Acute lymphoblastic leukemia (ALL) is the most common cancer of children, and current pediatric treatment protocols have resulted in 5-year overall survival rates up to 90% [1]. However, treatments often cause severe side effects [2]. In addition, relapsed pediatric patients have significantly reduced survival [2]. To address these issues, targeted therapies must be developed that are tailored to each individual case of ALL [3]. There are multiple potential therapeutic targets within pediatric ALL, but the IL-7Rα signaling pathways are particularly attractive. These pathways harbor genetic mutations at multiple signaling nodes in T-ALL and B-ALL [4]. Also, mutations of the IL-7Rα chain itself occur in ~10% of pediatric T-ALL cases [4]. These mutations enable homodimerization of IL-7Rα and lead to constitutive signaling, enabling increased cell survival and proliferation [5]. However, experiments in our lab showed that mutation of IL-7Rα alone was insufficient to cause transformation of primary T cells (W Li and SK Durum, unpublished results). We hypothesized that additional genetic lesions must collaborate with mutated IL-7Rα or its signaling pathway members to cause leukemia.
Pediatric patient data suggested at least three potential collaborative candidates: TLX3 (HOX11L2) expression, HOXA gene cluster overexpression, and mutations in NRAS and/or genes in its oncogenic pathways, the Raf/MEK/ERK and PI3K/Akt/mTOR pathways [5, 6]. These were considered potential collaborators because patients with mutated IL-7Rα were more likely to have TLX3 or HOXA subgroup leukemia [5]. In addition, concurrent genetic mutations in IL-7Rα and NRAS signaling pathways were quite common. For example, 10/24 T-ALL patients with NRAS/KRAS/NF1 pathway mutations and 13/37 Philadelphia chromosome-like BCP-ALL patients with NRAS pathway mutations had concurrent mutations in the IL-7Rα pathway [7, 8]. Concurrent NRAS and IL-7Rα pathway mutations also occurred in early T cell precursor leukemia and the T-ALL cell line DND41 [9, 10].
To assess these potential mutant IL-7Rα collaborations, we retrovirally transduced primary immature (CD4−CD8−) murine thymocytes, then injected these cells via tail vein into sub-lethally irradiated, 6–12-week-old, male and female Rag1−/− (B6.129S7-Rag1tm1Mom/J) mice, a strain that lacks mature T cells and B cells. We used thymocytes because T-ALL likely originates from these cells, and this technique has been reported [11]. Experiments were planned to include five animals per group based on previous experience suggesting this would be adequate to show differences between groups if any existed. Some groups were smaller than this because it was not possible to generate adequate cell numbers in culture. Non-parametric statistical tests were performed because there was heterogeneity of variance which was due to the biology of the response to experimental manipulations. Animal group allotment was not purposefully randomized. Animals that were sick due to causes other than the experimental manipulation (for example, tail trauma) and those that developed disease after 100 days were censored. The 100 day time-point was chosen during early experiments because one vector control animal developed leukemia at 110 days, an event that was considered unrelated to introduction of experimental oncogenes. Investigators and technicians were not blinded to experimental group for survival studies. Experiments were performed under the guidance of the Institutional Animal Care and Use Committee and the Institutional Biosafety Committee of the National Cancer Institute, Frederick, MD. As controls, thymocytes transduced with wild-type human IL-7Rα did not cause disease (Fig. 1a), and thymocytes transduced with mutant human IL-7Rα c.731_732insTTGTCCCAC alone (mutIL-7Rα-only) caused multisystemic inflammatory disease (Fig. 1a; Supplemental Fig. 1) of unknown mechanism, not leukemia.
