60. 10.1016/s0954-6111(03)00137-9 [DOI] [PubMed] [Google Scholar]
- 82.Atsuta J, Sterbinsky SA, Plitt J, Schwiebert LM, Bochner BS, Schleimer RP. Phenotyping and cytokine regulation of the BEAS-2B human bronchial epithelial cell: demonstration of inducible expression of the adhesion molecules VCAM-1 and ICAM-1. American journal of respiratory cell and molecular biology. 1997;17(5):571–82. 10.1165/ajrcmb.17.5.2685 [DOI] [PubMed] [Google Scholar]
- 83.Rada B, Leto TL. Pyocyanin effects on respiratory epithelium: relevance in Pseudomonas aeruginosa airway infections. Trends in microbiology. 2013;21(2):73–81. 10.1016/j.tim.2012.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bianchi SM, Prince LR, McPhillips K, Allen L, Marriott HM, Taylor GW, et al. Impairment of apoptotic cell engulfment by pyocyanin, a toxic metabolite of Pseudomonas aeruginosa. American journal of respiratory and critical care medicine. 2008;177(1):35–43. 10.1164/rccm.200612-1804OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jensen P, Bjarnsholt T, Phipps R, Rasmussen TB, Calum H, Christoffersen L, et al. Rapid necrotic killing of polymorphonuclear leukocytes is caused by quorum-sensing-controlled production of rhamnolipid by Pseudomonas aeruginosa. Microbiology (Reading, England). 2007;153(Pt 5):1329–38. 10.1099/mic.0.2006/003863-0 [DOI] [PubMed] [Google Scholar]
- 86.Horvat RT, Clabaugh M, Duval-Jobe C, Parmely MJ. Inactivation of human gamma interferon by Pseudomonas aeruginosa proteases: elastase augments the effects of alkaline protease despite the presence of alpha 2-macroglobulin. Infection and immunity. 1989;57(6):1668–74. 10.1128/IAI.57.6.1668-1674.1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Moraes TJ, Martin R, Plumb JD, Vachon E, Cameron CM, Danesh A, et al. Role of PAR2 in murine pulmonary pseudomonal infection. American journal of physiology Lung cellular and molecular physiology. 2008;294(2):L368–77. 10.1152/ajplung.00036.2007 [DOI] [PubMed] [Google Scholar]
- 88.Kida Y, Higashimoto Y, Inoue H, Shimizu T, Kuwano K. A novel secreted protease from Pseudomonas aeruginosa activates NF-kappaB through protease-activated receptors. Cellular microbiology. 2008;10(7):1491–504. 10.1111/j.1462-5822.2008.01142.x [DOI] [PubMed] [Google Scholar]
- 89.Casilag F, Lorenz A, Krueger J, Klawonn F, Weiss S, Häussler S. The LasB Elastase of Pseudomonas aeruginosa Acts in Concert with Alkaline Protease AprA To Prevent Flagellin-Mediated Immune Recognition. Infection and immunity. 2016;84(1):162–71. 10.1128/IAI.00939-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bardoel BW, van der Ent S, Pel MJ, Tommassen J, Pieterse CM, van Kessel KP, et al. Pseudomonas evades immune recognition of flagellin in both mammals and plants. PLoS pathogens. 2011;7(8):e1002206. 10.1371/journal.ppat.1002206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kasper M, Koslowski R, Luther T, Schuh D, Müller M, Wenzel KW. Immunohistochemical evidence for loss of ICAM-1 by alveolar epithelial cells in pulmonary fibrosis. Histochemistry and cell biology. 1995;104(5):397–405. 10.1007/BF01458134 [DOI] [PubMed] [Google Scholar]
- 92.Chan SC, Shum DK, Tipoe GL, Mak JC, Leung ET, Ip MS. Upregulation of ICAM-1 expression in bronchial epithelial cells by airway secretions in bronchiectasis. Respiratory medicine. 2008;102(2):287–98. 10.1016/j.rmed.2007.08.013 [DOI] [PubMed] [Google Scholar]
- 93.Tomassini JE, Graham D, DeWitt CM, Lineberger DW, Rodkey JA, Colonno RJ. cDNA cloning reveals that the major group rhinovirus receptor on HeLa cells is intercellular adhesion molecule 1. Proceedings of the National Academy of Sciences of the United States of America. 1989;86(13):4907–11. 10.1073/pnas.86.13.4907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gulraiz F, Bellinghausen C, Bruggeman CA, Stassen FR. Haemophilus influenzae increases the susceptibility and inflammatory response of airway epithelial cells to viral infections. FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 2015;29(3):849–58. 10.1096/fj.14-254359 [DOI] [PubMed] [Google Scholar]
- 95.Goffard A, Lambert V, Salleron J, Herwegh S, Engelmann I, Pinel C, et al. Virus and cystic fibrosis: rhinoviruses are associated with exacerbations in adult patients. Journal of clinical virology: the official publication of the Pan American Society for Clinical Virology. 2014;60(2):147–53. 10.1016/j.jcv.2014.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
BEAS-2B cells were stimulated for 24h with (A) 30 μL filtrate of the Early or Early ΔlasR strain or 20 ng/mL TNF-α; (B) starvation media (SM) +/- 30 μL SCFM medium; (C) 30 μL filtrates from six pairs of wild-type clinical isolates and isogenic lasR mutant. In (A) and (C), SCFM served as negative control (—dashed line) and 20 ng/mL TNF-α as positive control. (A+B) mICAM-1 levels were measured by flow cytometry and (B) AEC viability following filtrate stimulation was measured as the percentage of live cells (low eFluor 780) among all single cells by flow cytometry. The results are shown as mean ± SD of one representative experiment (from ≥ 2 independent experiments, each with biological triplicates). (D) Representative bacterial cultures of the Early, Early ΔlasR and Late strains, with pyocyanin (blue-green pigment) production only evident with the Early strain. Cultures were grown in SCFM, as used for filtrate production. *P < 0.05; **P < 0.01; ***P < 0.001.