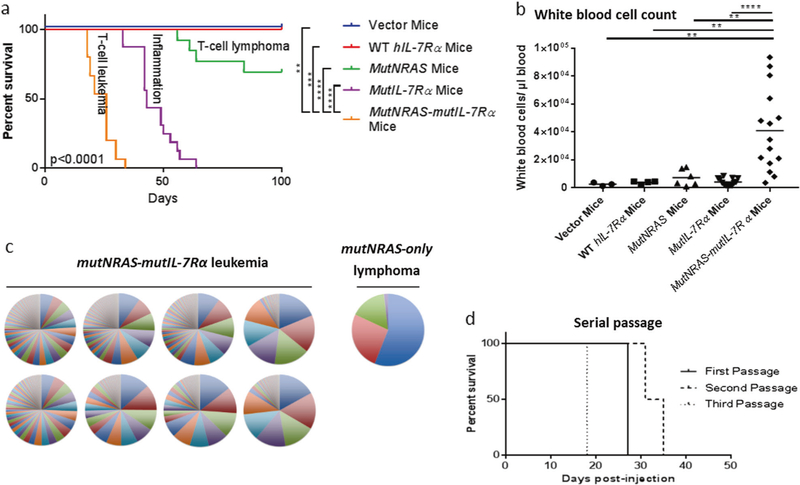
Fig. 1.
Mutant IL-7Rα combined with mutant NRas was sufficient to generate rapid-onset leukemia. Mice injected with mutNRas-mutIL-7Rα thymocytes developed clinical signs that required euthanasia more rapidly than control mice (a). MutNRas-mutIL-7Rα mice developed a circulating lymphoblastic leukocytosis (b). Splenic cells from mice injected with mutNRAS-mutIL-7Rα cells caused rapid-onset disease in serial recipients (d). Ligation-mediated PCR on splenic DNA to identify retroviral integration sites showed the mutNRas-mutIL-7Rα leukemias were markedly polyclonal in comparison to the oligoclonal MutNRas-only lymphoma. In this graph, each circle represents the splenic DNA (leukemic population) from a single mouse, and each wedge represents a unique integration site within the population. A single clone may have more than one integration site (c). Bars represent mean values. Survival statistical analysis used the Log-rank (Mantel-Cox) test comparing all groups as well as the mutNRas-mutIL-7Rα group to individual controls. Other statistical comparisons used the Mann–Whitney test comparing mutNRas-mutIL-7Rα cells to individual control groups. *p <0.05; **p <0.005; ***p ≤ 0.0005; ****p <0.0001. Survival and WBC count data include results from 1–3 independent experiments (vector N = 3; wild type N = 1, 4; mutNRas N = 1, 8, 4; mutIL-7Rα N = 3, 8, 5; mutNRas-mutIL-7Rα N = 2, 8, 5). Ligation-mediated PCR data are from a single experiment (mutNRas N = 1; mutNRas-mutIL-7Rα N = 8). Serial passage data are the best of four passage attempts (N = 5 recipients/ round of passage)
Only one of the five mice injected with thymocytes (sourced from 3–4-week-old C57BL/6 J female mice) transduced with both TLX3 and mutIL-7Rα (TLX3-mutIL-7Rα) developed lymphoma at Day 48 (Supplemental Fig. 2a). Neoplastic cells were GFP+(TLX3; immunohistochemistry (IHC) with Abcam ab6556 and flow cytometry autofluorescence), CD3+(IHC with Bio-Rad MCA1477) and CD4−CD8− (flow cytometry with Biolegend 100421, Biolegend 100737; data not shown).The remaining mice in this group developed multisystemic inflammatory disease similar to the mutIL-7Rα-only mice, presumably due to the subpopulation of mutIL-7Rα-only cells injected as a part of the unsorted cell population. The low penetrance and late occurrence of TLX3-mutIL-7Rα lymphoma suggested additional genetic lesion/s were required for the combination of TLX3 and mutIL-7Rα to cause cancer. Control TLX3-only cells did not cause disease.
All five mice injected with NUP98-HOXD13 transgenic thymocytes (sourced from a 10-week-old female C57BL/6-Tg(Vav1-NUP98/HOXD13)G2Apla/J mouse) transduced with mutIL-7Rα developed rapid-onset myeloid leukemia (Supplemental Fig. 2b). NUP98-HOXD13 thymocytes overexpress the Hoxα gene cluster [12]. NUP98-HOXD13-mutIL-7Rα myeloid cells caused a leukocytosis and infiltrated thymus, spleen, bone marrow, liver, lung, and lymph nodes (Supplemental Fig. 2c–e). Cells were GFP+ (mutIL-7Rα), CD11b+, CD3+, though neutrophilic morphology was not consistent with early thymic precursor ALL (Supplemental Fig. 2d and e and data not shown). Clonality analysis using ligation-mediated PCR to identify retroviral integration sites showed cell populations were oligoclonal suggesting in vitro and/or in vivo growth advantage of some clones (Supplemental Fig. 2f). Control NUP98-HOXD13-only cells did not cause disease.
Finally, thymocytes (sourced from 3–4-week-old C57BL/6 J female mice) transduced with both mutant IL-7Rα and mutant NRAS c.38G>A (G13D) caused full-penetrance, rapid-onset T cell lymphoblastic leukemia/lymphoma (Fig. 1a). MutNRas-mutIL-7Rα cells induced leukocytosis and infiltrated thymus, spleen, bone marrow, liver, and lung (Fig. 1b; Supplemental Figure 3a&b). Cells were GFP+ (MutNRas; IHC with Abcam ab6556 and flow cytometry autofluorescence), CD3+ (IHC with Bio-Rad MCA1477) Thy1.2+CD4+CD8+ or Thy1.2+CD8+TCRβ+ (Flow cytometry with Biolegend 105327, Biolegend 100421, Biolegend 100737, and Biolegend 109223, respectively; see Supplemental Figure 3b and data not shown). The neoplastic nature of these cells was further evident in a high rate of active cell cycling shown by ki67 (IHC with Abcam ab16667) expression (data not shown) and a leukemia-initiating cell frequency of at least 1 in 614 based on extreme limiting dilution analysis utilizing serial dilution of newly transduced thymocytes (Supplemental Table 1) [13]. In addition, serial passage of leukemic cells through three passages was successful in three of four attempts (Fig. 1d). Two of these serial passages were continued through eight and nine passages to generate cell lines. Of these, one cell line continued to express human IL-7Rα and GFP (mutNRas) and maintained an immunophenotype similar to the original leukemia (data not shown). This cell line was used in the drug study described below. Control mutNRas-only cells caused much later-onset, partially penetrant T cell lymphoma.
To further support the sufficiency of mutNRas and mutIL-7Rα to cause leukemia, we assessed clonality using ligation-mediated PCR to identify the retroviral integration sites. A sufficient combination would be expected to cause polyclonal leukemia composed of many different cells, each transduced with the combination of mutant genes. If, instead, additional mutations were necessary for leukemia, cells would be expected to be oligoclonal or monoclonal. The mutNRas-mutIL-7Rα leukemias were highly polyclonal in comparison to the oligoclonal mutNRas-only lymphoma and oligoclonal NUP98-HOXD13-mutIL-7Rα leukemias (Fig. 1c; Supplemental Fig. 2f).
Although mutNRas-mutIL-7Rα cell populations were polyclonal, there were some shared genes in the top 20 integration sites from each mouse, suggesting that differential growth of clones likely occurred both in vitro and in vivo (data not shown). Assessment of T cell receptor clonality showed variable clonality between animals, and there was evidence of clonal dominance in samples from the limiting dilution experiment (data not shown). On average, 5 of the 20 top integration sites in each animal impacted genes that were known tumor suppressors and/or oncogenes (data not shown) [14, 15]. It seems that, while the combination of mutant IL-7Rα and NRas is sufficient to induce T-ALL, cells can also be conferred growth advantage by additional genetic lesions. This is consistent with the observation that human T-ALL cases typically have more than two mutations [3]. Transcriptome analysis on cultured cells did not reveal the leukemogenic mechanism of mutNRas-mutIL-7Rα cells (data not shown).