(TIF)
(A) Caseinolytic activity in Early and Early ΔlasR filtrates (+/- heat treatment) was measured on skim milk agar plates. (B) Elastolytic activity in Early and Early ΔlasR filtrates (+/- heat treatment) was measured by Elastin-Congo Red assay. Correlation between (C) caseinolytic or (D) elastolytic activity of different P. aeruginosa filtrates and mICAM-1 levels on AEC stimulated with the respective filtrates. Results in (A) and (B) are shown as mean +SEM and are representative of ≥ 2 independent experiments, each with biological duplicates. In (C) and (D), each data point represents one wild-type or lasR mutant strain, with the X value displaying the mean ±SD ICAM-1 induction in one representative experiment (in biological triplicates) and the Y value displaying the mean ± SEM (C) caseinolytic or (D) elastolytic activity (two independent experiments, each with biological duplicates). The trendlines in (C) and (D) were calculated by linear regression. ND = not detectable. *P < 0.05; **P < 0.01; ***P < 0.001.
(TIF)
Caseinolytic (A+C) and elastolytic (B+D) activity of (A+B) PAO1-V and its isogenic protease mutants of lasA, lasB and aprA or (C+D) the Late strain complemented with lasA, lasB, aprA or prpL (T4P) was measured on skim milk agar plates and by Elastin-Congo Red assay, respectively. (E) rhICAM-1 was quantified by Western Blotting with a polyclonal anti rhICAM-1 antibody, following incubation for 24h with PBS (- control) or filtrates of PAO1-V and its isogenic protease mutants as indicated. (F) Adhesion of calcein-stained human primary neutrophils (green) after 2h of incubation in wells with or without AEC was analyzed by confocal imaging. Results in (A-D) are shown as mean ± SEM, with pooled data (n ≥ 3 biological replicates from ≥ 2 independent experiments). Results in (G) are representative of 2 independent experiments. Results in (F) are representative of 2 independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001.
(TIF)
(A) Representative image of the region of interest (ROI, green area) manually drawn to define the airway epithelium on mouse lung sections. (B) Representative image of an airway cross-section (20X objective, with digital magnification in the inset box) displaying high bronchial ICAM-1 expression (red) localized to the apical side the bronchial epithelium facing the lumen (arrows). The autofluorescence of the tissue was imaged in the green channel (Ex 488/ Em 518) and is shown both in grey (to better highlight the ICAM-1 signal) or green. (C) Total lung bacterial burden at 2 and 4 days p.i, in mice infected with the Early or Early ΔlasR strain. Results are pooled from two independent experiments. (D) Correlation between manual and automated scoring of the airway epithelial mICAM-1 fluorescence intensity. Each dot represents one distinct airway section analyzed. (E) ICAM-1 fluorescence intensity per ROI area in mice infected with the Early, Early ΔlasR or PBS control. Each dot represents one airway section. (F) Percentage of ICAM-1 positive airways in mice infected with the Early or Early ΔlasR strain at 2 and 4 days p.i. Results are shown as mean ± SEM (C), median ± IQR (E) or percentages (F). *P < 0.05; **P < 0.01; ***P < 0.001; n.s. P ≥ 0.05.
(TIF)
C57BL/6 mice were infected with the Early, Early ΔlasR or PBS control and sacrificed at 2 days p.i. (A) In BALF, the proportion of neutrophils was determined by Kwik-Diff staining. (B) Total BALF neutrophil counts, calculated by multiplying the proportion of neutrophils by the total number of live cells. Results are shown as medians (n = 2 mice in the control group, n ≥ 8 mice in infected groups). *P < 0.05; **P < 0.01; ***P < 0.001.
(TIF)
(DOCX)
(DOCX)
(DOCX)
Low protease production, the presence of a metallic sheen and low 3-oxo-C12-HSL signal level are characteristic phenotypes of LasR-deficient strains. NA not available.
(DOCX)
(XLSX)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.