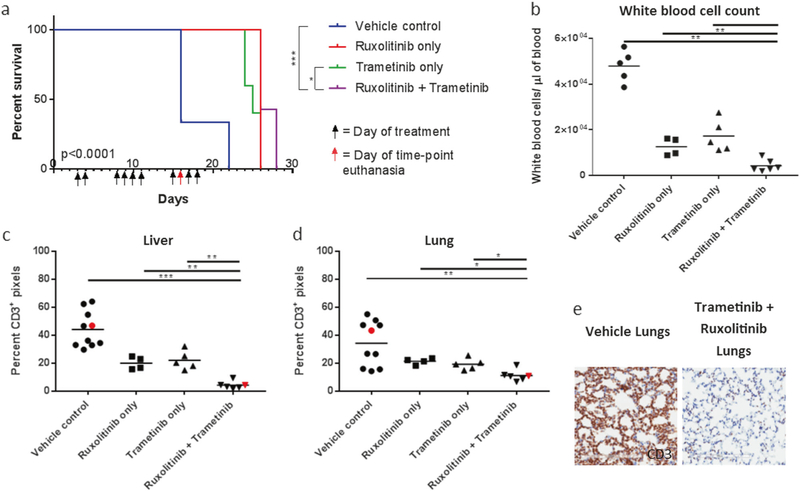
To assess whether targeted therapy could successfully treat mutNRas-mutIL-7Rα leukemia, we treated mice that had been injected with the mutNRas-mutIL-7Rα cell line described above. While targeting mutated RAS itself is notoriously difficult, targeting the downstream effector pathways Raf/MEK/ERK and PI3K/AKT/mTOR is more feasible [6]. Mutant IL-7Rα also signals through PI3K/AKT/mTOR as well as JAK1/STAT5 and can be targeted at multiple levels [4, 5]. Therefore, an optimal approach would include blockade of Raf/MEK/ERK, PI3K/AKT/mTOR, and JAK1/STAT5 pathways. However, humane considerations limited gavage frequency and number. Therein, we treated animals for 10 days with the MEK inhibitor Trametinib (targeting mutNRas signaling) and the JAK1 inhibitor Ruxolitinib (targeting mutIL-7Rα signaling) [4, 5, 7]. Technicians were blinded to treatment groups for this study. Treated mice survived significantly longer than untreated animals (Fig. 2a). At time-matched euthanasia, dual treatment significantly reduced disease progression, leading to lowered white blood cell counts, reduced spleen and liver weights, and decreased liver and lung infiltrates (Fig. 2b–e; Supplemental Figure 4a&b and data not shown).
Fig. 2.
Targeted therapy combining the JAK1 inhibitor Ruxolitinib with the MEK inhibitor Trametinib reduced disease progression and prolonged survival. Mice were treated using proper body weight dosing with Trametinib (0.39 mg/kg once a day) and Ruxolitinib (150 mg/kg twice a day) by gavage for 10 days. Dual therapy significantly improved survival (a). At time-matched euthanasia, dual therapy significantly reduced the white blood cell count (b) and tissue infiltrates as shown by digital image analysis of percent positive pixels from CD3 immunolabeled liver (c) and lung (d,e). In the graphs, red dots indicate animals whose tissues are pictured. Bars represent mean values. Scale bars = 200 μm. Survival analysis used the Log-rank (Mantel-Cox) test. Time-matched killing statistical analysis used the Mann-Whitney test comparing the dual therapy-treated mice to controls (*p < 0.05; **p < 0.005; ***p ≤ 0.0005). Data were from a single experiment (Vehicle-only N = 12; Ruxolitinib-only N = 9; Trametinib-only N = 10; dual treatment N = 13)
In conclusion, our data show that combined mutations in IL-7Rα and NRas appear sufficient to drive T-ALL formation. Targeted treatment of the resultant leukemia reduced leukemia progression and prolonged survival, consistent with previous reports [7]. Since concurrent mutations in the IL-7Rα and NRas pathways are relatively common in pediatric patients, these experiments could help to inform the development of targeted therapies.
Supplementary Material
Footnotes
Compliance with ethical standards
Conflict of interest S.K.D., W.L, and J.A.H. have applied for a patent titled “IL-7Rα antibodies for treating acute lymphoblastic leukemia.” (US patent application #62/238 612)
Electronic supplementary material The online version of this article (https://doi.org/10.1038/s41375-017-0001-0) contains supplementary material, which is available to authorized users.
References
- 1.Pui CH, Campana D, Pei D, Bowman WP, Sandlund JT, Kaste SC, et al. Treating childhood acute lymphoblastic leukemia without cranial irradiation. N Engl J Med 2009;360:2730–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nguyen K, Devidas M, Cheng SC, La M, Raetz EA, Carroll WL, et al. Factors influencing survival after relapse from acute lymphoblastic leukemia: a Children’s Oncology Group study. Leukemia. 2008;22:2142–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roberts KG, Mullighan CG. How new advances in genetic analysis are influencing the understanding and treatment of childhood acute leukemia. Curr Opin Pediatr 2011;23:34–40. [DOI] [PubMed] [Google Scholar]
- 4.Cramer SD, Aplan PD, Durum SK. Therapeutic targeting of IL-7Ralpha signaling pathways in ALL treatment. Blood. 2016; 128:473–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zenatti PP, Ribeiro D, Li W, Zuurbier L, Silva MC, Paganin M, et al. Oncogenic IL7R gain-of-function mutations in childhood T-cell acute lymphoblastic leukemia. Nat Genet 2011;43:932–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ward AF, Braun BS, Shannon KM. Targeting oncogenic Ras signaling in hematologic malignancies. Blood. 2012;120:3397–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cante-Barrett K, Spijkers-Hagelstein JA, Buijs-Gladdines JG, Uitdehaag JC, Smits WK, van der Zwet J, et al. MEK and PI3K-AKT inhibitors synergistically block activated IL7 receptor signaling in T-cell acute lymphoblastic leukemia. Leukemia 2016;30:1832–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roberts KG, Li Y, Payne-Turner D, Harvey RC, Yang YL, Pei D, et al. Targetable kinase-activating lesions in Ph-like acute lymphoblastic leukemia. N Engl J Med 2014;371:1005–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang J, Ding L, Holmfeldt L, Wu G, Heatley SL, Payne-Turner D, et al. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature. 2012;481:157–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Atak ZK, Gianfelici V, Hulselmans G, De Keersmaecker K, Devasia AG, Geerdens E, et al. Comprehensive analysis of transcriptome variation uncovers known and novel driver events in T-cell acute lymphoblastic leukemia. PloS Genet 2013;9:e1003997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Treanor LM, Zhou S, Janke L, Churchman ML, MaZ Lu T, et al. Interleukin-7 receptor mutants initiate early T cell precursor leukemia in murine thymocyte progenitors with multipotent potential. J Exp Med 2014;211:701–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin YW, Slape C, Zhang Z, Aplan PD. NUP98-HOXD13 transgenic mice develop a highly penetrant, severe myelodysplastic syndrome that progresses to acute leukemia. Blood. 2005;106:287–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu Y, Smyth GK. ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J Immunol Methods. 2009;347:70–8. [DOI] [PubMed] [Google Scholar]
- 14.Bushman F AllOnco. http://www.bushmanlab.org/links/genelists (2016). Accessed 19 December 2016.
- 15.Zhao M, Kim P, Mitra R, Zhao J, Zhao Z. TSGene 2.0: an updated literature-based knowledgebase for tumor suppressor genes. Nucleic Acids Res 2016;44:D1023–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.